Abstract
At the M2 terminal of the macrophage activation spectrum, expression of genes is regulated by transcription factors that include STAT6, CREB, and C/EBPβ. Signaling through β-adrenergic receptors drives M2 activation of macrophages, but little is known about the transcription factors involved. In the present study, we found that C/EBPβ regulates the signaling pathway between β-adrenergic stimulation and expression of Arg1 and several other specific genes in the greater M2 transcriptome. β-adrenergic signaling induced Cebpb gene expression relatively early with a peak at 1 hour post-stimulation, followed by peak Arg1 gene expression at 8 hours. C/EBPβ transcription factor activity was elevated at the enhancer region for Arg 1 at both 4 and 8 hours after stimulation but not near the more proximal promoter region. Knockdown of Cebpb suppressed the β-adrenergic-induced peak in Cebpb gene expression as well as subsequent accumulation of C/EBPβ protein in the nucleus, which resulted in suppression of β-adrenergic-induced Arg1 gene expression. Analysis of genome-wide transcriptional profiles identified 20 additional M2 genes that followed the same pattern of regulation by β-adrenergic- and C/EBPβ-signaling. Promoter-based bioinformatic analysis confirmed enrichment of binding motifs for C/EBPβ transcription factor across these M2 genes. These findings pinpoint a mechanism that may be targeted to redirect the deleterious influence of β-adrenergic signaling on macrophage involvement in M2-related diseases such as cancer.
Keywords: adrenergic receptor, bioinformatics, functional genomics, macrophage, transcription factor, tumor immunology
1. Introduction
Macrophages can take on many forms over space and time in the course of an inflammatory process [1]. Although the M1-M2 distinction continues to serve as a useful heuristic for the study of macrophage activation in the inflammatory process, macrophage biologists understand that these phenotypes are best understood as extremes on a spectrum constituted by several possible variations [2]. At the M2 extreme, induction of Arg1 by IL-4 requires transcription by STAT6 [3]. However, further research has shown that STAT6 is assisted by the transcription factor C/EBPβ, with both factors acting at an enhancer region ~3kb upstream of the Arg1 locus [4]-[5]. More recently, it was shown that C/EBPβ is regulated by cAMP and CREB. Deletion of CREB binding sites from the Cebpb promoter down-regulated expression of Arg1 and other M2-associated genes [6]. Moreover, stimulation of macrophages with cAMP increased C/EBPβ binding to the enhancer region for Arg1 [7], but it is possible that the effect of cAMP may also be mediated independently of CREB [8].
We previously found that transcription factor binding motifs for C/EBPβ were enriched among up-regulated genes in β-adrenergic-stimulated macrophages using a transcriptome-wide bioinformatic approach [9]. Ligation of β-adrenergic receptors activates the Gαs guanine nucleotide-binding protein to stimulate adenylyl cyclase synthesis of cAMP, which signals CREB in a PKA-dependent manner [10]. Our interest in β-adrenergic-stimulated macrophages stems from the finding that β-adrenergic signaling can promote mammary cancer progression, in part, by increasing macrophage accumulation in the primary tumor along with increased expression of Arg1 [11]. Also, several clinical studies have found reduced cancer progression in patients who are taking β-adrenergic antagonists (reviewed in [12]-[13]), although it is not clear from such studies that this protective effect is mediated through alterations in tumor-associated macrophages (TAMs). Nonetheless, these findings have spurred early phase clinical trials that seek to determine the utility of beta-blockers as an adjuvant treatment in cancer patients that can help prevent recurrence of malignancy [14]-[15]. Although Arg1 expression can also be upregulated in M1 macrophages [16], TAMs in several cancer types often exhibit M2-like properties [17]. There can however be substantial heterogeneity in phenotype, even within the same tumor [18]. Similarly, although β-adrenergic-stimulated macrophages did not fit cleanly into any one of the pre-defined categories along the M1-M2 spectrum in our previous research, the set of up-regulated genes in β-adrenergic-stimulated macrophages that exhibited enrichment of binding motifs for C/EBPβ were found to locate on the M2 side of the M1-M2 spectrum. However, it remains to be determined whether those genes were specifically or causally regulated by C/EBPβ. The present study addressed that issue by defining the extent to which C/EBPβ regulates the signaling pathway between β-adrenergic stimulation and specific genes in the greater M2 transcriptome.
2. Methods
2.1. Bone marrow-derived macrophage (BMDM) and RAW 264.7 cell culture models
Mice were handled under protocols approved by the Institutional Animal Use and Care Committee of the University of California, Los Angeles. Flushed bone marrow from female Balb/c mice (Charles River, 8–10 weeks) in RPMI-1640 with L-glutamine (Cellgro-Corning, Inc., #10–040-CV) was passed through a 30-μm cell strainer (Miltenyi, #130–041-407) and subjected to red blood cell lysis buffer (BD Biosciences, #555899). White blood cells were counted by hemocytometry and seeded at 1 × 106 cells per well in 1mL in 24-well polystyrene low attachment plates (Costar, #3473) for gene expression kinetics experiments or at 2.5 × 106 cells per well in 3mL in 6-well plates (Costar, #3471) for chromatin immunoprecipitation experiments. Bone marrow cells were cultured in RPMI-1640 with L-glutamine supplemented with 10% FBS (Atlanta Biologicals, #S11550H), 100 IU penicillin/mL, 100 μg streptomycin/mL (Cellgro-Corning, #30–002-CI), and 20 ng/mL of recombinant mouse M-CSF (Biolegend, #576402) in a humidified incubator at 37 °C, 5% CO2, for 7 days (media replenished after 2 and 5 days) before initiating β-adrenergic stimulation with the non-selective β-adrenergic agonist, isoproterenol (Sigma, #I2760) and/or the β-adrenergic antagonist, s-propranolol (Sigma, #P8688), dissolved in cell culture media as vehicle.
The RAW 264.7 macrophage-like cell line, which was derived the Balb/c mouse congenic substrain, BAB/14, was acquired from the European Collection of Authenticated Cell Cultures (ECACC) through its U.S. distributor (Millipore-Sigma, #91062702–1VL) and cultured in DMEM with high glucose, Glutamax, and pyruvate (Gibco, #10569044), with 10% FBS for 2 days before being seeded at 0.25 × 106 cells per well in 2mL in 6-well plates (Corning, #353046). Following 18–24 hours incubation after seeding, cells were transfected with Cebpb siRNA (Thermo Fisher Silencer Select, #s63860) or negative control siRNA (#4390843) at a concentration of 25nM, using Lipofectamine RNAiMAX transfection reagent (#13778075). Following 24 hours of transfection, cells were then stimulated with isoproterenol. We conducted extensive experimentation with BMDMs to try to knock down Cebpb but found that these primary cells were not amenable to efficient transfection with siRNA. As such, we focused our siRNA studies on RAW 264.7 cells, which were amenable to transfection with siRNA.
2.2. RT-qPCR
To quantify gene expression from BMDMs and RAW 264.7 cells, total RNA from RLT lysates was extracted (Qiagen RNeasy Mini Kit, #74104), cleared of contaminating DNA with on-column DNase digestion (Qiagen RNase-Free DNase Set, #79254), and quantified by spectrophotometry (NanoDrop ND-1000, Thermo Scientific). We did not include a control for the efficiency of on-column DNase digestion. Gene transcripts were examined by RT-qPCR with the CFX96 Touch Real-Time PCR Detection System (Bio-Rad), using one-step assay reagents (Qiagen Quantitect Probe RT-PCR, #204443) and TaqMan Gene Expression Assay primer-probes for mouse Cebpb and Arg1 (Thermo Fisher, Mm00843434_s1 and Mm00475988_m1, respectively). Following reverse transcription of RNA template for 30 minutes at 50°C, resulting product underwent an initial activation step at 94°C for 15 minutes followed by 50 amplification cycles of 15 seconds of strand separation at 94°C and 60 seconds of annealing and extension at 60°C. Triplicate determinations of each biological replicate were quantified by threshold cycle analysis of FAM fluorescence intensity using CFX Manager software (Bio-Rad), normalized to values of beta-actin mRNA amplified in parallel (Actb, #Mm00607939_s1).
2.3. Chromatin immunoprecipitation (ChIP) and qPCR
ChIP was performed on BMDMs using reagents from a commercially available kit (Active Motif, #53042). Isoproterenol-stimulated cells (or control cells) were removed from culture, fixed in 1% formaldehyde, and washed. Fixed cells were lysed via 1 flash freeze-thaw cycle followed by incubation with lysis buffer on ice and vortexing at highest speed with detergent. Chromatin was then sheared by immersion of a sonicator probe set to the highest allowable amplitude without inducing foam (Fisher Scientific Model 100 Sonic Dismembrator) to produce fragments ranging between 200 and 1200 bp in size. Input DNA was prepared from reverse cross-linked chromatin through phenol-chloroform extraction and analyzed for quality and quantity through spectrophotometry (Nanodrop). ChIP reactions began with overnight incubation of 20–35 μg of chromatin and 10 μg of antibody per reaction (Abcam, Anti-CEBP Beta antibody [1H7]-ChIP Grade, #AB15050). Antibody-bound protein/DNA complexes were then captured with Protein G agarose beads, and chromatin was eluted, reverse cross-linked, and DNA purified. DNA near the Arg1 locus was quantified with the CFX96 Touch Real-Time PCR Detection System (Bio-Rad), using Taqman Genotyping Master Mix (Thermo Fisher, #4371355) and TaqMan Copy Number primer-probes for mouse Arg1 and the enhancer region ~3kb upstream (NCBI GeneBank #AY074883.1) of the transcription start site for Arg1 (Thermo Fisher, Mm00735717_cn and custom assay ID ayforrev_cdkaj3j9, respectively; custom assay forward 5’-CAAGAAATACAGTTTACACATATGTCTGCAT-3’, reverse 5’-CCTTCTGCTCTCTGACTTCCTTATT, probe 5’-CTCGTCGGCTACCACCCTC-3’). Initial activation was at 95°C for 10 minutes, followed by 50 amplification cycles of 15 seconds of strand separation at 95°C and 60 seconds of annealing and extension at 60°C. Triplicate determinations of each biological replicate were quantified by threshold cycle analysis of FAM fluorescence intensity using CFX Manager software (Bio-Rad). ChIP-ed DNA was normalized to the amount of input DNA assayed in parallel. Positive and negative control antibodies and primer sets were utilized to test for defects in ChIP assay performance (Active Motif, #53027) and indicated that our chromatin preparation and immunoprecipitation procedures worked correctly (Supplementary Figure 1).
2.4. Flow cytometry
Flow cytometry was used to test for differences in expression of arginase I protein by BMDMs stimulated with isoproterenol. Following 8 and 12 hours of stimulation, cells were collected, treated with blocking solution (Innovex, #NB309), and then washed with cold FACS buffer (PBS, 2% FBS) before being fixed and permeabilized with an intracellular staining kit (Thermo Fisher, #88–8824-00). Cells were then incubated with fluorescence-conjugated monoclonal antibodies against arginase I, clone A1exF5 (Thermo Fisher, #17–3697-82) and washed. Cytometry was performed with a Becton Dickinson LSRFortessa X-20 SORP flow cytometer, using a 40mW solid state 640nm red laser to excite the antibody’s APC fluorophore. Forward and side scatter dot plot was used to identify intact cells versus debris. Side scatter and pulse width were used to exclude doublets from single cell measurement. Unlabeled cells were used to select the photomultiplier tube voltages required to detect baseline autofluorescence, and unlabelled cells from each experimental condition (control vs. isoproterenol) were measured. Relative quantification of arginase I protein for each sample was then accomplished by calculating the mean fluorescence intensity of a given sample’s fluorophore-positive cells, which were identified from orthogonal gating to that of unlabeled autofluorescent cells (no difference found in autofluorescence between conditions). Isotype control antibody-labeled cells (Thermo Fisher, #17–4321-81) from each experimental condition were also measured to test for difference in mean fluorescence intensity that was due to unspecific binding.
2.5. Liquid chromatography and tandem mass spectrometry (LC-MS/MS)
Liquid chromatography was used with tandem mass spectrometry to quantify arginase activity-dependent urea levels in BMDM cell culture media. Urea (Fisher Scientific, #BP169) was used to make calibrator standards in duplicate at concentrations of 1000, 500, 100, 50, 10 and 0 pg/uL. The urea chemical analogue, glycinamide (Sigma, #G6104), was used as an internal standard for all calibrators and samples. Samples were prepared in duplicate from cell culture media supernatants following centrifugation of cell culture solutions. Blank samples from unused cell culture media were also prepared. For each replicate of a calibration standard or sample, 100 μL was added to a microcentrifuge tube followed by 10 μL of internal standard at 10 mM and vortexed briefly. Protein was then precipitated by adding 400 μL of acetonitrile/water/formic acid (90/10/0.1, v/v/v) and vortexing before centrifugation at 16,000 × g for 5 minutes to pellet the precipitate. Supernatant (100 μL) was transferred to liquid chromatography (LC) injector vials. Aliquots of the replicate solutions (10 uL) were injected onto a HILIC column (Waters Acuity UPLC® BEH HILIC, 150 × 3 mm, 1.7 um particle size) equilibrated with 10% eluant A (0.1% formic acid in water) and 90% eluant B (0.1% formic acid in acetonitrile) and eluted (300 uL/min) with increasing concentration of eluant B (min/% B; 0/90, 4/90, 21/10, 24/10, 25/90, and 35/90). The effluent from the column was directed to an Agilent Jet Stream electrospray ionization (ESI) source connected to a triple quadrupole mass spectrometer (Agilent 6460) operating in the positive ion tandem mass spectrometric multiple reaction-monitoring (MRM) mode in which the intensity of the urea (m/z 61→44, retention time 4.1 min) and glycinamide (m/z 75→30, retention time 5.0 min) parent to fragment transitions were recorded using previously optimized instrument settings (fragmentor 50 and 35, collision energy 13 and 9, for urea and glycinamide, respectively). The amount of urea in each sample was then calculated by interpolation from the curves constructed using the data from the calibrator standards.
2.6. Immunofluorescent staining
For analysis of cellular C/EBPβ protein and transcription factor localization in nuclei, RAW 264.7 cells were cultured on glass cover slips in 6-well plates before fixation with 1% formaldehyde and permeabilization with 0.25% Triton X-100. Cells were treated with blocking solution (Innovex, #NB309) before overnight incubation at 4°C with antibodies against C/EBPβ (Abcam, Anti-CEBP Beta antibody [1H7]-ChIP Grade, #AB15050) at ¼00 dilution, followed by incubation with fluorescent Alexa 594-conjugated secondary antibody F(ab’)2 fragments (Thermo Fisher, #A-11020) at 1/500 dilution and DAPI nuclear counter stain solution (Molecular Probes #R37606). Coverslips were then mounted onto glass slides using antifade fluorescent mounting media (Agilent, #S302380–2). Fluorescence was visualized with a Zeiss inverted Axio Observer Z1 microscope and fluorescence filters, using a 20× Plan Apo objective with NA = 0.8. Digital images were acquired with the system’s AxioCam MRm monochromatic camera and Axiovision software (version 4.8.2.0), with gain and exposure time held constant across conditions of a given experiment. To quantify the relative amount of cellular C/EBPβ protein, 5 random images from each biological replicate were processed as 8-bit tagged image format files to accomplish in situ assay via pixel threshold analysis with ImageJ software [19]-[20]. To quantify relative amount of C/EBPβ localization in nuclei, JACoP plugin [21] was utilized, which calculates Mander’s Colocalization Coefficients [22], indicating the percentage of thresholded pixels in the blue channel that are occupied by corresponding thresholded pixels in the red channel.
2.7. Whole transcriptome analysis
Total RNA (2.5 μg per biological replicate) was converted to cDNA (Lexogen QuantSeq 3’ FWD) and sequenced using an Illumina HiSeq 4000 instrument in the UCLA Neuroscience Genomics Core Laboratory. Low-level sequencing data was mapped to the RefSeq mouse genome sequence and normalized to counts per million mapped reads (CPM) using the Illumina BaseSpace cloud computing platform. Gene expression data from the present study are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; accession No. GSE119707). Empirically-defined transcriptome-wide M2 genes from our previous study (9) were used as a reference set to identify M2 genes in the current study that exhibited up-regulation by β-adrenergic-signaling with inhibition by Cebpb knockdown. The reference set was defined as genes exhibiting an M2 diagnosticity score > 3 (i.e., > 3 standard deviations above the mean score, indicating greater expression of that gene by IL-4-stimulated M2 macrophages relative to IFN-γ-stimulated M1 macrophages; see reference (9) for full details). The orthogonal pattern of regulation was examined for M1 genes, using empiricallydefined M1 genes (i.e., diagnosticity score < −3) as a reference set from the same previous study. Genes constituting these M2 and M1 reference sets, with their corresponding diagnosticity scores, are listed in Dataset S1. Differential regulation was defined by ≥ 20% difference in mean log2-transformed CPM values in RAW 264.7 cells treated with isoproterenol vs. controls. Genes differentially expressed by isoproterenol were identified based on biological effect size (i.e., difference = mean isoproterenol – mean control) rather than statistical effect size (e.g., t statistic or P value), because previous research has shown that biological effect size-based criteria yield more replicable results than do statistical effect size criteria (e.g., t statistics, P values, or false discovery rate q values) [23]-[24]. To test for a significant difference in the prevalence of transcription factor binding motifs (TFBMs) for C/EBPβ in the promoters of genes that exhibited up-regulation or down-regulation by β-adrenergic-signaling, with inhibition or rescue by Cebpb knockdown, respectively, we conducted differential expression analysis with the Transcription Element Listening System (TELiS; www.telis.ucla.edu) as previously described [25]. The V$CEBPB_02 binding motif definition for C/EBPβ was retrieved from the TRANSFAC database. Analyses averaged results derived from nine parametric variations of promoter length (300 bp relative to RefSeq transcription start site, 600 bp, and 1000 bp to + 200) and target TFBM match stringency (MatSim = 0.80, 0.90, 0.95).
2.8. Statistical analysis
Data are reported as mean ± standard error of the mean. Analyses were conducted in the R environment, version 3.1.3 [26]. For Figure 1, Student’s t-test was used to analyze the effect of β-adrenergic stimulation on gene expression results at each time point in the context of a repeated measures univariate analysis of variance; factorial analysis of variance was used to examine the effects of isoproterenol and propranolol on gene expression results, with Tukey’s adjustment for multiple comparisons. For Figure 2, ChIP results, arginase I protein expression results, and urea results were analyzed with Student’s t-test. For Figure 3, factorial analysis of variance was used to examine the effects of β-adrenergic signaling and C/EBPβ inhibition on gene expression results and protein colocalization, with Tukey’s adjustment for multiple comparisons. For Figure 4, the mean value of all possible V$CEBPB_02 binding motif prevalence ratios was tested for significant deviation from a null population mean ratio of 1 with single-sample t-test for differentially-regulated genes vs. the reference genome in TELiS.
Figure 1.
Kinetic profile of β-adrenergic-induced Cebpb gene expression (A) and Arg1 gene expression (B) in BMDMs relative to initial control level at first time point. Abrogation of β-adrenergic-induced Cebpb gene expression at 1 hr (C) and Arg1 gene expression at 8 hrs (D) with the β-adrenergic antagonist, propranolol. Isoproterenol at 1μM. Propranolol at 10μM. Data represent mean ± SE of three independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05 vs. control at corresponding time point (A and B) or vs. no propranolol / no isoproterenol control condition (C and D).
Figure 2.
C/EBPβ binding near the Arg1 locus with subsequent arginase I protein expression and enzyme activity in BMDMs after β-adrenergic stimulation. Real-time qPCR of enhancer region DNA for Arg1 from anti-C/EBPβ immunoprecipitated chromatin at 4 hours (A) and 8 hours (B) after stimulation. Arginase I protein expression at 8–12 hours after stimulation (C) and arginase enzyme activity-dependent urea levels in culture media at 24 hours after stimulation (D). Isoproterenol at 1μM. Data for each ChIP time point in (A) and (B) represent mean ± SE of two independent experiments with three ChIP reactions per group in each experiment. Three experiments for (C) and two experiments for (D). **P < 0.01, *P < 0.05 vs. control.
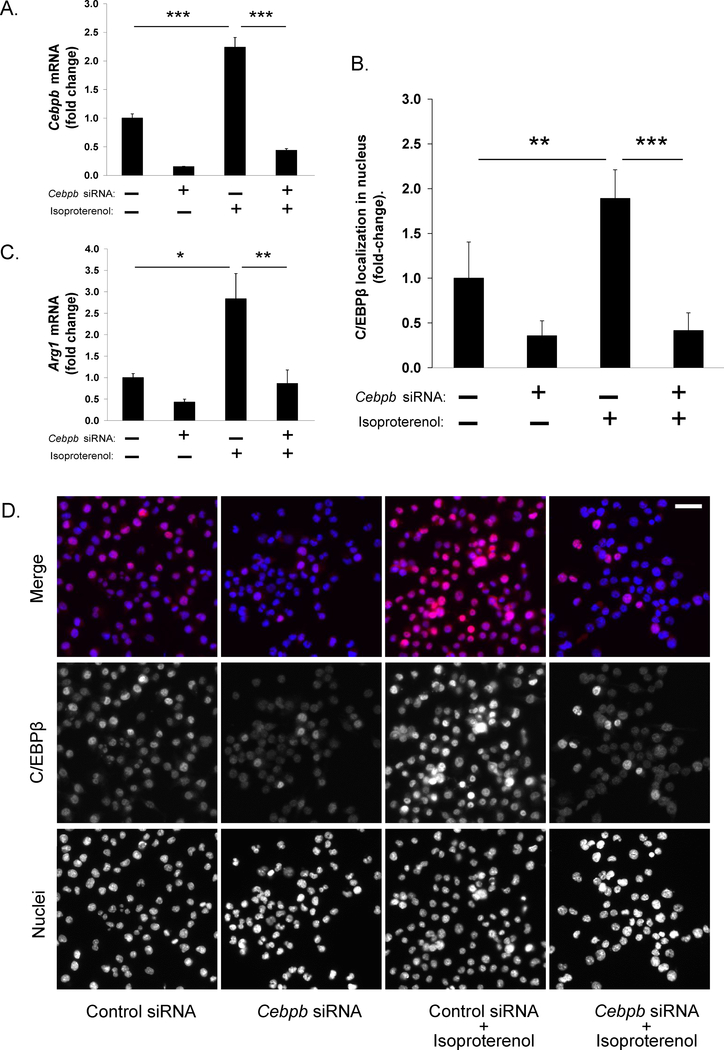
Figure 3.
Effects of C/EBPβ inhibition on β-adrenergic-induced transcription factor localization in the nucleus and Arg1 gene expression in RAW 264.7 cells. (A) Cebpb gene expression following 24 hours transfection with siRNA for Cebpb (or control siRNA) and 1 hour of stimulation with isoproterenol. (B) Percentage of nuclear stain co-localized with C/EBPβ stain at 4 hours following isoproterenol stimulation. (C) Arg1 gene expression following 24 hours transfection with siRNA for Cebpb (or control siRNA) and 8 hours of stimulation with isoproterenol. (D) Representative images of single channel gray scale and merged pseudocolored immunofluorescent staining across conditions; in merged images, blue signal is nucleus and red signal is C/EBPβ. Scale bar 100μm. Isoproterenol at 1μM. Data represent mean ± SE of three independent experiments. *** P < 0.001, **P < 0.01, *P < 0.05 between indicated pairs.
Figure 4.
C/EBPβ regulation of M2 and M1 genes modulated by β-adrenergic signaling in RAW 264.7 cells. (A) Isoproterenol up-regulated M2 genes that were suppressed by C/EBPβ inhibition in global gene expression profiles; red = up-regulation and blue = down-regulation, as mean fold change from control group fixed at 0 on log2 scale for each given gene. (B) TELiS differential expression analysis of transcription factor binding motif prevalence for C/EBPβ in M2 genes from panel A. (C) Isoproterenol down-regulated M1 genes that were rescued by C/EBPβ inhibition in global gene expression profiles. (D) TELiS differential expression analysis of transcription factor binding motif prevalence for C/EBPβ in M1 genes from panel B. *** P < 0.001 vs. reference genome.
3. Results
3.1. Kinetic profile of β-adrenergic-induced Cebpb and Arg1 gene expression
To investigate the connection between C/EBPβ and M2 gene expression in macrophages following β-adrenergic stimulation, we first determined the kinetics of isoproterenol-induced gene expression for C/EBPβ (Cebpb) and Arginase 1 (Arg1) in bone marrow-derived macrophages (BMDMs). Cebpb expression peaked at 1 hour (2.9-fold ± 0.2; P < 0.001) and remained significantly elevated above the control condition at 2 hours (2.0-fold ± 0.2; P < 0.001) and 4 hours (2.8-fold ± 0.2; P = 0.006) post stimulation (Figure 1A). In contrast, Arg1 expression did not peak until the 8-hour time point (6.3-fold ± 0.9; P < 0.001), although significant elevation above controls appeared as early as 2 hours post stimulation and remained elevated through 24 hours (P values < 0.05; Figure 1B). Blockade of β-adrenergic receptors with propranolol significantly abrogated peak Cebpb expression at 1 hour (mean suppression: 97% ± 2.5%; P < 0.001; Figure 1C) and peak Arg1 expression at 8 hours (mean suppression: 97% ± 1.9%; P < 0.001; Figure 1D). This temporal precedence of peak Cebpb expression before peak Arg1 expression suggested that β-adrenergic signaling might shift the macrophage transcriptome to the M2 side of the M1-M2 spectrum through its more immediate up-regulation of C/EBPβ. To further confirm that possibility, we conducted additional studies as follows.
3.2. C/EBPβ protein binding near the Arg1 locus after β-adrenergic stimulation
To determine whether β-adrenergic-induced Cebpb expression gives rise to greater level of C/EBPβ protein binding and transcription factor activity near the Arg1 locus at the later time points associated with peak expression, we conducted chromatin immunoprecipitation experiments at 4 and 8 hours after isoproterenol stimulation. Using real-time quantitative PCR of purified DNA from C/EBPβ-bound chromatin, we found that the Arg1 enhancer region was significantly more enriched in isoproterenol-treated cells vs. control cells at both 4 hours (2.6-fold ± 0.4; P = 0.009; Figure 2A) and 8 hours (2.3-fold ± 0.7; P = 0.031; Figure 2B). Subsequently, isoproterenol-treated cells exhibited significantly greater arginase I protein levels (8% ± 0.2%; P = 0.024; Figure 2C; Supplementary Figure 2) and marginally greater enzyme activity (12% ± 4%; P = 0.07; Figure 2D). In contrast, Arg1 DNA proper in C/EBPβ-bound chromatin exhibited no reliable increase at either 4 hours (P = 0.13) or 8 hours (P = 0.58) (Supplementary Figure 3). These results strongly suggested that β-adrenergic-induced C/EBPβ transcription factor activity up-regulates Arg1 expression and does so by binding the distal enhancer of Arg1 rather than its more proximal promoter area.
3.3. Effects of C/EBPβ inhibition on β-adrenergic-induced transcription factor localization in the nucleus
To further define β-adrenergic-induced C/EBPβ regulation of the M2 transcriptome, we conducted RNA interference experiments with RAW 264.7 cells. Twenty-four hours after transfection with Cebpb siRNA, cells were stimulated with isoproterenol for 1 hour. Similar to BMDMs, β-adrenergic signaling significantly increased Cebpb expression in RAW 264.7 cells (2.2-fold ± 0.2; P < 0.001; Figure 3A). Knockdown of Cebpb with siRNA (mean knockdown: 85%; Figure 3A) significantly inhibited β-adrenergic induction (mean suppression: 77% ± 2%; P < 0.001; Figure 3A). To determine the degree to which these results translated into suppression of C/EBPβ protein level and transcription factor localization in the nucleus, we used immunostaining to visualize C/EBPβ protein at 4 hours post stimulation. β-adrenergic signaling significantly increased C/EBPβ protein staining in the nucleus (1.9-fold ± 0.3; P = 0.006; Figure 3B,3D). Cebpb knockdown translated into knockdown of C/EBPβ protein (mean: 70%; Supplementary Figure 4) and significantly inhibited the β-adrenergic effect on C/EBPβ protein staining in the nucleus (mean suppression: 94% ± 22%; P < 0.001) and, in turn, suppressed β-adrenergic-induction of Arg1 expression at 8 hours post stimulation (mean suppression: 76% ± 17%; P = 0.002; Figure 3C). These results confirmed that β-adrenergic induction of Arg1 is regulated by C/EBPβ.
3.4. C/EBPβ regulation of M2 and M1 genes modulated by β-adrenergic signaling
To identify other β-adrenergic-induced M2 genes that are regulated by C/EBPβ, we screened RNAseq results from RAW 264.7 macrophages for previous empirically-defined M2 genes in the whole transcriptome (9) that followed the Arg1 pattern of up-regulation by β-adrenergic-signaling with inhibition by Cebpb knockdown. This analysis identified 20 additional M2 genes, including several that encode known protumoral mechanisms in tumor-associated macrophages (Figure 4A). TELiS differential expression analysis confirmed that transcription factor binding motifs for C/EBPβ (TRANSFAC V$CEBPB_02) were significantly enriched among these genes (mean fold difference [MFD] = 2.49, P < 0.001; Figure 4B). Because our previous study found that β-adrenergic signaling shifts the macrophage transcriptome to the M2 side of the M1-M2 spectrum, in part, through suppression of M1 transcripts (9), we also screened RAW 264.7 macrophages for empirically-defined M1 genes that exhibited the orthogonal pattern of Arg1 regulation (i.e., down-regulation by β-adrenergic signaling, with rescue of this effect by Cebpb knockdown). This latter screen identified eight M1 genes whose suppression by β-adrenergic signaling is regulated by C/EBPβ (Figure 4C). However, in contrast to the M2 geneset, TELiS differential expression analysis found no significant difference in prevalence of V$CEBPB_02 binding motifs among these M1 genes (MFD = 0.74, P = 0.27; Figure 4D). These results suggested that while suppression of M1 genes by C/EBPβ in β-adrenergic-stimulated macrophages is likely achieved through an indirect mechanism, C/EBPβ regulation of select M2 genes in β-adrenergic-stimulated macrophages likely involves direct transcription factor activity by the protein.
4. Discussion
These data confirm that β-adrenergic signaling induces an M2-like transcriptional profile in macrophages via induction of Cebpb gene transcription and subsequent recruitment of C/EBPβ protein to an array of M2-related gene loci, including Arg1. β-adrenergic signaling induced gene expression for C/EBPβ transcription factor (Cebpb) well in advance of peak Arg1 gene expression. Consequently, ChIP experiments showed that C/EBPβ transcription factor activity was elevated at the enhancer region for Arg1 during its peak expression, but not at the proximal promoter area. Additional C/EBPβ blocking experiments were performed through RNAi, which inhibited β-adrenergic induction of Cebpb, along with subsequent nuclear localization of C/EBPβ and Arg1 expression, further confirming that β-adrenergic induction of Arg1 is regulated by C/EBPβ. A screen of global gene expression results from RNAseq in the C/EBPβ blocking experiments identified 20 other β-adrenergic-induced M2 genes and eight β-adrenergic-suppressed M1 genes that are regulated by C/EBPβ, with bioinformatic indication that transcription factor binding motifs for C/EBPβ were over-represented in the M2 gene set but not in the M1 gene set. Together, these results show that C/EBPβ mediates the signaling pathway between β-adrenergic stimulation and expression of Arg1 and several other specific genes in the greater M2 transcriptome.
The M2 effects produced by β-adrenergic signaling in our study parallel the M2 effects produced by epinephrine and norepinephrine at concentrations ranging from 1nM to 1μM in a previous study [27]. That study also found that epinephrine and norepinephrine were acting through β2-adrenergic receptors rather than α-adrenergic receptors. Thus, given that both human [28] and rodent [29] stressor tasks have been shown to increase epinephrine into the nM range and norepinephrine levels to the near-μM range (0.1μM) in circulation, with presumably greater levels of norepinephrine in tissue microenvironments because of direct innervation (12), it seems plausible that the data from the present macrophage models may reflect how stress-induced β-adrenergic signaling could affect macrophage polarity in peripheral tissues.
C/EBPβ expression has been found to associate with greater progression in several tumor types, including colon [30], glioma [31], ovarian [32], and renal [33], although it is not clear that macrophages are the source in these cancer patient studies. Nonetheless, given its apparent involvement in malignancy, C/EBPβ has emerged as a target for the development of small-molecule inhibitors [34]-[35]. Similarly, early-phase clinical trials are beginning to target β-adrenergic receptors (14–15), given the sizeable body of pharmaco-epidemiological data that shows reduced cancer progression in patients who are taking β-adrenergic antagonists (reviewed in 12–13). The results from the current study underscore the notion that intervening at opportune points along the β-adrenergic-C/EBPβ pathway may inhibit malignancy by down-regulating M2 polarization in TAMs that can otherwise facilitate tumor progression.
Arginase I production by macrophages reduces the amount of L-arginine substrate available for nitric oxide production and, thus, is thought to reduce macrophage tumoricidal capacity [36]. However, the best defined mechanism by which arginase I has protumor effects may come from a line of studies that show arginase I enzyme activity depletes extracellular L-arginine, which in turn down-regulates expression of CD3ζ, cyclin D3, and cyclin dependent kinase 4 in neighboring T-cells, arresting them in the G0-G1 phase of the cell cycle and, thus, contributing to the well-known T-cell anergy that cancer immunotherapy seeks to reverse (reviewed in [37]). Interestingly, anti-tumor T-cell responses have also been found to be regulated by type I interferons from dendritic cells [38], which are up-regulated by lymphotoxin-β receptor (LTβR) signaling [39]. In this regard, LTβR signaling was recently found to enhance anti-tumor immunity in models of pancreatic, breast, and brain cancer [40]. Thus, the down-regulation of gene expression for the LTβR ligand subunit, Ltb, in the present study, alongside increased Arg1 expression, suggests the potential for a multifaceted immunosuppresive mechanism by which β-adrenergic-C/EBPβ signaling in TAMs could compliment direct inhibitory effects on T-cells by β-adrenergic signaling [41] in the promotion of tumor progression.
In addition to immunosuppression, other M2 properties contribute to malignant progression via tissue remodeling effects, which enable tumor angiogenesis, invasion, and metastasis (17). In the present study, we found that the β-adrenergic-C/EBPβ pathway up-regulates Csf2rb. This gene encodes the common beta chain of the receptor for GM-CSF, which has a well-established role for regulating matrix metalloproteinase (MMP) production by macrophages [42], and elevated MMP activity in the stroma surrounding breast cancer has been found to facilitate tumor invasion [43]. MMPs support malignant invasion and metastasis by degrading the extracellular matrix (ECM), which releases encapsulated cancer cells and facilitates neoangiogenesis and lymph-angiogenesis in the expanding tumor [44]-[45]. Thus, β-adrenergic-C/EBPβ signaling in macrophages may also contribute to cancer progression by promoting degradation of the ECM.
In addition to malignancy, these results may be relevant for other pathologies. For example, it is well-documented that β-adrenergic signaling primes the specialized population of macrophages in the brain, i.e., microglia, to respond with enhanced proinflammatory signaling molecules and bactericidal activity when stimulated with an immune challenge (reviewed in [46]). In turn, an amplified and prolonged neuroinflammatory response mediated by primed microglia is associated with cognitive and behavioral deficits and progressive neurodegenerative disease such as Alzheimer’s [47]. However, complicating the picture of neurodegeneration are recent reports that have found β-adrenergic signaling associates with a reduction in alpha-synuclein, the protein that accumulates to form Lewy bodies in the brains of most patients with Parkinson’s disease [48], with potential beneficial involvement of microglia in this effect (reviewed in [49],[50],[51]. Given the extensive role that C/EBPβ plays in myeloid cell development and function [52]), we speculate that this transcription factor may be involved in such effects. However, further research is needed to fully address the question.
It should be noted that extrapolation from murine in vitro models, like those in the present study, is necessarily limited. For several reasons, the genomic and protein markers that characterize a stimulated macrophage in one model may not always correlate well with phenotype in another macrophage model that uses the same stimulus (reviewed in [53]). Species differences between models can play a role in this. Compared to human macrophages, mouse macrophages can have faster response times to a given stimulus [54] and generally have higher sensitivity to the standard M1 stimulus, LPS (53). A larger factor affecting model differences may be the ontogeny of the macrophage model. For example, in vivo macrophages obtained from a participant’s specific tissue site have been shown to respond differently than the participant’s in vitro monocyte-derived macrophages when encountering the same stimulus [55],[56]. However, in the context of wound healing pathology (e.g., burn victims), which is characterized by M2 macrophage dynamics, circulating monocytes have been found to display M2 markers in parallel with macrophages situated at the site of tissue damage [57]. Nonetheless, further research with human macrophages is needed to determine the full relevancy of the present findings in mice.
In summary, the present study demonstrates that C/EBPβ mediates the signaling pathway between β-adrenergic stimulation and expression of Arg1 and several other elements of the M2 transcriptome. Although gene expression panels are asserted to be one of the clearest ways to distinguish the polarization state of macrophages (1), we note herein the caution that biological function cannot be definitively inferred from such profiles. Nonetheless, the current findings identify C/EBPβ signaling as a potential molecular target for inhibiting the effects of β-adrenergic signaling on macrophage activation in cancer and other macrophage-related diseases.
Supplementary Material
Highlights.
β-adrenergic-induced C/EBPβ transcription factor activity up-regulated Arg1 expression in macrophages
Knockdown of Cebpb inhibited β-adrenergic induction of C/EBPβ protein localization in the nucleus and subsequent Arg1 expression
Analysis of RNAseq results identified 20 additional M2 genes and eight M1 genes that were regulated by β-adrenergic-C/EBPβ signaling
Acknowledgments
This work was supported by the U.S. National Institutes of Health (K07CA188237; R01AG043404), the Australian National Health and Medical Research Council (APP1147498), and the UCLA Norman Cousins Center for PNI. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by NIH awards P30 CA016042 and 5P30 AI028697, and by the JCCC, the UCLA AIDS Institute, the David Geffen School of Medicine at UCLA, the UCLA Chancellor’s Office, and the UCLA Vice Chancellor’s Office of Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Murray PJ (2017) Macrophage Polarization. Annu Rev Physiol. 79,541–566 [DOI] [PubMed] [Google Scholar]
- [2].Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA (2014) Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 41,14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rutschman R, Lang R, Hesse M, Ihle JN, Wynn TA, Murray PJ (2001) Cutting edge: Stat6-dependent substrate depletion regulates nitric oxide production. J Immunol. 166, 2173–2177 [DOI] [PubMed] [Google Scholar]
- [4].Pauleau AL, Rutschman R, Lang R, Pernis A, Watowich SS, Murray PJ (2004) Enhancer-mediated control of macrophage-specific arginase I expression. J Immunol. 172, 7565–7573 [DOI] [PubMed] [Google Scholar]
- [5].Gray MJ, Poljakovic M, Kepka-Lenhart D, Morris SM Jr. (2005) Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene. 353,98–106 [DOI] [PubMed] [Google Scholar]
- [6].Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, Nerlov C (2009) A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A. 106, 17475–17480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sheldon KE, Shandilya H, Kepka-Lenhart D, Poljakovic M, Ghosh A, Morris SM Jr. (2013) Shaping the murine macrophage phenotype: IL-4 and cyclic AMP synergistically activate the arginase I promoter. J Immunol. 191, 2290–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Csóka B, Selmeczy Z, Koscsó B, Németh ZH, Pacher P, Murray PJ, Kepka-Lenhart D, Morris SM Jr, Gause WC, Leibovich SJ, Haskó G (2012) Adenosine promotes alternative macrophage activation via A2A and A2B receptors. FASEB J. 26, 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lamkin DM, Ho HY, Ong TH, Kawanishi CK, Stoffers VL, Ahlawat N, Ma JCY, Arevalo JMG, Cole SW, Sloan EK (2016) β-Adrenergic-stimulated macrophages: Comprehensive localization in the M1-M2 spectrum. Brain Behav Immun. 57, 338–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cole SW, Sood AK (2012) Molecular pathways: beta-adrenergic signaling in cancer. Clin Cancer Res. 18, 1201–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sloan EK, Priceman SJ, Cox BF, Yu S, Pimentel MA, Tangkanangnukul V, Arevalo JM, Morizono K, Karanikolas BD, Wu L, Sood AK, Cole SW (2010) The sympathetic nervous system induces a metastatic switch in primary breast cancer. Cancer Res. 70,7042–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cole SW, Nagaraja AS, Lutgendorf SK, Green PA, Sood AK (2015) Sympathetic nervous system regulation of the tumour microenvironment. Nat Rev Cancer. 15, 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hiller JG, Perry NJ, Poulogiannis G, Riedel B, Sloan EK (2018) Perioperative events influence cancer recurrence risk after surgery. Nat Rev Clin Oncol. 15, 205–218. [DOI] [PubMed] [Google Scholar]
- [14].Haldar R, Shaashua L, Lavon H, Lyons YA, Zmora O, Sharon E, Birnbaum Y, Allweis T, Sood AK, Barshack I, Cole S, Ben-Eliyahu S (2018) Perioperative inhibition of β-adrenergic and COX2 signaling in a clinical trial in breast cancer patients improves tumor Ki-67 expression, serum cytokine levels, and PBMCs transcriptome. Brain Behav Immun. 1591, 30187–30189 [DOI] [PubMed] [Google Scholar]
- [15].Knight JM, Kerswill SA, Hari P, Cole S,W, Logan BR, D’Souza A, Shah NN, Horowitz MM, Stolley MR, Sloan EK, Giles KE, Costanzo ES, Hamadani M, Chhabra S, Dhakal B, Rizzo JD (2018) Repurposing existing medications as cancer therapy: design and feasibility of a randomized pilot investigating propranolol administration in patients receiving hematopoietic cell transplantation. BMC Cancer. 18,593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, Basaraba RJ, König T, Schleicher U, Koo MS, Kaplan G, Fitzgerald KA, Tuomanen EI, Orme IM, Kanneganti TD, Bogdan C, Wynn TA,. Murray PJ (2008) Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 9, 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Biswas SK, Mantovani A(2010) Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 11, 889–896 [DOI] [PubMed] [Google Scholar]
- [18].Ostuni R, Kratochvill F, Murray PJ, Natoli G (2015) Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 36, 229–239. [DOI] [PubMed] [Google Scholar]
- [19].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9(7): 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hartig SM. Basic image analysis and manipulation in ImageJ. Curr Protoc Mol Biol. 2013;Chapter 14:Unit14.15. [DOI] [PubMed] [Google Scholar]
- [21].Bolte S, Cordelières FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 224,213–232 [DOI] [PubMed] [Google Scholar]
- [22].Manders EMM, Verbeek FJ, Aten JA (1993) Measurement of co-localization of objects in dual-colour confocal images. J Microsc. 169,375–382 [DOI] [PubMed] [Google Scholar]
- [23].Cole SW, Galic Z, Zack JA(2003) Controlling false-negative errors in microarray differential expression analysis: a PRIM approach. Bioinformatics. 19, 1808–1816 [DOI] [PubMed] [Google Scholar]
- [24].MAQC Consortium. (2010) The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarraybased predictive models. Nat. Biotechnol. 28, 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cole SW, Yan W, Galic Z, Arevalo J, Zack JA (2005) Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 21, 803–810 [DOI] [PubMed] [Google Scholar]
- [26].R Core Team. (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org [Google Scholar]
- [27].Grailer JJ, Haggadone MD, Sarma JV, Zetoune FS, Ward PA (2014) Induction of M2 regulatory macrophages through the b2-adrenergic receptor with protection during endotoxemia and acute lung injury. J Innate Immun. 6, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Greeson JM, Lewis JG, Achanzar K, Zimmerman E, Young KH, Suarez EC (2009) Stressinduced changes in the expression of monocytic beta2-integrins: The impact of arousal of negative affect and adrenergic responses to the Anger Recall Interview. Brain Behav. Immun. 23, 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Koolhaas JM, Bartolomucci A, Buwalda B, de Boer SF, Flügge G, Korte SM, Meerlo P, Murison R, Olivier B, Palanza P, Richter-Levin G, Sgoifo A, Steimer T, Stiedl O, vanDijk G, Wöhr M, Fuchs E (2011). Stress revisited: A critical evaluation of the stress concept. Neurosci Biobehav Rev. 35, 1291–1301. [DOI] [PubMed] [Google Scholar]
- [30].Rask K, Thörn M, Pontén F, Kraaz W, Sundfeldt K, Hedin L, Enerbäck S (2000). Increased expression of the transcription factors CCAAT-enhancer binding protein-beta (C/EBBeta) and C/EBzeta (CHOP) correlate with invasiveness of human colorectal cancer. Int J Cancer. 86, 337–343 [DOI] [PubMed] [Google Scholar]
- [31].Homma J, Yamanaka R, Yajima N, Tsuchiya N, Genkai N, Sano M, Tanaka R (2006) Increased expression of CCAAT/enhancer binding protein beta correlates with prognosis in glioma patients. Oncol Rep. 15, 595–601 [PubMed] [Google Scholar]
- [32].Sundfeldt K, Ivarsson K, Carlsson M, Enerbäck S, Janson PO, Brännström M, Hedin L (1999) The expression of CCAAT/enhancer binding protein (C/EBP) in the human ovary in vivo: specific increase in C/EBPbeta during epithelial tumour progression. Br J Cancer. 79, 1240–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Oya M, Horiguchi A, Mizuno R, Marumo K, Murai M (2003) Increased activation of CCAAT/enhancer binding protein-beta correlates with the invasiveness of renal cell carcinoma. Clin Cancer Res. 9, 1021–1027 [PubMed] [Google Scholar]
- [34].Jakobs A, Steinmann S, Henrich SM, Schmidt TJ, Klempnauer KH (2016) Helenalin Acetate, a Natural Sesquiterpene Lactone with Anti-inflammatory and Anti-cancer Activity, Disrupts the Cooperation of CCAAT Box/Enhancer-binding Protein β (C/EBPβ) and Co-activator p300. J Biol Chem. 291, 26098–26108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Coulibaly A, Haas A, Steinmann S, Jakob A, Schmidt TJ, Klempnauer KH (2018) The natural anti-tumor compound Celastrol targets a Myb-C/EBPβ-p300 transcriptional module implicated in myeloid gene expression. PLoS One. 10.1371/journal.pone.0190934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rath M, Müller I, Kropf P, Closs EI, Munder M (2014) Metabolism via Arginase or Nitric Oxide Synthase: Two Competing Arginine Pathways in Macrophages. Front Immunol. 5,532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Raber P, Ochoa AC, Rodríguez PC (2012) Metabolism of L-arginine by myeloid-derived suppressor cells in cancer: mechanisms of T cell suppression and therapeutic perspectives. Immunol Invest. 41,614–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF (2013) Type I interferon response and innate immune sensing of cancer. Trends Immunol. 34,67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Upadhyay V, Fu YX (2013) Lymphotoxin signalling in immune homeostasis and the control of microorganisms. Nat Rev Immunol. 13, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Allen E, Jabouille A, Rivera LB, Lodewijckx I, Missiaen R, Steri V, Feyen K, Tawney J, Hanahan D, Michael IP, Bergers G (2017) Combined antiangiogenic and anti-PD-L1 therapy stimulates tumor immunity through HEV formation. Sci Transl Med. 10.1126/scitranslmed.aak9679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nissen MD, Sloan EK, Mattarollo SR β-Adrenergic Signaling Impairs Antitumor CD8+ T-cell Responses to B-cell Lymphoma Immunotherapy. (2018) Cancer Immunol Res. 6, 98–109. [DOI] [PubMed] [Google Scholar]
- [42].Newby AC (2016) Metalloproteinase production from macrophages - a perfect storm leading to atherosclerotic plaque rupture and myocardial infarction. Exp Physiol. 101, 1327–1337 [DOI] [PubMed] [Google Scholar]
- [43].Basset P, Bellocq JP, Wolf C, Stoll I, Hutin P, Limacher JM, Podhajcer OL, Chenard MP, Rio MC, Chambon P (1990) A novel metalloproteinase gene specifically expressed in stromal cells of breast carcinomas. Nature. 348, 699–704 [DOI] [PubMed] [Google Scholar]
- [44].Alaseem A, Alhazzani K, Dondapati P, Alobid S, Bishayee A, Rathinavelu A (2017) Matrix Metalloproteinases: A challenging paradigm of cancer management. Semin Cancer Biol. 10.1016/j.semcancer.2017.11.008 [DOI] [PubMed] [Google Scholar]
- [45].Kim-Fuchs C, Le CP, Pimentel MA, Shackleford D, Ferrari D, Angst E, Hollande F, Sloan EK (2014) Chronic stress accelerates pancreatic cancer growth and invasion: a critical role for betaadrenergic signaling in the pancreatic microenvironment. Brain Behav Immun. 40, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Weber MD, Godbout JP, Sheridan JF (2017) Repeated Social Defeat, Neuroinflammation, and Behavior: Monocytes Carry the Signal. Neuropsychopharmacology. 42, 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Norden DM, Muccigrosso MM, Godbout JP (2015) Microglial priming and enhanced reactivity to secondary insult in aging, and traumatic CNS injury, and neurodegenerative disease. Neuropharmacology. 96(Pt A), 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mittal S, Bjørnevik K, Im DS, Flierl A, Dong X, Locascio JJ, Abo KM, Long E, Jin M, Xu B, Xiang YK, Rochet JC, Engeland A, Rizzu P, Heutink P, Bartels T, Selkoe DJ, Caldarone BJ, Glicksman MA, Khurana V, Schüle B, Park DS, Riise T, Scherzer CR (2017) β2-Adrenoreceptor is a regulator of the α-synuclein gene driving risk of Parkinson’s disease. Science. 357,891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].O’Neill E, Harkin A (2018) Targeting the noradrenergic system for anti-inflammatory and neuroprotective effects: implications for Parkinson’s disease. Neural Regen Res. 13,1332–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Butkovich LM, Houser MC, Tansey MG (2018) α-Synuclein and Noradrenergic Modulation of Immune Cells in Parkinson’s Disease Pathogenesis. Front Neurosci. 12,626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Magistrelli L, Comi C (2019) Beta2-Adrenoceptor Agonists in Parkinson’s Disease and Other Synucleinopathies. J Neuroimmune Pharmacol. 2019 January 7. doi: 10.1007/s11481-018-09831-0. [DOI] [PubMed] [Google Scholar]
- [52].Huber R, Pietsch D, Panterodt T, Brand K (2012). Regulation of C/EBPβ and resulting functions in cells of the monocytic lineage. Cell Signal. 24, 1287–96. [DOI] [PubMed] [Google Scholar]
- [53].Thomas AC, Mattila JT (2014) “Of mice and men”: Arginine metabolism in macrophages. Front Immunol. 5,479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Geelhaar-Karsch A, Schinnerling K, Conrad K, Friebel J, Allers K, Schneider T, Moos V (2013) Evaluation of arginine metabolism for the analysis of M1/M2 macrophage activation in human clinical specimens. Inflamm Res. 62,865–9. [DOI] [PubMed] [Google Scholar]
- [55].Weinberg JB, Misukonis MA, Shami PJ, Mason SN, Sauls DL,. Dittman WA, Wood ER, Smith GK, McDonald B, Bachus KE, Haney AF, Granger DL (1995) Human mononuclear phagocyte inducible nitric oxide synthase (iNOS): analysis of iNOS mRNA, iNOS protein, biopterin, and nitric oxide production by blood monocytes and peritoneal macrophages. Blood. 86,1184–95. [PubMed] [Google Scholar]
- [56].Hart PH, Bonder CS, Balogh J, Dickensheets HL, Donnelly RP, Finlay-Jones JJ (1999) Differential responses of human monocytes and macrophages to IL-4 and IL-13. J Leukoc Biol. 66,575–8. [PubMed] [Google Scholar]
- [57].Kobayashi M, Jeschke MG, Shigematsu K, Asai A, Yoshida S, Herndon DN, Suzuki F (2010) M2b monocytes predominated in peripheral blood of severely burned patients. J Immunol. 185,7174–9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.