Abstract
The thermally dimorphic fungus Histoplasma capsulatum is the causative agent of histoplasmosis, one of the most prevalent endemic mycosis in the Americas. In tropical regions, agro-ecosystems require organic matter replacement, therefore, the use of organic fertilizers has increased disregarding the fact that certain number of such fertilizers might be contaminated with the fungus, and with their handling resulting in human cases and even outbreaks of histoplasmosis. Additionally, in Colombia, chicken manure is the most common raw material used in the production of organic fertilizers. In this work, we reported the isolation of this fungus from chicken manure, and genetically compared with 42 clinical isolates. The genetically compared environmental isolates grouped together with the clinical ones. Our result suggests that chicken manure may be one of H. capsulatum infection sources. Also, the phylogenetic analyses done with other H. capsulatum isolates indicate that the Colombian isolates are widely distributed in the relational tree thus reveling towards the great genetic diversity among the H. capsulatum Colombian isolates.
Keywords: Agriculture, Environmental science, Earth sciences, Histoplasmosis, Organic fertilizers, Composted amendments, MLST technique, Hc100 nested PCR
1. Introduction
Histoplasmosis is a disease caused by the thermally dimorphic fungus Histoplasma capsulatum (H. capsulatum) widely found in all continents except the Antarctica. H. capsulatum infection occurs when a fungus contaminated area is disturbed, causing aerosolization and subsequent inhalation of infective hyphal fragments and microconidia. Once these particles are inhaled and reaching the alveoli, the body temperature stimulates a switch from the mycelium to the yeast form. Development of the disease depends on host factors such as immune system and lung conditions and on fungal factors such as virulence and the amount of inhaled infecting particles. This interaction gives rise to different clinical forms of the disease, ranging from an asymptomatic form to a severe disease that can be lethal (Kauffman, 2009; Knox and Hage, 2010; Scheel and Gómez, 2014).
H. capsulatum was first isolated from soil in 1945 by Emmons (Emmons, 1949), who observed an environmental association between the fungus and some animals such as bats and hens (Emmons, 1958; Emmons 1950). Later, other researchers, through the study of outbreaks, identified sources of infection and validated the relation between H. capsulatum with bats and birds manure (Ajello, 1964; McVeigh and Morton, 1965; Wheat et al., 1981; Morse et al., 1985; Storch et al., 1980; Sacks et al., 1986). These studies made it possible to delineate populations at risk of histoplasmosis, since most cases have been described in ecotourists, speleologists, archaeologists, construction workers, poultry farmers, chicken manure collectors, organic fertilizers producers, farmers, and in general, all whose handle organic fertilizers. For all the above, histoplasmosis has been defined as an occupational and recreational disease (Stobierski et al., 1996; Ashford et al., 1999; Litvintseva et al., 2015; Taylor et al., 2005; Jiménez et al., 2002; Morse et al., 1985; Negroni et al., 2010; Centers for Disease Control and Prevention, 2001; Ordóñez et al., 1997).
In Colombia, histoplasmosis reporting is not mandatory, hence the real prevalence of the disease is unknown. However, the rate of histoplasmosis in Colombia was calculated in 0,6 per 100.000 inhabitants, based on reported cases, statistics of the disease and some population parameters (Alvarez-Moreno et al., 2018). Also, outbreaks and cases have been described. Some of the outbreaks have been related the handling of organic fertilizers and its raw materials. Ordoñez et al., in 1997, made the compilation of 12 outbreaks occurred in the Colombian Andean Region, in 10 of those outbreaks the infection source was identified and in 2 the infection source were the handle of chicken manure contaminated with H. capsulatum (Ordóñez et al., 1997). Then in 2002, Jiménez et al. described an outbreak in a family who get infected after fertilizing a plant with a soil enriched with a H. capsulatum contaminated chicken manure (Jiménez et al., 2002). In Colombia, a total of 18 outbreaks of Histoplasmosis have been described, on those 415 people got expose and 188 (45.3%) were infected. The source of infection were mostly related with environmental sources especially caves visiting and chicken manure handling (Ordóñez et al., 1997; Jiménez et al., 2002; Alvarez-Moreno et al., 2018).
In the tropical agro-ecosystems the reconstitution of the organic matter is needed, then organic fertilizers are widely used. In Colombia, the most common raw material in the organic fertilizers production is chicken manure. Therefore, if H. capsulatum is found associated with chicken manure and also this is the principal raw material for organic fertilizers production in Colombia, then a lot of people may be exposed to the fungus.
In a previous work, we applied a protocol based on the Hc100 nested PCR to search for H. capsulatum in composted organic fertilizers, soils samples from caves and bird excretes. In that work, we were able to detected H. capsulatum DNA in 10% of the tested samples (Gómez et al., 2018). The present work was addressed to isolate H. capsulatum from Colombian environmental samples that tested positive by Hc100 nested PCR, in order to compare Colombian environmental and clinical isolates by using Multi-Locus Sequencing Typing (MLST).
Since the 1980, numerous studies have evaluated the genetic variation of H. capsulatum population by using typing techniques such as Restriction Fragment Long Polymorphism (RFLP) (Keath et al., 1992; Spitzer et al., 1989; Poonwan et al., 1998; Vincent et al., 1986), Variable Number of Tandem Repeats (VNTR) (Carter et al., 2001), Multi-Locus Sequence Typing (MLST) (Kasuga et al., 2003; Teixeira et al., 2016) and Whole Genome Sequencing (WGS) (Sepúlveda et al., 2017). These studies have consistently shown a strong association between phylogenetic clusters and geographic origin of the fungus. The currently accepted taxonomy indicates at least 8 distinct groups of H. capsulatum, based on MLST analysis of four single copy genes including fatty acid desaturase (ole), β tubulin 1 (tub1), ATP ribosylation factor (arf) and H antigen precursor (H-anti) (Kasuga et al., 2003; Teixeira et al., 2016). Nonetheless, as with many microorganisms, taxonomic status remains dynamic and even more recent research arguing H. capsulatum should be broken up into 4 completely separate species: H. mississippiense, H. ohiense, H. suramericanum and H. capsulatum sensu stricto (Sepúlveda et al., 2017).
H. capsulatum Colombian isolates have been analyzed in previous typing studies with 16 clinical ones. Interestingly, these isolates have shown marked differences among them in contrast to other H. capsulatum groups. The works by Kasuga et al. (1999, 2003), and Teixeira et al. (2016) showed a high degree of variability among the Colombian isolates by MLST (Kasuga et al., 1999; Kasuga et al., 2003; Teixeira et al., 2016). The work developed by Kasuga placed the Colombian isolates in Latin American clades (LAm) A, LAm B and 2 solitary lineages (Kasuga et al., 2003). Similarly, the work achieved by Teixeira et al. (2016) placed the Colombian isolates in the phylogenetic species LAm B1, LAm A1 and LAm A2 (Teixeira et al., 2016). Finally, Sepúlveda et al. (2017), evaluated two additional Colombian clinical isolates, MV3 and MZ5, which were classified into two different species: H. capsulatum sensu stricto and H. suramericanum, respectively (Sepúlveda et al., 2017). It is important to point out that these two last isolates are different from the 16 previously used.
In this work, the presence of H. capsulatum in the Colombian environment was achieved and the genetic analyses of both clinical and environmental Colombian isolates was done by MLST using the molecular markers used by Kasuga (Kasuga et al., 1999; Kasuga et al., 2003), the sequences of genes used for diagnosis purposes including the partial sequence of 100 KDa protein (Hc100) (Bialek et al., 2001), M antigen (Ohno et al., 2013) and two Sequences Characteristic Amplified Randomly (SCAR) (Frías De León et al., 2012).
2. Materials and methods
2.1. Search for H. capsulatum Colombian isolates from environmental sources
2.1.1. Environmental samples collection
The samples were send to the Grupo Interdisciplinario de Estudios Moleculares (GIEM), Chemistry Institute, Faculty of Exact and Natural Sciences, Universidad de Antioquia who has the permission from Instituto Colombiano Agropecuario (ICA) through the resolution number: 00005865 from May 19/2017 to perform the physical, chemical and microbiological analysis of organic fertilizers, soil amendments and nitrogen sources for organic fertilizers (S1 Appendix: ICA resolution number: 00005865).
The organic fertilizers were the compost products from an organic matter source (i.e., food scraps, pruning material, straw, or sawdust) and samples of nitrogen source were excrement from animals, primarily poultry or other birds. Between 500 and 1000 g of each environmental sample were send in a plastic zip lock bag and labeled with the date, and the type of sample (raw material, composted material, cave soil, bat excretes or chicken excretes). From the GIEM group the samples were sent to the Grupo de Micología Médica, School of Medicine, of the Universidad de Antioquia to be processed.
2.1.2. DNA extraction from environmental samples
The FastDNA SPIN Kit For Soil® (MP Biomedicals, Santa Ana, CA, USA) was used to extract DNA from environmental samples using the manufacturer's instructions with some modifications. Briefly, the extraction was performed using the supernatant obtained from the suspension of 10 g of sample in 30 ml of saline solution containing 0.001% Tween 80 and 0.1% antibiotics (gentamycin and penicillin; MK®. Cali, Valle, COL). The suspension was stirred vigorously for 1 min and allowed to settle for 20 min, this procedure was repeated twice. After the largest particles had settled, 300 μl of the supernatant was collected for DNA extraction. The other modification consisted of an increased contact time between the sample and kit reagents (Gómez et al., 2018).
2.1.3. Hc100 nested PCR assay
Two sets of specific primers targeting a fragment of the Hcp100 gene were used (Bialek et al., 2002; Porta et al., 1999). The conditions of the assay for organic fertilizers were described by Gómez et al. (2018) (Gómez et al., 2018). In the first reaction, 10μl of DNA was added to 50μl total reaction mix with a final concentration of 10 mM Tris - HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 1U Taq polymerase (Thermo Scientific, Ref: EP0402. Waltham, M A, USA), 0.2 mM of each primer (HcI-HcII), and 0.2mm deoxynucleoside triphosphate (Thermo Scientific, Ref: R0181. Waltham, M A, USA). The mixture for the nested PCR was similar, except using 2μl of the product of the first PCR and 0.2mM of the inner primers (HcIII-HcIV). Temperatures and times for the first reaction, containing external primers HcI-HcII were one cycle at 94 °C for 5 min; 35 cycles at 94 °C for 30s, 66 °C for 1 min, and 72 °C for 1 min, and then a final cycle of 72 °C for 5 min. For the second step, the reaction consisted of a cycle at 94 °C for 5 min; 35 cycles at 94 °C for 30s, 65 °C for 30s, and 72 °C for 1 min, and then a final extension cycle at 72 °C for 5 min. Synthesis of the primers was performed by Integrated DNA Technologies (IDT, Coralville, IA, USA) (Gómez et al., 2018).
2.1.4. Agarose gel electrophoresis
Agarose gels (Amresco, Ref.: N605-500G, Solon, OH, USA) prepared at 1.5% in Tris-borate EDTA buffer (TBE) were used to visualize the Hc100 nested PCR amplification products of the Hc100 nested PCR. Electrophoresis was performed for 40 min at 80 V with 10 μl of the PCR product and 5 μl of GelRed Nucleic Acid Gel stain (Biotum, Ref.: 41003, Hayward, CA, USA) in each lane. The bands were visualized and documented in a UV transilluminator (Doc Gel ™, BioRad. Hercules, CA, USA).
2.2. H. capsulatum culture recovery from environmental samples positive in the Hc100 nested PCR
The cultures were performed using the supernatant obtained from the suspension of 10 g of sample in 30 ml of sterile saline solution 0.85% containing 0.001% Tween 80 (Sigma, ref. P4780. St. Louis, MO, USA), 100 pg/ml oxytetracycline (MK®. Cali, Valle, COL), and 0.1% antibiotics (gentamycin and penicillin; MK®. Cali, Valle, COL). The suspension was stirred vigorously for 1 min and allowed to settle for 20 min, this procedure was repeated twice. After the largest particles had settled, the supernatant was collected and serial dilutions 1:10, 1:100 and 1:1000 were performed; from each dilution 200 μl were plated by duplicate in Mycosel agar (BBLTM; ref. 211462, Franklin Lakes, NJ, USA). Cultures were incubated at room temperature for 2 months and plates were visually inspected on days 5, 10, 15, 30, 45 and 60 in order to examine colonies with H. capsulatum appearance.
The colonies identified as H. capsulatum were cottony, raised, hard-edged, white, cream or light coffee and with slow growth. Each candidate colony was subcultured on a fresh Mycosel agar and the microscopic examination was done with Lactophenol blue to observe the characteristics indicative of H. capsulatum such as thin septate hyaline hyphae, thin wall microconidia and tuberous macroconidia. Colonies with these features were transformed into the yeast form using the procedure described in Gómez et al. (2018) (Gómez et al., 2018). The positive candidates were confirmed with sequencing of Hcp100 gene.
2.3. H. capsulatum mouse model recovery from environmental samples testing positive in the Hc100 nested PCR
2.3.1. Assays conducive to stablish the sensitivity of the mouse model
The efficacy of isolating H. capsulatum from the environmental sample was evaluated inoculating the mice with organic fertilizers samples contaminated with 3000 CFU/ml of H. capsulatum in its mycelial form (Gómez et al., 2019 in preparation manuscript). 1:10, 1:100 and 1:1000 dilutions were prepared from the contaminated sample supernatant and from each dilution, 2 mice were inoculated through the peritoneal cavity with 500 μl of the above inoculum. The animals maintenance and harvest are explained below.
2.3.2. Environmental samples preparation for mouse inoculation
Hc100 nested PCR positive samples were prepared by suspending 10 g of sample in 100 ml of saline solution containing 0.1% antibiotics (gentamycin and penicillin; MK®. Cali, Valle, COL). The suspension was stirred vigorously for 1 min and allowed to settle for 20 min. This procedure was repeated twice. After the last stirring, the suspension was allowed to settle 40 min; then, the supernatant was collected. The supernatant was diluted 1:10, 1:100 and 1:100 and each dilution was used for mice inoculation. The positive control was prepared for each environmental sample tested; this consisted in an aliquot of the supernatant mixed 1:1 with a suspension of 3000 CFU/ml of H. capsulatum in the mycelial form.
2.3.3. Maintenance and inoculation of mice
Specific pathogen-free male Balb/C mice (6–8 week old, 18–22 g weight) were obtained from the Laboratory Animal Center at the Corporación para Investigaciones Biológicas (CIB, Medellin, Colombia). Mice were housed in a caging system (RAIR HD Super Mouse 750TM Racks system, Lab Products, Inc. Seaford, DE, USA) equipped with high efficiency particulate air (HEPA) filters with controlled room temperature at 20–24 °C; and 12-h light/dark cycles, under sterile conditions and provided with sterilized food and water ad libitum.
Two mice were inoculated in the peritoneal cavity with 500μl of the supernatant for the 1:1, 1:10, 1:100 and 1:1000 dilutions of the positive samples and the positive control, using 10 mice to evaluate each environmental sample. The mice were treated with antibiotics (0.04 mg/ml gentamycin and 640 IU penicillin solution) one day before the challenge and 5 days following the sample inoculation (Zeidberg et al., 1952). After 17 days, mice were euthanized and the spleen, liver and lungs were extracted. The organs were macerated and cultured in Brain Hearth Infusion agar (BHI Agar, BBL TM; ref. 211065. Franklin Lakes, NJ, USA) supplemented with 1% glucose (Sigma, Ref. G5400. St. Louis, MO, USA), 0.001% L-cysteine (Sigma, ref. C - 7755. St. Louis, MO, USA) and 5% anticoagulated blood, then cultures were incubated at 37 °C in 5% CO2 atmosphere for two weeks and at room temperature in Mycosel agar for 2 months, in order to look for H. capsulatum compatible yeast and mold colonies, respectively.
2.3.4. Ethics statement
This study was performed according to recommendations of European Union, Canadian Council on Animal Care, and Colombian regulations (Law 84/1989, Resolution No. 8430/1993). The protocol was approved by the Ethics Research Committee at the CIB (Comité de Ética, electronic consultation on July 24th, 2014).
No specific permissions were required for the collection of the samples analyzed in this study. Also we did not perform field studies that involve endangered or protected species.
2.4. Colombian clinical isolates collection of H. capsulatum
2.4.1. H. capsulatum culture
A total of 42 human clinical isolates of H. capsulatum were obtained from the Corporación para Investigaciones Biológicas (CIB, Medellin, Colombia) (S2 Table). All isolates were transformed into the yeast form following the procedure were described in Gómez et al. (2018) (Gómez et al., 2018).
2.4.2. DNA extraction from yeast phase of H. capsulatum isolates
The phenol-chloroform-isoamyl alcohol method was used for DNA extraction from the H. capsulatum isolates in their yeast phase (Sambrook and Russell, 2001). The DNA concentration and quality were evaluated with a NanoDrop ND1000 spectrometer (Thermo Scientific, Wilmington, DE, USA) and 1% agarose gel electrophoresis, respectively.
2.5. Molecular epidemiology comparisons among the H. capsulatum isolates
2.5.1. MLST technique
The characteristics of the technique to compare H. capsulatum isolates were described by Kasuga et al. (1999) (Kasuga et al., 1999). The primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA). S3 Table describes the characteristics of each primer.
The standardized conditions for each MLST PCR assay and primers used are described on S3 Table.
2.5.2. Isolates comparison through other H. capsulatum genome regions
Given the difficulties associated with obtaining H. capsulatum isolates from environmental samples in microbiological culture, molecular diagnostic protocols originally designed to detect H. capsulatum DNA on clinical samples were standardized and applied to environmental samples. To fulfil this goal only the first round of the Hc100 Nested PCR and M antigen nested PCR assays were done given the larger size of the amplification product. Additionally, two PCR assays targeting two Sequences Characteristic Amplified Randomly (SCAR) in the H. capsulatum genome were also used. S3 Table describes the characteristics of each primer and the PCR reactions.
2.5.3. Sequences analyses
Bidirectional sequencing of the PCR amplification products was done using the chain termination method. This method used ABI 3730XL DNA sequencing technology with quality criteria QV20 (Macrogen Inc., Geumcheon-gu, Seoul, Korea). The obtained sequences were edited manually based on their chromatograms. The mold and complementary sequences were aligned and the consensus sequence was obtained using Geneious 11.0.2. Software. BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to verify that the sequenced PCR products belonged to H. capsulatum. The information of the 80 sequences was obtained from TreeBASE #1063, the matrix was separated gene by gene according to the length and the order reported by the author (Kasuga et al., 1999). Additionally, the sequences of 85 isolates were downloaded from NCBI using the access codes by the Batch Entrez program. Additionally, the sequences were aligned and concatenated using Geneious 11.0.2. software, and the matrix was analyzed using Maximum Likelihood analysis implemented in IqTree 1.4.4. software and branch fidelity was assessed via 10000 Ultra-fast boot. The phylogenetic tree obtained was visualized using FigTree v.1.4.3.
3. Results
3.1. Detection of H. capsulatum in environmental samples by Hc100 nested PCR
A total of 393 samples were collected between 2010 and 2017, of which 273 (69.5%) were composted fertilizers and organic amendments. A total of 120 (30.5%) samples did not have composting treatment. In the last group, 21 (5.3%) samples were from caves floors and/or bat droppings while a total of 99 (25.2%) were from bird depositions or chicken manure.
Out of the 393 environmental samples tested, a total of 39 (9.9%) were positive by Hc100 nested PCR. Of these positive samples, 21 (5.3%) were composted fertilizers and organic amendments, 17 samples (4.3%) were from bird feces or poultry manure and one (0.3%) from a cave (floor/bat droppings). Interestingly, we found higher positivity rates for samples without composting treatment 18/120 (15%) as compared with samples that had been previously composted 21/273 (7.7%).
3.2. Recovery of H. capsulatum in microbiological cultures from Hc100 nested PCR positive environmental samples
The 39 positive samples by the Hc100 nested PCR were seeded into Mycosel culture medium. However, only one sample taken from non-composted chicken manure produced viable H. capsulatum cultures. From this single environmental sample, three H. capsulatum colonies were obtained in different culture petri dishes, despite that probably those could be the same isolate recovered three times, the 3 colonies were used as different individuals for molecular analyses. Interestingly, while 2 of these isolates were morphologically similar, the third was distinct and did not fully switch to the yeast form. Abundant growth of bacteria and molds, such as Penicillium spp, Aspergillus spp, Trichoderma spp and Geotrichum spp were also observed in the H. capsulatum cultures from this environmental sample.
3.3. Recovery of H. capsulatum in the mouse model from Hc100 nested PCR positive environmental samples
In the experiments done to evaluate the sensitivity of the mouse model, H. capsulatum was recovered from mice inoculated with environmental samples artificially contaminated with 3000 CFU/ml of this fungus.
In the assays performed to obtain environmental isolates, the 39 positive samples by Hc100 nested PCR were also used to inoculate mice. From the mice challenged with each positive sample, around 9% of them died in the first 3 days after inoculation. Despite the use of broad-spectrum antibiotics in the sample preparation and the treatment of the mice with antibiotics before and after the inoculation, we were not able to obtain environmental isolates of H. capsulatum from the positive environmental samples.
3.4. MLST analysis of Colombian H. capsulatum isolates
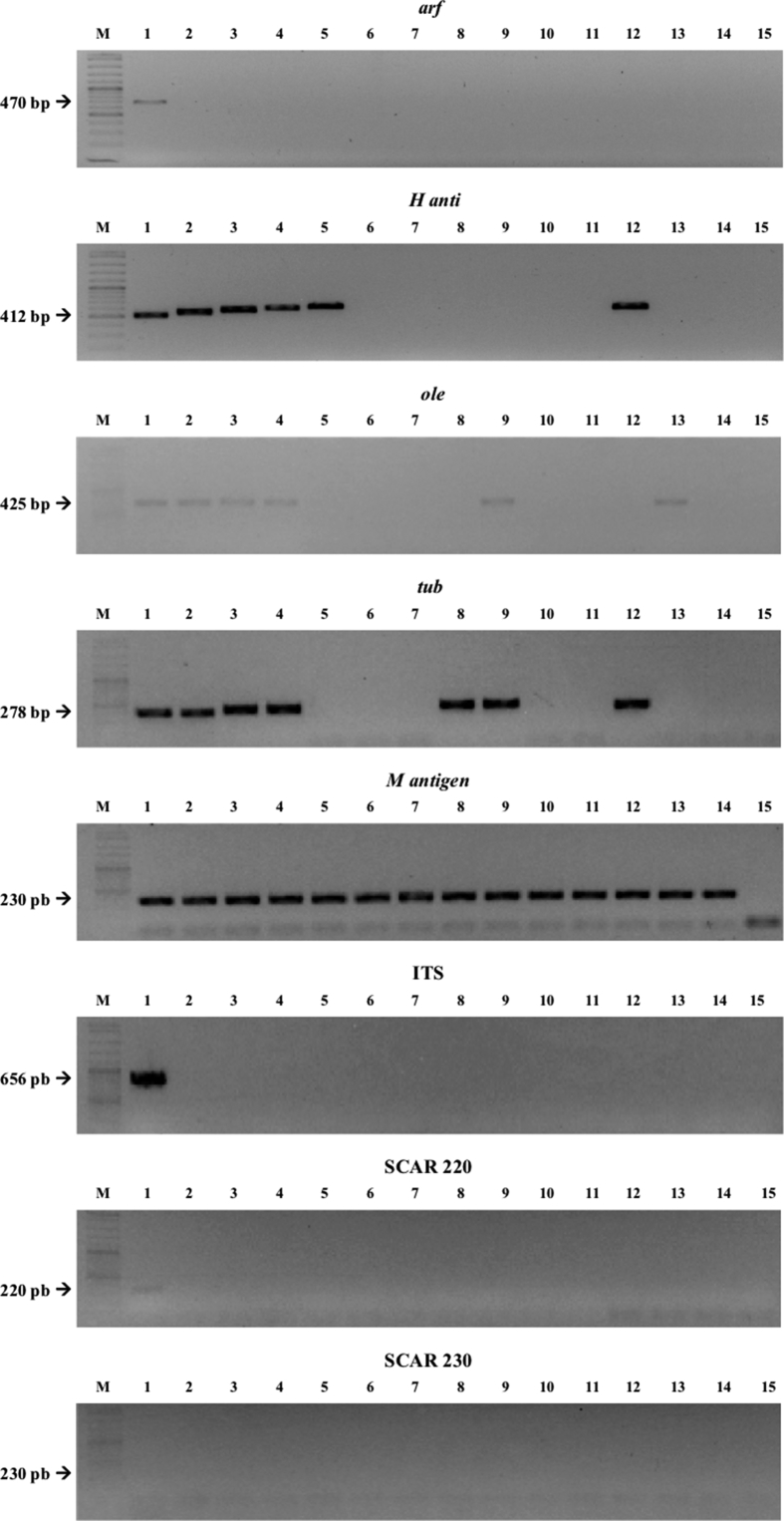
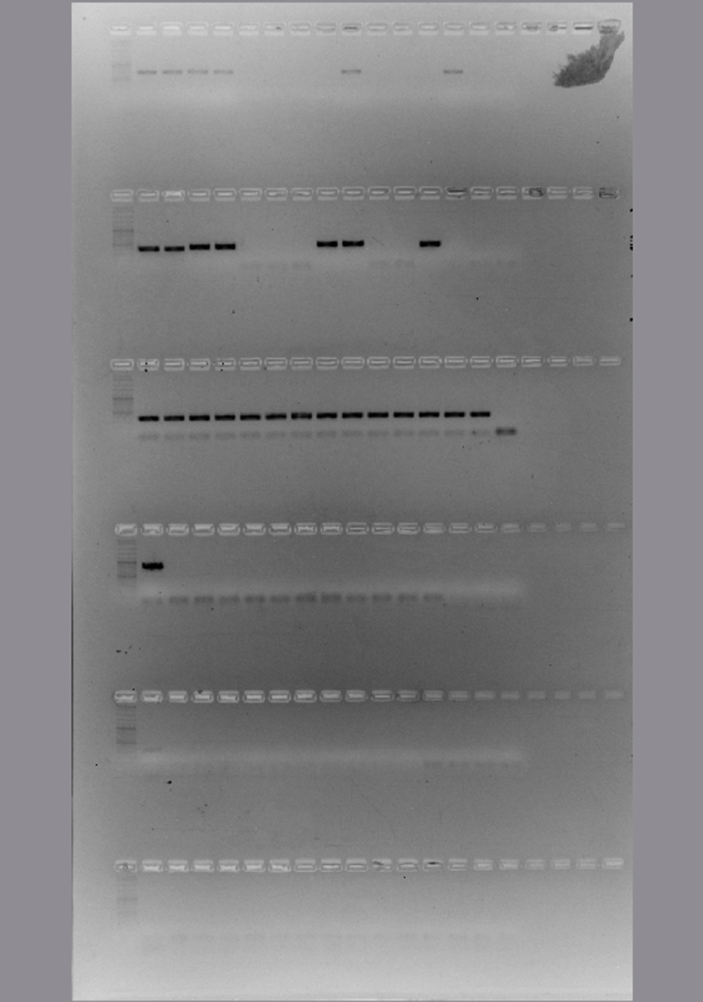
The conditions of each PCR protocol were standardized (S3 Table). Then the sensitivity, specificity and the ability to detect H. capsulatum DNA from environmental samples were evaluated (Fig. 1). All protocols, including both typing and diagnostic methods, were tested to detect H. capsulatum from environmental samples. The H-anti, ole, tub1 and M antigen PCR reactions showed an acceptable detection limit (Fig. 1, Lanes 1–7) with spiked samples with H. capsulatum DNA. In the PCR reactions done to check the specificity (Fig. 1, Lanes 8–13), none of these methods produced an amplicon from the 39 samples that were positive with the Hc100 nested PCR.
Fig. 1.
Detection limit, specificity and the ability to detect H. capsulatum DNA in environmental samples with the MLST analysis. Lane M: marker. Sensitivity assay with H. capsulatum DNA (from lane 1 to 7): lane 1: 2 ng/μl, lane 2: 1 ng/μl, lane 3: 500 pg/μl, lane 4: 200 pg/μl, lane 5: 100 pg/μl, lane 6: 50 pg/μl, lane 7: 20 pg/μl. Specificity assay with DNA from different fungi (from lane 8 to 13): lane 8: Paracoccidioides brasiliensis sensu stricto (Pb 18), lane 9: Paracoccidioides spp. (Pb 339), lane 10: Paracoccidioides restrepiensis (Pb 60855), lane 11: Coccidioides spp, lane 12: Emmonsia crecens, lane 13: Blastomyces dermatitidis, lane 14: DNA from a raw chicken manure positive by Hc100 nested PCR and culture in Mycosel, lane 15: PCR negative control with sterile water. Non edited figure can be reviewed in the supplemental material (S4, S5 and S6 Figures).
The standardized PCR protocols were used to obtain the sequences from all 45 Colombian isolates (42 human clinical and 3 environmental isolates) (S2 Table). After the PCR products were sequenced, the consensus sequence was obtained. The length for each locus was as follows: arf 478 bp, H-anti 413 bp, ole 428 bp, tub1 263 bp, Hcp100 209 bp, M antigen 276 bp, scar 220 208 bp, scar 230 262 bp, ITS 605 bp.
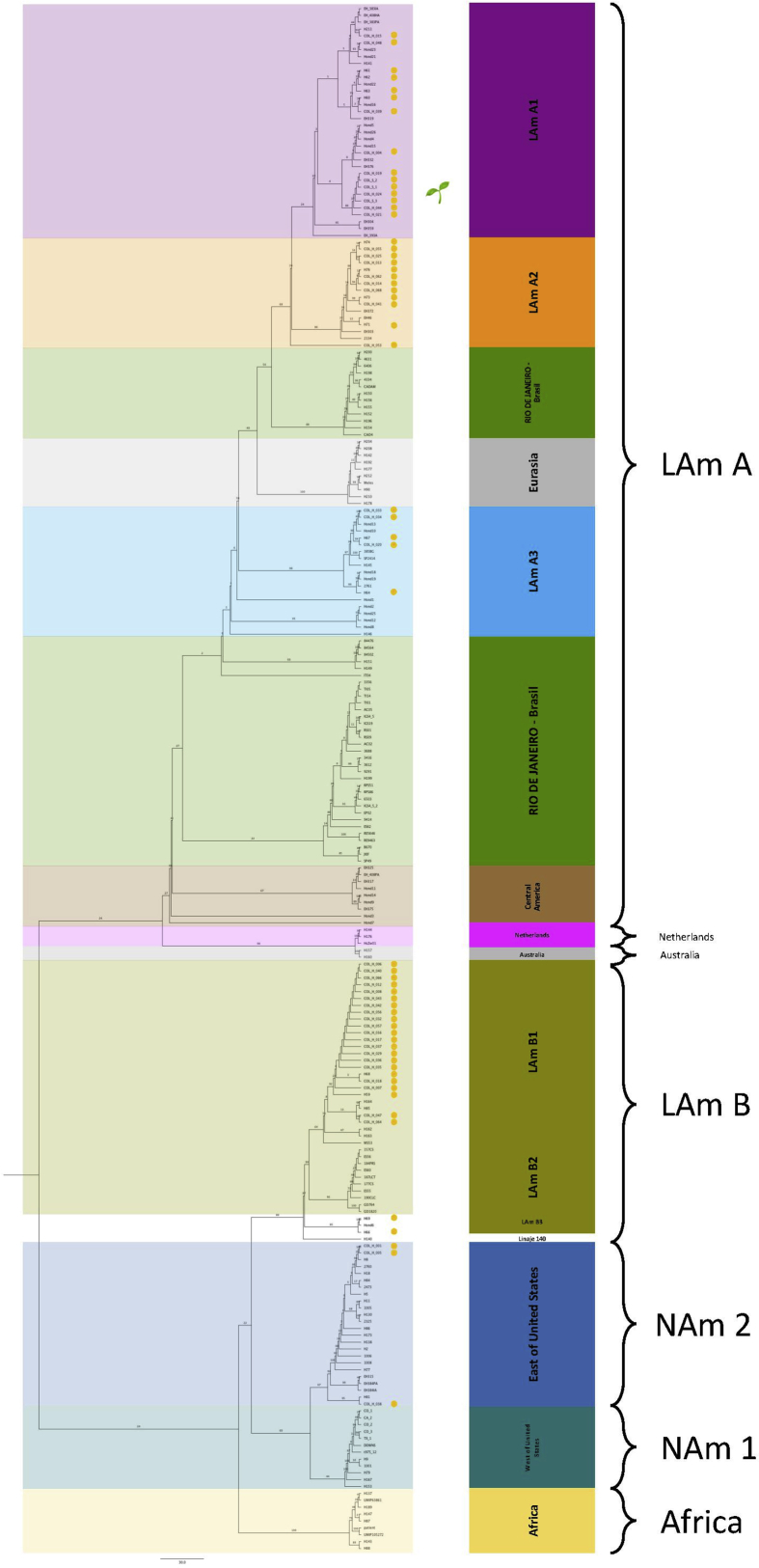
Subsequently, a matrix was constructed using the sequences of the genes in the following order: arf, H-anti, tub1 and ole. The matrix included a total of 225 sequences including the 45 obtained in this study and the other 180 from databases, had a length of 1582 bp and was partitioned by the type of sequence itself: CDS, exon or ITS. The evolutionary model was established as K2+G4 (Lemey et al., 2009). Seventeen phylogenetic groups were identified with similar distribution as reported by Kasuga et al., 2003 and Teixeira et al. (2016) (Kasuga et al., 2003; Teixeira et al., 2016) (Fig. 2).
Fig. 2.
Genetic comparison of H. capsulatum Colombian isolates. The groups were separated when bootstraps were over 70%. The contrast between our results and those of Teixeira et al. (2016) (Teixeira et al., 2016) (middle list) and of the Kasuga et al. (2003) (Kasuga et al., 2003) (external list), are marked using the same color pattern. Isolates from Colombia are shown with a yellow circle and they are distributed among LAm A2, LAm A1, LAm B1, LAm B2 and NAm 2 phylogenetic species. A leaf symbol shows the environmental isolates location; they have the same genetic information which lead to conclude the three colonies obtained from the same environmental same sample are the same strain.
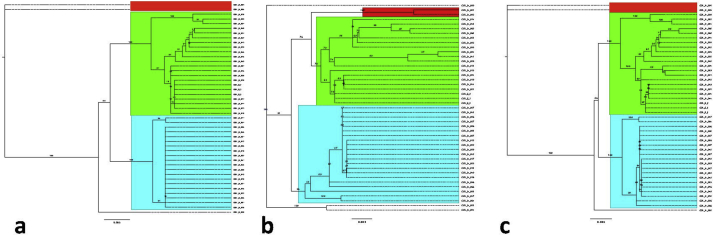
Additionally, three more matrices were built that only included the 45 Colombian isolates, because from those were obtained the sequences corresponding to Hcp100 gene, M antigen gene, SCAR 220, SCAR 230 and the ITS region. In these analyses, the first matrix included arf, H-anti, tub1 and ole. The second matrix included the sequences obtained using the PCR products of the diagnostic genes targets, those sequences were aligned in the order as shown above and a third matrix included all nine sequences. In all matrices, the genetic information of the three colonies from the environmental sample have the same genetic information showing that those are the same strain, additionally, they grouped together with the clinical isolates, proving their genetic homology. When the 3 different matrices were analyzed, the trees had the same topology, thus suggesting that the classical genes are adequate to achieve the genetic comparison of the H. capsulatum isolates (Fig. 3). According to the genes characteristics for MLST, the other regions of the genome tested had a lesser number of informative sites per gene than the classical targets (Table 1).
Fig. 3.
The phylogenetic analysis of the H. capsulatum Colombian isolates. All trees showed the same topology although they were built with different group of genes in their order, as follows: a. arf, H-anti, tub1 and ole. b. Hcp100 gene, M antigen gene, SCAR 220, SCAR 230 and the ITS region and c. Consensus tree for all 9 sequences arf, H-anti, tub1, ole, Hcp100 gene, M antigen gene, SCAR 220, SCAR 230 and the ITS region.
Table 1.
Other genome sequences evaluated for H. capsulatum genetic typing.
| Sequence | Length | Informative sites | Percentage informative sites |
|---|---|---|---|
| arf | 435 | 7 | 1,6 |
| H-anti | 373 | 6 | 1,6 |
| ole | 383 | 3 | 0,7 |
| tub1 | 218 | 3 | 1,3 |
| ITS | 608 | 4 | 0,6 |
| Hcp100 | 184 | 1 | 0,5 |
| M antigen | 276 | 7 | 2,5 |
| SCARS 220 | 180 | 5 | 2,7 |
| SCARS 230 | 152 | 1 | 0,6 |
For each sequence the bp length is described for analysis, as well as the quantity and the percentage of the informative sites.
4. Discussion
In this study through the phylogenetic analysis of over 3 environmental and 42 human clinical isolates, we have found that H. capsulatum isolates in Colombia show a particularly high degree of variation when compared to other geographic origins isolates. Also, the genetic comparison showed that the three colonies obtained from the same environmental sample are the same strain and grouped together with the clinical ones. These results suggest that chicken manure represents one of the H. capsulatum infection sources.
By means of the Hc100 nested PCR, we were able to detect H. capsulatum in 39 (9.9%) of the 393 of environmental samples studied. The fact that among the positive samples, 21.9% were found in non-composted samples compared to only 7.7% of those composted, suggests that a properly done composting process could reduce the risk of exposure to H. capsulatum when manipulating the final product of the organic fertilizer. This is because the composting process is a spontaneous decomposition of organic matter, mainly aerobic, in which mainly bacteria and fungi participate. The organic matter is transformed into a fertilizer free of toxins and pathogenic microorganisms due to the action of the saprophyte microorganisms. First, they are better competitors for space and nutrients and second due to their metabolic activity, they generate an increase in the temperature. These two actions reduce the pathogenic microorganisms population (Zucconi and De Bertoldi, 1987; Bernal et al., 2009; Gómez, 2006). In this sense, at the beginning of the composting process, there is a high risk infectious source, since we achieved the isolation of H. capsulatum from a raw/not composted chicken manure which happens to be the most commonly used raw material in organic fertilizers in Colombia (Bernal et al., 2009).
For the isolation of H. capsulatum from environmental samples, the mouse model has been considered the golden standard technique, but, in this study, we were unable to isolate the fungus using this model from the 39 samples previously marked as positives by the Hc100 nested PCR. This procedure is, additionally, time-consuming, expensive, requires training in handling the mice and according to results of previous studies, the success rates are between 0-50% (Emmons, 1949; Zeidberg et al., 1952; Hazen et al., 1956; Emmons, 1958; Ajello, 1964; Brandsberg et al., 1969; Disalvo et al., 1970; Moncada et al., 1989; Jiménez et al., 2002; Taylor et al., 2005; Huhn et al., 2005). Furthermore, we found that the high microbial background in our environmental samples leads to sepsis and subsequent high death rates, even thou the mice were treated with antibiotics before the challenge to prevent the sepsis. Moreover, the recovery of H. capsulatum using the mouse model requires that the fungus overcomes the saprophytic microorganisms, evades the immune system and establishes the infection in the mammalian host in order that it may be isolated from infected tissues (Emmons, 1949; Zeidberg et al., 1952; Hazen et al., 1956; Emmons, 1958; Ajello, 1964; Brandsberg et al., 1969; Disalvo et al., 1970; Moncada et al., 1989; Jiménez et al., 2002; Taylor et al., 2005; Huhn et al., 2005). One challenge associated with this method is that if the fungus has been in the environment for a long time without being exposed to a mammal host, the genes associated with pathogenicity may not be expressed. For example, when the α-(1–3)-glucan cell wall content has been studied in H. capsulatum, the fungus has been separated into chemotypes I and II; this classification has been correlated also with its virulence. Most virulent H. capsulatum isolates described are chemotype II, but can lose virulence spontaneously by successive passaging in microbiological cultures by loss of α-(1–3)-glucan in their cell wall (Rappleye et al., 2007; Sepúlveda et al., 2014; Holbrook and Rappleye, 2008). Nowadays, it is possible to compare isolates using the proteomic analysis. This approach could be used to elucidate genes related to pathogenicity by comparing gene expression and activity in environmental and clinical isolates (Kasap et al., 2017; Montagna et al., 2018; Jun et al., 2013). If the virulence genes have been not activated in the environmental isolates in the same form as in the clinical isolates, then the use of the mouse model for the isolation of H. capsulatum from environmental samples needs to be reconsidered as the gold standard technique.
In contrast to the above findings, we were able to isolate H. capsulatum by the direct culture in 1 of the 39 studied environmental samples previously marked as positives by the Hc100 nested PCR. In cases with high microbial backgrounds such as chicken manure samples, we were able to limit contamination by adding antibiotics to the culture media. Even though this method also had a low success rate, the recovery of just a few isolates proved to be very valuable for our study. Thus, we were able to demonstrate from our analysis of the 3 isolates from the non-composted chicken manure sample that viable fungus was present in the unprocessed materials. This has implications for worker safety in industrial contexts, and it is now required take actions regarding protective measures such as wearing high efficient respirators, gloves, boots, long sleeve shirts and showering at the end of the day. To follow these few recommendations will be best way to reduce the occurrence of occupational outbreaks of histoplasmosis (Lenhart et al., 2004). In order to promote the knowledge about histoplasmosis and how to avoid acquiring the infection with H. capsulatum, we wrote a booklet to teach people about the disease, its signs and symptoms, for those usually exposed and how to protect themselves (Gómez et al., 2017).
Due to the difficulties associated with obtaining isolates in culture from environmental samples, we tested the diagnostic PCR protocols used with clinical samples to obtain sequences directly from the environmental samples that could facilitate comparisons between clinical and environmental H. capsulatum isolates. However, none of the 8 PCR evaluated with the DNA obtained from the environmental sample positive with the Hc100 nested PCR produced an amplicon. Also, given that the trees obtained with the three different matrices analyzed had the same topology, the classical genes proposed by Kasuga et al. are adequate to achieve the genetic comparison of the H. capsulatum population (Bialek et al., 2002; Frías De León et al., 2012; Ohno et al., 2013; Kasuga et al., 1999; Kasuga et al., 2003; Matos Guedes et al., 2003).
Then, our successful recovery of three H. capsulatum isolates from the chicken manure sample enabled us to genetically compare these to the 42 human clinical isolates from Colombia, as well as to other isolates from around the world by the classical MLST (Kasuga et al., 1999; Kasuga et al., 2003). Our MLST analysis revealed that the isolates from chicken manure are most closely related to the clinical strains in Colombia. This finding implicates chicken manure as a source for exposure and infection by H. capsulatum. Previous studies of histoplasmosis outbreaks have used similar approaches and have also been able to establish associations between environmental and clinical H. capsulatum isolates (Keath et al., 1992; Kasuga et al., 2003; Teixeira et al., 2016).
Our work confirmed the high diversity of Colombian H. capsulatum isolates compared to isolates in other geographical regions (Teixeira et al., 2016; Kasuga et al., 2003; Sepúlveda et al., 2017). Although, it is not yet certain why there is such a high diversity, it is likely related to the ecological and geographical characteristics of Colombia. This country has a unique location that serves a crossing point for migratory birds and bats, which can carry and seed the fungus in the environment. This mechanism of dispersal has been demonstrated by studies of the bat Tadarida brasiliensis, whose range correlates well with the epidemiological map of histoplasmosis. This mechanism may explain the presence of H. capsulatum Colombian isolates in the clades that traditionally embrace isolates from other countries visited by bats in their migratory routes in the United States and Brazil. Furthermore, H. capsulatum has also been isolated from tissues and excreta from both birds and bats (González-González et al., 2012; Chávez-Tapia et al., 2005; Dias et al., 2011; Moncada et al., 1989; Bartlett et al., n.d.; Reyes-Montes et al., 2009; Tesh and Schneidau, 1966). Also, since tropical soils tend to be nutritionally poor, organic fertilizers made with chicken manure sources are commonly used, thus creating numerous possible habitats for H. capsulatum that overlaps with human activities. Our data implicates a mechanism of clinical infection originating from non-composted chicken manure sources. It is therefore likely that the risk of acquiring histoplasmosis can be reduced through complete composting process of chicken manure, before organic fertilizer will be handled by people. Risk may also be lower by the use of personal protection equipment when the people get exposed to risky activities like caves visiting, demolition or cleaning of old buildings or manipulation of excreta of birds and bats. Following these recommendations, histoplasmosis in individuals or populations could be prevented.
Declarations
Author contribution statement
Luisa Fernanda Gomez-Londoño: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Myrtha Arango-Arteaga, Juan Guillermo McEwen-Ochoa, Carlos Alberto Peláez-Jaramillo: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Oscar M Gomez-Guzman: Analyzed and interpreted the data; Wrote the paper.
Alejandra Zuluaga, Jose Miguel Acevedo-Ruiz: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Maria Lucia Taylor: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Maria Jimenez- Alzate: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Comité para el Desarrollo de la Investigación (CODI) project code 2014-1010 of Universidad de Antioquia, Medellín, Colombia and Departamento Administrativo de Ciencia, Tecnología e Innovación (Colciencias), Bogotá, Colombia, National Doctorate Program announcement 647 of 2014 to LFG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interest Statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at Genbank under accession numbers: ADP ribosylation factor (arf) MH338036 to MH338080, H antigen precursor (anti-H) MH122839 to MH122883, Delta-9 fatty acid desaturase (ole1) MH338126 to MH338170, Alpha-tubulin (tub1) MH338081 to MH338125, Hcp100 gene (100 kDa protein) MH122794 to MH122838, internal transcribed spacer (ITS) MH339542 to MH339586, SCAR markers 220 and 230 MH348521 to MH348610.
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2019.e02084.
Acknowledgements
The authors would like to acknowledge the members of the participant research groups for their feedback and improvement suggestions. We thank to Dra. Angela Restrepo and David Sexton for English editing of the manuscript. Also, thanks to Juan David Puerta for providing training and advising in handling the mice.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
ICA resolution number: 00005865.
List of the Histoplasma capsulatum Colombian isolates used in this work.
Table. Characteristics of the sequences and the structure of each primer and PCR reaction conditions. Reagents were as follow, the Deoxynucleoside triphosphates from Thomas Scientific, Swedesboro, NJ., the synthesis of the primers by Integrated DNA Technologies (IDT, Coralville, IA, USA), and the Taq polymerase form Thomas Scientific, Swedesboro, NJ.
S4 Fig (1).tiff.
Non edited figure of Detection limit, specificity and the ability to detect H. capsulatum DNA in environmental samples with arf and H anti.
S5 Fig.tiff.
Non edited figure of Detection limit, specificity and the ability to detect H. capsulatum DNA in environmental samples with ole, tub, M antigen, ITS, SCAR 220 and SCAR 230.
S6 Fig.tiff.
Non edited figure of Detection limit, specificity and the ability to detect H. capsulatum DNA in environmental samples with SCAR 220 and SCAR 230.
References
- Ajello L. “[Relationship of Histoplasma capsulatum with hens and other birds].” Boletin de La Oficina Sanitaria Panamericana. Pan Am. Sanitary Bur. 1964;56(3):232–236. http://www.ncbi.nlm.nih.gov/pubmed/14146787 [PubMed] [Google Scholar]
- Alvarez-Moreno Carlos Arturo, Cortes Jorge Alberto, Denning David W. Burden of fungal infections in Colombia. J. Fungi (Basel, Switzerland) 2018;4(2) doi: 10.3390/jof4020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford D a, Hajjeh R a, Kelley M.F., Kaufman L., Hutwagner L., McNeil M.M. Outbreak of histoplasmosis among cavers attending the National Speleological Society annual convention, Texas, 1994. Am. J. Trop. Med. Hyg. 1999;60(6):899–903. doi: 10.4269/ajtmh.1999.60.899. http://www.ncbi.nlm.nih.gov/pubmed/10403317 [DOI] [PubMed] [Google Scholar]
- Bartlett, P C, R J Weeks, and L Ajello. n.d. Decontamination of a Histoplasma capsulatum-infested bird roost in Illinois.” Arch. Environ. Health 37 (4): 221–223. http://www.ncbi.nlm.nih.gov/pubmed/7114902. [DOI] [PubMed]
- Bernal M.P., Alburquerque J.A., Moral R. Composting of animal manures and chemical criteria for compost maturity assessment. A review. Bioresour. Technol. 2009;100(22):5444–5453. doi: 10.1016/j.biortech.2008.11.027. [DOI] [PubMed] [Google Scholar]
- Bialek Ralf, Fischer J., Feucht Antje, Najvar Laura K., Dietz Klaus, Knobloch J., Graybill J.R. Diagnosis and monitoring of murine histoplasmosis by a nested PCR assay. J. Clin. Microbiol. 2001;39(4):1506–1509. doi: 10.1128/JCM.39.4.1506-1509.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek Ralf, Feucht Antje, Aepinus Christian, Just-Nübling Gudrun, Robertson Valerie J., Knobloch Jürgen, Rolf Hohle. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J. Clin. Microbiol. 2002;40(5):1644–1647. doi: 10.1128/JCM.40.5.1644-1647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsberg J.W., Weeks R.J., Hill W.B., Piggott W.R. A Study of Fungi Found in Association with Histoplasma Capsulatum: three bird roosts in S. E. Missouri, U.S.A. Mycopathologia et Mycologia Applicata. 1969;38(1):71–81. doi: 10.1007/BF02051677. http://www.ncbi.nlm.nih.gov/pubmed/5817912 [DOI] [PubMed] [Google Scholar]
- Carter D.A., Taylor J.W., Dechairo B., Burt A., Koenig G.L., White T.J. Amplified Single-Nucleotide Polymorphisms and a (GA)(n) Microsatellite Marker Reveal Genetic Differentiation between Populations of Histoplasma Capsulatum from the Americas. Fungal Genet. Biol. FG & B. 2001;34(1):37–48. doi: 10.1006/fgbi.2001.1283. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . Vol. 50. 2001. (Public Health Dispatch: Update: Outbreak of Acute Febrile Respiratory Illness Among College Students --- Acapulco, Mexico, March 2001). [Google Scholar]
- Chávez-Tapia Catalina Beatriz, Peña-Sandoval Gabriela, Rodríguez-Arellanes Gabriela, Reyes-Montes Maria del Rocio, Duarte-Escalante Esperanza, Taylor Maria Lucia. Aislamiento de Histoplasma Capsulatum En Los Murcielagos Desmodus Rotundos (No Migratorio) y Tadarida Brasiliensis (Migratorio de Larga Distancia): Primeros Registros En Mexico. Rev. Mex. Micol. 2005;20:61–70. [Google Scholar]
- Frías De León María Guadalupe, Arenas López Gabina, Taylor Maria Lucia, Acosta Altamirano Gustavo, Reyes-Montes María Del Rocío. Development of Specific Sequence-Characterized Amplified Region Markers for Detecting Histoplasma Capsulatum in Clinical and Environmental Samples. J. Clin. Microbiol. 2012;50(3):673–679. doi: 10.1128/JCM.05271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias M A Galvão, Zancopé Oliveira R.M., Giudice M.C., Netto H. Montenegro, Jordão L.R., Grigorio I.M., Rosa A.R. Isolation of Histoplasma Capsulatum from Bats in the Urban Area of São Paulo State, Brazil. Epidemiol. Infect. 2011;139(10):1642–1644. doi: 10.1017/S095026881000289X. [DOI] [PubMed] [Google Scholar]
- Disalvo Arthur F., Bigler William J., Ajello Libero, Johnson Joseph E., Palmer Joseph. Bat and Soil Studies for Sources of Histoplasmosis in Florida. Publ. Health Rep. (Washington, D.C. 1896) 1970;85(12):1063–1069. http://www.ncbi.nlm.nih.gov/pubmed/4991768 [PMC free article] [PubMed] [Google Scholar]
- Emmons C. Isolation of Histoplasma Capsulatum from Soil. Public Health Rep. (1896-1970) 1949;64(28):892–896. [PubMed] [Google Scholar]
- Emmons C.W. Histoplasmosis: Animal Reservoirs and Other Sources in Nature of Pathogenic Fungus, Histoplasma. Am. J. Publ. Health Nation's Health. 1950;40(4):436–440. doi: 10.2105/ajph.40.4.436. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1528481&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons C.W. Association of Bats with Histoplasmosis. Public Health Reports (Washington, D.C. : 1896) 1958;73(7):590–595. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1951700&tool=pmcentrez&rendertype=abstract [PMC free article] [PubMed] [Google Scholar]
- Gómez Raquel Barrena. 2006. Compostaje de Residuos Sólidos Orgánicos. Aplicación de Técnicas Respirométricas En El Seguimiento Del Proceso. [Google Scholar]
- Gómez Luisa F., Barrera Sebastian, Jiménez Maria del P., de Bedout Catalina, Torres Isaura P., Pelaéz Carlos, Acevedo Jose M., Taylor María L., Arango Myrtha. Primera ed. 2017. Histoplasmosis y Compostaje: ¿Estoy En Riesgo de Infectarme? Medellín. [Google Scholar]
- Gómez Luisa F., Torres Isaura P., Pilar Jiménez-A María Del, McEwen Juan Gmo, de Bedout Catalina, Peláez Carlos A., Acevedo José M., Taylor María L., Arango Myrtha. Detection of Histoplasma Capsulatum in Organic Fertilizers by Hc100 Nested Polymerase Chain Reaction and Its Correlation with the Physicochemical and Microbiological Characteristics of the Samples. Am. J. Trop. Med. Hyg. 2018;98(5):1303–1312. doi: 10.4269/ajtmh.17-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez Luisa F., Pérez Laura C., McEwen Juan G., Zuluaga Alejandra, Peláez Carlos A., Acevedo Jose M., Taylor María L., Arango Myrtha, Jiménez María del P. Capacity of Histoplasma capsulatum to survive the composting process. Appl. Environ. Soil Sci. J. 2019 in preparation. [Google Scholar]
- González-González A.E., Aliouat-Denis C.M., Carreto-Binaghi L.E., Ramírez J.A., Rodríguez-Arellanes G., Demanche C., Chabé M., Aliouat E.M., Dei-Cas E., Taylor M.L. An Hcp100 Gene Fragment Reveals Histoplasma Capsulatum Presence in Lungs of Tadarida Brasiliensis Migratory Bats. Epidemiol. Infect. 2012;140(11):1955–1963. doi: 10.1017/S0950268811002585. [DOI] [PubMed] [Google Scholar]
- Hazen E.L., Little G.N., Mordaunt V. Isolation of Histoplasma Capsulatum from Two Natural Sources in the Mohawk Valley; One the Probable Point Source of Two Cases of Histoplasmosis. Am. J. Publ. Health Nation's Health. 1956;46(7):880–885. doi: 10.2105/ajph.46.7.880. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1623868&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook Eric D., Rappleye Chad A. Histoplasma Capsulatum Pathogenesis: Making a Lifestyle Switch. Curr. Opin. Microbiol. 2008;11(4):318–324. doi: 10.1016/j.mib.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Huhn Gregory D., Austin Connie, Carr Mark, Heyer Diana, Boudreau Pam, Gilbert Glynnis, Terry Eimen. Two Outbreaks of Occupationally Acquired Histoplasmosis: More than Workers at Risk. Environ. Health Perspect. 2005;113(5):585–589. doi: 10.1289/ehp.7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez Roberto Alonso, Urán Martha Eugenia, de Bedout Catalina, Arango Myrtha, Tobón Angela Maria, Cano Luz Elena, Restrepo Angela. Outbreak of acute histoplasmosis in a family group: identification of the infection source. Biomedica : Revista Del Instituto Nacional de Salud. 2002;22(2):155–159. http://www.ncbi.nlm.nih.gov/pubmed/12152481 [PubMed] [Google Scholar]
- Jun He, Han Guangye, Chen Daiwen. Insights into enzyme secretion by filamentous fungi: comparative proteome analysis of trichoderma reesei grown on different carbon sources. J. Proteom. 2013;89(August):191–201. doi: 10.1016/j.jprot.2013.06.014. [DOI] [PubMed] [Google Scholar]
- Kasap Murat, Karadenizli Aynur, Akpınar Gürler, Uzuner Hüseyin, Ayimugu Abula, Karaosmanoğlu Kübra, Kadir Er. Doğanhan. Comparative analysis of proteome patterns of francisella tularensis isolates from patients and the environment. Curr. Microbiol. 2017;74(2):230–238. doi: 10.1007/s00284-016-1178-6. [DOI] [PubMed] [Google Scholar]
- Kasuga Takao, Taylor John W., White Thomas J. Phylogenetic Relationships of Varieties and Geographical Groups of the Human Pathogenic Fungus Histoplasma Capsulatum Darling. J. Clin. Microbiol. 1999;37(3):653–663. doi: 10.1128/jcm.37.3.653-663.1999. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=84508&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga Takao, White Thomas J., Koenig Gina, McEwen Juan, Restrepo Angela, Castañeda Elizabetha, Da Silva Lacaz Carlos. Phylogeography of the Fungal Pathogen Histoplasma Capsulatum. Mol. Ecol. 2003;12(12):3383–3401. doi: 10.1046/j.1365-294x.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- Kauffman Carol A. Histoplasmosis. Clin. Chest Med. 2009;30(2):217–225. doi: 10.1016/j.ccm.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Keath Elizabeth J., Kobayashi George S., Medoff Gerald. Typing of Histoplasma Capsulatum by Restriction Fragment Length Polymorphisms in a Nuclear Gene. J. Clin. Microbiol. 1992;30(8):2104–2107. doi: 10.1128/jcm.30.8.2104-2107.1992. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=265451&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox Kenneth S., Hage Chadi A. Histoplasmosis. Proc. Am. Thorac. Soc. 2010;7(3):169–172. doi: 10.1513/pats.200907-069AL. [DOI] [PubMed] [Google Scholar]
- Lemey Philippe, Salemi Marco, Vandamme Anne Mieke. In: The Phylogenetic Handbook: a Practical Approach to Phylogenetic Analysis and Hypothesis Testing. Lemey Philippe, Salemi Marco, Vandamme Anne-Mieke., editors. Cambridge University Press; Cambridge: 2009. [Google Scholar]
- Lenhart Steven W., Schafer Millie P., Mitchell Singal, Hajjeh Rana A. Centers for Disease Control and Prevention National; 2004. Histoplasmosis; Protecting Workers at Risk. [Google Scholar]
- Litvintseva Anastasia P., Brandt Mary E., Mody Rajal K., Lockhart Shawn R. Investigating fungal outbreaks in the 21st century. PLoS Pathog. 2015;11(5) doi: 10.1371/journal.ppat.1004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos Guedes Herbert Leonel De, Guimaraes Allan Jefferson, De Medeiros Muniz Mauro, Pizzini Claudia Vera, Hamilton Andrew John, Peralta Jose Mauro, Deepe George S., Zancope-Oliveira Rosely M. PCR assay for identification of histoplasma capsulatum based on the nucleotide sequence of the M Antigen. J. Clin. Microbiol. 2003;41(2):535–539. doi: 10.1128/JCM.41.2.535-539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVeigh Ilda, Morton Katherine. Nutritional Studies of Histoplasma Capsulatum. Mycopathol. Mycol. Appl. 1965;25(3):294–308. doi: 10.1007/BF02049917. http://www.ncbi.nlm.nih.gov/pubmed/5877181 [DOI] [PubMed] [Google Scholar]
- Moncada Luz Helena, Muñoz Javier, Pineda Fabio, Ferreira Gloria. Estudio de La Presencia de Histoplasma Capsulatum En La Tierra de 4 Cuevas Localizadas de La Región de Río Claro (Antioquia) Iatreia. 1989;2(3):195–200. https://aprendeenlinea.udea.edu.co/revistas/index.php/iatreia/article/view/3368/3130 [Google Scholar]
- Montagna Maria Teresa, De Donno Antonella, Caggiano Giuseppina, Serio Francesca, De Giglio Osvalda, Bagordo Francesco, D’Amicis R., Lockhart Shawn R., Cogliati Massimo. Molecular Characterization of Cryptococcus Neoformans and Cryptococcus Gattii from Environmental Sources and Genetic Comparison with Clinical Isolates in Apulia, Italy. Environ. Res. 2018;160(June 2017):347–352. doi: 10.1016/j.envres.2017.09.032. [DOI] [PubMed] [Google Scholar]
- Morse D.L., Gordon M.A., Matte T., Eadie G. An Outbreak of Histoplasmosis in a Prison. Am. J. Epidemiol. 1985;122(2):253–261. doi: 10.1093/oxfordjournals.aje.a114096. http://www.ncbi.nlm.nih.gov/pubmed/4014207 [DOI] [PubMed] [Google Scholar]
- Negroni R., Duré R., Ortiz Nareto Á., Arechavala A.I., Maiolo E.I., Santiso G.M., Iovannitti C., Ibarra-Camou B., Canteros C.E. Brote de Histoplasmosis En La Escuela de Cadetes de La Base Aérea de Morón, Provincia de Buenos Aires, República Argentina. Rev. Argent. Microbiol. 2010;42:254–260. doi: 10.1590/S0325-75412010000400003. [DOI] [PubMed] [Google Scholar]
- Ohno Hideaki, Tanabe Koichi, Umeyama Takashi, Kaneko Yukihiro, Yamagoe Satoshi, Miyazaki Yoshitsugu. Application of Nested PCR for Diagnosis of Histoplasmosis. J. Infect. Chemother. 2013;19(5):999–1003. doi: 10.1007/s10156-013-0548-2. [DOI] [PubMed] [Google Scholar]
- Ordóñez Nelly, Tobón Angela, Arango Myrtha, Angela Tabares, De Bedout Catalina, Gómez Beatriz, Castañeda Elizabeth, Restrepo Angela. Brotes de Histoplasmosis Registrados En El Área Andina Colombiana. Biomedica. 1997;17(2):105. [Google Scholar]
- Poonwan N., Imai T., Mekha N., Yazawa K., Mikami Y., Ando A., Nagata Y. Genetic Analysis of Histoplasma Capsulatum Strains Isolated from Clinical Specimens in Thailand by a PCR-Based Random Amplified Polymorphic DNA Method. J. Clin. Microbiol. 1998;36(10):3073–3076. doi: 10.1128/jcm.36.10.3073-3076.1998. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=105117&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta A., Colonna-Romano S., Callebaut I., Franco A., Marzullo L., Kobayashi G.S., Maresca B. An Homologue of the Human 100-KDa Protein (P100) Is Differentially Expressed by Histoplasma Capsulatum during Infection of Murine Macrophages. Biochem. Biophys. Res. Commun. 1999;254(3):605–613. doi: 10.1006/bbrc.1998.9894. [DOI] [PubMed] [Google Scholar]
- Rappleye Chad A., Eissenberg Linda Groppe, Goldman William E. Histoplasma Capsulatum Alpha-(1,3)-Glucan Blocks Innate Immune Recognition by the Beta-Glucan Receptor. Proc. Natl. Acad. Sci. U. S. A. 2007;104(4):1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Montes M.R., Rodríguez-Arellanes G., Pérez-Torres A., Rosas-Rosas A.G., Parás-García A., Juan-Sallés C., Taylor M.L. Identification of the Source of Histoplasmosis Infection in Two Captive Maras (Dolichotis Patagonum) from the Same Colony by Using Molecular and Immunologic Assays. Rev. Argent. Microbiol. 2009;41(2):102–104. http://www.ncbi.nlm.nih.gov/pubmed/19623900 [PubMed] [Google Scholar]
- Sacks Jeffrey J., Ajello Libero, Crockett Landis K. An Outbreak and Review of Cave-Associated Histoplasmosis Capsulati. Med. Mycol. 1986;24(4):313–327. doi: 10.1080/02681218680000471. [DOI] [PubMed] [Google Scholar]
- Sambrook Joseph, Russell David W. Molecular Cloning. third ed. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular Cloning. A Laboratory Manual; p. 2025. [Google Scholar]
- Scheel Christina M., Gómez Beatriz L. Diagnostic Methods for Histoplasmosis: Focus on Endemic Countries with Variable Infrastructure Levels. Curr. Trop. Med. Rep. 2014;78(April):129–137. doi: 10.1007/s40475-014-0020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepúlveda Victoria E., Williams Corinne L., Goldman William E., Sepulveda V.E., Williams Corinne L., Goldman William E. Comparison of Phylogenetically Distinct Histoplasma Strains Reveals Evolutionarily Divergent Virulence Strategies. MBio. 2014;5(4) doi: 10.1128/mBio.01376-14. e01376-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepúlveda Victoria E., Márquez Roberto, Turissini David A., Goldman William E., Matute Daniel R. Genome Sequences Reveal Cryptic Speciation in the Human Pathogen Histoplasma Capsulatum. MBio. 2017;8(6):1–23. doi: 10.1128/mBio.01339-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer E.D., Lasker B.A., Travis S.J., Kobayashi G.S., Medoff G. Use of Mitochondrial and Ribosomal DNA Polymorphisms to Classify Clinical and Soil Isolates of Histoplasma Capsulatum. Infect. Immun. 1989;57(5):1409–1412. doi: 10.1128/iai.57.5.1409-1412.1989. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=313291&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobierski M.G., Hospedales C.J., Hall W.N., Robinson-Dunn B., Hoch D., Sheill D.A. Outbreak of Histoplasmosis among Employees in a Paper Factory--Michigan, 1993. J. Clin. Microbiol. 1996;34(5):1220–1223. doi: 10.1128/jcm.34.5.1220-1223.1996. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=228985&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch G., Burford J.G., George R.B., Kaufman L., Ajello L. Acute Histoplasmosis. Description of an Outbreak in Northern Louisiana. Chest. 1980;77(1):38–42. doi: 10.1378/chest.77.1.38. [DOI] [PubMed] [Google Scholar]
- Taylor Maria Lucia, Ruíz-Palacios Guillermo M., Reyes-Montes María Del Rocío, Rodríguez-Arellanes Gabriela, Carreto-Binaghi Laura E., Duarte-Escalante Esperanza, Hernández-Ramírez Aurora. Identification of the Infectious Source of an Unusual Outbreak of Histoplasmosis, in a Hotel in Acapulco, State of Guerrero, Mexico. FEMS Immunol. Med. Microbiol. 2005;45(3):435–441. doi: 10.1016/j.femsim.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Teixeira Marcus de M., Patané José S.L., Taylor Maria L., Gómez Beatriz L., Theodoro Raquel C., Hoog Sybren de, Engelthaler David M., Zancopé-Oliveira Rosely M., Felipe Maria S.S., Barker Bridget M. Worldwide Phylogenetic Distributions and Population Dynamics of the Genus Histoplasma. PLoS Neglected Trop. Dis. 2016;10(6) doi: 10.1371/journal.pntd.0004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh R.B., Schneidau J.D. Experimental Infection of North American Insectivorous Bats (Tadarida Brasiliensis) with Histoplasma Capsulatum. Am. J. Trop. Med. Hyg. 1966;15(4):544–550. doi: 10.4269/ajtmh.1966.15.544. http://www.ncbi.nlm.nih.gov/pubmed/5941177 [DOI] [PubMed] [Google Scholar]
- Vincent R.D., Goewert R., Goldman W.E., Kobayashi G.S., Lambowitz a.M., Medoff G. Classification of Histoplasma Capsulatum Isolates by Restriction Fragment Polymorphisms. J. Bacteriol. 1986;165(3):813–818. doi: 10.1128/jb.165.3.813-818.1986. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=214500&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat L.J., Slama T.G., Eitzen H.E., Kohler R.B., French M.L., Biesecker J.L. A Large Urban Outbreak of Histoplasmosis: Clinical Features. Ann. Intern. Med. 1981;94(3):331–337. doi: 10.7326/0003-4819-94-3-331. http://www.ncbi.nlm.nih.gov/pubmed/7224378 [DOI] [PubMed] [Google Scholar]
- Zeidberg L.D., Ajello Libero, Dillon A., Runyon L.C. Isolation of Histoplasma Capsulatum from Soil. Am. J. Public Health Nation's Health. 1952;42(8):930–935. doi: 10.2105/ajph.42.8.930. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1526182&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucconi F de, De Bertoldi Marco. Compost: Production, Quality and Use. Elsevier; London: 1987. Compost Specifications for the Production and Characterization of Compost from Municipal Solid Waste; pp. 276–295. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ICA resolution number: 00005865.
List of the Histoplasma capsulatum Colombian isolates used in this work.
Table. Characteristics of the sequences and the structure of each primer and PCR reaction conditions. Reagents were as follow, the Deoxynucleoside triphosphates from Thomas Scientific, Swedesboro, NJ., the synthesis of the primers by Integrated DNA Technologies (IDT, Coralville, IA, USA), and the Taq polymerase form Thomas Scientific, Swedesboro, NJ.