Significance
Protein O-GlcNAcylation is a posttranslational modification essential for development. Recently, mutations in the O-GlcNAc transferase (OGT) substrate binding domain have been described that lead to intellectual disability, but the mechanisms underpinning pathogenesis remain to be explored. This work describes the first point mutation in the OGT catalytic domain leading to effects on O-GlcNAcylation and Host cell factor 1 processing in vitro and in a stem cell/Drosophila model, resulting in delayed neuronal differentiation. This establishes a potential link between OGT activity and intellectual disability.
Keywords: intellectual disability, O-GlcNAc, neurodevelopment
Abstract
O-GlcNAc transferase (OGT) is an X-linked gene product that is essential for normal development of the vertebrate embryo. It catalyses the O-GlcNAc posttranslational modification of nucleocytoplasmic proteins and proteolytic maturation of the transcriptional coregulator Host cell factor 1 (HCF1). Recent studies have suggested that conservative missense mutations distal to the OGT catalytic domain lead to X-linked intellectual disability in boys, but it is not clear if this is through changes in the O-GlcNAc proteome, loss of protein–protein interactions, or misprocessing of HCF1. Here, we report an OGT catalytic domain missense mutation in monozygotic female twins (c. X:70779215 T > A, p. N567K) with intellectual disability that allows dissection of these effects. The patients show limited IQ with developmental delay and skewed X-inactivation. Molecular analyses revealed decreased OGT stability and disruption of the substrate binding site, resulting in loss of catalytic activity. Editing this mutation into the Drosophila genome results in global changes in the O-GlcNAc proteome, while in mouse embryonic stem cells it leads to loss of O-GlcNAcase and delayed differentiation down the neuronal lineage. These data imply that catalytic deficiency of OGT could contribute to X-linked intellectual disability.
The O-linked β-N-acetyl-d-glucosamine (O-GlcNAc) transferase (OGT) and the hydrolase O-GlcNAcase (OGA) together orchestrate protein O-GlcNAcylation, a dynamic co/posttranslational modification cycling on thousands of nucleocytoplasmic proteins (1, 2). OGT catalyses the covalent attachment of the monosaccharide O-GlcNAc to serine or threonine. This modification has been suggested to affect transcription (3–6), translation (1), protein stability (7, 8), and subcellular localization (9, 10). O-GlcNAcylation plays a key role in regulating stress response (11–13), differentiation (14, 15), nutrient sensing (16, 17), and autophagy (18).
Genetic studies have highlighted the importance of OGT and protein O-GlcNAcylation in development. OGT is essential for embryonic stem cell viability (19, 20) and mouse embryonic development (21). Zebrafish lacking OGT function exhibit shortened body axis and smaller brains (22). The Drosophila melanogaster OGT encoded by the Polycomb group gene super sex combs (sxc) plays a critical role in establishing Drosophila segment identity (23, 24).
The O-GlcNAc modification is particularly abundant in the brain (25, 26), where it controls memory formation (27–29), circadian rhythm (30, 31), and appetite (32, 33). Numerous neuron-specific proteins are O-GlcNAcylated, such as the Microtubule-associated proteins tau (34) and CRMP2 (35), synaptic vesicle proteins (36), the transcriptional factor cyclic-AMP response element binding protein (CREB) (28), and the AMPA receptor GluA2 subunit (27). Growing evidence suggests that O-GlcNAcylation is essential for normal development and function of the mammalian nervous system (21, 33, 37–39).
Although O-GlcNAcylation has been long implicated in chronic metabolic diseases, such as type II diabetes mellitus, neurodegeneration, and cancer, its role in neurodevelopmental disorders has only recently become apparent. In the past year, a small number of missense mutations within the human OGT gene have been discovered in patients with X-linked intellectual disability (XLID) (40–43). Intellectual disability (ID) refers to a broad range of developmental disorders characterized by limited intellectual capacity, IQ below 70, and poor adaptive behavior with onset before the age of 18, affecting 1 to 3% of the population worldwide (44). Genetic factors are the major cause of this developmental condition involving over 700 ID genes (45). These genes encode for proteins that are required for neuronal development and activity, contributing to neuronal structure and function through several signaling pathways (46). Mutations in genes located on the X-chromosome account for ∼5 to 10% of all ID causative genes affecting predominantly male individuals (44, 47).
To date, XLID-associated mutations in the OGT gene have only been reported for male patients, causing developmental delay and severe cognitive disability. Accompanying clinical phenotypes were dysmorphic features, such as clinodactyly, eye abnormalities, and microcephaly. A common molecular trait of the missense XLID OGT variants is that they appear to retain substantial OGT catalytic activity. This is in agreement with these mutations all being localized to the C-terminal noncatalytic tetratricopeptide repeat (TPR) domain of OGT that is responsible for substrate specificity and scaffolding (40–43). In cultured cells, global O-GlcNAc levels on proteins were unaltered (41–43), and a decrease in OGA protein expression to compensate for the (presumed) moderate loss of OGT activity was only apparent in patient-derived fibroblast cells (43). In addition to its glycosyltransferase activity, mammalian OGT is involved in the unusual proteolytic maturation of Host cell factor 1 (HCF1) (48–50), a transcriptional cofactor implicated in cell cycle control (51), also identified as an ID gene (52–54). HCF1 is O-GlcNAc–modified and subsequently cleaved by OGT using the same active site for both enzymatic activities (49). Among XLID variant OGT models, a moderate effect on HCF1 processing was only detected for one of the mutations (41, 43).
From the currently available OGT XLID mutations, it remains unclear if the patient phenotypes observed are linked to changes in the O-GlcNAc proteome, loss of protein–protein interactions, or misprocessing of HCF1. Here, we characterize an OGT missense mutation in the catalytic domain as observed in female twins with ID and developmental delay. In vitro biochemical analysis of recombinant N567K OGT missense variant protein revealed that this mutation abrogates OGT and HCF1 proteolytic processing activity. Protein crystallography data explain the molecular basis of the observed loss of substrate binding. Introduction of this mutation into D. melanogaster by genome editing reveals global effects on the O-GlcNAc proteome. In mouse embryonic stem cells (mESCs) carrying this mutation, loss of OGA protein expression was observed, as well as defective HCF1 processing. The mutation caused defects in neurite outgrowth during neuronal differentiation. Taken together, our data provide evidence that loss of OGT activity could contribute to the XLID phenotype observed in the patients.
Results and Discussion
Monozygotic Twins with ID and Developmental Delay Possess a De Novo OGT Mutation.
Twin girls (twin 1 and twin 2) were born at 33-wk gestation by semielective caesarean section for maternal preeclampsia. Conception was unassisted. Twin 1 weighed 1,670 g at birth and required continuous positive airway pressure (CPAP) to aid breathing for 3 wk and supplemental oxygen for 58 d. Twin 2 weighed 2,150 g and needed CPAP for 2 d and supplemental oxygen for 20 d. Both babies were tube-fed initially but bottle feeding was established from 3 to 4 wk of age. The twins showed delay in reaching developmental milestones, especially in areas of speech and language development. By 4 y of age, twin 1 only used 3 to 4 single words; at 10 y she was able to put 2 to 3 words together. Language development of twin 2 was more advanced than her sister; at 4 y she was using short phrases and at 10 y she was speaking in sentences and able to recognize letters of the alphabet. Their movement and physical development were also affected: twin 1 by age 4 y and twin 2 by the age of 2 y were able to walk independently. However, twin 2 had significant gross motor difficulties and clumsiness at the age of 8 y. Both twins were diagnosed with cerebral visual impairment, twin 1 more severe than twin 2. They have attended an additional needs nursery and school.
The twins (Fig. 1 A and B) appeared to be affected by a similar developmental syndrome, including small nose, high arched palate, and fifth finger clinodactyly. Twin 1 has ataxic gait with knees bent, walking on toes with inverted feet. An MRI brain scan showed prominence of the lateral ventricles and an enlarged cistern magna in twin 1 at age 2 y, suggesting possible hypoplasia of the inferior cerebellar vermis. Although both siblings have kept generally good health, hypotonia was apparent in infancy and they required iron supplementation intermittently. Twin 1 had an adenoidectomy and treatment for gastro-esophageal reflux and constipation.
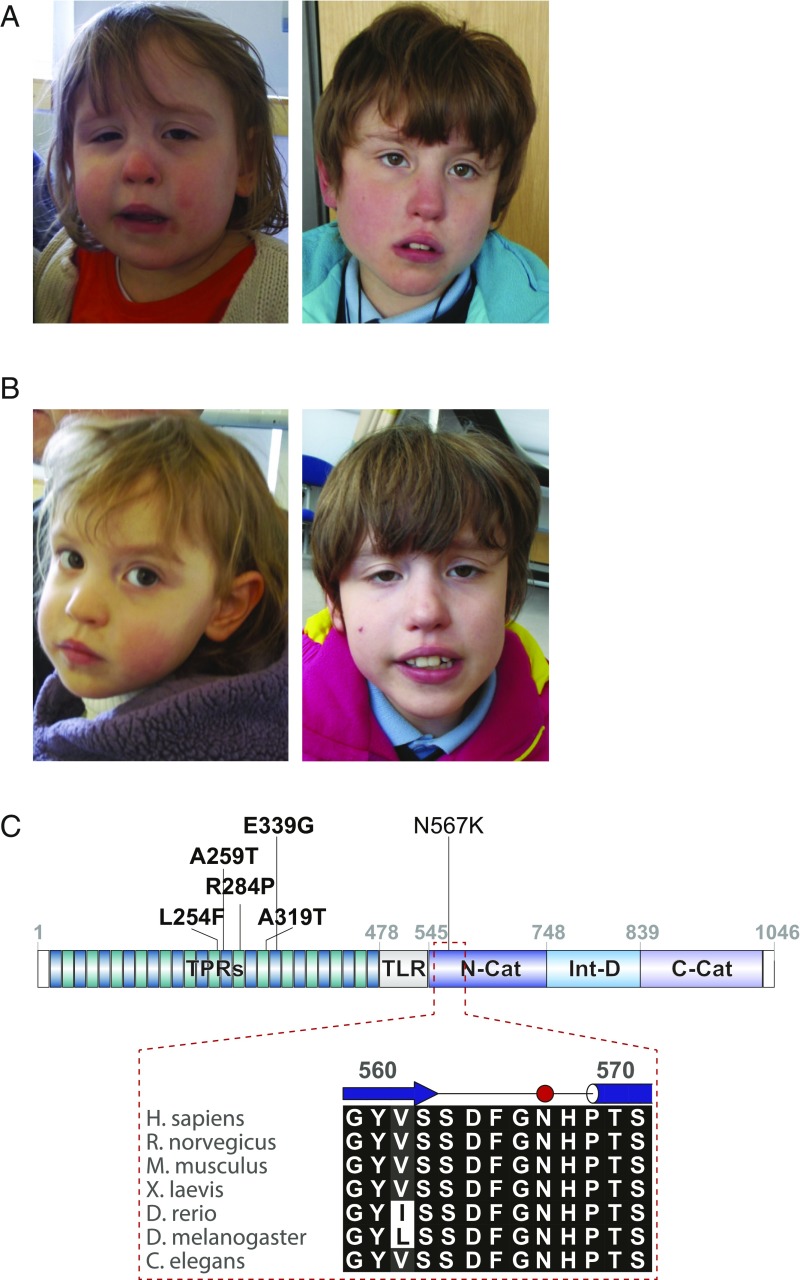
Fig. 1.
Clinical images from patients with the N567K mutation in the catalytic domain of OGT. (A) Picture of twin 1 showing dysmorphic features at age 4 y (Left) and 10 y (Right). (B) Picture of twin 2 showing dysmorphic features at age 4 y (Left) and 10 y (Right). (C) Schematic diagram of OGT highlighting the TPRs, TPR-like repeat, N-Cat, and C-Cat, and the site of N567K, as well as the previously identified XLID-associated mutations in OGT. The Inset shows the sequence alignment of the region encompassing the N567K mutation across the commonly used model organisms. N-Cat and C-Cat: N- and C-terminal lobes of OGT catalytic domain; TPR: tetratricopeptide repeat domain; TLR: tetratricopeptide repeat-like domain.
Sanger sequencing was performed as part of the Deciphering Developmental Disorders initiative, which currently lists genomic and clinical data from over 14,000 child patients with severe undiagnosed developmental disorders (55). This revealed a single pathogenic missense mutation in the OGT gene (X:70779215 T > A, N567K). The mutation was found absent in control population of 141,456 samples, comprising 125,748 exome sequences and 15,708 whole-genome sequences from unrelated individuals reported in the genome aggregation database, gnomAD (56). The mutation affected both monozygotic twins, whereas both of their parents were noncarriers. A 98:2 skew in X-inactivation was detected in both children using polymorphic markers that are differentially methylated on the active and inactive chromosome (SI Appendix, Fig. S1). However, being a de novo mutation, which 1 of the 2 chromosomes is predominantly inactivated remains unclear. Taking these data together, we have identified monozygotic twins with ID and developmental delay that possess a de novo OGT mutation.
The N567K Mutation Abrogates OGT Activity In Vitro.
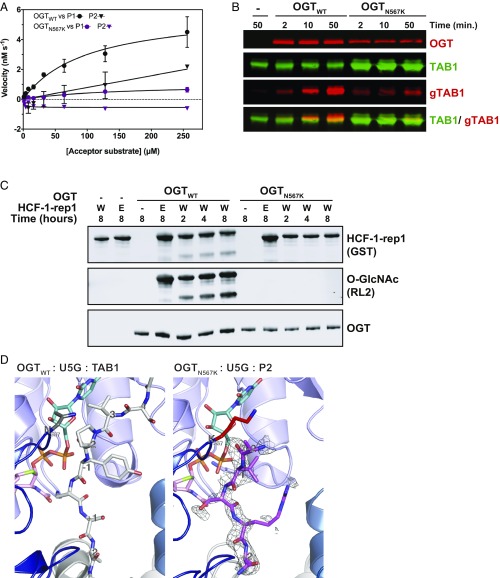
To understand the potential effects of the OGT N567K (OGTN567K) mutation, we next investigated changes in catalytic activity compared with full-length wild-type OGT (OGTWT) protein. Steady-state kinetics of OGT glycosyltransferase activity was measured against 2 acceptor peptide substrates, P1 (KKVPVSRA) and P2 (KKVAVSRA), that only varied at the −2 position adjacent to the O-GlcNAcylation site (57). OGTN567K activity was 12-fold reduced against P1 (Fig. 2A and Table 1) and not detectable against P2 (Fig. 2A). Next, OGT activity was measured against an intact protein substrate, TAK-1 binding protein (TAB1), where the reaction was followed with a TAB1 O-GlcNAc site-specific antibody (58). OGTN567K showed negligible glycosyltransferase activity (Fig. 2B).
Fig. 2.
The N567K mutation abrogates OGT activity due to disrupted acceptor binding site. (A) Michaelis–Menten kinetics of OGT glycosyltransferase activity against P1 and P2 peptides. (B) Immunoblots showing OGT glycosyltransferase activity against TAB1. (C) Immunoblots showing OGT glycosyltransferase and proteolytic activities against HCF1-rep1. HCF1-rep1: GST-tagged host cell factor-1 fragment containing the first PRO repeat. W: wild type. E: E1019Q. (D) Crystal structures of OGT ternary complexes of OGTWT and OGTN567K active site in complex with UDP-5S-GlcNAc (U5G, turquoise, orange atoms) and acceptor peptide. OGTWT is shown in complex with RB2 (Retinoblastoma-like protein 2 peptide, light gray carbon atoms) [PDB ID code: 5C1D (57)]; OGTN567K is shown with P2 peptide (pink carbon atoms) (PDB ID code 6IBO). K567 is shown in red. An unbiased Fo–Fc difference map before inclusion of the peptide is shown as a mesh.
Table 1.
Michaelis–Menten kinetics of OGT activity against P1 peptide substrate
| Enzyme | KM (nM⋅s−1) | kcat (s−1) | kcat/KM |
| OGTWT | 99 ± 19 | 119 ± 11 | 1.2 ± 0.3 |
| OGTN567K | 144 ± 63 | 20 ± 4 | 0.1 ± 0.2 |
OGT possesses a second catalytic activity in the form of promoting autocatalytic cleavage and activation of HCF1, which itself is an ID-associated gene (52). We next investigated the effects of the N567K mutation on OGT-mediated HCF1 processing. OGT proteolytic activity was measured using a GST-fusion construct of a minimal HCF1 fragment (HCF1-rep1) containing one of the PRO repeats that are known to be the targets of OGT-mediated proteolysis (49). Wild-type OGT not only promotes HCF1 proteolysis but was also able to hyperglycosylate this protein fragment (Fig. 2C). HCF1E1019Q, which lacks the key glutamate required for proteolytic processing, was not cleaved by wild-type OGT. Strikingly, even after 8 h of reaction time, OGTN567K was able to neither glycosylate nor cleave HCF1-rep1 (Fig. 2C). Taken together, the data show that the N567K mutation abrogates OGT activity.
The N567K Mutation Disrupts the OGT Acceptor Binding Site.
To understand the molecular basis of the observed loss of OGT catalytic activity, we next initiated structural characterization of OGTN567K. Inspection of the wild-type OGT structure reveals that Asn567 maps to a loop in the N-terminal lobe of the catalytic domain that is completely conserved through evolution from Caenorhabditis elegans to man (Fig. 1C). The Asn567 side chain borders the −2 subsite as part of the OGT acceptor substrate binding cleft (Fig. 2D). Therefore, we hypothesized that the N567K mutation would affect OGT acceptor substrate binding, without affecting the overall OGT fold. Recombinant OGTN567K was obtained from Escherichia coli using construct boundaries previously employed to crystallize OGTWT-substrate complexes (57). OGTN567K was then cocrystallized with a donor analog, UDP-5S-GlcNAc, and 2 OGT acceptor peptide substrates, P1 (KKVPVSRA) and P2 (KKVAVSRA), previously used for activity measurements (Fig. 2A). Only the condition containing P2, which contains an alanine at the −2 subsite, yielded crystals that diffracted to 2.3 Å and allowed structure solution by molecular replacement and refinement (Table 2). The OGTN567K structure superposed onto that of OGTWT with an rmsd of 0.4 Å across 675 Cα atoms, suggesting the mutation does not induce large conformational changes in the catalytic domain. Initial unbiased difference electron density maps revealed partial density corresponding to the acceptor peptide (Fig. 2D). Surprisingly, this only covered the P2 backbone in the +2 through to −2 subsites, suggesting that the N567K mutation may induce increased flexibility at the N-terminal tail of the acceptor peptide (Fig. 2D and SI Appendix, Fig. S2). In agreement with this, a comparative B-factor analysis suggested that the acceptor peptide bound to OGTN567K is substantially more flexible than that bound to OGTWT (SI Appendix, Fig. S2). Close inspection of the OGTN567K acceptor substrate binding cleft revealed that the protrusion caused by the N567K mutation partially occupies the position of the −2 proline frequently observed in OGT acceptor peptides (57, 59, 60). In summary, the N567K mutation appears to disrupt the OGT acceptor binding site.
Table 2.
X-ray diffraction data processing and refinement statistics of the OGTN567K ternary complex
| OGTN567K | |
| Data collection | |
| Space group | F222 |
| Cell dimensions | |
| α, β, γ (°) | 137.7, 150.6, 200.0 |
| A, B, C (Å) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 45.31–2.17 (2.17–2.23) |
| Rsym or Rmerge | 0.07 (0.91) |
| I/σI | 11.3 (1.3) |
| Completeness (%) | 100 (99) |
| Redundancy | 5.2 (5.3) |
| Refinement | |
| Resolution (Å) | 45.31–2.17 (2.17–2.23) |
| No. reflections | 286,887 (23,302) |
| Rwork/Rfree | 0.20/0.25 |
| No. atoms | |
| Protein | 5,530 |
| Nucleotide sugar | 39 |
| Peptide | 35 |
| Water | 328 |
| B-factors | |
| Protein | 53.2 |
| Nucleotide sugar | 46.6 |
| Peptide | 91.9 |
| Water | 51.3 |
| Rmsd | |
| Bond lengths (Å) | 0.02 |
| Bond angles (°) | 1.8 |
Numbers in brackets show the highest resolution bin.
The sxcN595K Mutation Leads to a Reduction of Protein O-GlcNAcylation in D. melanogaster.
Lack of functional OGT causes lethality and developmental arrest in most metazoan model organisms, including mice, frogs, zebrafish, and fruit flies (19, 22, 24, 61). To probe if the N567K mutation directly affects OGT catalytic activity in vivo, we exploited the genetically tractable system D. melanogaster, where phenotypes arisen due to null and hypomorphic alleles of sxc (the fly ogt ortholog) have been described (62, 63). Drosophila sxc null mutants (sxc1 and sxc6) die at the pharate adult stage with homeotic transformation defects (62). A hypomorph allele, sxcH537A, having substantially reduced OGT catalytic activity (64), shows significantly decreased protein O-GlcNAcylation levels (63) yet produce fertile offspring. Crucially, the fruit fly provides a platform to dissect the different effects of the OGT N567K mutation. In the fly, feedback between proteome O-GlcNAcylation levels and OGA/OGT protein/mRNA levels has not been observed. Furthermore, although Drosophila Hcf1 is heavily O-GlcNAc modified, proteolytic processing is catalyzed by the Taspase I protease instead of OGT (65).
We introduced the OGT N567K equivalent mutation (N595K) into D. melanogaster using a CRISPR/Cas9 gene-editing toolkit. Vasa::Cas9 embryos were injected in-house with a mixture of plasmids coding for the guide RNA and repair template DNA. In total, 100 potential candidate flies were screened exploiting restriction fragment-length polymorphism arising from the loss of a restriction digestion site introduced in parallel with the N595K mutation (SI Appendix, Fig. S3). We recovered 2 independent knockin lines (10.3 and 19.1) carrying the mutation, sxcN595K. Presence of the mutation was confirmed by sequencing the region ∼250 base pairs upstream and downstream from the mutation. Additionally, the lines were further validated by sequencing the full-length sxc mRNA. Both lines developed to adulthood without apparent homeotic defects. Transheterozygous sxc1/sxcH537A, sxc1/sxcN595K-19.1, and sxc1/sxcN595K-10.3 flies with presumably further reduced OGT activity were viable (SI Appendix, Fig. S4).
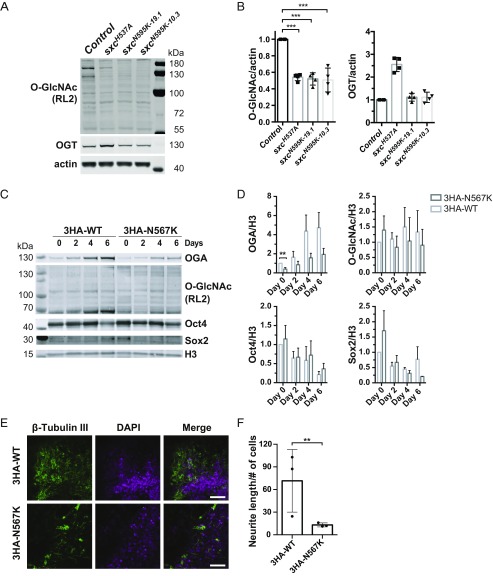
To probe the effect of the N595K mutation on OGT enzymatic activity and stability in vivo, we subjected adult fly head and embryo lysates to Western blotting with an O-GlcNAc antibody, RL2 (Fig. 3A and SI Appendix, Fig. S5). O-GlcNAc levels were reduced to levels comparable to those in the hypomorph sxcH537A flies (Fig. 3A and SI Appendix, Fig. S5). OGT protein levels are expressed at wild-type level (Fig. 3A and SI Appendix, Fig. S5). Taken together, phenotypic and molecular characterization of sxcN595K Drosophila lines revealed that the mutation leads to reduction of protein O-GlcNAcylation.
Fig. 3.
The N567K OGT mutation leads to altered O-GlcNAc homeostasis in D. melanogaster and neurite outgrowth defect in mESCs. (A) Immunoblot on 3- to 5-d-old adult Drosophila head lysates. (B) Quantification of O-GlcNAc modification on proteins and OGT protein level, normalized to actin signal. Protein O-GlcNAc modification is decreased in sxcN595K 19.1 (***P < 0.0001) and sxcN595K 10.3 (***P < 0.0001) lines to a comparable level found in the hypomorphic sxcH537A flies (***P < 0.0001). n = 4, mean ± SD. ANOVA with Tukey test. (C) Immunoblots on full cell lysate of 3HA-OGTWT and 3HA-OGTN567K mESCs showing OGA, Oct4, Sox2, Histone 3 (H3), and protein O-GlcNAcylation (RL2) levels. mESCs were differentiated for 0, 2, 4, and 6 d in N2B27 medium. (D) Quantification of immunoblots of OGA, Oct4, Sox2, and global protein O-GlcNAcylation in differentiated 3HA-OGTWT and 3HA-OGTN567K mESCs normalized to H3 signal. n = 4, mean ± SEM; **P = 0.012. Multiple t test using the Holm–Sidak method. (E) Immunofluorescence microscopy of 3HA-OGTWT and 3HA-OGTN567K mESCs after 8 d of neuronal differentiation in N2B27 medium. Cells were stained for tubulin III (green) and DAPI (magenta). (Scale bar, 50 μm.) (F) Quantification of neurite outgrowth. Total neurite length was measured per microscopic image by tracking β-tubulin III signal and then normalized to the number of nuclei (DAPI). (3HA-OGTWT, 108 to 800 nuclei per image; 3HA-OGTN567K, 260 to 740 nuclei per image) **P = 0.0079, Student t test, mean ± SD, n = 3.
The N567K Mutation Leads to a mESC Neuronal Differentiation Phenotype.
The twins carrying the N567K OGT mutation, and the previously published male patients carrying OGT mutations in the TPR domain, all suffered from neurodevelopmental delay. We next explored the early events of neuronal development by studying effects on pluripotency and differentiation in mESCs, a cellular system amenable to genetic modification. We generated male mESC lines expressing C-terminal triple hemagglutinin (3HA) -tagged version of wild-type OGT (3HA-OGTWT) and carrying the N567K mutation (3HA-OGTN567K) using 2 rounds of CRISPR/Cas9-mediated gene editing at the endogenous locus (SI Appendix, Figs. S6 and S7). At each step, at least 2 clones with the correct genotype were isolated and selected for further analysis.
Next, we examined whether neuronal differentiation was altered in 3HA-OGTN567K cells. Expression of pluripotency markers Sox2 and Oct4 was unaffected during the first 6 d of differentiation (Fig. 3 C and D). Neurite outgrowth was assessed after 8 d. Neurite length was significantly reduced in 3HA-OGTN567K cells compared with 3HA-OGTWT control cells immune-labeled against the neuron-specific β-tubulin III protein (Fig. 3 E and F). Thus, the N567K mutation leads to an mESC neuronal differentiation phenotype.
The N567K Mutation Leads to Misprocessing of HCF1.
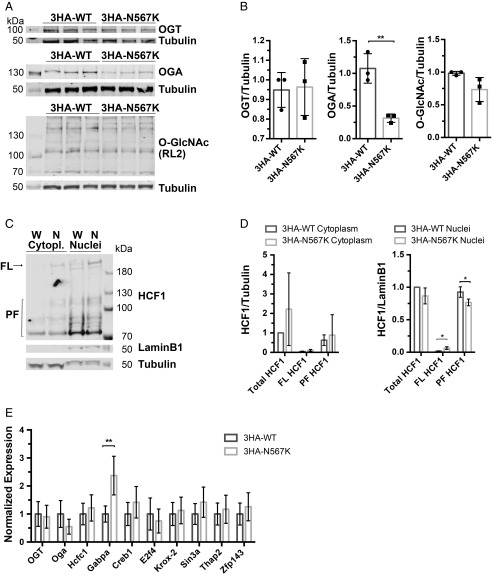
We next attempted to uncover the molecular mechanisms underpinning the observed differentiation phenotypes. We tested the levels of protein O-GlcNAcylation using Western blotting (Fig. 4A). Surprisingly, levels of protein O-GlcNAcylation in pluripotent mESCs appeared comparable in 3HA-OGTN567K and 3HA-OGTWT cells. Previous studies have shown that (an as yet to be discovered) feedback mechanism regulates OGA and OGT expression levels in response to protein O-GlcNAcylation levels (42, 43, 66, 67). Indeed, while OGT levels appeared to be unaltered, OGA levels were abrogated in 3HA-OGTN567K pluripotent mESCs (Fig. 4 A and B), presumably to compensate for reduced OGT activity, but appeared normal during differentiation (Fig. 3 C and D).
Fig. 4.
N567K mutation leads to defects in HCF1 processing and downstream gene expression in 3HA-OGTN567K pluripotent mESCs. (A) Immunoblots showing OGT, OGA, and protein O-GlcNAcylation (RL2) levels in 3HA-OGTWT and 3HA-OGTN567K undifferentiated mESCs. (B) Quantification of immunoblots of global protein O-GlcNAcylation, OGA, and OGT protein level in undifferentiated mESCs normalized to tubulin signal. n = 3, mean ± SD. Unpaired t test. **P = 0.0049. (C) 3HA-OGTN567K mESCs show impaired HCF1 proteolytic cleavage. Western blot analysis for HCF1, tubulin (cytoplasmic marker), and Laminin B1 (nuclear marker) indicates decreased level of proteolytic fragments of HCF1 in 3HA-OGTN567K compared with 3HA-OGTWT mESCs. (D) Quantification of immunoblots on nuclear and cytoplasmic fractions of undifferentiated 3HA-OGTN567K mESCs compared with 3HA-OGTWT mESCs control line showing HCF1 levels. HCF1 full-length (FL) and proteolytic fragments (PF) normalized to tubulin signal for the cytosolic fraction and to Lamin B1 signal for nuclear fraction. n = 3, mean ± SD. Multiple t test. *P = 0.0189 for FL and *P = 0.044 for PF. (E) qPCR analysis of gene expression of HCF1 targets in undifferentiated 3HA-OGTN567K and 3HA-OGTWT mESCs. Mean ± SEM, n = 3. **P = 0.002.
Next, we investigated the effects of the N567K mutation on the posttranslational maturation and cellular localization of HCF1 in pluripotent mESCs. Western blot analysis on cytoplasmic and nuclear fractions revealed an elevated ratio of full-length versus total HCF1 in the nuclear fractions of 3HA-OGTN567K compared with 3HA-OGTWT mESCs, while the levels of HCF1 proteolytic products were reduced (Fig. 4 C and D). This is in agreement with the impaired OGT-mediated proteolytic cleavage of HCF1 observed in vitro (Fig. 2C). HCF1 contributes to the regulation of transcriptional programs associated with controlling cell cycle and pluripotency (68, 69). Interestingly, the expression of some of the HCF1 interactors are also regulated by HCF1 itself (51, 66, 69). As an initial attempt to uncover possible candidate genes affected by HCF1 misprocessing, we investigated the expression levels of known HCF1 partners by qRT-PCR (Fig. 4E). The Ets transcription factor GA-binding protein subunit α (GABPA), a component of the nuclear respiratory factor-2 (NRF2) complex, showed a 2.3-fold increase in mRNA level in 3HA-OGTN567K cells compared with 3HA-OGTWT control samples (P = 0.002). GABPA/NRF2 regulates mitochondrial biogenesis and cell cycle progression (70, 71). Taking these data together, the OGT N567K mutation leads to misprocessing of HCF1 in mESCs.
Concluding Remarks
Previous studies have established a link between missense mutations in OGT and XLID in young males. These patients are hemizygous for the mutant OGT allele, with this genetic defect being present in all cells from early development to later life (41–43). However, residing exclusively in the OGT TPR domain, it was not clear whether these mutations led to the observed phenotypes through loss of glycosyltransferase activity, HCF1 misprocessing, or disruption of the OGT interactome (41–43). Here, we have presented female twins with XLID who are heterozygous for an N567K mutation in the OGT catalytic domain. Despite a wild-type ogt allele being present in all cells of these patients, its expression is arbitrarily silenced due to X chromosome inactivation at the late blastocyst stage of embryogenesis to compensate for gene dosage (72). Thus, these females represent an example of mosaic expression of an OGT XLID variant causing developmental abnormalities. Previous examples of female carriers of X-linked genetic disorders showed correlation between the X inactivation pattern and the severity of the condition (73–75). Hence, it is possible that the different etiology of ID in twins is the consequence of distinct spatial patterns and skew of X-inactivation.
The N567K variant of OGT retains negligible levels of catalytic activity. Thus, a possible mechanism underpinning the XLID phenotype is reduced protein O-GlcNAcylation on a subset of OGT target proteins at specific time points of development or differentiation. Previous genetic studies in mice have highlighted the vital role of OGT in both neuronal development and adult life (21, 33, 37–39). OGT is capable of transferring GlcNAc onto over 4,000 different substrate proteins; it possesses an acceptor-peptide specificity with a preference to a degenerated sequon motif (57, 76, 77). Our structural analysis has uncovered that the N567K mutation changes the surface of the substrate binding pocket, thereby altering substrate binding ability of the N567K OGT. Previously reported XLID associated mutations lie in the TPR domain of OGT (40–43, 78), so potentially influencing its scaffolding function or reducing O-GlcNAcylation on an as yet unknown, subset of substrates. The N567K mutation may affect the O-GlcNAc proteome globally yet leads to a similar clinical phenotype, suggesting it is the catalytic activity/dosage of OGT that is important.
We have identified one of the OGT substrates, HCF1, as a potential candidate that could at least partially explain some of the OGT N567K patient phenotypes. We showed that O-GlcNAcylation and proteolytic activation of HCF1 is significantly decreased, including in mESC nuclei. The HCF1 protein plays an important role in cell growth and cell cycle control through interacting with transcriptional complexes and epigenetic regulators (79–83). The HCF1 gene itself has been identified as an intellectual disability gene causing cobalamin type X deficiency, craniofacial abnormalities, and prenatal onset of microcephaly in the most extreme cases (52, 53, 84). Less-severe HCF1 mutations have been associated with ID and autism spectrum disorder without cobalamin deficiency (54). Patients with N567K OGT variant did not suffer from cobalamin deficiency and expressed a much milder form of congenital anomalies than the severe HCF1 group, suggesting that a degree of HCF1 activity is retained. Interestingly, we observed an increase in full-length HCF1 protein level and decreased level of the mature proteolytic fragment in nuclei, showing that HCF1 processing is affected in cellulo, albeit with retention of some activity. Surprisingly, the expression of the transcriptional factor GABPA downstream of HCF1 was increased indicating that the N567K mutation in OGT could translate into moderate molecular changes downstream of HCF1. The opposite effect, decreased expression of GABPA, was detected upon overexpression of the HCF1 N-terminal fragment (HCFN) (68). GABPA belongs to the E-26 family of DNA binding factors and regulates expression of several proteins required for mitochondrial DNA transcription and replication (85). Moreover, GABPA modulates the expression of genes involved in cell cycle control, apoptosis, differentiation, and hormonal regulation. Its importance in early embryonic development was demonstrated in knockout mouse studies (86). However, there is no established link between GABPA and neurodevelopment. Furthermore, expression of several other transcriptional factors targeted by HCF1—such as Creb1, E2F4, Krox-2, Sin3a, Thap2, and Zfp143—were found unaltered, suggesting that partial abrogation of HCF1 cleavage may affect just a small subset of targets, whose expression is influenced by the altered ratio of proteolytically cleaved and uncleaved HCF1.
While OGT is critical for mESC maintenance of pluripotency and differentiation, we have detected no difference at the level of pluripotency markers, Sox2 and Oct4, between 3HA-OGTWT and 3HA-OGTN567K mESCs. The pluripotency factor Sox-2 is O-GlcNAc modified at Ser248 (87) and this posttranslational modification is indispensable for sustaining pluripotency. However, its O-GlcNAc modification is not required for differentiation, with cells expressing an O-GlcNAc–deficient mutant version of Sox2 exhibited enhanced reprogramming ability (87). Furthermore, mESCs expressing O-GlcNAc–deficient Sox2 displayed changes in their transcriptional network, specifically increasing the expression of genes vital to maintain pluripotency (87). It is feasible that a similar mechanism is responsible to compensate for pluripotency defects induced by the N567K OGT mutation.
A Drosophila model of the N567K OGT mutation showed reduced protein O-GlcNAcylation. However, the abundance of O-GlcNAc–modified proteins was similar in 3HA-OGTWT and 3HA-OGTN567K mESCs. Interestingly, we observed decreased OGA protein levels as a compensatory regulatory mechanism responsible for maintaining protein O-GlcNAc levels in undifferentiated mESCs. Our finding is in line with previous data reported on patient-derived cell lines carrying XLID mutations in the TPR domain of OGT (42, 43). Although there is no evidence for a link between OGA and XLID, given the role of OGA in regulating transcription (88–91), it cannot be excluded that changes in OGA levels directly contribute to the XLID phenotype.
In agreement with previous studies implying a requirement for OGT function in normal neuron differentiation, morphology, and health (27, 37, 92–94), abnormal OGT activity in 3HA-OGTN567K cells resulted in reduced neurite outgrowth. These data suggest a possible mechanistic link between the mutation and the microcephaly and neurodevelopmental cognitive impairments observed in the patients.
In summary, we have shown that impaired catalytic activity of OGT leads to neurodevelopmental defects in humans, through combined downstream effects of impaired OGT activity and reduced HCF1 proteolytic processing. Further studies are required to define the mechanisms downstream of impaired OGT catalytic activity that affect neurodevelopment resulting in intellectual disability.
Materials and Methods
The study was approved by the University of Dundee Institutional Risk Assessment committee, and 1 of each participants’ parents provided written, informed consent. The DDD study has UK Research Ethics Committee (REC) approval (10/H0305/83, granted by the Cambridge South Research Ethic Committee, and GEN/284/12 granted by the Republic of Ireland REC).
X-Inactivation Analysis.
To determine if X chromosome inactivation occurred randomly or nonrandomly, the ratio of methylated and unmethylated maternal and paternal X chromosomes was determined. (SI Appendix, Fig. S1). Sites are methylated on the inactive X chromosome and unmethylated on the active X chromosome. The method utilizes the androgen receptor sequence that is encoded on the X chromosome (Xq11.2-q12) and contains a highly polymorphic polyglutamine (CAG) trinucleotide repeat in exon 1. There are 20 alleles of androgen receptor, with 11 to 31 repeats reported in the general population and 85 to 90% of patients are heterozygous for this locus, thus suitable for X-inactivation analysis. For experimental procedure, see SI Appendix, SI Materials and Methods.
Structure Solution.
Crystallization of truncated OGTWT/N567K (residues 323–1044) was performed as described previously (57) (for protein expression, purification, and crystallization, see SI Appendix, SI Materials and Methods). The structure was solved by molecular replacement using the structure for OGTWT [PDB ID code 5C1D (57)] as the search model. The resulting model was manually refined using Coot (95) and REFMAC (96), respectively. The editing and refinement of the model was iterated until it was in complete agreement with the data. Scaling and model building statistics can be seen in Table 2.
Enzyme Activity Assays.
Michaelis–Menten kinetics of OGT were measured using a fluorometric assay, as described previously (97), with the exception of reduced reaction volume of 25 µL and usage of 384-well plate. As acceptor substrate, P1 (KKVPVSRA) and P2 (KKVAVSRA) were used. Additional O-GlcNAcylation assays were performed on TAB1 protein (residues 7–420). Proteolytic assays were performed as described previously (43), except a noncleavable HCF1-rep1 fragment was used instead of OGA treatment as the negative control. For further details, see SI Appendix, SI Materials and Methods.
mESC Culture.
mESC AW2 line was acquired from the MRC Centre for Regenerative Medicine, Institute for Stem Cell Research, University of Edinburgh (98). mESCs were cultured in an undifferentiated state in 0.1% gelatin (wt/vol)-coated plates in GMEM BHK-21 supplemented with 10% FBS (vol/vol), 0.1 mM MEM nonessential amino acids, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol, and 100 units/mL LIF at 37 °C in 5% CO2. For differentiation assays, cells were cultured following a previously published protocol (99). Briefly, cells were cultured in DMEM and F12 media mixed at 1:1 ratio (vol/vol), supplemented with modified N2 (25 mg/mL insulin, 100 μg/mL apo-transferrin, 6 ng/mL progesterone, 16 μg/mL putrescine, 30 nM sodium selenite, and 50 μg/mL BSA fraction) and neurobasal supplement B27. Medium was renewed every 2 d. For generation of CRISPR/Cas9 lines, Immunocytochemistry, Western blotting, and qPCR analyses details are provided in SI Appendix, SI Materials and Methods.
Drosophila Husbandry, Microinjection, and Genetics.
Fly stocks were maintained on Nutri-Fly Bloomington Formulation fly food at 25 °C. Vasa::Cas9 (#51323), sxc1,bw1,sp1/SM5 (#3058) and Df(2R)BSC630/CyO (#25705) stocks were obtained from the Bloomington Drosophila Stock Centre. The sxc hypomorphic allele, sxcH537A, was described in ref. 63. The sxcN595K Drosophila lines were generated by CRISPR/Cas9 mutagenesis, as described in SI Appendix, SI Materials and Methods.
Western Blotting from Drosophila Samples.
Sample preparation of embryos and fly heads and Western blotting were performed as described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was funded by a Wellcome Trust Investigator Award (110061) (to D.M.F.v.A.). V.M.P. was supported by the SPRINT/MND-MS Scholarship (University of Edinburgh). The X-ray diffraction experiments were performed on beamline ID23-2 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. We are grateful to Local Contact at the ESRF for providing assistance for using beamline ID23-2. We acknowledge Dr. Louise A. Young for the aid in the X-inactivation analysis and Dr. Andrew Smith for the AW2 mouse embryonic stem cell line. We acknowledge the Deciphering Developmental Disorders study, presenting independent research commissioned by the Health Innovation Challenge Fund (grant no. HICF-1009-003), a parallel funding partnership between Wellcome and the Department of Health, and the Wellcome Sanger Institute (grant no. WT098051). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. The authors would also like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 6IBO).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900065116/-/DCSupplemental.
References
- 1.Zhu Y., et al. , O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol. 11, 319–325 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Zachara N., Akimoto Y., Hart G. W., The O-GlcNAc Modification (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2017). [PubMed] [Google Scholar]
- 3.Constable S., Lim J.-M., Vaidyanathan K., Wells L., O-GlcNAc transferase regulates transcriptional activity of human Oct4. Glycobiology 27, 927–937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamarre-Vincent N., Hsieh-Wilson L. C., Dynamic glycosylation of the transcription factor CREB: A potential role in gene regulation. J. Am. Chem. Soc. 125, 6612–6613 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Ranuncolo S. M., Ghosh S., Hanover J. A., Hart G. W., Lewis B. A., Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem. 287, 23549–23561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deplus R., et al. , TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu C. S., et al. , O-GlcNAcylation regulates EZH2 protein stability and function. Proc. Natl. Acad. Sci. U.S.A. 111, 1355–1360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambetta M. C., Müller J., O-GlcNAcylation prevents aggregation of the Polycomb group repressor polyhomeotic. Dev. Cell 31, 629–639 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Skorobogatko Y., et al. , O-linked β-N-acetylglucosamine (O-GlcNAc) site thr-87 regulates synapsin I localization to synapses and size of the reserve pool of synaptic vesicles. J. Biol. Chem. 289, 3602–3612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng C., et al. , Regulation of the hippo-YAP pathway by glucose sensor O-GlcNAcylation. Mol. Cell 68, 591–604.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Zachara N. E., Hart G. W., O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 1673, 13–28 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P., A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han C., et al. , O-GlcNAcylation of SIRT1 enhances its deacetylase activity and promotes cytoprotection under stress. Nat. Commun. 8, 1491 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andres L. M., et al. , Chemical modulation of protein O-GlcNAcylation via OGT inhibition promotes human neural cell differentiation. ACS Chem. Biol. 12, 2030–2039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang H., et al. , O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 11, 62–74 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Durning S. P., Flanagan-Steet H., Prasad N., Wells L., O-linked β-N-acetylglucosamine (O-GlcNAc) acts as a glucose sensor to epigenetically regulate the insulin gene in pancreatic beta cells. J. Biol. Chem. 291, 2107–2118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O., Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo B., et al. , O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 16, 1215–1226 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Shafi R., et al. , The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miura T., Nishihara S., O-GlcNAc is required for the survival of primed pluripotent stem cells and their reversion to the naïve state. Biochem. Biophys. Res. Commun. 480, 655–661 (2016). [DOI] [PubMed] [Google Scholar]
- 21.O’Donnell N., Zachara N. E., Hart G. W., Marth J. D., Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster D. M., et al. , O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev. Biol. 9, 28 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinclair D. A. R., et al. , Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gambetta M. C., Oktaba K., Muller J., Essential role of the glycosyltransferase Sxc/Ogt in polycomb repression. Science 325, 93–96 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Kreppel L. K., Blomberg M. A., Hart G. W., Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Lubas W. A., Frank D. W., Krause M., Hanover J. A., O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272, 9316–9324 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Taylor E. W., et al. , O-GlcNAcylation of AMPA receptor GluA2 is associated with a novel form of long-term depression at hippocampal synapses. J. Neurosci. 34, 10–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rexach J. E., et al. , Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol. 8, 253–261 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ardiel E. L., et al. , CaMK (CMK-1) and O-GlcNAc transferase (OGT-1) modulate mechanosensory responding and habituation in an interstimulus interval-dependent manner in Caenorhabditis elegans. bioRxiv:10.1101/115972 (10 March 2017).
- 30.Kim E. Y., et al. , A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 26, 490–502 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sui L., et al. , Clustered DNA damage induced by protons radiation in plasmid. Chin. Sci. Bull. 58, 3217–3223 (2013). [Google Scholar]
- 32.Dai C. L., Gu J. H., Liu F., Iqbal K., Gong C. X., Neuronal O-GlcNAc transferase regulates appetite, body weight, and peripheral insulin resistance. Neurobiol. Aging 70, 40–50 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Lagerlöf O., et al. , The nutrient sensor OGT in PVN neurons regulates feeding. Science 351, 1293–1296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold C. S., et al. , The microtubule-associated protein tau is extensively modified with O-linked N-acetylglucosamine. J. Biol. Chem. 271, 28741–28744 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Cole R. N., Hart G. W., Cytosolic O-glycosylation is abundant in nerve terminals. J. Neurochem. 79, 1080–1089 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Vosseller K., et al. , O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Francisco H., et al. , O-GLcNAc post-translational modifications regulate the entry of neurons into an axon branching program. Dev. Neurobiol. 69, 162–173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagerlöf O., Hart G. W., Huganir R. L., O-GlcNAc transferase regulates excitatory synapse maturity. Proc. Natl. Acad. Sci. U.S.A. 114, 1684–1689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang A. C., Jensen E. H., Rexach J. E., Vinters H. V., Hsieh-Wilson L. C., Loss of O-GlcNAc glycosylation in forebrain excitatory neurons induces neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 113, 15120–15125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouazzi H., Lesca G., Trujillo C., Alwasiyah M. K., Munnich A., Nonsyndromic X-linked intellectual deficiency in three brothers with a novel MED12 missense mutation [c.5922G>T (p.Glu1974His)]. Clin. Case Rep. 3, 604–609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selvan N., et al. , O-GlcNAc transferase missense mutations linked to X-linked intellectual disability deregulate genes involved in cell fate determination and signaling. J. Biol. Chem. 293, 10810–10824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaidyanathan K., et al. , Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J. Biol. Chem. 292, 8948–8963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willems A. P., et al. , Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J. Biol. Chem. 292, 12621–12631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bassani S., et al. , The neurobiology of X-linked intellectual disability. Neuroscientist 19, 541–552 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Neri G., Schwartz C. E., Lubs H. A., Stevenson R. E., X-linked intellectual disability update 2017. Am. J. Med. Genet. A 176, 1375–1388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boda B., Mas C., Muller D., Activity-dependent regulation of genes implicated in X-linked non-specific mental retardation. Neuroscience 114, 13–17 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Stevenson R. E., Schwartz C. E., Rogers R. C., Malformations among the X-linked intellectual disability syndromes. Am. J. Med. Genet. A 161A, 2741–2749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapuria V., et al. , Proteolysis of HCF-1 by Ser/Thr glycosylation-incompetent O-GlcNAc transferase:UDP-GlcNAc complexes. Genes Dev. 30, 960–972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazarus M. B., et al. , HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 342, 1235–1239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhuiyan T., Waridel P., Kapuria V., Zoete V., Herr W., Distinct OGT-binding sites promote HCF-1 cleavage. PLoS One 10, e0136636 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khurana B., Kristie T. M., A protein sequestering system reveals control of cellular programs by the transcriptional coactivator HCF-1. J. Biol. Chem. 279, 33673–33683 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Huang L., et al. , A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am. J. Hum. Genet. 91, 694–702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jolly L. A., et al. , HCFC1 loss-of-function mutations disrupt neuronal and neural progenitor cells of the developing brain. Hum. Mol. Genet. 24, 3335–3347 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Koufaris C., Alexandrou A., Tanteles G. A., Anastasiadou V., Sismani C., A novel HCFC1 variant in male siblings with intellectual disability and microcephaly in the absence of cobalamin disorder. Biomed. Rep. 4, 215–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Firth H. V., Wright C. F.; DDD Study , The deciphering developmental disorders (DDD) study. Dev. Med. Child Neurol. 53, 702–703 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Karczewski K. J., et al. , Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv:10.1101/531210 (30 January 2019).
- 57.Pathak S., et al. , The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol. 22, 744–750 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pathak S., et al. , O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. EMBO J. 31, 1394–1404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazarus M. B., et al. , Structural snapshots of the reaction coordinate for O-GlcNAc transferase. Nat. Chem. Biol. 8, 966–968 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schimpl M., et al. , O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis. Nat. Chem. Biol. 8, 969–974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kenwrick S., Amaya E., Papalopulu N., Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Dev. Dyn. 229, 289–299 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Ingham P. W., A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell 37, 815–823 (1984). [DOI] [PubMed] [Google Scholar]
- 63.Mariappa D., Ferenbach A. T., van Aalten D. M. F., Effects of hypo-O-GlcNAcylation on Drosophila development. J. Biol. Chem. 293, 7209–7221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mariappa D., et al. , Dual functionality of O-GlcNAc transferase is required for Drosophila development. Open Biol. 5, 150234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Capotosti F., Hsieh J. J.-D., Herr W., Species selectivity of mixed-lineage leukemia/trithorax and HCF proteolytic maturation pathways. Mol. Cell. Biol. 27, 7063–7072 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qian K., et al. , Transcriptional regulation of O-GlcNAc homeostasis is disrupted in pancreatic cancer. J. Biol. Chem., 10.1074/jbc.RA118.004709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Z., Tan E. P., VandenHull N. J, Peterson K. R., Slawson C., O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis. Front. Endocrinol. 5, 206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vogel J. L., Kristie T. M., The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 19, 683–690 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deléhouzée S., et al. , GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells 10, 717–731 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Jaworski A., Smith C. L., Burden S. J., GA-binding protein is dispensable for neuromuscular synapse formation and synapse-specific gene expression. Mol. Cell. Biol. 27, 5040–5046 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perdomo-Sabogal A., et al. , Human lineage-specific transcriptional regulation through GA-binding protein transcription factor alpha (GABPa). Mol. Biol. Evol. 33, 1231–1244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okamoto I., et al. , Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472, 370–374 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Allen R. C., Nachtman R. G., Rosenblatt H. M., Belmont J. W., Application of carrier testing to genetic counseling for X-linked agammaglobulinemia. Am. J. Hum. Genet. 54, 25–35 (1994). [PMC free article] [PubMed] [Google Scholar]
- 74.Ørstavik K. H., et al. , X chromosome inactivation in carriers of Barth syndrome. Am. J. Hum. Genet. 63, 1457–1463 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knudsen G. P. S., et al. , Increased skewing of X chromosome inactivation in Rett syndrome patients and their mothers. Eur. J. Hum. Genet. 14, 1189–1194 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Trinidad J. C., et al. , Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jochmann R., Holz P., Sticht H., Stürzl M., Validation of the reliability of computational O-GlcNAc prediction. Biochim. Biophys. Acta 1844, 416–421 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Gundogdu M., et al. , The O-GlcNAc transferase intellectual disability mutation L254F distorts the TPR helix. Cell Chem. Biol. 25, 513–518.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Machida Y. J., Machida Y., Vashisht A. A., Wohlschlegel J. A., Dutta A., The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem. 284, 34179–34188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu H., et al. , The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 30, 5071–5085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dejosez M., et al. , Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 24, 1479–1484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Julien E., Herr W., Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 22, 2360–2369 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rodriguez-Jato S., Busturia A., Herr W., Drosophila melanogaster dHCF interacts with both PcG and TrxG epigenetic regulators. PLoS One 6, e27479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu H.-C. C., et al. , An X-linked cobalamin disorder caused by mutations in transcriptional coregulator HCFC1. Am. J. Hum. Genet. 93, 506–514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bruni F., Polosa P. L., Gadaleta M. N., Cantatore P., Roberti M., Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication. J. Biol. Chem. 285, 3939–3948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ristevski S., et al. , The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol. Cell. Biol. 24, 5844–5849 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Myers S. A., et al. , SOX2 O-GlcNAcylation alters its protein-protein interactions and genomic occupancy to modulate gene expression in pluripotent cells. Elife 5, e10647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Akan I., Love D. C., Harwood K. R., Bond M. R., Hanover J. A., Drosophila O-GlcNAcase deletion globally perturbs chromatin O-GlcNAcylation. J. Biol. Chem. 291, 9906–9919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Z., et al. , O-linked N-acetylglucosamine (O-GlcNAc) transferase and O-GlcNAcase interact with Mi2β protein at theAγ-globin promoter. J. Biol. Chem. 291, 15628–15640 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Resto M., et al. , O-GlcNAcase is an RNA polymerase II elongation factor coupled to pausing factors SPT5 and TIF 1β. J. Biol. Chem. 291, 22703–22713 (2016). Correction in: J. Biol. Chem.292, 16524–16525 (2017). [DOI] [PMC free article] [PubMed]
- 91.Das S., et al. , O-GlcNAcylation of GLI transcription factors in hyperglycemic conditions augments Hedgehog activity. Lab. Invest. 99, 260– 270 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hwang H., Rhim H., Functional significance of O-GlcNAc modification in regulating neuronal properties. Pharmacol. Res. 129, 295–307 (2018). [DOI] [PubMed] [Google Scholar]
- 93.Fourneau J., Canu M. H., Cieniewski-Bernard C., Bastide B., Dupont E., Synaptic protein changes after a chronic period of sensorimotor perturbation in adult rats: A potential role of phosphorylation/O-GlcNAcylation interplay. J. Neurochem. 147, 240–255 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Tallent M. K., et al. , In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. J. Biol. Chem. 284, 174–181 (2009). [DOI] [PubMed] [Google Scholar]
- 95.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vagin A. A., et al. , REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2184–2195 (2004). [DOI] [PubMed] [Google Scholar]
- 97.Borodkin V. S., et al. , Bisubstrate UDP-peptide conjugates as human O-GlcNAc transferase inhibitors. Biochem. J. 457, 497–502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou X., et al. , Hes1 desynchronizes differentiation of pluripotent cells by modulating STAT3 activity. Stem Cells 31, 1511–1522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wongpaiboonwattana W., Stavridis M. P., Neural differentiation of mouse embryonic stem cells in serum-free monolayer culture. J. Vis. Exp., e52823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.