A recent report (1) presents the long-awaited confirmation of paternal inheritance of mtDNA in humans (2). Surprisingly, paternal transmission of mtDNA (1) follows a bimodal pattern: About half of the offspring show fairly uniform paternal/maternal heteroplasmy levels, while the rest do not inherit paternal mtDNA at all. This pattern resembles the inheritance of a dominant nuclear gene. The authors explain this pattern as permissive inheritance resulting from a faulty “gatekeeper” gene (1). However, 3 groups (3–5) instead suspect contamination with mtDNA nuclear pseudogenes (NUMTs), a notorious artifact (6).
Based on our vast NUMT experience, we support the authors’ response (7), asserting that NUMT artifact is unlikely (further explanation is given in Supporting Notes [SN] sections SN1 and SN2, see ref. 8). However, we also demonstrate that the authors’ dominant gatekeeper explanation is incorrect, because spermatozoa are functionally diploid (SN3) and thus are equally affected by the faulty gatekeeper. This leaves the quasi-Mendelian inheritance unexplained.
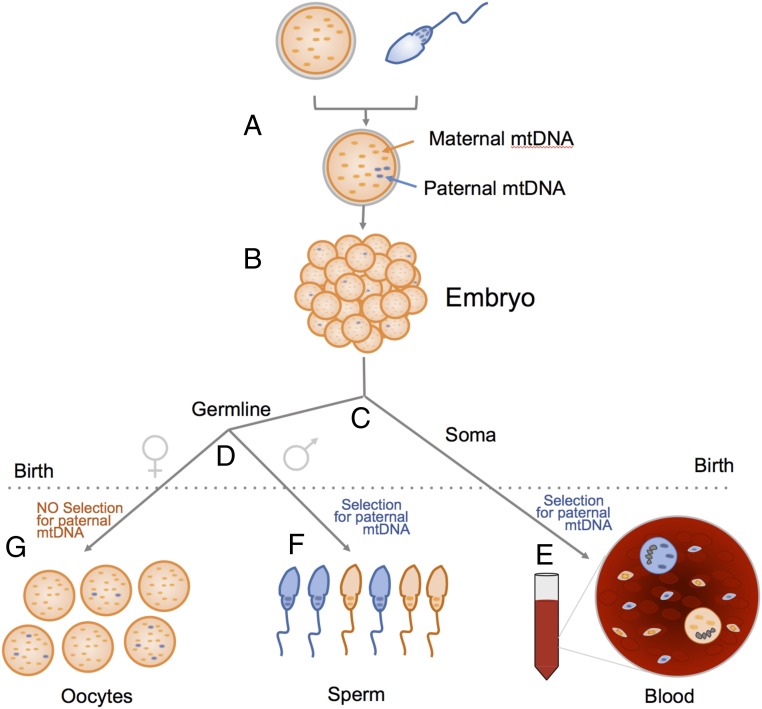
We offer an alternative explanation based on our analysis of the intracellular population dynamics of paternal mtDNA (Fig. 1). With a defective gatekeeper, a spermatozoon delivers <100 paternal mtDNA molecules (SN4) to the oocyte. In the beginning, cleavage of the embryo proceeds without mtDNA replication, and mtDNA molecules, including paternal ones, are randomly distributed among the blastomeres. Because of the low initial number of paternal molecules, some blastomeres are not “seeded” with paternal mtDNA (Fig. 1B) or later lose it due to intracellular “genetic drift.” Early replication of an mtDNA subpopulation offsets genetic drift, and ultimately about half of cells remain seeded with paternal mtDNA (SN7.2).
Fig. 1.
As paternal mtDNA (A) is distributed among blastomeres not all of them receive a copy (B). Later germline is parted from soma (C) and then the sex of the germline is set (D). After birth (dotted line), somatic and male germline cells exert selection in favor of the paternal mtDNA haplotype, so that somatic cells and spermatozoa end up as a paternal/maternal mtDNA mosaic. This explains somatic heteroplasmy (E) and quasi-Mendelian inheritance (F), respectively. In the female germline, there is no selection for the paternal haplotype (9), so oocytes keep low and variable proportions of paternal mtDNA (G) (SN7.3.2).
Because paternal inheritance includes reproducible, ∼1,000-times enrichment of paternal mtDNA, paternal mtDNA haplotype must have selective advantage (SN5). Competition between mtDNA haplotypes is well documented (9), and, as we show (SN6), is sufficiently strong to enrich the paternal haplotype to ∼100% in the “seeded” cell lineages. Nonseeded lineages naturally remain 100% maternal, so a mosaic of cells with mostly paternal and pure maternal mtDNA is created (Fig. 1E).
In mosaic somatic tissue, the heteroplasmy level equals the proportion of “paternal mtDNA” cells, which in turn is equal to the “seeding density,” that is, the proportion of embryonic cell lineages that received (and kept) paternal mtDNA.
Similarly, in male germline, selection for paternal mtDNA ultimately results in a mosaic of spermatozoa with paternal and maternal mtDNA (Fig. 1F). This explains the quasi-Mendelian bimodal inheritance, with a transmission rate equal to the “seeding density” of germline lineages. Female germline (no selection) follows a different path (Fig. 1G and SN7.3.2)
Intuitively, seeding density should depend on the initial number of paternal mtDNA, the timing of mtDNA replication initiation, and the dynamics of the intracellular population of mtDNA molecules. To investigate the combined effect of these factors quantitatively, we performed numerical simulations of mtDNA behavior (SN7.2). Simulations predict intermediate seeding densities and heteroplasmy levels in somatic and germ cells (SN7.3), in agreement with the observations (SN7.1).
In conclusion, despite apparent inconsistencies and some missing quality control (SN10), Luo et al. (1) have likely described genuine cases of paternal inheritance, and their seemingly impossible inheritance patterns are compatible with known mtDNA biology.
Footnotes
The authors declare no conflict of interest.
Data deposition: Supporting Notes for this letter are available at https://figshare.com/s/2c0d2508a0245b5e118f.
References
- 1.Luo S., et al. , Biparental inheritance of mitochondrial DNA in humans. Proc. Natl. Acad. Sci. U.S.A. 115, 13039–13044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz M., Vissing J., Paternal inheritance of mitochondrial DNA. N. Engl. J. Med. 347, 576–580 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Vissing J., Paternal comeback in mitochondrial DNA inheritance. Proc. Natl. Acad. Sci. U.S.A. 116, 1475–1476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lutz-Bonengel S., Parson W., No further evidence for paternal leakage of mitochondrial DNA in humans yet. Proc. Natl. Acad. Sci. U.S.A. 116, 1821–1822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salas A., Schönherr S., Bandelt H.-J., Gómez-Carballa A., Weissensteiner H., Extraordinary claims require extraordinary evidence in the case of asserted mtDNA biparental inheritance. bioRxiv:10.1101/585752 (25 March 2019). [DOI] [PubMed]
- 6.Hirano M., et al. , Apparent mtDNA heteroplasmy in Alzheimer’s disease patients and in normals due to PCR amplification of nucleus-embedded mtDNA pseudogenes. Proc. Natl. Acad. Sci. U.S.A. 94, 14894–14899 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo S., et al. , Reply to Lutz-Bonengel et al.: Biparental mtDNA transmission is unlikely to be the result of nuclear mitochondrial DNA segments. Proc. Natl. Acad. Sci. U.S.A. 116, 1823–1824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Annis S., et al. , Quasi-Mendelian paternal inheritance of mitochondrial DNA: A notorious artifact, or anticipated behavior? Figshare. https://figshare.com/s/2c0d2508a0245b5e118f. Deposited 5 March 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenuth J. P., Peterson A. C., Shoubridge E. A., Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 16, 93–95 (1997). [DOI] [PubMed] [Google Scholar]