Significance
Cryptic species complicate our ability to detect changes in biological communities that result from human-mediated shifts in ecosystem structure. We used DNA barcoding to compare the diversity of cryptic coralline algal assemblages in 2 alternative ecosystem states that differ in herbivore biomass and food chain length. We show that while coralline cover is greater in urchin-dominated sites (or “barrens”) subject to intense grazing than in kelp-dominated sites with low urchin densities, coralline communities in urchin barrens are significantly less diverse and dominated by 1 or 2 species. These findings illuminate ecological dynamics previously unexplored within this classic trophic cascade and show how identifying cryptic species can alter our interpretations of biological phenomena.
Keywords: cryptic species, coralline algae, trophic cascade, biodiversity
Abstract
Understanding how trophic dynamics drive variation in biodiversity is essential for predicting the outcomes of trophic downgrading across the world’s ecosystems. However, assessing the biodiversity of morphologically cryptic lineages can be problematic, yet may be crucial to understanding ecological patterns. Shifts in keystone predation that favor increases in herbivore abundance tend to have negative consequences for the biodiversity of primary producers. However, in nearshore ecosystems, coralline algal cover increases when herbivory is intense, suggesting that corallines may uniquely benefit from trophic downgrading. Because many coralline algal species are morphologically cryptic and their diversity has been globally underestimated, increasing the resolution at which we distinguish species could dramatically alter our conclusions about the consequences of trophic dynamics for this group. In this study, we used DNA barcoding to compare the diversity and composition of cryptic coralline algal assemblages at sites that differ in urchin biomass and keystone predation by sea otters. We show that while coralline cover is greater in urchin-dominated sites (or “barrens”), which are subject to intense grazing, coralline assemblages in these urchin barrens are significantly less diverse than in kelp forests and are dominated by only 1 or 2 species. These findings clarify how food web structure relates to coralline community composition and reconcile patterns of total coralline cover with the widely documented pattern that keystone predation promotes biodiversity. Shifts in coralline diversity and distribution associated with transitions from kelp forests to urchin barrens could have ecosystem-level effects that would be missed by ignoring cryptic species’ identities.
Changes in trophic dynamics can have widespread effects on the diversity and composition of communities (1–3). Although mounting evidence points to the prevalence of trophic downgrading across the world’s ecosystems (1, 2), we do not fully understand how diversity is distributed across communities (4, 5) or how it is influenced by large-scale shifts in food web structure (2, 3, 6). This knowledge gap is particularly wide for cryptic species whose existence may only be realized through modern DNA sequencing analyses (7). Cryptic species have caused researchers to rethink classic paradigms in ecology, such as the prevalence of generalist species (8, 9) and the mechanisms involved in ecological speciation (7, 10), but the importance of distinguishing cryptic species when discerning key ecological patterns remains unclear (11–13). If cryptic species are ecologically similar, then grouping species based on morphology or functional group may be a reasonable approach to studying biological distributions. However, if cryptic species differ ecologically, then species that look superficially similar may perform in fundamentally different ways across environmental gradients or in competitive interactions, and inconspicuous shifts in their distributions may significantly alter ecosystem structure.
In nearshore marine communities worldwide, shifts in trophic dynamics that result in high abundances of herbivorous sea urchins trigger dramatic shifts from biologically rich kelp forests to depauperate “urchin barrens” (14, 15). The collapse of kelp forests results in a transformation of reef function: Structural complexity and productivity are reduced (16), and biodiversity of many taxa is lost (17). However, calcified coralline algae (orders Corallinales, Hapalidiales, and Sporolithales), which may benefit from herbivore grazing pressure (18–22), often dominate these high herbivory environments and are thought to be ecological “winners” of this ecosystem state shift (14, 19–21, 23). Corallines are morphologically cryptic and, consequently, are often lumped together into a single vague category of crustose coralline algae (CCA) (e.g., refs. 23–25). Thus, we know little about the responses of coralline algal diversity and community composition to shifts in kelp forest regimes. If CCA abundance increases when kelp forests collapse, could this also reflect an increase in coralline diversity, providing an exception to the widely observed pattern (2) that keystone predation enhances biodiversity?
Herbivory is a key driver of macroalgal community dynamics (26, 27). Grazing can maintain coexistence and diversity of primary producers by decreasing the strength of competitive interactions (28–31) or even reversing them completely (31, 32), a paradigm that has been bolstered by empirical evidence in corallines (22, 28, 33). For example, thicker coralline crusts tend to outcompete thinner ones in the intertidal zone, but grazing can alter competitive outcomes by causing thinner species to overgrow themselves as they recover from disturbance, and effectively become thicker (32). If increased herbivory promotes coexistence, then we might expect urchin barrens with high urchin biomass to have greater coralline diversity than kelp forests, matching observed positive correlations between herbivore pressure and total coralline cover (18–22). However, the effect of herbivory on diversity depends upon intensity and context (26, 31), and herbivory in urchin barrens is unusually intense (14, 34). Thus, alternatively, herbivory in urchin barrens may reduce species coexistence by allowing herbivore-tolerant species to dominate and outcompete less tolerant species (26, 31, 35, 36). This latter hypothesis would suggest that increases in total “CCA” cover do not reflect patterns of diversity but, instead, emerge from the success of a small number of herbivore-tolerant species.
Coralline algae play essential roles in marine ecosystems and are ubiquitous in most rocky reef habitats (10, 23). Corallines cement coral reefs together (37), act as a substrate for algae and surf grass (38), facilitate kelp recruitment (39, 40), and chemically induce larval settlement of a wide phylogenetic range of invertebrates (41–45). However, our ability to effectively differentiate coralline species has been obscured by deceptively simple morphologies, rampant convergent evolution, and misunderstood phenotypic variation (46, 47). DNA barcoding techniques allow for the accurate identification of species and have demonstrated that some of the most common species under the morphological species concept in fact represent several to more than a dozen distinct genetic species (46, 48, 49). Widespread cryptic diversity is potentially problematic for ecologists because coralline species can differ in growth rate (43, 50), competitive ability (28, 32, 33), resistance to environmental stress (51, 52), and their effects on invertebrate recruitment (43, 53). Therefore, changes in the diversity of coralline assemblages could influence their contributions to ecosystem function, yet shifts could easily go undetected due to the cryptic nature of coralline taxa.
In the North Pacific, transitions from urchin barrens to kelp forests, and vice versa, can occur as a result of trophic cascades triggered by the local extirpation or recovery of sea otters that act as “keystone” predators, reducing the abundance of herbivorous urchins (17, 54). The loss of sea otters has occurred throughout the northeastern Pacific (14) and can lead to a 30- to 100-fold increase in macroalgal consumption rates (34). Here, we make use of a naturally occurring gradient in sea otter occupation (55) to examine how the species composition and diversity of coralline algae differ across 4 sites that range in sea urchin biomass and associated grazing pressure. We use DNA barcoding methods to identify coralline algal species and reliably quantify their diversity and abundance. We examine the distribution of species diversity across local scales and ask how the composition of coralline communities is influenced by the top-down control of sea otters on sea urchins and whether certain species, for example, those with thicker crusts (20, 32), show evidence of competitive advantage under heavy urchin grazing. We also test whether closely related species are distributed in similar ways across a gradient of top-down forcing. In doing so, we provide a taxonomically accurate look at the responses of cryptic coralline species to the pervasive shift between kelp-dominated and urchin-dominated ecosystem states.
Results
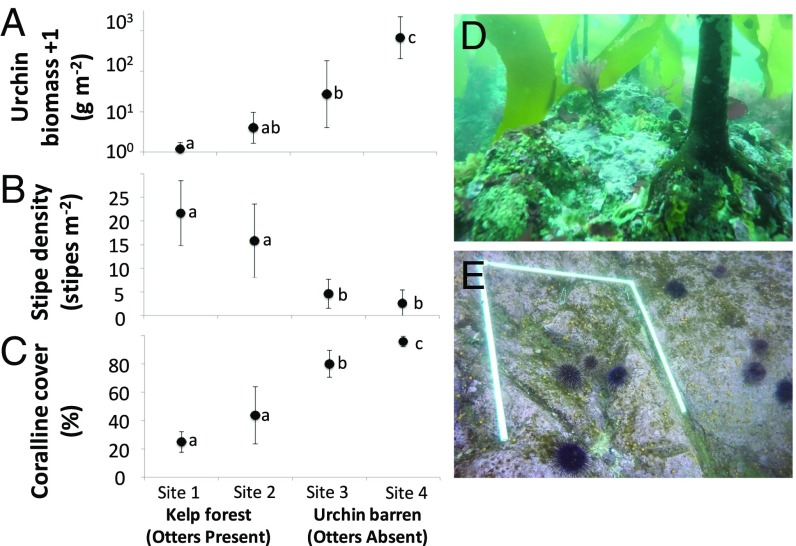
Sites without otters (i.e., urchin barrens) had higher urchin biomass, lower brown algal stipe densities (Laminariales and Desmarestiales), and higher coralline algal cover than otter-occupied sites (Fig. 1). Urchin biomass was significantly different among sites (Fig. 1A; ANOVA: degrees of freedom [df] = 3, F = 19.61, P < 0.0001), with the highest mean urchin biomass observed at the urchin barren sites, sites 3 and 4 (Tukey’s honest significant difference [HSD]: P < 0.001) (site 1 = 1.6 ± 1.6 g⋅m−2 [mean ± SE], site 2 = 36.7 ± 28.5 g⋅m−2, site 3 = 1,923.3 ± 720.5 g⋅m−2, site 4 = 2,032.1 ± 439.3 g⋅m−2; untransformed data are shown in SI Appendix, Fig. S1). Red urchins (Mesocentrotus franciscanus) made up more than 97% of urchin biomass at all sites, but green urchins (Strongylocentrotus droebachiensis) were rare at 3 of the 4 sites (site 1 = 3%, site 3 < 1%, site 4 < 1%). Kelp density was significantly different among sites (Fig. 1B; ANOVA: df = 3, F = 23.62, P < 0.0001): low in urchin barren sites (site 3 = 4.6 ± 1.5 stipes per square meter [mean ± SE], site 4 = 2.6 ± 1.4 stipes per square meter) and high in otter-occupied kelp forests (Tukey’s HSD: P < 0.001) (site 1 = 21.7 ± 3.5 stipes per square meter, site 2 = 15.8 ± 4.0 stipes per square meter). Percent cover of coralline algae was significantly different among sites (Fig. 1C; ANOVA: df = 3, F = 30.8, P < 0.0001), with significantly higher mean cover estimates recorded at urchin barren sites (Tukey’s HSD: P < 0.05) (site 1 = 24.9 ± 3.7% [mean ± SE], site 2 = 43.7 ± 10.3%, site 3 = 79.9 ± 4.9%, site 4 = 95.8 ± 1.9%). Coralline cover was significantly different between the 2 urchin barrens (Tukey’s HSD: P < 0.05) but not between the 2 kelp forests (Tukey’s HSD: P = 0.24).
Fig. 1.
Mean urchin biomass (A), mean adult stipe density of Laminariales and Desmarestiales (B), and total percent coralline cover (C) at kelp forest sites occupied by sea otters (site 1 = 34 y, site 2 = 18 y) and urchin barren sites not occupied by otters. Images from site 1 (D) and site 4 (E) are shown. Data are mean ± 95% confidence interval. Lowercase letters indicate significant differences between means as determined by using Tukey’s HSD post hoc test.
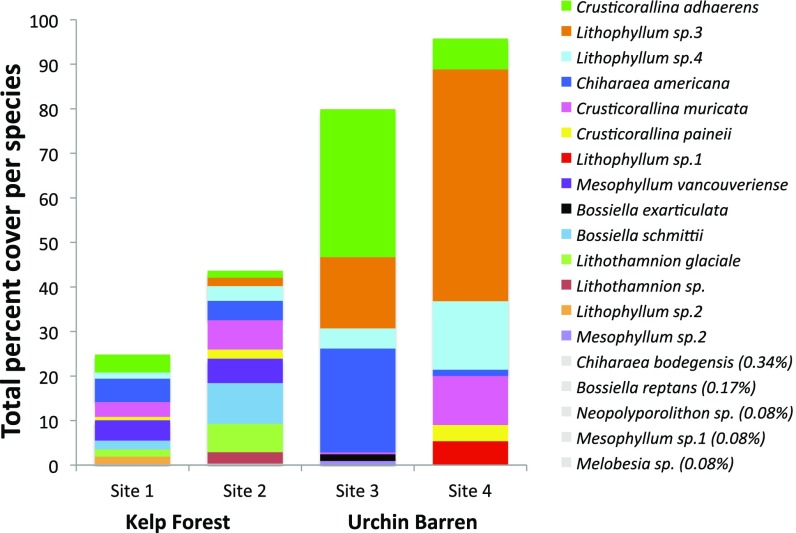
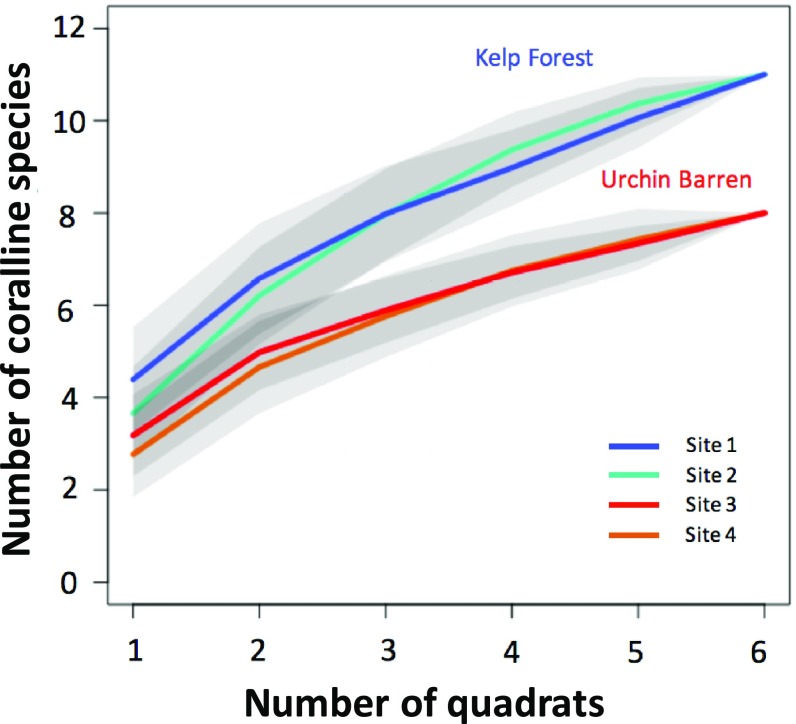
Across all sites, subtidal coralline algal assemblages were surprisingly diverse but diversity was lower in urchin barrens than in kelp forests. Using the psbA barcoding region, we identified 19 distinct genetic species of coralline algae (Fig. 2), some of which did not match any verified taxa in our database and are likely new to science, requiring taxonomic attention. Six species were shared between the 2 habitat types, 9 species were found only in kelp forests, and 5 species were found only at urchin barrens (Fig. 2). Across sites, there were fewer species detected in urchin barrens, with a total of 8 coralline species at each site, compared with 11 coralline species detected at each kelp forest site (Fig. 2). Although species accumulation curves suggest that further sampling may have revealed additional species in both habitats, species numbers increased more rapidly with additional sampling in kelp forest sites than in urchin barren sites (Fig. 3), suggesting that increased sampling might result in even greater discrepancies in species richness between kelp forest and urchin barren sites. Coralline assemblages were dominated by crustose corallines at all sites. Three of 4 articulated coralline species were found only at kelp forest sites and only in low abundance (<10%). One articulated species, Chiharaea americana, which has small, inconspicuous upright fronds (<2 mm tall) (56), was found in low abundances at 3 sites but was moderately abundant (23%) at just a single urchin barren site.
Fig. 2.
Stacked bar graphs showing the contribution of each genetic species (as determined by psbA) to total coralline percent cover across each site. Species observed with a total percent cover less than 0.4% are indicated in gray.
Fig. 3.
Accumulation curves for coralline algal species observed in kelp forests and urchin barrens. Sites are represented by different colors: site 1 (turquoise), site 2 (blue), site 3 (red), and site 4 (orange). Gray shading represents 95% confidence intervals.
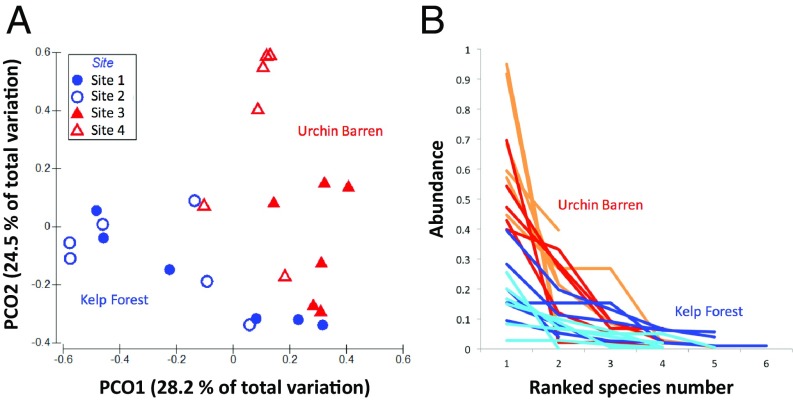
Urchin barren sites were dominated by only a few coralline species (Fig. 2), the identity of which differed between the 2 urchin barren sites. For example, Lithophyllum sp.3 dominated site 4, with 52% of total percent cover, whereas Crusticorallina adhaerens dominated site 3 with 33% of total percent cover. This pattern was reflected in the results of a permutational multivariate analysis of variance (PERMANOVA) on quadrat-level coralline community composition (Fig. 4A; PERMANOVA: df = 3, pseudo-F = 3.2013, P < 0.0001), which revealed that urchin barren sites were significantly different (post hoc pairwise comparison: P < 0.05) but the 2 kelp forest sites were similar (post hoc pairwise comparison: P = 0.62). Per quadrat, coralline species richness was not significantly different among habitats but there was a trend toward increased richness in quadrats measured at kelp forest sites (site 1: 3.7 ± 0.6, site 2: 4.3 ± 0.6, site 3: 3.2 ± 0.5, site 4: 2.8 ± 0.5; ANOVA: df = 1, F = 7.2, P = 0.12; SI Appendix, Table S1). Shannon diversity (site 1: 2.8 ± 0.4, site 2: 3.2 ± 0.4, site 3: 2.1 ± 0.3, site 4: 2.0 ± 0.3) and Simpson’s diversity (site 1: 2.5 ± 0.4, site 2: 2.8 ± 0.4, site 3: 1.9 ± 0.3, site 4: 1.8 ± 0.3) were significantly higher per quadrat at kelp forest sites (Shannon ANOVA: df = 1, F = 15.3, P < 0.05; Simpson’s ANOVA: df = 1, F = 37.3, P < 0.05; SI Appendix, Table S1), indicating that kelp forests were more even than urchin barrens. Rank abundance curves further demonstrate unevenness of urchin barrens relative to kelp forests (Fig. 4B).
Fig. 4.
(A) Principal coordinate (PCO) analysis of species composition in each quadrat; variation attributed to the top-ranking principal coordinates (PCO1 & PCO2) are shown. (B) Rank-abundance curve plotting coralline percent cover against species rank (based on abundance) for urchin barren and kelp forest sites. Each line represents a quadrat sampled. Sites are represented by different colors: site 1 (turquoise), site 2 (blue), site 3 (red), and site 4 (orange).
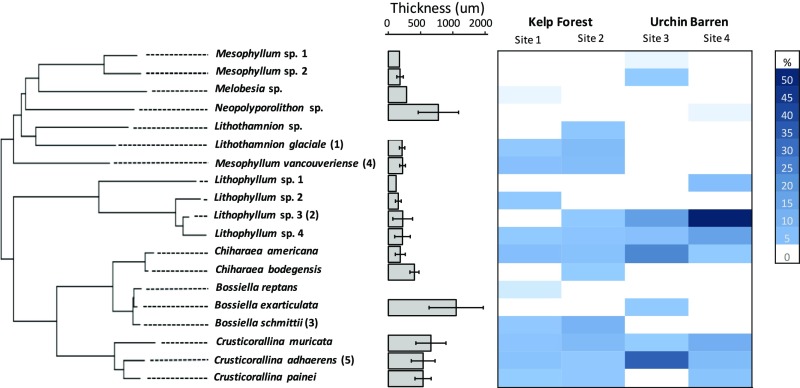
The distributions of coralline species across the herbivory gradient were not correlated with either crust thickness or phylogenetic relatedness (Fig. 5). To test for effects of phylogenetic signal and crust thickness on species distributions, we calculated differences in average abundance at sites with and without otters. Crust thickness did not significantly predict which species were found in each habitat type (linear model: F = 0.9354, df = 1 and df = 14, P = 0.3499). Moreover, there was no effect of site (ANOVA: F = 0.3581, df = 3, P = 0.7839) or total coralline cover (linear regression: F = 0.2587, df = 1 and df = 22, P = 0.6161) on community-weighted mean thickness (SI Appendix, Fig. S2). There was also no significant effect of phylogeny on differences in coralline species abundances at kelp forest and urchin barren sites (Blomberg’s K: K = 0.2198, P = 0.6609; Pagel’s λ: λ < 0.001, P = 1), and none of the sites that we assessed were significantly clustered or overdispersed phylogenetically, as determined by standard effect size (SES) of mean pairwise distance (MPD) and SES of mean nearest taxon distance (MNTD) (SI Appendix, Fig. S3). There was also no phylogenetic signal of crust thickness (Blomberg’s K: K = 0.469, P = 0.1311; Pagel’s λ: λ = 0.317, P = 0.3997) (Fig. 5). Supervised classification using randomForest analysis showed that 5 phylogenetically dissimilar species were the most informative in distinguishing kelp forests from urchin barrens (Fig. 5).
Fig. 5.
Phylogram of Corallinales from this study showing species relatedness (as inferred using an 856-bp region of psbA) plotted with crust thickness and site-level abundance. The heatmap shows site-level abundance of each coralline algal species sampled at each site. The numbers in parentheses indicate ranking of taxa that are most important in distinguishing kelp forests from urchin barrens based on supervised classification analysis. Lithothamnion glaciale was the most important discriminating species (mean decrease in accuracy = 0.0608, SD = 0.0313), followed by Lithophyllum sp.3, Bossiella schmittii, Mesophyllum vancouveriense, and C. adhaerens.
Discussion
Unmasking Cryptic Coralline Diversity.
Contrary to the simple conclusion that coralline algae are the “winners” in the transition from kelp forests to urchin barrens (e.g., refs. 14, 20, 21), our results indicate that the situation is more nuanced. Although abundance of corallines is greater at urchin barren sites, lumping species as “CCA” fails to capture the complex dynamics of coralline algal assemblages and how they are affected by the otter/urchin/kelp trophic cascade. In the absence of top-down control by sea otters, high urchin abundance and grazing pressure at rocky reef sites corresponded with lower coralline algal diversity and evenness despite higher total coralline cover. Our results show that while a few dominant coralline species strongly benefit from intense urchin herbivory, many coralline species decline or become excluded from urchin barren habitats. Multiple nonmutually exclusive hypotheses could explain these observed patterns. The loss of keystone predators, such as sea otters, can shift the landscape of competitive advantage for coralline algae: Without top-down predation, urchin herbivory is more intense and likely more homogeneous (34), in turn, favoring those coralline species that are tolerant of disturbance, while excluding species that are unable to withstand intense urchin grazing. Because coralline assemblages in the northeastern Pacific have evolved with otters present for millions of years (57), few species may be adapted to these environments of intense herbivory (34). Herbivore-mediated exclusions of algal species have been previously documented (e.g., refs. 18, 26, 58) and could explain the lower diversity present at urchin barren sites.
The transition from kelp forests to urchin barrens may also influence coralline communities through indirect processes. For example, otter-mediated heterogeneity in urchin grazing (34) may promote spatial variation in resources, limiting competitive advantage by allowing for differential success of species in varying microhabitats (59–61). In contrast, intense grazing pressure at urchin barrens could homogenize environmental factors such as light and flow (62–64), which could facilitate the dominance of a few species that are particularly successful under those conditions. Macroalgal canopy loss could also increase the importance of environmental factors in determining species distributions. For example, kelp cover can attenuate sunlight, preventing coralline bleaching (65), and can buffer pH (66), perhaps reducing the effects of ongoing ocean acidification on calcified organisms such as coralline algae (67, 68).
Regardless of the mechanism underlying these patterns, our results have broad implications for how we view cryptic species in ecological studies. Many studies lump phylogenetically similar cryptic species into a single category or inconsistently distinguish between species, and the common lumping of all CCA species into “CCA” is just an extreme example. Cryptic speciation is a widespread phenomenon in divergent groups such as insects (69), fungi (70), and embryophytes (71). As such, the importance of taxonomic resolution and the appropriateness of functional groups are widely debated topics in ecology (13, 72). In our study system, lumping all species has led to the interpretation that corallines are more successful when urchins are highly abundant (20, 23), but increasing taxonomic resolution suggests that coralline diversity is actually lost in these habitats. This demonstrates how improved resolution of cryptic species can illuminate previously hidden or underappreciated ecological patterns, which, in turn, raises new questions about the mechanisms that underlie ecological dynamics.
Dominance in Urchin Barrens Is Hard to Predict.
Previous work has suggested that crust thickness is a key predictor of competitive outcomes when grazers are present (20, 32). However, our results refute the generality of this conclusion by demonstrating that urchin barrens are not always composed of thicker species (Fig. 5 and SI Appendix, Fig. S2). For example, 1 urchin barren was dominated by a thick species (C. adhaerens) and another was dominated by a relatively thin Lithophyllum species (Fig. 3). Moreover, there was no significant correlation between crust thickness and species abundance in different rocky reef states. While crust thickness may determine competitive outcomes in controlled experiments (e.g., refs. 28, 32, 33), our results suggest this does not always scale up to natural community assemblages, at least not in high-herbivory subtidal systems. Instead, differences in physiology, including increased resistance to light stress (73), elevated growth rate (43), or perhaps species-specific recruitment characteristics, may play more significant roles in distributing species when kelp canopies are lost or recover from urchin grazing. The depth of conceptacles, cavities containing the reproductive structures of coralline algae, has also been hypothesized to correlate with herbivore resistance because deeper conceptacles may be protected from benthic grazers (19). We did not explicitly quantify conceptacle depth, but this hypothesis should be considered in future studies, particularly once the new species have been properly characterized and described. Because the identity of dominant species differed between urchin barren sites, site-level environmental conditions or stochasticity [e.g., lottery effects (74)] may play an important role in determining which species dominate in urchin barrens.
Coralline species were distributed across kelp forests and urchin barrens without any phylogenetic pattern. While phylogenetic signals are difficult to detect in low-diversity systems (75), our results clearly demonstrate that communities were composed of species from many different coralline lineages. Supervised classification analysis further revealed that 5 phylogenetically dispersed coralline species were the most informative in distinguishing urchin barrens and kelp forests (Fig. 5). This pattern suggests that responses to grazing are not phylogenetically conserved (76) but may, instead, involve several different species-specific or convergent strategies for resisting intense herbivory across taxa. Coralline algae are an ancient lineage (77), and convergent evolution is well noted across the diverse clade (20). For example, articulated corallines have evolved at least 3 times from crusts (77, 78), with reversals documented (46, 47), and variation in crust thickness has evolved repeatedly across the coralline phylogeny (Fig. 5). Convergent evolution tends to eliminate phylogenetic signal (79) and may explain why we observed no phylogenetic structure in assemblage composition.
While our results suggest convergence among distantly related lineages, they also provide evidence for divergence among closely related species. Divergent selection is a key evolutionary mechanism and leads to ecological differentiation among related taxa (80). Our data show that morphologically similar congeneric species (e.g., Lithophyllum spp. and Crusticorallina spp.), which are frequently lumped together in other studies, are ecologically different and are distributed differently across forested and deforested reefs (Fig. 5). For example, of 3 observed Crusticorallina species once thought to be a single entity (46), C. adhaerens was the only Crusticorallina species to dominate sites without otters (Fig. 2), suggesting that traits underlying species distributions may not be obvious in morphologically similar species. There is a clear need to use molecular identifications in future studies to compare the physiological and biotic tolerances of different coralline species and to tease apart the causes and consequences of cryptic species distributions in nature.
Implications Across the Seascape.
Enhancing the resolution at which we investigate coralline algal diversity could have important implications for understanding the community assembly and functioning of nearshore ecosystems. This may be particularly insightful across thousands of kilometers of coastline, where different drivers (e.g., sea otter recovery, fishing, urchin disease) trigger rocky reefs to shift between urchin-dominated barrens and macroalgae-dominated forests (14, 15). Coralline species differ in a number of ecological traits (43, 50–52), including their influence on recruitment of other organisms (43, 53); therefore, shifts in coralline diversity and composition at large geographic scales may affect the entire ecosystem. Shifts in coralline species that are necessary for macroalgal or invertebrate recruitment (25, 41, 43, 62, 81) could influence the composition of nearshore communities both inside and outside of urchin barrens. For example, retention of coralline species that urchin larvae prefer (25, 41) or the loss of coralline species necessary for kelp recruitment (40) could reinforce urchin barren states, contributing to the hysteresis observed in kelp forest/urchin barren regime shifts (14, 15, 53). Moreover, large-scale shifts in coralline species distributions could influence source-sink dynamics (82) across the seascape, and therefore further affect patterns of recruitment across entire coastlines. The challenge for future researchers will be to integrate physiological and ecological data for each cryptic species with molecularly confirmed field distributions to understand the drivers and downstream effects of shifting coralline community composition.
Samples and Methods
Field Surveys and Species Identification.
We conducted subtidal surveys (July 11–14, 2014) at 4 rocky reef sites: 2 kelp forests and 2 urchin barren sites on the central coast of British Columbia, Canada, where sea otter populations have been recovering (since around 1980) from extirpation due to the fur trade (83) (SI Appendix, Fig. S4). All methods were conducted by scuba diving. At each site, sea urchin density, sea urchin test diameter, and adult stipe density of kelps (Laminariales) and Desmarestialean algae (≥15 cm) were quantified in 18 stratified random 1-m2 quadrats (depths 3 to 12 m below mean low water) that spanned 6 30-m horizontal transects laid at 2 depth contours (details are provided in ref. 55). Sea urchin biomass was estimated from density and test diameter (84). Total coralline cover and the specific cover of genetically identified species were estimated from 6 0.25-m2 quadrats placed randomly near the first transect (8 to 13 m depth below mean low water) at each site. Although urchin biomass and kelp density were quantified across a larger area and wider range of depths (to provide representative reef-scale measures that accounted for spatial variability) than the coralline surveys, the same patterns were found when restricting analyses to just the first 2 spatially overlapping transects (SI Appendix, Fig. S5). Specimens were identified using amplification and sequencing of psbA, COI-5P, or rbcL genetic barcoding regions following Hind et al. (46) (SI Appendix, Dataset S1). The psbA sequence data were also used for phylogenetic inference.
Crust thickness measurements were mostly made on samples removed from the plots, when specimen size permitted. To increase sample size, additional measurements were made on herbarium samples from the British Columbia coast that had previously been identified using molecular sequence data. No crust thickness measurements were taken for Lithothamnion sp.1 since only very small fragments were obtained and no herbarium vouchers were available for measuring. Thus, Lithothamnion sp.1 and all articulated species were excluded from the analysis of crust thickness.
Statistical Analysis.
All analyses were conducted in R (R Core Team) using the packages “vegan” (85), “Phytools” (86), “picante” (87), and “randomForest” (88), except for the multivariate analyses that were conducted in PRIMER (89). Community-weighted mean thickness was calculated for each quadrat using the sum of the proportion of total coralline cover occupied by each species multiplied by its thickness. To determine whether closely related species were distributed similarly with respect to urchin grazing, we tested for the effect of phylogeny on community assembly at urchin barren and kelp forest sites. We first performed phylogenetic inference using an 856-bp alignment of psbA sequences aligned and edited in Geneious (90). A neighbor-joining tree was then inferred using a Tamura–Nei model in Geneious (90). Next, Blomberg et al.’s K (75) and Pagel’s λ (91) values were calculated to test for phylogenetic signal on species distributions and on crust thickness. To test for phylogenetic signal on species distributions, we calculated differences in average abundance at sites with and without otters (hereafter “habitat preference”). SESs of MPD and MNTD were also calculated to test for phylogenetic clustering or overdispersion relative to communities that were randomly generated but maintained species richness at the quadrat level. Detailed methods are available in SI Appendix, Extended Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Hakai Institute for their contributions and access to outstanding research facilities along the central coast of British Columbia. Special thanks to the following individuals: N. Roberts, O. Pontier, M. Vaughn, and B. Keeling for dive support; K. Demes for input on sampling logistics; C. Jensen for helping generate and analyze DNA sequence data; J. Lai for assistance acquiring GenBank accessions; and A. Houweling for measuring crust thickness. Thanks to Osprey Ranch for providing a peaceful and productive writing environment. The manuscript benefitted from fruitful conversations with M. O’Connor, P. Thompson, M. Whalen, and P. Gabrielson. Research funding was provided by the Hakai Institute, by Natural Sciences and Engineering Research Council (NSERC) Canadian Graduate Scholarships (to J.M.B. and S.S.), and by a CFI and Pew Fellowship (to A.K.S.), and by NSERC Discovery grants (to A.K.S. and P.T.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data accessibility statement data collected and analyzed in this study are available at https://github.com/martonelab/SubtidalCorallines2018.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900506116/-/DCSupplemental.
References
- 1.Jackson J. B. C., et al. , Historical overfishing and the recent collapse of coastal ecosystems. Science 293, 629–637 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Estes J. A., et al. , Trophic downgrading of planet Earth. Science 333, 301–306 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Terborgh J. W., Toward a trophic theory of species diversity. Proc. Natl. Acad. Sci. U.S.A. 112, 11415–11422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purvis A., Hector A., Getting the measure of biodiversity. Nature 405, 212–219 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Jetz W., McPherson J. M., Guralnick R. P., Integrating biodiversity distribution knowledge: Toward a global map of life. Trends Ecol. Evol. 27, 151–159 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Thompson R. M., et al. , Food webs: Reconciling the structure and function of biodiversity. Trends Ecol. Evol. 27, 689–697 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Bickford D., et al. , Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Smith M. A., Woodley N. E., Janzen D. H., Hallwachs W., Hebert P. D. N., DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc. Natl. Acad. Sci. U.S.A. 103, 3657–3662 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blair C. P., Abrahamson W. G., Jackman J. A., Tyrrell L., Cryptic speciation and host-race formation in a purportedly generalist tumbling flower beetle. Evolution 59, 304–316 (2005). [PubMed] [Google Scholar]
- 10.Fišer C., Robinson C. T., Malard F., Cryptic species as a window into the paradigm shift of the species concept. Mol. Ecol. 27, 613–635 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Hirst A. J., Influence of taxonomic resolution on multivariate analyses of arthropod and macroalgal reef assemblages. Mar. Ecol. Prog. Ser. 324, 83–93 (2006). [Google Scholar]
- 12.Konar B., Iken K., Influence of taxonomic resolution and morphological functional groups in multivariate analyses of macroalgal assemblages. Phycologia 48, 24–31 (2009). [Google Scholar]
- 13.Delić T., Trontelj P., Rendoš M., Fišer C., The importance of naming cryptic species and the conservation of endemic subterranean amphipods. Sci. Rep. 7, 3391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filbee-Dexter K., Scheibling R. E., Sea urchin barrens as alternative stable states of collapsed kelp ecosystems. Mar. Ecol. Prog. Ser. 495, 1–25 (2014). [Google Scholar]
- 15.Ling S. D., et al. , Global regime shift dynamics of catastrophic sea urchin overgrazing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20130269 (2015). [Google Scholar]
- 16.Teagle H., Hawkins S. J., Moore P. J., Smale D. A., The role of kelp species as biogenic habitat formers in coastal marine ecosystems. J. Exp. Mar. Biol. Ecol. 492, 81–98 (2017). [Google Scholar]
- 17.Estes J. A., Palmisano J. F., Sea otters: Their role in structuring nearshore communities. Science 185, 1058–1060 (1974). [DOI] [PubMed] [Google Scholar]
- 18.Johnson L. E., Paine R. T., Consistency in a marine algal-grazer interaction over multiple scales. J. Phycol. 52, 942–950 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Steneck R. S., “Adaptations of crustose coralline algae to herbivory: Patterns in space and time” in Paleoalgology, Toomey D. F., Nitecki M. H., Eds. (Springer, Berlin, 1985), pp. 352–366. [Google Scholar]
- 20.Steneck R. S., The ecology of coralline algal crusts: Convergent patterns and adaptative strategies. Annu. Rev. Ecol. Syst. 17, 273–303 (1986). [Google Scholar]
- 21.Lawrence J. M., On the relationships between marine plants and sea urchins. Oceanogr. Mar. Biol. Annu. Rev. 13, 213–286 (1975). [Google Scholar]
- 22.Paine R. T., Food webs: Linkage, interaction strength and community infrastructure. J. Anim. Ecol. 49, 667–685 (1980). [Google Scholar]
- 23.Steneck R. S., Dethier M. N., A functional group approach to the structure of algal-dominated communities. Oikos 69, 476–498 (1994). [Google Scholar]
- 24.Adey W. H., Macintyre I. G., Crustose coralline algae: A re-evaluation in the geological sciences. Bull. Geol. Soc. Am. 84, 883–904 (1973). [Google Scholar]
- 25.Baskett M. L., Salomon A. K., Recruitment facilitation can drive alternative states on temperate reefs. Ecology 91, 1763–1773 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Lubchenco J., Plant species diversity in a marine intertidal community: Importance of herbivore food preference and algal competitive abilities. Am. Nat. 112, 23–39 (1978). [Google Scholar]
- 27.Lubchenco J., Gaines S. D., A unified approach to marine plant-herbivore interactions. I. Populations and communities. Annu. Rev. Ecol. Syst. 12, 405–437 (1981). [Google Scholar]
- 28.Paine R. T., Ecological determinism in the competition for space: The Robert H. MacArthur Award Lecture. Ecology 65, 1339–1348 (1984). [Google Scholar]
- 29.Pacala S. W., Crawley M. J., Herbivores and plant diversity. Am. Nat. 140, 243–260 (1992). [DOI] [PubMed] [Google Scholar]
- 30.Gurevitch J., Morrison J. A., Hedges L. V., The interaction between competition and predation: A meta‐analysis of field experiments. Am. Nat. 155, 435–453 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Ishii R., Crawley M. J., Herbivore-induced coexistence of competing plant species. J. Theor. Biol. 268, 50–61 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Steneck R. S., Hacker S. D., Dethier M. N., Mechanisms of competitive dominance between crustose coralline algae: An herbivore-mediated competitive reversal. Ecology 72, 938–950 (1991). [Google Scholar]
- 33.McCoy S. J., Pfister C. A., Historical comparisons reveal altered competitive interactions in a guild of crustose coralline algae. Ecol. Lett. 17, 475–483 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Steinberg P. D., Estes J. A., Winter F. C., Evolutionary consequences of food chain length in kelp forest communities. Proc. Natl. Acad. Sci. U.S.A. 92, 8145–8148 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bentley S., Whittaker J. B., Effects of grazing by a chrysomelid beetle, Gastrophysa viridula, on competition between rumex obtusifolius and rumex crispus. J. Ecol. 67, 79–90 (1979). [Google Scholar]
- 36.Milchunas D. G., Sala O. E., Lauenroth W. K., A generalized model of the effects of grazing by large herbivores on grassland community structure. Am. Nat. 132, 87–106 (1988). [Google Scholar]
- 37.Steneck R. S., Adey W. H., The role of environment in control of morphology in Lithophyllum congestum, a Caribbean Algal Ridge Builder. Bot. Mar. 19, 197–216 (2009). [Google Scholar]
- 38.Turner T., Facilitation as a successional mechanism in a rocky intertidal community. Am. Nat. 121, 729–738 (1983). [Google Scholar]
- 39.Dayton P. K., Dispersion, dispersal, and persistence of the annual intertidal alga, Postelsia palmaeformis Ruprecht. Ecology 54, 433–438 (1973). [Google Scholar]
- 40.Barner A. K., Hacker S. D., Menge B. A., Nielsen K. J., The complex net effect of reciprocal interactions and recruitment facilitation maintains an intertidal kelp community. J. Ecol. 104, 33–43 (2016). [Google Scholar]
- 41.Rowley R. J., Settlement and recruitment of sea urchins (Strongylocentrotus spp.) in a sea-urchin barren ground and a kelp bed: Are populations regulated by settlement or post-settlement processes? Mar. Biol. 100, 485–494 (1989). [Google Scholar]
- 42.Gee J. M., Knight-Jones E. W., The morphology and larval behaviour of a new species of Spirorbis (Serpulidae). J. Mar. Biol. Assoc. U. K. 42, 641–654 (1962). [Google Scholar]
- 43.O’Leary J. K., et al. , Calcifying algae maintain settlement cues to larval abalone following algal exposure to extreme ocean acidification. Sci. Rep. 7, 5774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrigan J. F., “The planula larva of Pocillopora amicornis, lunar periodicity of swarming and substratum selection behavior,” PhD thesis, University of Hawaii at Manoa, Honolulu, HI (1972).
- 45.Carballo J. L., Ávila E., Enríquez S., Camacho L., Phenotypic plasticity in a mutualistic association between the sponge Haliclona caerulea and the calcareous macroalga Jania adherens induced by transplanting experiments. I: Morphological responses of the sponge. Mar. Biol. 148, 467 (2005). [Google Scholar]
- 46.Hind K. R., Gabrielson P. W., P Jensen C., Martone P. T., Crusticorallina gen. nov., a nongeniculate genus in the subfamily Corallinoideae (Corallinales, Rhodophyta). J. Phycol. 52, 929–941 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Hind K. R., Gabrielson P. W., Jensen C., Martone P. T., Evolutionary reversals in Bossiella (Corallinales, Rhodophyta): First report of a coralline genus with both geniculate and nongeniculate species. J. Phycol. 54, 788–798 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Hind K. R., Gabrielson P. W., Lindstrom S. C., Martone P. T., Misleading morphologies and the importance of sequencing type specimens for resolving coralline taxonomy (Corallinales, Rhodophyta): Pachyarthron cretaceum is Corallina officinalis. J. Phycol. 50, 760–764 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Gabrielson P. W., Hughey J. R., Diaz-Pulido G., Genomics reveals abundant speciation in the coral reef building alga Porolithon onkodes (Corallinales, Rhodophyta). J. Phycol. 54, 429–434 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Dethier M. N., Steneck R. S., Growth and persistence of diverse intertidal crusts: Survival of the slow in a fast-paced world. Mar. Ecol. Prog. Ser. 223, 89–100 (2001). [Google Scholar]
- 51.Noisette F., Egilsdottir H., Davoult D., Martin S., Physiological responses of three temperate coralline algae from contrasting habitats to near-future ocean acidification. J. Exp. Mar. Biol. Ecol. 448, 179–187 (2013). [Google Scholar]
- 52.Guenther R. J., Martone P. T., Physiological performance of intertidal coralline algae during a simulated tidal cycle. J. Phycol. 50, 310–321 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Breitburg D. L., Residual effects of grazing: Inhibition of competitor recruitment by encrusting coralline algae. Ecology 65, 1136–1143 (1984). [Google Scholar]
- 54.Estes J. A., Duggins D. O., Sea otters and kelp forests in Alaska: Generality and variation in a community ecological paradigm. Ecol. Monogr. 65, 75–100 (1995). [Google Scholar]
- 55.Stevenson C. F., Demes K. W., Salomon A. K., Accounting for size-specific predation improves our ability to predict the strength of a trophic cascade. Ecol. Evol. 6, 1041–1053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martone P. T., Lindstrom S. C., Miller K. A., Gabrielson P. W., Chiharaea and Yamadaia (Corallinales, Rhodophyta) represent reduced and recently derived articulated coralline morphologies. J. Phycol. 48, 859–868 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Vermeij G. J., et al. , The coastal North Pacific: Origins and history of a dominant marine biota. J. Biogeogr. 46, 1–18 (2019). [Google Scholar]
- 58.Harley C. D. G., Abiotic stress and herbivory interact to set range limits across a two-dimensional stress gradient. Ecology 84, 1477–1488 (2003). [Google Scholar]
- 59.Tylianakis J. M., et al. , Resource heterogeneity moderates the biodiversity-function relationship in real world ecosystems. PLoS Biol. 6, e122 (2008). [Google Scholar]
- 60.Hutchings M. L., John E. A., Stewart A. J. A., Eds., The Ecological Consequences of Environmental Heterogeneity: 40th Symposium of the British Ecological Society (Cambridge University Press, Vol. 40, 2000). [Google Scholar]
- 61.Estes J. A., Steinberg P. D., Predation, herbivory, and kelp evolution. Paleobiology 14, 19–36 (1988). [Google Scholar]
- 62.Reed D. C., Foster M. S., The effects of canopy shadings on algal recruitment and growth in a giant kelp forest. Ecology 65, 937–948 (1984). [Google Scholar]
- 63.Arkema K. K., Reed D. C., Schroeter S. C., Direct and indirect effects of giant kelp determine benthic community structure and dynamics. Ecology 90, 3126–3137 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Gaylord B., et al. , Spatial patterns of flow and their modification within and around a giant kelp forest. Limnol. Oceanogr. 52, 1838–1852 (2007). [Google Scholar]
- 65.Figueiredo M. A. D. O., Jones M. K., Norton T. A., Responses of crustose corallines to epiphyte and canopy cover. J. Phycol. 36, 17–24 (2000). [Google Scholar]
- 66.Frieder C. A., Nam S. H., Martz T. R., Levin L. A., High temporal and spatial variability of dissolved oxygen and pH in a nearshore California kelp forest. Biogeosciences 9, 3917–3930 (2012). [Google Scholar]
- 67.Ragazzola F., et al. , Ocean acidification weakens the structural integrity of coralline algae. Glob. Change Biol. 18, 2804–2812 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Kamenos N. A., et al. , Coralline algal structure is more sensitive to rate, rather than the magnitude, of ocean acidification. Glob. Change Biol. 19, 3621–3628 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaston K. J., The magnitude of global insect species richness. Conserv. Biol. 5, 283–296 (1991). [Google Scholar]
- 70.Geiser D. M., Pitt J. I., Taylor J. W., Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc. Natl. Acad. Sci. U.S.A. 95, 388–393 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fernandez C. C., Shevock J. R., Glazer A. N., Thompson J. N., Cryptic species within the cosmopolitan desiccation-tolerant moss Grimmia laevigata. Proc. Natl. Acad. Sci. U.S.A. 103, 637–642 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoeinghaus D. J., Winemiller K. O., Birnbaum J. S., Local and regional determinants of stream fish assemblage structure: Inferences based on taxonomic vs. functional groups. J. Biogeogr. 34, 324–338 (2007). [Google Scholar]
- 73.Melville A. J., Connell S. D., Experimental effects of kelp canopies on subtidal coralline algae. Austral Ecol. 26, 102–108 (2001). [Google Scholar]
- 74.Sale P. F., Coexistence of coral reef fishes—A lottery for living space. Environ. Biol. Fishes 3, 85–102 (1978). [Google Scholar]
- 75.Blomberg S. P., Garland T. Jr, Ives A. R., Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 57, 717–745 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Cavender-Bares J., Kozak K. H., Fine P. V. A., Kembel S. W., The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Aguirre J., Perfectti F., Braga J. C., Integrating phylogeny, molecular clocks, and the fossil record in the evolution of coralline algae (Corallinales and Sporolithales, Rhodophyta). Paleobiology 36, 519–533 (2010). [Google Scholar]
- 78.Johansen H. W., Coralline Algae: A First Synthesis (CRC Press, 1981). [Google Scholar]
- 79.Losos J. B., Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Schluter D., Ecological causes of adaptive radiation. Am. Nat. 148 (suppl), S40–S64 (1996). [Google Scholar]
- 81.Okamoto D. K., Stekoll M. S., Eckert G. L., Coexistence despite recruitment inhibition of kelps by subtidal algal crusts. Mar. Ecol. Prog. Ser. 493, 103–112 (2013). [Google Scholar]
- 82.Eriksson O., Regional dynamics of plants: A review of evidence for remnant, source-sink and metapopulations. Oikos 77, 248–258 (1996). [Google Scholar]
- 83.Nichol L. M., Watson J. C., Abernethy R., Rechsteiner E., Towers J., “Trends in the abundance and distribution of sea otters (Enhydra lutris) in British Columbia updated with 2013 survey results” (Fisheries and Oceans Canada, Ecosystems and Oceans Science, Rep. No. Fs70-5/2015-039E-PDF, 2015).
- 84.Burt J. M., et al. , Sudden collapse of a mesopredator reveals its complementary role in mediating rocky reef regime shifts. Proc Biol Sci 285, 20180553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oksanen J., et al. , vegan: Community Ecology Package, Version 2.5. https://cran.r-project.org/package=vegan/index,html. Accessed 8 September 2018.
- 86.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 87.Kembel S. W., et al. , Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 (2010). [DOI] [PubMed] [Google Scholar]
- 88.Breiman L., Cutler A., randomForest: Breiman and Cutler’s Random Forests for Classification and Regression, Version 4.6. https://cran.r-project.org/package=randomForest/index,html. Accessed 10 October 2018.
- 89.Clarke K. R., Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143 (1993). [Google Scholar]
- 90.Kearse M., et al. , Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pagel M., Inferring the historical patterns of biological evolution. Nature 401, 877–884 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.