Significance
We report that a previously uncharacterized protein genetically associated with cytoplasmic chemoreceptors of undefined function senses both iron and oxygen to mediate chemotaxis. Spectroscopic and crystallographic studies reveal an unusual di-iron center with remarkable oxygen-binding properties in this oxygen-binding di-iron protein (ODP). Moreover, the oxygen-bound state of ODP facilitates phosphate release from the histidine kinase CheA. This study defines a class of oxygen sensors and reveals functions for cytoplasmic chemoreceptors in diverse phyla of bacteria and archaea that include human pathogens and distantly related extremophiles. The presence of these systems in anaerobic bacteria of deep phylogeny suggests that they may represent early adaptations to oxic environments.
Keywords: oxygen sensor, chemoreceptor, phosphatase, signal transduction, molecular evolution
Abstract
Many bacteria contain cytoplasmic chemoreceptors that lack sensor domains. Here, we demonstrate that such cytoplasmic receptors found in 8 different bacterial and archaeal phyla genetically couple to metalloproteins related to β-lactamases and nitric oxide reductases. We show that this oxygen-binding di-iron protein (ODP) acts as a sensor for chemotactic responses to both iron and oxygen in the human pathogen Treponema denticola (Td). The ODP di-iron site binds oxygen at high affinity to reversibly form an unusually stable μ-peroxo adduct. Crystal structures of ODP from Td and the thermophile Thermotoga maritima (Tm) in the Fe[III]2-O22−, Zn[II], and apo states display differences in subunit association, conformation, and metal coordination that indicate potential mechanisms for sensing. In reconstituted systems, iron-peroxo ODP destabilizes the phosphorylated form of the receptor-coupled histidine kinase CheA, thereby providing a biochemical link between oxygen sensing and chemotaxis in diverse prokaryotes, including anaerobes of ancient origin.
During bacterial chemotaxis, methyl-accepting chemotaxis proteins (MCPs), or chemoreceptors, convey changes in the external chemical environment to a cytoplasmic phosphorelay system that ultimately directs rotation of the flagellar motor (1). Although canonical chemoreceptors contain transmembrane regions and periplasmic ligand-binding domains, several receptor classes found in such diverse organisms as Thermotoga maritima (Tm), Pseudomonas aeruginosa, Vibrio cholera, Rhodobacter sphaeroides, Bacillus subtilis, and Treponema denticola (Td) do not (2, 3). Nonetheless, these “soluble” receptors possess conserved regions known to interact with the primary chemotaxis histidine kinase CheA, as well as determinants needed to form the distinctive trimer-of-dimers assembly state characteristic of transmembrane receptors. Indeed, in several examined species, soluble receptors organize into hexagonal lattices bound to a baseplate of CheA and the adaptor protein CheW, much like their transmembrane counterparts (2). Nevertheless, what such receptors sense and how they convey signals to CheA is largely an open question.
A particular class of cytoplasmic receptor without a ligand-binding domain is found in the genomes of Thermotogota and is exemplified by the Tm receptor TM14. TM14 is a thermostable, soluble protein that deactivates CheA autophosphorylation activity and has a dimeric coiled-coil structure typical of MCPs, including a conserved tip that interacts with CheA and CheW (4). Where investigated, cytoplasmic receptors appear to respond to the metabolic state of the cell, but it is unknown what signals or ligands the receptors recognize (5). The tm14 gene is located adjacent to the tm13 gene, which codes for a protein that belongs to a superfamily of metalloenzymes, thus suggesting that the 2 proteins might interact physically or functionally.
Herein, we demonstrate that TM13 (hereafter named oxygen-binding di-iron protein [ODP]) exemplifies a distinct family of metalloproteins recruited to various bacterial and archaeal signal transduction pathways, including chemotaxis, to function as oxygen and iron sensors.
Results
ODP Represents a Distinct Di-Metalloprotein Family Associated with Signal Transduction.
ODP is recognized by the Lactamase_B Pfam model (PF00753), indicating that it is a member of the metallo-β-lactamase (MBL) superfamily, which is composed of functionally diverse enzymes that include β-lactam hydrolases that primarily have 1 or 2 zinc ions in their active sites (6). We found that ODP sequences form a distinct family within the MBL superfamily (SI Appendix, Fig. S1A), which is closely related to the family of flavodiiron proteins (FDPs) that serve as oxygen and/or nitric oxide reductases (7). In addition to distinction by sequence conservation, the ODP and FDP families associate their common metal-binding domain with different downstream protein domains. Whereas FDPs always contain a flavoprotein domain or other domains that support their enzymatic function (SI Appendix, Fig. S1 B and C), nearly 60% of all ODP genes in genomic datasets are either neighboring signal transduction genes or encode for a protein that contains a signal transduction domain (Fig. 1 and SI Appendix, Fig. S1 and Dataset S1). A conserved metal-binding site in FDPs and ODPs is strikingly similar, but different from the rest of the superfamily (SI Appendix, Fig. S1 and Dataset S2). Collectively, the results of genomic and protein sequence analyses suggest that ODPs are di-iron–binding domains recruited to signal transduction pathways in bacteria and archaea.
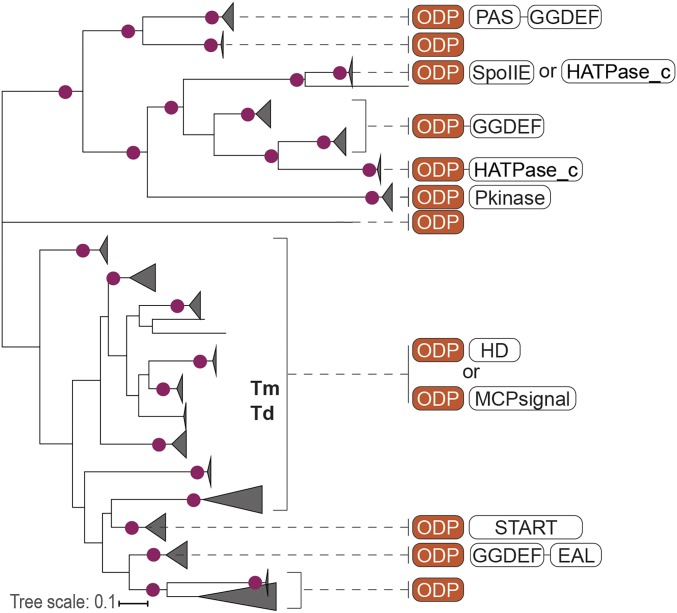
Fig. 1.
ODP is a family of metalloproteins associated with signal transduction in bacteria and archaea. A maximum likelihood phylogenetic tree of ODP representatives is shown, with gene neighborhoods and fusions to known signal transduction domains (Pfam nomenclature) depicted for each ODP branch. Nodes with bootstrap support >60 are marked by circles. Tm and Td indicate nodes containing TM13 orthologs from Tm and Td. Complete underlying data are shown as Dataset S1.
ODP Coevolves with Specific Cytoplasmic Chemoreceptors.
Database searches and genome and protein sequence analyses revealed that all closely related orthologs of TM13 that were identified in 8 bacterial and archaeal phyla come from genomes that also encode closely related orthologs of the TM14 chemoreceptor (SI Appendix, Fig. S2 and Dataset S1). In most cases, genes encoding ODP and the chemoreceptor were predicted to form a 2-gene operon (Dataset S1). Topology analysis of corresponding maximum-likelihood phylogenetic trees (SI Appendix, Fig. S2) provided further strong support for coevolution of ODP and the soluble chemoreceptor, thus suggesting that ODP might serve as input for the chemotaxis pathway and prompting its further analysis.
ODP Binds Iron.
Both Tm ODP and Td ODP were expressed in and purified from Escherichia coli (Ec). Td ODP is purple in color and has a visible absorbance spectrum characteristic of an Fe[III]2-O22− species (8) (Fig. 2A). Metal analysis by inductively coupled plasma atomic emission spectroscopy (ICP-AES) reveals that the protein binds iron in an ∼2:1 iron/subunit ratio, with no other metals being found in significant abundance (SI Appendix, Fig. S3A). Unlike Td ODP, Tm ODP is colorless after purification, and ICP-AES detected no bound metal ions. Lack of metal incorporation by recombinant Tm ODP is not surprising because Tm ODP originates from an extremophile (Tm grows at 90 °C, 2 atm), and thus may not display native behavior at room temperature (9).
Fig. 2.
Aerobic Td ODP contains a di-iron[III]-peroxo adduct. (A) UV/Vis spectroscopy of 480 μM ODP (ε590 = 1 mM−1⋅cm−1) demonstrates the loss of the characteristic di-Fe[III]-O22− bands (500 nm and 590 nm) under anaerobic conditions. The absorbance recovers upon reexposure to oxygen. Some active protein is lost during the 12-h anaerobic incubation (SI Appendix, Fig. S3D). (B) RR spectroscopy of peroxo-bound ODP produces vibrational bands indicative of a cis μ-1,2 iron-peroxo species (O-O [828 cm−1] and Fe-O [469 cm−1]) that downshifts upon 16O to 18O isotopic substitution. (C) Mössbauer spectroscopy confirms a cis μ iron-peroxo species (Left) in aerobic ODP and a di-ferrous state (Right) after oxygen release (∼70% di-ferrous state after 1 h). Data points (black dots) were fit (black line) to quadrupole doublets, with residuals shown as gray background lines.
Tm ODP was reconstituted with iron and zinc ions, and thermal melts were used to assess the relative extent of protein stabilization. Although Zn[II] and Fe[III] increase the melting temperature of the apo protein, Tm ODP is most stable upon reconstitution with Fe[II] (SI Appendix, Fig. S3B). Furthermore, oxidation of the Fe[II]-reconstituted Tm protein produces a color and absorbance spectrum similar to that of as-purified Td ODP (SI Appendix, Fig. S3E).
The ODP Oligomeric State Depends on Iron.
Multiangle light scattering (MALS) indicates that Td ODP forms a dimer in solution when the iron-peroxo adduct is present (60 kDa). However, the average molecular weight of the protein increases when the iron is removed (85 kDa). These data suggest that apo Td ODP undergoes a shift to a higher oligomeric state (SI Appendix, Fig. S4A). MALS experiments with apo and iron-reconstituted Tm ODP demonstrate that, like Td ODP, the average molecular weight of Tm ODP increases in the apo state. Specifically, apo Tm ODP forms a 120-kDa tetramer, while the metal-reconstituted protein is predominantly a dimer (SI Appendix, Fig. S4B). Thus, conversion to the apo forms of both Tm and Td ODP causes a transition from dimer to tetramer and decreases thermodynamic stability (SI Appendix, Figs. S3B and S4).
ODP Binds Oxygen as an Fe[III]2-Peroxo Species.
Purple ODP has a broad absorption band with peaks at 590 nm and 500 nm (Fig. 2A), indicative of a di-iron peroxo adduct (8, 10, 11). This spectrum is lost when Td ODP is reduced by dithionite but recovers upon titration with oxygenated buffer (SI Appendix, Fig. S3C). Resonance Raman (RR) spectroscopy of the oxygenated protein reveals oxygen-derived vibrational bands [ν(O-O) at 828 cm−1 and v(Fe-O) at 469 cm−1] that shift to lower energies (−45 cm−1 and −33 cm−1, respectively) upon isotopic substitution of 16O2 with 18O2 (Fig. 2B). Reduction of Td ODP with dithionite eliminates the isotope-sensitive bands (SI Appendix, Fig. S3I). The RR spectra closely resemble that of Fe[III]2(μ-O)(μ-η1:η1-O22−) biomimetic complexes, and thereby support the assignment of a symmetric cis μ-1,2 iron-peroxo adduct in ODP (8, 10, 11). The Mössbauer spectrum of ODP reconstituted with 57Fe indicates 2 electronically equivalent antiferromagentically coupled Fe[III] S = 5/2 centers that form a di-iron-peroxo species (δ = 0.58 mm⋅s−1, ∆EQ = 1.01 mm⋅s−1, Γ = 0.33 mm⋅s−1) (Fig. 2C and SI Appendix, Table S1). Notably, other proteins with cis μ-1,2 iron-peroxo adducts have optical transitions red-shifted relative to the 590-nm ODP charge-transfer band and also have ν(O-O) ∼ 30 to 40 cm−1 higher in energy than found for ODP (12). Spectral features like those displayed by ODP are often observed for asymmetric end-on η1 or η2 coordination of peroxide to 1 iron atom (12–14). However, there is considerable variability in these parameters; notably, the cis μ-1,2 iron-peroxo complex of human deoxyhyposine hydroxylase (hDOHH; discussed below) has RR and Mössbauer parameters similar to those of ODP (15) (SI Appendix, Table S1). Furthermore, biomimetic compounds that contain carboxylate bridges between iron atoms produce ultraviolet (UV)/visible (Vis) absorbance and RR spectra closely resembling those of Td ODP (8, 10). A single iron environment represented by the ODP Mössbauer parameters indicates near-symmetric coordination of the peroxo to the di-iron center.
Td ODP Oxygen Binding Is Reversible.
FDPs and other di-iron oxygen-binding proteins commonly generate iron-peroxo adducts to activate oxygen for redox reactions. In these cycles, a diferrous site is oxidized to a diferric state after substrate turnover. In the absence of substrate, the iron-peroxo species decays to an Fe[III]2 species (16). In both cases, reduction by a cofactor or coenzyme is required to reform the active Fe[II]2 state. Furthermore, the iron-peroxo adduct in these proteins is almost always short lived; reported “stable” adducts have a half-life (t1/2) on the order of seconds or minutes (16, 17). Unlike most other nonheme iron-peroxo proteins, ODP spontaneously releases oxygen from the iron center under anaerobic conditions to return to the Fe[II]2 state, which can again bind oxygen to reform the Fe[III]2-O22− adduct. Incubating oxygenated ODP under anaerobic conditions results in a steady decrease in absorbance at 590 nm that can be recovered by re-exposure to oxygen (Fig. 2A). Likewise, oxygen removal by either the glucose oxidase-catalase (GO-CAT) system (18, 19) or anaerobic incubation produces Mössbauer spectra indicative of an Fe[II]2 center (20) (Fig. 2C and SI Appendix, Figs. S3G and S8 and Table S1). In both cases, a fraction of the Fe[III]2-peroxo species remains in the samples. Deconvolution of the data indicates that the major species is di-Fe[II] S = 2 (δ = 1.20 mm⋅s−1, ∆EQ = 2.60 mm⋅s−1, Γ = 0.35 mm⋅s−1). Stable, reversible oxygen binding by ODP meets the requirements of an oxygen sensor.
To determine the rate of oxygen dissociation from the di-iron center, the GO-CAT system was used to scavenge released oxygen from Td ODP; spectral changes associated with O2 release were followed at 590 nm (SI Appendix, Fig. S3F). Conditions under which the rate of oxygen release did not depend on the concentration of GO-CAT gave a first-order off-rate constant (koff) of 0.002 ± 0.0004 s−1, or t1/2 ∼ 5.8 min (SI Appendix, Fig. S3F). Assuming that O2 binding by ODP has a typical on-rate constant (kon) in the range of 106 to 108 M−1⋅s−1, the dissociation constant (KD; koff/kon) is ∼2.0 to 0.02 nM, considerably lower than that of the di-iron oxygen carrier hemerythrin (Hr) (21–23). Under aerobic conditions, the steady-state stability of the Td ODP peroxy complex is extremely high and limited only by the degradation of the iron center and/or protein unfolding (t1/2 ∼ 50 h at 20 °C; SI Appendix, Fig. S3D). This stability contrasts with the majority of protein iron-peroxo adducts, which are short-lived intermediates. Several rare exceptions (14, 17) include hDOHH, mentioned above, which has a steady-state t1/2 ∼ 30 h at 20 °C (12).
ODP Mediates Chemotaxis Toward Iron and Away from Oxygen.
To test if ODP is responsible for sensing iron and oxygen as part of a chemotactic response, chemotaxis capillary assays were conducted in Td using oxygen and iron as candidate chemoeffectors (SI Appendix, Fig. S5A). An ODP knockout strain of Td (∆2498) was generated and compared with the behavior of wild-type (WT). RNA analysis indicates that the ODP gene is expressed under anaerobic growth conditions, but not in ∆2498 (SI Appendix, Fig. S5B).
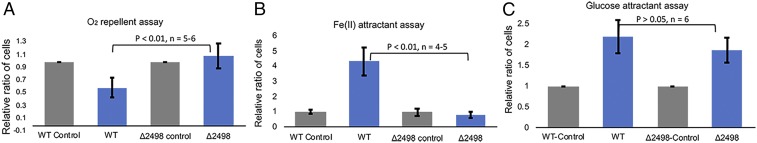
No significant difference was found between the WT and ∆2498 for chemotaxis to the known Td attractant glucose (24) (Fig. 3C). Notably, nearly all of the bacterial species that possess a receptor-coupled ODP homolog are anaerobes, and thus may contain a system for recognizing and avoiding oxygen. Indeed, although WT cells avoid oxygenated buffer, the ∆2498 mutants are deficient in this response (Fig. 3A). In addition, WT cells exhibit strong chemoattraction to Fe[II], whereas the ∆2498 mutant cells do not. Chemotaxis toward hemin, a previously reported Td iron source (25), was also impaired in the ∆2498 mutant (SI Appendix, Fig. S5C). Together, these data confirm that Td ODP is an oxygen and iron sensor that interfaces with the chemotaxis system of Td.
Fig. 3.
Capillary chemotaxis assays with Td confirm a role for ODP in sensing oxygen and iron. (A) Oxygenated buffer is a chemorepellent for Td WT but not ∆2498 (-ODP) cells. Only WT cells show less migration into capillaries when the buffer is oxygenated compared with anaerobic controls. (B) WT cells showed strong attraction to ferrous iron, but the ∆2498 cell line did not. (C) Both the WT and ∆2498 recognized glucose as a chemoattractant.
ODP Accelerates CheA Dephosphorylation.
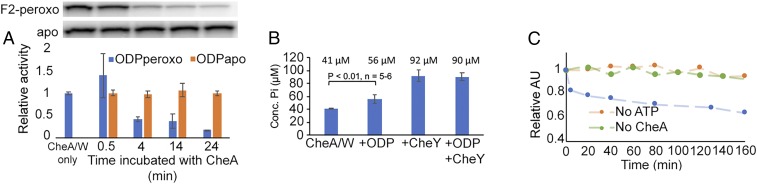
To explore the link between ODP and chemotaxis, Td CheA, CheW, CheY, and TM14 receptor were cloned; recombinantly expressed in Ec; and purified. Td CheA autophosphorylates in the presence of adenosine 5′-triphosphate (ATP), transfers phosphate to the response regulator CheY, and is inhibited by the TM14 homolog (SI Appendix, Fig. S6 A and B). To aid in assembly of the CheA/CheW complex, the TM14 homolog was artificially trimerized into its presumed trimeric oligomeric state by addition of a trimerization motif, as previously described (26) (SI Appendix, Fig. S6C). Surprisingly, addition of excess di-iron-peroxo ODP significantly decreases the relative amount of phosphorylated CheA (CheA-inorganic phosphate [Pi]) over time, whereas apo-ODP has no apparent effect (Fig. 4A). However, physical interaction between ODP and CheA could not be detected by pulldown or chemical cross-linking, suggesting a transient interaction between the proteins under these conditions.
Fig. 4.
Td ODP Fe[III]2-O22− affects CheA phosphorylation levels. (A) Autophosphorylation assays of CheA with γ-32P ATP show that iron-peroxo ODP decreases CheA-P but apo-ODP does not. CheA-P loss increases with time of incubation with ODP (quantification shown below the graph). (B) Amount of free phosphate formed by incubation of CheA/CheW and ATP increases with ODP Fe[III]2-O22− and CheY concentration (Conc.). (C) Fe[III]2-O22− adduct (as monitored in relative absorption units [AU] at 590 nm) depletes in the presence of CheA/CheW and ATP (blue), but not when either ATP (orange) or CheA (green) is removed.
The ability of ODP to decrease CheA-Pi levels could be due to an inhibition of autophosphorylation or an activation of phosphatase activity. To differentiate between these potential mechanisms, Pi was quantified in samples containing CheA and ATP in the absence and presence of di-iron-peroxo ODP or CheY. As expected, CheY increases phosphate release from CheA (Fig. 4B). Similarly, samples containing iron-peroxo ODP show an increase in [Pi] release compared with CheA and CheW alone (Fig. 4B), although not to the same extent as with CheY (Fig. 4B). Phosphate released from CheA by ODP and CheY matches that released by only CheY, indicating that under these conditions, CheY outcompetes ODP and ODP does not increase phosphate loss from CheY. To investigate whether the di-iron-peroxo species directly participates in phosphate hydrolysis, the stability of the peroxo adduct was monitored at 590 nm in the presence of CheA, CheW, and ATP. CheA only in the presence of ATP destabilized the ODP di-iron peroxo species (Fig. 4C). Free phosphate or ATP alone has little effect (SI Appendix, Fig. S3D). Therefore, the decay of the iron-peroxo adduct depends on CheA-Pi, a strong indication that phosphate release involves the di-iron-peroxo species.
Crystal Structures of Metal-Reconstituted Td ODP and Tm ODP.
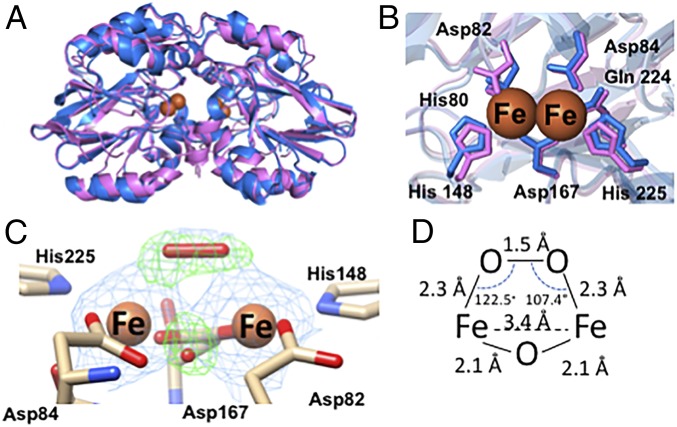
The 2.07-Å resolution crystal structure of Td ODP (6R9N) reveals a protein dimer with a di-iron site. Energy-dispersive X-ray spectroscopy (EDXS) of the crystals confirmed that iron is the only metal present (SI Appendix, Fig. S3H). As expected, Td ODP assumes the canonical β-lactam fold exhibited by β-lactamases and FDPs (Fig. 5A). Like most FDPs, the active site contains seven ligand residues: His80, Asp82, and His148 coordinate the iron closest to the dimer interface, whereas Asp84, Gln224, and His225 coordinate the other, and Asp167 bridges both metal ions (7, 27) (Fig. 5B). Most FDPs have a conserved histidine residue in place of the ODP residue Gln224 (28). Furthermore, the electron density around the active site reveals the presence of a bridging μ-oxo/hydroxo species and the cis μ-1,2 peroxo species that was identified by RR and Mössbauer spectroscopic techniques (Fig. 5 C and D). The bond lengths are consistent with those found in previously crystallized di-iron-peroxo proteins (17). Although the bridging peroxo species is not solvent-exposed, only minor conformational changes in the Val33-Tyr38 loop would be needed to expose it to solution or a substrate. With both oxygen and protein ligands, the coordination geometry of each iron atom is pseudo-octahedral, with the sixth coordination site occupied by the peroxo ligand. At this resolution, peroxide coordination (refined and unrestrained) is largely symmetric in both subunits (Fig. 5D and SI Appendix, Fig. S7D), consistent with results from spectroscopy (Fig. 2).
Fig. 5.
Crystal structures of Td and Tm ODP. (A) Overlay of the 2.07-Å resolution structure of iron-peroxo Td ODP (6R9N, blue) and the 2.0-Å resolution crystal structure of Zn-reconstituted Tm ODP (6QWO, magenta) (B) Di-iron centers of Td and Tm ODP conserve all metal-binding residues. (C) Di-iron site of Td ODP chain A contains a cis μ-1,2 iron-peroxo species and an oxo-bridge; 2Fo-Fc electron density map shown in blue at 2σ, and Fo-Fc omit map shown in green at 5.5σ. (D) Distances and angles of the iron-peroxo adduct are consistent with previously characterized cis-μ,1,2 iron-peroxo protein species and biomimetic compounds.
The di-iron-peroxo centers of the subunits are buried at the dimer interface with the peroxo moiety occluded from solvent by a ring of aromatic residues (F109, F195, Y199, and W106). Additionally, Gln81 and iron-ligand His148 coordinate a single ion that resides directly on the dimer axis and structurally couples the 2 active centers (SI Appendix, Fig. S5A). ICP-AES, several spectroscopic techniques (UV/Vis, RR, Mössbauer, and EDXS), and X-ray anomalous scattering did not identify an additional bound cation in the protein, whereas the coordination geometry suggests an anion, possibly chloride.
The iron complex of Tm ODP is unstable in most aerobic conditions; as such, Tm ODP was crystallized after reconstitution with Zn[II]. The 2.0-Å resolution zinc-reconstituted structure (6QWO) reveals a protein dimer nearly identical to that of Td ODP (Fig. 5A). However, unlike Td ODP, the Zn2+ form of Tm ODP does not display the interface ion that bridges the 2 histidine residues.
Crystal Structures of Apo ODP Suggest a Mechanism for Iron Sensing.
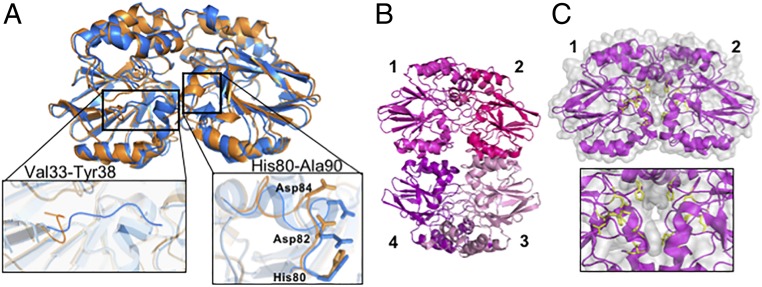
The 2.1-Å resolution crystal structure of apo Td ODP (6QNM) is similar to the iron-reconstituted structure with some exceptions. In the apo structure, an 11-residue loop (His80 to Ala90) that contains the metal ligands shifts and a surface-exposed loop that excludes the metal center from solvent (Val33 to Tyr38) displaces to expose the active site (Fig. 6A). The interface anion bridging the 2 active centers is also absent in the apo structure. Collectively, these structural changes result in solvent accessibility of the metal-binding pocket and the formation of a channel that includes the metal ligands at the dimer interface (SI Appendix, Fig. S7 B and C). The resulting increase in solvent accessibility and coupled conformational changes may allow for iron to access the metal-binding center.
Fig. 6.
Crystal structures of apo Td and Tm ODP. (A) Apo Td ODP (6QNM, orange) and iron-peroxo Td ODP (6R9N, blue) have similar conformations, with the exception of 2 small loops composed of residues Val33-Tyr38 and His80-Ala90. Movement of these loops in the apo form exposes the active site to solvent. (B) Apo Tm ODP crystallizes as a tetramer (6QRQ), consistent with molecular weight measurements by MALS. Subunits 1 and 2 or 3 and 4 compose the dimer found in the metal-bound structure. (C) Solvent exposure of the active site residues (yellow) increases in the apo structure, and Trp102 from the adjacent subunit moves into the metal-free center (SI Appendix, Fig. S7E).
Although apo Td ODP remains a dimer in crystals, the 2.56-Å resolution crystal structure of apo Tm ODP (6QRQ) is a tetramer, as observed in solution (Fig. 6B). The tetramer can be viewed as an end-to-end dimer that buries 1,109 Å2 additional surface of the metal-bound dimer. Therein, loop alterations create a solvent channel that exposes the metal-binding residues and also restructure the dimer interface and subunit orientation (Fig. 6C and SI Appendix, Fig. S7C). Furthermore, absence of the ions causes Trp102 from the adjacent subunit to insert into the metal-binding site (SI Appendix, Fig. S7E) and slightly displaces 2 helices (A49-V62 and V80-N93) at the tetramer interface; a loop from the adjacent subunit (F109-I117) moves to engage these helices. Such changes to the apo protein surface properties potentially enable engagement with partners.
Discussion
ODP is genetically linked to and coevolves with a nontransmembrane chemoreceptor that lacks a dedicated sensory domain, and this combined system senses oxygen and ferrous iron for chemotaxis responses. Td has been reported to survive in aerobic conditions by generating microaerobic environments, but it requires a source of iron for survival (29, 30). Here, we show that deletion of ODP in Td eliminates its ability to respond to iron and oxygen (Fig. 3). ODP is cytoplasmic, and therefore monitors the intracellular levels of these ligands. Iron binding induces conformational changes in ODP, which ultimately leads to changes in protein stability and oligomeric state. Furthermore, ODP reversibly binds oxygen with an affinity in the nanomolar range as an iron-peroxo adduct. Remarkably, iron-peroxo ODP destabilizes phosphorylated CheA.
The mechanism of ODP phosphatase activity is currently unclear. Although not homologous to ODP, purple acid phosphatases employ an Fe[II]-Fe[III] di-iron center (or Fe[II]-Zn[II] center), wherein both metal ions coordinate the phosphoryl ester and supply a bridging μ-OH, a pendant Fe[III]-OH(2), or an activated noncoordinating water molecule as a hydrolytic nucleophile to liberate phosphate (31). In ODP, the peroxo moiety would have to rearrange or be displaced to coordinate a phosphoryl group. The ODP metal center does not contain a bound hydroxide/water, but the μ-oxo/hydroxo could potentially act as a nucleophile. Further reduction of the μ-1,2 peroxo could also generate a pendant hydroxide. A cellular reductase or the coupled di-Fe[II] center of the ODP adjacent subunit may act as a source of electrons. It follows that multiple turnovers as a phosphatase may require ODP to be rereduced. If this is the case, the protein will likely be limited to a single reaction in vitro, which may explain why CheA dephosphorylation during steady-state autophosphorylation requires excess ODP.
Chemotaxis assays indicate that ODP functions as a chemoreceptor that senses iron as an attractant and oxygen as a repellent. A rationale for this activity pattern may involve at least 3 states of ODP participating in chemotaxis. In the apo state, ODP conformational changes in the subunits or tetramerization prevent interaction with the chemotaxis machinery. Upon iron binding, dimeric ODP then interacts with the kinase/receptor assembly (likely through TM14) and stimulates CheA autophosphorylation. In the presence of oxygen, dimeric ODP converts to the peroxo state, which will then destabilize CheA-Pi. Under such a scenario, metal-ion–driven conformational changes and active center oxygen activation both play a role in the ODP-sensing mechanism. An interaction between CheA and di-Fe[II] ODP has been challenging to observe, perhaps owing to the requirement of a functional hexagonal array of receptors, which is difficult to reconstitute under ex vivo anaerobic conditions.
In Ec, CheA activity increases in response to a repellent, whereas Bacillus subtilis (Bs) CheA activity increases in response to an attractant (32). Td and Tm CheA belong to the same evolutionary class (F1 [33]) as Bs CheA, whereas Ec CheA belongs to a different class (F7). Similarly, Td and Tm receptors belong to the same evolutionary class (44H [3]) as Bs receptors, whereas Ec receptors belong to another (36H). Furthermore, the unmodified receptors from Td and Tm deactivate CheA, which is a behavior consistent with an attractant-activating system like Bs (26, 34). Indeed, these considerations and data presented here support a model wherein Td/Tm CheA activity decreases with repellent (O2) and increases with attractant (Fe[II]).
Both prokaryotes and eukaryotes utilize nonheme iron proteins as oxygen and iron sensors (35, 36). The coupling of iron and oxygen sensing is advantageous because although Fe[II] is essential for cell survival, uptake of ferric iron is difficult and even deleterious (36). Nonetheless, other systems operate differently than ODP. Mammalian FBXL5 and Desulfovibrio vulgaris DcrH possess a di-iron Hr-like domain that undergoes rapid oxidation to a di-ferric state upon oxygen binding, which then induces conformational changes important for signaling (35). Like ODP, FBXL5 appears to modulate a posttranscriptional regulator in response to both oxygen and ferrous iron, and both moieties are required for activity (37). DcrH is a transmembrane MCP that contains an intracellular Hr-like domain to monitor the concentration of intracellular oxygen and modulate anaerotaxis (23). Oxygen accesses the di-iron site via a channel and then rapidly oxidizes Fe[II]. Unlike the Hr-like possessing proteins, ODP is not homologous to Hr and does not readily auto-oxidize to a ferric state. Hrs also reversibly bind oxygen as peroxide, but they form an end-on peroxo adduct instead of a cis μ-1,2 peroxo species (22). ODP is an example of a di-iron protein that couples oxygen and iron sensing through the reversible formation of a bridged Fe[III]2-peroxo species.
The geometry of the ODP cis μ-1,2 di-iron(III)-peroxo adduct is similar to that found in toluene monooxygenase (38), arylamine oxygenase (39), and deoxyhypusine hydroxylase (17). The μ-1,2 di-iron(III)-peroxo species have variable roles in the catalysis of these enzymes (40), as well as others (41), but often change coordination in conversion to more reactive species. ODP discourages such transitions and rather stabilizes the peroxo for conferring oxygen-binding signals to the chemotaxis machinery. FDPs do not form stable iron-peroxo species or reversibly bind oxygen, but they do conserve a hydrophobic pocket surrounding the metal centers. Unlike FDPs, ODP contains an iron-coordinating glutamine residue (Gln224) in place of a more typical His residue. Notably, Gln224 differentiates the metal coordination from typical FDPs; however, the Gln substitution alone may not explain the unusual oxygen-bonding properties of ODP. Substitution of the analogous His to Asn does not affect activity or stabilize an FDP iron-peroxo species (28). Thus, the oxygen-binding properties of ODP likely derive from the combined effects of the iron coordination geometry and protein environment.
Given its oxygen-sensing functions, the presence of ODP in an anaerobe such as Tm is intriguing. Tm, which inhabits hydrothermal vents and other environmental hot spots rich in Fe[II], belongs to a slowly evolving lineage with a deep phylogeny within bacteria (42, 43). Tm can tolerate small amounts of oxygen, and, in fact, the cell mounts several metabolic responses to aerobic exposure (43, 44). Oxidative stress up-regulates the transcription of several oxygen-scavenging proteins, and the primary oxygen consumer is the FDP and ODP homolog FprA (44, 45). Tm FprA has a di-iron domain with a similar fold and active site as ODP (with the exception of Gln224) (28). Likewise, Td also possesses an FDP shown to reduce oxygen and ameliorate oxygen sensitivity in vivo (46). For both species, it now appears that a chemotactic repellent response also aids survival when confronted with oxygen, and this system utilizes proteins that are homologous to oxygen-scavenging enzymes. Thus, the ODP-TM14 system may represent an ancient sensory system that evolved to position cells optimally in life-harboring environments of early Earth.
Materials and Methods
The materials and methods used in this study are described in detail in SI Appendix, Methods. Information includes bioinformatics, cloning, protein production and metal analysis, enzymatic assays, spectroscopy, crystallography, Td genetic manipulations, and chemotaxis assays.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R35GM122535 (to B.R.C.); NIH Grants R35GM131760 and R01DE024463 (to I.B.Z.); NIH Grants R01AI078958 and R01DE023080 (to C.L.); NIH Grant R35GM124908 (to K.M.L.); Grant DE-SC0013997 for Mössbauer spectroscopy; and a National Science Foundation (NSF) Graduate Research Fellowship 2014155468 (to A.R.M.). We thank Aaron Fleetwood, Avery Vilbert, Zach Maschmann, and T. K. Chua for help with experiments and analysis; and the Cornell High Energy Synchrotron Source (CHESS) and Northeast Collaborative Access Team (NE-CAT) at the Advanced Photon Source for access to data collection facilities. The CHESS is supported by NSF Award DMR-1332208 and NIH/National Institute of General Medical Sciences (NIGMS) Award GM-103485. The NE-CAT is supported by NIH/NIGMS Awards P30 GM124165 and S10 RR029205.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.Q. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6R9N, 6QWO, 6QNM, and 6QR2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904234116/-/DCSupplemental.
References
- 1.Hazelbauer G. L., Falke J. J., Parkinson J. S., Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem. Sci. 33, 9–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briegel A., et al. , Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. eLife 3, e02151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander R. P., Zhulin I. B., Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U.S.A. 104, 2885–2890 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollard A. M., Bilwes A. M., Crane B. R., The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry 48, 1936–1944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wadhams G. H., et al. , TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol. Microbiol. 46, 1211–1221 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Palzkill T., Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 1277, 91–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtz D. M., Jr, Flavo-diiron enzymes: Nitric oxide or dioxygen reductases? Dalton Trans. 37, 4115–4121 (2007). [Google Scholar]
- 8.Fiedler A. T., et al. , Spectroscopic and computational studies of (mu-oxo)(mu-1,2-peroxo)diiron(III) complexes of relevance to nonheme diiron oxygenase intermediates. J. Phys. Chem. A 112, 13037–13044 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Amico S., Marx J. C., Gerday C., Feller G., Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 278, 7891–7896 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Kodera M., et al. , Synthesis, characterization, and activation of thermally stable mu-1,2-peroxodiiron(III) complex. Inorg. Chem. 40, 4821–4822 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Zhang X., et al. , Structural and spectroscopic characterization of (μ-hydroxo or μ-oxo)(μ-peroxo)diiron(III) complexes: Models for peroxo intermediates of non-heme diiron proteins. J. Am. Chem. Soc. 127, 826–827 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Jasniewski A. J., Que L. Jr, Dioxygen activation by nonheme diiron enzymes: Diverse dioxygen adducts, high-valent intermediates, and related model complexes. Chem. Rev. 118, 2554–2592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roelfes G., et al. , End-on and side-on peroxo derivatives of non-heme iron complexes with pentadentate ligands: Models for putative intermediates in biological iron/dioxygen chemistry. Inorg. Chem. 42, 2639–2653 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Makris T. M., et al. , An unusual peroxo intermediate of the arylamine oxygenase of the chloramphenicol biosynthetic pathway. J. Am. Chem. Soc. 137, 1608–1617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vu V. V., et al. , Human deoxyhypusine hydroxylase, an enzyme involved in regulating cell growth, activates O2 with a nonheme diiron center. Proc. Natl. Acad. Sci. U.S.A. 106, 14814–14819 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krebs C., Bollinger J. M. Jr, Booker S. J., Cyanobacterial alkane biosynthesis further expands the catalytic repertoire of the ferritin-like ‘di-iron-carboxylate’ proteins. Curr. Opin. Chem. Biol. 15, 291–303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Z., et al. , Crystal structure of the peroxo-diiron(III) intermediate of deoxyhypusine hydroxylase, an oxygenase involved in hypusination. Structure 23, 882–892 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Efimov I., et al. , A simple method for the determination of reduction potentials in heme proteins. FEBS Lett. 588, 701–704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabian J., Simple method of anaerobic cultivation, with removal of oxygen by a buffered glucose oxidase-catalase system. J. Bacteriol. 89, 921 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stubna A., et al. , A structural and Mössbauer study of complexes with Fe(2)(μ-O(H))(2) cores: Stepwise oxidation from Fe(II)(μ-OH)(2)Fe(II) through Fe(II)(μ-OH)(2)Fe(III) to Fe(III)(μ-O)(μ-OH)Fe(III). Inorg. Chem. 43, 3067–3079 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Xiong J., et al. , The O(2) binding pocket of myohemerythrin: Role of a conserved leucine. Biochemistry 39, 8526–8536 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Wirstam M., Lippard S. J., Friesner R. A., Reversible dioxygen binding to hemerythrin. J. Am. Chem. Soc. 125, 3980–3987 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Isaza C. E., Silaghi-Dumitrescu R., Iyer R. B., Kurtz D. M. Jr, Chan M. K., Structural basis for O2 sensing by the hemerythrin-like domain of a bacterial chemotaxis protein: Substrate tunnel and fluxional N terminus. Biochemistry 45, 9023–9031 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Ruby J. D., Lux R., Shi W., Charon N. W., Dasanayake A., Effect of glucose on Treponema denticola cell behavior. Oral Microbiol. Immunol. 23, 234–238 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Xu X., Kolodrubetz D., Construction and analysis of hemin binding protein mutants in the oral pathogen Treponema denticola. Res. Microbiol. 153, 569–577 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Greenswag A. R., et al. , Preformed soluble chemoreceptor trimers that mimic cellular assembly states and activate CheA autophosphorylation. Biochemistry 54, 3454–3468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carfi A., et al. , The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 14, 4914–4921 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang H., et al. , Histidine ligand variants of a flavo-diiron protein: Effects on structure and activities. J. Biol. Inorg. Chem. 17, 1231–1239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X., Holt S. C., Kolodrubetz D., Cloning and expression of two novel hemin binding protein genes from Treponema denticola. Infect. Immun. 69, 4465–4472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai Y., Chu L., Novel mechanism for conditional aerobic growth of the anaerobic bacterium Treponema denticola. Appl. Environ. Microbiol. 74, 73–79 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oddie G. W., et al. , Structure, function, and regulation of tartrate-resistant acid phosphatase. Bone 27, 575–584 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Rao C. V., Kirby J. R., Arkin A. P., Design and diversity in bacterial chemotaxis: A comparative study in Escherichia coli and Bacillus subtilis. PLoS Biol. 2, E49 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wuichet K., Zhulin I. B., Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3, ra50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrity L. F., Ordal G. W., Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology 143, 2945–2951 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Taabazuing C. Y., Hangasky J. A., Knapp M. J., Oxygen sensing strategies in mammals and bacteria. J. Inorg. Biochem. 133, 63–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imlay J. A., The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson J. W., et al. , Structural and molecular characterization of iron-sensing hemerythrin-like domain within F-box and leucine-rich repeat protein 5 (FBXL5). J. Biol. Chem. 287, 7357–7365 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey L. J., Fox B. G., Crystallographic and catalytic studies of the peroxide-shunt reaction in a diiron hydroxylase. Biochemistry 48, 8932–8939 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Knoot C. J., Kovaleva E. G., Lipscomb J. D., Crystal structure of CmlI, the arylamine oxygenase from the chloramphenicol biosynthetic pathway. J. Biol. Inorg. Chem. 21, 589–603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acheson J. F., Bailey L. J., Brunold T. C., Fox B. G., In-crystal reaction cycle of a toluene-bound diiron hydroxylase. Nature 544, 191–195 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Yun D., et al. , (Mu-1,2-peroxo)diiron(III/III) complex as a precursor to the diiron(III/IV) intermediate X in the assembly of the iron-radical cofactor of ribonucleotide reductase from mouse. Biochemistry 46, 1925–1932 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Fitz-Gibbon S. T., House C. H., Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27, 4218–4222 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lakhal R., et al. , Oxygen uptake rates in the hyperthermophilic anaerobe Thermotoga maritima grown in a bioreactor under controlled oxygen exposure: Clues to its defence strategy against oxidative stress. Arch. Microbiol. 193, 429–438 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Le Fourn C., Fardeau M. L., Ollivier B., Lojou E., Dolla A., The hyperthermophilic anaerobe Thermotoga maritima is able to cope with limited amount of oxygen: Insights into its defence strategies. Environ. Microbiol. 10, 1877–1887 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Le Fourn C., et al. , An oxygen reduction chain in the hyperthermophilic anaerobe Thermotoga maritima highlights horizontal gene transfer between Thermococcales and Thermotogales. Environ. Microbiol. 13, 2132–2145 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Frederick R. E., et al. , Dioxygen and nitric oxide scavenging by Treponema denticola flavodiiron protein: A mechanistic paradigm for catalysis. J. Biol. Inorg. Chem. 20, 603–613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.