Significance
Legume crops are significant agriculturally and environmentally for their ability to form a symbiosis with specific soil bacteria capable of nitrogen fixation. However, nitrogen fixation is limited by the availability of the legume host’s bacterial partners in a given soil, and by strain variance in symbiotic effectiveness. In intensively managed agriculture systems, legume crops are provided specific inoculants; inoculation can fail if the added strains are unable to compete in soil with less symbiotically efficient endemic strains. Biogeographic insight is vital to understand what factors affect nitrogen fixation in legume crops and techniques to improve nitrogen fixation. Similarly, understanding the relationship between a legume crop’s symbionts in a geographic context can elucidate broader principles of microbial biogeography.
Keywords: microbial ecology, population genomics, integrative conjugative element, symbiosis, nitrogen fixation
Abstract
Although microorganisms are known to dominate Earth’s biospheres and drive biogeochemical cycling, little is known about the geographic distributions of microbial populations or the environmental factors that pattern those distributions. We used a global-level hierarchical sampling scheme to comprehensively characterize the evolutionary relationships and distributional limitations of the nitrogen-fixing bacterial symbionts of the crop chickpea, generating 1,027 draft whole-genome sequences at the level of bacterial populations, including 14 high-quality PacBio genomes from a phylogenetically representative subset. We find that diverse Mesorhizobium taxa perform symbiosis with chickpea and have largely overlapping global distributions. However, sampled locations cluster based on the phylogenetic diversity of Mesorhizobium populations, and diversity clusters correspond to edaphic and environmental factors, primarily soil type and latitude. Despite long-standing evolutionary divergence and geographic isolation, the diverse taxa observed to nodulate chickpea share a set of integrative conjugative elements (ICEs) that encode the major functions of the symbiosis. This symbiosis ICE takes 2 forms in the bacterial chromosome—tripartite and monopartite—with tripartite ICEs confined to a broadly distributed superspecies clade. The pairwise evolutionary relatedness of these elements is controlled as much by geographic distance as by the evolutionary relatedness of the background genome. In contrast, diversity in the broader gene content of Mesorhizobium genomes follows a tight linear relationship with core genome phylogenetic distance, with little detectable effect of geography. These results illustrate how geography and demography can operate differentially on the evolution of bacterial genomes and offer useful insights for the development of improved technologies for sustainable agriculture.
Biogeography studies the distribution of taxa and ecosystems in space and time and the factors that pattern those distributions. By observing global geographic patterns in plant and animal taxa and the ecosystems they comprise, 18th-century biologists contributed foundational insights to modern evolutionary biology and ecology. Biogeographic principles are less understood for microorganisms, despite the fact that they comprise the vast majority of life’s diversity.
For most of microbiology’s history, understanding the diversity and relatedness of microorganisms has come from studies of pure cultures, which produces a limited and biased view (1). Increasingly, studies examine diversity in microbial ecosystems interrogated through rRNA–gene surveys (2, 3), which allow high-throughput and relatively unbiased assessments of the composition of microbial ecosystems (4). These and related molecular genetic methodologies have begun to uncover biogeographic patterns. Multiple studies have shown that geographic distance between samples is less explanatory of microbial-taxa composition than factors such as pH (5, 6), temperature (7, 8), and salinity (9). The composition of atmospheric microbial communities has been shown to respond to weather (10), while marine microbial communities are structured by depth (11), southern versus northern hemisphere (12), and seasonally (2).
Despite these advances, methods that measure individual genomic features are unable to look confidently at patterns below the genera level and do not measure the explanatory factor by which endemism develops: evolutionary divergence. Whole-genome sequencing reveals the impact of horizontal genetic exchange. As little as 60% of genes in an individual bacterial genome are conserved across the entirety of its genospecies (13), even to the extent of microscale variation in nonhomologous cis-regulatory regions (14). This calls into question how organisms that exchange genes so regularly can form evolutionarily coherent groups. The inverse relationship of exchange frequency and phylogenetic relatedness may lead divergent genome groups to arise in microbial populations, but adaptive genes may cross between divergent populations (15, 16). Whole-genome data provide evidence for endemicity in microbial populations inhabiting island-like hot springs (17), as well as marine-distributed Vibrio cholerae (18). Conversely, photosynthetic marine Prochlorococcus genomes appear to be in equilibrium in genetic exchange across the Atlantic and Pacific oceans with the caveat that accessory genes may assort by ecological niche (19).
Because microbes leave no fossil record, placing observed biogeographic patterns and evolutionary events in microbial populations in time is complicated. Denef and Banfield (20) measured relative rates of recombination and mutation in metagenomes assembled from acid-mine drainage samples, but the geographic and temporal scales were limited to meters and decades, respectively. The well-studied legume–Rhizobium symbiosis provides a system to test hypotheses of bacterial population differentiation and biogeographic patterning on a global scale and over millennia-long time frames, in cases where the biogeography and domestication history of the legume host are well known.
Plants of the family Fabaceae (legumes) have evolved to form a highly specialized symbiosis with diverse Alphaproteobacteria and Betaproteobacteria, broadly referred to as rhizobia (21). Rhizobia provide the plant host with mineral forms of reduced atmospheric nitrogen in exchange for fixed carbon and shelter inside symbiosis-specific plant root nodules (22). Cross-kingdom signaling confers specificity to the symbiosis, such that different legume species generally partner only with circumscribed bacterial taxa and vice versa (23, 24), while gene transfer between related taxa can alter the symbiont’s host range (21).
Nitrogen availability is growth limiting in most agricultural systems (25). In highly managed agricultural systems, nitrogen is typically supplied as fertilizer from the fossil fuel-intensive Haber–Bosch process, accounting for 1 to 2% of global CO2 emissions (26). Legumes grown in rotation with cereal crops have been shown to contribute the equivalent of 30 to 100 kg N/ha—commensurate with agronomic recommendations for nitrogen fertilizer application (27). However, nitrogen fixation rates can vary by crop and geography (28), and the symbiosis is sensitive to environmental extremes (29). Even controlling for these factors, one still finds regional variability for the same crop grown under similar conditions in different locations (30), which may reflect differences in symbiont communities. Thus, legume crops often associate with bacterial strains that perform nitrogen fixation less efficiently than strains identified experimentally as optimal (31). Even in fields where commercial inoculants are provided, endemic rhizobia, present in the soil but inefficient with the legume crop, may outcompete the efficient inoculum in nodule formation (31–33). This has been termed the “competition problem” (31).
Root nodule formation is generally the result of an infection event by a single free-living rhizobial cell, making root nodules effectively clonal most often (34, 35). Inside of a nodule, rhizobial cells divide and endoreduplicate, resulting in many thousands of rhizobial genomes per plant cell (36). These factors enable accurate genome assemblies for discrete bacterial strains sampled as DNA directly from the environment, without culturing, which in cases where the natural history of a legume taxon is well understood can form the basis of hypothesis testing for the biogeographic constraints of its symbionts. Here, we focus on the biogeography of the legume crop chickpea and its nitrogen-fixing bacterial symbionts in the genus Mesorhizobium.
Chickpea (Cicer arietinum) originated in the fertile crescent between 10,000 and 12,000 y ago (37, 38), domesticated from the wild species Cicer reticulatum. C. reticulatum and its sister species Cicer echinospermum occur in contiguous but ecologically distinct ranges in modern-day southeastern Turkey (38). After domestication, chickpea was distributed throughout the Middle East and Mediterranean basin, reaching the Indian subcontinent a minimum of 4,000 y ago (37, 39) and Ethiopia between 2,000 and 3,000 y ago (37), with ensuing continuous cultivation. Genome analyses reveal a primary domestication bottleneck at the center or origin (38), and additional unique genetic bottlenecks and secondary diversification in both India and Ethiopia (39, 40). In the past century, chickpea cultivation was established in countries where modern, intensive agricultural practices predominate, including Canada, the United States, and Australia (37). The history of inoculum use differs substantially between these locations, being rare or absent among smallholder farmers of India and Ethiopia, and common in developed country scenarios. We sampled chickpea’s nitrogen-fixing rhizobial symbionts systematically across the crop’s global agricultural range, both ancient and recent, as well as the native range of its wild relatives. Our detailed understanding of chickpea’s biogeographic history gives us unparalleled ability to interpret patterns in the distribution and relationships of its symbionts.
Results and Discussion
Taxonomic Diversity of Bacterial Symbionts of Chickpea.
Nitrogen-fixing root nodules were collected from chickpea and its wild relatives across soil types, climates, growing seasons, agricultural methodologies, histories of cultivation, and multiple geographic scales (Dataset S1). Sampling consisted of a hierarchical scheme whereby multiple nodules were collected from a plant, multiple plants collected from a field, multiple fields within a region, and multiple regions within a country (Dataset S2). The countries we sampled span the vast majority of chickpea’s agricultural and natural range, including farms in North America, Australia, Morocco, Ethiopia, and India, and at wild ecological sites in the native range of southeastern Turkey. The identity and evolutionary relatedness of nodule bacteria were determined by genome sequencing (41–45), using a combination of pure cultures and metagenomics, with the goal of an unbiased and geographically representative sampling of in situ diversity. Metagenomic samples contained on average of 87.5% DNA from Mesorhizobium—the genus containing the known chickpea-nodulating rhizobia. In total, we obtained 805 genomes suitable for phylogenomic analyses (173 cultures and 632 metagenomes), and an additional 208 lower-quality genomes suitable for species assignment (Dataset S1).
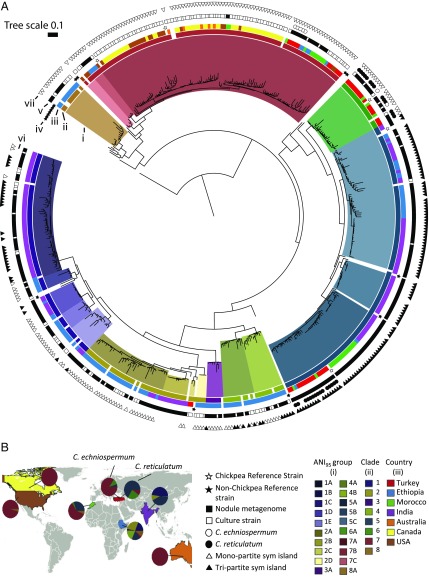
These bacteria occur throughout the full diversity of the genus Mesorhizobium, concentrated primarily in 10 phylogenetically broad clades, several of which contain strains diverse enough to constitute multiple distinct species (Fig. 1A, Dataset S3, and SI Appendix, Fig. S1). Pairwise average nucleotide identity (ANI) was calculated on 400 conserved single-copy marker genes (46) for all pairs of high-quality draft genomes, including reference strains that represent the phylogenetic breadth of Mesorhizobium (Dataset S1). Using 95% ANI (ANI95) as the lower boundary (47) circumscribed 36 distinct Mesorhizobium species, 28 of which are chickpea symbionts that include 20 previously unrecognized species. Many named Mesorhizobium species are misclassified from a genomic perspective (Dataset S3 and SI Appendix, Supplemental Text).
Fig. 1.
Phylogenetic relationships, species assignments, and geographic distribution of a global collection of chickpea’s Mesorhizobium symbiont. (A) Phylogenetic tree of Mesorhizobium cultures and root-nodule DNA extracts based on 400 single-copy marker genes (46). Concentric rings are (inner to outer): (i) 95% ANI cluster, (ii) major clade, (iii) country of collection, (iv) reference strain, (v) nodule metagenome or cultured strain, (vi) host of origin, and (vii) sym island structure. All strains originate from Cicer arietinum unless specified in ring (vi). Clades 9 and 10 are immediately basal to clade 6 and shown with greater clarity in SI Appendix, Fig. S1. The most abundant 20 species are shown, with details of 8 less abundant species given in Dataset S1. (B) Taxonomic composition of Mesorhizobium genomes from chickpea nodules for each country.
Geographic Patterns in Global Mesorhizobium Communities.
The diversity of chickpea mesorhizobia varies at different spatial scales. At a local scale, relatively few sites we sampled exhibited distinct and limited Mesorhizobium diversity. More often, divergent strains coexist, with strains from distinct Mesorhizobium clades occupying different nodules from plants within the same field, on an individual plant, or even individual nodules. Globally, individual agricultural fields typically contain 2 ANI95 groups forming nodules on chickpea. Rarefying to 4 plants sampled from a field—1 nodule per plant—we observe an average of 1.9 ANI95 groups per field in the 23 fields sampled at that depth or greater. We sampled 7 fields that contained 3 ANI95 and 1 field that contained 4 ANI95 groups. We estimate approximately one-third of individual chickpea plants are nodulated by 2 ANI95 groups (of 17 plants where we sequenced samples from 2 nodules, 6 were nodulated by Mesorhizobium strains from distinct ANI95 groups). Conversely, as described below, at a regional scale we document large differences in presence and abundance of chickpea’s distinct Mesorhizobium symbionts.
Chickpea’s wild ancestors show clear divergence in natural symbionts (Fig. 1A and SI Appendix, Fig. S2). In its native range, C. reticulatum—the crop’s immediate wild ancestor—nodulates with Mesorhizobium strains from ANI95 groups 5A—which contains the sequenced type strain for Mesorhizobium muleiense previously described to nodulate cultivated C. arietinum in China (48)—and 6A—containing Mesorhizobium mediterraneum, described to nodulate C. arietinum in Spain (49, 50). The distributions of groups 5A and 6A overlap at their centers of origins in southeastern Turkey, with both appearing at most sites where C. reticulatum is native (38). C. reticulatum’s sister species, C. echinospermum, nodulates primarily with strains from group 7A, containing M. ciceri. M. ciceri and M. mediterraneum were previously described as chickpea’s cognate rhizobial partners, but the type strains for each species were isolated from cultivated chickpea in Spain (49, 50). C. reticulatum and echinospermum occupy distinct geographies and soil types (38), suggesting that their differences in native Mesorhizobium symbionts reflect coadaptation to local host or environmental factors.
In regions where chickpea has been cultivated long-term under traditional agricultural practices, the crop’s predominant symbionts are distinct from those at the hosts’ center of origin and strongly structured by geography. Thus, the monophyletic group consisting of clades 1, 2, 3, and 4 is most abundant in sampled regions of India and Ethiopia, but not present in Morocco or chickpea’s native range of southeastern Turkey (Fig. 1A). The only named representative within this group occurs in clade 2, belonging to the species Mesorhizobium plurifarium, previously described to form nodules on tree and shrub legumes throughout the Old and New World tropics (51). These results suggest a pantropical distribution for this group, typically combined with characteristic local speciation. Strains from clade 5 are ubiquitous in chickpea fields sampled throughout Morocco, India, and Ethiopia. Phylogenetic diversity of these clade 5 groups is largely structured by geography, both within and among species, and is mostly distinct from clade 5 strains nodulating chickpea’s wild relative C. reticulatum in its native range (Fig. 1A). Similarly, strains in clade 7—which contains M. ciceri’s ANI95 group 7A—are globally disperse, largely structured by geography, and distinct from the phylogenetically coherent group of strains nodulating C. echinospermum in wild systems. Interestingly, a small number of M. mediterraneum strains (group 6A) were observed in chickpea nodules in Morocco (Fig. 1A) (and Ethiopia; Dataset S1), nesting phylogenetically within M. mediterraneum strains sampled from wild C. reticulatum.
In parts of the world where chickpea has been introduced recently and is typically grown with rhizobial inoculants (United States, Canada, Australia), nodules were exclusively occupied by strains closely related to but distinct from the inoculant (SI Appendix, Fig. S3), and further resolved from 7A genomes obtained from C. echinospermum nodules (Fig. 1A). This result contrasts with the diversity of Mesorhizobium genomes sampled from regions of long-standing chickpea cultivation, where inoculum use is absent or sparse, and where we observe a much broader range of Mesorhizobium ANI95 groups within and among the major centers of chickpea diversity (Fig. 1 A and B). Thus the Shannon diversity index (52, 53) of Mesorhizobium ANI95 groups is lower for nodules sampled from the US, Australia, or Canada, compared with that of Turkey, India, Ethiopia, or Morocco (Dataset S4). This result holds true whether comparing cultured genomes or both cultured and noncultured genomes, although we cannot exclude the possibility that sampled fields in North America and Australia might contain diversity not captured in isolation screens.
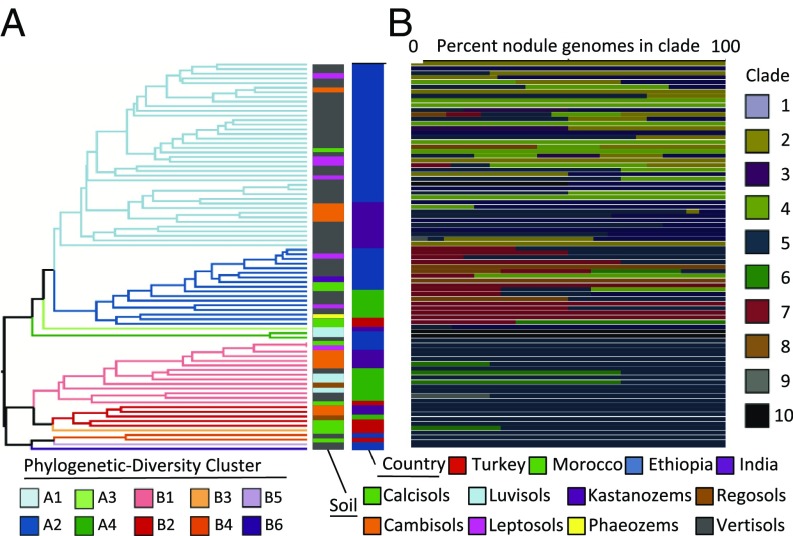
Chickpea’s nodule environment constitutes a homogeneous ecological niche with broad geographic distribution, providing an opportunity to assess biogeographic patterns of symbiosis and the ecological factors that structure them. To avoid possible bias imposed by culturing, we focused on 752 nodule metagenome samples collected from Turkey, Morocco, Ethiopia, and India. Across this distribution, we circumscribed 80 0.2 × 0.2° geographic cells (500 km2) (SI Appendix, Fig. S4), among which we calculated pairwise Mesorhizobium community similarity using the phylogenetically weighted Jaccard index (54, 55) (Fig. 2 and SI Appendix, Fig. S5). Most diversity clusters contain multiple Mesorhizobium clades (Fig. 2B and SI Appendix, Fig. S6), as has been observed for biogeographic patterns of marine picoplankton (56). Diversity clusters are broadly divided into 2 groups (apparent in Fig. 2A and in PC1 of SI Appendix, Fig. S5), driven by the predominance of clades 5 and 6 for diversity cluster B, and clades 1 to 4 and 7 for clusters A1 and A2, respectively (Fig. 2B and SI Appendix, Fig. S6). This division into A and B clusters correlates with latitude. The southernmost sampling sites are from Ethiopia, where 39 out of 43 sampling cells belong to A clusters (primarily A1). In India, samples were collected from 17 grid cells in both the north and south of the subcontinent, with stratification of A1 cells to the south and B cells to the north. The remaining B-cluster cells are from Turkey and Morocco, although both countries also contain cells from cluster A2 (Fig. 2 and SI Appendix, Figs. S4 and S6).
Fig. 2.
Diversity analysis and soil characteristics within sampled 500-km2 regions. (A) Hierarchical clustering of 0.2° × 0.2° grid cells by Mesorhizobium phylogenetic diversity (57, 58). (B) The horizontal colored bars indicate normalized taxon abundance of taxa within a cell, labeled according to country and predominant soil type. See SI Appendix, Table S17 for geographic coordinates of grids.
We used canonical correspondence analysis to test whether the observed variation in Mesorhizobium community composition across geographic space can be explained by climatic and soil variables, in particular soil type, soil pH, latitude, mean annual temperature, and mean annual precipitation. When tested individually, we found each environmental variable to explain a statistically significant portion of observed geographic variation in Mesorhizobium diversity, with soil type contributing the most (Table 1). We further performed forward selection analysis (57) of canonical analysis of principal coordinates (58) to control for correlation between these environmental variables, finding that soil pH does not significantly explain geographic variation in Mesorhizobium diversity, when accounting for the other included variables. This contrasts with previous findings for bulk soil microbial communities. Our forward selection model indicates that—in combination—soil type, latitude, precipitation, and temperature explain 27.6% of geographic variance in Mesorhizobium diversity. Variation in community composition along a north–south gradient was observed for Streptomyces in North American soils (59), explained as adaptations of divergent populations to differing temperatures (60). In the present case, variance partitioning reveals overlap in the contributions of latitude, precipitation, and soil genus (SI Appendix, Fig. S7 and Dataset S5), with temperature contributing predominantly independent of the other tested variables. This suggests the observed correspondence between latitude and diversity of chickpea-nodulating Mesorhizobium is largely a result of interactions between soil type, latitude, and precipitation. Even when accounting for correlations between explanatory variables, we found soil type to independently explain the largest portion of Mesorhizobium diversity variation (SI Appendix, Fig. S7 and Dataset S5), suggesting that the distributions of Mesorhizobium taxa are influenced by adaptation to soil conditions (Fig. 2A and SI Appendix, Figs. S8 and S9 A–D), with the largest split being between vertisols and other soil types. Vertisols are tropically distributed soils, providing further evidence that the latitudinal diversity gradient in chickpea’s global Mesorhizobium populations is best explained by soil factors, and that Mesorhizobium clades 1 to 4 may be tropically adapted.
Table 1.
Canonical analysis of principal coordinates partitioning variation in Mesorhizobium phylogenetic β-diversity among geographic grid cells by geographic and edaphic variables
| Geographic variable | R2 | P value | Confidence interval |
| Soil genus | 15.8 | <0.001*** | 12.5–20.0 |
| Mean annual precipitation | 9.54 | <0.001*** | 5.38–16.5 |
| Latitude | 11.4 | <0.001*** | 6.33–20.0 |
| Mean annual temperature | 5.26 | <0.002*** | 2.96–9.13 |
| Soil pH | 6.08 | <0.001*** | 3.39–10.5 |
Nucleotide-Level Versus Gene Content Variation in Global Chickpea-Mesorhizobium Genomes.
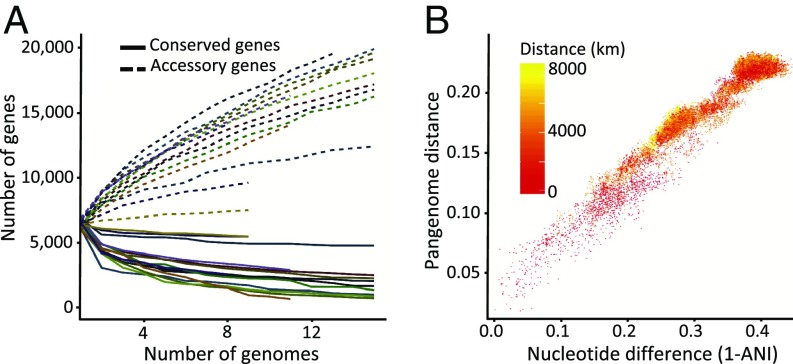
The total gene content of a given group of bacteria has come to be called the pangenome, consisting of genes conserved across the group (the core genome) and genes that are variable by strain (the accessory genome) (61). We compared the gene content of the genomes from each major and minor Mesorhizobium clade observed to nodulate chickpea as well as across the genus. Genomes comprised on average 6,552 predicted genes. Among a finished set of 15 phylogenetically representative strains, we find a strict core genome of 1,217 genes, with a total pangenome containing 41,874 genes. This is broadly comparable to the Prochlorococcus genus, which is estimated to have a global core genome of approximately 1,000 genes and a total pangenome of 84,872 genes (62). Among the larger set of high coverage draft genomes, we find 629 conserved orthologous groups present in greater than 95% of strains, with gene discovery likely limited by variation in genome assemblies. In total, we observed 171,982 orthologous groups of genes from chickpea-nodulating Mesorhizobium genomes. Using a 95% presence cutoff, core genome sizes within 20 chickpea-nodulating Mesorhizobium species from which we collected multiple genomes range from 1,051 to 2,856 genes, with an average of 1,979. The accessory genome size varies by clade but ranges from 17,912 to 38,028 genes when all identified strains are included in each clade. Comparing gene accumulation curves for the pangenome of each sampled Mesorhizobium species (Fig. 3A, SI Appendix, Fig. S10, and Dataset S6) reveals that even when controlling for background-genome phylogenetic distance (measured by ANI; Fig. 3B), Mesorhizobium species vary considerably in the size of core and accessory genomes, as well as the rates of accessory and core genome stabilization. Strikingly, sampling shows genomes from a single ANI95 group can share fewer than half of their genes even within single highly sampled fields, and that the accessory genome of such a geographically and phylogenetically defined group can exceed 15,000 distinct orthologous groups of genes (SI Appendix, Fig. S11). We estimated the exponent of the power law by which the pangenome of each adequately sampled ANI95 group grows with additional sampling (described by ref. 13), revealing that each Mesorhizobium pangenome sampled grows at a distinct rate but that each is open, meaning unlikely to reach saturation with additional sampling (Dataset S6).
Fig. 3.
Pangenome relationships in global Mesorhiozbium populations are driven by core genome evolution. (A) Pangenome gene accumulation curves for each 95% ANI group. The lines depict the average number of genes (core or accessory) present across rarefied genomes, with 10 replications, as the number of genomes increases. (B) Scatterplot depicting the portion of the pangenome shared by any 2 strains versus the nucleotide distance between those strains using 400 universal marker genes (Fig. 1A) (49), colored by geographic distance between those same pairs. Data include only nodule genome assemblies >90% complete.
The microbial pangenome reflects the ubiquity of horizontal gene transfer between distinct bacterial lineages (63). However, we observe a marked decrease of gene sharing between genomes as phylogenetic distance between genomes increases, irrespective of geographic distance. We performed multiple regressions on distance matrices (64) correlating pangenome dissimilarity and average nucleotide distance in 400 conserved marker genes (46). Across the full range of sampled genomes, we observed a strong positive correlation between the portion of genes shared between 2 genomes and their core genome nucleotide distance (Mantel r statistic: 0.9694; P < 0.001) (Fig. 3B). Similarly, clustering Mesorhizobium genomes by the presence or absence of genes in the genus-wide pangenome largely recapitulates the phylogeny calculated from sequence variation in conserved marker genes (SI Appendix, Fig. S12). This pattern corroborates predictions that genetic clusters can form even in light of horizontal gene transfer and agrees with prior observations that recombination rates decrease exponentially with nucleotide differences in homologous sequences (65, 66). Baltrus (67) interprets this in functional terms, as the cost of horizontal gene transfer. Irrespective of the mechanism, our observation that distinct Mesorhizobium species have characteristic core genomes, with genes from the core genome of 1 species often found in the accessory genome of other species, reveals species-level differentiation that is more pronounced with phylogenetic distance. Our results extend previous metagenomic studies in the marine cyanobacterium Prochlorococcus that found a similarly tight relationship between pairwise gene content distance and phylogenetic distance, but for which analysis of cis-relationships was restricted to metagenomic scaffolds rather than whole genomes (56).
Previous analyses reveal that geographic distance correlates with gene content distance in a variety of marine microbial species (68). However, this analysis does not take into account the effect of geography on microbial core genome relatedness. We find that geographic distance correlates significantly (Mantel r: 0.2242; P < 0.001) with gene content distance, but at a much lower level than phylogenetic distance (Mantel r: 0.9694; P < 0.001), which is lower than the correlation found by Nayfach et al. (68) for marine microorganisms. We similarly find that core genome phylogenetic distance correlates with geographic distance (Mantel r: 0.1674; P < 0.001), reflecting the geographic patterns in distributions of Mesorhizobium taxa described above. These results are consistent with the suggestion that phylogenetic relatedness primarily structures gene sharing between genomes, but that geographically close strains are more likely to share genes than distant strains of equal relatedness.
Chromosomal Structure of Chickpea Symbiosis Genes.
Symbiotic compatibility with chickpea appears to derive from horizontal transfer of symbiosis genes across diverse Mesorhizobium taxa, and transfer between strains is influenced by the evolutionary history of the background genome and the symbiosis genes themselves, as well as geography. Throughout Mesorhizobium diversity, all chickpea symbionts share a highly similar set of genes involved in nitrogen fixation and that are monophyletic relative to the species tree (SI Appendix, Fig. S13). In other Mesorhizobia, orthologous symbiosis genes occur in a ∼500-kb genome region that is horizontally transferred as an integrative conjugative element (ICE) (69–71) and that horizontal gene transfer is a driving force in the evolution of plant-commensal lifestyles in the bacterial order Rhizobiales (72). Recent work has also revealed that in some Mesorhizobium genomes the symbiosis island has a tripartite structure (73), excising and transferring from the genome as a single, circular DNA molecule, but undergoing recombination upon insertion and effectively dividing the ICE into 3 nonadjacent segments. We generated single-scaffold assemblies from 14 strains selected to represent most of the geographic and phylogenetic breadth of our sampled Mesorhizobium diversity, to identify the nature of the ICE conferring symbiotic specificity to chickpea. We find that chickpea’s Mesorhizobium symbionts can contain either monopartite (linear, nonrecombined elements) or tripartite symbiosis islands, and that this distinction has important impacts on the biogeographic distribution of the symbiosis island.
Tripartite symbiosis islands have been shown to insert into new genomes as a single element but to undergo 2 sequential, targeted chromosomal inversion events after insertion into the genome. Chromosomal insertion as well as subsequent genomic rearrangements each require a tyrosine recombinase enzyme to catalyze integration into distinct, conserved DNA motifs (attachment or att sites) (73, 74). Whole-genome alignments of finished Mesorhizobium genomes reveal a contiguous region of high nucleotide conservation that contains genes known to be involved in symbiosis (SI Appendix, Fig. S14 A and B). Depending on the strain, this region appears to constitute a monopartite symbiosis island or the α-region of the tripartite symbiosis island.
For most Mesorhizobium monopartite symbiosis islands, the att site resides within a tRNA gene. In 10 of the 14 Mesorhizobium single-scaffold genomes, the symbiosis island is inserted adjacent to 1 serine tRNA gene (with the same genomic position relative to a conserved ribosomal operon), with a tyrosine recombinase immediately downstream (SI Appendix, Fig. S14B). In each of these 10 genomes, this recombinase appears to be a highly conserved member of the same orthologous group, hereafter referred to as IntS1. No other tyrosine recombinase gene is conserved among these genomes. Haskett and colleagues (74) predicted that the chickpea symbiont Mesorhizobium ciceri strain ca181 possesses a tripartite symbiosis island and identified the symbiosis islands’ 3 putative integrase genes. We included the published genome for ca181 in our pangenome analysis and found that the genome does not contain a homolog of IntS1; instead, the IntS homolog identified by Haskett et al. belongs to a distinct orthogroup, hereafter called IntS2. Of the 4 genomes we sequenced where the evident primary symbiosis region did not integrate into the tRNA-ser, 3 possessed the same 3 symbiosis island integrases as ca181 (IntS2, IntG, and IntM) and did not contain a homolog of IntS1, suggesting that these 3 genomes possess a tripartite symbiosis island related to that of ca181 (SI Appendix, Fig. S15 A and B). The remaining genome (M6A.T.Cr.TU.016.01.1.1) possesses IntS1 and lacks homologs to ca181’s integrase genes, but the symbiosis island is not inserted at the same tRNA-ser. This genome’s symbiosis island appears distinct in other ways detailed below. We used the presence and absence of IntS1, IntS2, IntG, and IntM as markers to assign nodule-assembled Mesorhizobium genomes as possessing either tripartite and monopartite symbiosis islands, finding that out of 433 nodule assemblies, 200 likely possess a monopartite symbiosis island (based on the presence of IntS1 and absence of IntS2, IntG, and IntM) and 181 genomes likely contain a tripartite symbiosis island (1 or more of IntS2, IntG, and IntM, absence of IntS1).
Biogeography of the Chickpea-Symbiosis Islands.
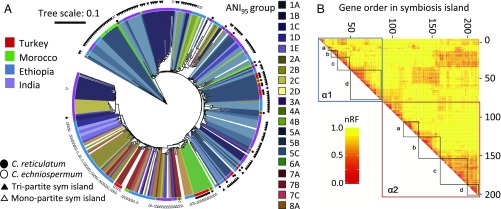
To evaluate the effects of geography, background genome phylogeny, and symbiosis island structure (tripartite versus monopartite) on the spread of the symbiosis island, we determined the conserved core of the symbiosis island, and concatenated alignments of each core symbiosis island gene in nodule-assembled genome drafts (SI Appendix, Supplemental Methods) and used this concatenated alignment to construct a symbiosis island phylogeny (Fig. 4A). We excluded cultured genomes to avoid the possibility of sampling biases imposed by culturing. Among these nodule-assembled Mesorhizobium genomes, we conducted Mantel correlation analyses between symbiosis island core phylogenetic distance and geographic distance, as well as background genome phylogenetic distance. Including all nodule-assembled Mesorhizobium genomes—regardless of symbiosis island type—we observe strong correlation between phylogenetic distance between genomes and phylogenetic distance between the symbiosis islands, but do not observe significant correlation between geographic distance and symbiosis island phylogenetic distance (Table 2). The effect of background genome phylogenetic distance on transfer of the symbiosis island is evident in the sym-core phylogeny as the clustering of primarily clade 5 symbiosis islands (Fig. 4A). Notably, for all clade 5 genomes where we were able to infer the structure of the symbiosis island, we predict these genomes possess a tripartite island (Figs. 1A and 4A). For most of the strains from outside of clade 5 predicted to also possess a tripartite symbiosis island, the symbiosis island core nests phylogenetically within the clade 5 symbiosis island group (as well as geographically circumscribed groups within clade 1 and clade 2). Conversely, the monopartite symbiosis island is broadly distributed through the total extent of Mesorhizobium diversity that we observe to nodulate chickpea, with the notable and evidently strict exception of clade 5. In addition, almost all strains from clade 6 (primarily from chickpea’s wild relatives in southeastern Turkey, as well as several strains from Morocco) cluster very closely phylogenetically. This group includes the finished genome whose symbiosis island is not inserted into the canonical monopartite att site in tRNA-ser, but which contains the characteristic monopartite IntS1, suggesting these genomes may contain a third type of chickpea symbiosis island of unknown arrangement.
Fig. 4.
The distribution of symbiosis island phylotypes is driven by ICE structure and geography, with frequent but patterned recombination. (A) Maximum-likelihood phylogenetic tree of genomes assembled from root nodules, inferred from concatenated alignments of 100 genes identified as core to the symbiosis island in all 14 PacBio assemblies (Dataset S6). Annotation rings are the same as in Figs. 1A and 2B (outside to inside: symbiosis island type, Cicer species, country, clade, and ANI95 group). (B) Heatmap of Robinson-Foulds distances calculated from maximum-likelihood phylogenetic tree comparisons using 10-gene sliding windows of 200 genes with >57% presence and syntenic in 14 PacBio symbiosis islands. α1 and α2 are the 2 conserved regions of the symbiosis island, highlighted in SI Appendix, Fig. S11B. Regulons of genes with related functions are noted: α1a, double-stranded DNA break repair; α1b, hypothetical proteins; α1c, genes involved in nod factor synthesis; α2d, genes involved in nitrogen fixation; α2a, type III secretion system and putative effectors; α2b, biofilm formation (including O-antigen, exopolysaccharide production, quorum-sensing genes, and the type II secretion system); α2c, conjugation (type IV secretion system, plasmid-transfer genes); α2d, cytochrome oxidases.
Table 2.
Mantel correlation tests between symbiosis island genetic distance and core genome phylogenetic distance and geographic distance
| Phylogenetic distance | Geographic distance | |||
| Island | Mantel | P value | Mantel | P value |
| All | 0.451 | <0.001*** | −0.011 | 0.713 |
| Tripartite | 0.341 | <0.001*** | 0.179 | <0.001*** |
| Monopartite | 0.142 | <0.001*** | 0.429 | <0.001*** |
These results suggest the tripartite and monopartite symbiosis islands have distinct phylogenetic distributions within the diversity of Mesorhizobium, and that this distinction is primarily responsible for the correlation between symbiosis island phylogenetic distance and background genome phylogenetic distance, with no detectable effect of geography at a global level. However, when we separately evaluate genomes assigned as possessing either monopartite or tripartite symbiosis islands, within each symbiosis island type, we observe significant correlations between symbiosis island phylogenetic distance and both phylogenetic distance as well as geographic distance (Table 2). In the case of tripartite symbiosis islands, the correlation coefficient for correlation with symbiosis island phylogenetic distance is higher for background genome phylogenetic distance than for geography (r = 0.3406 and 0.1792, respectively). Conversely, for monopartite symbiosis islands, the correlation with background genome phylogenetic distance is lower than that with geographic distance (r = 0.1422 and 0.4291, respectively), meaning that phylogenetically diverse strains that are geographically proximal are more likely to share a recently transferred monopartite symbiosis island, relative to phylogenetically close strains that are geographically distant.
Structure, Function, and Recombination within the Chickpea Symbiosis Island.
Although we infer the symbiosis island (tripartite and monopartite) to be transferred as a single ICE, we find evidence of significant additional gene flow among ICEs at rates higher than the background genome, with recombination structured by gene function. The conserved primary chickpea symbiosis island region varies in length from 352 to 564 kb (Dataset S7). Within this length, there are 2 regions of high nucleotide conservation and gene synteny. The region closer to the serine tRNA insertion site (in those strains where the symbiosis island is inserted in the tRNA-ser gene) contains genes involved in the type III and IV secretion system, as well as putative type III secreted effector genes. The second conserved region contains genes known to be involved in nitrogen fixation and biosynthesis of nod-factor—the signaling-molecule rhizobia produce to initiate nodulation with their cognate host. Outside of and between these 2 regions, the symbiosis island is highly variable both in terms of content and nucleotide sequence, with many annotated genes implicated in genomic transposition and recombination. Five of the 14 finished genomes contained a second type III secretion system located outside of the symbiosis island. In each case, genes from the nonsymbiotic type III secretion system (TTSS) display a phylogeny more similar to that of the background genome than of the symbiosis island (SI Appendix, Fig. S16).
We conducted pairwise whole-genome alignments between all pairs of single-scaffold PacBio Mesorhizobium genomes assembled for this study. Two of these genomes (M1D.F.Ca.ET.043.01.1.1 and M2A.F.Ca.ET.046.03.2.1) have highly similar monopartite symbiosis islands (SI Appendix, Fig. S17A), sharing almost 100% sequence identity throughout their length. The background genomes represent 2 distinct species of Mesorhizobium (ANI95 groups 1D and 2A). We infer conjugative transfer of the symbiosis island from a common source too recent for structural divergence, and indeed the strains originate from sites 16 km apart in northern Ethiopia. Both M1D.F.Ca.ET.043.01.1.1 and M2A.F.Ca.ET.046.03.2.1 possess a second, distinct and also highly conserved symbiosis island (SI Appendix, Fig. S17A). To quantify the number of chickpea-nodulating Mesorhizobium genomes that contain more than 1 symbiosis island, we used BLAST searches of nodC, finding that 4 additional draft genomes assembled from nodules—also from northern Ethiopia—contained 2 copies of nodC. Phylogenetic analysis reveals that all 6 secondary nodC genes are monophyletic within a broader nodC phylogeny, and widely diverged from nodC genes of the co-occurring chickpea symbiosis island (SI Appendix, Fig. S17B). Interestingly, each of these secondary nodC copies is truncated in the same location by the same mobile element (SI Appendix, Fig. S17C), suggesting that these symbiosis islands are nonfunctional, vestigial elements, derived from a common ancestral island and likely the same host plant, despite the fact that the background genomes represent 3 diverged Mesorhizobium specie (ANI95 groups 1C, 2A, and 5C).
Within the conserved regions of the primary symbiosis island, recombination rates appear higher than in the background genome. We constructed maximum-likelihood phylogenies from each conserved gene in the symbiosis island as well as from 400 universal, conserved single-copy nonsymbiosis marker genes. The average normalized Robinson-Foulds (nRF) distance between individual nonsymbiosis marker-gene trees and the concatenated nonsymbiosis marker-gene tree was 0.48, whereas between individual symbiosis genes and a concatenated consensus symbiosis gene tree was 0.8, indicating that phylogenies are more discordant within the symbiosis island than in the core genome. This phenomenon could result if phylogenetic signal is sufficiently low in symbiosis island genes that trees are divergent based on stochasticity, or could result if rates of recombination are higher within the symbiosis island than throughout the rest of the genome. To exclude the first hypothesis, we additionally calculated nRF values considering only branches with bootstrap support of 0.8 or greater—finding similar values. We also calculated nRF on trees for a subset of symbiosis genes using a broader set of genomes (all 14 PacBio genomes as well as 38 genomes collected from wild-Cicer nodules in southeastern Turkey) finding even greater phylogenetic discordance for symbiosis genes than when calculated only for PacBio genome assemblies alone (SI Appendix, Fig. S18).
We further performed pairwise comparisons between trees constructed from concatenated phylogenies of 10-gene sliding windows across the symbiosis island (Fig. 4B). Examining pairwise comparisons of phylogenetic trees constructed from individual symbiosis island genes, as well as between trees constructed from 10-gene sliding windows, reveals patterns of recombination and selection across the symbiosis island. Strikingly, adjacent genes often have higher phylogenetic concordance (low nRF) than comparisons among nonadjacent genes, with important exceptions detailed below. Adjacent genes do not uniformly give low-nRF signals, instead forming discreet blocks of phylogenetic concordance. Many of these blocks correspond to functional regulons of genes with known relevance to symbiotic nitrogen fixation, including nod factor synthesis, nitrogenase assembly, TTSS, biofilm formation, and bacterial conjugation. Similar patterns of low nRF are also observed for gene windows without known relevance to symbiosis, most prominently a string of hypothetical proteins of unknown function adjacent to nod factor synthesis genes, and a block of genes adjacent to the TTSS, which encodes 2-component response regulators among other functional categories. Comparisons of individual-gene trees identifies several symbiosis genes with low average nRF (<0.75) relative to all pairwise comparisons (0.896), including nodD—the transcriptional regulator of nod factor synthesis—and a gene predicted as part of the type II and IV secretion pseudopilus apparatus (SI Appendix, Fig. S19 and Dataset S8).
Comparisons of sliding window phylogenies also reveal interregulon patterns of phylogenetic concordance. In particular, the hypothetical proteins adjacent to nod factor synthesis genes have noticeably low nRF with genes in the nod factor synthesis cluster, suggesting these genes of unknown function may play a role in nod factor synthesis or other early-signaling processes. The large block of genes evidently involved in conjugation and plasmid transfer show phylogenetic concordance with adjacent genes that assemble as a cbb3-type cytochrome c oxidase toward the 3′-end of the symbiosis island. Cbb3-type cytochrome c oxidases play a role in improving respiration rates for aerobic proteobacteria in micro-oxic environments (such as a legume root nodule) and have been shown to be important for nitrogen fixation in Bradyrhizobium (75). The phylogenetic concordance between these genes and those involved in conjugation represents an evolutionary link between performing the symbiosis and transferring the symbiosis island, potentially suggesting further mechanisms of restricting symbiosis island transfer to other bacteria inhabiting root nodules. There are also 2 blocks of long-range phylogenetic concordance, between genes involved in nitrogen fixation with those involved in biofilm formation, as well as between genes involved in nod factor synthesis and those involved in conjugation.
Conclusion
Soil consistently appears among the most diverse microbial ecosystems that microbiologists have studied (76). This study demonstrates that Mesorhizobia are widely distributed in global agricultural soils, evincing the important ecological role of rhizobia. Furthermore, we observe biogeographic patterns in global populations of chickpea’s bacterial symbionts, despite the ubiquity of these taxa and the heterogeneity of soil environments.
The ancient domestication and distribution of the crop chickpea provide a natural experiment to evaluate the limitations of bacterial dispersal, range, and gene flow. We can hypothesize that the wild relatives of chickpea evolved specialized symbioses with distinct bacteria over the course of the plants’ hundred-thousand-year evolution and diversification (38). After chickpea was domesticated and subsequently spread to new locations, we envision 1 of 2 scenarios could have occurred in order for chickpea to continue symbiotic nitrogen fixation: first, that the crop began to partner with novel symbionts native to its new range; second, that the crops’ natural symbionts dispersed with chickpea. There are physical fossil and historical records that enable us to trace the history of chickpea’s domestication and distribution. No such similar evidence exists for chickpea’s bacterial symbionts, but the evolutionary history embedded in their genomes allows us to discriminate between these biogeographic scenarios. Furthermore, the unique biology of symbiotic nitrogen fixation allows us to systematically sample a set of related bacteria across a range of spatial scales.
This global hierarchical sampling scheme across the agricultural and ecological range of chickpea and its wild relatives enables us to analyze diversity of the plants’ symbiont communities to reconstruct their history as chickpea was domesticated and distributed. Phylogenetic analysis suggests that the bacteria responsible for nodule formation on chickpea throughout its natural and cultivated range are of the genus Mesorhizobium (SI Appendix, Fig. S1). This contrasts with some other legume systems for which N2-fixing symbionts often comprise multiple polyphyletic genera of bacteria, broadly known as rhizobia (21, 24). This analysis confirms that chickpea’s wild relatives did evolve for symbiosis with distinct bacterial partners, with distinct ecological ranges, and cross-compatible but phylogenetically differentiable genes for symbiosis. Outside of chickpea’s native range, we find evidence that a hybrid of the 2 predicted scenarios occurred: at present, across regions where chickpea has been cultivated without the intentional addition of specific symbionts, the majority of bacteria we observe to form root nodules are distinct phylogenetically from those that nodulate chickpea’s wild relatives. Furthermore, we find a gradient in Mesorhizobium diversity from north to south, and by soil type, providing evidence that the bacteria that dominate each location are likely adapted to the environmental conditions in those locations. Whole-genome alignments between chickpea’s symbionts’ reveal chromosomal genomes that are diverse at the nucleotide level and in terms of genome structure. Nevertheless, the genes associated with symbiosis in the diverse and locally adapted bacteria that nodulate chickpea outside the crop’s native range share high gene synteny and sequence-level resemblance to those found in chickpea’s natural symbionts in the crop’s native range. Together, this implies that chickpea’s coevolved symbionts dispersed along with the crop—the uniquely broad geographic distribution of strains clade 5A and its affinity with strains at wild chickpea’s center of origin may be a remnant of this dispersal—but were outcompeted in new locations by locally adapted bacteria that acquired symbiosis genes from the dispersed symbiont. This model suggests that adaptive genes can move through preexisting bacterial populations much faster than these genetically distinct populations can adapt to broad environmental changes.
One of the major questions in microbiology since the discovery of the pangenome is how can evolutionarily stable genetic clusters (e.g., species) of bacteria form if bacteria exchange genes so freely. Shapiro and Polz (77) suggest that because homologous recombination rates decline exponentially with nucleotide polymorphisms in homologous regions, genomes that are closely related in the background genome are also more likely to share genes through horizontal transfer. Our results corroborate this hypothesis for the broader Mesorhizobium pangenome, but also demonstrate that bacterial genomes possess mechanisms for fostering specific transfer across defined taxonomic lineages and that geographic factors influence this transfer. Haskett et al. (74) suggest 3 plausible selective advantages of tripartite ICEs. First, that the multiple attachment sites of the tripartite ICE afford a wider range of compatible background genomes. In contrast, our results indicate that the monopartite symbiosis island for chickpea has a broader phylogenetic distribution than the tripartite. Second, that the complex, sequential recombination reactions required to excise tripartite ICEs may aid persistence in host genomes in the absence of active stabilization (e.g., toxin/antitoxin systems), a hypothesis that our results are not structured to evaluate. Third, that monopartite ICEs may be unstable in populations with multiple ICEs competing for the same integration site, because the direct-repeat orientation of monopartite ICE attachment sites can lead to preferentially excised tandem ICE arrays, whereas a tripartite ICE will not be excised in the event of insertion of an invading monopartite ICE. In our results, we observe several instances of multiple symbiosis ICEs occupying the same Mesorhizobium genomes, and in each case, the symbiosis island for chickpea is monopartite rather than tripartite, consistent with the hypothesis of tripartite ICEs having selective advantage in ICE-competitive environments. Our results further suggest an intriguing corollary that the genomic backgrounds compatible with the tripartite symbiosis island are maladapted to successful integration and persistence by other symbiosis islands, in the sense of Baltrus (67). In particular, although we observe that the tripartite symbiosis island has integrated in genomes outside of clade 5, we never observe the monopartite symbiosis island in clade 5 genomes.
Our biogeographic understanding of chickpea—its domestication and distribution, and the effects that had on the genomes of its bacterial symbionts—is a powerful tool for discovering bacterial biogeography. The spread of the symbiosis ICE is a selective sweep in the microbe that originated at the crop’s center of origin. Its subsequent broad geographic distribution is the microbial genome’s analog of the chickpea crop’s domestication, evident as the increased diversity of compatible bacterial species especially at locations of long-standing secondary diversification in India and Ethiopia. Understanding the biogeography of chickpea’s nitrogen-fixing symbionts has important implications for the crop’s agricultural productivity. A common tool for increasing nitrogen fixation and yield in legume cropping systems is to inoculate the crop with specific bacterial strains, known to perform well with the crop under controlled conditions. Our observation that hybrid genotypes of the bacterium arise repeatedly and in parallel at sites of long-standing cultivation suggests that bacteria added as chickpea inoculants will be ecologically unstable over time. Thus, populations of bacteria, likely preexisting and adapted to local factors (e.g., soil), have the capacity to acquire the chickpea-compatible ICE and may ultimately outcompete the inoculant (69, 70). Published results suggest that nitrogen fixation can vary widely in controlled conditions based on the genomic background of the strain involved (78). Thus, it seems evident that researchers interested in providing optimally nitrogen-fixing strains with long-term stability in soil should therefore screen for adaptation to the intended soil environment in addition to nitrogen fixation performance.
Materials and Methods
A detailed description of the methods used in this study can be found in SI Appendix, Supplementary Materials and Methods.
Sample Collection and Processing.
Root nodules were sampled from the global agricultural and native range of chickpea and its closest wild relatives. Fresh or desiccated nodules were surface sterilized, crushed, and streaked onto YMA media for isolation of Mesorhizobium. Nodule samples from Turkey, Morocco, Ethiopia, and India were crushed in Qiagen Plant DNeasy extraction buffer AP1 and processed within 3 wk for DNA extraction.
Genome Sequencing.
DNA was prepared for whole-genome shotgun sequencing using Illumina’s Nextera XT library preparation kit (79), pooled and sequenced on the HiSeq 3000 or MiSeq platform. A subset of 14 cultures were selected for additional sequencing, high–molecular-weight DNA extracted and sequenced on the Pacific Biosciences RS II platform.
Genome Analyses.
Illumina genomic data from Mesorhizoibium cultures were assembled with SPADES (80). Root-nodule metagenomes were assembled and binned using a custom pipeline that included removing chickpea reads, assembling crude metagenome-wide contigs with metavelvet (81), mapping contigs to a reference database of phylogenetically representative Mesorhizobium genomes, and reassembling reads from Mesorhizobium contigs using SPADES (80). Genomes were annotated using prokka (82). Species phylogenies were constructed using the phylophlan pipeline (46). Biogeographic grid squares were clustered using the phylojaccard index implemented in the Biodiverse program (55). Phylojaccard distances between sampling grids was constrained to environmental variables using the capscale function in the R package vegan (83). Pangenome analyses were performed with Roary (84). Symbiosis island boundaries were inferred from whole-genome alignments of single-scaffold PacBio genome assemblies, and syntenic symbiosis genes assigned based on the pangenome of high-quality draft genomes. Sym-island phylogenies were inferred with RaxML (85) and phylogenetic incongruence calculated with the ete3 package (86).
Supplementary Material
Acknowledgments
We thank Dave Richter of the Sutter Basin Growers Co-op; Clarice Coyne, Rebecca McGee, and George Vandermark of Washington State University; Bunyamin Taran of University of Saskatchewan; as well as numerous smallholder farmers in Ethiopia, India, and Morocco, all for providing field samples. We acknowledge National Science Foundation Award IOS-1339346 (to D.R.C., E.J.B.v.W., and B.B.); US Agency for International Development (USAID) Award AID-OAA-A-14-00008 (to D.R.C., E.J.B.v.W., A.K., F.A., K.T., and A.F.). A.G. received support from the USAID Borlaug Fellows Program, and the University of California, Davis, Henry A. Jastro Graduate Research and Thompson Graduate-Student Research Assistantships.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All sequences reported in this paper have been deposited in the National Center for Biotechnology Information BioProject (accession no. PRJNA453501). A full list of biosample numbers is given in Datasets S1 and S7. Annotations are available at https://figshare.com/projects/Greenlon_Mesorhizobium_Biogeography/63542. Scripts and computational pipelines are available at https://github.com/alexgreenlon/meso_biogeo.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900056116/-/DCSupplemental.
References
- 1.Amann R. I., Ludwig W., Schleifer K.-H., Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ladau J., et al. , Global marine bacterial diversity peaks at high latitudes in winter. ISME J. 7, 1669–1677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barberán A., et al. , Continental-scale distributions of dust-associated bacteria and fungi. Proc. Natl. Acad. Sci. U.S.A. 112, 5756–5761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pace N. R., A molecular view of microbial diversity and the biosphere. Science 276, 734–740 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Fierer N., Jackson R. B., The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauber C. L., Hamady M., Knight R., Fierer N., Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fierer N., Carney K. M., Horner-Devine M. C., Megonigal J. P., The biogeography of ammonia-oxidizing bacterial communities in soil. Microb. Ecol. 58, 435–445 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Miller S. R., Strong A. L., Jones K. L., Ungerer M. C., Bar-coded pyrosequencing reveals shared bacterial community properties along the temperature gradients of two alkaline hot springs in Yellowstone National Park. Appl. Environ. Microbiol. 75, 4565–4572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozupone C. A., Knight R., Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U.S.A. 104, 11436–11440 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLeon-Rodriguez N., et al. , Microbiome of the upper troposphere: Species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc. Natl. Acad. Sci. U.S.A. 110, 2575–2580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delong E. F., et al. , Community genomics among microbial assemblages in the Ocean’ s interior. Science 311, 496–503 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Ghiglione J.-F., et al. , Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 109, 17633–17638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tettelin H., Riley D., Cattuto C., Medini D., Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 11, 472–477 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Oren Y., et al. , Transfer of noncoding DNA drives regulatory rewiring in bacteria. Proc. Natl. Acad. Sci. U.S.A. 111, 16112–16117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polz M. F., Alm E. J., Hanage W. P., Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 29, 170–175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumdicker F., Hess W. R., Pfaffelhuber P., The infinitely many genes model for the distributed genome of bacteria. Genome Biol. Evol. 4, 443–456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cadillo-Quiroz H., et al. , Patterns of gene flow define species of thermophilic Archaea. PLoS Biol. 10, e1001265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boucher Y., et al. , Local mobile gene pools rapidly cross species boundaries to create endemicity within global Vibrio cholerae populations. MBio 2, e00335-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coleman M. L., Chisholm S. W., Ecosystem-specific selection pressures revealed through comparative population genomics. Proc. Natl. Acad. Sci. U.S.A. 107, 18634–18639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denef V. J., Banfield J. F., In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science 336, 462–466 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Remigi P., Zhu J., Young J. P. W., Masson-Boivin C., Symbiosis within symbiosis: Evolving nitrogen-fixing legume symbionts. Trends Microbiol. 24, 63–75 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Friesen M. L., Widespread fitness alignment in the legume-Rhizobium symbiosis. New Phytol. 194, 1096–1111 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Masson-Boivin C., Giraud E., Perret X., Batut J., Establishing nitrogen-fixing symbiosis with legumes: How many Rhizobium recipes? Trends Microbiol. 17, 458–466 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Andrews M., Andrews M. E., Specificity in legume-rhizobia symbioses. Int. J. Mol. Sci. 18, E705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J., et al. , A high-resolution assessment on global nitrogen flows in cropland. Proc. Natl. Acad. Sci. U.S.A. 107, 8035–8040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen E. S., Hauggaard-Nielsen H., How can increased use of biological N2 fixation in agriculture benefit the environment? Plant Soil 252, 177–186 (2003). [Google Scholar]
- 27.Peoples M. B., Herridge D. F., Ladha J. K., Biological nitrogen fixation: An efficient source of nitrogen for sustainable agricultural production? Plant Soil 174, 3–28 (1995). [Google Scholar]
- 28.Peoples M. B., Herridge D. F., “Quantification of biological nitrogen fixation in agricultural systems” in Nitrogen Fixation: From Molecules to Crop Productivity (Springer, 2000), pp 519–524. [Google Scholar]
- 29.Zahran H. H., Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 63, 968–989 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herridge D. F., Peoples M. B., Boddey R. M., Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311, 1–18 (2008). [Google Scholar]
- 31.Triplett E. W., Sadowsky M. J., Genetics of competition for nodulation of legumes. Annu. Rev. Microbiol. 46, 399–428 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Streeter J. G., Failure of inoculant rhizobia to overcome the dominance of indigenous strains for nodule formation. Can. J. Microbiol. 40, 513–522 (1994). [Google Scholar]
- 33.Vlassak K. M., Vanderleyden J., Graham P. H., Factors influencing nodule occupancy by inoculant rhizobia. CRC Crit. Rev. Plant Sci. 16, 163–229 (1997). [Google Scholar]
- 34.Gage D. J., Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 184, 7042–7046 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gage D. J., Margolin W., Hanging by a thread: Invasion of legume plants by rhizobia. Curr. Opin. Microbiol. 3, 613–617 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Mergaert P., et al. , Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 5230–5235 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redden R. J., Berger J., “History and origin of chickpea” in Chickpea Breeding and Management, Yadav S. S., Redden R. J., Chen W., Sharma B., Eds. (CABI, Oxfordshire, UK: ), ed. 1, 2007), pp. 1–13. [Google Scholar]
- 38.von Wettberg E. J. B., et al. , Ecology and genomics of an important crop wild relative as a prelude to agricultural innovation. Nat. Commun. 9, 649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plekhanova E., et al. , Genomic and phenotypic analysis of Vavilov’s historic landraces reveals the impact of environment and genomic islands of agronomic traits. Sci. Rep. 7, 4816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varma Penmetsa R., et al. , Multiple post-domestication origins of kabuli chickpea through allelic variation in a diversification-associated transcription factor. New Phytol. 211, 1440–1451 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Greenlon A., Chang P. L., Cook D. R., Sequencing of a global collection of 1,315 chickpea nodulating Mesorhizobium strains. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA453501/. Deposited 14 January 2019.
- 42.Greenlon A., Mesorhizobium prokka genome annotations Figshare. https://figshare.com/projects/Greenlon_Mesorhizobium_Biogeography/63542. Deposited 10 May 2019.
- 43.Greenlon A., Mesorhizobium biogeograph R-scripts data. Figshare. https://figshare.com/projects/Greenlon_Mesorhizobium_Biogeography/63542. Deposited 10 May 2019.
- 44.Greenlon A., Rhizobiales-assigned draft genome orthology matrix. Figshare. https://figshare.com/projects/Greenlon_Mesorhizobium_Biogeography/63542. Deposited 10 May 2019.
- 45.Greenlon A., Alexgreenlon/meso_biogeo. Github. https://github.com/alexgreenlon/meso_biogeo. Deposited 10 May 2019.
- 46.Segata N., Börnigen D., Morgan X. C., Huttenhower C., PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 4, 2304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goris J., et al. , DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Zhang J. J., et al. , Mesorhizobium muleiense sp. nov., nodulating with Cicer arietinum L. Int. J. Syst. Evol. Microbiol. 62, 2737–2742 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Nour S. M., Cleyet-Marel J. C., Normand P., Fernandez M. P., Genomic heterogeneity of strains nodulating chickpeas (Cicer arietinum L.) and description of Rhizobium mediterraneum sp. nov. Int. J. Syst. Bacteriol. 45, 640–648 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Jarvis B. D. W., et al. , Transfer of Rhizobium loti, Rhizobium huakuii, Rhizobium ciceri, Rhizobium mediterraneum, and Rhizobium tianshanense to Mesorhizobium gen. nov. Int. J. Syst. Bacteriol. 47, 895–898 (1997). [Google Scholar]
- 51.Diouf F., et al. , Genetic and genomic diversity studies of Acacia symbionts in Senegal reveal new species of Mesorhizobium with a putative geographical pattern. PLoS One 10, e0117667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shannon C. E., A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 (1948). [Google Scholar]
- 53.Morris E. K., et al. , Choosing and using diversity indices: Insights for ecological applications from the German Biodiversity Exploratories. Ecol. Evol. 4, 3514–3524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leprieur F., et al. , Quantifying phylogenetic beta diversity: Distinguishing between “true” turnover of lineages and phylogenetic diversity gradients. PLoS One 7, e42760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laffan S. W., Lubarsky E., Rosauer D. F., Biodiverse, a tool for the spatial analysis of biological and related diversity. Ecography 33, 643–647 (2010). [Google Scholar]
- 56.Kent A. G., Dupont C. L., Yooseph S., Martiny A. C., Global biogeography of Prochlorococcus genome diversity in the surface ocean. ISME J. 10, 1856–1865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.ter Braak C. J. E., Verdonschot P. E. M., Canonical correspondence analysis and related multivariate methods in aquatic ecology. Aquat. Sci. 57, 255–289 (1995). [Google Scholar]
- 58.Anderson M. J., Willis T. J., Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 84, 511–525 (2003). [Google Scholar]
- 59.Choudoir M. J., Doroghazi J. R., Buckley D. H., Latitude delineates patterns of biogeography in terrestrial Streptomyces. Environ. Microbiol. 18, 4931–4945 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Choudoir M. J., Buckley D. H., Phylogenetic conservatism of thermal traits explains dispersal limitation and genomic differentiation of Streptomyces sister-taxa. ISME J. 12, 2176–2186 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R., The microbial pan-genome. Curr. Opin. Genet. Dev. 15, 589–594 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Biller S. J., Berube P. M., Lindell D., Chisholm S. W., Prochlorococcus: The structure and function of collective diversity. Nat. Rev. Microbiol. 13, 13–27 (2015). [DOI] [PubMed] [Google Scholar]
- 63.McInerney J. O., McNally A., O’Connell M. J., Why prokaryotes have pangenomes. Nat. Microbiol. 2, 17040 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Lichstein J. W., Multiple regression on distance matrices: A multivariate spatial analysis tool. Plant Ecol. 188, 117–131 (2006). [Google Scholar]
- 65.Shapiro B. J., Polz M. F., Microbial speciation. Cold Spring Harb. Perspect. Biol. 7, a018143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fraser C., Hanage W. P., Spratt B. G., Recombination and the nature of bacterial speciation. Science 315, 476–480 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baltrus D. A., Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Nayfach S., Rodriguez-Mueller B., Garud N., Pollard K. S., An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 26, 1612–1625 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sullivan J. T., Patrick H. N., Lowther W. L., Scott D. B., Ronson C. W., Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. U.S.A. 92, 8985–8989 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sullivan J. T., Ronson C. W., Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. U.S.A. 95, 5145–5149 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sullivan J. T., et al. , Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184, 3086–3095 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garrido-Oter R., et al. ; AgBiome Team , Modular traits of the Rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 24, 155–167.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haskett T. L., et al. , Assembly and transfer of tripartite integrative and conjugative genetic elements. Proc. Natl. Acad. Sci. U.S.A. 113, 12268–12273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haskett T. L., et al. , Evolutionary persistence of tripartite integrative and conjugative elements. Plasmid 92, 30–36 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Pitcher R. S., Watmough N. J., The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta Bioenerg. 1655, 388–399 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Fierer N., Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Shapiro B. J., Polz M. F., Ordering microbial diversity into ecologically and genetically cohesive units. Trends Microbiol. 22, 235–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elias N. V., Herridge D. F., Naturalised populations of mesorhizobia in chickpea (Cicer arietinum L.) cropping soils: Effects on nodule occupancy and productivity of commercial chickpea. Plant Soil 387, 233–249 (2015). [Google Scholar]
- 79.Illumina, Nextera XT DNA Sample Preparation Guide (Illumina, 2012).
- 80.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Namiki T., Hachiya T., Tanaka H., Sakakibara Y., MetaVelvet: An extension of velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res. 40, e155 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).24642063 [Google Scholar]
- 83.Dixon P., VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003). [Google Scholar]
- 84.Page A. J., et al. , Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.