Abstract
The innate immune defense of mammalian hosts relies on its capacity to detect invading pathogens and then directly eliminate them or help guide adaptive immune responses. Recognition of microbial DNA and RNA by pattern recognition receptors (PRRs) is central to the detection of pathogens by initiating cytokine-mediated innate immunity. In contrast, disturbance of this pathogen surveillance system can result in aberrant innate immune activation, leading to proinflammatory or autoimmune diseases. Among the many important PRRs are proteins of the retinoic acid-inducible gene-I (RIG-I)-like receptor (RLR) family as well as cyclic GMP-AMP synthase (cGAS), which detect viral RNA and DNA, respectively, within the host cell. Intriguingly, recent evidence has shown that “unmasked,” misprocessed, or mislocalized host-derived RNA or DNA molecules can also be recognized by RLRs or cGAS, thereby triggering antiviral host defenses or causing inflammation. Here, we review recent advances of endogenous nucleic acid recognition by RLRs and cGAS during viral infection and systemic proinflammatory/autoimmune disorders.
Keywords: innate immunity, interferon, RIG-I-like receptors, cGAS, viral infection, autoimmunity
Introduction
The innate immune system of vertebrates is an evolutionarily conserved surveillance apparatus that functions to detect microbial invasion or damaged “self,” thereby eliminating the threat and returning the cellular environment to homeostasis. Key mediators of innate immunity are germline-encoded receptors called pattern recognition receptors (PRRs) that detect conserved molecular patterns of foreign organisms (eg, viral or bacterial components such as nucleic acid) known as pathogen-associated molecular patterns (PAMPs) (Janeway and Medzhitov 2002).
Innate immune sensors fall into 2 major categories: First, membrane-bound sensors (eg, sensors of the Toll-like receptor and C-type lectin receptor families) recognize extracellular or membrane-encased PAMPs (Medzhitov and others 1997; Takeuchi and Akira 2010; Dambuza and Brown 2015). A second class of innate immune receptors, which are the focus of this review, recognize PAMPs within the host cell's environment, such as in the cytoplasm or nucleus (Chan and Gack 2016). The recognition of PAMPs by PRRs initiates innate immune signaling that ultimately results in the production of a multitude of host defense molecules, which together establish an antimicrobial milieu.
The recognition of viral RNA in the cytoplasm of infected cells is primarily mediated by members of the retinoic acid-inducible gene-I (RIG-I)-like receptor (RLR) family. Three RLR members have been identified: RIG-I (gene name: DDX58), melanoma differentiation-associated protein 5 (MDA5; gene name: IFIH1), and laboratory of genetics and physiology 2 (LGP2; gene name: DHX58) (Meylan and others 2006), the latter playing a regulatory role rather than functioning as a sensor protein. RIG-I and MDA5 harbor considerable similarities in domain structure and sequence; however, they recognize distinct types of viral RNA and hence different viral pathogens (Nakhaei and others 2009). RIG-I senses relatively short 5′ tri- or diphosphorylated (mostly blunt-ended) double-stranded RNA (dsRNA) or single-stranded RNA (ssRNA) species, which can be found in many negative-strand RNA viruses, including influenza A virus (IAV) and vesicular stomatitis virus (VSV), as well as positive-strand RNA viruses, such as Japanese encephalitis virus (Kato and others 2006; Loo and others 2008; Schlee 2013). Moreover, RIG-I has also been shown to sense poly-U or poly-UC sequence motifs (Saito and others 2008). In contrast, the optimal ligand for MDA5 is long dsRNA, or aggregates of dsRNA, as found, for example, in cells infected with certain picornaviruses. Furthermore, recent studies have shown that both RIG-I and MDA5 function in the detection of dengue virus (DENV), West Nile virus, and reoviruses (Loo and others 2008; Chan and Gack 2016).
Following the recognition of RNA ligands, RIG-I and MDA5 undergo various steps of activation, which include oligomerization, conformational changes, and a variety of post-translational modifications such as ubiquitination and dephosphorylation (Chiang and Gack 2017; van Tol and others 2017). Activated RIG-I and MDA5 trigger signal transduction through the adaptor protein mitochondrial antiviral signaling protein (MAVS; also known as Cardif, virus-induced signaling adapter [VISA], or interferon beta promoter stimulator protein 1 [IPS-1]), which is located on the outer membrane of mitochondria and at peroxisomes (Nakhaei and others 2009; Belgnaoui and others 2011). MAVS then recruits various innate signaling molecules, including the Ser/Thr kinases TANK-binding kinase 1 (TBK-1), IκB kinase-ɛ (IKKɛ), and IKKα/β/γ, which induce gene expression of various cytokines, including type I interferons (IFNs), as well as chemokines through the activation of transcription factors IFN-regulatory factor 3/7 (IRF3/7) and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). Secretion of IFN-α/β then leads to transcriptional induction of many IFN-stimulated genes (ISGs) via Janus kinase-signal transducers and actuators of transcription (JAK-STAT)-mediated signal transduction, which ultimately creates an antiviral state that impedes viral infection.
Cyclic GMP-AMP synthase (cGAS) is a cytosolic sensor that recognizes viral double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA), thereby eliciting innate immune defense against DNA viruses as well as some retroviruses (Cai and others 2014; Herzner and others 2015; Li and Chen 2018). Following DNA binding, cGAS undergoes a conformational change that leads to the synthesis of the second messenger cyclic GMP-AMP (cGAMP) from adenosine triphosphate (ATP) and guanosine triphosphate (GTP) (Chen and others 2016). cGAMP subsequently binds to the endoplasmic reticulum (ER)-resident membrane adaptor protein stimulator of IFN genes (STING), which leads to its activation and translocation from the ER to the Golgi apparatus. STING then activates the TBK-1/IRF3 and IKK/NF-κB axes, which ultimately triggers IFN- and ISG-dependent antiviral defenses (Barber 2014; Chen and others 2016).
Effective innate immune sensing is crucial for efficient production of type I IFNs and a rapid antiviral host response. However, dysregulated and overzealous production of type I IFNs (and also other cytokines or chemokines) can lead to inflammation and autoimmunity. Recent studies have shown that aberrant activation of RLR or cGAS signaling is associated with a variety of inflammatory/autoimmune diseases such as Aicardi-Goutières syndrome (AGS), Singleton-Merten syndrome (SMS), and systemic lupus erythematosus (SLE). Conceptually, these diseases arise from mutations in genes encoding these sensors and leading to their constitutive activation (so-called “gain-of-function” mutations) (reviewed in detail in Barrat and others 2016; Crowl and others 2017), or when these sensor proteins are activated by host-derived nucleic acids. The latter phenomenon is primarily due to loss-of-function mutations found in certain enzymes, in particular intracellular RNases or DNases, that are responsible for keeping the abundance of endogenous RNAs and DNAs at a minimal level (as described below).
Since the discovery of RLRs over a decade ago, much work has been done on defining the precise features of viral RNAs that allow their recognition by RIG-I and MDA5 (Sparrer and Gack 2015; Chan and Gack 2016; Schlee and Hartmann 2016). Intriguingly, a recent series of studies showed that RIG-I and MDA5 can also recognize host-derived RNAs. Conceptually, host RNA recognition by RLRs occurs when these RNAs are either mislocalized, incorrectly processed, or “unmasked” because of downregulation of proteins that usually bind to these RNAs. RLR activation by cellular RNA species has been implicated in the pathogenesis of proinflammatory and autoimmune diseases. Moreover, it has been demonstrated that RLR activation by host RNAs can also be beneficial to the host by mediating an antiviral response. Similar to host RNA recognition by RLRs, “self” recognition by cGAS can occur when host DNA (eg, genomic or mitochondrial DNA) is mislocalized, or because of loss-of-function mutations in genes that encode specific enzymes responsible for degrading or metabolizing host DNAs and keeping their levels at a minimum. In the following sections, we provide an overview of recent research on host nucleic acid sensing by RLRs and cGAS during viral infection and autoimmunity.
Recognition of Host RNA by RLRs During Viral Infection
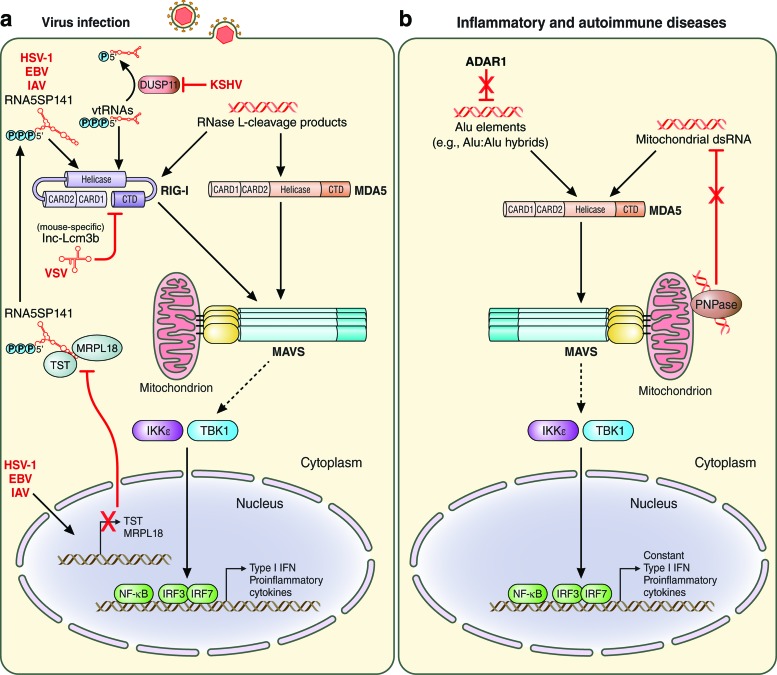
Recent studies have demonstrated that RLRs can recognize various types of endogenous RNAs to mediate an antiviral innate immune response. These host-derived RNAs are sensed by RLRs upon virus-induced “unmasking” from the proteins that normally bind to these RNAs, or after their incorrect processing and/or subcellular mislocalization. RIG-I protein affinity purification from herpes simplex virus type 1 (HSV-1)-infected cells followed by RNA deep sequencing identified that 5S ribosomal RNA pseudogene 141 (RNA5SP141) transcripts function as endogenous RIG-I ligands. 5S rRNA pseudogene transcripts are generated by RNA polymerase III (Pol III) and primarily found in the nucleus under normal (uninfected) conditions (Chiang and others 2018). The small amounts of RNA5SP141 transcripts found in the host cell cytoplasm were found to be bound by at least 2 RNA-binding proteins, Rhodanese (also known as thiosulfate sulfurtransferase, TST) and mitochondrial ribosomal protein L18 (MRPL18), thereby apparently preventing RNA5SP141 from binding to and activating RIG-I in uninfected cells (Fig. 1a). However, during HSV-1 infection, RNA5SP141 was found predominantly in the cytoplasm, where RIG-I resides, indicating relocalization of RNA5SP141 triggered by herpesvirus infection. Furthermore, HSV-1-induced host shutoff, which represents the massive downregulation of cellular transcripts and/or proteins by viral factors (eg, HSV-1 vhs protein, which is an RNase), led to markedly reduced protein abundance of TST and MRPL18, thereby liberating or “unmasking” RNA5SP141 and allowing its binding to RIG-I (Chiang and others 2018). Knockdown studies using siRNA or locked nucleic acid gapmers targeting RNA5SP141 in various human cell types, including primary human lung fibroblasts, showed that endogenous RNA5SP141 transcripts are important for proinflammatory cytokine responses not only to HSV-1 but also to the related Epstein-Barr virus (EBV), as well as IAV of the Orthomyxoviridae family (Chiang and others 2018). This study established that RNA5S pseudogene transcripts, whose functions are largely unknown in contrast to those of the parental 5S rRNA, serve as agonists of the sensor RIG-I to allow an antiviral or proinflammatory host response. Moreover, the data on RNA5SP141's role in RIG-I activation during IAV and EBV infections combined with previous studies that identified EBV- and IAV-derived RNAs as RIG-I agonists (Ablasser and others 2009; Chiu and others 2009; Baum and others 2010; Rehwinkel and others 2010) suggest that both viral and host-derived RIG-I ligands contribute to an effective host response to these viruses. Future studies will need to determine whether the detection of other viruses is mediated by RNA5SP141, or perhaps other RNA5S pseudogene transcripts.
FIG. 1.
Endogenous RNA recognition by RIG-I-like receptors. Recognition of endogenous RNAs by RIG-I and/or MDA5 during viral infection (a) or in inflammatory and autoimmune diseases (b). Specific RNA ligands are depicted as well as the signaling pathway initiated by RIG-I and MDA5 via their adaptor protein MAVS, which is localized at the mitochondrion. Activation of MAVS then leads to several downstream signaling events, including the activation of TBK-1, IKKɛ, IRF3, IRF7, and NF-κB. The detailed mechanisms of host ligand recognition by RLRs are described in the text. Solid lines indicate direct effects or signaling events. Dashed lines indicate signaling events that are indirect. Red lines indicate inhibitory effects. CARD, caspase activation and recruitment domain; CTD, carboxy-terminal domain; EBV, Epstein-Barr virus; HSV-1, herpes simplex virus type 1; IAV, influenza A virus; IKKɛ, IκB kinase-ɛ; IFN, interferon; IRF, IFN regulatory factor; KSHV, Kaposi's sarcoma-associated herpesvirus; MDA5, melanoma differentiation-associated protein 5; MAVS, mitochondrial antiviral signaling protein; NF-κB, nuclear factor-κB; P, phosphate; RIG-I, retinoic acid-inducible gene-I; TBK-1, TANK-binding kinase 1; VSV, vesicular stomatitis virus. Color images are available online.
A recent study showed that another type of cellular noncoding Pol III transcripts, called vault RNAs (vtRNAs), can be sensed by RIG-I during lytic reactivation of Kaposi's sarcoma-associated herpesvirus (KSHV) in patient-derived primary effusion lymphoma cells and the cell line iSLK.219, which is latently infected with recombinant KSHV (Zhao and others 2018) (Fig. 1a). In this case, it has been suggested that incorrect processing of these host RNAs allows for their recognition by RIG-I. This study showed that cellular DUSP11 (triphosphatase dual specificity phosphatase 11) removes 5′-triphosphate moieties from cellular vtRNAs during latent KSHV infection, thereby preventing detection of vtRNAs by RIG-I. During lytic reactivation of KSHV the expression of DUSP11 (both mRNA and protein levels) is downregulated, which leads to cytosolic accumulation of 5′-triphosphorylated vtRNAs and activation of RIG-I, which, in turn, restricts KSHV lytic infection (Zhao and others 2018). However, the physiological relevance of vtRNAs and their relative contribution to KSHV-mediated antiviral host responses remain to be determined. Moreover, a recent study suggested that short interspersed elements transcribed by Pol III could function as endogenous ligands of RIG-I and also MDA5 upon infection with murine gammaherpesvirus 68, leading to the activation of NF-κB in a MAVS-dependent manner (Mu and others 2016).
Several studies have demonstrated that, besides cellular Pol III transcripts, certain microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) can also serve as endogenous ligands of RIG-I. The expression of these types of host RNAs is induced upon viral infection, which allows for their detection by RIG-I. miRNA-136 was shown to be transcriptionally upregulated upon infection with the highly pathogenic H5N1 strain of IAV as well as VSV. miRNA-136 then binds to and activates RIG-I, which inhibits viral replication through production of IFN-β and interleukin-6 (Zhao and others 2015). In contrast to the described endogenous RIG-I agonists that mediate an antiviral/proinflammatory immune response, lncRNA-Lsm3b (which is mouse-specific) was shown to compete with viral RNA for RIG-I binding and to serve as a negative feedback inhibitory mechanism to dampen cytokine responses during the late stages of VSV, Sendai virus (SeV), and IAV infections (Jiang and others 2018) (Fig. 1a). Transcriptional upregulation of lnc-Lsm3b after VSV or SeV infection of mouse peritoneal macrophages directly correlated with decreased IFN gene expression. Furthermore, lnc-Lsm3b knock-out mice showed enhanced production of type I IFNs, lower viral burden, and reduced severity of pulmonary inflammation upon VSV infection compared with control mice. Mechanistically, lnc-Lsm3b sequesters monomers of RIG-I, thereby keeping this sensor in an inactive state to diminish innate immune responses during the late stages of infection.
Recent data indicated that mislocalized mitochondrial dsRNA can trigger MDA5-mediated innate immune responses. Loss of polynucleotide phosphorylase (PNPase), which is a critical component of the RNA degradosome, resulted in cytoplasmic accumulation of mitochondrial dsRNA, which subsequently activated MDA5 and led to the production of type I IFN and the upregulation of ISGs (Dhir and others 2018). Mice with a liver-specific knock-out of PNPase (Pnpt1HepKO mice) showed accumulation of dsRNA and enhanced transcript expression for IFN-β and ISGs in the liver compared with control animals. Although the majority of the experiments in this study were not performed in the context of viral infection, this study suggested that a similar mechanism of mitochondrial dsRNA mislocalization could also be relevant during infection with certain viruses such as encephalomyocarditis virus (EMCV), a picornavirus. Furthermore, it has been suggested that cellular NOP14 and GINS1 RNAs could serve as endogenous ligands of the sensor MDA5 to block the lytic reactivation of KSHV (Zhao and others 2018). However, future studies will need to determine the physiological relevance of these RNAs in the activation of MDA5-mediated antiviral responses. Moreover, 2′,5′-linked oligoadenylate synthetase-dependent ribonuclease RNase L generates small RNA cleavage products from self-RNA upon infection with certain viruses, such as EMCV and SeV (Malathi and others 2007). In fact, RNase L-cleavage products were the first cellular RNAs identified to activate RIG-I and MDA5 and trigger host antiviral immune responses (Malathi and others 2007) (Fig. 1a).
In conclusion, these recent findings on innate immune sensing of host RNA during viral infection revealed that unmasking or inappropriate processing of noncoding Pol III transcripts, or transcriptional upregulation of other types of noncoding RNAs (eg, certain miRNAs), allow for the recognition of these RNAs by RLRs. Interestingly, these endogenous RNA ligands share most, but not necessarily all, molecular features that have previously been found in viral RNAs to be important for triggering RLR activation. For example, while most of the described host-derived RIG-I ligands have a 5′-triphosphate moiety and dsRNA stretches, several of them are, however, not blunt-ended, which suggests that endogenous RLR ligands do not strictly adhere to all criteria of “classic” agonists, which have often been studied using in vitro experimental systems. Furthermore, mislocalized cellular dsRNA that accumulates in the cytoplasm due to organelle damage can also be recognized by the RLR machinery, activating antiviral and proinflammatory host responses. Lastly, while most host-derived RLR ligands identified so far activate innate immunity, as one would expect, some host-derived RNAs may also dampen antiviral and/or proinflammatory responses as a means of negative feedback inhibition, as it has been shown for lnc-Lsm3b.
Recognition of Host RNA by RLRs in Autoimmunity
RLR activation by endogenous RNAs has also been implicated in inappropriate upregulation of type I IFN (and also other cytokines) commonly associated with a number of inflammatory and autoimmune diseases. Recent studies have reported that gain-of-function mutations in the IFIH1 gene (which encodes MDA5) lead to constant type I IFN signaling because of stronger MDA5 binding to RNA and/or hypersensitivity to self-RNA ligands, resulting in certain forms of AGS, SMS, or SLE (Rice and others 2014; Rutsch and others 2015). In contrast, loss-of-function mutations in the genes of specific enzymes involved in RNA metabolism can also lead to autoimmune diseases through aberrant RLR activation by certain host RNAs. Stetson and others reported that biallelic mutations in the gene SKIV2L, which regulates RNA turnover as part of the cytosolic RNA exosome, lead to the generation of endogenous immunostimulatory RNAs, which ultimately triggers RIG-I-mediated IFN production in trichohepatoenteric syndrome patients (Eckard and others 2014). Furthermore, Adar1 deficiency leads to the accumulation of retroelements, such as Alu:Alu hybrids, in the cytoplasm, which are then recognized by MDA5, resulting in an overzealous proinflammatory response (Rice and others 2012; Ahmad and others 2018; Chung and others 2018) (Fig. 1b). Mouse models deficient in Adar1, or encoding editing-deficient Adar1 mutant variants, established that dysregulated RNA editing caused MDA5-driven autoimmunity (Mannion and others 2014; Liddicoat and others 2015; Pestal and others 2015). Similarly, a recent study reported that patients harboring hypomorphic mutations in PNPT1 (encoding PNPase) display accumulation of mitochondrial dsRNA coupled with upregulation of MDA5-mediated innate immune responses (Dhir and others 2018) (Fig. 1b).
Collectively, these findings suggested that increased avidity of RLRs to self-ligands caused by gain-of-function mutations in RLR genes, or aberrant accumulation of cytosolic RNA due to loss-of-function mutations in genes encoding specific enzymes that metabolize or degrade host RNAs, can lead to abnormal RLR activation and thereby autoimmune diseases.
Recognition of Cellular DNA by cGAS During Viral Infection
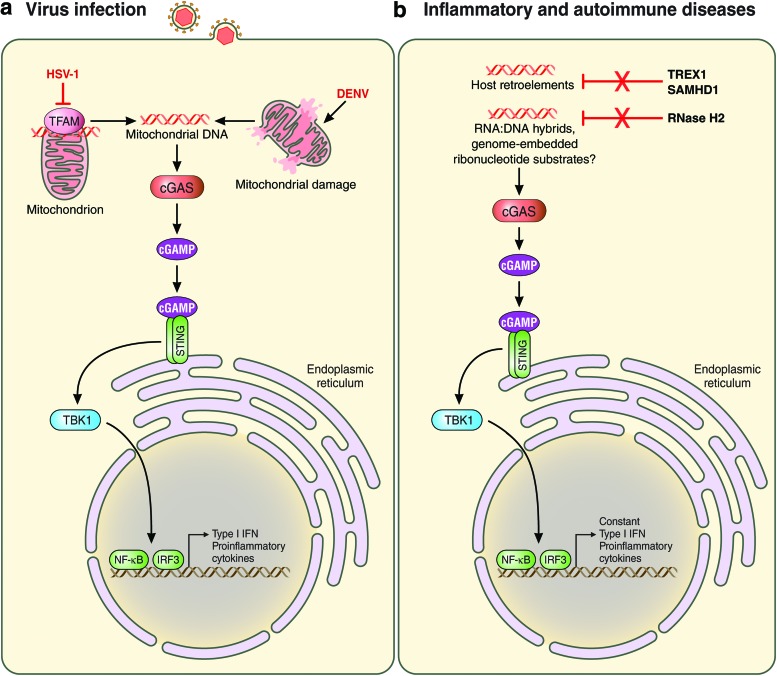
Recent evidence has indicated that mislocalized cellular DNA can be sensed by the cGAS–STING pathway during virus infection. Infection with DENV, a positive-strand RNA virus of the Flaviviridae family, induces mitochondrial membrane alterations, which triggers the release of mitochondrial DNA (mtDNA) into the cytoplasm and subsequent cGAS activation in various cell types, including primary human monocyte-derived dendritic cells (Aguirre and others 2017) (Fig. 2a). Similarly, mtDNA was shown to activate cGAS-mediated antiviral innate immunity during HSV-1 infection. HSV-1 infection induced mtDNA stress through loss of the mtDNA-binding protein TFAM (transcription factor A, mitochondrial), and TFAM deficiency induced the release of mtDNA into the cytoplasm where it is sensed by cGAS (West and others 2015) (Fig. 2a). Furthermore, mtDNA leakage has been implicated in cGAS-mediated immunity triggered by apoptotic stimuli. Apoptosis is mediated through the activation of members of the caspase family of proteases and further regulated by Bcl-2-family proteins (both pro- and anti-apoptotic ones), which control the formation of the Bax/Bak channel at the mitochondrial outer membrane. However, recent studies demonstrated that an unexpected caspase-inhibited apoptosis mechanism triggered Bax/Bak-dependent mtDNA leakage and thereby activation of innate immunity. In caspase-9-deficient mice (Casp9−/−) as well as caspase-9 and/or Bax/Bak knock-out primary mouse fibroblasts, mtDNA was shown to accumulate in the cytoplasm, leading to the activation of type I IFN production via cGAS and STING (Rongvaux and others 2014; White and others 2014). Enhanced innate immunity by this mechanism of mtDNA leakage could confer an antiviral state as Casp9−/− mice had better survival rates following EMCV infection compared with control animals. Taken together, these results suggest that mislocalization of mtDNA following virus-induced mitochondrial alterations, or apoptotic stimuli, can trigger cGAS–STING-mediated host immunity.
FIG. 2.
Endogenous DNA recognition by cGAS. Recognition of endogenous DNA species by cGAS during viral infection (a) or in inflammatory and autoimmune diseases (b). Following the recognition of cellular DNA, cGAS produces the second messenger cGAMP, which then binds to and activates STING at the ER. STING then recruits and activates the kinase TBK-1, leading to IRF3 activation. In addition, NF-κB is activated. The detailed mechanisms of host DNA recognition by cGAS are described in the text. Solid lines indicate direct effects or signaling events. Red lines indicate inhibitory events. cGAMP, cyclic GMP-AMP; cGAS, cyclic GMP-AMP synthase; DENV, dengue virus; ER, endoplasmic reticulum; STING, stimulator of IFN genes. Color images are available online.
Recognition of Cellular DNA by cGAS in Autoimmunity
cGAS-mediated sensing of DNA (pathogen-derived or cellular) is required for effective antiviral host defense against a number of pathogens; however, aberrant activation of the cGAS–STING pathway has also been linked to autoimmune and inflammatory disorders (Fig. 2b). Recent research has shown that recessive mutations in genes involved in DNA metabolism are responsible for improper cGAS–STING signaling. For instance, certain mutations in the gene that encodes 3′ repair exonuclease 1 (TREX1, previously called DNase III) cause the accumulation of ssDNA from endogenous retroelements, leading to a cell-intrinsic autoimmune response in AGS and chilblain lupus patients (Crow and others 2006a; Yang and others 2007; Stetson and others 2008).
Furthermore, genetic mutations in the genes encoding the 3 subunits of the RNase H2 enzyme complex, RNASEH2A, RNASEH2B, or RNASEH2C, lead to reduced RNase H2 function and thereby higher levels of endogenous RNA–DNA hybrids or possibly also retroelements. These endogenous ligands stimulate aberrant type I IFN production via the cGAS–STING signaling axis in AGS (Crow and others 2006b). Studies in mice supported the notion that cGAS–STING-mediated autoimmunity is triggered by endogenous ligands that are usually degraded by RNase H2. Rnaseh2b (A174T/A174T) and Rnaseh2a (G37S/G37S) knock-in mice exhibited AGS-like symptoms such as chronic IFN/ISG production due to the accumulation of host nucleic acid species that activated cGAS–STING signaling (Mackenzie and others 2016; Pokatayev and others 2016).
Furthermore, it has been demonstrated that Samhd1 deficiency induces cGAS–STING-dependent autoimmunity (Rice and others 2009; Maelfait and others 2016). Certain mutations in the Samhd1 gene induce the generation of host retroelements such as long interspersed elements 1, which can cause AGS (Zhao and others 2013) (Fig. 2b). Moreover, in mice that lack lysosomal DNase II, inefficiently digested self-DNA leaks from the endosome/lysosome into the cytosol where it activates cGAS and STING, resulting in an abnormal production of multiple cytokines, including type I IFNs and tumor necrosis factor α (Ahn and others 2012). Notably, to date, no mutations have been identified in the gene that encodes lysosomal DNase II in humans.
In summary, these studies established that loss-of-function mutations in genes encoding enzymes that play crucial roles in DNA metabolism result in the accumulation of certain host dsDNA species, or RNA–DNA hybrids, which ultimately induces inappropriate innate immune signaling via the cGAS–STING axis.
Concluding Remarks
Over the past decades, ample progress has been made toward defining the molecular features that allow innate immune sensors to discriminate between cellular and pathogen-derived RNA or DNA. Whereas in most cases cellular nucleic acid recognition leads to proinflammatory or autoimmune diseases and thus has deleterious effects on the host organism, intriguing new research indicates that sensing of cellular RNA or DNA can also mount an antiviral response. In this case, recognition of “self” represents an indirect mechanism of pathogen detection wherein the mammalian host senses viral manipulation of host RNA/DNA metabolism, or virus-induced changes in the subcellular localization of host nucleic acids.
Intriguingly, the recognition of host RNA or DNA recently has also been found to play an important role in cancer. For example, in breast cancer cells, RIG-I recognizes unmasked noncoding RNA RN7SL1 (a Pol III transcript), which is released by stromal cells via exosomes and induces robust cytokine production, resulting in therapy resistance and cancer progression (Nabet and others 2017). Similarly, cellular RNA species, including endogenous retrovirus transcripts that are upregulated by DNA methyltransferase inhibitor treatment, an effective cancer therapy, are detected by MDA5 and cause cytokine responses in ovarian cancer and melanoma (Chiappinelli and others 2015). Moreover, recent studies have shown that DNA damage and senescence induced by ionizing irradiation lead to cytoplasmic chromatin fragments, which can activate the cGAS–STING pathway (Deng and others 2014; Dou and others 2017).
Many questions remain in this new and rapidly evolving field of “self” nucleic acid sensing. Just as viral ligands for PRRs have been extensively characterized, the molecular features of host RNAs and DNAs that trigger RLR or cGAS activation need to be elucidated. Furthermore, it will be important to understand the precise subcellular localization of host-derived PRR ligands before and after viral infection. New technologies such as intracellular RNA fluorescent in situ hybridization of endogenous ligands combined with live-cell imaging could be utilized to understand in precise detail when and where these host RNAs are bound to RLRs. Furthermore, for host RNAs that are transcriptionally induced by viral infection or other stimuli, single-cell transcriptomics could be applied to understand the kinetics of their production as well as their expression in infected versus bystander cells. Most importantly, while several studies have shown that certain host RNAs/DNAs can bind to intracellular sensors during infection or other pathological conditions, in many cases the physiological relevance and contribution of these endogenous ligands to the activation of innate immunity and/or pathogenesis still need to be established. Moreover, in light of recent findings that showed that RIG-I can bind to the 3′-UTR of certain mRNAs, such as the mRNA that encodes NF-κB (Zhang and others 2013), the full range of cellular RNA species that can bind (and potentially activate) RLRs remains to be determined. As of yet, self-RNAs recognized by other intracellular PRRs, such as LGP2, have yet to be described, which represents an exciting new avenue for future research. A sound understanding of the molecular mechanisms by which intracellular sensors recognize endogenous nucleic acid may lead to novel therapeutic approaches for infectious diseases, autoimmune disorders, and cancer.
Acknowledgments
Current research in Gack's laboratory is supported by the National Institutes of Health grants R01 AI087846, R01 AI127774, and R21 AI118509.
Author Disclosure Statement
No competing financial interests exist.
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol 10(10):1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, Fredericks AC, Tripathi S, Zhu T, Pintado-Silva J, Webb LG, Bernal-Rubio D, Solovyov A, Greenbaum B, Simon V, Basler CF, Mulder LC, Garcia-Sastre A, Fernandez-Sesma A. 2017. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2:17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang CZ, Hur S. 2018. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172(4):797 e13–810 e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Gutman D, Saijo S, Barber GN. 2012. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A 109(47):19386–19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. 2014. STING-dependent cytosolic DNA sensing pathways. Trends Immunol 35(2):88–93 [DOI] [PubMed] [Google Scholar]
- Barrat FJ, Elkon KB, Fitzgerald KA. 2016. Importance of nucleic acid recognition in inflammation and autoimmunity. Annu Rev Med 67:323–336 [DOI] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A 107(37):16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui SM, Paz S, Hiscott J. 2011. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol 23(5):564–572 [DOI] [PubMed] [Google Scholar]
- Cai X, Chiu YH, Chen ZJ. 2014. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell 54(2):289–296 [DOI] [PubMed] [Google Scholar]
- Chan YK, Gack MU. 2016. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol 14(6):360–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun L, Chen ZJ. 2016. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 17(10):1142–1149 [DOI] [PubMed] [Google Scholar]
- Chiang C, Gack MU. 2017. Post-translational control of intracellular pathogen sensing pathways. Trends Immunol 38(1):39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JJ, Sparrer KMJ, van Gent M, Lassig C, Huang T, Osterrieder N, Hopfner KP, Gack MU. 2018. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat Immunol 19(1):53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Budhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Merghoub T, Chan TA, Baylin SB, Strick R. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162(5):974–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138(3):576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR, Rice CM. 2018. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172(4):811 e14–824 e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T. 2006a. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet 38(8):917–920 [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. 2006b. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet 38(8):910–916 [DOI] [PubMed] [Google Scholar]
- Crowl JT, Gray EE, Pestal K, Volkman HE, Stetson DB. 2017. Intracellular nucleic acid detection in autoimmunity. Annu Rev Immunol 35:313–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambuza IM, Brown GD. 2015. C-type lectins in immunity: recent developments. Curr Opin Immunol 32:21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, Huang X, Gajewski TF, Chen ZJ, Fu YX, Weichselbaum RR. 2014. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41(5):843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A, Crow YJ, Rice GI, Duffy D, Tamby C, Nojima T, Munnich A, Schiff M, de Almeida CR, Rehwinkel J, Dziembowski A, Szczesny RJ, Proudfoot NJ. 2018. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560(7717):238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, Simithy J, Lan Y, Lin Y, Zhou Z, Capell BC, Xu C, Xu M, Kieckhaefer JE, Jiang T, Shoshkes-Carmel M, Tanim K, Barber GN, Seykora JT, Millar SE, Kaestner KH, Garcia BA, Adams PD, Berger SL. 2017. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550(7676):402–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckard SC, Rice GI, Fabre A, Badens C, Gray EE, Hartley JL, Crow YJ, Stetson DB. 2014. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol 15(9):839–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kubler K, Wittmann S, Gramberg T, Andreeva L, Hopfner KP, Mertens C, Zillinger T, Jin T, Xiao TS, Bartok E, Coch C, Ackermann D, Hornung V, Ludwig J, Barchet W, Hartmann G, Schlee M. 2015. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol 16(10):1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA, Jr., Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216 [DOI] [PubMed] [Google Scholar]
- Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, Wang W, Liu S, Liu W, Ma Y, Zhang L, Cao X. 2018. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173(4):906 e13–919 e13 [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441(7089):101–105 [DOI] [PubMed] [Google Scholar]
- Li T, Chen ZJ. 2018. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 215(5):1287–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349(6252):1115–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 82(1):335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Lettice L, Tarnauskaite Z, Reddy K, Dix F, Revuelta A, Abbondati E, Rigby RE, Rabe B, Kilanowski F, Grimes G, Fluteau A, Devenney PS, Hill RE, Reijns MA, Jackson AP. 2016. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J 35(8):831–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelfait J, Bridgeman A, Benlahrech A, Cursi C, Rehwinkel J. 2016. Restriction by SAMHD1 limits cGAS/STING-dependent innate and adaptive immune responses to HIV-1. Cell Rep 16(6):1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M, Jr., Silverman RH. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448(7155):816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellaker C, Vesely C, Ponting CP, McLaughlin PJ, Jantsch MF, Dorin J, Adams IR, Scadden AD, Ohman M, Keegan LP, O'Connell MA. 2014. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 9(4):1482–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388(6640):394–397 [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J, Karin M. 2006. Intracellular pattern recognition receptors in the host response. Nature 442(7098):39–44 [DOI] [PubMed] [Google Scholar]
- Mu X, Ahmad S, Hur S. 2016. Endogenous retroelements and the host innate immune sensors. Adv Immunol 132:47–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, Benci JL, DeMichele AM, Tchou J, Marcotrigiano J, Minn AJ. 2017. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell 170(2):352 e13–366 e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhaei P, Genin P, Civas A, Hiscott J. 2009. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol 21(4):215–222 [DOI] [PubMed] [Google Scholar]
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. 2015. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ sevelopment. Immunity 43(5):933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokatayev V, Hasin N, Chon H, Cerritelli SM, Sakhuja K, Ward JM, Morris HD, Yan N, Crouch RJ. 2016. RNase H2 catalytic core Aicardi-Goutieres syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. J Exp Med 213(3):329–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140(3):397–408 [DOI] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. 2009. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 41(7):829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, Casarano M, Chouchane M, Cimaz R, Collins AE, Cordeiro NJ, Dale RC, Davidson JE, De Waele L, Desguerre I, Faivre L, Fazzi E, Isidor B, Lagae L, Latchman AR, Lebon P, Li C, Livingston JH, Lourenco CM, Mancardi MM, Masurel-Paulet A, McInnes IB, Menezes MP, Mignot C, O'Sullivan J, Orcesi S, Picco PP, Riva E, Robinson RA, Rodriguez D, Salvatici E, Scott C, Szybowska M, Tolmie JL, Vanderver A, Vanhulle C, Vieira JP, Webb K, Whitney RN, Williams SG, Wolfe LA, Zuberi SM, Hur S, Crow YJ. 2014. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet 46(5):503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanelli M, De Laet C, de Lonlay P, del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla A, Heiberg A, Kawaguchi M, Kumar R, Lin JP, Lourenco CM, Male AM, Marques W, Jr., Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe WG, Vanderver A, Vassallo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, McLaughlin PJ, Keegan LP, O'Connell MA, Lovell SC, Crow YJ. 2012. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet 44(11):1243–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, Taniguchi T, Shadel GS, Chen ZJ, Iwasaki A, Flavell RA. 2014. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159(7):1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsch F, MacDougall M, Lu C, Buers I, Mamaeva O, Nitschke Y, Rice GI, Erlandsen H, Kehl HG, Thiele H, Nurnberg P, Hohne W, Crow YJ, Feigenbaum A, Hennekam RC. 2015. A specific IFIH1 gain-of-function mutation causes Singleton-Merten syndrome. Am J Hum Genet 96(2):275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454(7203):523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M. 2013. Master sensors of pathogenic RNA–RIG-I like receptors. Immunobiology 218(11):1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Hartmann G. 2016. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol 16(9):566–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrer KM, Gack MU. 2015. Intracellular detection of viral nucleic acids. Curr Opin Microbiol 26:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134(4):587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140(6):805–820 [DOI] [PubMed] [Google Scholar]
- van Tol S, Hage A, Giraldo MI, Bharaj P, Rajsbaum R. 2017. The TRIMendous role of TRIMs in virus-host interactions. Vaccines (Basel) 5(3):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. 2015. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520(7548):553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, Huang DC, Kile BT. 2014. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159(7):1549–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE. 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 131(5):873–886 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Liu ZX, Sun YP, Zhu J, Lu SY, Liu XS, Huang QH, Xie YY, Zhu HB, Dang SY, Chen HF, Zheng GY, Li YX, Kuang Y, Fei J, Chen SJ, Chen Z, Wang ZG. 2013. Rig-I regulates NF-kappaB activity through binding to Nf-kappab1 3′-UTR mRNA. Proc Natl Acad Sci U S A 110(16):6459–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, Cheung LE, Wang G, Kazazian HH, Jr., Yu XF. 2013. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep 4(6):1108–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Zhu J, Zhou H, Zhao Z, Zou Z, Liu X, Lin X, Zhang X, Deng X, Wang R, Chen H, Jin M. 2015. Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Sci Rep 5:14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ye X, Dunker W, Song Y, Karijolich J. 2018. RIG-I like receptor sensing of host RNAs facilitates the cell-intrinsic immune response to KSHV infection. Nat Commun 9(1):4841. [DOI] [PMC free article] [PubMed] [Google Scholar]