Abstract
Background
Cetaceans exhibit an exceptionally wide range of body size, yet in this regard, their genetic basis remains poorly explored. In this study, 20 body-size-related genes for which duplication, mutation, or deficiency can cause body size change in mammals were chosen to preliminarily investigate the evolutionary mechanisms underlying the dramatic body size variation in cetaceans.
Results
We successfully sequenced 20 body-size-related genes in six representative species of cetaceans. A total of 46 codons from 10 genes were detected and determined to be under strong positive selection, 32 (69.6%) of which were further found to be under radical physiochemical changes; moreover, some of these sites were localized in or near important functional regions. Interestingly, positively selected genes were well matched with body size evolution: for small cetaceans, strong evidence of positive selection was detected at ACAN, OBSL1, and GRB10, within which mutations or duplications could cause short stature; positive selection was found in large cetaceans at CBS and EIF2AK3, which could promote growth, and at the PLOD1 gene, within which mutations could cause tall stature. Importantly, relationship analyses revealed that the evolutionary rate of CBS was positively related to body length and body mass with statistical significance. Additionally, we identified 32 cetacean-specific amino acid changes in 10 genes.
Conclusions
This is the first study to investigate the molecular basis of dramatic body size variation in cetaceans. Our results provide evidence of the positive selection of several body-size-related genes in cetaceans, as well as divergent selection between large or small cetaceans, which suggest cetacean body size variation possibly associated with these genes. In addition, cetacean-specific amino acid changes might have played key roles in body size evolution after the divergence of cetaceans from their terrestrial relatives. Overall, the evolutionary pattern of these body-size-related genes could provide new insights into genetic mechanisms for the body size variation in cetaceans.
Electronic supplementary material
The online version of this article (10.1186/s12862-019-1461-9) contains supplementary material, which is available to authorized users.
Keywords: Cetacean, Body size, Body-size-related genes, Positive selection
Background
Fossils have revealed that cetaceans (whales, dolphins, and porpoises) originated 56–53 Ma from terrestrial artiodactyl ancestors [1]. Extant cetaceans can be subdivided into two suborders (Odontoceti and Mysticeti) within a wide range of marine habitats from coastal to oceanic and from tropical to polar [2]. As a result of lifestyle changes, cetaceans presented many extreme physiological and morphological adaptations among mammals, such as the loss of hindlegs, forelimbs that changed into flippers, lack of hair coats, thick blubber, feeding transition from herbivorous to carnivorous, and the loss of nearly all taste receptors, as well as the development of underwater sensory systems [3–6]. Benefiting from advances in cetacean whole genome sequencing projects, the molecular mechanisms underlying those major evolutionary changes, e.g., thickening of blubber, loss of hair, feeding habit transitions, brain size enlargements, and hypoxia tolerance [7–12], are beginning to be understood.
One of the most remarkable external changes in cetacean evolution is the dramatically wide range of body size, from less than 2 m in length and less than 50 kg in weight for porpoises to over 30 m and more than 150 tons for blue whales (Balaenoptera musculus), with more than a 20-fold increase in body length and a 3500-fold increase in body mass. Cetacean body size seems to have increased over evolutionary time, which follows Cope’s rule [13]. Moreover, the evolutionary rate of body mass in cetaceans exceeds that of terrestrial mammals perhaps due to “aquatic weightlessness”, migratory behaviour, and selection related to thermoregulation and feeding ecology in an aquatic environment [14]. Among these factors, temperature was reported to affect the body size of cetaceans, in agreement with Bergmann’s rule that animals living in colder climates are generally larger than those living in warmer regions [15]. For example, the largest living animal, the blue whale, frequents Arctic and Antarctic waters in the respective summer seasons and moves to the warmer subtropics and tropics during winters. By contrast, dolphins and porpoises, generally much smaller than whales, e.g., vaquita Phocoena sinus, the smallest cetacean, is only 1.4 m in body length and 42 kg in body mass and usually lives close to shore in shallow water. Large body size has been demonstrated to have many advantages, such as enhancing predation success, suitability for a more generalist diet, and increasing tolerance to extreme environments, but it also has some weaknesses such as being more susceptible to extinction and having lower fecundity [16]. However, the genetic bases of dramatic body size variation in cetaceans remain poorly explored.
Body size is a typical quantitative or complex trait that shows continuous variation [17]. Previous studies have reported that many discrete genes are involved in individual development, genetic diseases, or body size regulation. It has been reported that some genes involved in promoting growth or mutations in genes could cause tall stature (e.g., gigantism) and overgrowth, such as aryl hydrocarbon receptor interacting protein (AIP), cystathionine β - synthase (CBS), Natriuretic peptide receptor 2 (NPR2), nuclear receptor binding SET domain protein 1 (NSD1), lysyl hydroxylase 1 (PLOD1, also LH1), pleomorphic adenomagene 1 (PLAG1), translation initiation factor2-α kinase 3 (EIF2AK3), G-protein-coupled receptor 101 (GPR101), N-acetylgalactosamine-6-sulfate-sulfatase (GALNS), multiple endocrine neoplasia type I (MEN1), and mediator complex subunit 12 (MED12), which have been called “tall stature-related genes”. In contrast, some genes involved in inhibiting growth or mutations in genes result in short stature (e.g., dwarfism), including aggrecan (ACAN), obscurin-like 1 (OBSL1), growth factor receptor-bound protein 10 (GRB10), pituitary specific transcription factor 1 (PIT-1), Kir inward rectifier potassium channels (KCNJ2), Noggin (NOG), cyclin-dependent kinase inhibitor 1B (CDKN1B), and glypican-3 (GPC3), which have been called “short stature-related genes”. In addition, some genes, such as Fibrillin-1 (FBN1), have been associated with both overgrowth and dwarfism, depending on the kinds of changes that occurred on them; mutations in this gene have been described in Marfan syndrome, which is characterized by tall stature and arachnodactyly, whereas the TB5 mutations in this gene were responsible for short stature phenotypes. These genes, due to their association with body size, are termed body-size-related genes in the present study, and detailed function information is listed in (see Additional file 1: Table S1).
In the present study, coding sequences of the above 20 candidate genes were examined in cetaceans of both large and small body size to explore the evolutionary patterns and their association with the morphological variables of body size. Our results provide evidence of the positive selection and cetacean-specific amino acid changes of body-size-related genes in cetaceans, as well as divergent selection between large or small cetaceans, which suggested body size variation in cetaceans possibly associated with these genes.
Results
Positive selection of body-size-related genes in cetaceans
We successfully sequenced 20 body-size-related genes in six representative species of cetaceans: Omura’s baleen whales Balaenoptera omurai, striped dolphins Stenella coeruleoalba, pantropical spotted dolphins S. attenuata, common dolphins Delphinus delphis, Risso’s dolphins Grampus griseus, and dwarf sperm whales Kogia simus. Newly sequenced genes (GenBank accession nos. MH729659-MH729778) covered at least 80% of the full CDS. In addition, the orthologous genes of the 20 body-size-related genes were downloaded from another 10 cetacean species from their published database (see Additional file 1: Table S2). Preliminary alignment of gene sequences showed no frame shift mutations or premature stop codons. The one-ratio model, which assumes that one ratio occurs across the phylogenetic tree, showed that the ω ratios of 20 body-size-related genes ranged from 0.009 to 0.332 (see Additional file 1: Table S3), which indicates that strong purifying selection acts on these genes to constrain their important functions in body size development. A pair of site models (M8 VS M8a), implemented in the CODEML program of PAML 4.7 [18], was further used to test positively selected sites in the cetacean-only dataset. The likelihood ratio test (LRT) showed that M8 fitted the data better than M8a at 10 genes (i.e., ACAN, AIP, CDKN1B, EIF2AK3, FBN1, MED12, MEN1, NPR2, NSD1, and OBSL1), with 65 codons identified to be under positive selection using the BEB approach with posterior probabilities ≥0.85 (Table 1). Significant evidence of positive selection was further corroborated by the other two ML methods (FEL and REL) implemented in Datamonkey. Sixteen codons from 7 genes (ACAN, AIP, CDKN1B, EIF2AK3, FBN1, MED12, and NPR2) and 61 codons from 8 genes (ACAN, AIP, EIF2AK3, FBN1, MED12, MEN1, NSD1, and OBSL1) were examined to be under positive selection by REL and FEL, respectively (Table 1). A total of 46 positively selected sites were thus identified in 10 genes (i.e., ACAN, AIP, CDKN1B, EIF2AK3, FBN1, MED12, MEN1, NPR2, NSD1, OBSL1) by at least two ML methods and were thus regarded as robust candidates for positively selected sites. Thirty-two (69.2%) of them were found to have radical amino acid changes detected using a complementary protein-level approach implemented in TreeSAAP (see Additional file 1: Table S4), which provided additional evidence for positive selection in cetaceans.
Table 1.
Genes and sites inferred to be under positive selection using three ML methods
| Genes | PAML | Datamonkey | ||||||
|---|---|---|---|---|---|---|---|---|
| -Ln(M8a) | -Ln(M8) | -2△Lnl (p-value) | ω value | Positively selected sites (P ≥ 85%)a | FEL (p < 0.1)b | REL (p > 50)c | % of Sites d | |
| ACAN | (16 sequences) | |||||||
| (2022aa) | 13,517.793 | 13,357.328 | 320.930 (<0.001) | 11.434 | 9, 360, 703, 806, 975, 980, 985, 992, 1002, 1068, 1100, 1185, 1270, 1411, 1532, 1626, 1710, 1734, 1944 | 9, 146, 360, 407, 703, 980, 985 | 9, 146, 219, 360, 396, 407, 686, 703, 975, 980, 985, 1016, 1100, 1567, 1734, 1750, 1944 | 0.544(11) |
| AIP | (14 sequences) | |||||||
| (330aa) | 1859.035 | 1855.058 | 7.953 (<0.005) | 4.488 | 43, 45, 78, 86, 131, 218 | 43, 131 | 43, 45, 56, 78, 131, 301 | 1.212(4) |
| CDKN1B | (16 sequences) | |||||||
| (198aa) | 1015.965 | 1013.544 | 4.841 | 21.427 | 162 | 162 | 0.505(1) | |
| EIF2AK3 | (16 sequences) | |||||||
| (964aa) | 5985.966 | 5976.756 | 18.42 (<0.001) | 3.978 | 154, 358, 558, 628, 677, 897 | 154 | 154, 677, 795, 897 | 0.311(3) |
| FBN1 | (15 sequences) | |||||||
| (2871aa) | 15,888.036 | 15,872.839 | 30.393 (<0.001) | 3.387 | 300, 2443, 2696, 2699, 2701, 2737, 2741 | 1946, 2701, 2741 | 120, 1175, 1946, 2336, 2443, 2696, 2699, 2701, 2737, 2741, 2775 | 0.244(7) |
| MED12 | (16 sequences) | |||||||
| (2050aa) | 9844.493 | 9840.242 | 8.502 (<0.005) | 12.298 | 312, 1621 | 1621 | 1621 | 0.049(1) |
| MEN1 | (16 sequences) | |||||||
| (610aa) | 3657.161 | 3628.394 | 57.534 (<0.001) | 7.403 | 526, 536, 542, 594 | 394, 459, 475, 526, 542 | 0.328(2) | |
| NPR2 | (16 sequences) | |||||||
| (1047aa) | 5716.770 | 5687.387 | 58.825 (<0.001) | 14.294 | 325 | 325 | 0.096(1) | |
| NSD1 | (16 sequences) | |||||||
| (2696aa) | 13,297.254 | 13,276.127 | 42.253 (<0.001) | 4.399 | 111, 652, 964, 978, 1144, 1493, 1518, 1853, 2236, 2264, 2335, 2411 | 111, 652, 964, 978, 1493, 1518, 1853, 2236, 2264, 2335, 2411 | 0.408(11) | |
| OBSL1 | (16 sequences) | |||||||
| (1729aa) | 10,250.581 | 10,242.647 | 15.868 (<0.001) | 4.348 | 638, 1242, 1395, 1423, 1428, 1564, 1617 | 592, 638, 1242, 1395, 1428, 1617 | 0.289(5) | |
Note: The codons identified to be under positive selection by at least two or three ML methods are marked in bold and "bold and Italic", respectively.
aCodons identified by M8 model in PAML using a Bayes Empirical Bayes (BEB) analysis with posterior probabilities ≥85%
bCodons detected by FEL implemented in Datamonkey web server with significance levels of 0.1
cCodons determined by REL in Datamonkey web server with Bayes factors >50
dNo. of sites indicate positively selected sites identified by both ML method
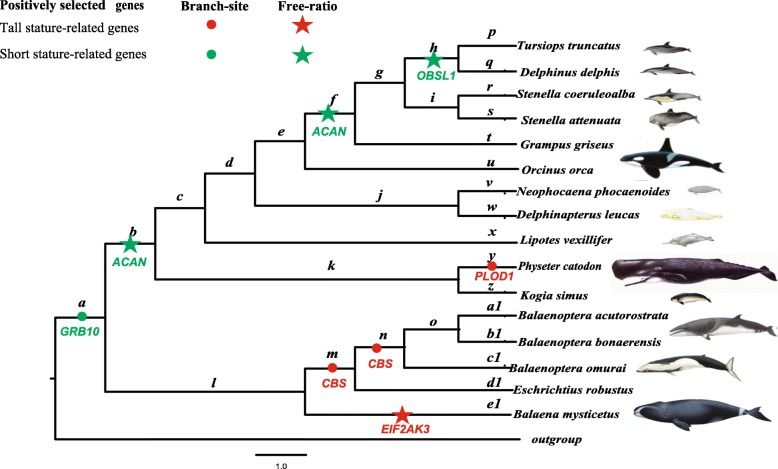
To test whether positive selection was restricted to some specific lineages, the free-ratio and branch-site models were used in our analysis. LRT revealed that the free-ratio model fitted the data better than the one-ratio model at 11 genes (i.e., ACAN, AIP, EIF2AK3, FBN1, GALNS, GRB10, MED12, MEN1, OBSL1, PLAG1, and PLOD1) (Additional file 1: Table S3). Specifically, evidence of positive selection was determined along the lineage to the last common ancestor (LCA) of Odontoceti and LCA of G. griseus at ACAN, the LCA of T. truncatus and D. delphis at OBSL1, as well as the terminal branch of B. mysticetus at EIF2AK3 (Fig. 1). Moreover, the more stringent branch-site model revealed that positive selection was identified along the LCA of Balaenopteridae and LCA of E. robustus at CBS, the LCA of cetaceans at GRB10, as well as the lineage leading to Physeter catodon at PLOD1 after FDR correction for multiple tests. In addition, four codons were identified to be under positive selection by the BEB procedure (Fig. 1).
Fig. 1.
Evidence of positive selection across the phylogeny of cetaceans identified by the free-ratio and branch-site models. Positive selection across the cetacean phylogeny is marked with different colours: tall stature-related genes (red), short stature-related genes (green)
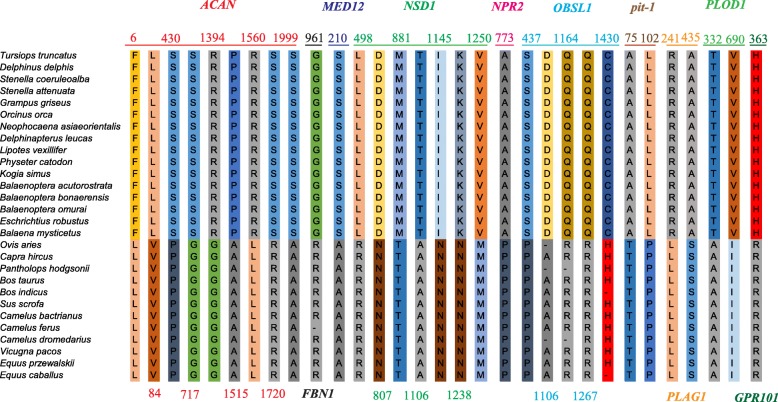
We classified representative cetaceans into small cetaceans and large cetaceans to explore whether cetaceans with divergent body size evolved under different evolutionary pressure. The ω values estimated for small cetaceans were almost twice as high as those in the large cetaceans at four genes, i.e., FBN1, GRB10, NPR2 and NSD1 (see Additional file 1: Table S5). Additionally, compared with terrestrial mammals, 32 cetacean-specific amino acid changes were identified at 10 genes (ACAN, FBN1, GPR101, MED12, NPR2, NSD1, OBSL1, PIT-1, PLAG1, PLOD1) (Fig. 2).
Fig. 2.
Special sites of cetaceans when compared with related terrestrial mammals
Spatial distribution of positively selected sites and cetacean-specific sites in protein structures
To gain insight into the functional significance of the putative positively selected sites and cetacean-specific sites, a total of 46 radical amino acid changes subjected to positive selection and 32 cetacean-specific sites were mapped onto the 3D structures of the corresponding proteins. It was observed that many positively selected sites and cetacean-specific sites were located in or close to the functional regions (see Additional file 2: Figure S1). For example, one positively selected site of AIP (site 78) was located in N-terminus immunophilin-like domains, and missense mutation of several sites in this domain were reported to cause gigantism (e.g., V49 M, R56C, K58 N, E84K). In addition, radical change of amino acids was detected at residue 162 by two ML methods in the CDKN1B gene. 3D analysis showed that the site was located in the nuclear localization signal, which may contribute to its recognition with cytosolic nuclear transport receptors and that radical change in site 162 may help the protein in cell nuclei be more efficient and have the function of negative regulation of cell growth. Several radical changes subject to positive selection of ACAN, such as 975, 980, 985, were located in the chondroitin sulfate-rich domain (CS1), which is composed of repeats of nineteen amino acids and provides the aggrecan with its high anionic charge. For cetacean-specific sites, site 435 of PLAG1 was located in the C-terminal serine-rich transactivation domain, which possesses a phosphorylation site and raises the possibility that phosphorylation may regulate the transactivation capacity of PLAG1. Another gene, OBSL1, also showed some cetacean-specific sites, among them, site 437 located in fibronectin-like (Fn3) domains, sites 1106 and 1164 located in immunoglobulin (Ig-like 11) and site 1267 located in the Ig-like 12 domain.
Association between gene evolution and morphological variables
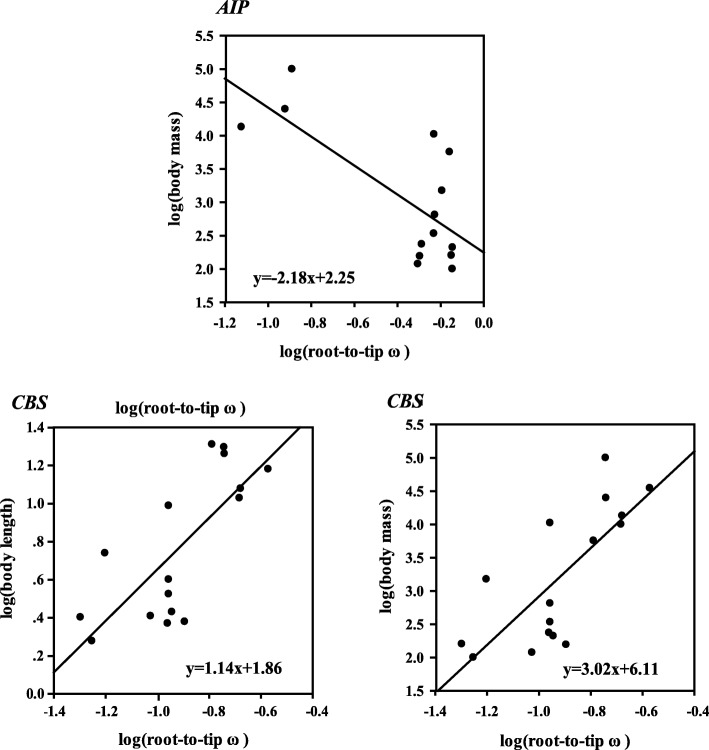
It has been regarded that tests for statistical association between genes and normal variation in phenotypes is a good strategy for examining the phenotypic consequences of a signature of ongoing selection [19]. We performed phylogenetic generalized least squares (PGLS) regressions to explicitly address the link between evolutionary rate of each gene under positive selection and body length/body mass. Regression analyses revealed significant positive association between log (root-to-tip ω) and log (body length) at the CBS (R2 = 0.551, P = 0.014) and between log (root-to-tip ω) and log (body mass) at the CBS (R2 = 0.561, P = 0.017). However, a significant negative association between log (root-to-tip ω) and log (body mass) was tested at the AIP gene (R2 = 0.522, P = 0.004, Fig. 3), whereas no such association was detected for other genes under positive selection (see Additional file 1: Table S6).
Fig. 3.
Regression analyses between root-to-tip ω and morphological variables (body length and body mass)
Discussion
As is well known, modern cetaceans show an exceptionally wide range of body size, including the largest animal ever to live on the earth, the blue whale, and some small dolphins and porpoises, with a 20-fold body size difference and a 3500-fold mass difference. Previous studies have revealed clear evidence that the ancestor of odontocetes and mysticetes had a body size similar to that of the adult living bottlenose dolphin [20]. During the adaptive radiation, body size increased in mysticetes, whereas odontocetes exhibited a general decrease in body size (except for the sperm whale with increased body size), which was regarded to be related to their special dietary strategy, diving depth, temperature, and other factors [20, 21]. It is widely accepted that body size variation during cetacean evolution is the product of rapid divergence into new adaptive zones. Our study showed clear evidence that different evolutionary patterns of body-size-related genes, investigated in cetacean clades with contrasting body size, might contribute to an understanding of body size evolution at the molecular level.
Positive selection in cetacean clades with contrasting body size
A total of 46 codons from 10 body-size-related genes were detected to be under positive selection by at least two ML methods, 32 (69.2%) of which were further found to be under radical physiochemical changes; some of these sites were localized in or near the important functional regions of proteins. Interestingly, positive selection of body-size-related genes identified in cetacean clades were well matched with their contrasting body size. Positive selection detected at ACAN, OBSL1 and GRB10, whose mutations or duplications could cause short stature in mice, could provide insights into body decrease along lineages of small cetaceans, whereas positive selection at CBS, PLOD1 and EIF2AK3 in large cetacean genes could be explained by their role in promoting growth and causing body increase and tall stature.
Strong evidence of positive selection was found to act on the ACAN gene in odontocetes, which has a relatively smaller body size. Three codons (975, 980, 985) especially under positive selection were located in the chondroitin sulfate-rich domain (CS1), which is composed of repeats of nineteen amino acids and provides the aggrecan with its high anionic charge. In addition, some sites were found to be under radical change in the amino acid property. For example, position 985 of ACAN was identified to have a change in the “alpha-helical tendencies” property that would lead to a more rigid alpha helical form, which would help provide a more stable lipid raft composition [22]. Therefore, we speculated that the amino change of this site might play a crucial effect for the structure and function of protein. Significant positive selection in ACAN was subsequently examined in the common ancestor branch of Odontoceti, which was characterized by decreases in body mass [14]. ACAN encodes aggrecan, a proteoglycan in the extracellular matrix of the growth plate and other cartilaginous tissues, which is essential for cartilage structure. ACAN gene mutation could cause bulldog dwarfism in Dexter cattle due to the absence of normal aggrecan [23]. For humans, a single base pair insertion in ACAN resulted in spondyloepiphyseal dysplasia type Kimberly, characterized by shortened limbs and trunk [24]. Taken together, positive selection identified in cetaceans at the ACAN gene suggested this gene were possibly associated with restricting the body size increase during the evolution of small cetaceans.
Similar results were also found in the GRB10, a growth suppressor. It was found that the evolutionary rate of GRB10 for small cetaceans was almost twice as high as that of large whales. More importantly, branch-site analysis revealed significant positive selection in the ancestor of cetaceans that was estimated to evolutionarily decrease body mass [14]. Previous experiments confirmed that overexpression of GRB10 in human or transgenic mice could cause postnatal growth restriction [25] and that loss of its function in mice resulted in foetal and placental overgrowth [26]. The higher evolutionary rate of GRB10 in small cetaceans and positive selection in the lineages with decreased body mass would thus help to restrict body size growth. OBSL1 is a cytoskeletal adaptor protein, and mutation of this gene was confirmed to cause the primordial growth disorder 3-M syndrome in humans [27]. Similarly, positive selection acting on the OBSL1 was identified in the last common ancestor of both small dolphins, i.e., the bottle-nosed dolphin and the common dolphin, which implies that the evidence of positive selection in this gene might play roles in restricting the body size increase of small cetaceans.
In contrast, significant positive selection was identified in the large whales at the CBS, PLOD1, and EIF2AK3 genes that might promote body increase. Positive selection acting on the CBS gene was detected in both branch m, with large body length up to 15 m and body mass up to 35,000 kg, and branch n, with large body length up to 12 m and body mass up to 135,000 kg (Fig. 1). Moreover, our association analyses showed that the CBS evolutionary rate was significantly positively related to body length and body mass. This result is consistent with the fact that CBS-deficient (Cbs−/−) mice displayed severe growth retardation [28]. Interestingly, evidence of positive selection was also determined in the lineage leading to the largest toothed whale, i.e., the sperm whale (20.5 m in length and 57,000 kg in mass) in the PLOD1 gene, which has been previously shown to cause the Nevo syndrome, clinically characterized by increased growth, kyphosis, a prominent forehead, and other factors [29]. Previous studies showed that loss-of-function mutations in EIF2AK3 resulted in Wolcott-Rallison Syndrome in humans and that EIF2AK3-deficient (Perk−/−) mice also exhibited skeletal dysplasias at birth and postnatal growth retardation [30, 31]. Notably, significant positive selection in the EIF2AK3 gene was examined in the lineages leading to the bowhead whale (19.8 m in length and 100,000 kg in mass), which suggests that this gene may contribute to the tremendous body size of the bowhead whale. Notably, the function of these body-size-related genes used in our study are forecasted according to mutations from human diseases and experiment of knock-out mice. Considering these genes are pleiotropic, we should do functional experiment in future to test whether these positive selected sites paly a key role in cetacean body size variation.
Cetacean-specific amino acid change provides evidence of adaptive evolution
Compared with terrestrial mammals, 32 cetacean-specific amino acid changes in 10 genes were identified; 26 of them (81.2%) were identified to be under radical changes, and some of them were also located in or close to important functional regions. For example, one cetacean-specific amino acid site (773) identified to be under radical change at the NPR2 gene was located in the kinase homology domain of the NPR2. It was reported that the NPR2 gene positively regulates longitudinal bone growth. Loss-of-function mutations of NPR2 cause short stature, and, conversely, gain-of-function mutations are associated with an overgrowth disorder [32]. For ACAN, nine cetacean-specific amino acid changes were found and six of them were identified to be under radical changes. Among them, site 717 was located in the KS domain, which could enhance the load-bearing capability of aggrecan in cartilage and may contribute to tissue development. Sites 1394, 1426, 1515, 1560 and 1720 were located in the CS2 domain, and negatively charged chondroitin sulfate chains in this domain account for the major function of aggrecan as a structural proteoglycan. Note that seven positively selected sites (i.e., 1100, 1411, 1532, 1626, 1710, 1734, 1944) were also located in this CS2 domain. In another example, one cetacean-specific acid site (75) was identified in the PIT-1 gene, which is a transcription factor responsible for anterior pituitary development and pituitary-specific gene expression [33]. Mutations in this gene were first observed in Snell and Jackson dwarf mice because of pituitary hormone deficiency (CPHD). Previous studies have shown that mutations in codon 75 (T → A) could increase its interaction with the LHX3 gene, which encodes LIM homeodomain class transcription factors that have important roles in pituitary and nervous system development. Interestingly, an A (alanine) is present in all cetaceans, whereas a T (threonine) is present in other terrestrial mammals. Notably, change in site 76 (P → L) results in CPHD1, which reduces transactivation capacity on the GH1 gene, increases the functional binding on the GH1 promoter, and increases the interaction with ELK1, LHX3 and PITX1. Therefore, our results imply that these special sites might play key roles in body size evolution after the divergence of cetaceans from their terrestrial relatives. Of course, further investigation of the site-directed mutagenesis of these important sites is necessary in the future to confirm their role in body size evolution.
Conclusions
Cetaceans show an exceptionally wide range of body size, which has been regarded to be related to their special dietary strategy, diving depth, temperature, and other factors. In this paper we present the first systematic investigation of 20 body size-related genes of representative cetacean lineages. Our results provide evidence of the positive selection of several body-size-related genes in cetaceans, as well as divergent selection between large and small cetaceans, which suggest their contribution to body size variation in cetaceans. Moreover, cetacean-specific amino acid changes might have played key roles in body size evolution after the divergence of cetaceans from their terrestrial relatives. Overall, the evolutionary pattern of these body-size-related genes could provide new insights into genetic mechanisms for body size variation in cetaceans. Importantly, some crucial codons detected in this study, including positively selected sites and cetacean-specific sites, provide a basis for function tests in the future.
Methods
Body-size-related genes acquisition
The coding sequences of 20 body-size-related genes were first screened and downloaded from the genomes of ten cetacean species: bottlenose dolphins T. truncatus, killer whales Orcinus orca, baiji Lipotes vexillifer, Yangtze finless porpoises Neophocaena asiaeorientalis, beluga whales Delphinapterus leucas, sperm whales Physeterm acrocephalus, bowhead whales Balaena mysticetus, minke whales B. acutorostata, Antarctic minke whales B. bonaerensis, and grey whales Eschrichtius robustus (see Additional file 1: Table S2). Low-quality and/or low-integrity gene sequences were further confirmed by searching from genome sequences of relevant cetaceans using bottlenose dolphin genes as queries to the BLAST (basic local alignment search tool) algorithm. Then, we amplified and sequenced these 20 genes in another six cetacean species: B. omurai, S. coeruleoalba, S. attenuata, common dolphins D. delphis, G. griseus, and K simus. The six cetacean samples were collected from dead individuals in the wild; no ethics statement is required for such occasions. Genomic DNA extraction, polymerase chain reaction (PCR) amplification, and sequencing were conducted as described in Xu et al. [11]. The PCR primers are listed in (see Additional file 3: Table S7).
The orthologous genes of each candidate gene were also downloaded from twelve terrestrial relatives: cows Bos taurus, zebu cattle Bos indicus, sheep Ovis aries, goats Capra hircus, Tibetan antelopes Pantholops hodgsonii, camels Camelus ferus, bactrian camels Camelus bactrianus, Arabian camels Camelus dromedarius, alpacas Vicugna pacos, pigs Sus scrofa, Przewalski’s horses Equus przewalskii, and horses Equus caballus (Additional file 1: Table S2). For genomic DNA, intron-exon boundaries were recognized from strict conserved splice signals (GT/AG), and the exons of each gene were concatenated according to the coding sequences of the known relative species. The nucleotide sequences and their deduced amino acid sequences of each gene were aligned using Muscle in MEGA6 [34] and verified by visual inspection (see Additional file 4).
Molecular evolutionary analysis
Comparisons of nonsynonymous (dN)/synonymous (dS) substitution ratios have become a useful means for quantifying the impact of natural selection on molecular evolution [35, 36]. Values of ω < 1, = 1, and > 1 correspond to purifying selection, neutral evolution, and positive selection, respectively. The codon-based maximum likelihood models implemented in the CODEML program of PAML 4.7 [18] were applied to estimate the ω values. To identify the probabilities of sites under positive selection in each gene, two pairs of site models: M8a (beta & ω2 = 1) versus M8 (beta & ω2 > 1), for which ω could vary among sites were implemented in the cetacean-only dataset. LRT statistic (2△L), which approximates a chi-square distribution, was used to compare nested likelihood models. Positively selected sites were identified using BEB analysis with posterior probabilities of ≥0. 85. Considering that ω values estimated by PAML models only allow for variation in the nonsynonymous substitution rate, and positively selected sites were further evaluated by fixed-effect likelihood (FEL) and random effect likelihood (REL) models implemented in the Datamonkey web server, which incorporated variation in the rate of synonymous substitution [37]. The REL approach allows variation in nonsynonymous and synonymous rates across sites according to a predefined distribution, and the FEL method directly estimates nonsynonymous and synonymous substitution rates at each site [38]. Therefore, sites with a significance level < 0.1 for FEL or a Bayes factor > 50 for REL were regarded as candidates for selection.
To test whether positively selected sites were limited to a specific lineage, the branch-site model and free-ratio model implemented in the CODEML program of PAML 4.7 were used in the all-mammals dataset. The branch-site model can detect positive selection at specific sites along a specific branch [39], whereas the free-ratio model allows an independent ω value for each branch [18]. A false discovery rate (FDR) correction for multiple tests was conducted in the branch-site model analysis [40]. BEB analysis was also used to test positively selected sites with posterior probabilities of ≥0.85 in the branch-site model. Three starting ω values (0.4, 1, and 2) were used to check for the existence of multiple local optima.
To determine if different selection pressures were acting on cetacean clades with contrasting body size, we divided sixteen cetacean species into large-bodied cetaceans (body length > 6 m), including bowhead whales, minke whales, Antarctic minke whales, Omura’s baleen whales, grey whales, sperm whales and killer whales, and small cetaceans (such as bottlenose dolphins, baiji and finless porpoises) according to the recommendation of Weber [41]. Branch models that allow different branches to have different ω, the so-called ‘two ratio’ and ‘three ratio’ models, were implemented in CodeML and were used to evaluate the ω values between the large and small cetacean groups. First, we set up foreground (particular lineages of interest) and background lineages (the remaining lineages). The one-ratio model, which enforces the same ω ratio for all lineages, was compared with the two-ratio model that allows one ω ratio for all branches of cetaceans and another for all terrestrial mammal branches. Moreover, we used the three-ratio model, which assumes that large cetaceans, small cetaceans and terrestrial mammals have independent ω values, to investigate whether different selective pressures were imposed on different types of cetaceans. All nested models were compared using LRTs.
To detect significant physicochemical amino acid changes in each gene, we used the algorithm implemented in the TreeSAAP 3.2 software package [42], which measures the selection based on 31 structural and biochemical amino acid property changes. According to the change in specific physicochemical properties, the magnitudes of non-synonymous changes are classified into eight categories from conservative (1–3) to very radical substitutions (6–8). A z-score was calculated for each category. Only significant positive z-scores in radical substitutions were considered to be affected by positive destabilizing selection.
Mapping of positively selected sites and cetacean-specific sites onto protein structures
To gain insights into the functional significance of the putative positively selected sites and cetacean-specific sites, we mapped these sites onto the crystal structures. The three-dimensional (3D) structures of each gene subject to positive selection and cetacean-specific sites were constructed by using I-TASSER server [43], a hierarchical protein structure modelling approach based on the secondary structure enhanced profile–profile threading alignment and the iterative implementation of the TASSER program. Additionally, the functional information of genes under positive selection was derived from UniProt (http://www.uniprot.org/).
Association analysis between gene evolution and phenotypes
To explore the potential relationships between the evolutionary rate (ω) of genes and body size phenotypes, we used the root-to-tip ω, which includes more evolutionary history of a locus as the evolutionary rate according to the method suggested by Montgomery et al. [44]. The branch models were used to estimate the average ω from the ancestral cetacean to each terminal species implemented in the CODEML program in PAML 4.7 [18]. The phenotypic traits, including body length and body mass of representative species of cetaceans, were collected from previously published data (see Additional file 5: Table S8). Phylogenetic generalized least squares (PGLS) regression was used to analyse the relationship between log-transformed (root-to-tip ω) and each log-transformed morphological variable. The lambda (λ) value estimated by the maximum likelihood method was used as a quantitative measure of phylogenetic signals [45]. All statistical analyses were performed using R 3.4.3 in the package Caper [46, 47].
Additional files
Table S1. Twenty body-size-related genes and their functions. Table S2. Sequence data used in this study, including taxonomy and accession numbers or Emsemble ID. Table S3. One-ratio model, Free-ratio model and Branch-site model analysis in 20 body-size-related genes. Table S4. Radical amino acid sites under positive selection detected by PAML, Datamonkey and TreeSAAP. Table S5. Log likelihood and omega values estimates under different branch models according contracting body length of cetaceans. Table S6. Association analysis between gene evolution and phenotypes. (DOCX 107 kb)
Figure S1. Radical amino acid changes in selected sites and cetacean-special sites mapped on the three-dimensional structure of body-size-related genes. Sites marked with red balls stand for robust sites under selection and yellow balls stand for cetacean-special sites. The figures were created using PyMOL (http://www.pymol.org). (PDF 2961 kb)
Table S7. Primer sets used to amplify the coding regions of body-size-related genes in this study (XLSX 25 kb)
Alignment sequences of 20 body-size related genes used for this study. These files are Fast format, please use MEGA or Clustal to open them. (ZIP 133 kb)
Table S8. Previously reported body size information for cetaceans. (DOCX 17 kb)
Acknowledgements
A special thank you is due to Mr. Xinrong Xu for collecting samples for many years. We also thank Dr. Ran Tian for her technical support.
Abbreviations
- 3D
three-dimensional
- ACAN
aggrecan
- AIP
aryl hydrocarbon receptor interacting protein
- BEB
Bayes Empirical Bayes
- BLAST
basic local alignment search tool
- CBS
cystathionine β – synthase
- CDKN1B
cyclin-dependent kinase inhibitor 1B
- dN
nonsynonymous substitution
- dS
synonymous substitution
- EIF2AK3
translation initiation factor2-α kinase 3
- FBN1
Fibrillin-1
- FDR
false discovery rate
- FEL
fixed effects likelihood
- GALNS
N-acetylgalactosamine-6-sulfate-sulfatase
- GPC3
glypican-3
- GPR101
G-protein-coupled receptor 101
- GRB10
growth factor receptor-bound protein 10
- KCNJ2
Kir inward rectifier potassium channels
- LRT
likelihood ratio test
- MED12
mediator complex subunit 12
- MEN1
multiple endocrine neoplasia type I
- NOG
Noggin
- NPR2
Natriuretic peptide receptor 2
- NSD1
nuclear receptor binding SET domain protein 1
- OBSL1
obscurin-like 1
- PCR
polymerase chain reaction
- PGLS
phylogenetic generalized least squares
- PIT-1
pituitary specific transcription factor 1
- PLAG1
pleomorphic adenomagene 1
- PLOD1 (also LH1)
lysyl hydroxylase 1 gene
- REL
random effect likelihood
Authors’ contributions
SX and GY conceived the project and designed the experiments. YS, YL1, XS, YL2, and DY performed the molecular evolution analysis. YL1 amplified these genes in representative species. YS and SX wrote the manuscript, and SX and GY improved the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Programme of Research and Development, Ministry of Science and Technology (Grant number 2016YFC0503200 to GY and SX); the Key Project of the National Natural Science Foundation of China (NSFC) (grant no. 31630071 to GY), the NSFC (grant nos. 31570379, 31772448 to SX); and the Priority Academic Program Development of Jiangsu Higher Education Institutions to GY and SX. These funding bodies played no role in study design, data collection, analysis, interpretation of data, and in writing the manuscript.
Availability of data and materials
All the data supporting our findings are contained within the manuscript and in the supplemental files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yingying Sun and Yanzhi Liu contributed equally to this work.
Contributor Information
Yingying Sun, Email: sunyingying718@163.com.
Yanzhi Liu, Email: 18260029208@163.com.
Xiaohui Sun, Email: sxh1991a8a9@163.com.
Yurui Lin, Email: linyurui0214@163.com.
Daiqing Yin, Email: 15928035490@163.com.
Shixia Xu, Email: xushixia78@163.com.
Guang Yang, Email: gyang@njnu.edu.cn.
References
- 1.Thewissen J, Cooper LN, George JC, Bajpai S. From land to water: the origin of whales, dolphins, and porpoises. Evol Educ Outreach. 2009;2:272–288. doi: 10.1007/s12052-009-0135-2. [DOI] [Google Scholar]
- 2.Zhou X, Xu S, Yang Y, Zhou K, Yang G. Phylogenomic analyses and improved resolution of Cetartiodactyla. Mol Phylogenet Evol. 2011;61:255–264. doi: 10.1016/j.ympev.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Pauly D, Trites A, Capuli E, Christensen V. Diet composition and trophic levels of marine mammals. ICES J Mar Sci. 1998;55:467–481. doi: 10.1006/jmsc.1997.0280. [DOI] [Google Scholar]
- 4.Fordyce RE, Barnes LG, Miyazah N. General aspects of the evolutionary history of whales and dolphins. Island Arc. 1994;3(4):373–391. doi: 10.1111/j.1440-1738.1994.tb00121.x. [DOI] [Google Scholar]
- 5.Reidenberg J. Anatomical adaptations of aquatic mammals. Anat Rec A. 2007;290:507–513. doi: 10.1002/ar.20541. [DOI] [PubMed] [Google Scholar]
- 6.Zhu K, Zhou X, Xu S, Sun D, Ren W, Zhou K, Yang G. The loss of taste genes in cetaceans. BMC Evol Biol. 2014;14:218. doi: 10.1186/s12862-014-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Chen Z, Xu S, Ren W, Zhou K, Yang G. ‘Obesity’is healthy for cetaceans? Evidence from pervasive positive selection in genes related to triacylglycerol metabolism. Sci Rep. 2015;5:14187. doi: 10.1038/srep14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Zhang Z, Sun Y, Li J, Xu S, Yang G. Comparative genomics analyses of alpha-keratins reveal insights into evolutionary adaptation of marine mammals. Front Zool. 2017;14(1):41. doi: 10.1186/s12983-017-0225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Xu S, Du K, Huang F, Chen Z, Zhou K, Ren W, Yang G. Evolution of digestive enzymes and RNASE1 provides insights into dietary switch of cetaceans. Mol Biol Evol. 2016;33(12):3144–3157. doi: 10.1093/molbev/msw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian R, Wang Z, Niu X, Zhou K, Xu S, Yang G. Evolutionary genetics of hypoxia tolerance in cetaceans during diving. Genome Biol Evol. 2016;8(3):827–839. doi: 10.1093/gbe/evw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu S, Chen Y, Cheng Y, Yang D, Zhou X, Xu J, Zhou K, Yang G. Positive selection at the ASPM gene coincides with brain size enlargements in cetaceans. Proc Roy Soc B. 2012;279(1746):4433–4440. doi: 10.1098/rspb.2012.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu S, Sun X, Niu X, Zhang Z, Tian R, Ren W, Zhou K, Yang G. Genetic basis of brain size evolution in cetaceans: insights from adaptive evolution of seven primary microcephaly (MCPH) genes. BMC Evol Biol. 2017;17:206. doi: 10.1186/s12862-017-1051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker J, Meade A, Pagel M, Venditti C. Adaptive evolution toward larger size in mammals. Proc Natl Acad Sci U S A. 2015;112(16):5093–5098. doi: 10.1073/pnas.1419823112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montgomery SH, Geisler JH, McGowen MR, Fox C, Marino L, Gatesy J. The evolutionary history of cetacean brain and body size. Evolution. 2013;67(11):3339–3353. doi: 10.1111/evo.12197. [DOI] [PubMed] [Google Scholar]
- 15.Blackburn TM, Gaston KJ, Loder N. Geographic gradients in body size: a clarification of Bergmann’s rule. Divers Distrib. 1999;5(4):165–174. doi: 10.1046/j.1472-4642.1999.00046.x. [DOI] [Google Scholar]
- 16.Hone DW, Benton MJ. The evolution of large size: how does Cope's rule work? Trends Ecol Evol. 2005;20(1):4–6. doi: 10.1016/j.tree.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Kemper KE, Visscher PM, Goddard ME. Genetic architecture of body size in mammals. Genome Biol. 2012;13(4):244. doi: 10.1186/gb-2012-13-4-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 19.Mekel-Bobrov N, Lahn BT. Response to Comments by Timpson et al. and Yu et al. Science. 2007;317(5841):1036b. doi: 10.1126/science.1143658. [DOI] [Google Scholar]
- 20.Pyenson ND, Sponberg SN. Reconstructing body size in extinct crown Cetacea (Neoceti) using allometry, phylogenetic methods and tests from the fossil record. J Mamm Evol. 2011;18(4):269–288. doi: 10.1007/s10914-011-9170-1. [DOI] [Google Scholar]
- 21.Slater GJ, Price SA, Santini F, Alfaro ME. Diversity versus disparity and the radiation of modern cetaceans. Proc Biol Sci. 2010;277(1697):3097–3104. doi: 10.1098/rspb.2010.0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burkin DJ, Kim JE, Gu M, Kaufman SJ. Laminin and alpha7beta1 integrin regulate agrin-induced clustering of acetylcholine receptors. J Cell Sci. 2000;113(16):2877–2886. doi: 10.1242/jcs.113.16.2877. [DOI] [PubMed] [Google Scholar]
- 23.Cavanagh JA, Tammen I, Windsor PA, Bateman JF, Savarirayan R, Nicholas FW, et al. Bulldog dwarfism in Dexter cattle is caused by mutations in ACAN. Mamm Genome. 2007;18(11):808–814. doi: 10.1007/s00335-007-9066-9. [DOI] [PubMed] [Google Scholar]
- 24.Gleghorn L, Ramesar R, Beighton P, Wallis G. A mutation in the variable repeat region of the aggrecan gene (AGC1) causes a form of spondyloepiphyseal dysplasia associated with severe, premature osteoarthritis. Am J Hum Genet. 2005;77(3):484–490. doi: 10.1086/444401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blagitko N, Mergenthaler S, Schulz U, Wollmann HA, Craigen W, Eggermann T, et al. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue-and isoform-specific fashion. Hum Mol Genet. 2000;9(11):1587–1595. doi: 10.1093/hmg/9.11.1587. [DOI] [PubMed] [Google Scholar]
- 26.Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc Natl Acad Sci U S A. 2003;100(14):8292–8297. doi: 10.1073/pnas.1532175100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson D, Murray PG, Sud A, Temtamy SA, Aglan M, Superti-Furga A, et al. The primordial growth disorder 3-M syndrome connects ubiquitination to the cytoskeletal adaptor OBSL1. Am J Hum Genet. 2009;84(6):801–806. doi: 10.1016/j.ajhg.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akahoshi N, Kamata S, Kubota M, Hishiki T, Nagahata Y, Matsuura T, et al. Neutral aminoaciduria in cystathionine β-synthase-deficient mice, an animal model of homocystinuria. Am J Physiol Renal Physiol. 2014;306(12):F1462–F1476. doi: 10.1152/ajprenal.00623.2013. [DOI] [PubMed] [Google Scholar]
- 29.Giunta C, Randolph A, Al‐Gazali LI, Brunner HG, Kraenzlin ME, Steinmann B. Nevo syndrome is allelic to the kyphoscoliotic type of the Ehlers–Danlos syndrome (EDS VIA) Am J Med Genet A. 2005;133A(2):158–164. doi: 10.1002/ajmg.a.30529. [DOI] [PubMed] [Google Scholar]
- 30.Zhang P, McGrath B, Sa L, Frank A, Zambito F, Reinert J, et al. The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22(11):3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta S, McGrath B, Cavener DR. PERK (EIF2AK3) regulates proinsulin 767 trafficking and quality control in the secretory pathway. Diabetes. 2010;768 59(8):1937–1947. doi: 10.2337/db09-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasques GA, Arnhold IJ, Jorge AA. Role of the natriuretic peptide system in normal growth and growth disorders. Horm Res Paediatr. 2014;82(4):222–229. doi: 10.1159/000365049. [DOI] [PubMed] [Google Scholar]
- 33.Pernasetti F, Milner RD, al Ashwal AA, de Zegher F, Chavez VM, Muller M, et al. Pro239Ser: a novel recessive mutation of the pit-1 gene in seven middle eastern children with growth hormone, prolactin, and thyrotropin deficiency. J Clin Endocrinol Metab. 1998;83(6):2079–2083. doi: 10.1210/jcem.83.6.4901. [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2759. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura M. The neutral theory of molecular evolution. New York: Cambridge University Press; 1983. [Google Scholar]
- 36.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15(12):496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pond SLK, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21(10):2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 38.Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22(12):2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 40.Anisimova M, Yang Z. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol Biol Evol. 2007;24(5):1219–1228. doi: 10.1093/molbev/msm042. [DOI] [PubMed] [Google Scholar]
- 41.Weber PW, Howle LE, Murray MM, Reidenberg JS, Fish FE. Hydrodynamic performance of the flippers of large-bodied cetaceans in relation to locomotor ecology. Mar Mammal Sci. 2014;30(2):413–432. doi: 10.1111/mms.12040. [DOI] [Google Scholar]
- 42.Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics. 2003;19(5):671–672. doi: 10.1093/bioinformatics/btg043. [DOI] [PubMed] [Google Scholar]
- 43.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montgomery SH, Capellini I, Venditti C, Barton RA, Mundy NI. Adaptive evolution of four microcephaly genes and the evolution of brain size in anthropoid primates. Mol Biol Evol. 2011;28:625–638. doi: 10.1093/molbev/msq237. [DOI] [PubMed] [Google Scholar]
- 45.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 46.Orme D, Freckleton RP, Thomas G, Petzoldt T, Fritz SA. Caper: comparative analyses of phylogenetics and evolution in R. 2011. [Google Scholar]
- 47.R Development Core Team . R: A Language and Environment for Statistical Computing. 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Twenty body-size-related genes and their functions. Table S2. Sequence data used in this study, including taxonomy and accession numbers or Emsemble ID. Table S3. One-ratio model, Free-ratio model and Branch-site model analysis in 20 body-size-related genes. Table S4. Radical amino acid sites under positive selection detected by PAML, Datamonkey and TreeSAAP. Table S5. Log likelihood and omega values estimates under different branch models according contracting body length of cetaceans. Table S6. Association analysis between gene evolution and phenotypes. (DOCX 107 kb)
Figure S1. Radical amino acid changes in selected sites and cetacean-special sites mapped on the three-dimensional structure of body-size-related genes. Sites marked with red balls stand for robust sites under selection and yellow balls stand for cetacean-special sites. The figures were created using PyMOL (http://www.pymol.org). (PDF 2961 kb)
Table S7. Primer sets used to amplify the coding regions of body-size-related genes in this study (XLSX 25 kb)
Alignment sequences of 20 body-size related genes used for this study. These files are Fast format, please use MEGA or Clustal to open them. (ZIP 133 kb)
Table S8. Previously reported body size information for cetaceans. (DOCX 17 kb)
Data Availability Statement
All the data supporting our findings are contained within the manuscript and in the supplemental files.