Abstract
Identifying a common set of genes that mediate host-microbial interactions across populations and species of mammals has broad relevance for human health and animal biology. However, the genetic basis of the gut microbial composition in natural populations remains largely unknown outside of humans. Here, we used wild house mouse populations as a model system to ask three major questions: (1) Does host genetic relatedness explain inter-individual variation in gut microbial composition? (2) Do population differences in the microbiota persist in a common environment? (3) What are the host genes associated with microbial richness and the relative abundance of bacterial genera? We found that host genetic distance is a strong predictor of the gut microbial composition as characterized by 16S amplicon sequencing. Using a common garden approach, we then identified differences in microbial composition between populations that persisted in a shared laboratory environment. Finally, we used exome-sequencing to associate host genetic variants with microbial diversity and relative abundance of microbial taxa in wild mice. We identified 20 genes that were associated with microbial diversity or abundance including a macrophage-derived cytokine (IL12a) that contained three nonsynonymous mutations. Surprisingly, we found a significant overrepresentation of candidate genes that were previously associated with microbial measurements in humans. The homologous genes that overlapped between wild mice and humans included genes that have been associated with traits related to host immunity and obesity in humans. Gene-bacteria associations identified in both humans and wild mice suggest some commonality to the host genetic determinants of gut microbial composition across mammals.
Keywords: Microbiome, Coevolution, GWAS, Metagenomics, Mammals, Mus musculus
Introduction
Host-associated microbial communities play an important role in health and fitness (McFall-Ngai et al., 2013). Compositional and functional variation in the gut microbiota has been linked to a variety of diseases in humans and lab mouse models including obesity, inflammatory bowel disease, and autism (e.g. Hsiao et al., 2013; Marchesi et al., 2007; Turnbaugh et al., 2006). Links between the gut microbiota and fitness-related traits have also been reported in wild mammals including traits related to digestion, immunity, and behavior (Suzuki, 2017). Therefore, understanding the mechanisms governing the maintenance and function of gut microbial communities is important in medicine and animal biology more broadly.
Host genetics may play an important role in structuring gut microbial communities. For example, genome-wide markers have been associated with overall differences in the microbiome (i.e. beta-diversity) in humans (Blekhman et al., 2015; J. Ma et al., 2014). Twin studies have shown that monozygotic twins tend to have more similar microbial composition compared to dizygotic twins (Goodrich, Davenport, Beaumont, et al., 2016; Goodrich et al., 2014). Mouse knockout experiments have identified genes involved in immunity, metabolism, and behavior that affect the gut microbiota (Spor, Koren, & Ley, 2011). Mouse quantitative trait locus (QTL) mapping studies have also identified multiple genomic regions associated with the relative abundance of different microbial taxa (Benson et al., 2010; Leamy et al., 2014; McKnite et al., 2012; Org et al., 2015; Wang et al., 2015).
In human populations, microbiome genome-wide association studies (mGWAS) have identified specific candidate genes associated with natural variation of the gut microbiota (Blekhman et al., 2015; Bonder et al., 2016; Davenport et al., 2015; Goodrich, Davenport, Beaumont, et al., 2016; Knights et al., 2014; Turpin et al., 2016; Wang et al., 2016), and a few gene-bacteria associations have been replicated in multiple human populations (Goodrich, Davenport, Waters, Clark, & Ley, 2016; Hall, Tolonen, & Xavier, 2017). However, this approach has not been used to look for gene-bacteria associations in wild mammals. Genes identified from human mGWAS are often compared with those identified in laboratory mice, but gene-bacteria associations identified in a controlled laboratory environment may differ from those in a complex natural environment. In fact, the function and composition of the gut microbiota in lab mice are known to differ from those of their wild relatives (Rosshart et al., 2017). Samples from a wild population would provide an opportunity for mGWAS that is more directly comparable to human mGWAS.
Wild house mice (Mus musculus domesticus) are globally distributed and live in a wide range of environments in association with humans (Phifer-Rixey & Nachman, 2015). The house mouse is a powerful model because it is possible to disentangle variables using experimental manipulation (Wang et al., 2015, 2014) and assess the functions of the microbiome using germ-free mice (Rosshart et al., 2017). Previous work has shown that geographic and genetic distances (Linnenbrink et al., 2013), diet as measured by stable isotopes (Wang et al., 2014), reproductive status, body size, age, viral and parasite infection status (Weldon et al., 2015), gut regions (Suzuki & Nachman, 2016), and altitude (Suzuki, Martins, & Nachman, 2018) are associated with compositional differences in the gut microbiota of wild house mice. However, there have been no previous efforts to identify specific genes underlying compositional variation in the gut microbiota of wild mice.
Here, we characterize natural variation in the gut microbiota of wild house mice sampled from five populations along an environmental gradient in eastern North America and identify specific host genes associated with gut microbial composition. First, we show that genetic distance correlates with microbial composition (i.e. beta-diversity), both within and between populations. Second, using a common garden experiment, we show that differences in the gut microbiota among wild populations persist in the laboratory, suggesting that they are not driven by the environment such as diet. Third, we identify genome-wide gene-bacteria associations in wild mice using the complete exome sequences of all mice. Finally, we document significant overlap between the genes associated with microbiota variation in mice and genes associated with microbiota variation in humans.
Materials and Methods
Sample collection
We collected a total of 50 adult house mice (Mus musculus domesticus) from five populations in eastern North America during the summer of 2012 (Phifer-Rixey et al., 2018) and generated 80 lab-reared individuals in the laboratory for a common garden experiment (see below). For wild-caught individuals, we collected ten mice each from each of five populations: Florida (FL), Georgia (GA), Virginia (VA), Pennsylvania (PA), and New Hampshire - Vermont (NH-VT) (summarized in Table S1). Sherman live traps were used with peanut butter and oats as bait. Each mouse was caught a minimum of 500m from all other mice to avoid sampling close relatives. Animals were kept in Sherman traps, euthanized by cervical dislocation, and all tissues and external measurements were collected within 24 hours after capture. Cecum and liver were stored in liquid nitrogen in the field and then stored in a deep freezer (−80°C) until sequencing. Diet was inferred using carbon (δ13C) and nitrogen (δ15N) stable isotopes from mouse hair following the protocol of Suzuki and Nachman (2016). Climate data were inferred using the first two principal components (Table S2) calculated from 19 climatic variables downloaded from WorldClim database (Hijmans, Cameron, Parra, Jones, & Jarvis, 2005) and the R package “dismo”.
For the common garden experiment, we captured live animals close to the northern and southern populations using Sherman live traps during summer, 2013 from Saratoga Springs, NY and Gainsville, FL. Within each location, animals were collected from at least 10 sites that were a minimum of 500m apart. Animals were housed in the laboratory at 23°C with a light cycle of 10 hours dark and 14 hours light. Teklad Global food (18% Protein Rodent Diet) was fed ad libitum. Wild-caught mice from the same population were paired to produce F1 offspring. Ten independent crosses were conducted and offspring were housed with their own littermates. Body weight and fresh fecal samples were collected from 40 individuals per population representing four adults (two females and two males) each from the 10 independent crosses of wild-caught founders. Fecal samples were stored in −80°C until sequencing. Detailed information of lab-reared animals is in Table S3. All procedures involving animals were reviewed and approved by the IACUC at the University of Arizona (07–004) and at the University of California Berkeley (R361–0514).
16S rRNA gene sequencing
We extracted DNA from cecal and fecal samples from wild-caught and lab-reared mice, respectively. Although these sample types are different, fecal samples are known to closely match individual differences in cecal samples in wild house mice (Suzuki & Nachman, 2016). We followed the DNA extraction protocol described in Suzuki and Nachman (2016). Briefly, we added a bead-beating step before step 4 in the protocol from QIAamp DNA stool Minikit (Qiagen). The V4 region of the 16S rRNA gene was amplified and multiplexed using the standard primers and barcodes described in (Caporaso et al., 2012), and the samples were sequenced on two lanes of 150bp pair-end Illumina MiSeq at the Next Generation Sequencing Core Facility at Argonne National Laboratory. Negative controls were included in every set of amplifications. To avoid potential lane bias, the same DNA aliquots of six samples from the first lane (MPR108, MPR114, MPR120, MPR135, MPR138, and MPR144) were run on the second lane. Lane 1 included all the wild-caught populations and Lane 2 included all the lab-reared populations plus these six controls.
Mouse exome data
We used exome data from Phifer-Rixey et al. (2018). That study is based on the same individuals used in this study. Briefly, DNA was extracted from frozen liver, kidney, or spleen. Genomic libraries were enriched for mouse exons using a NimbleGen in-solution capture array (SeqCap EZ) and sequenced using 100bp pair-end Illumina HiSeq2000. After quality filtering and SNP discovery, we further filtered the SNPs to only include those with a minor allele frequency of 5% or greater. This resulted in 279,278 SNPs. Each SNP was annotated to a single gene or multiple genes using Variant Effect Predictor in Ensembl. Additional details of quality filtering and SNP discovery are given in Phifer-Rixey et al. (2018).
16S data processing
We processed all of the 16S data in QIIME version 1.9.0 (Caporaso et al., 2010). The forward reads were demultiplexed and quality-filtered using default parameters using split_libraries_fastq.py. Chimeric sequences were removed using USEARCH 6.1 (Edgar, Haas, Clemente, Quince, & Knight, 2011). A subsampled open-reference OTU picking approach (pick_open_reference_otus.py) was employed with default parameters. OTUs at 97% similarity were generated using UCLUST (Edgar, 2010) and taxa were assigned based on the Greengene database 13.8 (DeSantis et al., 2006). To remove sequence errors and very rare OTUs, OTUs with <10 reads across all samples were removed. A phylogenetic tree was created using FastTree (Price, Dehal, & Arkin, 2009). The OTU table was rarefied to an even depth of 5,000 reads. Two samples (FL08M1 and FL08M2) were removed from all analyses due to low sequence reads (<200 reads).
Despite rarefying the reads to equal depth for all samples, the OTU counts were consistently higher in lane 1 compared to lane 2 for the six control samples (Fig. S1). This lane bias is likely due to the greater average sequence depth of lane 2 (68,196 reads per sample) compared to lane 1 (13,918 reads per sample) resulting in an excess of rare OTUs in lane 2. To account for this, we removed rare OTUs from lane 2 to normalize the OTU counts between lanes before rarefaction (i.e. OTUs with a relative abundance less than 8.0×10−6 were removed) (Fig. S1). Since lane 1 included all wild-caught mice and lane 2 included all lab-reared mice, all conclusions derived from comparisons within these groups (essentially all major conclusions; see Results) were not affected by lane bias. Moreover, conclusions drawn from comparisons between wild-caught and lab-reared mice remained the same with or without correcting for lane bias (see Results). The OTU table corrected for lane bias was used for all analyses presented below.
Statistical analysis
We calculated beta-diversity measurements (i.e. Bray-Curtis dissimilarity, Binary-Sorensen-Dice, Unweighted- and Weighted-UniFrac distances) among all individuals using beta_diversity.py in QIIME. Pairwise distances for geography (km) were calculated based on GPS coordinates of the sampling locations. Pairwise distances for host genomes were calculated based on the exome data (~280,000 SNPs) using ngsDist (Vieira, Lassalle, Korneliussen, & Fumagalli, 2016) which takes into account uncertainty of the genotype calls. We used Mantel tests to test for correlations between beta-diversity and eight predictor variables (genetic distance, geographic distance, body weight, BMI, diet (δ13C and δ15N), climate PC1, and climate PC2) among all 50 wild-caught mice. We used Partial Mantel tests to ask whether host genetic distance is independently associated with Bray-Curtis dissimilarity while controlling for the effects of other variables. We also made comparisons among individuals within populations using Spearman’s rho correlation. Specifically, we compared Bray-Curtis dissimilarity and geographic distance and we also compared Bray-Curtis dissimilarity and genetic distance. We also assessed the effects of geography while controlling for genetics (and the effects of genetics while controlling for geography) by comparing the residuals in a covariate regression with Bray-Curtis dissimilarity. Similarly, to test for correlations between beta-diversity and body size measurements, Spearman’s rho correlation with residuals between body size and latitude was used to control for the known effect of latitude on body size (Table S4).
We calculated Bray-Curtis dissimilarity separately among mice within the northern-wild population (NH-VT), the southern-wild population (FL), the northern-lab population (NY), and the southern-lab population (FL). Similarly, Bray-Curtis dissimilarity was calculated separately between the two wild populations and between the two lab populations. We tested whether Bray-Curtis dissimilarity between population comparisons were significantly greater than within population comparisons in both wild-caught and lab-reared individuals using Wilcoxon permutation tests based on 9999 Monte-Carlo resampling with the “Wilcox_test” function in the R package “coin”. We calculated alpha-diversity using alpha_diversity.py and relative abundances of bacterial taxa using summarize_taxa.py using the rarefied OTU table. We used phylogenetic diversity (Faith, 1992) as an alpha-diversity measurement and we focused on the relative abundances of 17 bacterial genera that were present in at least 50% of the individuals and had an average relative abundance of >1% across all individuals. The motivation for selecting a small set of common bacterial genera is both statistical (e.g. to minimize 0 values and multiple testing) and biological (e.g. narrower taxonomic groups [e.g. genera] have been shown to be associated with host genomic regions better than broader taxonomic groups [e.g. classes or phyla] (Benson et al., 2010)). Wilcoxon tests and Kruskal-Wallis tests were used for all pairwise and group comparisons unless otherwise stated.
Wild mouse mGWAS and overlap with human mGWAS
We used a multivariate linear mixed model for association tests in GEMMA (version 0.94) using the exome (279,278 SNPs). The phylogenetic diversity measure and the relative abundances of 17 bacterial genera described above were Box-Cox transformed following Goodrich et al. (2016) using the “PowerTransform” function in the R package “car”. A multiple linear regression was used on the transformed microbial measurements to regress out the covariates including population structure, latitude, and hidden factors. We calculated population structure using SNPRelate (version 1.10.2) and used the first four genetic principal components (which together explain 18.5% of the genetic variation). Latitude was also used as a covariate to control for bacteria that vary latitudinally (Thompson et al., 2017). Hidden factors were calculated to account for experimental cofounders and batch effects by inferring 10 cofounders in PEER (Stegle, Parts, Durbin, & Winn, 2010). We also accounted for relatedness by using a relatedness matrix estimated in GEMMA. Manhattan plots and QQ-plots were generated using the R package “qqman”. To control for false discovery, q-values were calculated using the R package “qvalue” based on Likelihood ratio p-values. Significant SNPs were called at a q-value < 0.1. The functions of nonsynonymous SNPs were predicted using mouse genome version GRCm38 in SIFT (Sim et al., 2012).
To test for overlap of bacteria-associated genes identified in this study and previously published human mapping studies, we identified significant SNPs with a q-value < 0.2 to increase power. We compiled SNPs and genes that were associated with microbial measurements (e.g. relative abundance of taxa, alpha- or beta-diversity) in seven human mGWAS studies (Table S5). To test whether the overlap of candidate genes in humans and mice was significant, we used a hypergeometric test with the “phyper” function in R. We used the total number of possible human-mouse orthologous genes in our exome (19,100) based on the Ensembl database (Sep 2017). To test whether the proportion of significant gene-bacteria associations was greater in the candidate gene set compared to all genes using the entire exome, we used a chi-square test with Yate’s correction.
To calculate effect sizes and to ask whether different genotypes have significantly different relative abundances of taxa or alpha-diversity measurements, we used ANOVA. Residuals after covariate regression (i.e. population structure, latitude, and hidden factors) on the box-cox transformed microbial measurements were used for the analyses. We used a sign test to ask whether the directionality of genotype-bacteria associations within populations deviates significantly from the expected 50:50 ratio.
Results
Host genetic distance and body size are associated with compositional variation in the gut microbiota
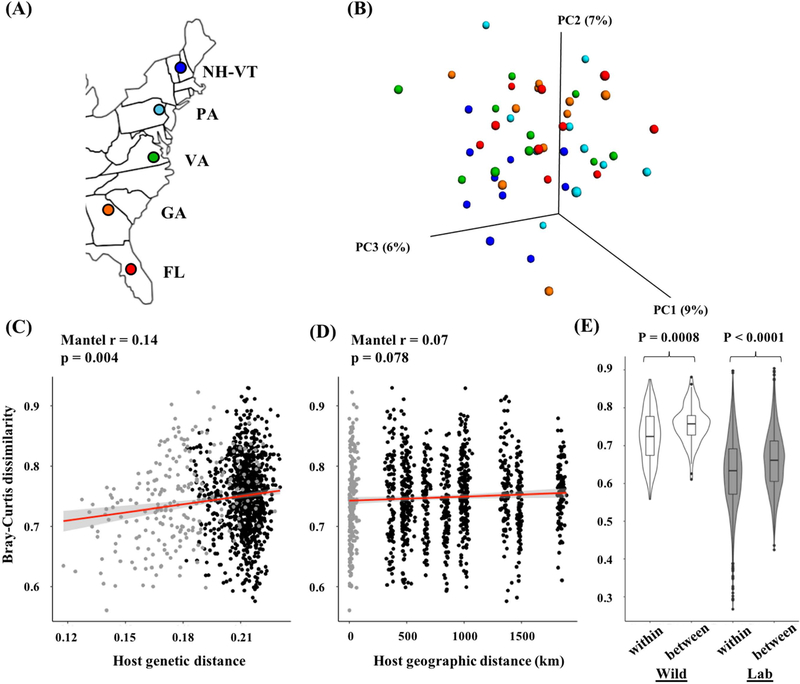
We found significant differences in gut microbial communities among five populations of house mice in eastern North America (Fig. 1A&B) based on Bray-Curtis dissimilarity (ADONIS, R2 = 0.095, p = 0.04), Binary-Sorensen-Dice (R2 = 0.099, p = 0.002), and unweighted UniFrac distance (R2 = 0.097, p = 0.003), but not based on weighted UniFrac distance (R2 = 0.087, p = 0.34). While these results are consistent with findings in European populations of house mice (Linnenbrink et al., 2013), the population differences observed in the present study were relatively small. To understand how host genetic and environmental factors contribute to variation in gut microbial communities across all samples, we measured correlations between Bray-Curtis dissimilarity and eight putative predictor variables using Mantel tests (Table 1). We found that Bray-Curtis dissimilarity was significantly correlated with both host genetic distance and body mass index (BMI) after correcting for multiple tests (Mantel r = 0.14, p = 0.004 and Mantel r = 0.25, p < 0.0001, respectively) (Table 1 and Fig. 1C). Diet (δ13C), climate PC1, and geographic distance (Fig. 1D) also showed weak correlations with Bray-Curtis dissimilarity, but they were not significant (Table 1). Diet measurements (δ13C and δ14N) did not vary among populations (Fig.S1) or by latitude (Table S4). The overall results were similar using other beta-diversity measurements (Table S6).
Figure 1.
Population differences and host genetic distance are associated with compositional variation in the gut microbiota. (A) Sampling locations of five house mouse populations. (B) PCoA plot of Bray-Curtis dissimilarity. The color corresponds to populations in Fig. 1A. Populations show weak, but significant clustering (ADONIS R2=0.095, p=0.04). (C) A significant positive correlation between microbial distance and host genetic distance (calculated by ngsDist) and (D) a non-significant correlation between microbial distance and geographic distance using all individuals. Correlations for within-population comparisons (grey points) and between-population comparisons (black dots) are shown. (E) Violin boxplots of Bray-Curtis dissimilarity within population comparisons (within) and between population comparisons (between) of the most northern (NH-VT) and southern (FL) populations in the wild (white bars) and lab (gray bars). P-values are Wilcoxon permutation tests.
Table 1.
Correlations between microbial beta-diversity and predictor variables using Mantel test.
| Predictor variables | n | Bray-Curtis dissimilarity |
|
|---|---|---|---|
| Mantel r | p-value | ||
| Genetic distance (ngsDist) | 50 | 0.14 | 0.004 |
| Geographic distance (km) | 50 | 0.07 | 0.078 |
| Body weight (g) | 50 | 0.08 | 0.232 |
| BMI (g/mm2) | 50 | 0.25 | <0.001 |
| Diet (δ13C) | 50 | 0.14 | 0.044 |
| Diet (δ15N) | 50 | 0.04 | 0.67 |
| ClimatePC1 | 50 | 0.10 | 0.066 |
| ClimatePC2 | 50 | 0.01 | 0.913 |
Bolded values show significance below Bonferroni corrected p-value = 0.05/8 = 0.0063
The observed correlation between microbial distance and genetic distance was independent of other variables including geographic distance (Table S7). First, there was no pattern of genetic isolation-by-distance among these populations (Fig. S2). Second, the correlation between host genetic distance and Bray-Curtis dissimilarity remained significant after controlling for seven predictor variables, including geographic distance, using Partial Mantel tests (Table S7). When comparisons were made between individuals within populations, both host genetic distance and geographic distance showed significant correlations with Bray-Curtis dissimilarity (Fig. S3). Consistent with the results among populations, we found that the correlation between host genetic distance and Bray-Curtis dissimilarity within populations remained significant after controlling for geographic distance using residuals of covariate regression (Spearman’s rho = 0.26, p < 0.0001). In contrast, we found that the correlation between geographic distance and Bray-Curtis dissimilarity did not remain significant after controlling for genetic distance (Spearman’s rho = 0.06, p = 0.38). These results suggest that host genetics and/or vertical transmission have stronger effects on the gut microbiota than the geographic distance between individuals.
The correlation between BMI and Bray-Curtis dissimilarity also remained significant after controlling for geographic distance (partial mantel r = 0.25, p < 0.001). This association is interesting because BMI and body weight vary clinally with latitude (Table S4), a pattern consistent with Bergmann’s rule (Bergmann, 1847) and presumably reflecting thermoregulatory adaptation (Lynch, 1992; Phifer-Rixey et al., 2018). Moreover, at higher latitudes, individuals tended to have a greater ratio of Firmicutes to Bacteroidetes, which is associated with obesity in humans (Ley, Turnbaugh, Klein, & Gordon, 2006) and in laboratory mice (Ley et al., 2005; Turnbaugh et al., 2006) (Fig. S4). We also identified various microbial taxa that correlated with latitude and body size after accounting for latitude (Fig. S5). For example, the phylum Proteobacteria was the taxon whose relative abundance was most positively correlated with latitude (rho = 0.443, p < 0.01). The phylogenetic diversity of the microbial community and the relative abundance of genus Odoribacter were most positively correlated with body weight after correcting for latitude (rho = 0.410, p < 0.01 and rho = 0.412, p < 0.01, respectively).
Population differences in the microbiota persist in a common laboratory environment
We found a significant difference in Bray-Curtis dissimilarity among wild mice in comparisons within versus between populations from the ends of the transect (NH-VT and FL) (Fig. 1E), indicating that mice from these populations harbor compositionally distinct microbial communities. To test whether population differences in the microbiota were driven by environmental differences, we collected live animals close to the most northern and southern populations (NY and FL) and conducted a common garden experiment. Twenty unrelated wild mice were collected from each population and returned to the lab (10 males, 10 females). For each population, we created 10 crosses between wild-caught parents to produce 40 offspring which were reared under identical conditions. The lab-born mice showed major shifts in alpha-diversity and in the relative abundances of bacterial phyla and genera compared to the wild-caught animals (Table S8). For example, alpha-diversity measurements and the relative abundances of Firmicutes and Proteobacteria significantly decreased, and the relative abundance of Bacteroidetes significantly increased in lab-reared animals compared to wild-caught animals (Phylogenetic diversity and Shannon index, p < 0.001; all phyla, p <0.001) (Table S8). Although there is a potential for batch effects since the microbiota of wild-caught and lab-reared individuals were sequenced on separate lanes, the same results were obtained with or without correcting for lane bias (Shannon index, p < 0.001; all phyla, p < 0.001). Interestingly, diet as assessed by stable isotopes showed some overlap between wild and lab mice (Fig. S6). The lab diet was significantly different from that of the wild as assessed by δ15N (lab mean: 6.5, wild mean: 7.1, p < 0.05 ) but not as assessed by δ13C (lab mean: −19.0, wild mean: 19.1, p = 0.12) (Fig. S6).
Despite the dramatic shifts in the microbiota from the wild to the lab environment (Fig. S7), population differences in the microbiota persisted among lab-reared offspring (Fig. 1E). Compositional differences in the microbiota both within and between populations were reduced in the laboratory setting compared to the wild (Fig. 1E). However, the microbial community composition of lab populations was more similar, on average, to the wild populations from which they came than to the wild populations at the other end of the transect (Wilcoxon permutation test, P = 0.029, Fig. S8). Overall, these results indicate that environmental differences alone (e.g. diet, temperature, etc.) cannot fully explain the population differences in the microbiota. The observed population differences are consistent either with a role for host genetics or simply with vertical transmission shaping the variation of the gut microbiota in wild mice.
Identification of genetic loci underlying gut microbiota variation in wild mice
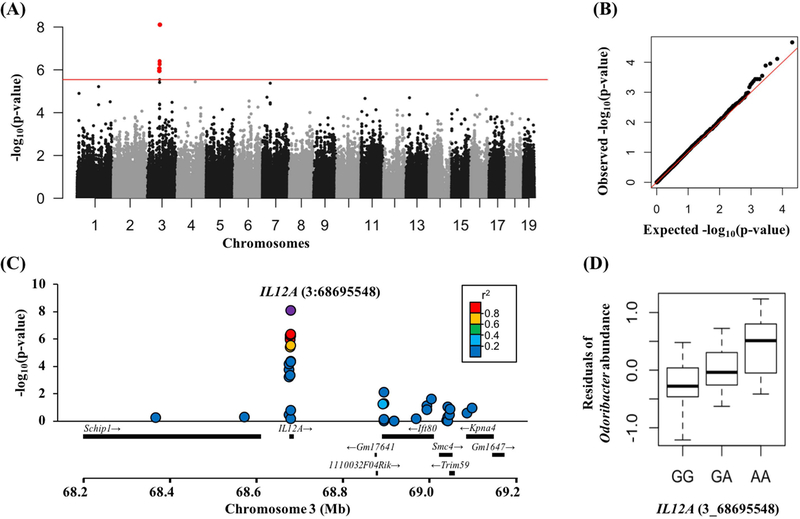
To identify host genes contributing to differences in the gut microbiota, we conducted a mGWAS using ~280,000 SNPs identified from sequencing the complete exomes of the 50 wild-caught mice (Phifer-Rixey et al., 2018). We searched for associations between host genetic variation and the relative abundances of 17 bacterial genera that were common (an average relative abundance of > 1% and present in > 50% of all individuals). We also searched for associations between host genetic variation and alpha-diversity represented by phylogenetic diversity. Analyses were done using Multivariate Linear Mixed Models in GEMMA while controlling for host population structure, relatedness, latitude, and hidden factors. Among the 18 bacterial measurements, two bacteria genera (Odoribacter and Bacteroides) and phylogenetic diversity showed significant associations with host genetic loci (Table 2). Across all tests, we identified a total of 24 SNPs in 20 genes that passed a genome-wide significance threshold (q-value < 0.1). Although none of the GO terms were significantly overrepresented after false discovery correction, the top three GO terms include mRNA transcription (Mier1, p-value = 0.006), protein lipidation (Zdhhc7, p-value = 0.04), and nucleobase-containing compound transport (Slc35d1, p-value = 0.07).
Table 2.
Loci significantly associated with the relative abundances of bacterial taxa or phylogenetic diversity (q-value < 0.1) in wild mouse mGWAS.
| Chr | Bp | Annotated gene(s) | Associated microbial measurements | P-values1 | Q-values | Effect size (%)2 | Missense variant3 |
|---|---|---|---|---|---|---|---|
| 3 | 68695209 | Il12a | Odoribacter | 8.41E–07 | 0.042 | 23.1 | Thr - Ser |
| 3 | 68695333 | 1.10E–06 | 0.045 | 19.9 | Ser - Met * | ||
| 3 | 68695379 | 5.37E–07 | 0.033 | 18.0 | - | ||
| 3 | 68695382 | 5.37E–07 | 0.033 | 18.0 | - | ||
| 3 | 68695502 | 4.13E–07 | 0.033 | 22.2 | - | ||
| 3 | 68695548 | 7.79E–09 | 0.002 | 24.4 | Gly - Ser | ||
| 3 | 86138475 | Snord73a, Rnu73b, Rps3a1 | Bacteroides | 1.77E–06 | 0.054 | 18.1 | - |
| 3 | 86138574 | 6.90E–07 | 0.028 | 18.4 | - | ||
| 3 | 86138625 | 2.00E–06 | 0.054 | 29.8 | - | ||
| 4 | 103170679 | Mier1, Slc35d1 | Phylogenetic diversity | 2.19E–06 | 0.092 | 5.4 | - |
| 4 | 89692441 | Dmrta1 | Bacteroides | 4.73E–07 | 0.023 | 27.9 | - |
| 5 | 90490831 | Afp | Bacteroides | 1.27E–08 | 0.002 | 27.0 | - |
| 5 | 90490846 | 1.27E–08 | 0.002 | 27.0 | - | ||
| 5 | 90491657 | 8.67E–08 | 0.007 | 17.5 | - | ||
| 6 | 121221243 | Tuba8, Gm15856 | Bacteroides | 4.19E–06 | 0.086 | 23.4 | - |
| 6 | 121222841 | 3.03E–06 | 0.074 | 23.3 | - | ||
| 6 | 128374454 | Foxm1, Tex52 | Phylogenetic diversity | 4.09E–07 | 0.051 | 33.3 | - |
| 6 | 128374521 | 4.09E–07 | 0.051 | 33.3 | - | ||
| 6 | 128374742 | 9.63E–07 | 0.055 | 34.0 | - | ||
| 8 | 120092803 | Zdhhc7, Gm20388, Gm15898 | Bacteroides | 3.38E–07 | 0.021 | 38.2 | - |
| 8 | 13142468 | Cul4a | Bacteroides | 3.56E–06 | 0.079 | 22.5 | - |
| 8 | 16358320 | Csmd1 | Phylogenetic diversity | 1.09E–06 | 0.055 | 17.0 | - |
| 11 | 3132802 | Sfi1, Pisd-ps1 | Phylogenetic diversity | 8.39E–07 | 0.055 | 12.0 | - |
| 13 | 33671503 | Serpinb6d | Bacteroides | 1.38E–06 | 0.048 | 31.7 | - |
Likelihood ratio p-values.
ANOVA R2 values. Residuals after covariate regression (i.e. Genetic PC1–4 and Latitude) on the box-cox transformed relative abundance of bacterial taxa was used.
Amino acid changes are shown; aCt/aGt (Threnine/Serine), aCg/aTg (Serine/Methionine), and Ggc/Agc (Glycine/Serine).
indicate significant deleterious changes predicted by SIFT (SIFT score < 0.05).
Although we attempted to account for population structure in identifying these genes using GEMMA, observed gene-bacteria associations might still be driven by differences among populations that are not fully accounted for by the model. To further account for population structure, we first looked at associations within individual populations and then examined whether the direction of the association was consistent among populations. Overall, most of the within-population genotype-bacteria comparisons showed the same direction as the all-population comparisons (37 out of 43 comparisons, sign test p-value < 0.0001, Table S9). Moreover, 20 of these 37 comparisons were individually significant (ANOVA, p-value < 0.05) despite the fact that these tests are underpowered with only 10 individuals per population (Table S9). Together, the results suggest that the observed genotype-bacteria associations are unlikely to be explained by population structure.
Among the 20 genes that were associated with bacterial measurements, the interleukin 12a gene (IL12a) included a SNP with the lowest p-value across all tests in this study (Table 2). IL12 is a cytokine that plays a key role in innate and adaptive immunity by activating natural killer cells and regulating differentiation of T cells (Trinchieri, 1998). We identified six SNPs in IL12a that were significantly associated with the relative abundance of Odoribacter after accounting for population structure, latitude, and hidden factors as covariates (Fig. 2A-D, Table 2). Three of these six SNPs were nonsynonymous changes, and one of these (3_68695333) was predicted in silico to be deleterious (SIFT score = 0.002) (Table 2).
Figure 2.
Results of mGWAS in wild mice. (A) Manhattan plot of Odoribacter. Six SNPs in IL12A on chromosome 3 are highlighted. Red line shows the genome-wide cut-off of the likelihood ratio test p-value (q-value < 0.1). (B) Quantile-quantile plot of Odoribacter p-values. The red diagonal line represents the expected distributions of p-values. (C) Zoom in plot around the SNP (3:68695548) that has the lowest p-value (represented by purple). Each dot is a SNP and colors represent pairwise linkage disequilibrium measures (r2) between the SNP (3:68695548) and SNPs within the surrounding 1 Mb window. r2 was calculated based on genotype allele counts using PLINK. (D) Box-plot of Odoribacter abundance and IL12A genotypes. A missense SNP in IL12A gene has significantly different abundances of Odoribacter (ANOVA R2 = 0.224, p = 0.0014). Residual values were used for the y-axis controlling for population structure, latitude, and hidden factors using covariate regression. Whiskers indicate highest and lowest values.
Homologous genes underlie gut microbiota variation in humans and mice
A common set of genes may underlie host-bacterial interactions across diverse mammals. To test this idea, we asked whether there was significant overlap between the genes underlying variation in the microbiota of mice and humans using two different approaches. First, we compiled genes that were associated with microbial measurements (e.g. relative abundance of taxa, alpha- or beta-diversity) in seven different human mGWAS. This comprised a set of 469 genes with one-to-one mouse-human orthologs (Table S5). We then conducted association analyses in GEMMA using this set of 469 genes in mice and found that 10 were significantly associated with one or more bacterial measurements (q-value < 0.1) (Table S10). This fraction of genes showing associations (10 out of 469 = 2.13%) is significantly greater than the fraction discovered in the initial analysis using all genes (20 out of 21,954 = 0.09%) suggesting that mouse mGWAS hits are overrepresented among genes previously identified in human mGWAS (Chi-square test with Yate’s correction p < 0.0001).
Second, we asked how many genes overlapped between the 469 genes identified in human mGWAS and the 20 mouse-human orthologous genes that were identified in the mouse mGWAS. Using the genome-wide cut-off of q-value < 0.1, there was only one gene, Csmd1 that overlapped between these sets, and this degree of overlap was marginally not significant (hypergeometric test p = 0.06). However, when we made the genome-wide cut-off less stringent (q-value < 0.2), we identified 96 mouse-human orthologous genes (Table S11) and eight genes overlapped with 469 human candidate genes (Table S12). The number of overlapped genes identified at q-value < 0.2 was greater than expected by chance (hypergeometric test p = 0.0006). Not surprisingly, all eight genes were also identified by the candidate gene approach mentioned above (Table S10 & S12). Among the eight homologous overlapping genes between human and mouse mGWAS, all show expression in the brain of mice and humans (Table S12) and some have been associated with phenotypes related to obesity and immunity in other human GWAS.
Discussion
Understanding how mammalian hosts maintain the composition and function of the gut microbiota remains a major challenge in microbial ecology and biomedical research. We showed that differences in host genetics and body mass were significantly associated with compositional differences in the microbiota of wild mice. We did not find significant associations between the microbiota and diet as inferred by carbon and nitrogen stable isotopes. However, fine-scale dietary differences that are not reflected in isotope measurements might also contribute to variation in the wild mouse gut microbiota. In other studies, host genetic distance (Blekhman et al., 2015; J. Ma et al., 2014), body size measurements (e.g. Ley et al., 2006; Turnbaugh et al., 2009), and diet (David et al., 2014) have all been associated with differences in the gut microbial composition in humans. We found that mouse populations from higher latitudes had a greater ratio of Firmicutes/Bacteroidetes, a pattern also seen among human populations (Suzuki & Worobey, 2014). This is interesting since a higher ratio of Firmicutes/Bacteroidetes is known to be associated with obesity in humans (Ley et al., 2006) and in laboratory mice (Ley et al., 2005; Turnbaugh et al., 2006). The mice sampled here conform to Bergmann’s rule with mice from higher latitudes exhibiting larger body sizes than mice from lower latitudes (Lynch, 1992; Phifer-Rixey et al., 2018). Further experiments, including transplants into gnotobiotic mice, would be useful for testing the role of the gut microbiome, if any, in adaptive host body size variation.
To test whether population differences in the gut microbiota were due to the environment or to host genetics, we identified population differences in the gut microbiota that persisted in a common laboratory environment. Consistent with previous studies of wild mice raised in captivity (Wang et al., 2015, 2014), we observed a decrease in alpha-diversity and in the relative abundances of Firmicutes and Proteobacteria, and an increase in the relative abundance of Bacteroidetes, in lab-reared animals compared to wild-caught animals. Despite the changes in gut microbial composition that occurred when progeny of wild mice were raised in the lab, population differences persisted among the lab-reared mice. Moreover, the gut microbiota of lab-reared mice resembled the gut microbiota of the population of origin more than of the population at the other end of the transect. Similar patterns have also been found in other wild-derived mouse strains where population differences in the field were maintained in captivity for over 10 generations (Moeller, Suzuki, Phifer-Rixey, & Nachman, 2018). These observations suggest that environmental differences alone (e.g. diet, temperature, etc.) cannot explain the population differences in the microbiota. Instead, host genetics and/or vertical transmission must partly account for the observed population differences in the gut microbiota of wild mice. Experiments allowing the exchange of microbes between individuals from different populations could be used to directly test the effect of host genotype on the gut microbiota.
To explore the genetic basis of the gut microbiota, we identified both novel and previously known gene-bacteria associations in wild mice using a genome-wide mapping approach. The top association identified in this study was between the relative abundance of Odoribacter and a non-synonymous SNP in IL12a, a cytokine that is involved in innate and adaptive immunity (Trinchieri, 1998). The up-regulation of IL12a production has been linked to Crohn’s disease in humans (Parronchi et al., 1997) and mucosal inflammation in mice (Z. Liu et al., 2001). Furthermore, a recent study in humans demonstrated that inflammatory cytokine responses are associated with microbial taxa composition, metagenomic functional profiles, and microbial metabolites (Schirmer et al., 2016). Interestingly, the relative abundance of Odoribacter was significantly correlated with tumor necrosis factor alpha (TNF-α) (Schirmer et al., 2016), which is another macrophage-derived cytokine that interacts with IL12a in mediating inflammatory responses in mammals (X. Ma, 2001). These observations lend further support to the role of IL12a in mediating host-microbial interactions in wild mice.
Finally, a significantly greater number of genes overlapped between human mGWAS and mouse mGWAS than expected by chance, including genes related to the nervous system, immunity, and obesity. For example, a SNP in Csmd1 is associated with alpha-diversity (i.e. phylogenetic diversity) in mice and showed the lowest p-value among the eight human-mouse overlapping genes. Csmd1 is highly expressed in the central nervous system and in epithelial tissue and is involved in regulating the development of the central nervous system (Kraus et al., 2006). In humans, Csmd1 is associated with beta-diversity of the gut microbiota (Wang et al., 2016), obesity-related traits (Comuzzie et al., 2012; Irvin et al., 2011; C. T. Liu et al., 2013), parasite infection status (Deng et al., 2013), and antibody response to smallpox vaccine (Ovsyannikova et al., 2012). Similarly, Gpr158 is also highly expressed in mouse and human brains and is associated with bacterial taxa in the order Clostridiales in both wild mice and humans (Goodrich, Davenport, Beaumont, et al., 2016). Gpr158 has been associated with variation in energy expenditure in a native American population that has a high prevalence of obesity (Piaggi et al., 2017). These results suggest host genes related to the nervous system, immunity, and obesity may underlie gut microbial variation across diverse mammalian species.
In conclusion, we presented evidence that the host genome affects gut microbial composition within and between populations of wild mice using field observations and laboratory experiments. Gene-bacteria associations identified in wild mice and humans using similar mapping methods are strong candidates for genes influencing the mammalian gut microbial composition in a natural environment. Replicating these results in independent populations of wild mice and validating the functions of candidate SNPs in wild-derived inbred mice would further strengthen the observed gene-bacteria associations.
Supplementary Material
Acknowledgements
We thank Andrew Moeller, Mohamed Noor, the members of the Nachman lab, and three anonymous reviewers for discussions and comments on the manuscript. This work was supported by NIH grants (RO1 GM074245, RO1 GM127468) to MWN and an NSF grant (1501646) to TAS.
Footnotes
Data Accessibility Statement
All museum specimens (skins and skulls) were prepared and have been deposited in the mammal collections of the Museum of Vertebrate Zoology at the University of California, Berkeley with ancillary data uploaded to ARCTOS (see Table S1 for accession numbers). All 16S rRNA sequence data were uploaded to European Molecular Biology Laboratory, European Nucleotide Archive (ENA) database (PRJEB32701). The mouse exome sequence data are available under NCBI SRA (PRJNA397406) as described in (Phifer-Rixey et al., 2018).
References
- Benson AK, Kelly S. a, Legge R, Ma F, Low SJ, Kim J, … Pomp D (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proceedings of the National Academy of Sciences of the United States of America, 107(44), 18933–18938. 10.1073/pnas.1007028107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C (1847). Uber die verhaltnisse der warmeokonomie der thiere zu ihrer grosse. Göttinger Studien. Pt, 1, 595–708. [Google Scholar]
- Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, … Clark AG (2015). Host genetic variation impacts microbiome composition across human body sites. Genome Biology, 16(1), 191 10.1186/s13059-015-0759-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, … Zhernakova A (2016). The effect of host genetics on the gut microbiome. Nature Genetics, 48(11), 1407–1412. 10.1038/ng.3663 [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, … Knight R (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5), 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters W. a, Berg-Lyons D, Huntley J, Fierer N, … Knight R (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal, 1–4. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed]
- Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, & Butte NF (2012). Novel Genetic Loci Identified for the Pathophysiology of Childhood Obesity in the Hispanic Population. PLoS ONE, 7(12). 10.1371/journal.pone.0051954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, & Gilad Y (2015). Genome-Wide Association Studies of the Human Gut Microbiota. PloS ONE, 10(11), e0140301 10.1371/journal.pone.0140301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, … Turnbaugh PJ (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature, 505(7484), 559–563. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Sabino EC, Cunha-Neto E, Ribeiro AL, Ianni B, Mady C, … Seielstad M (2013). Genome wide association study (GWAS) of chagas cardiomyopathy in trypanosoma cruzi seropositive subjects. PLoS ONE, 8(11), 4–10. 10.1371/journal.pone.0079629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, … Andersen GL (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 72(7), 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26(19), 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, & Knight R (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27(16), 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP (1992). Conservation evaluation and phylogenetic diversity. Biological Conservation, 61(1), 1–10. 10.1016/0006-3207(92)91201-3 [DOI] [Google Scholar]
- Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, … Ley RE (2016). Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host and Microbe, 19(5), 731–743. 10.1016/j.chom.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Davenport ER, Waters JL, Clark AG, & Ley RE (2016). Cross-species comparisons of host genetic associations with the microbiome. Science, 352(6285), 29–32. 10.1126/science.aad9379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, … Ley RE (2014). Human genetics shape the gut microbiome. Cell, 159(4), 789–799. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AB, Tolonen AC, & Xavier RJ (2017). Human genetic variation and the gut microbiome in disease. Nature Reviews Genetics, 18(11), 690–699. 10.1038/nrg.2017.63 [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones G, & Jarvis A (2005). Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol, 25, 1965–1978. 10.1002/joc.1276 [DOI] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, … Mazmanian SK (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell, 155(7), 1451–1463. 10.1016/j.cell.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin MR, Wineinger NE, Rice TK, Pajewski NM, Kabagambe EK, Gu CC, … Arnett DK (2011). Genome-wide detection of allele specific copy number variation associated with insulin resistance in african americans from the hyperGEN study. PLoS ONE, 6(8), 9–11. 10.1371/journal.pone.0024052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, … Xavier RJ (2014). Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Medicine, 6, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD, … Holers VM (2006). CSMD1 Is a Novel Multiple Domain Complement-Regulatory Protein Highly Expressed in the Central Nervous System and Epithelial Tissues. The Journal of Immunology, 176(7), 4419–4430. 10.4049/jimmunol.176.7.4419 [DOI] [PubMed] [Google Scholar]
- Leamy LJ, Kelly S. a, Nietfeldt J, Legge RM, Ma F, Hua K, … Pomp D (2014). Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biology, 15(12), 552 10.1186/s13059-014-0552-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, & Gordon JI (2005). Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America, 102(31), 11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, & Gordon JI (2006). Microbial ecology: human gut microbes associated with obesity. Nature, 444(7122), 1022–1023. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Linnenbrink M, Wang J, Hardouin E. a, Künzel S, Metzler D, & Baines JF (2013). The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Molecular Ecology, 22, 1904–1916. 10.1111/mec.12206 [DOI] [PubMed] [Google Scholar]
- Liu CT, Monda KL, Taylor KC, Lange L, Demerath EW, Palmas W, … Fox CS (2013). Genome-Wide Association of Body Fat Distribution in African Ancestry Populations Suggests New Loci. PLoS Genetics, 9(8), e1003681 10.1371/journal.pgen.1003681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Geboes K, Heremans H, Overbergh L, Mathieu C, Rutgeerts P, & Ceuppens JL (2001). Role of interleukin-12 in the induction of mucosal inflammation and abrogation of regulatory T cell function in chronic experimental colitis. Eur J Immunol, 31(5), 1550–60. [DOI] [PubMed] [Google Scholar]
- Lynch C (1992). Clinal variation in cold adaptation in Mus domesticus: verification of predictions from laboratory populations. American Naturalist, 139(6), 1219–1236. Retrieved from http://www.jstor.org/stable/2462338 [Google Scholar]
- Ma J, Coarfa C, Qin X, Bonnen PE, Milosavljevic A, Versalovic J, & Aagaard K (2014). mtDNA haplogroup and single nucleotide polymorphisms structure human microbiome communities. BMC Genomics, 15(1), 257 10.1186/1471-2164-15-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X (2001). TNF-α and IL-12:a balancing act in macrophage functioning. Microbes and Infection, 3(2), 121–129. 10.1016/S1286-4579(00)01359-9 [DOI] [PubMed] [Google Scholar]
- Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, … Wang Y (2007). Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. Journal of Proteome Research, 6(2), 546–551. 10.1021/pr060470d [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, … Wernegreen JJ (2013). Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences, 110(9), 3229–3236. 10.1073/pnas.1218525110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnite AM, Perez-Munoz ME, Lu L, Williams EG, Brewer S, Andreux PA, … Ciobanu DC (2012). Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS ONE, 7(6), e39191 10.1371/journal.pone.0039191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Suzuki TA, Phifer-Rixey M, & Nachman MW (2018). Transmission modes of the mammalian gut microbiota. Science, 362(6413), 453–457. 10.1126/science.aat7164 [DOI] [PubMed] [Google Scholar]
- Org E, Parks BW, Joo JWJ, Emert B, Schwartzman W, Kang EY, … Lusis AJ (2015). Genetic and environmental control of host-gut microbiota interactions. Genome Research, 25(10), 1558–1569. 10.1101/gr.194118.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Kennedy RB, O’Byrne M, Jacobson RM, Pankratz VS, & Poland GA (2012). Genome-wide association study of antibody response to smallpox vaccine. Vaccine, 30(28), 4182–4189. 10.1016/j.vaccine.2012.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parronchi P, Romagnani P, Annunziato F, Sampognaro S, Becchio a, Giannarini L, … Romagnani S (1997). Type 1 T-helper cell predominance and interleukin-12 expression in the gut of patients with Crohn’s disease. The American Journal of Pathology, 150(3), 823–832. [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M, Bi K, Ferris KG, Sheehan MJ, Lin D, Mack KL, … Nachman MW (2018). The genomic basis of environmental adaptation in house mice. PLoS Genetics, 14(9), e1007672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M, & Nachman MW (2015). Insights into mammalian biology from the wild house mouse Mus musculus. ELife, 2015(4), 1–13. 10.7554/eLife.05959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaggi P, Masindova I, Muller YL, Mercader J, Wiessner GB, Chen P, … Baier LJ (2017). A Genome-Wide Association Study Using a Custom Genotyping Array Identifies Variants in GPR158 Associated With Reduced Energy Expenditure in American Indians. Diabetes, 66(8), 2284–2295. 10.2337/db16-1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, & Arkin AP (2009). Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Molecular Biology and Evolution, 26(7), 1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, … Rehermann B (2017). Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell, 1–14. 10.1016/j.cell.2017.09.016 [DOI] [PMC free article] [PubMed]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, … Xavier RJ (2016). Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell, 167(4), 1125–1136. 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, & Ng PC (2012). SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Research, 40(W1), W452–W457. 10.1093/nar/gks539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, & Ley R (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nature Reviews. Microbiology, 9(4), 279–290. 10.1038/nrmicro2540 [DOI] [PubMed] [Google Scholar]
- Stegle O, Parts L, Durbin R, & Winn J (2010). A bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Computational Biology, 6(5), 1–11. 10.1371/journal.pcbi.1000770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki TA (2017). Links between Natural Variation in the Microbiome and Host Fitness in Wild Mammals. Integrative and Comparative Biology, 57(4), 756–769. 10.1093/icb/icx104 [DOI] [PubMed] [Google Scholar]
- Suzuki TA, Martins FM, & Nachman MW (2018). Altitudinal variation of the gut microbiota in wild house mice. Molecular Ecology, (October), 1–13. 10.1111/mec.14905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki TA, & Nachman MW (2016). Spatial heterogeneity of gut microbial composition along the gastrointestinal tract in natural populations of house mice. PLoS ONE, 11(9), 1–15. 10.1371/journal.pone.0163720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki TA, & Worobey M (2014). Geographical variation of human gut microbial composition. Biology Letters, 10(2), 20131037–20131037. 10.1098/rsbl.2013.1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, … Knight R (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature, 551(7681), 457–463. 10.1038/nature24621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G (1998). Interleukin-12: a cytokine at the interface of inflammation and immunity. Advances in Immunology, 70(2), 83–243. https://doi.org/98427913 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, … Gordon JI (2009). A core gut microbiome in obese and lean twins. Nature, 457(7228), 480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald M. a, Magrini V, Mardis ER, & Gordon JI (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 444(7122), 1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Turpin W, Espin-garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, … Croitoru K (2016). Association of host genome with intestinal microbial composition in a large healthy cohort. Nature Genetics, 48(11), 1413–1417. 10.1038/ng.3693 [DOI] [PubMed] [Google Scholar]
- Vieira FG, Lassalle F, Korneliussen TS, & Fumagalli M (2016). Improving the estimation of genetic distances from Next-Generation Sequencing data. Biological Journal of the Linnean Society, 117(1), 139–149. 10.1111/bij.12511 [DOI] [Google Scholar]
- Wang J, Kalyan S, Steck N, Turner LM, Harr B, Künzel S, … Baines JF (2015). Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nature Communications, 6, 6440 10.1038/ncomms7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Linnenbrink M, Künzel S, Fernandes R, Nadeau M-J, Rosenstiel P, & Baines JF (2014). Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proceedings of the National Academy of Sciences of the United States of America, 111(26), E2703–10. 10.1073/pnas.1402342111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Thingholm LB, Skiecevic J, Rausch P, Kummen M, Holm K, … Franke A (2016). Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nature Genetics, 48(11), 1396–1406. 10.1038/ng.3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon L, Abolins S, Lenzi L, Bourne C, Riley EM, & Viney M (2015). The gut microbiota of wild mice. PLoS ONE, 10(8), 1–15. 10.1371/journal.pone.0134643 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.