Abstract
During industrial fermentation, Streptomyces clavuligerus F613-1 simultaneously produces primary product clavulanic acid (CA) and cephamycin C. The cephamycin C biosynthetic gene cluster and pathway have been basically elucidated and the CcaR positive regulator was found to control the cephamycin genes expression. However, additional mechanisms of regulation cannot be excluded. The BB341_RS13780/13785 gene pair in S. clavuligerus F613-1 (annotated as SCLAV_2960/2959 in S. clavuligerus ATCC27064) encodes a bacterial two-component system (TCS) and were designated as CepRS (for cephamycin regulator/sensor). CepRS significantly affects cephamycin C production but only slightly affects CA production. To further understand the regulation of cephamycin C biosynthesis, the cepRS genes were deleted from S. clavuligerus F613-1. The deletion mutant resulted in decreased cephamycin C production but had no phenotypic effects. Real-time quantitative polymerase chain reaction analysis revealed that CepRS regulates the expression of most genes involved in cephamycin C biosynthesis, with electrophoretic mobility shift assays showing that CepR interacts with the cefD-cmcI intergenic region. These results demonstrate that the CepR response regulator serves as a transcriptional activator of cephamycin C biosynthesis, which may provide an approach for metabolic engineering methods for CA production by S. clavuligerus F613-1 in future.
Electronic supplementary material
The online version of this article (10.1186/s13568-019-0844-z) contains supplementary material, which is available to authorized users.
Keywords: Streptomyces clavuligerus, CepRS, Cephamycin C, Clavulanic acid
Introduction
The actinomycete Streptomyces clavuligerus produces a variety of secondary metabolites, including cephamycin C (7-methoxy-3ʹ-carbamoyl-deacetyl-cephalosporin C) and clavulanic acid (CA). CA is a broad-spectrum inhibitor of β-lactamases produced by penicillin- and cephalosporin-resistant bacteria (Paradkar 2013; Saudagar et al. 2008). S. clavuligerus F613-1 is used industrially to produce the β-lactamase inhibitor CA. Cephamycin C, which is also present in the fermentation broth, is considered undesirable when CA is produced (Jin et al. 2015; Qin et al. 2017).
Genes responsible for cephamycin C and CA biosynthesis are clustered in the genome, forming a β-lactam supercluster (Ward and Hodgson 1993). The cephamycin C biosynthetic gene cluster mainly includes ten genes: pcd, lat, pcbAB, pcbC, cefD, cefE, cmcI, cmcJ, cefF, and cmcH. The four genes pcd, lat, pcbAB, pcbC are involved in the early stages of cephamycin C biosynthesis. pcd and lat are involved in the formation of α-aminoadipic. pcbAB and pcbC are involved in the formation of isopenicillin N. cefD and cefE are involved in the intermediate stages of cephamycin C biosynthesis, and are responsible for forming deacetylcephalosporin C. The four genes cmcI, cmcJ, cefF, and cmcH are involved in the late stages of cephamycin C biosynthesis and are responsible for the specific C-7 methoxylation and carbamoylation steps (Coque et al. 1995a, b; Enguita et al. 1996).
Secondary metabolites are usually produced at very low levels, indicating the existence of mechanisms that tightly control their biosynthesis (Chater 1993). In fact, the biosynthesis of both cephamycin C and CA is very precisely regulated. Disruption of the biosynthetic or regulatory genes of the cephamycin C/CA supercluster indicated a connection between the biosynthetic pathways of these β-lactam compounds via a regulatory cascade that includes both pleiotropic and pathway-specific regulators (Liras et al. 2008; Martín and Liras 2010). Regulatory gene ccaR, located in the cephamycin C biosynthetic gene cluster, encodes protein product CcaR, which appears to be a key Streptomyces antibiotic regulatory protein-type positive regulator. Deletion of ccaR completely block the production of both cephamycin C and CA (Pérez-Llarena et al. 1997; Santamarta et al. 2002, 2011). Recently, examination of the DNA-binding characteristics of CcaR revealed that it binds to the lat, cefF, cefD-cmcI, and ccaR promoter regions in the cephamycin C biosynthetic gene cluster (Kyung et al. 2001; Santamarta et al. 2011). In addition, CcaR affects CA production by binding to the claR, ceaS2 and oppA1 promoters and, therefore, controlling the transcription of claR and consequently of the genes ceaS2 and bls2 (Álvarez-Álvarez et al. 2014; Santamarta et al. 2011).
The genome of S. clavuligerus F613-1 encodes 47 paired two-component systems (TCSs), amongst which, CagRS has been characterized as a global regulator of both primary and secondary metabolism (Fu et al. 2019). To further study the regulation mechanism of TCSs on the secondary metabolism, we constructed a series of single-gene and double-gene knockout strains. Among them, we found that the biosynthesis of cephamycin C was significantly decreased in the cepRS mutant stain, while the biosynthesis of clavulanic acid was slightly affected. The CepRS TCS remains uncharacterized in strain F613-1, although CepR is annotated as the response regulator and CepS as the histidine kinase. The cepRS genes are located 1.38 Mb away from the pcbC gene of the cephamycin C gene cluster. Sequence analysis indicated that CepS belongs to the sub-family of histidine kinases and CepR belongs to the LuxR family of transcriptional regulators. The LuxR family proteins are approximately 250 amino acids long and include two functional domains: an amino-terminal AHL (N-acyl homoserine lactone) binding domain and a carboxy-terminal DNA-binding domain (Nasser and Reverchon 2007). LuxR family response regulators are common in two-component systems (Zhang et al. 2018), however, many of the LuxR family response regulators are orphan response regulators (Fuqua 2006; Patankar and González 2009; Subramoni et al. 2011; Subramoni and Venturi 2009). Here, we show that CepRS affects cephamycin C production and that CepR interacts with the cefD-cmcI bidirectional promoter. In addition, we found that CepRS has limited effects on CA production. Our results may provide new insights into the regulatory network of the cephamycin C biosynthesis in S. clavuligerus.
Materials and methods
Bacterial strains and growth conditions
All the strains and plasmids used in this study are listed in Additional file 1: Table S1. Culturing of Escherichia coli, S. clavuligerus and its derivative strains were performed as described previously (Fu et al. 2019; Qin et al. 2017). In this study, for phenotypic analysis, S. clavuligerus F613-1 (CGMCC NO. 12830) and its mutant strains were cultured at 25 °C and 50–60% relative humidity on BSCA (1.5% (w/v) malt extract, 0.3% (w/v) tryptone, 0.4% (w/v) glucose, and 2.0% (w/v) agar powder, pH 7.5) and TSA (3% (w/v) tryptone soya broth, and 2.0% (w/v) agar powder, pH 7.2) plates.
To examine cephamycin C production, strain F613-1 and its derivatives were grown at 25 °C and a relative humidity of 50–60% on TSA plates for 9 days. Agar blocks were collected at days 1, 3, 5, 7, and 9 post-inoculation to determine cephamycin C concentrations. For CA fermentation assays, the culture conditions were performed as described previously (Fu et al. 2019).
Primers and DNA manipulation
All the primers used in the construction of the ΔcepRS mutant and complementation strains, in the confirmation of the transconjugants, and in the electrophoretic mobility shift assays (EMSAs) are listed in Additional file 1: Table S2. The DNA manipulation method were performed as described previously (Fu et al. 2019).
Construction and complementation of a cepRS null mutant and overexpression strain
The cepRS genes were knocked out as described previously (Fu et al. 2019). In this study, the upstream homologous arm of the cepRS fragment was amplified by PCR using primers cepRS L-F/R. The downstream homologous arm of the cepRS fragment was PCR-amplified using primers cepRS R-F/R. All the recombinant plasmids were verified by DNA sequencing. Conjugation was performed using S. clavuligerus F613-1 and E. coli ET12567/pUZ8002 as described previously (Fu et al. 2019; Kieser et al. 2000; Sambrook 1989). The ΔcepRS mutant strain was confirmed by PCR analysis using the cepRS V-F/R primer pair.
For cepR gene deletion, the upstream homologous arm of the cepR fragment was amplified by PCR using primers cepRS L-F/R. The downstream homologous arm of the cepR fragment was PCR-amplified using primers cepR R-F/R. The mutant strain was confirmed by PCR analysis using the cepR V-F/R primer pair. For cepS gene deletion, the upstream homologous arm of the cepS fragment was amplified by PCR using primers cepS L-F/R. The downstream homologous arm of the cepS fragment was PCR-amplified using primers cepRS R-F/R. The ΔcepR and ΔcepS mutant strains were confirmed by PCR analysis using the cepRS V-F/R primer pair.
For complementation, primers cepRScom-F and cepRScom-R were used to amplify a fragment containing the cepRS coding sequence and its native promoter, which was then cloned into BamHI/XbaI-digested pSET152, generating recombinant plasmid pSET-cepRS. Plasmids pSET152 and pSET-cepRS were introduced separately into the ΔcepRS mutant by conjugation to generate ΔcepRS-pSET152 and ΔcepRScom strains. The ΔcepRScom complemented mutant strain was confirmed by PCR analysis using primer pair cepRS V-F/R.
For cepRS overexpression, primers cepRS-F/R were used to amplify a fragment containing the cepRS coding sequence, which was then cloned into pHLY12, generating recombinant plasmid pHLY-cepRS. Plasmids pHLY-cepRS were introduced into the ΔcepRS mutant by conjugation to generate cepRS overexpression strain ΔcepRS-cepRS. The ΔcepRS-cepRS complemented mutant strain was confirmed by PCR analysis using primer pair cepRS V-F/R.
Measurement of cell growth and cephamycin C production
Determination of cell growth was performed as described previously (Yu et al. 2012). Briefly, strain F613-1 and the derivative strains were cultured on TSA solid plates covered with cellophane and incubated at 25 °C. For dry weight measurement, cultures were harvested at days 1, 3, 5, 7, and 9 post-inoculation and then dried for 4 h at 80 °C.
Cephamycin C concentrations were determined by agar diffusion bioassay as described previously (Leite et al. 2016). Cephalosporin C zinc salt (J & K Scientific Ltd, CAS #59,143–60-1) was used as a standard, and cephamycin C concentration was measured as “total cephalosporins” present in the sample. E. coli ESS 2235, which is highly sensitive to β-lactam antibiotics, was used as the test organism and was cultured for 24 h in LB medium at 37 °C. All samples were treated with penicillinase (Beckton Dickinson, cat. no.: 215,331) to remove penicillin. For biomass measurement and analysis of cephamycin C production, all assays were performed three biological replicates independently. Statistical analysis was analyzed using IBM SPSS Statistics V19.0 software.
HPLC analysis of CA production
CA concentrations were analyzed by HPLC analysis as previously described (Fu et al. 2019; Jin et al. 2015; Qin et al. 2017). Briefly, the concentration of CA in the SCF fermentation medium was detected by HPLC using an Inertsil ODS-3 4.6 mm × 150 mm, 5 μm column, and using clavulanate lithium as the standard for quantification. All HPLC assays were performed three biological replicates independently. Statistical analysis was analyzed using IBM SPSS Statistics V19.0 software. Biomass was also measured prior to determination of CA production using aliquots (1 g) of fermentation supernatant centrifuged at 6000×g to collect mycelia.
Real-time quantitative polymerase chain reaction (RT-qPCR) analysis
To determine the expression levels of the cephamycin C biosynthesis genes, spores (2 × 106 CFU/ml) from wild-type strain F613-1 and the ΔcepRS deletion mutant were plated onto TSA medium covered with cellophane and incubated at 25 °C for 72 h. To examine the expression levels of the CA biosynthesis genes, mycelia from S. clavuligerus strains F613-1 and ΔcepRS were harvested from SCF fermentation broth at 72 h post-inoculation. Total RNA isolation and RT-qPCR procedures were performed as described previously (Fu et al. 2017; Qin et al. 2017). Relative transcript levels were normalized to those of the 16S rRNA gene. For RT-qPCR assays, experiments were performed three biological replicates independently. Statistical analysis was analyzed using IBM SPSS Statistics V19.0 software.
Expression and purification of His-tagged CepR
cepR was amplified using primer pair cepRHis-F/R (Additional file 1: Table S2) before being cloned into vector pMD18T, generating the intermediate recombinant plasmid pMD18T-cepR. Following DNA sequence confirmation, cepR was released from pMD18T-cepR and cloned into the pET-15bto obtain pET-cepR. Finally, pET-cepR was introduced into E. coli BL21(DE3). His-tagged CepR expression was induced and the protein was purified as previously described [5, 6]. The purity of His-tagged CepR was determined by 10% SDS-PAGE.
EMSAs
DNA fragments corresponding to the intergenic regions of the cephamycin C biosynthetic gene cluster were amplified using F613-1genomic DNAas template (primers are listed in Additional file 1: Table S2). EMSAs were performed as previously described (Fu et al. 2019; Zhang et al. 2015).
Results
Disruption of cepRS leads to a significant reduction in cephamycin C production by S. clavuligerus F613-1
CepRS is one of the uncharacterized two-component systems in S. clavuligerus F613-1. To investigate the role of the individual components of the CepRS system, each gene was mutated separately in S. clavuligerus F613-1. The ΔcepR strain, which lacks the coding sequence for the cepR response regulator gene, and the ΔcepS strain, which lacks the coding sequence for the cepS sensor kinase, were constructed (Additional file 1: Figure S1A). Phenotypic differences were not obvious noted for ΔcepR or ΔcepS compared with F613-1 on BSCA solid medium (Figure S1B). While cephamycin C production was reduced for both ΔcepR and ΔcepS strains (Additional file 1: Figure S1C), suggesting that CepR and CepS are critical for cephamycin C production.
To further characterize the function of CepRS, a cepRS null mutant strain, ΔcepRS, and a complemented mutant strain, ΔcepRScom, were constructed and confirmed by PCR (Fig. 1). No phenotypic differences were noted for ΔcepRS relative to parental strain F613-1 on BSCA and TSA solid media (Additional file 1: Figure S1B and data not shown). Interestingly, agar diffusion assays revealed that cepRS double gene deletion resulted in a significant reduction of cephamycin C biosynthesis in ΔcepRS compared with that of F613-1 despite similar biomass values (Fig. 2). The cephamycin C production was almost restored in the complementation strain ΔcepRScom (Fig. 2b). The deletion of cepRS was complemented by introduction of cepRS genes (Additional file 1: Figure S1A and Fig. 2), suggesting that the CepRS mainly contributes to the regulation of cephamycin C biosynthesis and not affect phenotype of F613-1.
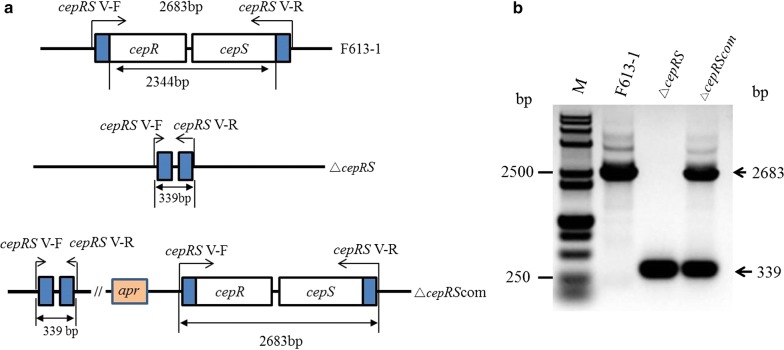
Fig. 1.
Verification of cepRS mutant strain ΔcepRS. a Schematic representation outlining the construction of the cepRS deletion and complementation strains. b Polymerase chain reaction-based verification of the ΔcepRS and ΔcepRScom strains. M: DNA marker
Fig. 2.
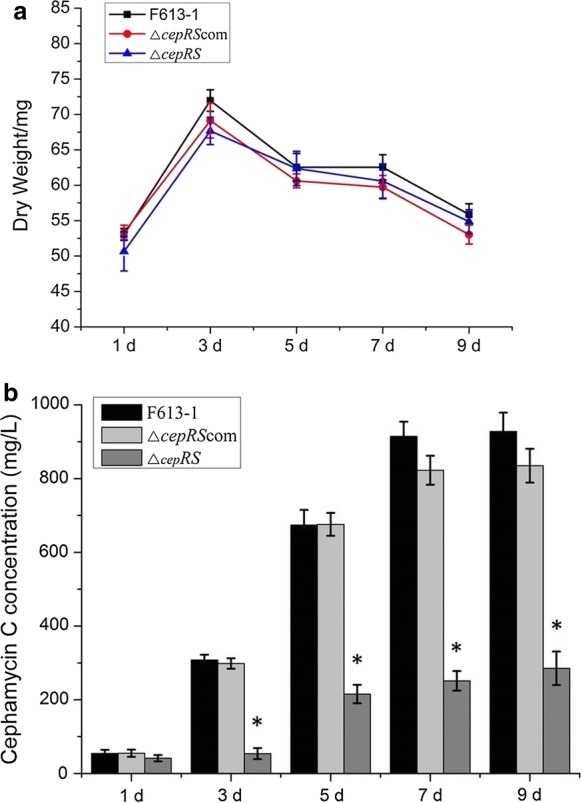
Biomass and cephamycin C concentration analyses of Streptomyces clavuligerus strains F613-1, ΔcepRS, and ΔcepRScom on TSA medium. a Growth curves of strains F613-1, ΔcepRS, and ΔcepRScom grown on TSA plates. Samples for biomass analysis were harvested at days 1, 3, 5, 7, and 9 post-inoculation. Data are the mean ± standard deviation of three separate experiments. b Concentrations of cephamycin C produced by S. clavuligerus strains F613-1, ΔcepRS, and ΔcepRScom strains. *Statistically significant difference (P < 0.05) between ΔcepRS and F613-1 at the same time point
In addition, no phenotypic differences were noted for cepRS overexpression strain ΔcepRS-cepRS relative to the parental strain F613-1 on BSCA solid media (Additional file 1: Figure S4A). The bioassay data indicated that cephamycin C production was slightly increased in ΔcepRS-cepRS compared with F613-1 (Additional file 1: Figure S4B).
Over all, the similar phenotypes and decreased cephamycin C production displayed by ΔcepR, ΔcepS, and ΔcepRS support the prediction that CepR and CepS form a paired TCS and indicate that CepRS is important for regulating cephamycin C production.
cepRS regulates the expression of most of the cephamycin C biosynthetic genes
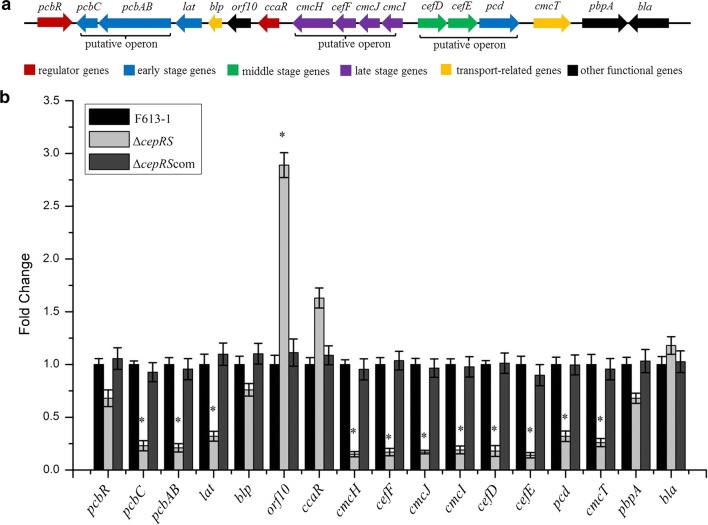
Sequence analysis of the S. clavuligerus F613-1 genome revealed that this cluster is essentially identical to that of S. clavuligerus NRRL27064 (Liras et al. 2008; Cao et al. 2016). As shown in Fig. 3a, three putative operons (pcbAB-pcbC, cmcI-cmcJ-cefF-cmcH, and cefD-cefE-pcd) were predicted in the gene cluster based on intervals between the ORFs. Because the CepRS TCS appears to significantly affect cephamycin C production, RT-qPCR analysis was performed to gain further insights into the mechanisms of CepRS regulation of cephamycin C biosynthesis. As shown in Fig. 3b, the expression of pcbR, blp, ccaR, pbpA, and bla was not significantly affected by disruption of CepRS; however, the expression of uncharacterized gene orf10 was significantly increased in ΔcepRS compared with that in F613-1. Further, the expression levels of all of the biosynthetic genes pcbC, pcbAB, lat, cefD, cefE, cmcI, cmcJ, cefF, and cmcH were significantly reduced in the ΔcepRS mutant compared with those in F613-1. The RT-qPCR data revealed that expression of genes in the early (pcbC, pcbAB and lat), middle (cefD and cefE), and late (cmcI, cmcJ, cefF, cmcH) stages of cephamycin C biosynthesis was reduced in ΔcepRS compared with F613-1, which is consistent with the agar diffusion assay results showing that cephamycin C levels were reduced in the ΔcepRS mutant compared with wild-type strain F613-1. In addition, the RT-qPCR data revealed that the effect on expression level of cephamycin C biosynthetic genes was not significantly affected by cepRS overexpression compared with F613-1 (Additional file 1: Figure S4C). Overall, the above data suggest that CepRS positively regulates the expression of most of the cephamycin C biosynthetic genes.
Fig. 3.
Comparison of gene expression. a Schematic diagram of the cephamycin C biosynthetic gene cluster. b The expression of genes belonging to the cephamycin C biosynthetic gene cluster was examined by real-time quantitative PCR analysis using RNA extracted from Streptomyces clavuligerus strains F613-1, ΔcepRS and ΔcepRScom. Results were normalized against the expression of the 16S rRNA gene and are shown as fold change over the F613-1 control. Data are the mean ± standard deviation of three independent biological experiments. *Compared with F613-1, P < 0.05
CepR specifically binds to the promoters of cefD-cmcI
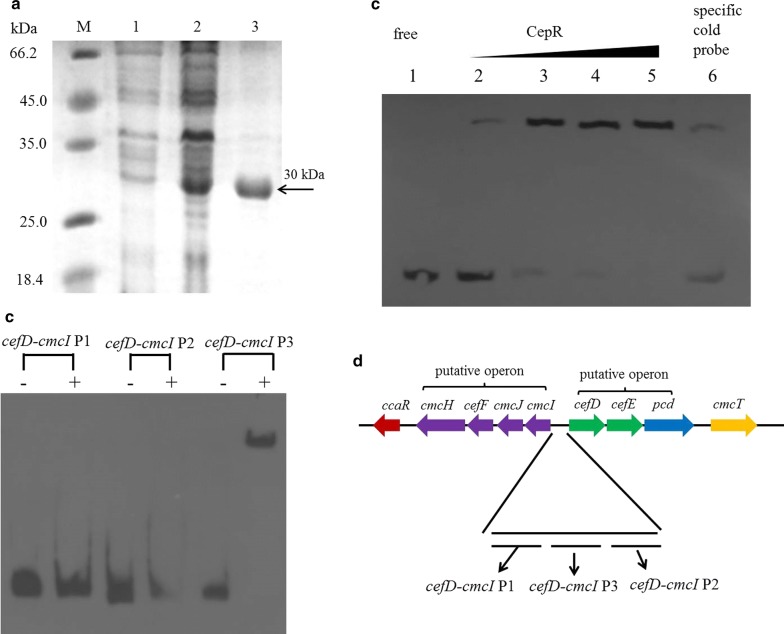
The above data indicated that the CepRS TCS affects the expression of 12 genes in the cephamycin C biosynthetic gene cluster. Therefore, we speculated that response regulator CepR may interact directly with the promoters of the affected genes. To investigate the targets of CepR, we amplified from the intergenic regions upstream of the genes different DNA fragments to use them as probes in EMSAs: 196 bp upstream region of pcbAB, two upstream regions (330 bp and 313 bp respectively) of lat, two upstream regions (148 bp and 193 bp respectively) of orf10, two upstream regions (117 bp and 208 bp respectively) of ccaR, three intergenic regions (208 bp, 315 bp and 318 bp respectively) of cefD-cmcI, 149 bp upstream region of cmcT, 210 bp upstream region of pbpA. Recombinant 6 × His-CepR was expressed and purified for EMSAs (Fig. 4a). When incubated with the purified His-tagged CepR, obvious shifting was only observed with probe containing intergenic region P3 of cefD-cmcI (Fig. 4b), indicating that CepR interacts with cefD-cmcI intergenic region P3. However, shifting was not observed with probes containing the intergenic regions of other genes in the cephamycin C biosynthetic gene cluster. Further EMSA experiments showed that shifting was almost abrogated by the addition of excess specific unlabeled probe P3 (Additional file 1: Fig. 4c, lanes 2–6), indicating that binding of CepR to the cefD-cmcI P3 intergenic region was specific. These in vitro results suggested that CepR interacts directly with the promoters of two critical genes (cefD and cmcI) necessary for cephamycin C biosynthesis. However, the cefD-cmcI intergenic region lies between the cmcI-cmcJ-cefF-cmcH and cefD-cefE-pcd operons (Fig. 4d), indicating that response regulator CepR may directly regulate the expression of all seven genes (cmcI, cmcJ, cefF, cmcH, cefD, cefE, and pcd).
Fig. 4.
Binding of CepR to the cefD-cmcI promoter regions. a Expression and purification of His-tagged CepR analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Lane 1, control sample. Lane 2, His-tagged CepR induced by the addition of IPTG. Lane 3, purified fusion protein. Lane M, protein marker. b Electrophoretic mobility shift assay analysis of the binding of His-tagged CepR to the promoter regions of cefD-cmcI. The P1, P2, and P3 cefD-cmcI intergenic regions were 208 bp, 315 bp, and 318 bp, respectively. The probes were incubated either with no protein ( − ) or with 3.0 μg of CepR ( + ).1.0 µg polydI/dC was used as a competitor. c A fixed amount of probe P3 was incubated with no CepR (Lane 1), 0.5–3.0 µg of CepR (Lanes 2–5), or 3.0 μg of CepR and 100-fold excess of unlabeled specific probe (Lane 6). 1.0 µg polydI/dC was used as a competitor. d Positions of P1–P3 cefD-cmcI intergenic regions in the cephamycin C biosynthetic gene cluster
Discussion
CepRS affects cephamycin C production but not cell growth
A bacterial TCS consists of a sensor histidine kinase and a response regulator (Fu et al. 2019). TCSs mediate adaptation to changing environments and are reported to be involved in a variety of bacterial cellular responses, including morphological development, biofilm development, sporulation, osmoregulation, chemotaxis, photosynthesis, antibiotic production, and pathogenicity (Bijlsma and Groisman 2003; Fu et al. 2019; Hutchings et al. 2004; Mikkelsen et al. 2011; Ogura and Tanaka 2002). TCSs are highly abundant in Streptomyces species and reportedly affect the production of antibiotics such as CA (Fu et al. 2019). Here, we presented a typical and paired TCS (CepRS) in S. clavuligerus, bioinformatics analysis revealed that CepS is a histidine kinase containing histidine phosphotransfer domain and ATPase domain, while CepR is a response regulator containing phosphorylation domain and DNA binding domain, function analysis showed that mutation of cepRS, or each gene individually, leads to a significant reduction in cephamycin C production but does not affect phenotype or biomass. ΔcepS, ΔcepR, and the double gene deletion mutant ΔcepRS has similar effect on the cephamycin C production, indicating CepRS is a paired TCS and works together. We further determined that CepRS regulates the expression of multiple genes critical for cephamycin C biosynthesis, most notably pcbC, pcbAB, lat, cmcH, cefF, cmcJ, cmcI, cefD, cefE, pcd, and cmcT. Therefore, CepRS may be a specific TCS for the regulation of cephamycin C biosynthesis, while the functions of many other TCSs remain unclear.
Possible mechanisms for CepRS regulation of cephamycin C biosynthesis
In the cephamycin C biosynthetic gene cluster of S. clavuligerus, a strong bidirectional promoter located between cefD and cmcI could give rise to two polycistronic transcripts: cefD-cefE-pcd and cmcI-cmcJ-cefF-cmcH (Santamarta et al. 2011). Our RT-qPCR data revealed that the cefD-cefE-pcd and cmcI-cmcJ-cefF-cmcH genes in S. clavuligerus strain F613-1 likely form two polycistronic transcripts because the expression levels of the grouped genes are almost identical (Fig. 3). In addition, RT-qPCR data also revealed that CepR negatively regulated the resistance gene orf10. The orf10 expression level was affected by CcaR (Santamarta et al. 2011), also affected by pimM (Martínez-Burgo et al. 2019), the presence of regulators CcaR, PimM and CepR imply that the biosynthesis of cephamycin C is intricately regulated.
Additionally, the cmcI-cefD bidirectional promoter was characterized as a target of CepR in S. clavuligerus F613-1 (Fig. 4). Therefore, CepR may simultaneously control the expression of early- (pcd), middle- (cefD, cefE), and late-stage (cmcI, cmcJ, cefF, cmcH) genes of the cephamycin C biosynthetic pathway. In addition, CcaR reportedly also binds to the cmcI-cefD bidirectional promoter (Santamarta et al. 2002); however, our RT-qPCR data revealed that CepRS does not affect ccaR transcription, indicating that CepRS and CcaR may independently regulate cephamycin C production. Further, our RT-qPCR data revealed that expression of pcbC, pcbAB, and lat was also significantly reduced in ΔcepRS compared with that in F613-1. CepR could not bind to the intergenic fragments used in the EMSA assays of the three genes: pcbC, pcbAB, and lat; however, a binding sequence located upstream of probes used in this work cannot be excluded. It has been reported that the response regulator AfsR binds to afsS after phosphorylation in Streptomyces coelicolor (Lee et al. 2002). But we found that the response regulator CepR did not bind to promoter region of cepS when we analyzed the binding targets of CepR in EMSA.
The pathway specific regulator CcaR belongs to the family of SARP proteins and positively regulate the production of cephamycin C (Pérez-Llarena et al. 1997; Santamarta et al. 2011, 2002). CcaR were reported to be bind to the cefD-cmcI intergenic region (Santamarta et al. 2011). Like CcaR, CepR also positively regulate the production of cephamycin C and could interact with cefD-cmcI intergenic region (P3). A direct repeat sequence GGCGGTCGATCGGCGGT in P3 region was found by hand, which might be a CepR box, while the significance of these sequences will remain unclear until footprinting experiments confirm their binding to CepR. In addition, the presence of two positive regulators CcaR and CepR suggesting that the biosynthesis of cephamycin C has a complex regulation mechanism.
CepRS does not affect clavulanic acid biosynthesis
Because genes involved in the biosynthesis of cephamycin C and CA form a supercluster (Ward and Hodgson 1993), we also examined whether CepRS affects CA production in S. clavuligerus F613-1. HPLC was carried out to analyze the concentration of CA in ΔcepRS and F613-1 in SCF fermentation medium (Additional file 1: Figure S2). HPLC results revealed that there are no obvious differences in CA concentrations between strains ΔcepRS and F613-1 (Additional file 1: Figure S2). There are three clusters of genes involved in CA biosynthesis in S. clavuligerus: the CA biosynthetic gene cluster, the clavam gene cluster, and the paralogue gene cluster (Fu et al. 2019). RT-qPCR-based analysis of the effects of CepRS on the expression of the CA biosynthetic genes failed to identify any significant changes in the expression of these genes in the ΔcepRS mutant compared with wild-type strain F613-1 (Additional file 1: Figure S3). The RT-qPCR data are also consistent with HPLC results showing no obvious differences in CA concentrations between strains ΔcepRS and F613-1. Therefore, these findings suggest that CepRS mainly regulates the production of cephamycin C and has little effect on CA production. As such, it may be possible to increase CA production by modifying CepRS, providing an approach for metabolic engineering efforts for CA production by S. clavuligerus F613-1 in future.
Additional files
Additional file 1: Table S1. Plasmids and strains used in this study. Table S2. Primers used in this study. Figure S1. (A) Polymerase chain reaction-based verification of the ΔcepS and ΔcepR strains. M: DNA marker. (B) Phenotype of F613-1, △cepRS, △cepR, △cepS and △cepRS on BSCA solid media. (C) Ceph C (cephamycin C) obtained in F613-1, △cepRS, △cepR and △cepS strains on TSA solid medium. *, statistically significant difference (P < 0.05) between △cepRS, △cepR, △cepS and F613-1 at the same time point. Figure S2. Biomass and clavulanic acid (CA) concentration of F613-1, △cepRS and the complemented strain. A: Growth curves of F613-1, △cepRS and △cepRScom in SCF fermentation medium. Samples for growth curve analysis were harvested at five time points (1, 3, 5, 7 and 9 d). Data are the mean ± SD of three independent experiments. B: Analysis of the change of CA concentration during fermentation. Data are the mean ± SD of three independent biological experiments. Figure S3. Expression of genes in the CA biosynthetic gene cluster were examined by RT-qPCR between F613-1 (black bars), △cepRS (grey bars) and △cepRScom (dark grey bars). Results were normalized for 16S rRNA gene content and are shown as fold change over the F613-1 control. Data are the mean ± SD of three independent biological experiments. Figure S4. A: Phenotype of F613-1, △cepRS, △cepRScom and △cepRS-cepRS (over-expression strain) on BSCA solid media. B: Cephamycin C obtained in F613-1, △cepRS and △cepRS-cepRS (over-expression strain) strains on TSA solid medium. *, compared with F 613-1, P < 0.05. Data are the mean ± SD of three independent biological experiments. C: Expression of genes in the cephamycin C biosynthetic gene cluster were examined by RT-qPCR between F613-1 (black bars), △cepRS (grey bars) and △cepRS-cepRS (over-expression strain) (dark grey bars). Results were normalized for 16S rRNA gene content and are shown as fold change over the F613-1 control. *, compared with F613-1, P < 0.05. Data are the mean ± SD of three independent biological experiments.
Acknowledgements
Not applicable.
Abbreviations
- TCS
two-component system
- CepRS
cephamycin regulator/sensor
- CA
clavulanic acid
- EMSA
electrophoretic mobility shift assay
- HPLC
high performance liquid chromatography
- RT-qPCR
real-time quantitative polymerase chain reaction
Authors’ contributions
GC designed the work and RQ executed the experiments. JF analyzed the data and prepared the manuscript draft, submission and revised the manuscript. GZ and CZ carried out the interpretation of data, drawing up figures and statistical analysis. PZ and NK helped in data analysis. XQ contributed to the experimental design, manuscript preparation. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31700065), the Shandong Provincial Natural Science Foundation (No. ZR2017BC040), and the Innovation Project of the Shandong Academy of Medical Sciences (No. 201604).
Availability of data and materials
Please contact author for data requests.
Ethics approval and consent to participate
No animal or human subjects were used in this work.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiafang Fu and Ronghuo Qin have contributed equally
Contributor Information
Jiafang Fu, Email: mefujiafang@163.com.
Ronghuo Qin, Email: qinronghuo@139.com.
Gongli Zong, Email: zonggongli.mf@163.com.
Chuanqing Zhong, Email: zhongchuanqing@sdjzu.edu.cn.
Peipei Zhang, Email: zhangpei-yes@163.com.
Ni Kang, Email: kangni15891448943@163.com.
Xiaoyu Qi, Email: qixiaoyu2006@sdjzu.edu.cn.
Guangxiang Cao, Phone: +86 531 8291 9606, Email: caozhong0402@163.com.
References
- Álvarez-Álvarez R, Rodríguez-García A, Santamarta I, Pérez-Redondo R, Prieto-Domínguez A, Martínez-Burgo Y, Liras P. Transcriptomic analysis of Streptomyces clavuligerus DeltaccaR::tsr: effects of the cephamycin C-clavulanic acid cluster regulator CcaR on global regulation. Microb Biotechnol. 2014;7(3):221–231. doi: 10.1111/1751-7915.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma JJ, Groisman EA. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 2003;11(8):359–366. doi: 10.1016/S0966-842X(03)00176-8. [DOI] [PubMed] [Google Scholar]
- Cao G, Zhong C, Zong G, Fu J, Liu Z, Zhang G, Qin R. Complete genome sequence of Streptomyces clavuligerus F613–1, an industrial producer of clavulanic acid. Genome Announc. 2016;4(5):e01020. doi: 10.1128/genomeA.01020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater KF. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- Coque JJ, Enguita FJ, Martin JF, Liras P. A two-protein component 7 alpha-cephem-methoxylase encoded by two genes of the cephamycin C cluster converts cephalosporin C to 7-methoxycephalosporin C. J Bacteriol. 1995;177(8):2230–2235. doi: 10.1128/jb.177.8.2230-2235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque JJ, Perez-Llarena FJ, Enguita FJ, Fuente JL, Martin JF, Liras P. Characterization of the cmcH genes of Nocardia lactamdurans and Streptomyces clavuligerus encoding a functional 3′-hydroxymethylcephem O-carbamoyltransferase for cephamycin biosynthesis. Gene. 1995;162(1):21–27. doi: 10.1016/0378-1119(95)00308-S. [DOI] [PubMed] [Google Scholar]
- Enguita FJ, Liras P, Leitao AL, Martin JF. Interaction of the two proteins of the methoxylation system involved in cephamycin C biosynthesis* Immunoaffinity, protein cross-linking, and fluorescence spectroscopy studies. J Biol Chem. 1996;271(52):33225–33230. doi: 10.1074/jbc.271.52.33225. [DOI] [PubMed] [Google Scholar]
- Fu J, Qin R, Zong G, Liu C, Kang N, Zhong C, Cao G. The CagRS Two-Component System Regulates Clavulanic Acid Metabolism via Multiple Pathways in Streptomyces clavuligerus F613–1. Front Microbiol. 2019;10:244. doi: 10.3389/fmicb.2019.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Zong G, Zhang P, Zhao Z, Ma J, Pang X, Cao G. XdhR negatively regulates actinorhodin biosynthesis in Streptomyces coelicolor *M145. FEMS Microbiol Lett. 2017;364(22):1. doi: 10.1093/femsle/fnx226. [DOI] [PubMed] [Google Scholar]
- Fuqua C. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J Bacteriol. 2006;188(9):3169–3171. doi: 10.1128/JB.188.9.3169-3171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings MI, Hoskisson PA, Chandra G, Buttner MJ. Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2) Microbiology. 2004;150(Pt 9):2795–2806. doi: 10.1099/mic.0.27181-0. [DOI] [PubMed] [Google Scholar]
- Jin X, Cao G, Zhang X, Chen Y, Wang L, Zhong C. Studies on the formation and synthetic mechanism of related substance G in potassium clavulanate production. Braz J Pharm Sci. 2015;51(1):77–83. doi: 10.1590/S1984-82502015000100008. [DOI] [Google Scholar]
- Kieser T, Chater KF, Buttner MJ, Hopwood DA (2000) Practical Streptomyces Genetics: a laboratory Manual
- Kyung YS, Hu WS, Sherman DH. Analysis of temporal and spatial expression of the CcaR regulatory element in the cephamycin C biosynthetic pathway using green fluorescent protein. Mol Microbiol. 2001;40(3):530–541. doi: 10.1046/j.1365-2958.2001.02386.x. [DOI] [PubMed] [Google Scholar]
- Lee PC, Umeyama T, Horinouchi S. afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2) Mol Microbiol. 2002;43(6):1413–1430. doi: 10.1046/j.1365-2958.2002.02840.x. [DOI] [PubMed] [Google Scholar]
- Leite CA, Cavallieri AP, Baptista AS, Araujo ML. Dissociation of cephamycin C and clavulanic acid biosynthesis by 1,3-diaminopropane in* Streptomyces clavuligerus. FEMS Microbiol Lett. 2016;363(1):215. doi: 10.1093/femsle/fnv215. [DOI] [PubMed] [Google Scholar]
- Liras P, Gomez-Escribano JP, Santamarta I. Regulatory mechanisms controlling antibiotic production in Streptomyces clavuligerus. J Ind Microbiol Biot. 2008;35(7):667–676. doi: 10.1007/s10295-008-0351-8. [DOI] [PubMed] [Google Scholar]
- Martín JF, Liras P. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol. 2010;13(3):263–273. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Martínez-Burgo Y, Santos-Aberturas J, Rodriguez-Garcia A, Barreales EG, Tormo JR, Truman AW, Reyes F, Aparicio JF, Liras P. Activation of secondary metabolite gene clusters in Streptomyces clavuligerus by the PimM regulator of Streptomyces natalensis. Front Microbiol. 2019;10:580. doi: 10.3389/fmicb.2019.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, Sivaneson M, Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ Microbiol. 2011;13(7):1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- Nasser W, Reverchon S. New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Anal Bioanal Chem. 2007;387(2):381–390. doi: 10.1007/s00216-006-0702-0. [DOI] [PubMed] [Google Scholar]
- Ogura Mitsuo. Recent progress in Bacillus subtilis two-component regulation. Frontiers in Bioscience. 2002;7(1-3):d1815. doi: 10.2741/ogura. [DOI] [PubMed] [Google Scholar]
- Paradkar A. Clavulanic acid production by Streptomyces clavuligerus: biogenesis, regulation and strain improvement. J Antibiot. 2013;66(7):411–420. doi: 10.1038/ja.2013.26. [DOI] [PubMed] [Google Scholar]
- Patankar AV, González JE. Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev. 2009;33(4):739–756. doi: 10.1111/j.1574-6976.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- Pérez-Llarena FJ, Liras P, Rodríguez-García A, Martín JF. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both beta-lactam compounds. J Bacteriol. 1997;179(6):2053–2059. doi: 10.1128/jb.179.6.2053-2059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R, Zhong C, Zong G, Fu J, Pang X, Cao G. Improvement of clavulanic acid production in Streptomyces clavuligerus F613–1 by using a claR-neo reporter strategy. Electron J Biotechnol. 2017;28:41–46. doi: 10.1016/j.ejbt.2017.05.002. [DOI] [Google Scholar]
- Sambrook JEFF, J. Maniatis. (1989) Molecular cloning: a laboratory manual, 2nd ed.
- Santamarta I, López-García MT, Kurt A, Nardiz N, Álvarez-Álvarez R, Pérez-Redondo R, Martín JF, Liras P. Characterization of DNA-binding sequences for CcaR in the cephamycin–clavulanic acid supercluster of Streptomyces clavuligerus. Mol Microbiol. 2011;81(4):968–981. doi: 10.1111/j.1365-2958.2011.07743.x. [DOI] [PubMed] [Google Scholar]
- Santamarta I, Rodríguez-García A, Pérez-Redondo R, Martín JF, Liras P. CcaR is an autoregulatory protein that binds to the ccaR and cefD-cmcI promoters of the cephamycin C-clavulanic acid cluster in Streptomyces clavuligerus. J Bacteriol. 2002;184(11):3106–3113. doi: 10.1128/JB.184.11.3106-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudagar PS, Survase SA, Singhal RS. Clavulanic acid: a review. Biotechnol Adv. 2008;26(4):335–351. doi: 10.1016/j.biotechadv.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Subramoni S, González JF, Johnson A, Pechy-Tarr M, Rochat L, Paulsen I, Loper JE, Keel C, Venturi V. Bacterial subfamily of LuxR regulators that respond to plant compounds. Appl Environ Microbiol. 2011;77(13):4579–4588. doi: 10.1128/AEM.00183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni S, Venturi V. LuxR-family 'solos': bachelor sensors/regulators of signalling molecules. Microbiology. 2009;155(Pt 5):1377–1385. doi: 10.1099/mic.0.026849-0. [DOI] [PubMed] [Google Scholar]
- Ward JM, Hodgson JE. The biosynthetic genes for clavulanic acid and cephamycin production occur as a 'super-cluster' in three Streptomyces. FEMS Microbiol Lett. 1993;110(2):239–242. doi: 10.1111/j.1574-6968.1993.tb06326.x. [DOI] [PubMed] [Google Scholar]
- Yu Z, Zhu H, Dang F, Zhang W, Qin Z, Yang S, Tan H, Lu Y, Jiang W. Differential regulation of antibiotic biosynthesis by DraR-K, a novel two-component system in Streptomyces coelicolor. Mol Microbiol. 2012;85(3):535–556. doi: 10.1111/j.1365-2958.2012.08126.x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Teng K, Wang J, Zhang Z, Zhang J, Sun S, Li L, Yang X, Zhong J. CerR, a single-domain regulatory protein of the LuxR family, promotes cerecidin production and immunity in Bacillus cereus. Appl Environ Microbiol. 2018;84:5. doi: 10.1128/AEM.02245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhao Z, Li H, Chen XL, Deng Z, Bai L, Pang X. Production of the antibiotic FR-008/candicidin in Streptomyces sp FR-008 is co-regulated by two regulators, FscRI and FscRIV, from different transcription factor families. Microbiology. 2015;161(Pt 3):539–552. doi: 10.1099/mic.0.000033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Plasmids and strains used in this study. Table S2. Primers used in this study. Figure S1. (A) Polymerase chain reaction-based verification of the ΔcepS and ΔcepR strains. M: DNA marker. (B) Phenotype of F613-1, △cepRS, △cepR, △cepS and △cepRS on BSCA solid media. (C) Ceph C (cephamycin C) obtained in F613-1, △cepRS, △cepR and △cepS strains on TSA solid medium. *, statistically significant difference (P < 0.05) between △cepRS, △cepR, △cepS and F613-1 at the same time point. Figure S2. Biomass and clavulanic acid (CA) concentration of F613-1, △cepRS and the complemented strain. A: Growth curves of F613-1, △cepRS and △cepRScom in SCF fermentation medium. Samples for growth curve analysis were harvested at five time points (1, 3, 5, 7 and 9 d). Data are the mean ± SD of three independent experiments. B: Analysis of the change of CA concentration during fermentation. Data are the mean ± SD of three independent biological experiments. Figure S3. Expression of genes in the CA biosynthetic gene cluster were examined by RT-qPCR between F613-1 (black bars), △cepRS (grey bars) and △cepRScom (dark grey bars). Results were normalized for 16S rRNA gene content and are shown as fold change over the F613-1 control. Data are the mean ± SD of three independent biological experiments. Figure S4. A: Phenotype of F613-1, △cepRS, △cepRScom and △cepRS-cepRS (over-expression strain) on BSCA solid media. B: Cephamycin C obtained in F613-1, △cepRS and △cepRS-cepRS (over-expression strain) strains on TSA solid medium. *, compared with F 613-1, P < 0.05. Data are the mean ± SD of three independent biological experiments. C: Expression of genes in the cephamycin C biosynthetic gene cluster were examined by RT-qPCR between F613-1 (black bars), △cepRS (grey bars) and △cepRS-cepRS (over-expression strain) (dark grey bars). Results were normalized for 16S rRNA gene content and are shown as fold change over the F613-1 control. *, compared with F613-1, P < 0.05. Data are the mean ± SD of three independent biological experiments.
Data Availability Statement
Please contact author for data requests.