Abstract
The spontaneous self-assembly of α-synuclein (α-syn) into aggregates of different morphologies is associated with the development of Parkinson’s disease. However, the mechanism behind the spontaneous assembly remains elusive. The current study shows a novel effect of phospholipid bilayers on the assembly of the α-syn aggregates. Using time-lapse atomic force microscopy, it was discovered that α-syn assembles into aggregates on bilayer surfaces, even at the nanomolar concentration range. The efficiency of the aggregation process depends on the membrane composition, with the greatest efficiency observed for of 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS). Importantly, assembled aggregates can dissociate from the surface, suggesting that on-surface aggregation is a mechanism by which pathological aggregates may be produced. Computational modeling revealed that dimers of α-syn assembled rapidly, through the membrane-bound monomer on POPS bilayer, due to an aggregation-prone orientation of α-syn. Interaction of α-syn with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) leads to a binding mode that does not induce a fast assembly of the dimer. Based on these findings, we propose a model in which the interaction of α-syn with membranes plays a critical role initiating the formation of α-syn aggregates and the overall aggregation process.
Keywords: α-synuclein, amyloid aggregation, lipid bilayer, Parkinson’s disease, time-lapse AFM, computer modeling
1. Introduction
Self-assembly of amyloidogenic peptides and proteins is associated with the development of a large number of neurodegenerative diseases, including Parkinson’s disease (PD). PD is associated with the presence of cytosolic inclusions, named Lewy bodies (LBs), that contain amyloid-type fibrils typically localized to presynaptic terminals. [1] These fibrils are assembled from the α-synuclein (α-syn) protein, which can spontaneously self-assemble into amyloid aggregates in solution. [2] In addition to PD, α-syn is associated with the development of several other neurodegenerative diseases, including LB dementia and Alzheimer’s disease; however, the aggregation mechanism leading to the disease-prone state remains elusive. [3–5].
The self-assembly process of amyloid proteins, including α-syn, has led to the amyloid cascade hypothesis (ACH), which posits that the spontaneous assembly of amyloidogenic polypeptides is the key feature defining the disease state. [6–8] Numerous data support the ACH hypothesis, and it remains the theory on which in vitro and in vivo studies related to the molecular mechanisms of amyloid aggregation are performed. However, there is a serious complication in translating the current knowledge on amyloid aggregation in vitro to the aggregation process in vivo. Namely, the concentrations of amyloidogenic polypeptides dramatically differ in vitro compared to in vivo. For example, the critical concentration for the spontaneous aggregation of α-syn in vitro is in the high micromolar range, in stark contrast to the picomolar concentration range for α-syn in the cerebrospinal fluid (CSF). [9] While α-syn is indeed an abundant protein in presynaptic vesicles, evidence is missing that proves the disease state is initiated in this segment of neurons. The immunohistochemical stains of LB dementia and PD brain samples show accumulation of aggregated α-syn in the entire neuronal body, where the α-syn concentration would resemble the average concentration in the brain, ~10 nM. [10] Furthermore, we discovered a novel aggregation pathway that essentially eliminates the disparity between in vivo and in vitro concentrations found in ACH; in the presence of a surface, using time-lapse atomic force microscopy (AFM), we observed the spontaneous assembly of amyloid beta (Aβ) peptides and α-syn proteins in the nanomolar concentration range. [11] Based on our findings, we hypothesized that a similar on-surface aggregation mechanism is possible on membrane surfaces.
Monomeric α-syn interacts with membranes by binding to phospholipid molecules as part of its normal function. [12, 13] Past reports suggest that the normal membrane-binding function of α-syn is related to regulation of synaptic vesicle trafficking. [12–14] The interaction of α-syn with lipid membranes has been shown to elicit changes in the protein structure. [13, 15–19] The α-syn protein has also been shown to undergo accelerated aggregation when incubated in the presence of phospholipid vesicles at high protein-to-lipid ratios. [20–22] Additionally, the ability of aggregates to induce changes in the properties of cellular membrane is considered a mechanism underlying the development of PD. [23] One current model posits that neurotoxic effects are associated with membrane permeability, and cell death is mediated by interactions with oligomeric α-syn. [24–29] The key role that membrane-induced α-syn aggregation plays in neurotoxicity, [22] potentially via a mechanism involving membrane permeabilization, [30–33] is in line with our recent finding that surface interactions dramatically facilitate the aggregation process of amyloidogenic proteins, including α-syn. [11] Together, these findings suggest that self-assembly on the surface of cellular membranes is the mechanism by which potentially neurotoxic oligomers are assembled at physiologically relevant protein concentrations. [11].
In the current study, the above hypothesis is tested by direct visualization of α-syn aggregation on the surfaces of supported lipid bilayers (SLBs) using time-lapse AFM. [34] We demonstrate that SLBs catalyze the aggregation of α-syn at concentrations as low as 10 nM, which corresponds to the concentration range of the CSF. [9] The aggregation kinetics were also found to be dependent on the SLB composition, being considerably greater for a bilayer composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) than 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC). The assembled aggregates were not strongly bound to the surface and were capable of spontaneous dissociation into solution. Importantly, the self-assembly process did not cause damage to the surface, as no defects were detected after the aggregates dissociated. Computational modeling suggested that α-syn monomers change conformation upon interaction with the bilayers, and these interactions are dependent on the composition of the bilayer. The conformations of α-syn after binding to POPS dramatically facilitated assembly into the dimer, a property that is in contrast with POPC and in line with experimental data.
2. Results
2.1. Experimental AFM studies of α-syn aggregation on lipid bilayers
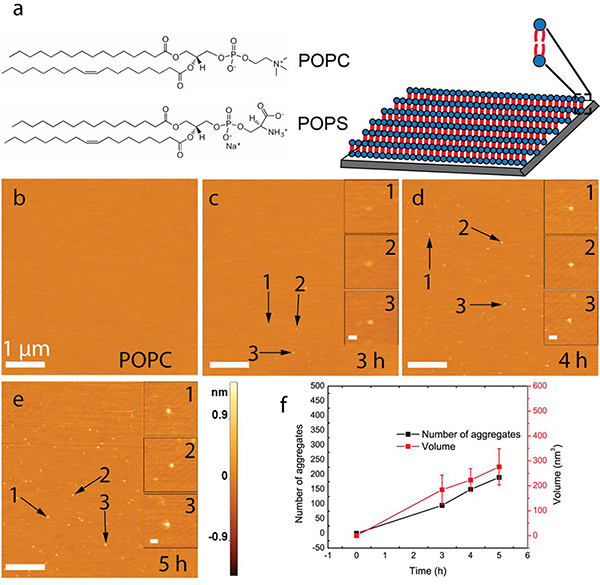
Lipid bilayers were assembled on the surface of freshly cleaved mica to directly visualize interactions between the protein and bilayer over many hours with AFM (see Methods section for details). Based on their prevalence in neuronal cellular membranes, the following three types of bilayers were used, as shown in Figure 1a: POPC, POPS, and an equimolar mixture of the two. Time-lapse AFM studies require that the bilayers are stable during the entire multi-hour AFM experiments. Additionally, the surface needs to be smooth to allow for detection of protein aggregates while they are formed. We developed a procedure to assemble POPC and POPS bilayers, with no defects, over areas of at least 4 μm × 4 μm (Fig. 1b and Fig. S1). Each surface was tested for smoothness and stability prior to AFM studies for the α-syn-membrane interactions, as illustrated in the AFM topographic image in Fig. S1. Only similarly smooth surfaces were used to conduct time-lapse AFM experiments.
Figure 1. Time-lapse AFM images to characterize α-syn aggregation on the POPC supported lipid bilayer (SLB).
(a) POPC and POPS were used in the present study, and their chemical structures are shown (left). Schematic of an SLB on freshly cleaved mica (right) is shown only for displaying the model for the SLB; it does not indicate any phase of the bilayer. (b) The image of the POPC SLB surface immediately after buffer exchange with 10 nM α-syn solution. (c)-(e) Images of the SLB surface taken at time points after adding the protein. Insets show zoomed images of three representative aggregates. (f) A graphical illustration of the evolution of aggregate quantity and mean volume with respect to time. Data are shown as the mean ± SD. The scale bar in panels b-e is 1 μm, while scale bars for the insets are 80 nm, and the Z-scale is shown to the right of panel e.
2.2. α-Syn aggregation on the POPC bilayer
First, we determined how the presence of a POPC bilayer affects the α-syn aggregation. A 10 nM α-syn solution was exchanged with the buffer on a smooth POPC SLB and the bilayer was visualized by time-lapse AFM imaging. Aggregation of α-syn on supported POPC bilayers was investigated over a period of five hours, as shown in Figure 1. Further, Figure 1b shows the POPC surface immediately after exchange of buffer with an α-syn solution at a concentration of 10 nM. Aggregates appeared after three hours of incubation, indicated by arrows in Figure 1c. More aggregates appeared over the next two hours of observation, accompanied by an increase in their sizes (volumes), as shown in Figures 1d and 1e. The number and volume of aggregates were measured and revealed that both parameters gradually increase over time, as shown in Figure 1f. Note that 10 nM solution of α-syn incubated under the same conditions but in the absence of a phospholipid bilayer or mica surface (Fig. S2 a and b) did not show a large number of aggregates. Importantly, this suggests the formation of α-syn dimers due to S-S bonds between C-terminal Cys residues are unlikely because the histograms are essentially identical; the volume of the protein remains indistinguishable from its monomeric form. [35] For further verification, on-surface aggregation (on aminopropyl silatrane, APS- functionalized mica) experiment was performed in presence and absence of dithiothreitol (DTT) (Fig. S2c). The volume of the α-syn aggregates remains essentially similar in both situations, thereby indicating that the on-surface formation of dimers through terminal Cys residues is not likely in our experimental setup.
These results were also compared with the aggregation of α-syn on APS-functionalized mica in which the catalytic effect of surfaces was initially demonstrated. [11] These data are shown in Figure S3. Aggregates formed on APS-mica appeared after three hours of incubation, which was also the case for aggregates formed on the POPC bilayer (Fig. S3e). However, qualitatively, aggregation on APS was less efficient compared to POPC, as measured by the number and size of aggregates (Fig. S3h and Fig. S4).
2.3. α-Syn aggregation on the POPS bilayer
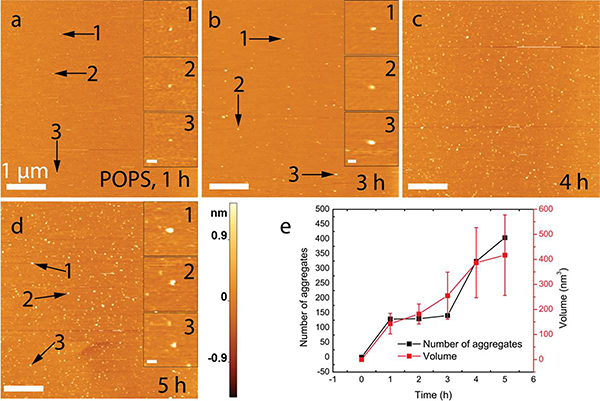
Next we tested how the composition of the bilayer affects the aggregation of α-syn by performing experiments with POPS bilayers. Both POPC and POPS bilayers are composed of hydrocarbon chains, but POPS has a serine head group that renders the surface negatively charged at physiological pH, while POPC has a net neutral charge. AFM images, shown in Figure 2, demonstrate that aggregates appear after one hour of incubation with POPS (Fig. 2a), and the surface is densely covered with α-syn aggregates after five hours incubation (Figs. 2b–2d). Quantitative analysis demonstrated the number of aggregates and their size increase over time (Fig. 2e). Histograms for the volume distribution at each time point are shown in Figure S4 (for more data, see Table 1). Compared with POPC, aggregation on POPS bilayers was accelerated, with the first detectable aggregates appearing after one hour on POPS bilayers compared with three hours on POPC bilayers. Additionally, at the end of the 5 h experiment, the number of aggregates was more than double for POPS (n = 404) compared to that of POPC (n = 190). Furthermore, aggregates formed in the presence of POPS had a larger mean volume (417 nm3 for POPS vs. 276 nm3 for POPC; Fig. S4 and Table 1).
Figure 2. Enhanced aggregation kinetics of α-Syn when applied to a supported POPS bilayer.
(a-d) AFM images acquired at time-points after buffer exchange with 10 nM protein solution. Insets show zoomed images of three representative aggregates for the selected time-points. (e) A graph showing the time-dependent evolution of aggregate quantity and mean volume. The data are shown as the mean ± SD. The scale bar in panels (a-d) is 1 μm, while scale bars for the insets are 80 nm, and the Z-scale is shown to the right of panel d.
Table 1.
Summary of results from three independent experiments. The table shows the volume and number of aggregates on POPC, POPS, and POPC:POPS surfaces at different time intervals.
| POPC | POPS | POPC/POPS | |||||
|---|---|---|---|---|---|---|---|
| Volume (nm3) | Number of aggregate measured | Volume (nm3) | Number of aggregate measured | Volume (nm3) | Number of aggregate measured | ||
| Experiment 1 | 1 h | -- | -- | 143±41 | 129 | -- | -- |
| 2 h | -- | -- | 182±40 | 130 | 137±37 | 82 | |
| 3 h | 184±59 | 95 | 255±94 | 141 | 187±59 | 176 | |
| 4 h | 223±46 | 149 | 387±140 | 324 | 248±87 | 291 | |
| 5 h | 276±73 | 190 | 417±161 | 404 | 312±116 | 372 | |
| Experiment 2 | 1 h | -- | -- | 123±48 | 116 | -- | -- |
| 2 h | -- | -- | 169±42 | 120 | 112±48 | 71 | |
| 3 h | 159±53 | 86 | 227±91 | 272 | 176±73 | 166 | |
| 4 h | 198±45 | 132 | 331±126 | 307 | 209±96 | 272 | |
| 5 h | 228±85 | 172 | 363±165 | 383 | 289±136 | 351 | |
| Experiment 3 | 1 h | -- | -- | 143±53 | 125 | -- | -- |
| 2 h | -- | -- | 212±47 | 140 | 157±46 | 91 | |
| 3 h | 179±82 | 106 | 284±95 | 153 | 200±67 | 187 | |
| 4 h | 240±41 | 161 | 414±142 | 343 | 277±83 | 283 | |
| 5 h | 319±96 | 206 | 460±163 | 425 | 347±113 | 356 | |
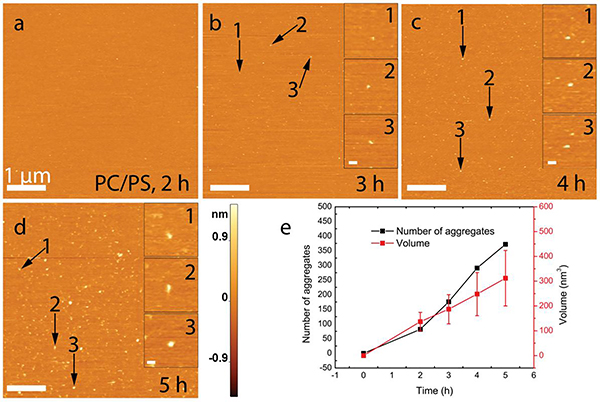
α-Syn aggregation on the POPS:POPC bilayer
The effect of the bilayer composition on the aggregation of α-syn was also examined by using an equimolar mixture of POPC and POPS; the results are shown in Figure 3. For the mixture, aggregates appeared after two hours (Fig. 3a), and their number increase over time (Figs. 3a–3d). Quantitative analysis showed that the number and size of the aggregates grew monotonically over time (Fig. 3e). Although the number of aggregates on POPC:POPS bilayer was close to that of POPS (372 vs 404), the average size of aggregates was much smaller (see panel ‘Experiment 1’ in Table 1; mean volume value of 312 nm3 on POPC:POPS versus 417 nm3 on POPS). Furthermore, the first appearance of aggregates on the equimolar mixture of the POPC:POPS SLB occurred one hour later than for the POPS SLB. The volume distributions at each time point for the POPC:POPS SLB is shown in Figure S4 and Table 1. Direct comparison of aggregates on the different bilayer surfaces reveals that the general aggregation propensity follows this order: POPS > POPC:POPS > POPC (Table 1). Changes in the volume and number of aggregates assembled on the POPC, POPS and POPC:POPS bilayer surfaces are graphically shown in Figure S5.
Figure 3. The aggregation of α-syn on a POPC:POPS SLB.
(a) An AFM image of the initial bilayer immediately after the exchange. (b-d) Corresponding images taken at 1-hour time intervals. Zoomed images of three representative aggregates are shown on the right side of images. (e) A line plot showing the time-dependent evolution of the quantity and mean volume of aggregates. Data are shown as the mean ± SD. The scale bar in panels a-d is 1 μm, while scale bars for the insets are 80 nm, and the Z-scale is shown to the right of panel d.
2.4. Time-lapse observation of the on-surface dynamics of α-syn aggregates
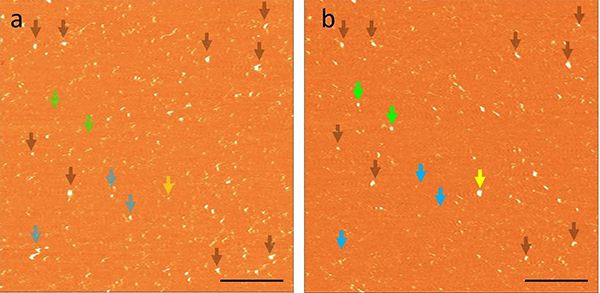
The data from the time-lapse experiments allows us to follow the dynamics of individual aggregates. Figure 4 shows scans of the same area taken during a 30-minute interval on the POPS surface. The aggregates are highlighted with different colored arrows to indicate different types of aggregate dynamics. New aggregates appearing in panel B are highlighted with green arrows. Aggregates that did not change between panels A and B are marked with transparent black arrows. One aggregate on panel A dramatically increased in size in panel B and is highlighted with a yellow arrow. Interestingly, several aggregates highlighted in blue in panel A are not found in panel B, suggesting these aggregates spontaneously dissociated from the surface during the 30-minute interval. Importantly, the surface remained smooth, indicating that no damage occurs to the bilayer surface following dissociation of the aggregates.
Figure 4. The dynamics of α-syn aggregates on a POPS SLB.
(a-b) AFM image captured after 6 h and 6.5 h. Aggregates highlighted with black arrows are features that did not change between frames and demonstrate the absence of drift. Blue arrows in panel a correspond to aggregates that have dissociated in panel b. New aggregates that appeared in panel b are highlighted with green. A growing aggregate is highlighted in yellow. Scale bars are 500 nm.
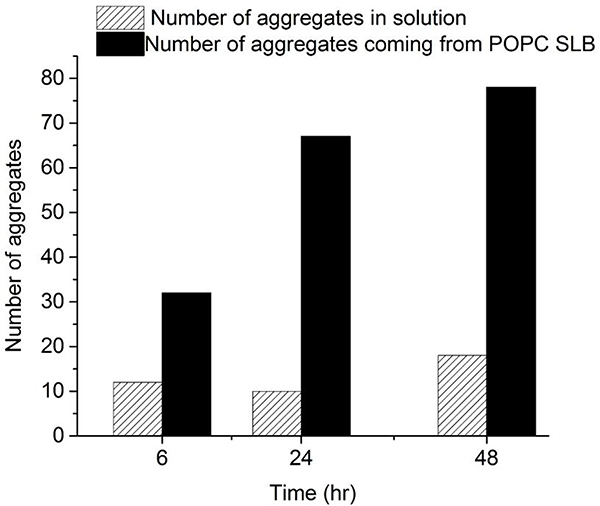
The same spontaneous dissociation was also observed for aggregates assembled in presence of POPC bilayers (Fig. S6). Thus, aggregates assembled on the surface were able to dissociate back into solution, suggesting that aggregates should appear in the bulk solution above the bilayer. This assumption was tested by directly measuring the time-dependent accumulation of α-syn aggregates in the bulk solution above the bilayer surface. To this end, aliquots were taken from the bulk solution, deposited on mica, imaged with AFM, and analyzed for the presence of aggregates. AFM images in Figure S7 clearly demonstrate the accumulation of aggregates in the bulk solution; these data are quantitatively shown in Figure 5. The presence of the bilayers produced significantly more aggregates (solid black bars) compared with the control (dashed bars). These results support the conclusion that aggregates assembled on the lipid bilayer can dissociate into the bulk solution. Note that a similar effect was observed in our recent study, [11] in which the assembly of α-syn aggregates on a mica surface was studied.
Figure 5. Accumulation of α-syn aggregates in solution above the POPC SLB.
A 10 nM α-syn solution was incubated in the presence of a POPC SLB. Aliquots of 5 μl of the solution were taken out at different time points (6 h, 24 h, 48 h) and analyzed by AFM. Solid black bars show the number of aggregates that appeared in the bulk solution above the POPC SLB at different times. In a parallel experiment, a 10 nM α-syn solution was incubated in a test tube; an aliquot of 5 μl was taken out at similar time-points and analyzed by AFM to test aggregation in the absence of a POPC SLB (striped bars). Aggregates were counted in 2 μm × 2 μm AFM images.
2.5. Computational modeling of the interaction of α-syn with lipid bilayers
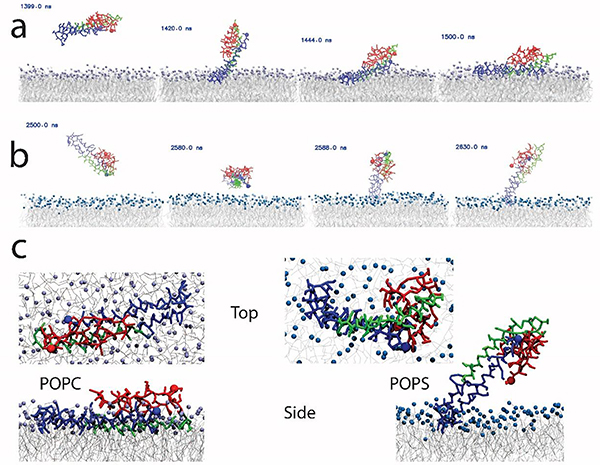
To obtain insight into the underlying molecular mechanism behind the aggregation of α-syn on the bilayer surface, molecular dynamics (MD) simulations were performed to identify interactions of α-syn with the POPC and POPS bilayers. Briefly, a monomer of α-syn was placed 6 nm above the center of a 13 nm × 13 nm bilayer patch (512 lipids), and interactions with the bilayer were then simulated. A few selected snapshots illustrating the dynamics of the interaction of α-syn with the POPC bilayer are shown in Figure 6a. The set of data for the interaction with POPC is assembled as a movie (Movie S1). According to Figure 6a, α-syn initially bound to the POPC membrane through its N-terminal segment (frame (ii)). Over time, the length of the protein segment in contact with the POPC surface increased, so that the non-Aβ component (NAC) segment approached the surface as well (frames (iii-iv)). Graphically, this change in binding is illustrated by the kymograph shown in Figure S8a and b for POPC and POPS, respectively. In fact, α-syn can undergo multiple association-dissociation events, as evidenced by the fluctuations in the number of contacts over time (Fig. S8c). Eventually (after ~1.5 μs, seen as a jump in the graph), the protein strongly interacted with the bilayer and remained bound to the surface until the end of the simulation. Throughout the simulation, the end-to-end distance and the radius of gyration of the α-syn molecule experienced minor fluctuations (Figs. S8d and e).
Figure 6. Molecular dynamics (MD) simulations of the interaction of α-syn with lipid bilayers reveal distinct conformations.
(a-b) The results show stable binding of α-syn to POPC (a) and POPS (b) bilayers; time-resolved stability is presented in Figure S8. (c) Top and side view snapshots are given for the last frame, at 4 μs, of the MD simulations for POPC and POPS, left and right respectively. The α-syn N-terminal segment is colored blue; the NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are colored grey, while the POPC and POPS head groups are colored purple and blue, respectively.
A similar analysis was performed for α-syn interactions with a POPS bilayer. A few selected frames are shown in Figure 6b, and the full set of data for the interaction is assembled as Movie S2. Similar to the data obtained for the POPC bilayer, the N-terminal segment of α-syn bound to the membrane surface, but unlike POPC, the interaction with POPS was limited to a short central region (G36-K58) of the protein, graphically illustrated in Figure S8b. As a result, the protein remained extended out of the plane of the POPS surface.
To account for the structural diversity reported for α-syn, we performed MD simulations using two additional conformations of α-syn, obtained from all-atom discrete MD (DMD) simulations [36] and from coarse-grained simulations using a random coil criterion (see methods), Figure S9a (i) and (ii) respectively. The DMD structure is similar to the previous structure, but with a more extended α-helical region as well as a more flexible C-terminal; these two factors contribute to the rapid interaction with both POPC and POPS, Figure S9b(i) and c(i) respectively. However, unlike the previous simulations with POPC, the α-syn monomer only interacts with the bilayer through a small section of the N-terminus and assumes an orientation similar to that for POPS (on Figure 6), Figure S9b(i) and Movie S3. Interactions with POPS are initiated through the same N-terminus region as interactions with POPC but later grow to include the entire N-terminal region as well as the NAS, Figure S9c(i) and Movie S4. These interaction lead to the insertion of the monomer into the bilayer interfacial region with the C-terminal region extending out of the bilayer, Movie S5.
The α-syn conformation obtained through coarse-grained simulation is largely unstructured and has a compact shape and, unlike previous simulations, rarely interacts with POPC (Figures S9a(ii) and b(ii)). Similar decrease in interaction is seen in presence of POPS, and although initial interactions happen through the same N-terminal region (residues 30–50) neither the full N-terminal region nor the NAC region remain in contact with the membrane as previous (Figure S9c(ii)). To determine the underlying cause, we performed all-atom simulations using the same initial structure and the results are shown in Figure S10. In presence of POPC, the α-syn monomer sporadically interacts with the bilayer and undergoes structural change, initially adopting small helical segments in the N-terminal region before gaining small β-strands, as well as changes shape, becoming more elongated, Movie S6 and S7. On the other hand, in presence of POPS, once the monomer interacts with the bilayer, through the N-terminal region, it stays connected and undergoes similar change as was seen on POPC, Movie S8. Furthermore, the monomer orientation on POPS is similar to that on Figure 6.
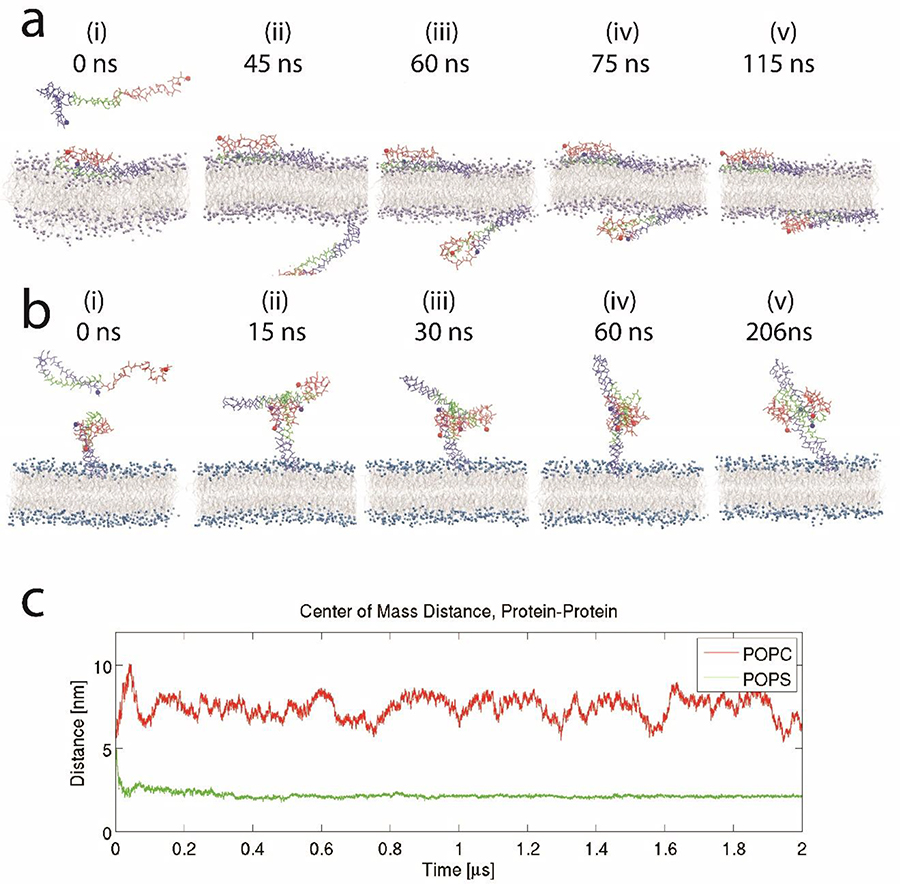
The interaction of membrane-bound α-syn was then modeled using a second free-α-syn molecule; the results are shown in Figure 7. Frame (i) in Figure 7a shows the second protein floating around the α-syn bound to the POPC surface, but later (frame ii) it moves away from the bound protein and binds to the other side of the bilayer, gradually increasing the number of segments interacting with the bilayer, as shown in frames (iii) to (iv). An animation of the dynamics is assembled as Movie S9.
Figure 7. Results of molecular dynamics (MD) simulations showing interaction between free and membrane-bound α-syn.
(a) Binding of a free α-syn to the POPC membrane; the free α-syn traverses through the periodic boundary to the inner leaflet and stably binds; the mode of interaction is similar to the initial α-syn interaction shown in Figure 6a. (b) On the POPS membrane, the free α-syn rapidly binds membrane-bound α-syn through NAC-NAC and NAC-C-terminal interactions. (c) Center of mass distance between the two α-syn molecules for the POPC and POPS systems. For POPC, after the transition through the periodic boundary, the fluctuations in the center of mass distance reflect the diffusion of the proteins in the XY-plane. The α-syn N-terminal segment is colored blue; the NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are colored grey, while the POPC and POPS head groups are colored purple and blue, respectively.
Similar MD simulations with the POPS bilayer produced entirely different results compared with POPC. As described in Figure 7b, a free protein, shown in frame (i), rapidly binds to membrane-bound extended α-syn, and the dimer is rapidly formed, after only 15 ns, shown in frame (ii). These events occurred due to interactions of the NAC segments of the two proteins as well as by interactions of the NAC-C-terminal. The proteins in frame (iii) rearranges into a parallel orientation with an extended NAC-NAC interaction interface (frames (iv)-(v)), and the dimer remains stable for the remainder of the simulation. The center of mass (CoM) distance plot, Figure 7c, further corroborates these observations, with the distance for POPS quickly stabilizing to approximately 2.5 nm, while the distance for POPC is large and fluctuates around 7 nm, which is equal to the thickness of the bilayer plus contributions from the position on the bilayer surface (2D diffusion). This dynamic process is presented as Movie S10. Furthermore, geometric analysis of the proteins (Fig. S11) demonstrates that interactions within the dimer (on POPS) primarily occurs between the NAC and C-terminal segments of the membrane-bound protein and the second α-syn molecule.
3. Discussion
Overall, our studies demonstrate that phospholipid bilayers catalyze the aggregation of α-syn at conditions where no aggregates are assembled in bulk solution. The aggregation process was directly observed using time-lapse AFM, which showed that the number and size of aggregates increase gradually with the incubation time. The efficient assembly of aggregates on phospholipid bilayers is in line with other studies, [37–39] which reported accelerated formation of α-syn fibrils on phospholipid vesicles, although much larger concentrations of α-syn were typically used. Moreover, our studies reveal a number of important features for the self-assembly process catalyzed by the membrane bilayers.
The aggregation efficiency is dependent on the phospholipid composition, with the general aggregation propensity conforming to the following order: POPS > POPC:POPS > POPC (Table 1 and Fig. S5). POPS is an anionic phospholipid, which suggests electrostatic interactions between the negatively charged surface and the positively charged lysine-rich segment of α-syn can contribute to the catalytic effect this surface, which is in line with previously reported data. [23] In turn, this initial interaction increases the local concentration of the protein on the bilayer surface and accelerates the aggregation process. Importantly, previously reported findings suggest that the levels of anionic lipids in the brain increase with age [40] and that the ratio of acidic to zwitterionic phospholipids further increases in the PD brain. [41] Based on these data and our observations, we hypothesize that amyloidogenic aggregates of α-syn assemble on cellular membranes and that the membrane composition is the factor that controls the aggregation process. [22, 33] Recent studies are in line with our observation, where the role of membrane composition has been identified as a major factor for different types of amyloid proteins towards disease development. The specific role of GM1 ganglioside as a binding target of α-syn and, at the same time fibril formation inhibitor, along with the other results (reviewed in [42]) reveal the importance of membrane composition for PD neuropathology. The presence of GM1 ganglioside has been shown to also induce formation of toxic Aβ42 oligomers [43]. Additionally, interactions of GM1 with HypF-N oligomers has been shown to lead the cytotoxicity[44].
Computer modeling provides insights into differences for α-syn interactions with POPC and POPS bilayers. According to Figures 6a and 6b, α-syn binds differently to both surfaces. In the case of POPC, α-syn initially binds using its N-terminal segment, but it makes multiple contacts with the surface by different segments “spreading” on it. On the contrary, α-syn remains bound to POPS surface by its N-terminal segment “popping-out” from the surface over entire simulation period. Similar interactions through the N-terminal region are seen for different initial conformations of the α-syn monomer (Figure S9). However, while interactions with POPC do occur, these conformations show more extended interactions with the POPS surface. Common for all initial conformations of α-syn is the orientation of the monomer on the bilayer surface, with interactions and binding primarily occurring through the residues of the N-terminal region and leaving the C-terminal oriented away from the surface and freely accessible. As a result of this arrangement, a second monomer can rapidly find the membrane-bound α-syn monomer and form a stable dimer (Fig. 7b and movie S9). No such rapid interactions were detected for α-syn bound to the POPC surface (Fig. 7a and movie S10). The POPC-bound monomer remains undetected by the second monomer as it passes the periodic boundary and binds to the surface on the other leaflet, far from the first monomer. Note that the initial distances between the monomers for POPC and POPS simulations were the same. The different modes of α-syn interaction with lipid bilayers is in line with past studies [17, 45–48] that examined α-syn binding to lipid vesicles. Interestingly, an extended conformation with exposed NAC region has been reported, [46] which is similar to the conformation of α-syn bound to POPS in our study. Thus, the binding modes of α-syn to POPC and POPS can help explain differences in the aggregation propensity for the protein on these two bilayers.
It has been widely discussed in literature that interactions of amyloid proteins, including α-syn, with lipid bilayers are accompanied by changes in the bilayer structure and even the disruption of the bilayer. [15, 49, 50] The formation of channel-like features assembled by amyloid protein oligomers has also been reported. [26, 27] However, in the present study, we did not observe such changes in the bilayers. As shown in Figure 4, aggregates highlighted with blue dissociated and did not appear in frame B; however, no damage to the bilayer surface is seen in the images prior to and after dissociation of the aggregate. An explanation for this difference may be the concentration of the α-syn protein. For example, α-syn pores were previously obtained [26] using α-syn concentrations at six orders of magnitude greater than in our experiments. Note as well, these pores were observed on bilayers assembled from a mixture of lipids and protein (20:1 weight ratio). We used a very different approach - our bilayers were assembled prior to addition of α-syn. This setup is designed to better mimic the in vivo interaction of α-syn with the cell membrane. In experiments by others using preassembled bilayers, the α-synuclein concentration was also six orders of magnitude greater than that of our studies. [51] As a result, micrometer-sized clusters were observed that led to membrane damage. An additional explanation for the discrepancy could be the presence of defects on the bilayer. For example, α-syn aggregates are reported to bind packing defects, and induce lateral expansion of lipid molecules that progress further to bilayer remodeling by insertion of α-syn into the headgroup region. [52] In our study, we developed a procedure by which the bilayers remain defect-free during the entire time-lapse experiment (Fig. S1). This model is in line with the data of Chaudhary and coworkers, [53] in which homogeneous bilayers remain intact despite the formation of α-syn oligomers.
We also find that the self-assembly of aggregates on the membrane bilayers is a dynamic process. The aggregates grow gradually, and some of them can dissociate from the surface into the bulk solution (Fig. 4). This process leads to the accumulation of aggregates in the solution, as supported by direct measurements (Fig. 5). Thus on-surface aggregation is the mechanism by which amyloid aggregates are assembled regardless of very low concentration of the protein and delivered to the solution. This is the surface mediated catalysis of amyloid aggregation that may have important biological significance. Note that at our experimental conditions (low concentration of α-syn) the surface assembled aggregates are oligomers, which are considered to be the most neurotoxic amyloid aggregates.
4. Conclusion
In conclusion, our studies reveal the ability of membrane surfaces to catalyze the assembly of α-syn aggregates at such low concentrations that no aggregation in bulk solution is observed. Importantly, the aggregation catalysis of the bilayer surfaces depends on the phospholipid type. Computational modeling revealed different modes of interactions between α-syn and bilayers that may explain the elevated catalytic activity of POPS compared with POPC surface. The aggregates assembled on the membrane surfaces can dissociate into solution suggesting that on-membrane aggregation is the mechanism by which the overall aggregation process may not only be initiated, but rather the aggregation process is further maintained by the on-membrane production of amyloid aggregates. The observation that aggregation occurs at low concentrations of α-syn, which eliminates potential need for the high concentration of the protein, as it is required for experiments in vitro. Altogether, based on the data obtained we hypothesize that the membrane aggregation is the mechanism by which the disease-prone state is initiated. We also posit that the change of the membrane composition to one that facilitates the on-surface aggregation process is a factor that may contribute to triggering of the disease state. Although this hypothesis is supported by the data on the age-related change of the membrane composition towards increase POPS concentration, [40, 41] direct data are needed to prove the proposed model.
5. Experimental Section
Materials
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) were purchased from Avanti Polar Lipids, Inc, (Alabaster, AL); Chloroform ((> 99.5%, Sigma–Aldrich Inc.); dry bath incubator (Fisher Scientific); a 10 mM pH 7.4 sodium phosphate buffer (PBS, NaH2PO4•H2O: Na2HPO4 = 1:3.4 without additional salt) was prepared and filtered through a disposable Millex-GP syringe filter unit (0.22 μm) before use. Deionized (DI) water (18.2 MΩ, 0.22 μm pore size filtered, APS Water Services Corp., Van Nuys, CA) was used for all experiments. Muscovite mica (Asheville Schoonmaker Mica Co., Newport News, VA). 1-(3-aminopropyl)silatrane was synthesized as previously described, [54] ImmEdge hydrophobic barrier pen (Vector Laboratories, Inc. Burlingame, CA); Aron alpha industrial glue (Toagosei America, West Jefferson, OH); S/P Brand Bev-L-Edge micro glass slides (Allegiance Healthcare Corporation, McGaw Park, IL).
Preparation of α-syn solution
Wild-type A140C α-syn in which the C-terminal alanine was replaced with a cysteine was prepared as described previously. [55] α-Syn solutions were freshly prepared by dissolving 0.4 to 0.8 mg of the lyophilized powder in 200 μL water (the pH was adjusted to 11 with NaOH), in the presence of 1 μL of 1 M dithiothreitol (DTT) to break disulfide bonds, followed by the addition of 300 μL of 10 mM sodium phosphate buffer (pH 7.4). The solution was filtered through an Amicon filter with a molecular weight cutoff of 3 kDa at 14,000 rpm for 15 min. The filtration was repeated three times to completely remove free DTT. The concentration of the stock solutions was determined by spectrophotometry (Nanodrop® ND-1000, Wilmington, DE) using the molar extinction coefficients 1280 cm−1·M−1 and 120 cm−1·M−1 for tyrosine and cysteine at 280 nm, respectively. In general, freshly prepared stock solutions were used for all the experiments. No significant dimerization was observed when 10 nM protein was kept incubated for 6 h in solution (Fig. S2).
Preparation of APS-mica
Freshly cleaved mica strips (5.0 × 1.5 cm, L×W) were immersed in plastic cuvettes containing 167 μM APS solution for 30 min, [35, 55] followed by rinsing with deionized water and drying in argon flow. The APS-mica was stored in a vacuum chamber for use over the following few weeks. [56] The APS-mica strips were cut into ~1.5 × 1.5 cm pieces and glued to a glass slide for sample preparation.
Preparation of SLBs
We followed a published protocol with minor modifications. [57] Lipid powder (25 mg) was first dissolved in 1 mL chloroform to make a 25 mg/mL stock solution. The stock solution was aliquoted and stored at −20°C. The aliquoted solution (20 μL) was thawed and brought to room temperature before it was blow dried with a gentle argon flow and vacuum desiccated overnight. Next, a 1 mL solution of 10 mM sodium phosphate buffer (pH 7.4) was injected into the glass container to make a 0.5 mg/mL solution, unless a different concentration was stated. The solution was sonicated (Branson 1210, Branson Ultrasonics, Danbury, CT) until mostly clear to obtain small unilamellar vesicles. A mica piece mounted on a glass slide was then prepared for experiments by drawing around the perimeter of the mica with a hydrophobic pen to prevent overflow. A range of 60 μL to 80 μL of the lipid solution was deposited onto the freshly cleaved mica surface. The lipid solution was incubated at 60°C while buffer was supplied periodically. After 1 h, excess lipids were washed off by extensive gentle exchange of the lipid solution with buffer. The resulting SLB was kept in buffer and imaged.
In situ time-lapse AFM imaging in liquid
AFM imaging was conducted on an MFP-3D (Asylum Research, Santa Barbara, CA). Tapping mode was used. A MSNL cantilever (Bruker, Santa Barbara, CA) with a nominal spring constant of 0.1 N/m was used for imaging in liquid. The resonance frequency varied between 7 kHz to 9 kHz.
SLBs were scanned at different magnifications to establish homogeneity before the final scan size was set to 5 μm × 5 μm and experiments commenced at 1 Hz scan rate. After the bilayer surface was characterized, the buffer was exchanged with a solution containing 10 nM monomeric α-synuclein. For an area of 1 μm x1 μm this approximately yields ~106 phospholipid molecules and ~10 molecules of α-syn in the volume (1 μm × 1 μm ×1 μm) above the area. Images were acquired with 512 pixels per line resolution.
Buffer was injected periodically to keep the sample at a constant volume. 10 mM sodium phosphate, pH 7.4 has been used in the time-lapse experiments, except in one situation, where 1 mM DTT has been incorporated in the imaging buffer to find out the effect of DTT (Fig. S2c). For in situ time-lapse AFM experiments, the images were acquired at different time points. Between images, the AFM tip was placed on idle (electronically retracted, approximately 4 μm above the scan area) to ensure that it exerted minimum influence on the sample.
Data analysis
All AFM images were flattened and then processed using FemtoScan Online software (Advanced Technologies Center, Moscow, Russia). The features on the bilayer surfaces were visually inspected and analyzed using “Grain Analysis” in the software. The aggregate volumes were arranged in histograms and fit with a Gaussian distribution using OriginPro software (OriginLab, Northampton, MA), yielding mean ± SD values. Scatter plots of number/volume against incubation times were drawn with the Origin software.
Statistical analysis was performed using OriginPro; comparison was performed at each time-point using the pooled data from three separate experiments and using ANOVA followed by Tukey’s range test to determine significance differences.
Coarse-grained molecular dynamics simulations
Lipid bilayers of POPC and POPS were prepared using the insane.py script (available at http://md.chem.rug.nl) using the MARTINI2.2refP [58] force field together with the polarizable water [59] model. The MARTINI force field is widely used for computer simulations of membranes [60] and is also suitable for simulations of relatively large proteins such as α-synuclein. The initial bilayer was constructed using 512 lipids in total placed randomly in a bilayer structure with 40 water molecules per lipid and 150 mM NaCl. After energy minimization using the steepest decent algorithm, the bilayers were simulated as an NPT ensemble for 500 ns using a 20 fs integration time step. The simulation employed periodic boundary conditions with a semi-isotropic pressure coupling using the Parrinello-Rahman barostat at 1 bar with a 12 ps coupling constant. The temperature was kept at 300 K using the velocity rescaling algorithm. Electrostatic interactions were calculated using the particle mesh Ewald algorithm, with a real space cutoff of 1.1 nm. All simulations were performed using the 2016 version of the GROMACS suite of programs. [61] Only the final frame of each bilayer simulation was used for further simulations.
Several studies have shown interactions between α-syn and POPC [62–64] but spectroscopic studies did not show a significant change in secondary structure of α-syn during interaction with the bilayer. Due to this, we selected three starting conformations of α-syn: 1) micelle-bound α-syn (PDB ID: 1XQ8), 2) a conformation obtained from extensive all-atom DMD simulations, [36] and 3) a conformation obtained by running 2 μs coarse-grained MD simulation using MARTINI and the 1XQ8 α-syn structure with all backbone definitions set to random coil.
α-Synuclein-membrane interactions were then simulated using the relaxed bilayers and coarse-grained α-syn structures, generated using the martinize.py script and the all-atom PDB structures. The coarse-grained α-syn structure was placed at a CoM distance of 6 nm from the bilayer core in a parallel orientation (along the long protein axis) to the bilayer. The system was then solvated in a box of 13×13×18 nm3 water and 150 mM NaCl. The simulation procedure was the same as previously described for bilayers alone.
Simulations with membrane-bound and additional free α-syn were conducted using the last frame of the previous simulations and adding another α-syn at a CoM distance of 6 nm from the membrane-bound α-syn. Orientation of the free α-syn was parallel to the bilayer using the same protein conformation as the initial α-syn-bilayer simulation. Simulations for both POPC and POPS were carried out for 2 μs each using the previously described parameters.
All-atom molecular dynamics simulations
To evaluate the change in α-syn secondary structure during interaction with the bilayers, we performed all-atom MD simulations using the α-syn conformation obtained by running 2 μs coarse-grained MD simulation using MARTINI and POPC and POPS bilayers, consisting of 512 lipid molecules each, generated using CHARMM-GUI. [65] The bilayers were constructed with 40 TIP3P waters [66] per lipid, neutralized and kept at 150 mM ionic concentration using Na+ and Cl− counterions, and converted to AMBER format using the lipid17 force field (an extension and refinement of lipid14 [67]). After which 150 ns NPT MD simulations were performed using a 2 fs integration time step. The simulations employed periodic boundary conditions with a semi-isotropic pressure coupling at 1 bar, a constant temperature of 300 K, non-bonded interactions truncated at 10 Å, and electrostatic interactions treated using particle-mesh Ewald [68]. Simulations were performed using the Amber16 package [69]. These relaxed bilayers were then used to investigate α-syn interactions.
To investigate the interaction of α-syn monomer with the bilayers, we extracted the POPC and POPS bilayers from the final frame of the pure bilayer simulations, added α-syn at 6 nm from the bilayer core, solvated with TIP3P water, neutralized with NaCl counter ions, and maintained a final NaCl concentration of 150 mM. Proteins were described using the Amber ff99SB-ILDN force field. [70] The systems were then simulated as an NPT ensemble for 50 ns (using the same parameters as previous bilayer simulations) before being submitted to the special purpose supercomputer Anton2 for long production runs. Simulations on Anton2 employed the multigrator algorithm and treated electrostatics using the Gaussian split Ewald method.
Supplementary Material
Supplementary Movie 1. Coarse-grained MD simulation of α-syn interaction with a POPC lipid bilayer. Stable binding of the α-syn protein to the bilayer is observed. The α-syn N-terminal segment is colored blue, NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are in grey, while the POPC head groups are in purple.
Supplementary Movie 9. Coarse-grained MD simulation of a free α-syn interacting with a membrane bound α-syn on a POPC bilayer. The binding of α-syn to the bilayer is shown. Α-syn N-terminal segment is colored blue, NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are in grey, while the POPC head groups are in purple.
Supplementary Movie 10. Coarse-grained MD simulation of a free α-syn interacting with a membrane bound α-syn on a POPS bilayer. The binding of α-syn to the membrane-bound α-syn is shown. α-syn N-terminal segment is colored blue, NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are in grey, while the POPS head groups are in blue.
Supplementary Movie 2. Coarse-grained MD simulation of α-syn interaction with a POPS lipid bilayer. The stable binding event of α-syn to the bilayer is shown. α-syn N-terminal segment is colored blue, NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are in grey, while the POPS head groups are in blue.
Supplementary Movie 3. Coarse-grained MD simulation of interaction of α-syn monomer, initial conformation from DMD simulations, and POPC bilayer. Interaction with and binding to the POPC surface is observed. Blue backbone denotes the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Lipid tails are in grey, while the POPC head groups are in purple.
Supplementary Movie 4. Coarse-grained MD simulation of interaction of α-syn monomer, initial conformation from DMD simulations, and POPS bilayer. Conversion from a bound extended orientation to a parallel (to the bilayer surface) orientation is observed, with majority of N-terminal region going to the surface of the bilayer. Blue backbone denotes the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Lipid tails are in grey, while the POPS head groups are in blue.
Supplementary Movie 5. Coarse-grained MD simulation of interaction of α-syn monomer, initial conformation from DMD simulations, and POPS bilayer. Stable interaction and insertion of the N-terminal and NAC regions in the bilayer interfacial region is observed while the C-terminal region stays off the surface and flexible. Blue backbone denotes the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Lipid tails are in grey, while the POPS head groups are in blue.
Supplementary Movie 6. All-atom MD simulation of interaction of α-syn monomer, initial conformation from coarse-grained simulations, and POPC bilayer. Initial interactions happen through N-terminal region followed by residues in the NAC- and C-terminal regions. Large blue sphere denotes the N-terminus Cα, the small blue sphere the C-terminal Cα, blue backbone the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Gold spheres mark the P atoms of the lipid heads. Interacting protein residues and lipid head groups are depicted in licorice style, with protein residues being in a wider shape.
Supplementary Movie 7. All-atom MD simulation of interaction of α-syn monomer, initial conformation from coarse-grained simulations, and POPC bilayer. Interaction with the bilayer is observed, initially through the N-terminal region followed by the NAC-region; which is then converted to an extended conformation with only N-terminal region interacting, as observed in coarse-grained simulations. Large blue sphere denotes the N-terminus Cα the small blue sphere the C-terminal Cα, blue backbone the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Gold spheres mark the P atoms of the lipid heads. Interacting protein residues and lipid head groups are depicted in licorice style, with protein residues being in a wider shape.
Supplementary Movie 8. All-atom MD simulation of interaction of α-syn monomer, initial conformation from coarse-grained simulations, and POPS bilayer. Stable interaction with the bilayer is observed, through the N-terminal region; initially in a compact conformation which then extends with C-terminal region moving away from the bilayer surface, similar to observations from coarse-grained simulations. Large blue sphere denotes the N-terminus Cα, the small blue sphere the C-terminal Cα, blue backbone the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Gold spheres mark the P atoms of the lipid heads. Interacting protein residues and lipid head groups are depicted in licorice style, with protein residues being in a wider shape.
Acknowledgements
The work at the University of Nebraska Medical Center (UNMC) was supported by grants from the National Institutes of Health to Y.L.L. (R01 GM096039, R01GM118006 and R21 NS101504). J.C.R. was supported by the Branfman Family Foundation. M.H. was partially supported by the UNMC Graduate Fellowship. The computational modeling was partially performed using resources at the Holland Computing Center of the University of Nebraska, which receives support from the Nebraska Research Initiative. Anton 2 computer time was provided by the Pittsburgh Supercomputing Center (PSC) through Grant R01GM116961 from the National Institutes of Health. The Anton 2 machine at PSC was generously made available by D.E. Shaw Research. The authors thank Melody A. Montgomery for the professional editing of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Goedert M, Spillantini MG, Del Tredici K, Braak H, 100 years of Lewy pathology, Nat Rev Neurol, 9 (2013) 13–24. [DOI] [PubMed] [Google Scholar]
- [2].Luth ES, Bartels T, Dettmer U, Kim NC, Selkoe DJ, Purification of alpha-synuclein from human brain reveals an instability of endogenous multimers as the protein approaches purity, Biochemistry, 54 (2015) 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO, Structural conversion of neurotoxic amyloid-beta(1–42) oligomers to fibrils, Nat Struct Mol Biol, 17 (2010) 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Friedrich RP, Tepper K, Ronicke R, Soom M, Westermann M, Reymann K, Kaether C, Fandrich M, Mechanism of amyloid plaque formation suggests an intracellular basis of Abeta pathogenicity, Proc Natl Acad Sci U S A, 107 (2010) 1942–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ross CA, Poirier MA, Protein aggregation and neurodegenerative disease, Nat Med, 10 Suppl (2004) S10–17. [DOI] [PubMed] [Google Scholar]
- [6].Hardy J, An ‘anatomical cascade hypothesis’ for Alzheimer’s disease, Trends Neurosci, 15 (1992) 200–201. [DOI] [PubMed] [Google Scholar]
- [7].Hardy JA, Higgins GA, Alzheimer’s disease: the amyloid cascade hypothesis, Science, 256 (1992) 184–185. [DOI] [PubMed] [Google Scholar]
- [8].Karran E, Mercken M, De Strooper B, The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics, Nat Rev Drug Discov, 10 (2011) 698–712. [DOI] [PubMed] [Google Scholar]
- [9].Wang Y, Shi M, Chung KA, Zabetian CP, Leverenz JB, Berg D, Srulijes K, Trojanowski JQ, Lee VM, Siderowf AD, Hurtig H, Litvan I, Schiess MC, Peskind ER, Masuda M, Hasegawa M, Lin X, Pan C, Galasko D, Goldstein DS, Jensen PH, Yang H, Cain KC, Zhang J, Phosphorylated alpha-synuclein in Parkinson’s disease, Sci Transl Med, 4 (2012) 121ra120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Landeck N, Hall H, Ardah MT, Majbour NK, El-Agnaf OMA, Halliday G, Kirik D, A novel multiplex assay for simultaneous quantification of total and S129 phosphorylated human alpha-synuclein, Molecular Neurodegeneration, 11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Banerjee S, Hashemi M, Lv Z, Maity S, Rochet JC, Lyubchenko YL, A novel pathway for amyloids self-assembly in aggregates at nanomolar concentration mediated by the interaction with surfaces, Sci Rep, 7 (2017) 45592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Diao J, Burre J, Vivona S, Cipriano DJ, Sharma M, Kyoung M, Sudhof TC, Brunger AT, Native alpha-synuclein induces clustering of synaptic-vesicle mimics via binding to phospholipids and synaptobrevin-2/VAMP2, Elife, 2 (2013) e00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Davidson WS, Jonas A, Clayton DF, George JM, Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes, J Biol Chem, 273 (1998) 9443–9449. [DOI] [PubMed] [Google Scholar]
- [14].Venda LL, Cragg SJ, Buchman VL, Wade-Martins R, alpha-Synuclein and dopamine at the crossroads of Parkinson’s disease, Trends Neurosci, 33 (2010) 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jo E, McLaurin J, Yip CM, St George-Hyslop P, Fraser PE, alpha-Synuclein membrane interactions and lipid specificity, J Biol Chem, 275 (2000) 34328–34334. [DOI] [PubMed] [Google Scholar]
- [16].Eliezer D, Kutluay E, Bussell R Jr., Browne G, Conformational properties of alpha-synuclein in its free and lipid-associated states, J Mol Biol, 307 (2001) 1061–1073. [DOI] [PubMed] [Google Scholar]
- [17].Bodner CR, Dobson CM, Bax A, Multiple tight phospholipid-binding modes of alpha-synuclein revealed by solution NMR spectroscopy, J Mol Biol, 390 (2009) 775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hellstrand E, Grey M, Ainalem M-L, Ankner J, Forsyth VT, Fragneto G, Haertlein M, Dauvergne M-T, Nilsson H, Brundin P, Linse S, Nylander T, Sparr E, Adsorption of α-Synuclein to Supported Lipid Bilayers: Positioning and Role of Electrostatics, ACS Chemical Neuroscience, 4 (2013) 1339–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D, Membrane-Bound α-Synuclein Forms an Extended Helix: Long-Distance Pulsed ESR Measurements Using Vesicles, Bicelles, and Rodlike Micelles, J Am Chem Soc, 130 (2008) 12856–12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee H-J, Choi C, Lee S-J, Membrane-bound a-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form., J. Biol. Chem, 277 (2002) 671–678. [DOI] [PubMed] [Google Scholar]
- [21].Galvagnion C, Buell AK, Meisl G, Michaels TC, Vendruscolo M, Knowles TP, Dobson CM, Lipid vesicles trigger alpha-synuclein aggregation by stimulating primary nucleation, Nat Chem Biol, 11 (2015) 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ysselstein D, Joshi M, Mishra V, Griggs AM, Asiago JM, McCabe GP, Stanciu LA, Post CB, Rochet JC, Effects of impaired membrane interactions on alpha-synuclein aggregation and neurotoxicity, Neurobiology of disease, 79 (2015) 150–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pfefferkorn CM, Jiang Z, Lee JC, Biophysics of alpha-synuclein membrane interactions, Biochim Biophys Acta, 1818 (2012) 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Butterfield SM, Lashuel HA, Amyloidogenic Protein-Membrane Interactions: Mechanistic Insight from Model Systems, Angewandte Chemie International Edition, 49 (2010) 5628–5654. [DOI] [PubMed] [Google Scholar]
- [25].Green JD, Kreplak L, Goldsbury C, Li Blatter X, Stolz M, Cooper GS, Seelig A, Kistler J, Aebi U, Atomic force microscopy reveals defects within mica supported lipid bilayers induced by the amyloidogenic human amylin peptide, J Mol Biol, 342 (2004) 877–887. [DOI] [PubMed] [Google Scholar]
- [26].Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R, Amyloid ion channels: a common structural link for protein-misfolding disease, Proc Natl Acad Sci U S A, 102 (2005) 10427–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stockl MT, Zijlstra N, Subramaniam V, alpha-Synuclein oligomers: an amyloid pore? Insights into mechanisms of alpha-synuclein oligomer-lipid interactions, Mol Neurobiol, 47 (2013) 613–621. [DOI] [PubMed] [Google Scholar]
- [28].Dante S, Hauss T, Brandt A, Dencher NA, Membrane fusogenic activity of the Alzheimer’s peptide A beta(1–42) demonstrated by small-angle neutron scattering, J Mol Biol, 376 (2008) 393–404. [DOI] [PubMed] [Google Scholar]
- [29].Viennet T, Wordehoff MM, Uluca B, Poojari C, Shaykhalishahi H, Willbold D, Strodel B, Heise H, Buell AK, Hoyer W, Etzkorn M, Structural insights from lipid-bilayer nanodiscs link alpha-Synuclein membrane-binding modes to amyloid fibril formation, Commun Biol, 1 (2018) 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Reynolds NP, Soragni A, Rabe M, Verdes D, Liverani E, Handschin S, Riek R, Seeger S, Mechanism of membrane interaction and disruption by alpha-synuclein, J Am Chem Soc, 133 (2011) 19366–19375. [DOI] [PubMed] [Google Scholar]
- [31].Comellas G, Lemkau LR, Zhou DH, George JM, Rienstra CM, Structural intermediates during alpha-synuclein fibrillogenesis on phospholipid vesicles, J Am Chem Soc, 134 (2012) 5090–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lee JH, Hong CS, Lee S, Yang JE, Park YI, Lee D, Hyeon T, Jung S, Paik SR, Radiating amyloid fibril formation on the surface of lipid membranes through unit-assembly of oligomeric species of alpha-synuclein, PLoS One, 7 (2012) e47580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ysselstein D, Dehay B, Costantino IM, McCabe GP, Frosch MP, George JM, Bezard E, Rochet JC, Endosulfine-alpha inhibits membrane-induced alpha-synuclein aggregation and protects against alpha-synuclein neurotoxicity, Acta neuropathologica communications, 5 (2017) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lv Z, Banerjee S, Zagorski K, Lyubchenko YL, Supported Lipid Bilayers for Atomic Force Microscopy Studies, Methods Mol Biol, 1814 (2018) 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lv Z, Krasnoslobodtsev Alexey V., Zhang Y, Ysselstein D, Rochet J-C, Blanchard Scott C., Lyubchenko Yuri L., Direct Detection of α-Synuclein Dimerization Dynamics: Single-Molecule Fluorescence Analysis, Biophys J, 108 (2015) 2038–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang Y, Hashemi M, Lv Z, Williams B, Popov KI, Dokholyan NV, Lyubchenko YL, High-speed atomic force microscopy reveals structural dynamics of alpha-synuclein monomers and dimers, J Chem Phys, 148 (2018) 123322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Galvagnion C, Brown JW, Ouberai MM, Flagmeier P, Vendruscolo M, Buell AK, Sparr E, Dobson CM, Chemical properties of lipids strongly affect the kinetics of the membrane-induced aggregation of alpha-synuclein, Proc Natl Acad Sci U S A, 113 (2016) 7065–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Galvagnion C, Buell AK, Meisl G, Michaels TC, Vendruscolo M, Knowles TP, Dobson CM, Lipid vesicles trigger alpha-synuclein aggregation by stimulating primary nucleation, Nature chemical biology, 11 (2015) 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Perni M, Galvagnion C, Maltsev A, Meisl G, Müller MBD, Challa PK, Kirkegaard JB, Flagmeier P, Cohen SIA, Cascella R, Chen SW, Limboker R, Sormanni P, Heller GT, Aprile FA, Cremades N, Cecchi C, Chiti F, Nollen EAA, Knowles TPJ, Vendruscolo M, Bax A, Zasloff M, Dobson CM, A natural product inhibits the initiation of α-synuclein aggregation and suppresses its toxicity, Proceedings of the National Academy of Sciences, 114 (2017) E1009–E1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Giusto NM, Salvador GA, Castagnet PI, Pasquare SJ, Ilincheta de Boschero MG, Age-associated changes in central nervous system glycerolipid composition and metabolism, Neurochem Res, 27 (2002) 1513–1523. [DOI] [PubMed] [Google Scholar]
- [41].Riekkinen P, Interaction Between Dopamine and Phospholipids, Archives of Neurology, 32 (1975) 25. [DOI] [PubMed] [Google Scholar]
- [42].Ledeen RW, Wu G, Gangliosides, α-Synuclein, and Parkinson’s Disease, 156 (2018) 435–454. [DOI] [PubMed] [Google Scholar]
- [43].Cascella R, Evangelisti E, Bigi A, Becatti M, Fiorillo C, Stefani M, Chiti F, Cecchi C, Nacmias B, Soluble Oligomers Require a Ganglioside to Trigger Neuronal Calcium Overload, Journal of Alzheimer’s Disease, 60 (2017) 923–938. [DOI] [PubMed] [Google Scholar]
- [44].Oropesa-Nuñez R, Seghezza S, Dante S, Diaspro A, Cascella R, Cecchi C, Stefani M, Chiti F, Canale C, Interaction of toxic and non-toxic HypF-N oligomers with lipid bilayers investigated at high resolution with atomic force microscopy, Oncotarget, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bodner CR, Maltsev AS, Dobson CM, Bax A, Differential phospholipid binding of alpha-synuclein variants implicated in Parkinson’s disease revealed by solution NMR spectroscopy, Biochemistry, 49 (2010) 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shvadchak VV, Yushchenko DA, Pievo R, Jovin TM, The mode of alpha-synuclein binding to membranes depends on lipid composition and lipid to protein ratio, FEBS Lett, 585 (2011) 3513–3519. [DOI] [PubMed] [Google Scholar]
- [47].Lokappa SB, Suk JE, Balasubramanian A, Samanta S, Situ AJ, Ulmer TS, Sequence and membrane determinants of the random coil-helix transition of alpha-synuclein, J Mol Biol, 426 (2014) 2130–2144. [DOI] [PubMed] [Google Scholar]
- [48].Fusco G, De Simone A, Gopinath T, Vostrikov V, Vendruscolo M, Dobson CM, Veglia G, Direct observation of the three regions in alpha-synuclein that determine its membrane-bound behaviour, Nat Commun, 5 (2014) 3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yip CM, McLaurin J, Amyloid-beta peptide assembly: a critical step in fibrillogenesis and membrane disruption, Biophys J, 80 (2001) 1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yip CM, Darabie AA, McLaurin J, Abeta42-peptide assembly on lipid bilayers, J Mol Biol, 318 (2002) 97–107. [DOI] [PubMed] [Google Scholar]
- [51].Iyer A, Petersen NO, Claessens MM, Subramaniam V, Amyloids of alpha-synuclein affect the structure and dynamics of supported lipid bilayers, Biophys J, 106 (2014) 2585–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ouberai MM, Wang J, Swann MJ, Galvagnion C, Guilliams T, Dobson CM, Welland ME, alpha-Synuclein senses lipid packing defects and induces lateral expansion of lipids leading to membrane remodeling, J Biol Chem, 288 (2013) 20883–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chaudhary H, Iyer A, Subramaniam V, Claessens MM, alpha-Synuclein Oligomers Stabilize Pre-Existing Defects in Supported Bilayers and Propagate Membrane Damage in a Fractal-Like Pattern, Langmuir, 32 (2016) 11827–11836. [DOI] [PubMed] [Google Scholar]
- [54].Rauscher S, Gapsys V, Gajda MJ, Zweckstetter M, de Groot BL, Grubmuller H, Structural Ensembles of Intrinsically Disordered Proteins Depend Strongly on Force Field: A Comparison to Experiment, J Chem Theory Comput, 11 (2015) 5513–5524. [DOI] [PubMed] [Google Scholar]
- [55].Krasnoslobodtsev AV, Peng J, Asiago JM, Hindupur J, Rochet JC, Lyubchenko YL, Effect of spermidine on misfolding and interactions of alpha-synuclein, PLoS One, 7 (2012) e38099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shlyakhtenko LS, Gall AA, Lyubchenko YL, Mica functionalization for imaging of DNA and protein-DNA complexes with atomic force microscopy, Methods Mol Biol, 931 (2013) 295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mingeot-Leclercq MP, Deleu M, Brasseur R, Dufrene YF, Atomic force microscopy of supported lipid bilayers, Nature protocols, 3 (2008) 1654–1659. [DOI] [PubMed] [Google Scholar]
- [58].de Jong DH, Singh G, Bennett WF, Arnarez C, Wassenaar TA, Schafer LV, Periole X, Tieleman DP, Marrink SJ, Improved Parameters for the Martini Coarse-Grained Protein Force Field, J Chem Theory Comput, 9 (2013) 687–697. [DOI] [PubMed] [Google Scholar]
- [59].Yesylevskyy SO, Schafer LV, Sengupta D, Marrink SJ, Polarizable water model for the coarse-grained MARTINI force field, PLoS Comput Biol, 6 (2010) e1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Marrink SJ, Tieleman DP, Perspective on the Martini model, Chemical Society Reviews, 42 (2013) 6801. [DOI] [PubMed] [Google Scholar]
- [61].Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E, GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 1–2 (2015) 19–25. [Google Scholar]
- [62].Rhoades E, Ramlall TF, Webb WW, Eliezer D, Quantification of α-Synuclein Binding to Lipid Vesicles Using Fluorescence Correlation Spectroscopy, Biophys J, 90 (2006) 4692–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jiang Z, de Messieres M, Lee JC, Membrane Remodeling by α-Synuclein and Effects on Amyloid Formation, J Am Chem Soc, 135 (2013) 15970–15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Middleton ER, Rhoades E, Effects of Curvature and Composition on α-Synuclein Binding to Lipid Vesicles, Biophys J, 99 (2010) 2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: a web-based graphical user interface for CHARMM, J Comput Chem, 29 (2008) 1859–1865. [DOI] [PubMed] [Google Scholar]
- [66].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys, 79 (1983) 926–935. [Google Scholar]
- [67].Dickson CJ, Madej BD, Skjevik ÅA, Betz RM, Teigen K, Gould IR, Walker RC, Lipid14: The Amber Lipid Force Field, J Chem Theory Comput, 10 (2014) 865–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Darden T, York D, Pedersen L, Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems, The Journal of Chemical Physics, 98 (1993) 10089–10092. [Google Scholar]
- [69].Case VBDA, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Gohlke H, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A, Lee TS, LeGrand S, Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C, Salomon-Ferrer R, Seabra G, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X and Kollman PA, AMBER 16, University of California, San Francisco, 2016. [Google Scholar]
- [70].Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE, Improved side-chain torsion potentials for the Amber ff99SB protein force field, Proteins, 78 (2010) 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1. Coarse-grained MD simulation of α-syn interaction with a POPC lipid bilayer. Stable binding of the α-syn protein to the bilayer is observed. The α-syn N-terminal segment is colored blue, NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are in grey, while the POPC head groups are in purple.
Supplementary Movie 9. Coarse-grained MD simulation of a free α-syn interacting with a membrane bound α-syn on a POPC bilayer. The binding of α-syn to the bilayer is shown. Α-syn N-terminal segment is colored blue, NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are in grey, while the POPC head groups are in purple.
Supplementary Movie 10. Coarse-grained MD simulation of a free α-syn interacting with a membrane bound α-syn on a POPS bilayer. The binding of α-syn to the membrane-bound α-syn is shown. α-syn N-terminal segment is colored blue, NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are in grey, while the POPS head groups are in blue.
Supplementary Movie 2. Coarse-grained MD simulation of α-syn interaction with a POPS lipid bilayer. The stable binding event of α-syn to the bilayer is shown. α-syn N-terminal segment is colored blue, NAC region is in green, and the C-terminal segment is in red. N- and C-terminal residues are highlighted with a sphere. Lipid tails are in grey, while the POPS head groups are in blue.
Supplementary Movie 3. Coarse-grained MD simulation of interaction of α-syn monomer, initial conformation from DMD simulations, and POPC bilayer. Interaction with and binding to the POPC surface is observed. Blue backbone denotes the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Lipid tails are in grey, while the POPC head groups are in purple.
Supplementary Movie 4. Coarse-grained MD simulation of interaction of α-syn monomer, initial conformation from DMD simulations, and POPS bilayer. Conversion from a bound extended orientation to a parallel (to the bilayer surface) orientation is observed, with majority of N-terminal region going to the surface of the bilayer. Blue backbone denotes the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Lipid tails are in grey, while the POPS head groups are in blue.
Supplementary Movie 5. Coarse-grained MD simulation of interaction of α-syn monomer, initial conformation from DMD simulations, and POPS bilayer. Stable interaction and insertion of the N-terminal and NAC regions in the bilayer interfacial region is observed while the C-terminal region stays off the surface and flexible. Blue backbone denotes the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Lipid tails are in grey, while the POPS head groups are in blue.
Supplementary Movie 6. All-atom MD simulation of interaction of α-syn monomer, initial conformation from coarse-grained simulations, and POPC bilayer. Initial interactions happen through N-terminal region followed by residues in the NAC- and C-terminal regions. Large blue sphere denotes the N-terminus Cα, the small blue sphere the C-terminal Cα, blue backbone the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Gold spheres mark the P atoms of the lipid heads. Interacting protein residues and lipid head groups are depicted in licorice style, with protein residues being in a wider shape.
Supplementary Movie 7. All-atom MD simulation of interaction of α-syn monomer, initial conformation from coarse-grained simulations, and POPC bilayer. Interaction with the bilayer is observed, initially through the N-terminal region followed by the NAC-region; which is then converted to an extended conformation with only N-terminal region interacting, as observed in coarse-grained simulations. Large blue sphere denotes the N-terminus Cα the small blue sphere the C-terminal Cα, blue backbone the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Gold spheres mark the P atoms of the lipid heads. Interacting protein residues and lipid head groups are depicted in licorice style, with protein residues being in a wider shape.
Supplementary Movie 8. All-atom MD simulation of interaction of α-syn monomer, initial conformation from coarse-grained simulations, and POPS bilayer. Stable interaction with the bilayer is observed, through the N-terminal region; initially in a compact conformation which then extends with C-terminal region moving away from the bilayer surface, similar to observations from coarse-grained simulations. Large blue sphere denotes the N-terminus Cα, the small blue sphere the C-terminal Cα, blue backbone the N-terminal region, green backbone the NAC region, and red backbone the C-terminal region of the α-syn molecule. Gold spheres mark the P atoms of the lipid heads. Interacting protein residues and lipid head groups are depicted in licorice style, with protein residues being in a wider shape.