SUMMARY
Severe obesity is a rapidly growing global health threat. Although often attributed to unhealthy lifestyle choices or environmental factors, obesity is known to be heritable and highly polygenic – the majority of inherited susceptibility is related to the cumulative impact of many common DNA variants. Here, we derive and validate a new polygenic predictor comprised of 2.1 million common variants to quantify this susceptibility and test this predictor in >300,000 individuals ranging from middle age to birth. Among middle-aged adults, we observe a 13 kg gradient in weight and a 25-fold gradient in risk of severe obesity across polygenic score deciles. In a longitudinal birth cohort, we note minimal difference in birthweight across score deciles, but a significant gradient emerged in early childhood and reached 12 kg by age 18 years. This new approach to quantify inherited susceptibility to obesity using affords new opportunities for clinical prevention and mechanistic assessment.
eTOC Blurb
A genome-wide polygenic score quantifies inherited susceptibility to obesity, integrating information from 2.1 million common genetic variants to identify adults at risk of severe obesity.
INTRODUCTION
Severe obesity, defined as body mass index (BMI) ≥ 40 kg/m2, is a rapidly growing public health issue already afflicting 8% of American adults (Flegal et al., 2016; NHLBI Expert Panel, 1998). Although present in less than 1% of the population in middle-income countries such as India and China, prevalence of severe obesity in these countries has increased more than 100-fold over the last three decades and shows no signs of slowing (NCD Risk Factor Collaboration, 2017). Individuals with severe obesity are often stigmatized due to the commonly-held belief that their condition results primarily from unhealthy lifestyle choices (Tomiyama et al., 2018). However, obesity is known to be heritable, suggesting that inborn DNA variation confers increased susceptibility in some individuals and protection in others (Elks et al., 2012; Maes et al., 1997; Whitaker et al., 1997; Yang et al., 2015).
Inherited susceptibility to obesity can, in rare cases, be attributed to a large-effect mutation that perturbs energy homeostasis or fat deposition (Barsh et al., 2000). For example, genetic inactivation of the melanocortin 4 receptor (MC4R) gene is associated with obesity in both mouse models and humans (Farooqi et al., 2003; Huszar et al., 1997; Vaisse et al., 1998; Yeo et al., 1998). However, for the vast majority of severely obese individuals, no such monogenic mutation can be identified (Larsen et al., 2005; Stutzmann et al., 2008; Vaisse et al., 2000). Their genetic susceptibility may instead result from the cumulative impact of numerous variants with individually modest effect – a ‘polygenic’ model. This paradigm is similar to other complex diseases in which polygenic inheritance, involving many common genetic variants, accounts for the majority of inherited susceptibility (Golan et al., 2014; International Schizophrenia et al., 2009; Visscher et al., 2012; Yang et al., 2011; Zhu et al., 2015).
A recently published genome-wide association study (GWAS) quantified the relationship between each of 2.1 million common genetic variants and BMI in over 300,000 individuals (Locke et al., 2015). None of the individual variants accounts for a large proportion of the phenotype. The strongest association was noted for a common variant at the FTO locus: the risk allele was associated with a statistically robust, but clinically modest, increase in weight of approximately 1 kilogram per inherited risk allele. Obtaining meaningful predictive power thus requires aggregating information from many common variants into a polygenic score (Chatterjee et al., 2016; Khera et al., 2018a). However, previous efforts to create an effective polygenic score for obesity have had only modest success (Loos and Janssens, 2017).
Here, we use recently developed computational algorithms and large datasets to derive, validate and test a robust polygenic predictor of BMI and obesity. This genome-wide polygenic score (GPS) integrates all available common variants into a single quantitative measure of inherited susceptibility. It identifies a subset of the adult population that is at substantial risk of severe obesity – in some cases equivalent to rare monogenic mutations – and others that enjoy considerable protection. The GPS is associated with only minimal differences in birthweight, but it predicts clear differences in weight during early childhood and profound differences in weight trajectory and risk of developing severe obesity in subsequent years.
RESULTS
In order to create a GPS, we obtained the average effects for each of 2,100,302 genetic variants on BMI from the largest published GWAS study of obesity to date (Locke et al., 2015). We used a recently developed computational algorithm to reweight each variant according to the effect size and strength of statistical significance observed in the prior GWAS, the degree of correlation between a variant and others nearby, and a tuning parameter that denotes the proportion of variants with non-zero effect size (Vilhjalmsson et al., 2015). Because the best choice of this tuning parameter is difficult to know a priori, a range of 5 values was tested as previously recommended (Vilhjalmsson et al., 2015).
We set out to validate these 5 scores—and to choose the best score for further analysis—by testing their ability to predict measured BMI in a validation dataset of 119,951 middle-aged adult participants of the UK Biobank. The UK Biobank enrolled participants aged 40 to 69 years from across the United Kingdom and allows for linkage of measurements such as BMI to extensive genetic data (Bycroft et al., 2018; Sudlow et al., 2015). Within this dataset we estimated the heritability of BMI explained by common variants to be 23.4% using a recently developed approach (Bulik-Sullivan et al., 2015), consistent with prior estimates ranging from 17 to 27% (Yang et al., 2015; Yang et al., 2011; Zhu et al., 2015).
Each of the five candidate GPSs was strongly associated with observed BMI (p < 0.0001), with similar correlation coefficients ranging from 0.283 to 0.292 (See also Table S1). Nearly identical results were obtained after adjustment of each of the candidate GPSs for genetic background, as assessed by principal components of ancestry (Table S2). We selected the best score, with correlation of 0.292, to take forward into four testing datasets below. Additional details of GPS derivation and validation are provided in Figure 1 and the STAR Methods.
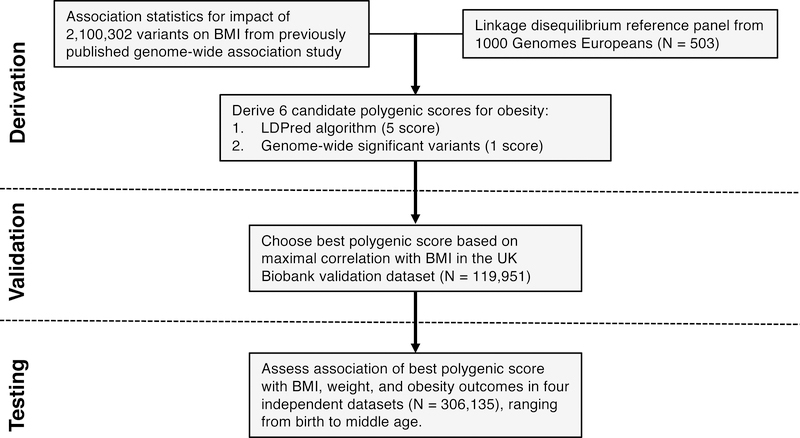
Figure 1. Derivation, validation, and testing of a genome-wide polygenic score for obesity.
A genome-wide polygenic score (GPS) for obesity was derived by starting with two independent datasets: first, a list of 2,100,302 common genetic variants and estimated impact of each on BMI from a large GWAS study (Locke et al., 2015), and second, genetic information from 503 individuals of European ancestry from the 1000 Genomes Study used to measure ‘linkage disequilibrium,’ the correlation between genetic variants (The 1000 Genomes Project Consortium, 2015). Candidate GPSs were derived using the LDPred computational algorithm, a Bayesian approach to calculate a posterior mean effect for all variants based on a prior (effect size and statistical significance in the previous GWAS) and subsequent shrinkage based on linkage disequilibrium (Vilhjalmsson et al., 2015). The five candidate LDPred scores vary with respect to the tuning parameter ρ (that is, the proportion of variants assumed to be causal), as previously recommended. A sixth polygenic score was derived based on only the 141 independent variants that had achieved genome-wide levels of statistical significance in the previous GWAS. The optimal GPS was chosen based on maximal correlation with BMI in the UK Biobank validation dataset (N = 119,951 Europeans), and subsequently tested in multiple independent testing datasets of 306,135 individuals. See also Tables S1, S2, and S3.
Our GPS of 2,100,302 variants had substantially greater predictive power than a sixth polygenic score comprised of only the 141 independent variants that had reached genome-wide levels of statistical significance in the prior GWAS. Within the 119,951 participants in the validation dataset, correlation with BMI for this 141-variant score was only 0.133. This lower strength of association using fewer variants is consistent with earlier studies, where predictors of up to 97 variants had a relatively low correlation with measured BMI, ranging from 0.01 to 0.12 (Belsky et al., 2013; Hung et al., 2015; Li et al., 2010; Sandholt et al., 2010).
Having derived and validated a new polygenic predictor which considerably outperformed earlier scores, we explored the predictive power of the GPS on BMI, weight, and severe obesity in 306,135 individuals of four independent testing datasets, spanning the age spectrum from middle age to time of birth (Table 1).
Table 1.
Genome-wide polygenic score for obesity assessed in four independent testing datasets.
| UK Biobank | Partners Healthcare | Framingham Offspring / CARDIA | Avon Longitudinal Study of Parents and Children | |
|---|---|---|---|---|
| N Participants | 288,016 | 6,536 | 3,722 | 7,861 |
| Study Design | Cross-sectional | Case-control | Longitudinal | Longitudinal |
| Age Range | 40—69 years | ≥ 18 years | 18—40 years | Birth |
| Female sex | 55% | 61% | 48% | 49% |
| Outcomes | Weight, Severe obesity, Bariatric surgery, Cardiometabolic diseases, Mortality | Bariatric surgery | Incident severe obesity (27 years median follow-up) | Weight at birth and subsequent visits (0—18 years) |
CARDIA – Coronary Artery Risk Development in Young Adults
Polygenic susceptibility to weight and severe obesity in middle age
We determined the extent to which the GPS predicted weight and severe obesity in a testing dataset of 288,016 middle-aged participants of the UK Biobank (independent of the 119,951 validation dataset participants studied above). Participant mean age was 57 years and 55% were female. Mean weight was 78.1 kilograms and mean BMI was 27.4 kg/m2. 23.9% of the participants were obese (BMI ≥ 30 kg/m2) and 1.8% met criteria for severe obesity.
The GPS approximated a normal distribution in the population (Figure S1). The correlation of the GPS and observed BMI was 0.29, identical to the UK Biobank validation dataset. Correlations were similar when participants were stratified into 5-year age bins, ranging from 0.28 to 0.31 (Table S3).
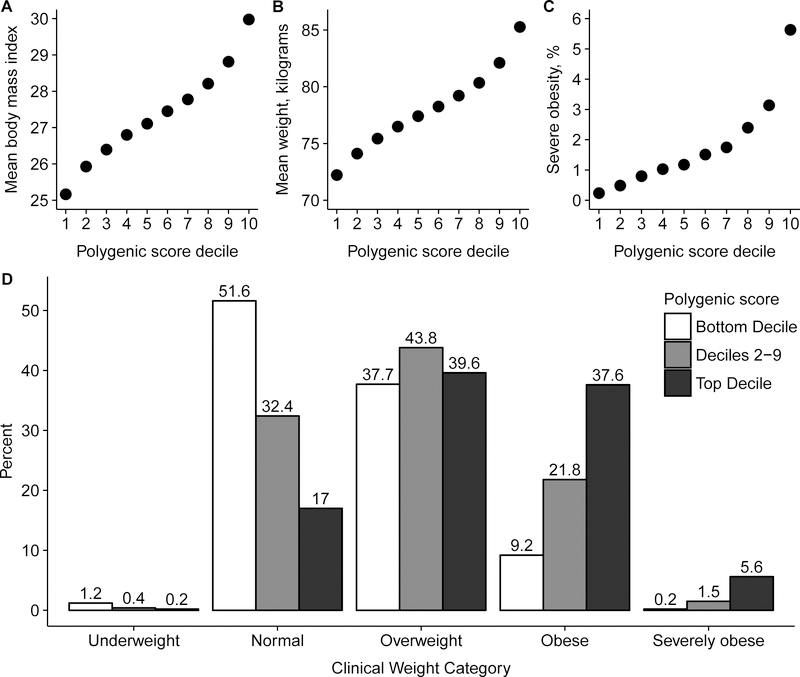
We next stratified the population according to GPS decile and found a striking gradient with respect to BMI, weight, and prevalence of obesity (Figure 2A–2C). For example, average BMI was 30.0 kg/m2 for those in the top decile of the GPS and 25.2 kg/m2 for those in the bottom decile, a difference of 4.8 kg/m2 (p < 0.0001). Similarly, average weight was 85.3 kilograms for those in the top decile versus 72.2 kilograms for those in the bottom decile, a difference of 13.0 kilograms (p < 0.0001). 43.2% of those in the top decile were obese versus 9.5% of those in the bottom decile (Figure S2). Severe obesity was present in 1,621 of 28,784 (5.6%) of those in the top decile of the GPS versus 69 of 28,834 (0.2%) of those in the bottom decile, corresponding to a 25-fold gradient in risk of severe obesity (p < 0.0001).
Figure 2. Relationship of genome-wide polygenic score distribution with body mass index, weight, and severe obesity.
288,016 middle-aged UK Biobank participants were binned into 10 deciles according to the polygenic score. Body mass index (Panel A), weight (Panel B), and prevalence of severe obesity (Panel C) each increased across deciles of the polygenic score (p < 0.0001 for each). Significant differences in clinical categories of obesity were noted (Panel D) when participants were stratified into three categories – bottom decile, deciles 2–9, and top decile. Underweight refers to BMI < 18.5 kg/m2, normal as 18.5 to 24.9 kg/m2, overweight as 25.0 to 29.9 kg/m2, obesity as 30.0 to 39.9 kg/m2, and severe obesity as ≥ 40 kg/m2 (NHLBI Expert Panel, 1998). See also Figures S1, S2, and S3.
Despite the strength of these associations, polygenic susceptibility to obesity is not deterministic. Among those in the top decile of the GPS, 83% were overweight or obese but 17% had BMI within the normal range and 0.2% were underweight (Figure 2D). These results were nearly identical after adjustment of the GPS for genetic background, as assessed by principal components of ancestry (Figure S3).
High polygenic score common among those with extreme obesity
Traditional analyses of rare genetic mutations are performed by comparing heterozygous mutation carriers to noncarriers. An important example is the p.Tyr35Ter premature stop codon present in 0.02% of the population and typically inherited as a shared haplotype with the p.Asp37Val missense mutation, which has been previously shown to completely inactivate MC4R activity in in vitro functional assays (Larsen et al., 2005; Xiang et al., 2006). A recent analysis linked this variant to an average weight increase of 7 kilograms (Turcot et al., 2018).
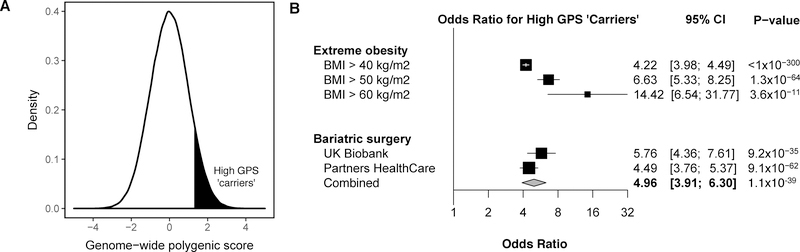
We sought to mimic this approach using the GPS by labeling the top decile of the GPS distribution as ‘carriers’ and those in the remainder of the distribution as non-carriers (Figure 3A). The 10% of the population who carried ‘high GPS’ demonstrated an average BMI that was 2.9 kg/m2 higher and weight 8.0 kilograms higher than noncarriers (p < 0.0001 for both comparisons). Results were similar when high GPS carriers were compared to individuals within the middle quintile of the score distribution instead of the bottom 90% of the distribution, with difference in BMI and weight of 2.6 kg/m2 and 7.4 kilograms, respectively.
Figure 3. Association of high genome-wide polygenic score with extreme obesity and bariatric surgery.
We considered the top 10% of the distribution as a ‘carrier’ of high genome-wide polygenic score (GPS), represented by the shading in panel A, and compared risk of obesity-related outcomes to the remaining 90% of the distribution. The x-axis represents the polygenic score, with values scaled to a mean of 0 and standard deviation 1 to facilitate interpretation. In Panel B, the relationship of high GPS to extreme obesity and treatment with bariatric surgery was quantified using logistic regression.
CI – confidence interval
Furthermore, the magnitude of risk conferred by high GPS increased at more extreme levels of observed obesity. The proportion of high GPS carriers was 9.7% among individuals with BMI < 40 kg/m2; 31% among the 5,232 individuals with BMI ≥ 40 kg/m2; 42.3% among the 331 individuals with BMI ≥ 50 kg/m2; and 61.5% among the 26 individuals with BMI ≥ 60 kg/m2. Compared with the remainder of the GPS distribution, high GPS was associated with a 4.2, 6.6, and 14.4-fold increased risk of BMI ≥ 40, 50, and 60 kg/m2, respectively (Figure 3B).
Another indicator of extreme obesity involves individuals who undergo treatment with bariatric surgery, acknowledging that factors in addition to severity of obesity contribute to the decision to move forward with an invasive procedure to assist with weight loss. We identified 208 such participants in the UK Biobank testing dataset, of whom 81 (38.9%) carried high GPS. This finding was replicated among 714 severely obese patients treated with bariatric surgery within the Partners HealthCare System (Hatoum et al., 2013; Karlson et al., 2016). 238 of these 714 (33%) patients carried high GPS. A combined analysis of the 922 bariatric surgery participants noted high GPS in 319 (34.6%). Compared with remainder of the distribution, high GPS was associated with a 5.0-fold increased risk of severe obesity treated with bariatric surgery (Figure 3B).
High polygenic score associated with increased risks for cardiometabolic disease and mortality
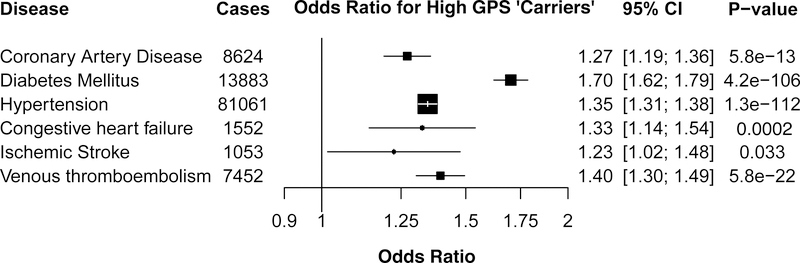
Beyond severe obesity, individuals in the UK Biobank who carried high GPS were at increased risk for six common cardiometabolic diseases, including a 28% increased risk of coronary artery disease, a 72% increased risk for diabetes mellitus, 38% increased risk for hypertension, 34% increased risk for heart failure, 23% increased risk for ischemic stroke, and 41% increased risk for venous thromboembolism (p < 0.05 for each; Figure 4).
Figure 4. Association of high genome-wide polygenic score with cardiometabolic diseases.
The relationship of high genome-wide polygenic score (GPS), defined as the top decile of the score distribution, with the prevalence of six cardiometabolic diseases was determined in a logistic regression model within the UK Biobank testing dataset of 288,016 participants.
CI – confidence interval
We next determined the relationship between high polygenic score and all-cause mortality. Death following enrollment occurred in 8,102 (2.8%) participants over a median follow-up of 7.1 years, including 940 (3.3%) of those in the top decile of the polygenic score distribution and 7,162 (2.8%) in the remainder of the distribution (p < 0.0001). In a survival analysis that additionally included time to death in the statistical model, high polygenic score was associated with a 19% increased risk of incident mortality (p < 0.0001).
Polygenic score identifies 1.6% of the population with BMI increase similar to a monogenic mutation
Rare inactivating mutations in the MC4R gene are among the most common monogenic mutations for obesity (Farooqi et al., 2003; Stutzmann et al., 2008; Vaisse et al., 2000), but few prior studies have analyzed gene sequencing data and performed clinical grade variant classification in a large population of unascertained adults.
We performed whole exome sequencing of 6,547 UK Biobank participants, identifying 24 rare (allele frequency < 1%) protein-altering variants in the MC4R gene. A total of 54 of the 6,547 individuals (0.8%) harbored one of these variants. Average BMI of these 54 individuals was 30.8 kg/m2 versus 28.4 kg/m2 in the remainder of the population, a difference of 2.4 kg/m2 (95% confidence interval [CI] 1.0 to 3.7; p = 0.001).
Given that the majority of rare missense mutations have little or no functional impact on protein function (Boyko et al., 2008; Yampolsky et al., 2005), a clinical laboratory geneticist on our team who was blinded to participant phenotypes classified each of the 24 observed MC4R variants according to current clinical guidelines (Richards et al., 2015), integrating information from population allele frequency data, computational prediction and conservation scores, functional assay data, and prior reports of the variant segregating with obesity. 4 of these 24 variants met these clinical criteria as pathogenic or likely pathogenic for monogenic obesity, including the p.Tyr35Ter premature stop codon noted above, an inactivating frameshift mutation (p.Phe280AlafsX12), and two missense mutations – p.Arg165Gln and p.Glu61Lys – previously shown to segregate with obesity in family studies and impair receptor activity in functional assays. A summary of the evidence used to classify each of the 24 variants is provided in Table S4.
A total of 9 of the 6,547 individuals harbored one of the 4 pathogenic MC4R variants, corresponding to a prevalence of 0.14% (95% confidence interval [CI] 0.06 to 0.26%). Subsequent unblinding of phenotype information revealed that average BMI of these 9 carriers was 32.5 kg/m2 as compared to 28.4 kg/m2 in the remainder of the population, a difference of 4.1 kg/m2 (95% CI 0.8 to 7.3; p = 0.02). However, consistent with recent observations of incomplete penetrance in an adult population (Turcot et al., 2018), only one of the 9 carriers was severely obese. An additional 3 were obese, and the remaining 5 were overweight but not obese.
We hypothesized that individuals in the extreme of the GPS distribution might have an increase in BMI that approaches or exceeds the 4.1 kg/m2 increase noted for carriers of pathogenic MC4R mutations, and tested progressively more extreme tails of the distribution. The top 1.6% of the GPS distribution had a mean BMI 4.1 kg/m2 higher than the remaining 98.4% – 31.4 versus 27.3 kg/m2, and 9.1% of these individuals were severely obese.
Young adults risk’ of developing severe obesity varies according to polygenic score
Although only a small minority of individuals are severely obese in early adulthood, the prevalence increases rapidly over subsequent decades (NCD Risk Factor Collaboration, 2017). We hypothesized that the GPS might predict who would go on to develop severe obesity during the transition from young adulthood to middle age. We analyzed data from the Framingham Offspring and Coronary Artery Risk Development in Young Adults (CARDIA) studies, in which participants were weighed at an initial baseline assessment and at additional study visits over the subsequent decades (Feinleib et al., 1975; Friedman et al., 1988). We identified 3,722 young adult participants – none of whom were severely obese at time of baseline assessment – in whom GPS calculation was possible. Mean age at baseline assessment was 28.0 years, 48% were female, and mean BMI was 24.2 kg/m2. These individuals were weighed at up to 8 subsequent visits over a median follow-up of 27 years to determine incidence of severe obesity.
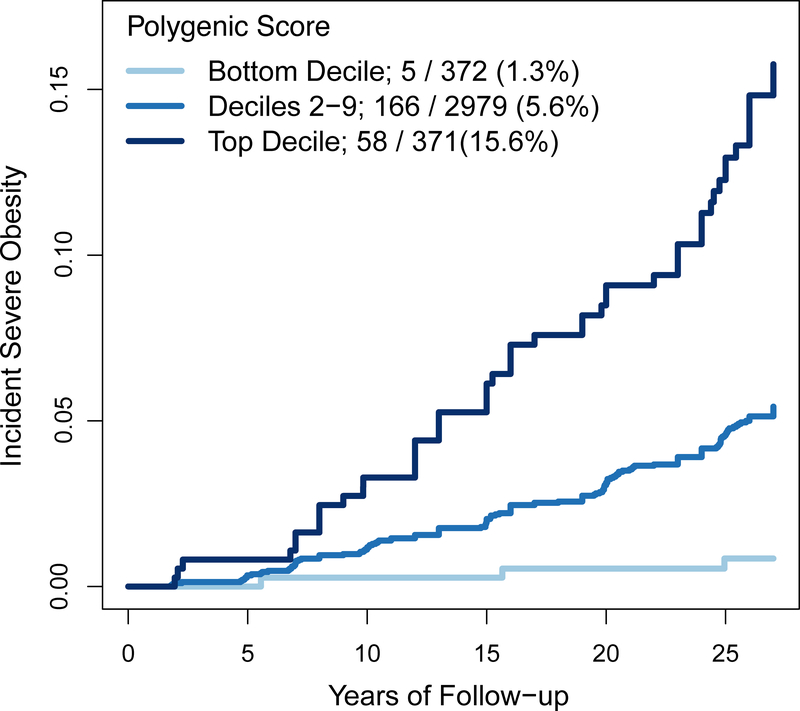
Among individuals in the top decile of the GPS, 58 of 371 (15.6%) went on to develop severe obesity as compared with 5.6% of those in deciles 2—9 (Figure 5). By contrast, among those in the lowest decile, only 5 of 372 (1.3%) individuals went on to develop severe obesity.
Figure 5. Association of GPS with incident severe obesity among young adults.
Among 3,722 young adults in the Framingham Offspring and Coronary Artery Risk Development in Young Adults studies, individuals were stratified based on their GPS into three categories – bottom decile, deciles 2–9, and top decile. Incident severe obesity is plotted according to GPS category over a median follow-up of 27 years (p < 0.0001 for each between-group comparison).
Impact of polygenic susceptibility emerges in early childhood
Given the gradients in weight and severe obesity observed in adulthood, we next posed the question – at what age does this gradient first start to emerge? We explored this question in a birth cohort from the United Kingdom, the Avon Longitudinal Study of Parents and Children (ALSPAC); (Boyd et al., 2013; Fraser et al., 2013). The ALSPAC study recruited pregnant mothers in the United Kingdom between 1991 and 1992, and followed offspring with serial weight assessments from time of birth to age 18 years. We identified 7,861 participants with both weight and genotyping array data available for analysis.
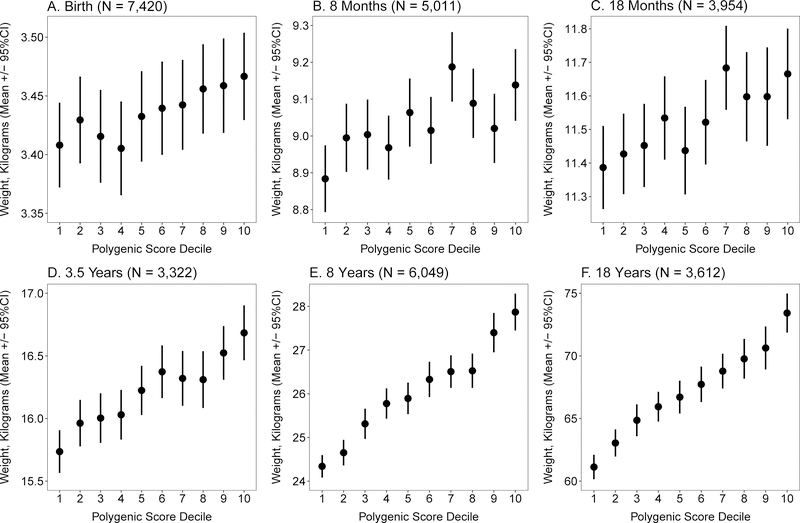
The GPS was associated with only small differences in birthweight: the mean was 3.47 kilograms for those in the top decile vs. 3.41 kilograms for those in the bottom decile, a difference of 0.06 kilograms (p = 0.02) (Figure 6A–F). By age 8 years, the difference increased to 3.5 kilograms (p < 0.0001), with mean weight 27.9 versus 24.3 kilograms. By age 18 years, the difference reached 12.3 kilograms (p < 0.0001). Strikingly, this weight difference between top and bottom GPS deciles at age 18 years (12.3 kilograms) was comparable to that seen in participants in the UK Biobank at mean age 57 years (13.0 kilograms).
Figure 6. Association of obesity GPS decile with weight from birth to 18 years.
Within the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort, 7,861 participants were stratified according to decile of the GPS distribution. Average weight and 95% confidence interval within each decile is displayed at 6 representative ages; corresponding sample size for number of participants with follow-up weight available at each time point is provided (Panels A-F). P-value for linear trend across deciles was 0.003 at birth (A) and < 0.0001 at all subsequent ages. See also Figure S4 and Figure S5.
We observed similar results after converting participants’ weights to z-scores – the number of standard deviations a child’s weight differs from a population and age-specific normative value (Figure S4). The difference in z-score between the top and bottom deciles was 0.11 for birthweight (p = 0.03), but this gradient had increased to 0.75 by 8 years and 0.90 by 18 years (p < 0.0001).
We modeled the trajectories of weight from birth to 18 years, stratifying individuals according to the top decile of the GPS distribution, deciles 2–9, and the bottom decile. This longitudinal analysis confirmed a separation in weight that starts in early childhood and continues to diverge into adulthood (Figure S5).
DISCUSSION
We describe a systematic approach to derive and validate a GPS, incorporating information from 2.1 million common genetic variants, to predict polygenic susceptibility to obesity and tested the polygenic score in 306,135 participants from four cohorts. The GPS accurately predicted striking differences in weight, severe obesity, cardiometabolic disease and overall mortality in middle-aged adults, with the extreme of the GPS distribution inheriting susceptibility to obesity equivalent to rare monogenic mutations in MC4R. The score had minimal association with birthweight, but it was strongly associated with a gradient in weight that started to emerge in early childhood and even larger differences in weight and severe obesity in subsequent decades.
The GPS far outperformed a score based only on the 141 variants most strongly associated with BMI, consistent with the highly polygenic nature of BMI and obesity. For example, in a direct comparison in 119,951 individuals, we observe a correlation with BMI of 0.29 for the GPS as compared with 0.13 with the 141-variant score. This improved performance using a genome-wide set of common variants was anticipated by a prior theoretical projection study based on early GWAS results and an analysis that indicated minimal ‘missing heritability’ of BMI when accounting for the full range of observed genetic variation (Chatterjee et al., 2013; Yang et al., 2015). Here, we use a recently developed computational algorithm that explicitly models the correlation structure between variants in calculating variant weights(Vilhjalmsson et al., 2015). This algorithm has been shown to outperform prior methods for a range of complex traits including cardiovascular disease, type 2 diabetes, and educational attainment (Khera et al., 2018a; Lee et al., 2018).
The ability to quantify inborn susceptibility using genome-wide polygenic scores is likely to be generalizable across a broad range of complex diseases, contingent upon availability of a large discovery GWAS, independent validation and testing datasets, and the heritability of a given disease explained by common variants (Torkamani et al., 2018). Predictive power will likely continue to improve in coming years as a function of larger discovery GWAS studies and improved computational algorithms that integrate functional genomics annotation, variant-variant interactions, and rare large-effect variants into the predictive model (Chatterjee et al., 2016; Zhang et al., 2018).
We note that both a pathogenic MC4R mutation and the extreme of the GPS distribution predisposed individuals to a BMI 4.1 kg/m2 higher than the remainder of the population. However, despite an identical effect size, we estimate that extreme GPS has a prevalence an order of magnitude higher than pathogenic MC4R mutations – 1.6% versus 0.14% respectively.
Both extreme polygenic score and pathogenic MC4R mutations demonstrate incomplete penetrance – not all carriers manifest severe obesity. This observation is consistent with recent large-scale gene sequencing studies across a broad range of complex diseases, including diabetes, cardiovascular disease, and breast cancer (Flannick et al., 2013; Khera et al., 2016; Manickam et al., 2018). Additional studies of large unascertained populations are needed to determine whether a larger effect size for pathogenic MC4R mutations is noted among children or young adults – as has been suggested in prior reports (Farooqi et al., 2003; Stutzmann et al., 2008) – and the extent to which a favorable polygenic background can explain the absence of obesity noted among many mutation carriers.
Genetic risk predictors have important potential implications for clinical medicine, because they identify individuals at risk before the condition has manifested. For example, individuals with high polygenic score for heart attack derive the greatest benefit from preventive medications such as cholesterol lowering therapy and those with the highest polygenic scores for breast cancer may benefit from earlier and more intensive mammography screening (Natarajan et al., 2017; Pharoah et al., 2008).
Although the average BMI has increased substantially across populations, so too has the variability within any given population – suggesting that an increasingly obesogenic environment may have led to preferential ‘unmasking’ of inherited susceptibility among those with highest genetic risk (Smith, 2016; Yanovski and Yanovski, 2018). For example, prior studies suggest that the impact of unhealthy diet, physical activity, and sedentary behavior on BMI are most pronounced in those with a genetic predisposition (Qi et al., 2014; Qi et al., 2012; Tyrrell et al., 2017). The ability to identify high-risk individuals from the time of birth may facilitate targeted strategies for obesity prevention with increased impact or cost-effectiveness. Given that the weight trajectories of individuals in different GPS deciles start to diverge in early childhood, such interventions may have maximal impact when employed early in life.
The GPS may also accelerate research insights into the molecular and physiological basis of severe obesity. Traditional research approaches have compared the physiology of severely obese individuals to lean controls. However, it is difficult to draw inferences from such studies, since observed differences might be either a cause or a consequence of severe obesity. The GPS permits identification of individuals, from the time of birth, who inherit high susceptibility and before clinical disease is manifest. Careful study of individuals at the extremes of a GPS distribution might uncover new causal risk factors or pathways underlying disease. For example, healthy individuals with high polygenic score for heart attack were enriched for higher blood pressure, increased cholesterol levels, and increased rates of type 2 diabetes – each of these is a well-known and modifiable clinical risk factor (Khera et al., 2018b). Similarly, clinical and multi-omic profiling of those at the extremes of a GPS distribution for obesity may uncover the contributions and molecular correlates of pathways related to appetite regulation, fat storage, and microbiome perturbation and might enable identification of clinically relevant subtypes of severe obesity that most benefit from a given pharmacologic or behavioral intervention.
Individuals who maintain normal weight despite an unfavorable GPS – or develop severe obesity despite a favorable GPS – may be of particular interest. The discordance between polygenic susceptibility and clinical phenotype in these individuals could result from a disproportionate influence of environment, the effect of a rare, large-effect mutation not captured by the polygenic score, or other undetermined factors.
Finally, a clear understanding of the genetic predisposition to obesity may help to destigmatize obesity among patients, their health care providers, and the general public.
We anticipate that our approach to constructing a robust GPS predictor of obesity will generalize across a range of common diseases, raising both important opportunities and potential challenges for clinical medicine. First, the cohorts studied here were of European ancestry – future studies are needed to extend this approach across additional ancestral background and ensure equitable implementation into clinical practice. Second, rare monogenic mutations for conditions such as obesity can sometimes be treated by precise targeting of the perturbed pathway (Kuhnen et al., 2016). Whether polygenic risk can be disaggregated into driving pathways within each individual in a similar fashion remains uncertain (Khera and Kathiresan, 2017). Lastly, additional work is needed to optimize genetic risk disclosure and to test whether this disclosure can improve disease prevention or treatment.
STAR METHODS
LEAD CONTACT FOR REAGENT AND RESOURCE SHARING
Sekar Kathiresan, MD, Center for Genomic Medicine, Massachusetts General Hospital, 185 Cambridge Street, CPZN 5.821A, Boston, MA 02114, skathiresan1@mgh.harvard.edu
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Study cohorts
The UK Biobank is a large observational study that enrolled 502,617 individuals aged 40 to 69 years of age from across the United Kingdom beginning in 2006 (Sudlow et al., 2015). We identified 407,969 individuals of European ancestry with genotyping array and BMI data available. Individuals in the UK Biobank underwent genotyping with one of two closely related genotyping arrays consisting of over 800,000 genetic markers scattered across the genome (Bycroft et al., 2018). Additional genotypes were imputed centrally using the Haplotype Reference Consortium panel version 1.1, the UK10K panel, and the 1000 Genomes panel. To analyze individuals with a relatively homogenous ancestry and owing to small percentages of non-British individuals, the present analysis was restricted to the white British ancestry individuals. This subpopulation was constructed centrally using a combination of self-reported ancestry and genetic confirmation by principal components of ancestry. Additional exclusion criteria included outliers for heterozygosity or genotype missing rates, discordant reported versus genotypic sex, putative sex chromosome aneuploidy, or withdrawal of informed consent, derived centrally as previously reported (Bycroft et al., 2018).
The genome-wide polygenic score was validated within 119,951 participants of the UK Biobank Phase 1 validation dataset, and subsequently tested in the remaining 288,016 participants. Avoidance of sample overlap between the validation and testing datasets prevents test statistic inflation (Vilhjalmsson et al., 2015).
Whole exome sequencing was performed at the Broad Institute of MIT and Harvard (Cambridge, MA, USA) in a subset of 6,552 UK Biobank participants. Libraries were constructed as previously reported (Fisher et al., 2011) and sequenced on an Illumina HiSeq sequencer with the use of 151 bp paired-end reads. In-solution hybrid selection was performed using the Illumina Nextera Exome Kit. Aligned non-duplicate reads were locally realigned and base qualities were recalibrated using Genome Analysis Toolkit software (McKenna et al., 2010; Van der Auwera et al., 2013). Variants were jointly called using Genome Analysis Toolkit HaplotypeCaller software. We removed samples with contamination > 10% (N = 0), samples with < 80% of target bases at 20X coverage (N = 3), samples with discordance between self-reported and genetic sex (N = 0), and samples with discordant reported versus genotypic sex (N = 2). Mean target coverage among the remaining 6,547 samples was 75X, and 91.4% of target bases were captured at >20X sequencing depth. The subset of rare (allele frequency < 1%) variants in the melanocortin 4 receptor gene (MC4R; Ensemble transcript ID: ENST00000299766) were narrowed to those meeting American College of Medical Genetics and Genomics (ACMG)/Association of Molecular Pathology (AMP) pathogenic or likely pathogenic criteria by an American Board of Genetics and Genomics (AMBGG)-certified clinical laboratory geneticist within the Partners HealthCare Laboratory for Molecular Medicine (Boston, MA, USA) who was blinded to any phenotype information (Richards et al., 2015). Among the 24 rare coding variants analyzed, mean sequencing depth across the 6,547 variants was 80X with a genotype missingness rate of 0%. See also Table S4.
The Partners HealthCare system case-control cohort was assembled using 718 individuals with severely obese individuals of European ancestry who underwent Roux-en-Y gastric bypass surgery in the Partners HealthCare System (Hatoum et al., 2013). 5,822 controls were derived from a population of European participants of the Partners HealthCare Biobank (Karlson et al., 2016). Control participants were excluded if they had undergone bariatric surgery or had a Charlson comorbidity index >3 (Charlson et al., 1987). Samples were imputed to the Haplotype Reference Consortium panel version 1.1 using the Michigan Imputation Server (Das et al., 2016; McCarthy et al., 2016).
The Framingham Offspring Study is a prospective cohort study that recruited 5,124 individuals beginning in 1971 (Feinleib et al., 1975). We identified 2,177 young adults aged 18 to 40 years with available data on BMI. BMI was assessed at baseline and during six subsequent visits to ascertain incident severe obesity. Individuals with severe obesity at baseline or missing data from subsequent visits were excluded.
The Coronary Artery Risk Development in Young Adults (CARDIA) Study is a prospective cohort study of 5,115 black and white participants beginning in 1985 (Friedman et al., 1988). We analyzed 1,545 white participants aged 18 to 30 years at time of enrollment. BMI was assessed at baseline and up to 8 subsequent visits to ascertain incident severe obesity. Individuals with missing baseline BMI, severe obesity at baseline, and pregnant females were excluded.
The Avon Longitudinal Study of Parents and Children (ALSPAC) is a prospective birth cohort study investigating factors that influence normal childhood development and growth (Boyd et al., 2013; Fraser et al., 2013). Briefly, 14,541 pregnant women resident in a defined area of the South West of England, with an expected delivery date of April 1, 1991 and December 31, 1992 were enrolled to the cohort. Of these, 13,988 live-born children who were still alive 1 year later have been followed-up to date with regular questionnaires and clinical measures, providing behavioral, lifestyle and biological data. For the present analysis, up to 7,861 participants with both weight and genotyping array data available were included. Weight was assessed at subsequent visits up to age 18 years. Z-scores were computed using the Growth Analyzer RCT program (https://growthanalyser.org/software/growth-analyser-rct/), with age-specific reference weights derived from the United Kingdom/Northern Ireland reference population. The study website (http://www.bristol.ac.uk/alspac/researchers/our-data/) contains details of all the data that are available through a fully searchable data dictionary and variable search tool.
Informed consent and study approval
Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committee and written informed consent was obtained from both the parent/guardian and, after the age of 16, children provided written assent. For the three remaining cohorts, informed consents were obtained by investigators of each study and analysis approved by the Institutional Review Board of Partners HealthCare (Boston, MA).
METHOD DETAILS
Polygenic score derivation and validation
Polygenic scores provide a quantitative metric of an individuals inherited risk based on the cumulative impact of many common (minor allele frequency ≥1%) variants. Weights are generally assigned to each genetic variant according to the strength of its association with a given trait (effect estimate). Individuals are scored based on how many risk alleles they have for each variant (e.g. 0, 1, 2 copies) included in the polygenic score.
For our score derivation, we used summary statistics from a recent genome-wide association study (GWAS) for body mass index (BMI) including up to 339,224 individuals and a linkage disequilibrium reference panel of 503 European samples from 1000 Genomes phase 3 version 5 (Locke et al., 2015; The 1000 Genomes Project Consortium, 2015). DNA polymorphisms with ambiguous strand (A/T or C/G) were removed from the score derivation.
5 candidate polygenic scores were derived using the LDPred computational algorithm (Vilhjalmsson et al., 2015). This Bayesian approach calculates a posterior mean effect size for each variant based on a prior and subsequent shrinkage based on the extent to which this variant is correlated with similarly associated variants in the reference population. The underlying Gaussian distribution additionally considers the fraction of causal (i.e., non-zero effect sizes) markers via a tuning parameter, ρ. Because ρ is unknown for any given disease, a range of ρ values, the fraction of causal variants, was used – 1, 0.3, 0.1, 0.03, 0.01. A sixth score was derived with variants restricted to those meeting genome-wide levels of statistical significance (p< 5 × 10−8) using the linkage disequilibrium-based clumping procedure in PLINK version 2.0 (Chang et al., 2015). The algorithm identifies a list of independent (r2 < 0.2) variants with this level of statistical significance.
The 6 candidate polygenic scores were calculated in a validation dataset of 119,951 participants of European ancestry of the UK Biobank Phase I validation dataset. More than 99% of variants in the genome-wide polygenic scores were available for scoring purposes in the validation dataset with sufficient imputation quality (INFO > 0.3); Table S1. The polygenic score with the strongest correlation with observed BMI in the validation dataset was determined based on Pearson correlation, and the best score carried forward into subsequent analyses in independent testing datasets. A sensitivity analysis that included adjustment for genetic background was performed as described previously (Khera et al., 2018b). In brief, we fit a linear regression model using the first ten principal components of ancestry to predict polygenic score. The residual from this model was used to create an ancestry-corrected polygenic score.
UK Biobank Phenotypes
Within the UK Biobank, severe obesity was defined as BMI ≥ 40 kg/m2 (NHLBI Expert Panel, 1998). Additional phenotypes with respect to disease status and bariatric surgery were ascertained via linkage to data based on self-report in an interview with a trained nurse, and diagnosis and procedure codes within the electronic health record. Bariatric surgery status was ascertained based on having a OPCS-4 primary procedure code for Roux-en-Y gastric bypass, sleeve gastrectomy, or duodenal switch procedure (G28.1–5, G31.2, G 32.1, G33.1, G71.6).
With respect to additional classification of prevalent cardiometabolic diseases in UK Biobank participants, coronary artery disease ascertainment was based on a composite of myocardial infarction or coronary revascularization. Myocardial infarction was based on self-report or hospital admission diagnosis, as performed centrally by the UK Biobank. This included individuals with International Classification of Diseases (ICD)-9 codes of 410.X, 411.0, 412.X, 429.79 or ICD-10 codes of I21.X, I22.X, I23.X, I24.1, I25.2 in hospitalization records. Coronary revascularization was assessed based on an OPCS4 coded procedure for coronary artery bypass grafting (K40.1–40.4, K41.1–41.4, K45.1–45.5) or coronary angioplasty with or without stenting (K49.1–49.2, K49.8–49.9, K50.2, K75.1–75.4, K75.8–75.9). Diabetes mellitus ascertainment was based on a composite of self-report, use of insulin, ICD-9 codes of 250.X or ICD-10 codes of E10.X, E11.X, E12.X, E 13.X, E14.X in hospitalization records. Hypertension ascertainment was based on self-report, ICD-9 codes of 40.X or ICD-10 codes of I10, I11.X, I 12.X, I13.X, or I15.X in hospitalization records. Heart failure was ascertained based on self-report, ICD-9 codes of 425.4, 428.0, 428.1, 428.9 or ICD-10 codes of I11.0, I13.0, I13.2, I25.5, I42.X in hospitalization records. Ischemic stroke was ascertained centrally based on self-report or hospitalization admission diagnosis of ICD-9 codes 430, 431, 434, or 436 and ICD-10 codes of I60, I61, I63, I64. Venous thromboembolism was diagnosed based on self-report, ICD-9 codes of 415.1, 451.1, ICD-10 codes of I26.X, I80.X, I82.X in the hospitalization records or insertion of an IVC filter or open thrombectomy of lower-extremity veins in procedure registries.
QUANTIFICATION AND STATISTICAL ANALYSIS
We estimated the heritability of BMI based on common variation within the validation set of the UK Biobank composed of 119,951 European individuals. We used previously recommended parameters suggested for heritability assessment using LD-score regression (Bulik-Sullivan et al., 2015). In brief, we tested for an association between 1,163,095 common variants that were well-imputed and available in HapMap3 (minor allele frequency > 0.01, imputation INFO > 0.9) and BMI using a linear regression model adjusted for age, sex, genotyping array, and the first 10 principal components of ancestry. We then estimated heritability using the resulting association statistics and a linkage disequilibrium reference panel of individuals of European ancestry from the 1000 Genomes Study (The 1000 Genomes Project Consortium, 2015).
Within each of the four testing cohorts, genotyping array data was imputed and a genome-wide polygenic score calculated for each individual. Scores were generated by multiplying the genotype dosage of each risk allele for each variant by its respective weight, and then summing across all variants in the score. Incorporating genotype dosages accounts for uncertainty in genotype imputation. Scoring was done using the PLINK2 software program (Chang et al., 2015). Within the UK Biobank, participants were stratified according to decile of the GPS. Average weight and prevalence of severe obesity was determined within each decile. The relationship of high polygenic score, defined as the top decile of the GPS, with severe obesity and treatment with bariatric surgery was next determined in both the UK Biobank and Partners HealthCare system cohorts using logistic regression.
Associations of high polygenic score with severe obesity, bariatric surgery, and six cardiometabolic diseases, were determined using logistic regression models. Association with incident all-cause mortality was determined using a Cox regression model survival analysis.
The incidence of severe obesity among young adults according to GPS category was assessed in the Framingham Offspring and CARDIA studies using an unadjusted Kaplan-Meier survival analysis.
Within the ALSPAC cohort, individuals were stratified according to GPS decile and mean weights determined within each of 6 representative ages. P-values for linear trend were assessed using GPS decile as a predictor of observed weight at each age. Linear spline multi-level models were used to examine the association between the polygenic score and change in weight from birth to 18 years. Multi-level models estimate the mean trajectories of weight while accounting for non-independence of repeated measures within individuals, change in scale and variance of measures over time, differences in the number and timing of measurements between individuals (using all available data from all eligible participants under a missing-at-random assumption) (Howe et al., 2016; Tilling et al., 2014). Linear splines allow know points to be fitted at different ages to derive periods of change that are approximately linear. All participants with at least one measure of weight were included under a missing-at-random assumption to minimize selection bias in trajectories estimated using linear spline multi-level models (with two levels: measurement occasion and individual). Knot points were placed at ages 1, 8 and 15 years based on the distribution and longitudinal pattern of weight measures between birth and 18 years. All trajectories were models in MLwiN version 3.01 (UoBc, 2017) called from Stata version 15 using the “runmlwin” command (UoBc, 2016).
Statistical analyses were conducted using R version 3.4.3 software (The R Foundation) and Stata version 15.
DATA AND SOFTWARE AVAILABILITY
The genome-wide polygenic score validated and tested here will be made available to the research community prior to publication at: http://www.broadcvdi.org/informational/data.
Supplementary Material
Figure S1. Distribution of the genome-wide polygenic score for obesity, Related to Figure 2.
The distribution of the genome-wide polygenic score (GPS) in 288,016 participants of the UK Biobank testing dataset is displayed. The x-axis represents the polygenic score, with values scaled to a mean of 0 and standard deviation 1 to facilitate interpretation.
Figure S2. Relationship of genome-wide polygenic score distribution with obesity, Related to Figure 2
288,016 middle-aged UK Biobank participants were binned into 10 deciles according to the polygenic score, with significant differences in the prevalence of obesity (body mass index ≥ 30 kg/m2) noted across deciles of the polygenic score (p < 0.0001).
Figure S3. Relationship of genome-wide polygenic score distribution with body mass index, weight, and severe obesity, including adjustment for principal components of ancestry, Related to Figure 2
A sensitivity analysis was performed after adjustment of the genome-wide polygenic score for genetic background, as assessed by the first ten principal components of ancestry. 288,016 middle-aged UK Biobank participants were binned into 10 deciles according to this ancestry-adjusted genome-wide polygenic score. Body mass index (Panel A), weight (Panel B), and prevalence of severe obesity (Panel C) each increased across deciles of the polygenic score (p < 0.0001 for each). Significant differences in clinical categories of obesity were noted (Panel D) when participants were stratified into three categories – bottom decile, deciles 2–9, and top decile. Underweight refers to BMI < 18.5 kg/m2, normal as 18.5 to 24.9 kg/m2, overweight as 25.0 to 29.9 kg/m2, obesity as 30.0 to 39.9 kg/m2, and severe obesity as ≥ 40 kg/m2.
Figure S4. Association of obesity GPS decile with weight z-scores from birth to 18 years, Related to Figure 6
Within the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort, 7,861 participants were stratified according to decile of the GPS distribution. Average z-score and 95% confidence interval within each decile is displayed at 6 representative ages; corresponding sample size for number of participants with follow-up weight available at each time point is provided (Panels A-F). P-value for linear trend across deciles was 0.003 at birth (A) and < 0.0001 at all subsequent ages.
Figure S5. Association of GPS with longitudinal weight trajectory from birth to 18 years, Related to Figure 6
Among 7,861 participants of the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort, individual were stratified based on their GPS into three categories – bottom decile, deciles 2–9, and top decile. Longitudinal weight trajectories from birth to 18 years were modeled using linear spline multilevel models with knot points.
Table S4. Variant Classification of Melanocortin 4 Receptor (MC4R) gene identified by whole exome sequencing, Related to STAR Methods.
Melanocortin 4 Receptor (MC4R) gene variants identified by exome sequencing of 6,547 UK Biobank participants were classified according to current clinical guidelines (Richards et al., 2015) to identify those that with allele frequency < 0.01 that were pathogenic or likely pathogenic for obesity. Variants are described based on ‘chromosome:position:reference allele>alternate allele’ formatting, with chromosome positions based on the GRCh37 genome assembly.
HIGHLIGHTS.
A genome-wide polygenic score can quantify inherited susceptibility to obesity
Polygenic score impact on weight emerges early in life and increased into adulthood
Impact of polygenic score can be similar to a rare, monogenic obesity mutation
High polygenic score is a strong risk factor for severe obesity and associated diseases
ACKNOWLEDGEMENTS
This work was supported by a K08 award from the National Human Genome Research Institute (1K08HG0101), a Junior Faculty Research Award from the National Lipid Association, a BroadIgnite grant from the Broad Institute of MIT and Harvard (to A.V.K.), funding from the Wellcome Trust (202802/Z/16/Z), the University of Bristol NIHR Biomedical Research Centre (S- BRC-1215-20011), and the MRC Integrative Epidemiology Unit (MC_UU_12013/3 to N.J.T.), a RO1 award the National Heart Lung and Blood Institute (HL127564 to S.K.) and the Ofer and Shelly Nemirovsky Research Scholar Award from Massachusetts General Hospital (to S.K.).
Exome sequencing of UK Biobank participants was supported by a UM1 award from the National Human Genome Research Institute (HG008895; to E.S.L. and S.K.).
The Partners HealthCare System bariatric surgery cohort analysis was supported by grants from the National Institutes of Health (DK088661, DK090956, and DK040561), Merck Research Laboratories, and Ethicon Endo-Surgery (all to L.M.K.).
The Coronary Artery Risk Development in Young Adults Study (CARDIA) is supported by contracts HHSN268201300025C, HHSN268201300026C, HHSN268201300027C, HHSN268201300028C, HHSN268201300029C, and HHSN268200900041C from the National Heart, Lung, and Blood Institute (NHLBI), the Intramural Research Program of the National Institute on Aging (NIA), and an intra-agency agreement between NIA and NHLBI (AG0005). Genotyping and imputation were funded as part of the Gene Environment Association Studies (GENEVA) through grants U01-HG004729, U01-HG04424, and U01-HG004446 from the National Human Genome Research Institute. This manuscript has been reviewed and approved by CARDIA for scientific content.
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and Wellcome (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. The ALSPAC analysis is the work of the authors, and Drs. Kaitlin Wade and Professor Nicholas Timpson will serve as guarantors for the contents of this paper.
Footnotes
DECLARATION OF INTERESTS
A.V.K. and S.K. are listed as co-inventors on a patent application for the use of polygenic scores to determine risk and guide therapy, and have received consultant fees from Color Genomics (Burlingame, CA). E.S.L serves on the Board of Directors for Codiak BioSciences and Neon Therapeutics, and serves on the Scientific Advisory Board of F-Prime Capital Partners and Third Rock Ventures; he is also affiliated with several non-profit organizations including serving on the Board of Directors of the Innocence Project, Count Me In, and Biden Cancer Initiative, and the Board of Trustees for the Parker Institute for Cancer Immunotherapy. He has served and continues to serve on various federal advisory committees.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barsh GS, Farooqi IS, and O’Rahilly S (2000). Genetics of body-weight regulation. Nature 404, 644–651. [DOI] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Sugden K, Williams B, Houts R, McCarthy J, and Caspi A (2013). Development and evaluation of a genetic risk score for obesity. Biodemography Soc Biol 59, 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, and Davey Smith G (2013). Cohort Profile: the ‘children of the 90s’--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 42, 111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, et al. (2008). Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet 4, e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics, C., Patterson N, Daly MJ, Price AL, and Neale BM (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, and Lee JJ (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson ME, Pompei P, Ales KL, and MacKenzie CR (1987). A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40, 373–383. [DOI] [PubMed] [Google Scholar]
- Chatterjee N, Shi J, and Garcia-Closas M (2016). Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet 17, 392–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, and Park JH (2013). Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet 45, 400–405, 405e401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, et al. (2016). Next-generation genotype imputation service and methods. Nat Genet 48, 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elks CE, den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos RJ, and Ong KK (2012). Variability in the heritability of body mass index: a systematic review and meta-regression. Front Endocrinol (Lausanne) 3, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, and O’Rahilly S (2003). Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med 348, 1085–1095. [DOI] [PubMed] [Google Scholar]
- Feinleib M, Kannel WB, Garrison RJ, McNamara PM, and Castelli WP (1975). The Framingham Offspring Study. Design and preliminary data. Prev Med 4, 518–525. [DOI] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. (2011). A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 12, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick J, Beer NL, Bick AG, Agarwala V, Molnes J, Gupta N, Burtt NP, Florez JC, Meigs JB, Taylor H, et al. (2013). Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nat Genet 45, 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegal KM, Kruszon-Moran D, Carroll MD, Fryar CD, and Ogden CL (2016). Trends in Obesity Among Adults in the United States, 2005 to 2014. JAMA 315, 2284–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, Henderson J, Macleod J, Molloy L, Ness A, et al. (2013). Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol 42, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR Jr., Liu K, and Savage PJ (1988). CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 41, 1105–1116. [DOI] [PubMed] [Google Scholar]
- Golan D, Lander ES, and Rosset S (2014). Measuring missing heritability: inferring the contribution of common variants. Proc Natl Acad Sci U S A 111, E5272–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum IJ, Greenawalt DM, Cotsapas C, Daly MJ, Reitman ML, and Kaplan LM (2013). Weight loss after gastric bypass is associated with a variant at 15q26.1. Am J Hum Genet 92, 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe LD, Tilling K, Matijasevich A, Petherick ES, Santos AC, Fairley L, Wright J, Santos IS, Barros AJ, Martin RM, et al. (2016). Linear spline multilevel models for summarising childhood growth trajectories: A guide to their application using examples from five birth cohorts. Stat Methods Med Res 25, 1854–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CF, Breen G, Czamara D, Corre T, Wolf C, Kloiber S, Bergmann S, Craddock N, Gill M, Holsboer F, et al. (2015). A genetic risk score combining 32 SNPs is associated with body mass index and improves obesity prediction in people with major depressive disorder. BMC Med 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, et al. (1997). Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88, 131–141. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, and Sklar P (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson EW, Boutin NT, Hoffnagle AG, and Allen NL (2016). Building the Partners HealthCare Biobank at Partners Personalized Medicine: Informed Consent, Return of Research Results, Recruitment Lessons and Operational Considerations. J Pers Med 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. (2018a). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, Lichtman JH, D’Onofrio G, Mattera JA, Dreyer RP, et al. (2018b). Whole Genome Sequencing to Characterize Monogenic and Polygenic Contributions in Patients Hospitalized with Early-Onset Myocardial Infarction. Circulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, and Kathiresan S (2017). Is Coronary Atherosclerosis One Disease or Many? Setting Realistic Expectations for Precision Medicine. Circulation 135, 1005–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Won HH, Peloso GM, Lawson KS, Bartz TM, Deng X, van Leeuwen EM, Natarajan P, Emdin CA, Bick AG, et al. (2016). Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J Am Coll Cardiol 67, 2578–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnen P, Clement K, Wiegand S, Blankenstein O, Gottesdiener K, Martini LL, Mai K, Blume-Peytavi U, Gruters A, and Krude H (2016). Proopiomelanocortin Deficiency Treated with a Melanocortin-4 Receptor Agonist. N Engl J Med 375, 240–246. [DOI] [PubMed] [Google Scholar]
- Larsen LH, Echwald SM, Sorensen TI, Andersen T, Wulff BS, and Pedersen O (2005). Prevalence of mutations and functional analyses of melanocortin 4 receptor variants identified among 750 men with juvenile-onset obesity. J Clin Endocrinol Metab 90, 219–224. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Karlsson Linner R, et al. (2018). Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 50, 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhao JH, Luan J, Luben RN, Rodwell SA, Khaw KT, Ong KK, Wareham NJ, and Loos RJ (2010). Cumulative effects and predictive value of common obesity-susceptibility variants identified by genome-wide association studies. Am J Clin Nutr 91, 184–190. [DOI] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. (2015). Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJF, and Janssens A (2017). Predicting Polygenic Obesity Using Genetic Information. Cell Metab 25, 535–543. [DOI] [PubMed] [Google Scholar]
- Maes HH, Neale MC, and Eaves LJ (1997). Genetic and environmental factors in relative body weight and human adiposity. Behav Genet 27, 325–351. [DOI] [PubMed] [Google Scholar]
- Manickam K, Buchanan AH, Schwartz MLB, Hallquist MLG, Williams JL, Rahm AK, Rocha H, Savatt JM, Evans AR, Ledbetter DH, et al. (2018). Exome Sequencing–Based Screening for BRCA½ Expected Pathogenic Variants Among Adult Biobank Participants. JAMA Network Open 1, e182140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, et al. (2016). A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48, 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, Sartori S, Fuster V, Reilly DF, Butterworth A, et al. (2017). Polygenic Risk Score Identifies Subgroup With Higher Burden of Atherosclerosis and Greater Relative Benefit From Statin Therapy in the Primary Prevention Setting. Circulation 135, 2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCD Risk Factor Collaboration (2017). Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet 390, 2627–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NHLBI Expert Panel (1998). Clinical Guidelines on the Identification, Evaluation, and Treatment of Overweight and Obesity in Adults--The Evidence Report. National Institutes of Health. Obes Res 6 Suppl 2, 51S–209S. [PubMed] [Google Scholar]
- Pharoah PD, Antoniou AC, Easton DF, and Ponder BA (2008). Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med 358, 2796–2803. [DOI] [PubMed] [Google Scholar]
- Qi Q, Chu AY, Kang JH, Huang J, Rose LM, Jensen MK, Liang L, Curhan GC, Pasquale LR, Wiggs JL, et al. (2014). Fried food consumption, genetic risk, and body mass index: gene-diet interaction analysis in three US cohort studies. BMJ 348, g1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Chu AY, Kang JH, Jensen MK, Curhan GC, Pasquale LR, Ridker PM, Hunter DJ, Willett WC, Rimm EB, et al. (2012). Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med 367, 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17, 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandholt CH, Sparso T, Grarup N, Albrechtsen A, Almind K, Hansen L, Toft U, Jorgensen T, Hansen T, and Pedersen O (2010). Combined analyses of 20 common obesity susceptibility variants. Diabetes 59, 1667–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GD (2016). A fatter, healthier but more unequal world. Lancet 387, 1349–1350. [DOI] [PubMed] [Google Scholar]
- Stutzmann F, Tan K, Vatin V, Dina C, Jouret B, Tichet J, Balkau B, Potoczna N, Horber F, O’Rahilly S, et al. (2008). Prevalence of melanocortin-4 receptor deficiency in Europeans and their agedependent penetrance in multigenerational pedigrees. Diabetes 57, 2511–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. (2015). UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12, e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilling K, Macdonald-Wallis C, Lawlor DA, Hughes RA, and Howe LD (2014). Modelling childhood growth using fractional polynomials and linear splines. Ann Nutr Metab 65, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyama AJ, Carr D, Granberg EM, Major B, Robinson E, Sutin AR, and Brewis A (2018). How and why weight stigma drives the obesity ‘epidemic’ and harms health. BMC Med 16, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Wineinger NE, and Topol EJ (2018). The personal and clinical utility of polygenic risk scores. Nat Rev Genet 19, 581–590. [DOI] [PubMed] [Google Scholar]
- Turcot V, Lu Y, Highland HM, Schurmann C, Justice AE, Fine RS, Bradfield JP, Esko T, Giri A, Graff M, et al. (2018). Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet 50, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell J, Wood AR, Ames RM, Yaghootkar H, Beaumont RN, Jones SE, Tuke MA, Ruth KS, Freathy RM, Davey Smith G, et al. (2017). Gene-obesogenic environment interactions in the UK Biobank study. Int J Epidemiol 46, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UoBc, C.f.M.M. (2016). runmlwin: Stata module for fitting multilevel models in the MLwiN software.
- UoBc, C.f.M.M. (2017). MLwiN Version 3.01.
- Vaisse C, Clement K, Durand E, Hercberg S, Guy-Grand B, and Froguel P (2000). Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest 106, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisse C, Clement K, Guy-Grand B, and Froguel P (1998). A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet 20, 113–114. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11 10 11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhjalmsson BJ, Yang J, Finucane HK, Gusev A, Lindstrom S, Ripke S, Genovese G, Loh PR, Bhatia G, Do R, et al. (2015). Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet 97, 576–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, and Yang J (2012). Five years of GWAS discovery. Am J Hum Genet 90, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker RC, Wright JA, Pepe MS, Seidel KD, and Dietz WH (1997). Predicting obesity in young adulthood from childhood and parental obesity. N Engl J Med 337, 869–873. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Litherland SA, Sorensen NB, Proneth B, Wood MS, Shaw AM, Millard WJ, and Haskell-Luevano C (2006). Pharmacological characterization of 40 human melanocortin-4 receptor polymorphisms with the endogenous proopiomelanocortin-derived agonists and the agouti-related protein (AGRP) antagonist. Biochemistry 45, 7277–7288. [DOI] [PubMed] [Google Scholar]
- Yampolsky LY, Kondrashov FA, and Kondrashov AS (2005). Distribution of the strength of selection against amino acid replacements in human proteins. Hum Mol Genet 14, 3191–3201. [DOI] [PubMed] [Google Scholar]
- Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AA, Lee SH, Robinson MR, Perry JR, Nolte IM, van Vliet-Ostaptchouk JV, et al. (2015). Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 47, 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, et al. (2011). Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet 43, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovski SZ, and Yanovski JA (2018). Toward Precision Approaches for the Prevention and Treatment of Obesity. JAMA 319, 223–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GS, Farooqi IS, Aminian S, Halsall DJ, Stanhope RG, and O’Rahilly S (1998). A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat Genet 20, 111–112. [DOI] [PubMed] [Google Scholar]
- Zhang M, Lykke-Andersen S, Zhu B, Xiao W, Hoskins JW, Zhang X, Rost LM, Collins I, Bunt MV, Jia J, et al. (2018). Characterising cis-regulatory variation in the transcriptome of histologically normal and tumour-derived pancreatic tissues. Gut 67, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Bakshi A, Vinkhuyzen AA, Hemani G, Lee SH, Nolte IM, van Vliet-Ostaptchouk JV, Snieder H, LifeLines Cohort S, Esko T, et al. (2015). Dominance genetic variation contributes little to the missing heritability for human complex traits. Am J Hum Genet 96, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Distribution of the genome-wide polygenic score for obesity, Related to Figure 2.
The distribution of the genome-wide polygenic score (GPS) in 288,016 participants of the UK Biobank testing dataset is displayed. The x-axis represents the polygenic score, with values scaled to a mean of 0 and standard deviation 1 to facilitate interpretation.
Figure S2. Relationship of genome-wide polygenic score distribution with obesity, Related to Figure 2
288,016 middle-aged UK Biobank participants were binned into 10 deciles according to the polygenic score, with significant differences in the prevalence of obesity (body mass index ≥ 30 kg/m2) noted across deciles of the polygenic score (p < 0.0001).
Figure S3. Relationship of genome-wide polygenic score distribution with body mass index, weight, and severe obesity, including adjustment for principal components of ancestry, Related to Figure 2
A sensitivity analysis was performed after adjustment of the genome-wide polygenic score for genetic background, as assessed by the first ten principal components of ancestry. 288,016 middle-aged UK Biobank participants were binned into 10 deciles according to this ancestry-adjusted genome-wide polygenic score. Body mass index (Panel A), weight (Panel B), and prevalence of severe obesity (Panel C) each increased across deciles of the polygenic score (p < 0.0001 for each). Significant differences in clinical categories of obesity were noted (Panel D) when participants were stratified into three categories – bottom decile, deciles 2–9, and top decile. Underweight refers to BMI < 18.5 kg/m2, normal as 18.5 to 24.9 kg/m2, overweight as 25.0 to 29.9 kg/m2, obesity as 30.0 to 39.9 kg/m2, and severe obesity as ≥ 40 kg/m2.
Figure S4. Association of obesity GPS decile with weight z-scores from birth to 18 years, Related to Figure 6
Within the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort, 7,861 participants were stratified according to decile of the GPS distribution. Average z-score and 95% confidence interval within each decile is displayed at 6 representative ages; corresponding sample size for number of participants with follow-up weight available at each time point is provided (Panels A-F). P-value for linear trend across deciles was 0.003 at birth (A) and < 0.0001 at all subsequent ages.
Figure S5. Association of GPS with longitudinal weight trajectory from birth to 18 years, Related to Figure 6
Among 7,861 participants of the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort, individual were stratified based on their GPS into three categories – bottom decile, deciles 2–9, and top decile. Longitudinal weight trajectories from birth to 18 years were modeled using linear spline multilevel models with knot points.
Table S4. Variant Classification of Melanocortin 4 Receptor (MC4R) gene identified by whole exome sequencing, Related to STAR Methods.
Melanocortin 4 Receptor (MC4R) gene variants identified by exome sequencing of 6,547 UK Biobank participants were classified according to current clinical guidelines (Richards et al., 2015) to identify those that with allele frequency < 0.01 that were pathogenic or likely pathogenic for obesity. Variants are described based on ‘chromosome:position:reference allele>alternate allele’ formatting, with chromosome positions based on the GRCh37 genome assembly.
Data Availability Statement
The genome-wide polygenic score validated and tested here will be made available to the research community prior to publication at: http://www.broadcvdi.org/informational/data.