Abstract
Antigen presentation by classical MHC class I molecules to CD8+ T cells is a central aspect of the adaptive immune response. Here, we describe methods to monitor antigen presentation using the model ovalbumin Kb-binding peptide, SIINFEKL. SIINFEKL genetically incorporated into viral or cellular source proteins can be used to precisely probe various aspects of antigen presentation, including the kinetics of peptide generation, MHC class I surface stability, and presentation efficiency following pharmacological and genetic manipulations including genome wide and high throughput drug screening.
Keywords: MHC-I, MHC class I, SIINFEKL, SL8, H-2Kb, DRiPs
1. Introduction
Antigen presentation by MHC class I molecules is a core function of adaptive immunity. CD8+ T cell immunosurveillance of cell surface MHC enables the immune system to monitor the cellular translatome to eradicate virus infected and cancerous cells. Conversely, dysregulation of antigen presentation can contribute to a myriad of autoimmune diseases.
All cell types, with the exception of erythrocytes, constitutively express class I molecules on their surfaces. The business end of the molecule contains a peptide, typically 8–11 amino acids, bound to a groove in the extracellular domain. This elegant mode of peptide display affords the T-cell repertoire a molecular window into the translatome of all cells. T-cells are activated by cells that escape self-tolerance mechanisms. These include (1) pathogen-derived peptides, (2) mutated peptides stemming from genetic abnormalities (termed neoepitopes), and (3) unmutated peptides which stem from alternative translation events that do not occur during thymic selection.
Research into the class I-CD8+ T cell system spans multiple fields, including genomics (as class I genes are the most polymorphic genes in most jawed vertebrate species), medicine (organ and bone marrow transplants), virology, and many aspects of basic cell biology. Viruses have played a key role in understanding virtually all aspects of the system, starting from the discovery of MHC restriction by Zinkernagel and Doherty. It has been clear for decades that viral peptides on surface class I molecules are generated at similar timeframes as their source proteins, even though the source proteins exhibit half-lives on the order of days ([1], see also Fig. 2a). This led to the hypothesis that defective ribosomal products (DRiPs) are an important source of peptides [2]. DRiPs represent improper translation products that stem from any number of missteps in the synthesis of polypeptide from mRNAs, including improper splicing, tRNA misacylation, alternative start codons, frameshifting, premature termination, stop codon read-through, or failure of the nascent full-length polypeptide to achieve a stable conformation.
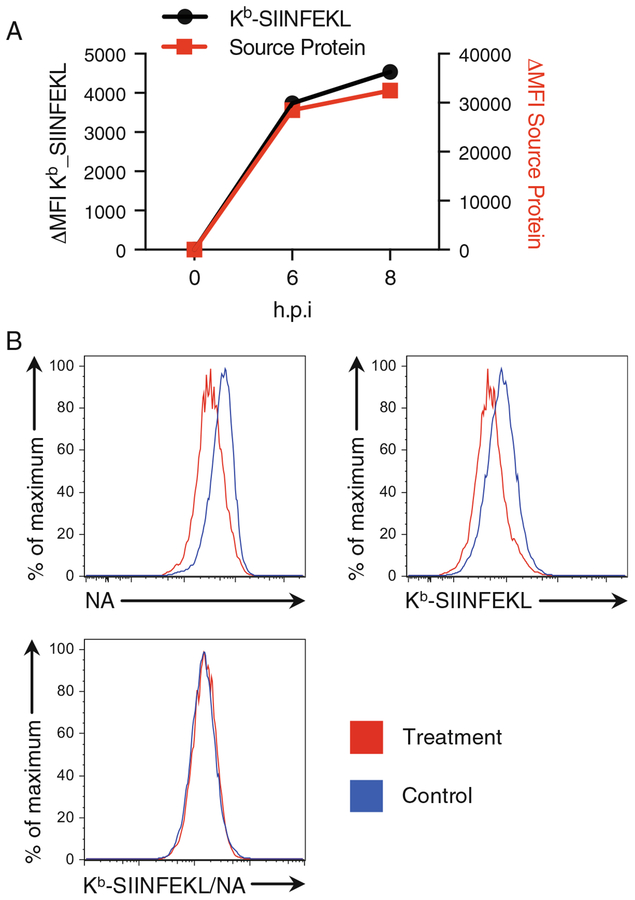
Fig. 2.
Kb-SIINFEKL presentation on 293Kb cell surface after IAV infection. SIINFEKL is inserted into the stalk region of neuraminidase (NA). Source protein (NA) and Kb-SIINFEKL levels were analyzed as in Subheading 3.3. (a) Time course showing Kb-SIINFEKL expression paralleled NA expression, indicating SIINFEKL generation from DRiPs. (b) At 8 hpi, in response to a treatment, expression levels of both NA and Kb-SIINFEKL were downregulated, while the ratio (peptide presentation efficiency) was unchanged
Recent remarkable advances in T cell-based immunotherapy has rejuvenated the antigen processing field. Resistance to immunotherapies has reinforced the concept of tumor immunoevasion/immunoediting including down regulation of antigen presentation [3, 4]. Cancer-associated neoepitopes that avoid central and peripheral tolerance are an important area of research, with mass spectrometry [5] and ribosome profiling [6] filling in the gaps between DNA/RNA sequencing and T-cell immunology.
Here we describe basic experiments to study antigen presentation using the model peptide antigen SIINFEKL from ovalbumin. This system originated with Mike Bevan’s desire to avoid viruses at all costs and find a model antigen that came in an inexpensive bottle from Sigma. Following a report that internal virion proteins can be presented to T cells following virus mediated delivery to the cytosol [7], Bevan and colleagues showed that pinocytosed ovalbumin (Ova) was presented to T cells if it was released from pinosomes into the cytosol by osmotic lysis of pinosomes [8]. A key to this discovery is that SIINFEKL is on the high end of class I binding peptides (10 nM binding to Kb) and is efficiently liberated from Ova by proteasomes and trimmed for class I binding. Propelling Kb-SIINFEKL as the most studied complex in vitro and in vivo are the availability Kb-SIINFEKL specific reagents: B3Z hybridoma cells [9], OT-I TCR transgenic mice [10], and the 25D1.16 monoclonal antibody [11], which enables precise quantitation of complexes by flow cytometry. Although SIINFEKL is criticized as being an unusual peptide, its affinity for class I and efficiency of generation is similar to many immunodominant viral peptides [12]. Antibodies for other class I/peptide complexes are available, but few, if any, give as high a signal to noise ratio as 25D1.16, which under the best conditions, rivals T cells in sensitivity [11, 13].
2. Materials
2.1. Cell Culture
Cell lines of interest—must contain H-2Kb or will need to create H-2Kb-expressing line.
Appropriate tissue culture media for cell lines being used (e.g., DMEM with 10% FBS).
Phosphate-buffered saline (PBS) −/−.
Cell dissociation reagent if adherent cells are used, such as0.05% trypsin, TrypLE (ThermoFisher 12563011) or Versene (PBS–EDTA).
2.2. Transfection
Sleeping Beauty H-2Kb expression vector if making stable cells, such as Addgene #111623.
Sleeping Beauty 100× vector if making stable cells; Addgene #34879 according to [14, 15].
Expression vector containing SIINFEKL, such as Addgene #111624.
Transfection reagent, such as GenJet version II.
DMEM without FBS.
6-well tissue culture plates.
2.3. Viral Infection
BSS–BSA: filtered sterilized balanced salt solution (BSS) containing 0.1% bovine serum albumin or suitable medium for viral infection.
Virus of choice encoding SIINFEKL.
Appropriate tissue culture media for cell lines being used.
Tube rotator in incubator, with tube angle set at an angle that allows maximum mixing of samples without spilling.
2.4. Acid Stripping and Brefeldin A Experiments
Citric acid buffer (pH 3.0): 46.5 mL 0.1 M citric acid, 3.5 mL 0.1 M sodium citrate; dilute to 100 mL with 50 mL water.
PBS.
Stock of brefeldin A at 3 mg/mL in methanol.
2.5. Antibodies and Antibody Labeling
25D1.16 antibody directly conjugated with fluorophore (e.g., Alexa Fluor 647 antibody labeling kit, Invitrogen A20186).
Antibodies against viral proteins directly conjugated with fluorophores (e.g., Alexa Fluor 488 antibody labeling kit, Invitrogen A20181).
Secondary fluorescent antibodies of choice, if necessary.
2.6. Cells and Antibody Staining
BSS–BSA as above, or suitable blocking/staining buffer for flow cytometry.
Fixation buffer: 1% paraformaldehyde (PFA) in PBS.
Permeabilization buffer: BSS–BSA containing 0.1% saponin.
Round bottom 96-well plates.
2.7. FACS Analysis
Flow cytometer equipped with the appropriate lasers and filter sets.
Appropriate analytical software such as FlowJo.
3. Methods
3.1. General Considerations
Culture cells of interest according to ATCC or standard culturing procedures. Adherent cells should be passaged before they reach confluence. Typical passage procedure includes removing growth medium, washing cells gently with PBS−/−, and trypsinizing at 37 °C, followed by neutralization of the trypsin with fresh growth medium. Count cells and replate according to cell line instructions.
Cells for studying SIINFEKL presentation must contain endogenous H-2Kb or must be engineered to express a H-2Kb transgene. If cells need to be first generated, simply use our deposited H-2Kb Addgene vector #111623 in combination with the optimized Sleeping Beauty transposase Addgene #34879 according to [15]. H-2Kb can associate with human β2–microglobulin (β2m), so there is no need to also express the mouse β2m gene in a human cell line.
To study antigen presentation efficiency, we recommend the use of cytosolic fluorescent source proteins containing SIINFEKL in an optimal position. See our deposited example Addgene vector #111624, which expresses Venus fluorescent protein followed by LEQLE-SIINFEKL-TEW-stop (see Notes 1 and 2).
The 25D1.16 antibody is used to quantitate Kb-SIINFEKL complexes on the cell surface. We find it best to conjugate this antibody directly to a fluorophore, in particular if it will be used in combination with other antibodies for staining purposes (see Note 3).
3.2. Measuring Antigen Presentation Efficiency by Fluorescent SIINFEKL-Containing Plasmid Reporters
- To test the effect on antigen presentation of a drug, knockdown, knockout, temperature, or other treatment, transient transfection of a reporter gene may be sufficient (see Note 4). This bypasses any effect that a virus might have on translation, cell viability, or antigen presentation which may be considerable. Follow the steps below for a standard transient transfection experiment.
- Plate cells the day before transfection on a 6-well plate such that they are between 60% and 80% confluent on the day of transfection.
- Numerous transfection agents can be used, such as Lipofectamine 2000, but we typically use GenJet version II and follow the manufacturer’s protocol, as below. Be sure to include separate wells for appropriate controls such as nontransfected and mock transfected cells.
- Change medium to fresh growth medium ~20–30 min prior to transfection; use 2 mL per well of a 6-well dish.
- Place 1 μg of the SIINFEKL reporter gene vector into 50 μL of DMEM (no FBS) (do not use Opti-MEM when using GenJet version II transfection reagent).
- Add 3 μL of GenJet version II into a separate 50 μL of DMEM (no FBS) (do not use Opti-MEM when using GenJet version II transfection reagent).
- Mix the entire GenJet solution into the tube of DNA by gentle trituration.
- Incubate at room temperature for 10 min.
- Add the 100 μL solution dropwise to the well of cells and swirl the medium slightly to ensure even spread.
- Change medium 5–6 h later to fresh, prewarmed growth medium.
- Antigen presentation can be tested typically either 24 or 48 h post-transfection by flow cytometry with conjugated 25D1.16 antibody.
- Harvest cells using as mild of detachment as possible. We typically wash cells in PBS, followed by either PBS–EDTA (Versene) or with TrypLE, though standard trypsin solutions can be used if necessary.
- Neutralize the TrpLE/trypsin in full growth medium and move cells to standard 1.5 mL Eppendorf tubes.
- Centrifuge at 300 × g for 4 min at 4 °C and remove supernatant.
- Wash cells into BSS–BSA or alternative blocking buffer and transfer ~1 × 106 cells to a 96-well round bottom plate, 200 μL total (plates are used to scale up experiment, though staining can also be done in tubes).
- Centrifuge at 300 × g for 4 min at 4 °C and remove supernatant.
- Resuspend cells in an appropriate dilution of 25D1.16 in BSS–BSA or blocking buffer (see Note 3).
- Cover the plate to protect from light, and subject to slight shaking at 4 °C for 30 min.
- Centrifuge at 300 × g for 4 min at 4 °C and remove the supernatant.
- Wash cells in 200 μL of BSS–BSA or blocking buffer and repeat the centrifugation and wash steps 3×.
- Resuspend cells in ~300 μL of blocking buffer and move to flow cytometry tubes for analysis.
- Analyze cells on a flow cytometer with channels for forward scatter, side scatter, GFP/Venus, and 25D1.16 label channel. Gate for live cells, gate away doublets, and examine the 25D1.16 staining as a function of GFP expression. The SIINFEKL presentation should be proportional to source protein expression (Fig. 1). If it is not, this is either biologically interesting or an artifact that needs troubleshooting.
Antigen presentation efficiency (Kb-SIINFEKL per source protein) can be calculated by the “derived parameter” function in FlowJo (see Note 5). Enter the 25D1.16 channel, the division sign, and the Venus channel to create this new parameter which can be applied to all samples. It helps to bring the axis to “logarithmic” and the expand the axis from 0.001 to 1000. The mean of this parameter can then be quantified for each sample.
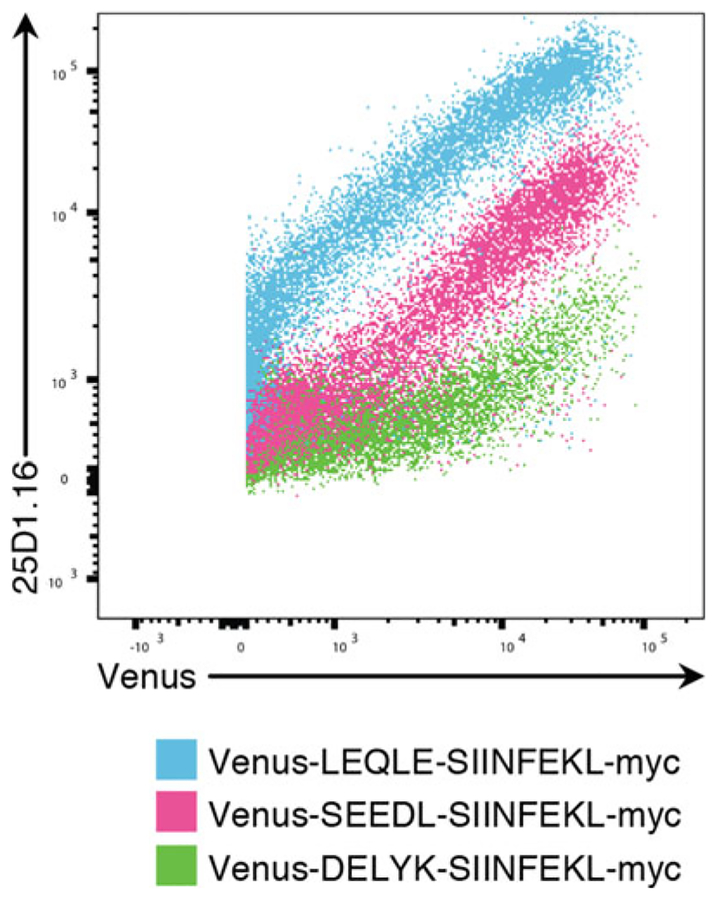
Fig. 1.
293Kb cells were transfected with the indicated plasmid reporters according to Subheading 3.2, step 1. and analyzed by 25D1.16 staining as in Subheading 3.2, step 2. The N-terminal flanking residues just prior to the SIINFEKL peptide can modulate antigen presentation efficiency by ~100×. DELYK is the nature end to many commonly used fluorescent proteins, and as such should not be used immediately upstream of the SIINFEKL peptide
3.3. Measuring Antigen Presentation Efficiency by SIINFEKL-Containing Viral Reporters
- Antigen presentation from viral source protein can be measured after infecting cells with recombinant viruses containing SIINFEKL in the coding sequence (see Note 6). Several time points can be taken after viral infection to assess the kinetics of antigen presentation (Fig. 2a, see also Note 7). Recombinant influenza A virus (IAV) is used in the example described below.
- Wash suspension cells or detached adherent cells with PBS. The total cell number is governed by the number of time points taken. Generally, for each time point, start with 5 × 105 cells (You will lose up to 50% cells during processing, depending on the cell line (sticky and proapoptotic are worst) and your experimental skills.). 105 cells at the end is plenty for accurate quantitation.
- Infect cells (1 × 107 cells/mL) at a multiplicity of infection (MOI) of 10 in BSS–BSA at 37 °C for 1 h in a culture tube. A mock-infected sample is prepared in parallel. Additional infected samples are prepared to serve as single color controls and fluorescence minus one controls for flow cytometry analysis.
- Wash cells by adding PBS, centrifuging at 300 × g for 4 min, and resuspend cells in culture medium at 106 cells/mL (see Note 8).
- Take an aliquot of cells at 4, 6, and 8 h post infection (hpi).
- Wash and resuspend cells with ice-cold 200 μL BSS–BSA, transfer cells to one well of a 96-well round bottom plate and keep on ice until antibody staining and flow cytometry analysis (see Note 9).
- The use of conjugated 25D1.16 antibody is as described above. Antibodies against viral proteins can be conjugated with fluorophores according to manufacturer’s protocols and used along with conjugated 25D1.16 to assess the level of viral source protein expression and thus calculate antigen presentation efficiency (Fig. 2b). One step staining can be conducted with antibodies against viral surface protein (HA, NA, M2) and 25D1.16 as described in Subheading 3.2, step 2. If necessary, staining of intracellular viral proteins is performed after 25D1.16 staining following the steps described below.
- Fix cells with freshly made 1.6% PFA at room temperature for 20 min (protect from light).
- Wash cells by adding PBS, centrifuging the plate at ~700 × g for 1 min at 4 °C, and gently flick the plate (Find an immunologist to show you the flick if you are new to this) to remove liquid without disturbing the pelleted cells.
- Wash cells 2× by adding BSS–BSA, centrifuging the plate at ~700 × g for 1 min at 4 °C, and gently flick plate to remove liquid without disturbing the pelleted cells.
- Permeabilize cells by resuspending cells in permeabilization solution containing antibodies for intracellular staining.
- Cover the plate to protect from light, and gently shake at 4 °C for 30 min.
- Wash cells 3× in 200 μL permeabilization buffer and run cells on the flow cytometer (see Note 10).
Single color compensation should be conducted when using multiple colors in flow cytometry. Because viral infection often changes the auto fluorescence level of cells, the use of fluorescence minus one controls should be considered. The best negative control is cells infected with an identical virus that does not encode SIINFNEKL, or for the ultimate control, encodes a non-class I binding variant of SIINFEKL (there are many).
3.4. Kinetic Analyses of Kb-SIINFEKL or Total MHC Class I Presentation
- To examine the stability of cell surface class I molecules, export of class I molecules from the cis-Golgi can be blocked by treating cells with brefeldin A blockade [16]. Typically, this is monitored kinetically, with Kb-SIINFEKL complexes stained at different time points post-brefeldin A treatment (see Note 11).
- Prepare cells to be tested as in Subheading 3.2 or 3.3. Drug treatments, knockouts or knockdowns, or any other manipulation of the cells should be included in the experimental setup.
- If necessary, harvest cells using as mild of detachment as possible. We wash cells in PBS, followed by either Versene or with TrypLE, though standard trypsin solutions can be used if necessary.
- Neutralize the TrpLE/trypsin/Versene in full growth medium and move cells to a conical tube.
- Centrifuge at 300 × g for 4 min at 4 °C and remove supernatant.
- Resuspend cells into fresh growth medium containing 6 μg/mL brefeldin A. (A few cell lines (e.g., MDCK cells) are resistant to brefeldin A) The volume can be adjusted based on total cell number and how many time-points are to be analyzed.
- Place the cells (now in suspension in a conical tube) on a slow rotator in an incubator to prevent cell attachment (see Note 8).
- Wait 10 min for cells to reach the correct temperature, and then pipet out the first timepoint (t = 0), typically 5 × 105 or 1 × 106 cells. Place this aliquot of cells on ice.
- Continue to remove aliquots of cells for the desired time course—generally every 1–1.5 h for a total of 6–10 h should yield sufficient data to appreciate kinetic differences among samples.
- At the last time point, pellet all tubes by 300 × g for 4 min 4 °C centrifugation and proceed with 25D1.16 staining as described.
- Quantify the mean 25D1.16/Venus derived parameter as a function of time.
- To assess the kinetics of peptide generation and presentation, an acid stripping step can be performed to remove preexisting class I peptide complexes on cell surface and measure the recovery over time. The readout can be overall class I expression on cell surface using pan-class I, allomorph specific or peptide class I specific reagents (see Note 11).
- Resuspend PBS washed cells in ice-cold citric acid buffer at 1 × 107 cells/mL for 2 min (see Note 12).
- Add >20× volumes of PBS or medium to neutralize acid (see Note 13).
- Centrifuge cells immediately at ~700 × g for 1 min at 4 °C, and remove supernatant.
- Wash cells twice more with PBS.
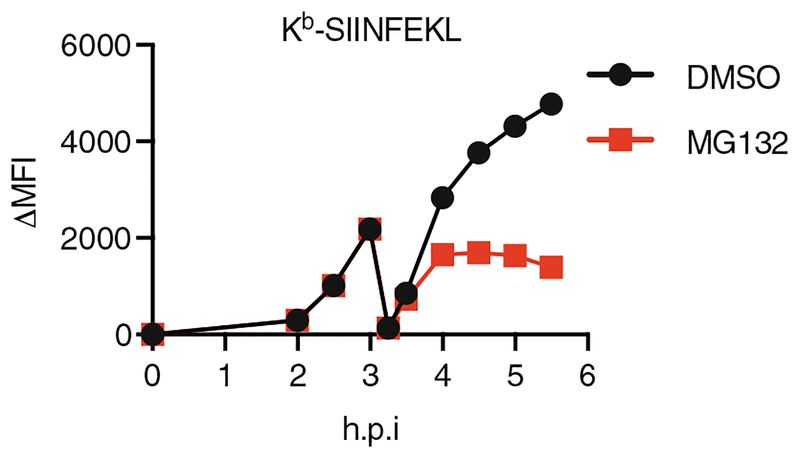
- Resuspend cells in cell culture medium and culture cells at 37 °C as in Subheading 3.4, step 1. At this point, drug treatments such as MG-132 can be included to assess the recovery of Class I as a function of the desired perturbation (Fig. 3).
- Take an aliquot of cells for antibody staining and flow cytometry analysis as Subheading 3.4, step 1.
- Quantify the mean 25D1.16/source protein derived parameter as a function of time.
Fig. 3.
Kb-SIINFEKL presentation on 293Kb cell surface after virus infection and acid wash. At 3 hpi, acid wash was conducted as in Subheading 3.4, step 2. Kb-SIINFEKL recovered slower in response to the proteasome inhibitor MG132
4. Notes
Our recommendation of a cytosolic fluorescent reporter such as Venus-LEQLE-SIINFEKL-TEW is only a guideline—source proteins can be anything detectable by flow cytometry. Additionally, antigen presentation from specific organelle-targeted proteins can be easily studied by the creative use of source proteins and permeabilization.
The location of SIINFEKL is critical in the design of a custom vector. In particular, upstream N-terminal flank residues can drastically alter antigen presentation efficiency [17]. SIINFEKL appended to the C-terminus of GFP or Venus, for example, will lead to very low Kb-SIINFEKL expression (Fig. 1). We prefer to use the LEQLE natural Ova flanking sequence, which has shown the highest efficiency in our hands. The C-terminal flanking residues appear to less important, and in fact SIINFEKL-stop constructs present sometimes even slightly better than C-terminally flanked SIINFEKL, likely due to one less proteolytic cleavage step needed in the presentation pathway.
We recommend using a bright fluorophore for the labeling of 25D1.16 antibody, and we follow the protein labeling kits by the manufacturer’s protocol. If sensitivity is of the essence, the use of longer wavelength fluorophores can exploit the lower cellular autofluorescence in the red and far red wavelengths. Calculate the moles of dye per mole of antibody to ensure the labeling was successful. Most importantly, the antibody must be titrated, typically in the range of 1:100–1:5000, in a pilot experiment before proceeding with real test samples to establish the optimal signal to noise and also to save time and money in producing antibody conjugates.
The experiment described here represents a straightforward transient transfection, which may be useful enough for a quick analysis of a particular treatment or genetic disruption. One should consider creating stable cell clones, which can be easily generated with our Addgene vector #111624 in conjunction with the Sleeping Beauty transposase [14, 15]. This standardizes expression of the source protein and eliminates transfection differences, toxicities associated with transfection reagents, etc. If examining presentation from an endogenously translated protein, SIINFEKL can be knocked into the genome using CRISPR/Cas9 techniques.
The derived parameter calculation in FlowJo is essential for understanding how antigen presentation is different across treatments, cells, knockdowns, etc. This is a true measurement of antigen presentation efficiency because it measures SIINFEKL presentation as a function of source protein across all cells on a per-cell basis. Thus, even small effects in the derived parameter can be highly significant. See Fig. 2b for an example of how both Kb-SIINFEKL and its source protein decrease, such that the ratio is unchanged.
Similar to plasmid reporters, flanking sequences also affect the efficiency of SIINFEKL presentation from viral reporters. In addition, the insertion of SIINFEKL in the viral coding sequence can impact virus fitness, with its effect ranging from unmeasurable to catastrophic. The effect of SIINFEKL insertion on viral protein detection via antibody also needs to be considered and assessed in a pilot experiment.
A pilot experiment can be performed to determine the appropriate time points and MOI. The time point taken to assess Kb-SIINFEKL on cell surface depends on virus and cells used. For vaccinia virus infected cells, we can observe signal over background as early as 2 hpi. For IAV infected cells, we typically take measurements starting from 4 hpi (MOI = 10), though peptide can be generated in the first hour [18]. A lower MOI can be used to avoid excessive cell death if Kb-SIINFEKL is measured after overnight culture.
To facilitate taking aliquots for time course experiments, we dissociate adherent cells. Viral infection and subsequent cell culture is conducted in culture tubes. We rotate tubes at a moderate speed that allows general mixing and aeration without vigorous vibrating. A thorough cell mixing is conducted by triturating cell cultures when time point samples are taken. While dissociating cells is pragmatic, it might alter antigen presentation. Ultimately, antigen presentation will have to studied in situ in living animals. While the day is coming no doubt, there are remaining technical hurdles.
We use BSA to block nonspecific antibody binding. Fc block should be used for cell lines with Fc receptors. In our experiments, dead cells are gated out by FSA/SSA. Live/dead dyes can also be used when necessary.
Secondary staining can be conducted if necessary. If the primary antibodies are raised in the same species, samples need to be stained separately. As a result, “derived parameter” function cannot be used to calculate antigen presentation efficiency on a per cell basis.
For kinetics experiments, it is not necessary to specifically look at Kb-SIINFEKL complexes via the 25D1.16 antibody. One can also use antibodies to specific HLA allotypes, or the W6/32 antibody to track overall class I levels.
The time to acid strip surface MHC-I peptides is cell type dependent. A pilot experiment should be performed for each cell type. An optimal treatment decreases surface class I level without excessive damage to cells. The majority of cells should recover in subsequent cell culture. The stripping is typically in the range of 1–5 min.
After acid treatment, cell tend to clump together. It is crucial to add PBS immediately, and repeatedly triturate to ensure a complete and homogeneous mixture to stop the reaction uniformly.
Acknowledgments
The authors are supported by the Division of Intramural NIAID.
References
- 1.Esquivel F, Yewdell JW, Bennink JR (1992) RMA/S cells present endogenously synthesized cytosolic proteins to class I-restricted cytotoxic T lymphocytes. J Exp Med 175:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yewdell JW, Anton LC, Bennink JR (1996) Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J Immunol 157(5):1823–1826 [PubMed] [Google Scholar]
- 3.Challa-Malladi M, Lieu YK, Califano O, Holmes AB, Bhagat G, Murty VV, Dominguez-Sola D, Pasqualucci L, Dalla-Favera R (2011) Combined genetic inactivation of beta2-Microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell 20(6):728–740. 10.1016/j.ccr.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, Birkbak NJ, Veeriah S, Van Loo P, Herrero J, Swanton C (2017) Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171(6):1259–1271.e1211. 10.1016/j.cell.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassani-Sternberg M, Braunlein E, Klar R, Engleitner T, Sinitcyn P, Audehm S, Straub M, Weber J, Slotta-Huspenina J, Specht K, Martignoni ME, Werner A, Hein R, HB D, Peschel C, Rad R, Cox J, Mann M, Krackhardt AM (2016) Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 7:13404 10.1038/ncomms13404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, Weissman JS, Fuchs E (2017) Translation from unconventional 5′ start sites drives tumour initiation. Nature 541(7638):494–499. 10.1038/nature21036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yewdell JW, Bennink JR, Hosaka Y (1988) Cells process exogenous proteins for recognition by cytotoxic T lymphocytes. Science 239 (4840):637–640 [DOI] [PubMed] [Google Scholar]
- 8.Moore MW, Carbone FR, Bevan MJ (1988) Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell 54:777–785 [DOI] [PubMed] [Google Scholar]
- 9.Karttunen J, Sanderson S, Shastri N (1992) Detection of rare antigen-presenting cells by the lacZ T-cell activation assay suggests an expression cloning strategy for T-cell antigens. Proc Natl Acad Sci U S A 89:6020–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke SR, Barnden M, Kurts C, Carbone FR, Miller JF, Heath WR (2000) Characterization of the ovalbumin-specific TCR transgenic line OT-I: MHC elements for positive and negative selection. Immunol Cell Biol 78(2):110–117. 10.1046/j.1440-1711.2000.00889.x [DOI] [PubMed] [Google Scholar]
- 11.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN (1997) Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity 6(6):715–726 [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Anton LC, Bennink JR, Yewdell JW (2000) Dissecting the multifactorial causes of immunodominance in class I-restricted T cell responses to viruses. Immunity 12(1):83–93 [DOI] [PubMed] [Google Scholar]
- 13.Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW (2003) Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18(3):343–354 [DOI] [PubMed] [Google Scholar]
- 14.Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, VandenDriessche T, Ivics Z, Izsvak Z (2009) Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41(6):753–761. 10.1038/ng.343 [DOI] [PubMed] [Google Scholar]
- 15.Kowarz E, Loscher D, Marschalek R (2015) Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J 10(4):647–653. 10.1002/biot.201400821 [DOI] [PubMed] [Google Scholar]
- 16.Yewdell JW, Bennink JR (1989) Brefeldin A specifically inhibits presentation of protein antigens to cytotoxic T lymphocytes. Science 244(4908):1072–1075 [DOI] [PubMed] [Google Scholar]
- 17.del Val M, Schlicht HJ, Ruppert T, Reddehase MJ, Koszinowski UH (1991) Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell 66:1145–1153 [DOI] [PubMed] [Google Scholar]
- 18.Zanker D, Waithman J, Yewdell JW, Chen W (2013) Mixed proteasomes function to increase viral peptide diversity and broaden antiviral CD8+ T cell responses. J Immunol 191(1):52–59. 10.4049/jimmunol.1300802 [DOI] [PMC free article] [PubMed] [Google Scholar]