Abstract
Chemically induced dimerization (CID) systems, in which two proteins dimerize only in the presence of a small molecule ligand, offer versatile tools for small molecule sensing and actuation. However, only a handful of CID systems exist and creating one with the desired sensitivity and specificity for any given ligand is an unsolved problem. Here, we developed a combinatorial binders-enabled selection of CID (COMBINES-CID) method broadly applicable to different ligands. We demonstrated a proof-of-principle by generating nanobody-based heterodimerization systems induced by cannabidiol with high ligand selectivity. We applied the CID system to a sensitive sandwich ELISA-like assay of cannabidiol in body fluids with a detection limit of ~0.25 ng/mL. COMBINES-CID provides an efficient, cost-effective solution for expanding the biosensor toolkit for small molecule detection.
Keywords: chemically induced dimerization, combinatorial nanobody library, cannabidiol, phage display
Graphical Abstract
Since the concept of using CID systems as a molecular switch was introduced by Schreiber, Crabtree, and coworkers in 1993,1 a few naturally occurring CID systems, such as rapamycin-inducible FK506 binding protein (FKBP)/FKBP-rapamycin binding domain (FRB),2 gibberellin-inducible GAI/GID1 complexes,3 and their derivatives,1, 4–5 have been engineered as genetically encoded biosensors to dissect or manipulate transcription and translation regulation, signaling and metabolic pathways, and other biological processes.6–8 Moreover, CID offers a new intriguing mechanism for in vivo and in vitro small molecule detection; for example, CID proteins can be genetically fused with reporter tags for optical or transcriptional readouts of metabolite concentrations, or serve as affinity reagents for sandwich enzyme-linked immunosorbent assay (ELISA)-like assays applicable to the point-of-care testing of small-molecule targets, such as drugs, toxins, and pollutants.
Despite their widespread use, creating CID systems for new ligands is, so far, an unsolved problem. Existing methods, such as animal immunization,9 in vitro selection,10–11 and computational design,12 can generate protein binders, such as antibodies, that function via binary protein‒ligand interactions; however, it is difficult to obtain protein pairs that only form a ternary complex in the presence of a ligand. Some methods created CID by chemical linking of two ligands that independently bind to the same or different proteins,1, 4–5, 13–15 or by selecting antibodies against an existing protein-ligand complex (e.g., a B-cell lymphoma family protein (BCL-xL)–ABT-737),16 where the bound ligand shows a large solvent-exposed moiety for the antibody recognition to ensure the specificity of ligand-induced dimerization. However, these methods are all limited by the choice of ligands.
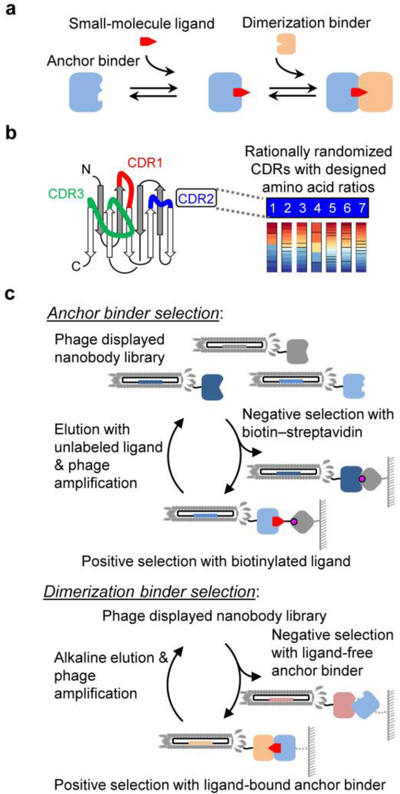
Here we propose a COMBINES-CID method to select CID proteins for any given ligand—an ‘anchor binder’ that first binds to a ligand, and a ‘dimerization binder’ that only binds to the anchor binder‒ligand complex not the unbound anchor binder (Figure 1a). This method is based on the in vitro selection of vastly diverse protein binder libraries, such as combinatorial antibody libraries,17 which can be selected against virtually any epitope. In this work, we focus on a single-domain antibody (or nanobody), a 12–15 kDa functional antibody fragment from camelid comprising a universal scaffold and three variable complementarity-determining regions (CDRs) (Figure 1b).18 We reasoned that the three CDR loops might form a binding pocket with adaptable sizes for small-molecule epitopes.19–20 Of note, unlike a rigid binding site, the flexible CDR loops might undergo conformational changes upon the ligand binding,19, 21 providing a basis for the selection of conformationally selective binders22 only recognizing ligand-bound anchor binders. A stepwise phage-display screening strategy was devised to first obtain anchor binders which are then used as baits to select dimerization binders (Figure 1c).
Figure 1.
(a) Ligand-induced dimerization of an anchor and a dimerization binders. (b) Schematic of the generation of a synthetic nanobody combinatorial library. (c) Overview of the COMBINES-CID method.
As a proof-of-principle, cannabidiol (CBD), a non-psychoactive phytocannabinoid with many medical uses,23 was chosen as the ligand. Unlike large, polar, or charged ligands that might be easier targets for binder selection, CBD is hydrophobic and smaller than most ligands in all existing CID systems. Thus, CBD provides a rigorous test of our method. Other CID engineering methods5–7, 13–16 tend to generate or use relatively large ligands; for example, a FKBP homo-dimerization ligand, FK1012, is a conjugated dimer of tacrolimus with molecular weight of 1,564 daltons.1 However, smaller ligands are often preferred for the use in biological and clinical applications.24–25
To enhance the success of selection, we prepared a high-quality nanobody library with high protein diversity and stability. A combinatorial gene library, designed with a thermally stable nanobody scaffold and three rationally randomized CDRs, was chemically synthesized by a trinucleotide mutagenesis technology,26 similarly as previously described.27 The synthetic DNA library of ~1012 sequences was subcloned and transformed into Escherichia coli to produce phage-displayed nanobodies (Supplementary Methods). The quality of the phage library was assessed by Sanger and deep sequencing. Approximately 74% of the clones were found within the designed sequences. 39,289,832 out of 41,458,478 merged 2×150 bp paired-end reads were found to be unique (Figure S1) and the library diversity was estimated to be 1.23 to 7.14×109 by an empirical Bayesian statistical method.28 The amino acid distributions of CDRs were close to the expected ratios (Figure S2).
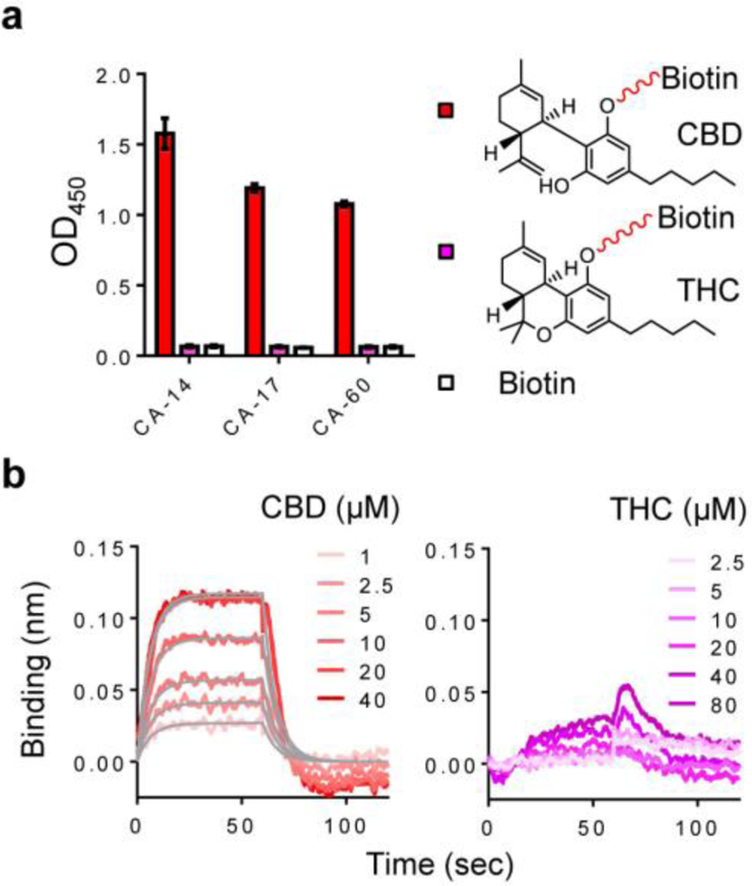
To obtain CBD anchor binders (CA), the library was screened using biotinylated CBD as bait. Phage displayed nanobodies were captured by biotinylated CBD bound to streptavidin-coated magnetic beads and eluted by unlabeled CBD (Figure 1c). After six rounds of selection, three unique clones were found from 96 randomly picked clones. Single-phage ELISA showed that all had specific binding to CBD‒biotin‒streptavidin but minimal binding to its structural analog, tetrahydrocannabinol (THC)‒biotin‒streptavidin (Figures 2a and S3). The binding selectivity suggests that the minor structural difference in CBD (i.e., the open ring with a phenolic hydroxyl group) is key to the molecular recognition. Interestingly, CDRs in the three nanobodies have little or no sequence similarity (Table S1).
Figure 2. Anchor binder analysis.
(a) ELISA of three anchor binders against biotinylated CBD and THC immobilized on streptavidin-coated plates. Biotin was used as a control. Data represent mean values of 3 measurements; error bars, standard deviation. (b) BLI sensorgrams of CA-14 with unlabeled CBD or THC. CA-14 was immobilized on Super Streptavidin biosensors. CBD binding sensorgrams (red curves) were modelled using a global fit (grey lines).
To confirm that the nanobody binding only requires CBD, and not the linker between CBD and biotin or the streptavidin, we measured the binding affinity using unlabeled CBD. Anchor-binder hits were expressed in E. coli periplasm and purified by nickel-affinity and size-exclusion chromatography (SEC). Purified nanobodies were enzymatically biotinylated (Figure S4a–b) and immobilized on Super Streptavidin biosensors to quantify binding affinities to CBD or THC by Bio-Layer Interferometry (BLI) (Figure S5a). All hits showed high specificity to CBD; dissociation constants (KDs) were measured to be 6.0 (CA-14), 26.4 (CA-17), and 3.5 (CA-60) μM (Figures 2b and S5b–c). The binding affinities are relatively weak compared with those in natural CID complexes; for example, the KDs of FKBP–rapamycin and GyrB–coumermycin complexes were reported to be ~0.2 and ~100 nM, respectively.29–30 Although the ligand-binding affinity of nanobodies can be further improved by engineering the binding sites,19 we hypothesized that even a relatively weak anchor binder–ligand complex could form a stable ternary CID complex via the cooperative interaction with a dimerization binder. To test this, CA-14 with the highest protein yield (> 5 mg per liter of culture) was selected for the dimerization binder (DB) selection.
To select specific ligand-induced dimerization, the library was subjected to negative and positive screening using CBD-free and bound CA-14 as baits, respectively (Figure 1c). After four rounds of selection, 24 unique clones out of 384 were identified and all showed CBD concentration-dependent binding to CA-14 by titration ELISA (Figure S6). They also showed minimal or no binding to CA-14 in the absence of CBD. Four clones with diverse CDR sequences (Table S1) and relatively high protein yields (> 2.5 mg per liter of culture) were purified for further characterization.
With the purified DB nanobodies in hand, we first sought to determine their binding affinities to the CA-14–CBD complex. In a three-component system, CID binders often exhibit more complex binding behaviors than their two-component counterparts,31 for example, CBD might directly bind to dimerization binders, which can complicate the affinity measurement. To assess this possibility, the four nanobodies were biotinylated (Figure S4c) and immobilized on streptavidin biosensors to measure the interaction with CBD by BLI. All showed minimal or no interaction with CBD (Figure S7). Moreover, the binding-dissociation dynamics of the CA-14–CBD complex can also affect the measurement of dimerization binder affinities. To address this, the affinity was measured by a BLI assay in which the biosensor-immobilized dimerization binders were interacted with CA-14 preequilibrated with varied concentrations of CBD (Figure S8a). The concentrations of the CA-14–CBD complex in equilibrium were calculated based on input CA-14 and CBD concentrations (Supplementary Note 1). The CA-14-CBD complex concentration were approximated to be constant because only a minimal fraction of the complex bound to the biosensor tips during the BLI assay. The result confirms that CA-14 only interacts with the dimerization binders in the presence of CBD (Figure 3a). The KDs of the four dimerization binders varied from 56.4 nM to 19 μM (Table S2). CA-14 and DB-21 appear to form the most stable CID complex due to a relatively slow dissociation rate (koff = 6.17×10−4/s; Table S2), and the binding and dissociation rates appear to be sensitive to pH and ion strength changes (Figure S9). All binders showed a high ligand specificity and the dimerization was not induced by THC (Figure S8b).
Figure 3. CBD-induced nanobody dimerization.
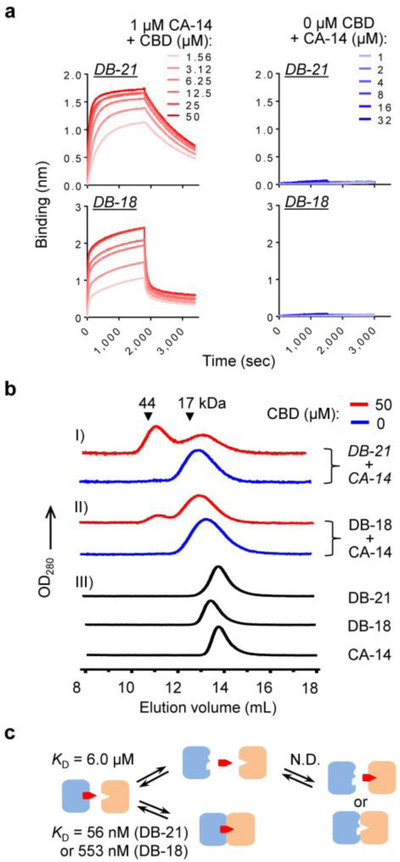
(a) BLI sensorgrams of DB-21 and DB-18 immobilized on streptavidin biosensors binding to CA-14 preequilibrated with different concentrations of CBD. (b) SEC analysis of CBD-induced heterodimerization. I) 5 μM (each) DB-21 and CA-14 and II) 10 μM (each) DB-18 and CA-14, in the presence or absence of CBD, were crosslinked by 50 μM bis-N-succinimidyl-(pentaethylene glycol) ester for 30 min at room temperature before SEC. III) 30 μM non-crosslinked DB-21, DB18, and CA-14 were separately analyzed. Elution volumes of protein standards are marked by triangles. Chromatograms in different groups are shown with different Y scales. (c) Measured KDs of the ternary complex formation. N.D., not detectable or too weak to be determined.
Next, we confirmed the CBD-induced heterodimerization of the two most stable CID systems (CA-14–DB-21 and CA-14–DB-18) by analytical SEC. As expected, the anchor and dimerization binders by themselves were detected exclusively as monomers, but after adding CBD, SEC peaks corresponding to the heterodimers were observed (Figure 3b). These results demonstrate a nanobody-based CID binding mechanism (Figures 3c), which can be explained by a cooperative binding model of the three-component binding equilibrium (Supplementary Note 1). In brief, this model suggests that CBD works as a bridge between the two binders, and the dimerization binders stabilize the anchor binder–ligand complex by cooperative binding, similar to the rapamycin CID system.31 Additionally, the CBD-bridged CID model is supported by Rosetta-based ligand docking calculations showing that the solvent-exposed phenolic hydroxyl group of anchor-bound CBD, where biotin was attached in the ligand during the anchor binder selection (Figure 2a), might be involved in the dimerization recognition (Supplementary Note 2).
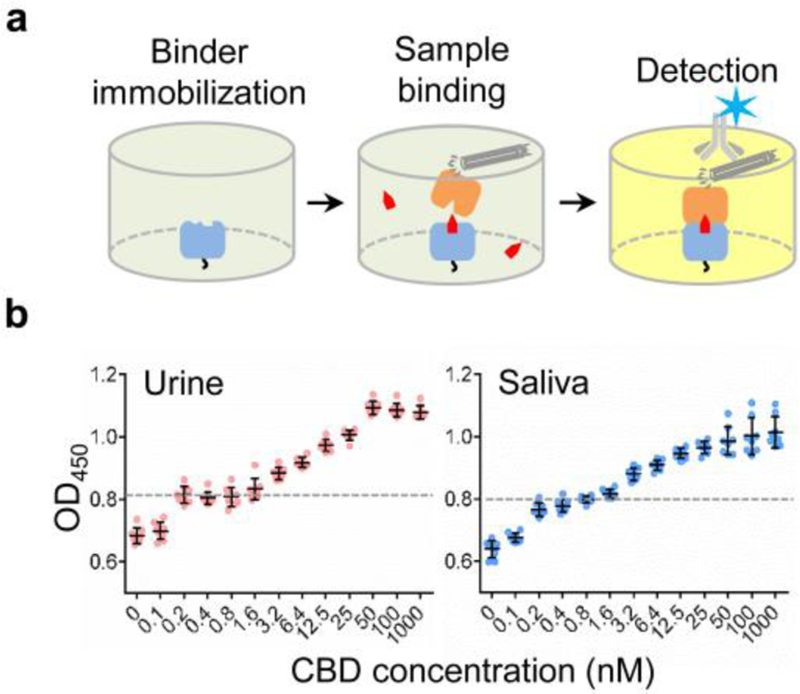
Lastly, to demonstrate the application in small molecule detection, we applied the CA-14–DB-21 dimerization system to a sandwich ELISA-like assay (Figure 4a) and a split luciferase assay32 (Figure S10). Small molecule detection often relies on complex, time- and resource-consuming technologies, such as mass spectrometry. In contrast, antibody-based approaches, such as ELISAs, can be performed with minimal training and easily interpretable results available within a few hours. Traditional ELISA-based small molecule detection requires the use of ligand-binding antibodies, which are difficult to obtain by animal immunization because small molecules, by themselves, are non-immunogenic and can only elicit antibodies upon conjugation to protein carriers. Moreover, ELISA requires labeling of ligands—i.e., chemical linking of ligands or their competitive inhibitors to solid supports or reporter molecules—and thus, are not generally applicable to different ligands. However, the CID-based detection only requires the labeling of CID binders. By analyzing CBD-spiked urine and saliva samples from three healthy donors, the assay showed a broad detection range with Limit of Detection (LoD) of ~0.8 nM or ~0.25 ng/mL (Figures 4b and S11), which meets requirements for the diagnostic applications.33 The dose-dependent dimerization is corroborated by the luciferase assay with a NanoLuc complementation reporter (Figure S10).
Figure 4. Sandwich ELISA-like detection of CBD.
(a) CA-14 was immobilized on a plate and the CBD-induced binding of phage-displayed DB-21 was detected by a horseradish peroxidase-conjugated antibody. (b) The detection of CBD spiked in urine and saliva samples. Limits of detection (LoDs = meanblank + 3×(standard deviation of the blank, n=8)) for urine and saliva samples were determined to be ~0.8 nM (~0.25 ng/mL).
In summary, COMBINES-CID enables the efficient selection of vastly diverse combinatorial binders to obtain ligand-induced dimerization systems. We demonstrated for the first time that stable, specific CID systems can be constructed with synthetic nanobodies. Our method is applicable to select protein binders with other immunoglobulin, non-immunoglobulin, or computationally designed scaffolds. The selection is cost-effective and fast (Figure S12) since the same binder library can be used for different ligands and binders with desired qualities were often obtained without in vitro affinity maturation. Together, our results show that this method can generate CID systems for a challenging ligand like CBD, thus suggesting that COMBINES-CID is a suitable method to address the unmet challenge in in vitro and in vivo small molecule sensing.8
Supplementary Material
ACKNOWLEDGMENT
We thank D. Baker and L. Stewart for providing biotinylated CBD and THC, M. Dinh, J. Lee, and M. Buerger for the assist on binder screening, Huang Z. for the help on BLI assay, Xue W. for the ligand docking, and D. Durnam for editing the manuscript. This work was supported by the University of Washington Innovation Award (to L.G. and F.D.), a grant from the U.S. National Institutes of Health (1R35GM128918 to L.G.), and a startup fund of the University of Washington (to L.G.). H.J. is supported by a Washington Research Foundation undergraduate fellowship.
ABBREVIATIONS
- COMBINES
combinatorial binders-enabled selection
- CID
chemically induced dimerization
- CBD
cannabidiol
- THC
tetrahydrocannabinol
- CA
CBD anchor
- DB
dimerization binder
- CDR
complementarity-determining region
- ELISA
enzyme-linked immunosorbent assay
- BLI
bio-layer interferometry
- SEC
size-exclusion chromatography
- LoD
Limit of Detection
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: https://doi.org/10.1021/jacs.9b03522
General information about materials and methods, chemical synthesis and characterization, nanobody library construction and selection, binder characterization, binding data analysis, and structural modeling.
A provisional patent related to this work has been filed by the University of Washington.
REFERENCES
- (1).Spencer DM; Wandless TJ; Schreiber SL; Crabtree GR, Controlling signal transduction with synthetic ligands. Science 1993, 262, 1019–1024. [DOI] [PubMed] [Google Scholar]
- (2).Rivera VM; Clackson T; Natesan S; Pollock R; Amara JF; Keenan T; Magari SR; Phillips T; Courage NL; Cerasoli F; Holt DA; Gilman M, A humanized system for pharmacologic control of gene expression. Nat. Med 1996, 2, 1028–1032. [DOI] [PubMed] [Google Scholar]
- (3).Miyamoto T; DeRose R; Suarez A; Ueno T; Chen M; Sun TP; Wolfgang MJ; Mukherjee C; Meyers DJ; Inoue T, Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nat. Chem. Biol 2012, 8, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ho SN; Biggar SR; Spencer DM; Schreiber SL; Crabtree GR, Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature 1996, 382, 822–826. [DOI] [PubMed] [Google Scholar]
- (5).Belshaw PJ; Ho SN; Crabtree GR; Schreiber SL, Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc. Natl. Acad. Sci. U. S. A 1996, 93, 4604–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Fegan A; White B; Carlson JCT; Wagner CR, Chemically controlled protein assembly: techniques and applications. Chem. Rev 2010, 110, 3315–3336. [DOI] [PubMed] [Google Scholar]
- (7).DeRose R; Miyamoto T; Inoue T, Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 2013, 465, 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Stanton BZ; Chory EJ; Crabtree GR, Chemically induced proximity in biology and medicine. Science 2018, 359, eaao5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hunter MM; Margolies MN; Ju A; Haber E, High-affinity monoclonal antibodies to the cardiac glycoside, digoxin. J. Immunol 1982, 129, 1165–1172. [PubMed] [Google Scholar]
- (10).Bradbury ARM; Sidhu S; Dubel S; McCafferty J, Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol 2011, 29, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chen G; Hayhurst A; Thomas JG; Harvey BR; Iverson BL; Georgiou G, Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS). Nat. Biotechnol 2001, 19, 537–542. [DOI] [PubMed] [Google Scholar]
- (12).Tinberg CE; Khare SD; Dou JY; Doyle L; Nelson JW; Schena A; Jankowski W; Kalodimos CG; Johnsson K; Stoddard BL; Baker D, Computational design of ligand-binding proteins with high affinity and selectivity. Nature 2013, 501, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Farrar MA; AlberolaIla J; Perlmutter RM, Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature 1996, 383, 178–181. [DOI] [PubMed] [Google Scholar]
- (14).Erhart D; Zimmermann M; Jacques O; Wittwer MB; Ernst B; Constable E; Zvelebil M; Beaufils F; Wymann MP, Chemical Development of Intracellular Protein Heterodimerizers. Chemistry & Biology 2013, 20, 549–557. [DOI] [PubMed] [Google Scholar]
- (15).Ballister ER; Aonbangkhen C; Mayo AM; Lampson MA; Chenoweth DM, Localized light-induced protein dimerization in living cells using a photocaged dimerizer. Nat. Commun 2014, 5, 5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Hill ZB; Martinko AJ; Nguyen DP; Wells JA, Human antibody-based chemically induced dimerizers for cell therapeutic applications. Nat. Chem. Biol 2018, 14, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lerner RA, Combinatorial antibody libraries: new advances, new immunological insights. Nat. Rev. Immunol 2016, 16, 498–508. [DOI] [PubMed] [Google Scholar]
- (18).Muyldermans S, Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem 2013, 82, 775–797. [DOI] [PubMed] [Google Scholar]
- (19).Fanning SW; Horn JR, An anti-hapten camelid antibody reveals a cryptic binding site with significant energetic contributions from a nonhypervariable loop. Protein Sci. 2011, 20, 1196–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zavrtanik U; Luken J; Loris R; Lah J; Hadzi S, Structural basis of epitope recognition by heavy-chain camelid antibodies. J. Mol. Biol 2018, 430, 4369–4386. [DOI] [PubMed] [Google Scholar]
- (21).Al Qaraghuli MM; Ferro VA, Analysis of the binding loops configuration and surface adaptation of different crystallized single-domain antibodies in response to various antigens. J. Mol. Recognit 2017, 30, e2592. [DOI] [PubMed] [Google Scholar]
- (22).McMahon C; Baier AS; Pascolutti R; Wegrecki M; Zheng SD; Ong JX; Erlandson SC; Hilger D; Rasmussen SGF; Ring AM; Manglik A; Kruse AC, Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol 2018, 25, 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Izzo AA; Borrelli F; Capasso R; Di Marzo V; Mechoulam R, Non-psychotropic plant cannabinoids: new therapeutic opportunities from an ancient herb. Trends Pharmacol. Sci 2009, 30, 515–527. [DOI] [PubMed] [Google Scholar]
- (24).Wu CY; Roybal KT; Puchner EM; Onuffer J; Lim WA, Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 2015, 350, aab4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Di Stasi A; Tey SK; Dotti G; Fujita Y; Kennedy-Nasser A; Martinez C; Straathof K; Liu E; Durett AG; Grilley B; Liu H; Cruz CR; Savoldo B; Gee AP; Schindler J; Krance RA; Heslop HE; Spencer DM; Rooney CM; Brenner MK, Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med 2011, 365, 1673–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Virnekas B; Ge LM; Pluckthun A; Schneider KC; Wellnhofer G; Moroney SE, Trinucleotide phosphoramidites: ideal reagents for the synthesis of mixed oligonucleotides for random mutagenesis. Nucleic Acids Res. 1994, 22, 5600–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Moutel S; Bery N; Bernard V; Keller L; Lemesre E; de Marco A; Ligat L; Rain JC; Favre G; Olichon A; Perez F, NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. eLife 2016, 5, e16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Daley T; Smith AD, Predicting the molecular complexity of sequencing libraries. Nat. Methods 2013, 10, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Banaszynski LA; Liu CW; Wandless TJ, Characterization of the FKBP.rapamycin.FRB ternary complex. J. Am. Chem. Soc 2005, 127, 4715–4721. [DOI] [PubMed] [Google Scholar]
- (30).Gormley NA; Orphanides G; Meyer A; Cullis PM; Maxwell A, The interaction of coumarin antibiotics with fragments of the DNA gyrase B protein. Biochemistry 1996, 35, 5083–5092. [DOI] [PubMed] [Google Scholar]
- (31).Douglass EF; Miller CJ; Sparer G; Shapiro H; Spiegel DA, A comprehensive mathematical model for three-body binding equilibria. J. Am. Chem. Soc 2013, 135, 6092–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Dixon AS; Schwinn MK; Hall MP; Zimmerman K; Otto P; Lubben TH; Butler BL; Binkowski BF; Machleidt T; Kirkland TA; Wood MG; Eggers CT; Encell LP; Wood KV, NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol 2016, 11, 400–408. [DOI] [PubMed] [Google Scholar]
- (33).Schwope DM; Milman G; Huestis MA, Validation of an enzyme immunoassay for detection and semiquantification of cannabinoids in oral fluid. Clin. Chem 2010, 56, 1007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.