Abstract
Current clinical practice relies on clinical history to determine the time since stroke onset (TSS). Imaging-based determination of acute stroke onset time could provide critical information to clinicians in deciding stroke treatment options such as thrombolysis. Patients with unknown or unwitnessed TSS are usually excluded from thrombolysis, even if their symptoms began within the therapeutic window. In this work, we demonstrate a machine learning approach for TSS classification using routinely acquired imaging sequences. We develop imaging features from the magnetic resonance (MR) images and train machine learning models to classify TSS. We also propose a deep learning model to extract hidden representations for the MR perfusion-weighted images and demonstrate classification improvement by incorporating these additional deep features. The cross-validation results show that our best classifier achieved an area under the curve of 0.765, with a sensitivity of 0.788 and a negative predictive value of 0.609, outperforming existing methods. We show that the features generated by our deep learning algorithm correlate with MR imaging features, and validate the robustness of the model on imaging parameter variations (e.g., year of imaging). This work advances magnetic resonance imaging (MRI) analysis one step closer to an operational decision support tool for stroke treatment guidance.
Keywords: Deep learning, autoencoder, acute ischemic stroke, stroke onset time, MR perfusion imaging
I. Introduction
WITH approximately 795,000 new cases each year, stroke is the fifth leading cause of death and the primary cause of long-term disability in the United States [1]. Acute stroke treatments focus on restoring blood flow to hypoperfused regions to minimize infarction (i.e., tissue death). Intravenous (IV) tissue plasminogen activator (tPA) remains the dominant thrombolytic treatment for acute stroke, with a strict time usage guideline (no more than 4.5 hours from witnessed stroke symptom onset, i.e., time-since-stroke (TSS) A < 4.5hrs) due to the increased risk of hemorrhage when administered beyond that time interval. Mechanical thrombectomy (clot retrieval) is an alternative or adjunct therapy to IV tPA, yet its optimal a treatment time window remains uncertain [2]. Although IV tPA administration is the most common clinical therapy in most stroke centers, about 30% of the population cannot receive IV tPA because of unknown TSS (e.g., wake-up strokes or unwitnessed strokes). These patients are ineligible for tPA treatment despite the fact that their strokes may have actually occurred within the treatment window [3].
Previous work has argued for administering tPA based on a “tissue clock” determined via image analysis [4]. Studies are underway to investigate the use of a simple imaging feature, a mismatch pattern between magnetic resonance (MR) diffusion weighted imaging (DWI) and fluid attenuated inversion recovery (FLAIR) imaging, to estimate TSS. This method is based on the fact that the ischemic tissue is nearly immediately visible in DWI at stroke onset whereas it takes 3–4 hours for the ischemic tissue to appear in FLAIR [5]–[8]. The mismatch pattern is known as “DWI-FLAIR mismatch,” which is defined as the presence of visible acute ischemic lesions on DWI with no traceable hyperintensity in the corresponding region on FLAIR imaging (Fig. 1). Several clinical trials are ongoing to evaluate this mismatch method and determine if it is a suitable technique to apply on unwitnessed acute stroke patients in clinical settings [9]–[11]. While this is the current state-of-the-art method for determining eligibility for thrombolytic therapy in cases of unknown TSS, computing mismatch using MR imaging is a difficult task that requires extensive training and for which clinician inter-observer agreement has been found to be only moderate [12], [13]. Most of the previous studies [6]–[8] reported that the mismatch method could only achieve a specificity of 0.60 to 0.80 with a moderate sensitivity of 0.5 to 0.6 and a moderate negative predictive value (NPV) of 0.2 to 0.5. One study reported an area under the receiver operator curve (AUC) of 0.58 [8]. The preliminary work of the DWI-FLAIR mismatch method demonstrates a potential opportunity for using image analysis to classify TSS. However, the mismatch method may be too stringent, and therefore miss individuals who could benefit from thrombolytic therapy [14]. A recent study has shown that lesion water uptake obtained from Computed Tomography (CT) images may estimate TSS more accurately than MRI [15], yet CT research on TSS classification is limited. Furthermore, many stroke centers skip CT imaging and obtain only MRI prior to stroke intervention to save time and because it provides more information for clinical diagnosis.
Fig. 1.
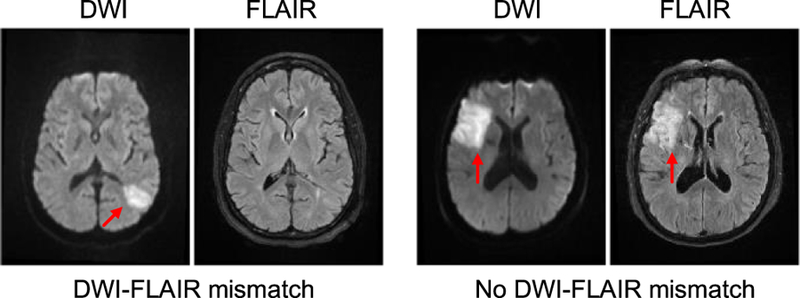
Examples of DWI-FLAIR mismatch. LEFT: presence of DWI-FLAIR mismatch (TSS = 1hr); RIGHT: absence of DWI-FLAIR mismatch (TSS = 8hrs). Hyperintensities are indicated by the red arrows.
Machine learning models have been applied widely and can achieve good classification performance for problems in the healthcare domain because of their ability to learn and utilize patterns from data to make predictions. In particular, recent developments in an area of machine learning, deep learning [16], have drawn significant research interest because of the technique’s ability to automatically learn feature detectors specific to the data for classification, achieving state-of-the-art performance in challenging medical imaging problems (e.g., brain tumor segmentation [17], high-resolution histological segmentation [18], organ classification [19], retinal image anomaly detection [20], etc.).
Predictive models have been made in attempt to predict stroke patient outcomes (e.g., mortality) using basic imaging features (e.g., lesion volume) [21], [22]. While much work has been done in predicting stroke patient outcome and treatment response, there is limited work in determining TSS using MR perfusion-weighted images (PWIs). These images may contain information that encodes TSS [23]–[25]. Ho, et al. [26] have previously shown the potential of classifying TSS from MR images using only a simple feature (i.e., mean intensity value) in a dataset of 105 patients. In the current work, we built off of this preliminary analysis by developing a set of hundreds of imaging features and analyzed the performance on a larger acute stroke patient dataset. We developed a deep learning algorithm based on an autoencoder architecture [27] to extract latent representative imaging features (i.e., deep features) from PWIs and evaluate the effectiveness of classifiers with and without the deep features to classify TSS.
In summary, the main contributions of this work are:
We developed a set of imaging features from the MR images (DWI, ADC, and FLAIR) and the perfusion parameter maps (derived from the PWIs) and compared five machine learning models on TSS classification using these imaging features.
We proposed a deep learning model with training patch coupling strategies to learn latent deep features from four-dimension (4-D) PWIs that can be used in TSS classification.
We compared our proposed machine learning models (with and without deep features) and show that the deep features improve TSS classification and the models outperform the DWI-FLAIR mismatch method.
This work is the illustration of machine learning models on TSS classification using imaging features derived from the MR images and the perfusion parameter maps. The results show that imaging features derived from stroke images can be predictive of TSS, demonstrating a possible alternative to DWI-FLAIR mismatch, which is known to be difficult to evaluate consistently. This work represents a step towards an operational decision support tool for guiding acute stroke treatment.
II. MATERIALS AND IMAGE PREPROCESSING
A. Patient Cohort and Imaging Data
Under institutional review board (UCLA IRB#18–000329) approval, a total of 181 patient MR images were examined from the University of California-Los Angeles picture archiving and communication system (PACS) between December 2011 and December 2017. The inclusion criteria were all patients with: 1) acute ischemic stroke due to middle cerebral artery (MCA) occlusion; 2) a recorded time of observed stroke symptom onset; 3) a recorded time of initial pretreatment imaging; and 4) a complete MR imaging sequence set (PWI, FLAIR, DWI, and ADC). The presence of a DWI-FLAIR mismatch was determined [28] by an expert neuroradiologist (Dr. S. El-Saden) using Medical Image Processing, Analysis, and Visualization (MIPAV) software [29], following the published protocol [8]. The presence of a DWI-FLAIR mismatch was labeled 1. The absence of a DWI-FLAIR mismatch was labeled 0. Patients’ TSS was calculated by subtracting the time at which the stroke symptoms were first observed from the time at which the first imaging was obtained. We followed the existing DWI-FLAIR TSS classification task [8] to binarize the TSS into two classes: positive (<4.5hrs) and negative (≥4.5hrs). After applying the inclusion criteria, a total of 131 patients were retrieved for the analysis (85 positive class; 46 negative class). This cohort subset was used to build the models for TSS classification. The patient characteristics are summarized in Table I.
TABLE I.
ISCHEMIC STROKE PATIENT COHORT CHARACTERISTICS
| Patients (n = 131) | |
|---|---|
| Demographics | |
| Age | 72.9±13.9 |
| Gender | 72 females |
| Clinical Presentation | |
| Time since stroke (continuous) | 256±247 minutes |
| NIHSS† | 10.1±7.87 |
| Atrial fibrillation | 37 |
| Hypertension | 87 |
| Stroke location (hemisphere) | |
| Left | 65 |
| Right | 66 |
| Classification Label | |
| Time since stroke (binary) | <4.5hrs (85); ≥4.5hrs (46) |
NIHSS = NIH Stroke Scale International; scale: 0 (no stroke symptoms) −42 (severe stroke)
All patients underwent MRI using a 1.5 or 3 Tesla echo planar MR imaging scanner (Siemens Medical Systems); scanning was performed with 12-channel head coils. The PWIs were acquired using a repetition time (TR) range of 1,490 to 2,640 ms and an echo time (TE) range of 23 to 50 ms. The pixel dimension of the PWIs varied from 1.72 × 1.72 × 6.00 to 2.61 × 2.61 × 6.00 mm. The FLAIR images were acquired using a TR range of 8,000 to 9,000 ms and a TE range of 88 to 134 ms. The pixel dimension of the FLAIR images varied from 0.688 × 0.688 × 6.00 to 0.938 × 0.938 × 6.50 mm. The DWI/ADC images were acquired using a TR range of 4,000 to 9,000 ms and a TE range of 78 to 122 ms. The pixel dimension of the DWI images varied from 0.859 × 0.859 × 6.00 to 1.85 × 1.85 × 6.50 mm. We note that in MR imaging, each brain voxel has three spatial dimensions for three axes (x-, y-, z-). We ignore the z-dimension in data generation (i.e., patch creation) due to slice thickness. Thus, our notation is simplified as we may denote the size of a “voxel” as 1 × 1 only.
B. Image Preprocessing
Intra-patient registration of pre-treatment PWIs, DWI, ADC and FLAIR images was performed with a six degree of freedom rigid transformation using FMRIB’s Linear Image Registration Tool (FLIRT) [30]. Through the registration, each voxel in the PWI, DWI, and ADC images was made to correspond to the same anatomical location in FLAIR. Gaussian filters with a size of 2.35 mm full width at half maximum (FWHM) were applied to remove spatial noise. Skulls and different tissue type masks (e.g., cerebrospinal fluid (CSF), gray/white matter) were identified using Statistical Parametric Mapping 12 (SPM12) [31]. CSF was excluded from this analysis. Perfusion parameter maps were generated using block-circulant singular value decomposition (bSVD) as provided by the sparse perfusion deconvolution toolbox (SPD) [32]; the arterial input function (AIF) was generated by the ASIST-Japan Perfusion mismatch analyzer (PMA) [33]. All DWI, ADC, and FLAIR intensity values were standardized to zero-mean and unit-variance globally on a brain-by-brain basis. The standardized images were used in the feature generation step for TSS classification.
III. METHODS
Inspired by the extensive research work in other medical domains (e.g., lung nodule detection [34]), in which hundreds of hand-crafted imaging features were defined for classification, we propose to train machine learning models with imaging features derived from MR images and perfusion parameter maps to classify TSS. This section is divided into four parts: A. Imaging Feature Generation describes the set of baseline imaging features and deep learning features for TSS classification; B. Machine Learning Models for TSS Classification describes the details of using machine learning models for TSS classification; C. Experimental Setup describes the implementation details of the deep learning models, the machine learning model training configuration; D. Evaluation describes the evaluation analysis and the metrics. The overview of the TSS classification is shown in Figure 2.
Fig. 2.
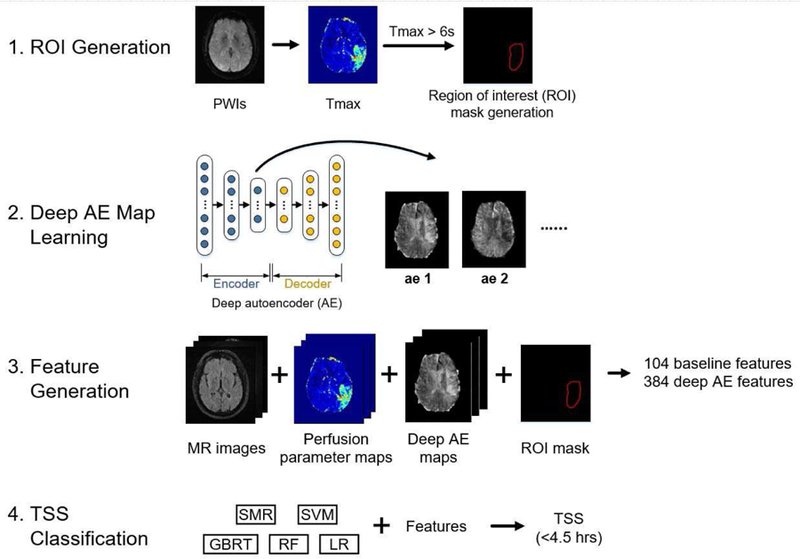
The overview of the TSS classification. The classification involved four steps: (1) ROI generation, (2) deep AE map learning, (3) feature generation, and (4) TSS classification. The ROI generation step defines the region of interest (Tmax > 6s) for generating imaging features. The deep AE map learning step generates new feature maps using deep autoencoders (AEs). The feature generation step includes imaging feature generation from the MR images, the perfusion parameter maps and the deep AE maps, resulting in a set of 104 baseline imaging features and 384 deep AE features. The TSS classification step trains five machine learning models with the imaging features to classify TSS<4.5 hrs.
A. Imaging Feature Generation
PWIs are spatio-temporal imaging data (4-D) that show the flow of a gadolinium-based contrast bolus into and out of the brain over time. They contain concentration time curves (CTCs) for each brain voxel, that describe the flow of the contrast (i.e. signal intensity change) over time. The global arterial input function (AIF) describes the contrast input to the vasculature (within a voxel) at a certain time t and it is defined in the MCA [35]. Perfusion parameter maps [36] can be derived from the AIF and CTCs, including cerebral blood volume (CBV), cerebral blood flow (CBF), mean transit time (MTT), time-to-peak (TTP), and time-to-maximum (Tmax). Briefly, CBV describes the total volume of flowing blood in a given volume of a voxel and CBF describes the rate of blood delivery to the brain tissue within a volume of a voxel. CBV and CBF are used to derive MTT, which represents the average time it takes the contrast to travel through the tissue volume of a voxel. TTP is the time required for the CTC to reach its maximum, which approximates the time needed for the bolus to arrive at the voxel with delay caused by brain vessel narrowing or obstruction. Tmax is the time point where the contrast residue function reaches its maximum, which approximates the true time needed for the bolus to arrive at the voxel.
We proposed and compared two ways to generate the imaging features for TSS classification. The first way was to generate imaging features from the MR images and the perfusion parameter maps, in which descriptive statistical features (e.g., mean) were defined. The second way was to generate imaging features directly from the 4-D PWIs. We proposed to use a deep autoencoder to learn hidden representation of every CTC within the PWIs. After transforming the CTCs into a number of hidden features using the trained autoencoders, we then aggregated these hidden representations into new feature maps that indicated hidden characteristics of the stroke tissue. The descriptive statistical features could then be generated from these new feature maps for TSS classification.
The imaging feature generation involved three parts: (1) region of interest generation, (2) baseline imaging feature generation, and (3) deep imaging feature generation.
1). Region of Interest Generation:
Generating imaging features based on entire brain MR images may be less descriptive to the stroke pathophysiology and less predictive of TSS because often stroke occurs in only one cerebral hemisphere. Therefore, we first needed to define the regions of interest (ROIs) to generate the imaging features. Specifically, the ROIs were defined by Tmax>6s, which captures both the dead tissue core and the salvageable tissue that can possibly be saved by intervention aimed at restoring blood flow [37]. The largest connected region in which Tmax>6s on the stroke hemisphere was used as the ROI mask.
2). Baseline Imaging Feature Generation:
The imaging features are summarized in Table II. The baseline imaging features were generated from the MR images (DWI, ADC, and FLAIR), the perfusion parameter maps (CBV, CBF, MTT, and TTP), and the Tmax>6s ROI mask. There are two major types of imaging features: descriptive statistics and the morphological features. Descriptive statistics included the maximum, minimum, median, mean, standard deviation, and variance of the intensity/parameter value within the ROI. Relative value (i.e., the ratio between the value of interest and the corresponding value on the contralateral side of the brain) has been shown to be predictive in stroke tissue outcome prediction [38], and therefore relative statistics (e.g., relative maximum) were also included as part of the descriptive statistics. Relative statistics of ADC-to-FLAIR and DWI-to-FLAIR were included, as inspired by the DWI-FLAIR mismatch method. This resulted in a set of 96 baseline descriptive features. Morphological features [34] were calculated using the ROI mask, including area, volume, circularity, and sphericity. Two shape features [39] were included: the ratio between the volume of the ROI and its bounding box (BE), and the ratio between the lesion surface area and the lesion volume (SV). The maximum and minimum diameter of the ROI mask were also included. This resulted in a set of 8 baseline morphological features. In total, a set of 104 baseline imaging features were generated. All the features were standardized independently to zero mean with a standard deviation of 1 for TSS classification.
TABLE II.
IMAGING FEATURES TYPES FOR TSS CLASSIFICATION
| Type | Features | Sources |
|---|---|---|
| Descriptive Statistics (n=96 for baseline features) (n=384 for deep features) | (Relative†) maximum, (relative) minimum, (relative) median, (relative) mean, (relative) standard deviation, (relative) variance | DWI, ADC, FLAIR, CBV, CBF, MTT, TTP, deep feature maps |
| Morphological Features (n=8 for baseline features) | Area, volume, circularity, sphericity, the ratio between the volume of the ROI and the bounding box. the ratio between the lesion surface area and the lesion volume, maximum diameter, minimum diameter | Tmax > 6s ROI mask |
Relative = the ratio between the value of interest and the value in its contralateral side of the brain
3). Deep Imaging Features Generation:
We hypothesized that a deep learning approach can automatically learn feature detectors to extract latent features from PWIs that can improve TSS classification. We implemented a deep autoencoder (deep AE) that is based on a stacked autoencoder [27] to learn the hidden features from PWIs (Fig. 3). Each PWI voxel CTC at location i , with a size of 1 × t (t = time for perfusion imaging), is transformed by the deep AE into K new feature representations that can represent complex voxel perfusion characteristics (in this work, the optimal value of K for AE reconstruction is determined by cross-validation). The learning of these features is automatic, and it is achieved by the hierarchical feature detectors, which are sets of weights that are learned in training via backpropagation. The deep AE consists of an encoder and a decoder. The encoder consists of two components: 1) an input layer; and 2) fully-connected layers, in which input neurons are fully-connected to each previous layer’s output neuron. The encoder is connected to the decoder, which follows reversely the same layer patterns of the encoder. The encoder output (i.e., the middle layer output of the deep AE) is the set of K new feature representations. Each new feature representation of all CTCs is aggregated to form a new feature map, known as “AE feature map” (Fk):
| (1) |
where I is the set of pixels in a PWI. In total, there are K new AE feature maps for a PWIs. New AE deep imaging features (descriptive statistics) were then generated from the AE features maps following the same procedure as described in 2) Baseline Imaging Feature Generation.
Fig. 3.

Deep AE feature generation. Training patches (with a size of 3 × 3 × 64) were randomly generated from PWIs. Each patch was coupled with an extra patch (AIF only, contralateral only, or AIF+contralateral) and the combined matrix was unrolled into a 1D vector that would be fed into the deep network. The proposed deep AE consisted of an encoder and decoder, in which the encoder output would be the new compact representation for the input. The encoder outputs of all PWI voxels were aggregated into the final deep AE feature maps. A ROI mask (Tmax>6s) was then applied to the new feature maps to generate the imaging features (descriptive statistics). Note that the input z-dimension is not included.
The proposed deep AE is trained via an unsupervised learning procedure in which the decoder output is the reconstruction of the encoder input. The network is optimized to obtain weights, Θ, that minimize the binary cross-entropy loss between the input, I , and the reconstructed output, , across the samples with size n [40]:
| (2) |
4). Training Input Patch Coupling and Generation:
Previous work [41], [42], suggests that regional information corresponding to a voxel’s surroundings improves classification in the MR images. Therefore, a small region (8 neighboring voxels) was included with each training voxel, leading to a size of 3×3 ×t (width × height × time; the z-dimension is omitted; t = 64 in our dataset), where the center of the patch is the voxel of interest for the deep AE feature learning. Previous work showed that patch coupling in voxel-wise stroke classification could improve the learning of hidden features, yielding better performance [43]. Therefore, we proposed three approaches for patch coupling: (1) training patch with global AIF patch; (2) training patch with its corresponding contralateral patch, which could be used as a matched control (reference) to improve feature learning [44]; (3) training patch with both the AIF patch and the contralateral patch. Each training patch (with the coupled patch(es)) was then unrolled from a size of 3 × 3 × t × p ( p = 2 or p = 3, depending on the number of coupled patches) into a 1-D vector. The 1-D data were used to train the deep AEs, which consisted of the fully-connected layers. Three different deep AEs were optimized for the three patch coupling methods. In total, 105,000 training data were generated by sampling randomly and equally from all the patient PWIs to train the deep AEs.
B. Machine Learning Models for TSS Classification
We constructed and compared the performance of five machine learning methods for binary TSS classification (TSS<4.5hrs or TSS≥4.5hrs): logistic regression (LR), random forest (RF), gradient boosted regression tree (GBRT), support vector machine (SVM), and stepwise multilinear regression (SMR). Briefly, LR is a probabilistic classification model in which binary label probabilities are found by fitting a logistic function of feature values [45]. RF is an ensemble learning method in which a multitude of decision trees are randomly constructed and the classification is based on the mode of the classes output by individual trees [46]. GBRT is an ensemble learning method similar to RF, in which a multitude of decision trees are randomly generated, yet these trees are added to the model in a stage-wise fashion based on their contribution to the objective function optimization [47]. SVM is a supervised learning classification algorithm that constructs a hyperplane (or set of hyperplanes) in a higher dimensional space for classification [48]. SMR is a stepwise method for adding and removing features from a multilinear model based on their statistical significance (e.g., F-statistics) to improve model performance [49]. In addition to the five machine learning models, we also trained four popular end-to-end convolutional neural networks (CNNs) to classify TSS. The input to the CNNs were the stacked images (the MR images + the perfusion parameter maps) and the output was the TSS classification. The details of the CNN implementation are described in the supplementary materials (S.1).
C. Experimental Setup
1). Autoencoder configurations and implementations details:
We optimized the deep AE using Adam, which computes adaptive learning rates during training and has demonstrated superior performance over other optimization methods [50]. An early-stopping strategy was applied to improve the learning of deep AE weights and prevent overfitting, where the training would be terminated if the performance did not improve over five consecutive epochs (maximum number of training epochs: 50). The deep AE was implemented in Torch7 [40], and the training was done on two NVIDIA Titan X GPUs and an NVIDIA Tesla K40 GPU. Ten-fold patient-based cross-validation was performed to determine the optimal deep AE architectures, including the number of encoder hidden layers (from 1–3) and the number of hidden units (factor of 4, 8, 16, 32).
2). Machine Learning Model Training:
The LR, RF, and SVM were developed using the Python Scikit-learn library [51]. The SMR and GBRT were developed using MATLAB and the XGBoost library [52] respectively. Different model hyperparameters (e.g., a LR’s hyperparameter, C) contribute differently to the classification and different machine learning methods may not perform equally on the same feature set. Evaluating model performance without hyperparameter tuning may lead to decreased predictive power due to over-fitting, especially on small and imbalanced datasets. Therefore, we performed nested ten-fold cross-validation for all five classifier evaluation to avoid classification bias [53]. Briefly, an outer ten-fold cross-validation was performed to obtain the overall classifier performance. Within each outer fold (in which a validation fold was held out), an inner ten-fold cross-validation was performed first to determine the optimal model hyperparameters using the training data (i.e., the nine folds), and then the model was trained with the optimal hyperparameters and applied to the validation fold. The details of the optimal model hyperparameter determination are described in the supplementary materials (S.2).
D. Evaluation
1). ROI Sensitivity Analysis:
To investigate the effect of ROI generation on classification, we explored the impact of two additional Tmax cutoff values [37], [54]. One is Tmax>4s, which is a softer cutoff value that may include normal brain tissue; one is Tmax>8s, which is a stricter cutoff value that captures only the severe hypoperfused stroke region. We followed the same experimental procedures to extract the imaging features from the ROIs generated by the two new cutoff values and evaluated their performance.
2). Feature Correlation Analysis:
A question one may ask is the correlation of the new deep features to the baseline imaging features. Recently, deep learning has been criticized as a “black-box” approach [55] that yields state-of-the-art performance, yet the classification mechanism is unclear. To understand what the deep features represented, we proposed an approach based on the correlation analysis. First, we calculated the correlation between the deep AE features and the baseline imaging features. Then, for each baseline imaging feature, the most correlated deep AE feature was identified. For each identified deep AE feature, the top five correlated baseline imaging features were obtained. All correlations were calculated using Pearson correlation [56].
3). TSS Subgroup Classification Analysis:
The dataset was created from the patient imaging exams obtained from 2011 to 2017. Changes in MR image acquisition parameters (e.g., field strength) across any years may impact the classifier performance [57]. We explored the impact of two image-related variations, magnetic field strength and year of imaging acquisition, on the TSS classification. For the field strength, we performed two-fold cross-validation to evaluate the classifiers on TSS classification, i.e., trained on a data subset with one field strength (e.g., 1.5T) and evaluated on a data subset with another field strength (e.g., 3T). For the year of imaging, we trained the classifiers with the data collected from 2011–2014 and evaluated the models with the data collected from 2015–2017. This evaluation was meant to explore whether the model still performed well on the newer data when training on the older data.
4). Metrics:
We computed the area under the ROC curve (AUC), which is a classifier’s probability of predicting an outcome better than chance, for all five classifiers. To determine if the performance of the models significantly differed, we used the Hanley and McNeil significant test [58] with the improved covariance calculation [59] to compare the model AUCs. We also computed the model AUCs using the method published by Ho, et al. in 2017 [26] trained with our dataset. Sensitivity, specificity, F1-score, positive predictive value (PPV), and negative predictive value (NPV) were calculated for the DWI-FLAIR mismatch method. Given the DWI-FLAIR mismatch method specificity, the performance (sensitivity, F1-score, PPV, and NPV) was calculated for the machine learning classifiers and compared against the DWI-FLAIR mismatch method.
IV. RESULTS
A. TSS Classification
The optimal AE model architectures (number of layers, number of hidden units) for three types of coupling patch were determined (Table III). All three optimal AE architectures had 32 hidden units (AE1 to AE32) in the middle layer (i.e., 32 deep feature maps), with mean square error (MSE) of at least 40% smaller than the average MSE of all of the trained AEs. These optimal AE models were used to generate deep feature maps from the patient PWIs, in which the ROI masks were applied to generate the deep AE features. The classifiers were trained with three different groups of features: (1) the baseline imaging (BI) features (96 descriptive statistics and 8 morphological features); (2) the deep AE features (384 descriptive statistics); (3) the baseline and deep AE features. The AUCs of the classifiers are depicted in Table IV.
TABLE III.
OPTIMAL AE ARCHITECTURE FOR EACH COUPLING TYPE
| Coupling Patch Type | Optimal AE Architecture (# of hidden units/layer) | Optimal MSE (Average Deep AE MSE) |
|---|---|---|
| AIF patch only | 1152–192-32–32-192–1152 | 0.606 (1.54) |
| Contralateral patch only | 1152–288-32–32-288–1152 | 1.16(1.95) |
| AIF + Contralateral patch | 1728–288-32–32-288–1728 | 1.06 (4.49) |
TABLE IV.
THE AUCS OF CLASSIFIERS ON TSS CLASSIFICATION BOLD INDICATED THE HIGHEST AUC FOR A GIVEN CLASSIFIER
| Classifier | Ho, et al. (2017) [26] | No AE | AIF coupling patch only | Contralateral coupling patch only | AIF + contralateral patch | |||
|---|---|---|---|---|---|---|---|---|
| BI† | AE¥ | BI+AEΩ | AE | BI+AE | AE | BI+AE | ||
| LR | 0.574 | 0.618 | 0.650 | 0.658 | 0.647 | 0.676 | 0.710 | 0.765* |
| RE | 0.624 | 0.640 | 0.650 | 0.669 | 0.662 | 0.682 | 0.592 | 0.690 |
| GBRT | 0.567 | 0.608 | 0.590 | 0.570 | 0.676 | 0.674 | 0.612 | 0.670 |
| SVM | 0.669 | 0.636 | 0.477 | 0.736 | 0.605 | 0.666 | 0.600 | 0.746* |
| SMR | 0.683 | 0.661 | 0.574 | 0.707 | 0.650 | 0.677 | 0.705 | 0.730 |
BI = Models were trained with the baseline imaging features (94 descriptive statistics and 8 morphological features)
AE = Models were trained with the deep AE features (384 descriptive statistics generated from deep AE feature maps)
BI + AE = Models were trained with the baseline and deep AE features
With the baseline imaging features alone, all classifiers (LR, RF, GBRT, SVM, and SMR) achieved an AUC of at least 0.6 on TSS classification. With the deep AE features alone, most classifiers also achieved an AUC of at least 0.6, showing that the proposed deep AEs extracted hidden features in PWIs which are predictive of TSS. With the combination of baseline imaging features and deep features, all classifiers (except the GBRT trained with AIF coupling patch) showed improvement in AUC (compared to when using only the baseline imaging features). Among all the patch coupling methods, deep features generated from the AIF + contralateral coupling method improved TSS classification in most of all classifiers, e.g., LR has the best AUC with the AIF + contralateral patches (0.765 vs. 0.658 vs. 0.676). Both LR and SVM had significantly better AUCs (p-value=0.003 and p-value=0.024 respectively) with the features from the AIF + contralateral coupling than with the features from only the baseline imaging. Comparing to the method published by Ho et al. in 2017, all classifiers (AIF + contralateral patches) performed better using the current method.
Figure 4 shows the ROCs of the classifiers trained with the baseline features and the deep features (generated from the deep AE with the AIF + contralateral coupling patch), and the neuroradiologist performance using the DWI-FLAIR mismatch method. Three classifiers (LR, SMR, and SVM) achieved higher sensitivity (while having the same specificity) than the DWI-FLAIR mismatch method with the addition of the deep features, demonstrating the ability of using imaging features with machine learning models to classify TSS. Among all the classifiers, the LR trained with baseline imaging features and the deep features performed the best, with an AUC of 0.765. Comparing to the mismatch method, LR achieved higher sensitivity (0.788 vs 0.694), F1-score (0.788 vs 0.728), NPV (0.609 vs 0.519), and PPV (0.788 vs 0.766) while maintaining same specificity (0.609). Therefore, LR with the baseline imaging features and the deep AE features was determined to be the most suitable classifier for the TSS classification.
Fig. 4.

The ROCs of different classifiers trained with both the baseline imaging features and the deep features (generated from the deep AE with the AIF + contralateral coupling patch). The red cross indicated the neuroradiologist classification using the DWI-FLAIR mismatch method.
B. Example of Classification
Figure 5 shows the TSS classification example of the optimal LR classifier, trained with both the baseline and deep AE features (generated from the AIF + contralateral coupling patch). The classifier was able to classify patients with clear mismatch between DWI and FLAIR (Figure 5, patient #1 and patient #2). In cases where the mismatch was not clear, the classifier was able to correctly classify some cases (patient #3), but occasionally resulted in misclassifications (patient #4 and patient #5).
Fig. 5.

Examples of TSS classification of the optimal LR classifier trained with both the baseline and deep AE features. Patient #1 (deep white matter infarct) and patient #2 (cortical infarct) were correctly classified as having TSS<4.5hrs and showed clear mismatch between DWI and FLAIR. In patient #3, the mismatch between DWI and FLAIR was less obvious, but the classifier still classified correctly. Patient #4 was misclassified because there was a visible mismatch between DWI and FLAIR images, but clinical history determined TSS to be > 4.5hrs. In patient #5, the infarct was more conspicuous on DWI but essentially matched on FLAIR, and was also misclassified.
C. ROI Sensitivity Analysis
We performed the ROI sensitivity analysis on the deep AE feature maps generated from the AIF + contralateral coupling patch. Table V shows the TSS classification results. We observed that the deep AE features were still able to improve the performance of almost every classifier and threshold combination. The only exception was SMR with Tmax>8s, but the difference was not statistically significant (p-value=0.089). These results show that the deep AE feature generation is robust across ROI generation thresholds. Among all the cutoff values, Tmax>6s provided ROIs that resulted in optimal performance in all classifiers with baseline and deep AE features.
TABLE V.
THE AUCS OF CLASSIFIERS (WITH AIF + CONTRALATERAL PATCH) ON TSS CLASSIFICATION IN ROI SENSITIVITY ANALYSIS
| Classifier | Tmax>4s | Tmax>6s¥ | Tmax>8s | |||
|---|---|---|---|---|---|---|
| BI† | BI+AEΩ | BI | BI+AE | BI | BI+AE | |
| LR | 0.520 | 0.690* | 0.618 | 0.765* | 0.622 | 0.651 |
| RE | 0.667 | 0.678 | 0.640 | 0.690 | 0.610 | 0.666 |
| GBRT | 0.607 | 0.650 | 0.608 | 0.670 | 0.618 | 0.644 |
| SVM | 0.479 | 0.649* | 0.636 | 0.746* | 0.624 | 0.683 |
| SMR | 0.494 | 0.591 | 0.661 | 0.730 | 0.696 | 0.624 |
BI = Models were trained with the baseline imaging features (94 descriptive statistics and 8 morphological features)
BI + AE = Models were trained with the baseline and deep AE features
The result is obtained from Table IV
D. Feature Correlation Analysis
Table VI shows several examples of the deep AE feature correlation to the baseline imaging features. It is interesting to observe that different deep AE features correlated well with certain categories of baseline imaging features. For example, the AE8 feature correlated well with the time-related baseline imaging features (TTP and MTT), whereas the AE7 feature correlated well with the morphological baseline imaging features (e.g., area). Some deep AE feature (e.g., AE16) correlated well to an image type (e.g., ADC). The correlation analysis demonstrates that the deep AE features capture a variety of complex representations (i.e., shape, morphology) that led to better TSS classification.
TABLE VI.
FEATURE CORRELATION BETWEEN THE DEEP AE FEATURES AND THE BASELINE IMAGING FEATURES
| Rank | AE8 relative minimum | AE7 Relative max | AE16 variance | AE23 Relative variance |
|---|---|---|---|---|
| 1 | TTP relative minimum | Area | ADC variance | DWI relative maximum |
| 2 | TTP minimum | Maximum diameter | ACD-FLAIR relative mean | DWI variance |
| 3 | MTT relative minimum | Volume | ADC-FLAIR Relative variance | FLAIR relative maximum |
| 4 | TTP maximum | Minimum diameter | MTT variance | DWI-FLAIR relative variance |
| 5 | DWI minimum | TTP minimum | ADC mean | SV |
| Interpretation | Time-related | Morphology-related | ADC-related | DWI-related |
E. TSS Subgroup Classification Analysis
The TSS subgroup classification result is shown in Table Three out of five classifiers showed improvement with the addition of deep AE features on the field strength subgroup analysis, and four out of five classifiers showed improvements on the year of imaging subgroup analysis. We observed that the SMR (with the baseline imaging features and the deep AE features) did not perform well (AUC=0.488) in the year of imaging subgroup analysis. We suspect that this may be due to the nature of the SMR feature selection mechanism, where small feature set could be selected and led to poor performance. Overall, we could still observe the improvement of the TSS classification with the additional new imaging features. The subgroup analysis shows that the classifiers were robust to both field strength and year of image acquisition.
V. DISCUSSION
Determining stroke onset time independent of patient history is a challenging and important task for better stroke evaluation and stroke treatment decision-making. The DWI-FLAIR mismatch method is the current state-of-the-art method that can provide clinicians with insight into stroke onset time based on observable mismatch patterns between DWI and FLAIR. One study reported the DWI-FLAIR mismatch method could achieve an AUC of 0.58 in a data set of 194 ischemic stroke patients [8]. A clinical trial showed that using this method is safe (i.e., no increased risk of hemorrhage) in selecting patients whose stroke onset time is unknown for IV tPA treatment [11]. Yet, this method suffers from its simplicity, i.e., the mismatch pattern between DWI and FLAIR may not capture all patients in whom TSS<4.5hrs [14], which can lead to a misclassification. In this work, we proposed a classification framework (defining ROIs, generating features, and training classifiers) for TSS classification. To generate useful information with a limited number of patients (n=131) and high dimensional data (4-D PWIs), we proposed to first use an autoencoder with several patch-coupling strategies to learn voxel-wise hidden representations. We then aggregated these hidden representations to generate new feature maps, which can be used to generate new AE features. Using this approach, we developed new imaging features from routinely acquired MR imaging sequences, perfusion parameter maps, and deep AE feature maps that capture information predictive of TSS. Our results show that this machine learning approach can potentially serve as an improved alternative to the DWI-FLAIR mismatch method. The proposed methodology may also be applied to other medical imaging data (e.g., cardiac PWIs).
With only the baseline imaging features, the best classifier (SMR) can achieve an AUC of 0.661 on TSS classification (Table IV). This indicates that the machine learning models capture signal changes from the MR images and the perfusion parameter maps that are predictive of TSS. One possible signal is the change of the perfusion parameter value (e.g., CBV) over time within the ischemic stroke regions, previously demonstrated in animal studies [24], [25]. This also shows that the enriched baseline imaging feature set improves the TSS classification, in which previous work [26] showed a limited performance (AUC<0.700) with a single mean intensity value feature. There is an interesting observation that the deep AE feature maps generated from the AIF + contralateral coupling input are more predictive than the deep AE feature maps generated from the AIF coupling input or the contralateral coupling input. This supports our hypothesis that the AIF patch provides the base for the initial bolus setting (e.g., how fast the bolus is injected) whereas the contralateral patch provides a matched control for the healthy brain concentration time curve. We also observe that adding the deep imaging features (from the AIF + contralateral coupling) could improve the best classifier (LR) by at least an AUC of 0.1, and the correlation shows that 4 out of 10 top-10 correlated are the deep AE features (supplementary material Table SII). These observations suggest that the deep AE features are important for improving the TSS classification. Compared to the best proposed model, the best end-to-end CNN had a lower AUC (0.575 vs. 0.765, p-value = 0.0001; supplementary material Table SI). We suspect that the low performance may be due to the limited training data, a common problem with medical datasets, and a large number of trainable weights (>100,000).
Deep learning approaches have been criticized as “black-box”, in which the learning and the classification mechanism are too complicated and difficult to understand, engendering doubt in medical applicability because clinical decision making is ideally evidence-based [60]. In this work, we interpreted the complex deep AE features via a correlation analysis and found that some deep AE features correlated well with some baseline intuitive imaging features (e.g., morphology and time). This is an important first step because it shows clinicians what these deep AE features may represent, helping them to understand more about how the classifiers make the classification and why they can achieve better performance. The next important step will be the visualization [26], which may bring further insight into TSS classification, like highlighting the important brain regions that drive a specific classification. Through a comprehensive visualization tool, clinicians may then be able to associate clinical reasoning (e.g., the location and the strength of the highlighted signals) with the TSS classification, making the deep learning approach more intuitive and therefore integral to the medical decision-making process.
Our study does have some limitations. The machine learning models were trained and validated on only the MR images. Our next step will be collecting the Computed Tomographic (CT) perfusion images and validating the robustness of our model on the new imaging modality. CT perfusion imaging is cheaper, faster and more readily available than MRI and could become the imaging modality of choice for acute ischemic stroke patients if TSS analysis on CT images was accurate and independent of clinical history. Finally, we did not consider clinical variables (e.g., age) in our classification, which may further improve the TSS classification. We plan to explore the existing machine learning models, incorporating clinical variables for TSS classification in the future.
VI. CONCLUSION
In this work, we developed new imaging features from MR images, perfusion parameter maps, and deep AE feature maps, and showed that they can be utilized by machine learning models to classify TSS. We showed that the best machine learning model can outperform the current state-of-the-art DWI-FLAIR mismatch method. We also proposed a correlation method to interpret the deep AE features and demonstrated that our proposed classification method is robust to variations in imaging acquisition. The method proposed here provides a foundation to utilize deep learning and machine learning techniques in TSS classification, which could ultimately provide decision-making guidance for clinicians in acute stroke intervention treatment.
Supplementary Material
ACKNOWLEDGMENT
Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R01NS100806. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional funding support was provided by a UCLA Radiology Department Exploratory Research Grant 16–0003, and an NVIDIA Academic Hardware Grant.
For example, “This work was supported in part by the U.S. Department of Commerce under Grant BS123456”.
Footnotes
This paragraph of the first footnote will contain the date on which you submitted your paper for review. It will also contain support information, including sponsor and financial support acknowledgment.
Contributor Information
King Chung Ho, Departments of Bioengineering and Radiological Sciences, and the Medical Imaging Informatics (MII) Group, University of California, Los Angeles, CA 90024 USA.
William Speier, Department of Radiological Sciences and the Medical Imaging Informatics (MII) Group, University of California, Los Angeles, CA 90024 USA.
Haoyue Zhang, Departments of Bioengineering and Radiological Sciences, and the Medical Imaging Informatics (MII) Group, University of California, Los Angeles, CA 90024 USA.
Fabien Scalzo, Departments of Neurology and Computer Science, University of California, Los Angeles, CA 90024 USA.
Suzie El-Saden, Department of Radiological Sciences and the Medical Imaging Informatics (MII) Group, University of California, Los Angeles, CA 90024 USA.
Corey W. Arnold, Department of Radiological Sciences and the Medical Imaging Informatics (MII) Group, University of California, Los Angeles, CA 90024 USA.
References
- [1].Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, and others, “Heart disease and stroke statistics—2018 update: a report from the American Heart Association,” Circulation, vol. 137, no. 12, pp. e67–e492, 2018. [DOI] [PubMed] [Google Scholar]
- [2].Furlan AJ, “Endovascular Therapy for Stroke — It’s about Time,” N. Engl. J. Med, pp. 1–3, 2015. [DOI] [PubMed] [Google Scholar]
- [3].Moradiya Y and Janjua N, “Presentation and outcomes of ‘wake-up strokes’ in a large randomized stroke trial: analysis of data from the International Stroke Trial,” J. Stroke Cerebrovasc. Dis, vol. 22, no. 8, pp. e286–e292, 2013. [DOI] [PubMed] [Google Scholar]
- [4].Buck D, Shaw LC, Price CI, and Ford GA, “Reperfusion therapies for wake-up stroke: systematic review.,” Stroke, vol. 45, no. 6, pp. 1869–75, June 2014. [DOI] [PubMed] [Google Scholar]
- [5].Thomalla G, Rossbach P, Rosenkranz M, Siemonsen S, Krützelmann A, Fiehler J, and Gerloff C, “Negative fluid-attenuated inversion recovery imaging identifies acute ischemic stroke at 3 hours or less,” Ann. Neurol, vol. 65, no. 6, pp. 724–732, 2009. [DOI] [PubMed] [Google Scholar]
- [6].Thomalla G, Cheng B, Ebinger M, Hao Q, Tourdias T, Wu O, Kim JS, Breuer L, Singer OC, and Warach S, “DWI-FLAIR mismatch for the identifi cation of patients with acute ischaemic stroke within 4 · 5 h of symptom onset ( PRE-FLAIR ): a multicentre observational study,” Lancet Neurol, vol. 10, no. 11, pp. 978–986, 2011. [DOI] [PubMed] [Google Scholar]
- [7].Ebinger M, Galinovic I, Rozanski M, Brunecker P, Endres M, and Fiebach JB, “Fluid-attenuated inversion recovery evolution within 12 hours from stroke onset: A reliable tissue clock?,” Stroke, vol. 41, no. 2, pp. 250–255, 2010. [DOI] [PubMed] [Google Scholar]
- [8].Emeriau S, Serre I, Toubas O, Pombourcq F, Oppenheim C, and Pierot L, “Can Diffusion-Weighted Imaging–Fluid-Attenuated Inversion Recovery Mismatch (Positive Diffusion-Weighted Imaging/Negative Fluid-Attenuated Inversion Recovery) at 3 Tesla Identify Patients With Stroke at < 4.5 Hours?,” Stroke, vol. 44, no. 6, pp. 1647–1651, 2013. [DOI] [PubMed] [Google Scholar]
- [9].Koga M, Toyoda K, Kimura K, Yamamoto H, Sasaki M, Hamasaki T, Kitazono T, Aoki J, Seki K, Homma K, Sato S, and Minematsu K, “THrombolysis for Acute Wake-up and unclear-onset Strokes with alteplase at 0.6 mg/kg (THAWS) Trial,” Int. J. Stroke, vol. 9, no. 8, pp. 1117–1124, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Thomalla G, Fiebach JB, Østergaard L, Pedraza S, Thijs V, Nighoghossian N, Roy P, Muir KW, Ebinger M, Cheng B, Galinovic I, Cho TH, Puig J, Boutitie F, Simonsen CZ, Endres M, Fiehler J, and Gerloff C, “A multicenter, randomized, double-blind, placebo-controlled trial to test efficacy and safety of magnetic resonance imaging-based thrombolysis in wake-up stroke (WAKE-UP),” Int. J. Stroke, vol. 9, no. 6, pp. 829–836, 2014. [DOI] [PubMed] [Google Scholar]
- [11].Schwamm L, “MR WITNESS: A Study of Intravenous Thrombolysis With Alteplase in MRI-Selected Patients (MR WITNESS),” ClinicalTrials.gov, 2011. .
- [12].Ziegler A, Ebinger M, Fiebach JB, Audebert HJ, and Leistner S, “Judgment of FLAIR signal change in DWI-FLAIR mismatch determination is a challenge to clinicians,” J. Neurol, vol. 259, no. 5, pp. 971–973, 2012. [DOI] [PubMed] [Google Scholar]
- [13].Galinovic I, Puig J, Neeb L, Guibernau J, Kemmling A, Siemonsen S, Pedraza S, Cheng B, Thomalla G, Fiehler J, and Fiebach JB, “Visual and region of interest-based inter-rater agreement in the assessment of the diffusion-weighted imaging-fluid-attenuated inversion recovery mismatch,” Stroke, vol. 45, no. 4, pp. 1170–1172, 2014. [DOI] [PubMed] [Google Scholar]
- [14].Odland A, Særvoll P, Advani R, Kurz MW, and Kurz KD, “Are the current MRI criteria using the DWI-FLAIR mismatch concept for selection of patients with wake-up stroke to thrombolysis excluding too many patients?,” Scand. J. Trauma. Resusc. Emerg. Med, vol. 23, p. 22–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Minnerup J, Broocks G, Kalkoffen J, Langner S, Knauth M, Psychogios MN, Wersching H, Teuber A, Heindel W, Eckert B, and others, “Computed tomography–based quantification of lesion water uptake identifies patients within 4.5 hours of stroke onset: A multicenter observational study,” Ann. Neurol, vol. 80, no. 6, pp. 924–934, 2016. [DOI] [PubMed] [Google Scholar]
- [16].LeCun Y, Bengio Y, and Hinton G, “Deep learning,” Nature, vol. 521, no. 7553, pp. 436–444, 2015. [DOI] [PubMed] [Google Scholar]
- [17].Davy A, Havaei M, Warde-Farley D, Biard A, Tran L, Jodoin P-M, Courville A, Larochelle H, Pal C, and Bengio Y, “Brain Tumor Segmentation With Deep Neural Networks,” Med. Image Anal, vol. 35, p. 1–2017. [DOI] [PubMed] [Google Scholar]
- [18].Li J, Sarma KV, Ho KC, Gertych A, Knudsen BS, and Arnold CW, “A Multi-scale U-Net for Semantic Segmentation of Histological Images from Radical Prostatectomies,” in AMIA Annual Symposium Proceedings, 2017. [PMC free article] [PubMed] [Google Scholar]
- [19].Shin HC, Orton MR, Collins DJ, Doran SJ, and Leach MO, “Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4D patient data,” IEEE Trans. Pattern Anal. Mach. Intell, vol. 35, no. 8, pp. 1930–1943, 2013. [DOI] [PubMed] [Google Scholar]
- [20].Schlegl T, Seeböck P, Waldstein SM, Schmidt-Erfurth U, and Langs G, “Unsupervised anomaly detection with generative adversarial networks to guide marker discovery,” in International Conference on Information Processing in Medical Imaging, 2017, pp. 146–157. [Google Scholar]
- [21].Ho KCKC, Speier W, El-Saden S, Liebeskind DSDS, Saver JLJL, Bui AAAAT, and Arnold CWCW, “Predicting discharge mortality after acute ischemic stroke using balanced data.,” AMIA Annu. Symp. Proc, vol. 2014, pp. 1787–96, 2014. [PMC free article] [PubMed] [Google Scholar]
- [22].Vogt G, Laage R, Shuaib A, and Schneider A, “Initial lesion volume is an independent predictor of clinical stroke outcome at day 90: An analysis of the Virtual International Stroke Trials Archive (VISTA) database,” Stroke, vol. 43, no. 5, pp. 1266–1272, 2012. [DOI] [PubMed] [Google Scholar]
- [23].Thomalla G and Gerloff C, “Treatment Concepts for Wake-Up Stroke and Stroke with Unknown Time of Symptom Onset,” Stroke, vol. 46, no. 9, pp. 2707–2713, 2015. [DOI] [PubMed] [Google Scholar]
- [24].Mcleod DD, Parsons MW, Levi CR, Beautement S, Buxton D, Roworth B, and Spratt NJ, “Establishing a rodent stroke perfusion computed tomography model,” Int. J. Stroke, vol. 6, no. 4, pp. 284–289, 2011. [DOI] [PubMed] [Google Scholar]
- [25].Murphy BD, Chen X, and Lee T-Y, “Serial changes in CT cerebral blood volume and flow after 4 hours of middle cerebral occlusion in an animal model of embolic cerebral ischemia,” Am. J. Neuroradiol, vol. 28, no. 4, pp. 743–749, 2007. [PMC free article] [PubMed] [Google Scholar]
- [26].Ho KC, Speier W, EL-Saden S, and Arnold WC, “Classifying Acute Ischemic Stroke Onset Time using Deep Imaging Features,” in AMIA Annual Symposium Proceedings, 2017. [PMC free article] [PubMed] [Google Scholar]
- [27].Vincent P, Larochelle H, Lajoie I, Bengio Y, and Manzagol P-A, “Stacked Denoising Autoencoders: Learning Useful Representations in a Deep Network with a Local Denoising Criterion,” J. Mach. Learn. Res, vol. 11, no. 3, pp. 3371–3408, 2010. [Google Scholar]
- [28].Tourdias T, Renou P, Sibon I, Asselineau J, Bracoud L, Dumoulin M, Rouanet F, Orgogozo JM, and Dousset V, “Final cerebral infarct volume is predictable by MR imaging at 1 week.,” AJNR. Am. J. Neuroradiol, vol. 32, no. 2, pp. 352–8, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McAuliffe MJ, Lalonde FM, McGarry D, Gandler W, Csaky K, and Trus BL, “Medical image processing, analysis and visualization in clinical research,” in Computer-Based Medical Systems, 2001. CBMS 2001. Proceedings. 14th IEEE Symposium on, 2001, pp. 381–386. [Google Scholar]
- [30].Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, and Matthews PM, “Advances in functional and structural MR image analysis and implementation as FSL,” Neuroimage, vol. 23, no. SUPPL. 1, pp. 208–219, 2004. [DOI] [PubMed] [Google Scholar]
- [31].Ashburner J, Barnes G, Chen C, Daunizeau J, Flandin G, Friston K, Kiebel S, Kilner J, Litvak V, Moran R, and others, “SPM12 Manual The FIL Methods Group (and honorary members),” Wellcome Trust Cent. Neuroimaging, London, UK, 2014. [Google Scholar]
- [32].Fang R, Chen T, and Sanelli PC, “Towards robust deconvolution of low-dose perfusion CT?: Sparse perfusion deconvolution using online dictionary learning,” Med. Image Anal, vol. 17, no. 4, pp. 417–428, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kudo K, Christensen S, Sasaki M, Østergaard L, Shirato H, Ogasawara K, Wintermark M, and Warach S, “Accuracy and reliability assessment of CT and MR perfusion analysis software using a digital phantom,” Radiology, vol. 267, no. 1, pp. 201–211, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Messay T, Hardie RC, and Rogers SK, “A new computationally efficient CAD system for pulmonary nodule detection in CT imagery,” Med. Image Anal, vol. 14, no. 3, pp. 390–406, 2010. [DOI] [PubMed] [Google Scholar]
- [35].Calamante F, “Arterial input function in perfusion MRI: A comprehensive review,” Prog. Nucl. Magn. Reson. Spectrosc, vol. 74, pp. 1–32, 2013. [DOI] [PubMed] [Google Scholar]
- [36].Ho KC, Scalzo F, Sarma VK, EL-Saden S, and Arnold WC, “A Temporal Deep Learning Approach for MR Perfusion Parameter Esti mation in Stroke,” in International Conference of Pattern Recognition, 2016. [Google Scholar]
- [37].Olivot JM, Mlynash M, Thijs VN, Kemp S, Lansberg MG, Wechsler L, Bammer R, Marks MP, and Albers GW, “Optimal tmax threshold for predicting penumbral tissue in acute stroke,” Stroke, vol. 40, no. 2, pp. 469–475, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Park H-I, Cha J-K, Kang M-J, Kim D-H, Yoo N-T, Choi J-H, and Huh J-T, “Reduced rCBV ratio in perfusion-weighted MR images predicts poor outcome after thrombolysis in acute ischemic stroke,” Eur. Neurol, vol. 65, no. 5, pp. 257–263, 2011. [DOI] [PubMed] [Google Scholar]
- [39].Frindel C, Rouanet A, Giacalone M, and Cho T, “Validity of Shape as a Predictive Biomarker of Final Infarct Volume in Acute Ischemic Stroke,” Stroke, vol. 46, pp. 976–981, 2015. [DOI] [PubMed] [Google Scholar]
- [40].Collobert R, Kavukcuoglu K, and Farabet C, “Torch7: A Matlab-like Environment for Machine Learning,” BigLearn, NIPS Work; , pp. 1–6, 2011. [Google Scholar]
- [41].Scalzo F, Hao Q, Alger JR, Hu X, and Liebeskind DS, “Regional prediction of tissue fate in acute ischemic stroke,” Ann. Biomed. Eng, vol. 40, no. 10, pp. 2177–2187, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ho KC, Scalzo F, Sarma VK, El-Saden S, and Arnold WC, “A Temporal Deep Learning Appraoch for MR Perfusion Parameter Estimation in Stroke,” in International Conference of Pattern Recognition, 2016. [Google Scholar]
- [43].Ho KC, Scalzo F, Sarma K, El-Saden S, Bui A, and Arnold C, “A Novel Bi-Input Convolutional Neural Network for Deconvolution-Free Estimation of Stroke MR Perfusion Parameters,” in 2016 Annual Meeting of the Radiological Society of North America, 2016. [Google Scholar]
- [44].V Campbell BC, Christensen S, Levi CR, Desmond PM, Donnan GA, Davis SM, and Parsons MW, “Cerebral blood flow is the optimal CT perfusion parameter for assessing infarct core,” Stroke, vol. 42, no. 12, pp. 3435–3440, 2011. [DOI] [PubMed] [Google Scholar]
- [45].Bewick V, Cheek L, and Ball J, “Statistics review 14: Logistic regression,” Crit. care, vol. 9, no. 1, p. 112, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Breiman L, “Random forests,” Mach. Learn, vol. 45, no. 1, pp. 5–32, 2001. [Google Scholar]
- [47].Friedman JH, “Greedy function approximation: a gradient boosting machine,” Ann. Stat, pp. 1189–1232, 2001. [Google Scholar]
- [48].Cortes C and Vapnik V, “Support-vector networks,” Mach. Learn, vol. 20, no. 3, pp. 273–297, 1995. [Google Scholar]
- [49].Draper NR and Smith H, Applied regression analysis John Wiley & Sons, 2014. [Google Scholar]
- [50].Kingma DP and Ba JL, “Adam: a Method for Stochastic Optimization,” Int. Conf. Learn. Represent. 2015, pp. 1–15, 2015. [Google Scholar]
- [51].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, and others, “Scikit-learn: Machine learning in Python,” J. Mach. Learn. Res, vol. 12, no. Oct, pp. 2825–2830, 2011. [Google Scholar]
- [52].Chen T and Guestrin C, “XGBoost?: Reliable Large-scale Tree Boosting System,” arXiv, pp. 1–6, 2016. [Google Scholar]
- [53].Krstajic D, Buturovic LJ, Leahy DE, and Thomas S, “Cross-validation pitfalls when selecting and assessing regression and classification models,” J. Cheminform, vol. 6, no. 1, p. 10, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zaro-Weber O, Moeller-Hartmann W, Heiss W-D, and Sobesky J, “Maps of time to maximum and time to peak for mismatch definition in clinical stroke studies validated with positron emission tomography,” Stroke, vol. 41, no. 12, pp. 2817–2821, 2010. [DOI] [PubMed] [Google Scholar]
- [55].Yosinski J, Clune J, Nguyen A, Fuchs T, and Lipson H, “Understanding Neural Networks Through Deep Visualization,” Int. Conf. Mach. Learn. - Deep Learn. Work. 2015, p. 12, 2015. [Google Scholar]
- [56].Grech R, Galvin PL, Power S, O’Hare A, Looby S, Brennan P, and Thornton J, “Outcome prediction in acute stroke patients considered for endovascular treatment: a novel tool,” Interv Neuroradiol, vol. 20, no. 1591–0199 (Print), pp. 312–324, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mayerhoefer ME, Szomolanyi P, Jirak D, Materka A, and Trattnig S, “Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: an application-oriented study,” Med. Phys, vol. 36, no. 4, pp. 1236–1243, 2009. [DOI] [PubMed] [Google Scholar]
- [58].Hanley JA and McNeil BJ, “A method of comparing the areas under receiver operating characteristic curves derived from the same cases.,” Radiology, vol. 148, no. 3, pp. 839–43, 1983. [DOI] [PubMed] [Google Scholar]
- [59].Hanley JA and Hajian-Tilaki KO, “Sampling variability of non-parametric estimates of the areas under receiver operating characteristic curves: an update,” Acad. Radiol, vol. 4, no. 1, pp. 49–58, 1997. [DOI] [PubMed] [Google Scholar]
- [60].Ribeiro MT, Singh S, and Guestrin C, “Why should i trust you?: Explaining the predictions of any classifier,” in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016, pp. 1135–1144. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


