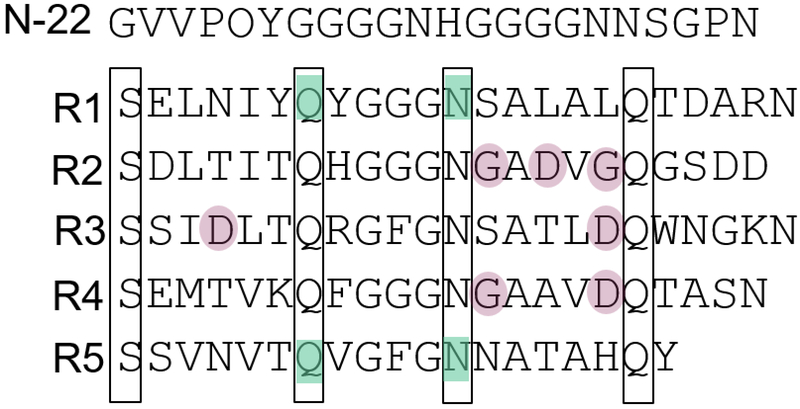Abstract
The discovery of intrinsic disorderness in proteins and peptide regions has given a new and useful insight into the working of biological systems. Due to enormous plasticity and heterogeneity, intrinsically disordered proteins or regions in proteins can perform myriad of functions. The flexibility in disordered proteins allows them to undergo conformation transition to form homopolymers of proteins called amyloids. Amyloids are highly structured protein aggregates associated with many neurodegenerative diseases. However, amyloids have gained much appreciation in recent years due to their functional roles. A functional amyloid fiber called curli is assembled on the bacterial cell surface as a part of the extracellular matrix during biofilm formation. The extracellular matrix that encases cells in a biofilm protects the cells and provides resistance against many environmental stresses. Several of the Csg (curli specific genes) proteins that are required for curli amyloid assembly are predicted to be intrinsically disordered. Therefore, curli amyloid formation is highly orchestrated so that these intrinsically disordered proteins do not inappropriately aggregate at the wrong time or place. The curli proteins are compartmentalized and there are chaperone-like proteins that prevent inappropriate aggregation and allow the controlled assembly of curli amyloids. Here we review the biogenesis of curli amyloids and the role that intrinsically disordered proteins play in the process.
Keywords: Intrinsically disordered proteins, phase transition, functional amyloids, curli, biofilms
1. The dynamics of unstructured proteins
Until recently, there was an assumption that protein function must be tightly linked to a definable globular structure [1]. Hence, the “sequence-structure-function paradigm” that posits the globular structure is determined by the amino acid sequence, which in turn dictates the function of the protein, is well-conceived and accepted [1]. However, the advent of new technologies and discoveries has introduced the idea that some proteins are functional in a ‘natively disordered’ state [2]. Natively disordered or intrinsically disordered proteins (IDPs) are classically defined as proteins that lack a stable definitive globular structure [3]. IDPs exist in a dynamic ensemble of conformations that enables them to functionally engage with multiple substrates [4]. Some proteins have discrete domains or sequences that are intrinsically disordered, called IDRs (intrinsically disordered regions) [5]. It is now well established that IDPs and IDRs contribute to various cellular biological processes [4].
Pontius and Berg were the first to link protein disorder and function when they identified a functional IDR in the C-terminus of hnRNP A1[6]. Pontius proposed that IDRs may have higher utility since they can switch the conformations rapidly and can bind to multiple partners [6]. Later, bioinformatic analyses found that ‘disorderness’ varies among proteins, but can be classified as the following: 1) ‘molecular switches’ that fold upon binding, 2) ‘morphers’ that vary in structure based on their partners, 3) ‘dormant disorder’ where disorder is triggered by interacting with other proteins or molecules, and 4) ‘fuzzies’ that remain flexible even after binding [3]. The hypothesis that particular amino acid sequences beget disorder was tested by developing predictors to predict fold based on amino acid sequences [7]. The results from these predictors suggested that peptide chains with less hydrophobic and more charged amino acids are likely to remain unfolded or disordered[8]. IDPs are typically rich in amino acids like M, K, R,S, Q, P, E and significantly depleted in W, Y, F, V, L, C [9]. Therefore, combinations of certain amino acids allow IDPs to remain in a dynamically unfolded state [4][10].
The astonishing dynamics possessed by IDPs gives them the ability to switch between structures in a very short time scale [9]. It is a challenge to structurally characterize IDPs using conventional biochemical and biophysical techniques like X-ray crystallography [11]. Web based prediction software like PONDR, DISOPRED, GlobPlot, and FoldIndex have been successful in identifying the propensity of intrinsic disorderness in a protein sequence [12]. Bioinformatic tools like molecular dynamic (MD) simulations (CAMPRI and Rosetta) have been crucial to understand the structural dynamics of IDPs [13]. Nuclear magnetic resonance (NMR) has emerged as a powerful probe that has been widely and successfully used to map flexibility, dynamics and binding mechanism in IDPs under both in vitro and cell based environments [14]. New and more sophisticated techniques like single molecule atomic force microscopy and single molecule FRET (fluorescence resonance energy transfer) have been recently implemented to understand the conformational polymorphism in IDPs [15].
2. Role of IDPs in phase transition
Owing to low complexity and high net charge IDPs and IDRs are most suitable to undergo liquid-liquid phase transition to confine certain biological molecules and reactions in compartments within nucleus or cytoplasm and therefore making proteinaceous membrane less organelles (PMLO) [16,17]. PMLOs are formed in response to environmental changes and are the result of regulated liquid-liquid phase transition [16,17] which is driven by the primary sequence of the protein components [18-20]. The current knowledge on protein component in PMLO is limited. However, recent studies indicate that the presence of IDPs or IDRs may be a prerequisite for formation of formation of dynamic, cell-size dependent PMLOs [16].The highly dynamic nature of PMLOs allows them to participate in a wide range of molecular interactions [21]. A recent review by Darling et al provides a comprehensive list of IDPs and IDRs involved in PMLO formation [21]. The review also highlighted presence of disordered proteins linked to PMLOs in different organisms suggesting IDPs/IDRs to be an evolutionary conserved factor [21]. IDPs are also involved in phase transition and formation of membrane-less organelles in plants [22]. Finally, IDPs that phase transition can drive the formation of structured homoaggregates or amyloids associated with neurodegenerative diseases [23-26].
3. IDPs and amyloid formation (disease related and functional)
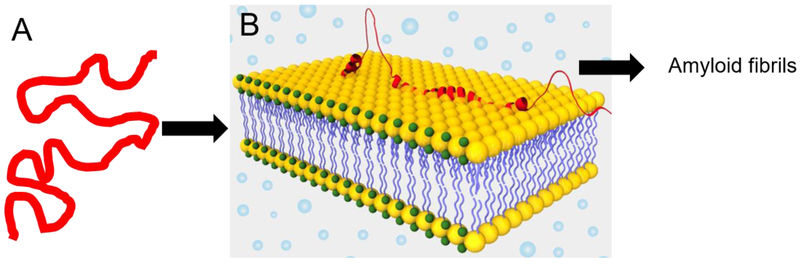
The conformational dynamics of IDPs may act as double edged sword where it allows the polypeptide chain to perform various functions but is also poised to collapse to form “molten globules” that lead to oligomer and amyloid formation [25]. Amyloids are highly ordered cross β-sheet structures of proteins that are commonly associated with neurodegenerative disorders like Alzheimer’s, Parkinson’s and Huntington’s disease [27]. Some natively folded, globular proteins become denatured prior to forming amyloid [28]. However, owing to intrinsic disorderedness, IDPs are prone to amyloid aggregation. α-Synuclein (α-syn) is one such IDP that has a high level of structural plasticity and undergoes conformation transitions upon binding to different substrates [29]. Amyloid formation by α-synuclein is linked with pathophysiology of Parkinson’s disease [30]. As illustrated in figure 1, disordered α-synuclein undergoes membrane-induced conformational switch to helical structure. The membrane bound helical α-synuclein has higher propensity to aggregate and form β-sheet rich amyloids [31].
Figure 1:
Conformational switch of α-synuclein (AS; an archetypal IDP) induced by lipid bilayer. A. Native unfolded structure of AS in solution. B. Predominantly helical structure in the presence of lipid bilayer which is prone to aggregate and form amyloid fibrils.
Although readily recognized as the product of protein misfolding and aggregation, some amyloids are beneficial for cells [32]. These so called ‘functional’ amyloids can be found in all walks of life from bacteria to mammals [33]. Functional amyloids made by bacteria are an important constituent of the extracellular matrix that protects cells from environmental stressors [34]. Cytoplasmic Polyadenylation Element Binding (CPEB) from sea snails, and its homolog in Drosophila, are functional amyloids that help with memory consolidation [35,36]. Humans and other mammals possess homologs of CPEB that are putatively involved in similar functions [37]. One of the fascinating ways of utilizing amyloid scaffold is to store peptide hormones in exocrine and endocrine glands. Upon receiving appropriate signals, the amyloid form of hormones gets disaggregate to release the peptides [38].
The subunit proteins that make up functional amyloid fibers are often intrinsically disordered, thus making the monomers prone to oligomerization and aggregation [39]. Therefore, the biogenesis of functional amyloids must be spatially and temporarily regulated such that potential cellular toxicity is mitigated [40]. Certain functional amyloids transition from monomer to amyloid polymer efficiently enough to limit the accumulation of oligomeric intermediates [41]. For instance, the human functional amyloid Pmel forms ordered amyloid aggregates at a much faster rate compared to disease-associated proteins like Aβ and α-synuclein [42]. Other functional amyloids have adapted ‘nucleator’ proteins that provide an amyloid-like scaffold which can be used as a folding template to accelerate amyloid formation [43]. Dedicated chaperone proteins can also prevent amyloid formation at the wrong time or the wrong place within the cell [44-47].
For the remainder of the review we will discuss the functional amyloid systems with a focus on the bacterial amyloid called curli. The curli amyloid assembly system provides many elegant examples of how cells can control disordered and highly aggregation prone proteins.
4. Curli proteins in Escherichia coli: the disordered form
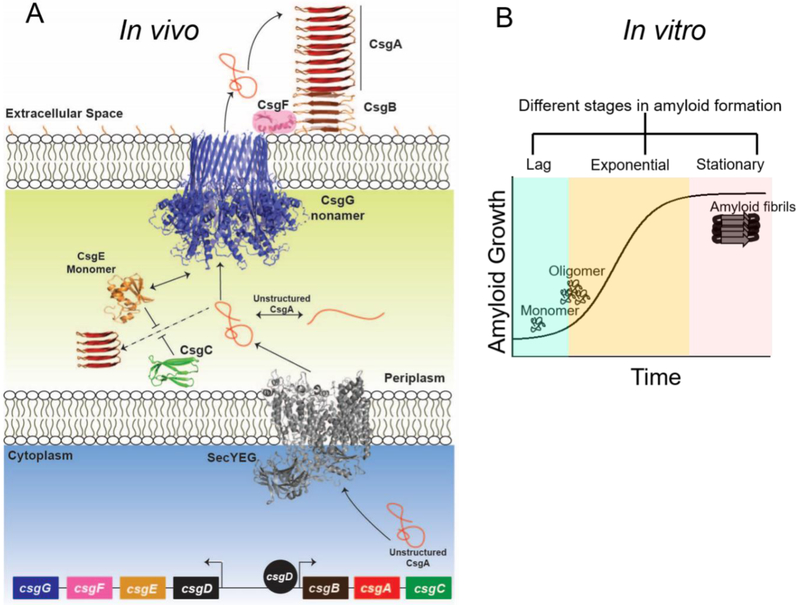
Bacteria are generally found in sessile communities called biofilms, where individual cells can be adhered to other cells or to a surface by biopolymers assembled on the cell surface [48]. Since the extracellular space will be exposed to environmental stresses, it is not surprising that the protein polymers in the extracellular matrix (ECM) are often amyloids [34]. The functional amyloid fiber (called curli) found in the extracellular matrix of E. coli are heteropolymers of two proteins, CsgA and CsgB [49].CsgA and CsgB are IDPs with flexible and aggregation prone regions [49]. In a test tube, purified CsgA and CsgB exist transiently in a random coil conformation, but then aggregate into ordered amyloid fibers [50].Inside the cell an efficient chaperone network helps to discourage CsgA and CsgB aggregation, so that CsgA and CsgB can be secreted out of the cell where they polymerize into curli amyloid fibers [44,46]. The csgA and csgB genes chaperoned and secreted to the cell surface with the help of proteins encoded on a divergently transcribed operon, csgDEFG [34]. Figure 2A illustrates how Csg proteins coordinate to assemble curli amyloid fibers on the bacterial cell surface. The next sections are dedicated to the biogenesis of curli on cell surface and the regulatory machinery that keeps a check on intracellular aggregation.
Figure 2:
The biogenesis model of curli in vivo and in vitro. A. Controlled translocation of disordered monomeric CsgA and CsgB on cell surface. CsgA self assembles into fibrils on nucleator CsgB amyloids. B. In vitro amyloid formation of CsgA follows a nucleation-dependent aggregation pattern.
4.1. The major curli subunit; CsgA:
The major curli subunit CsgA, is secreted to the cell surface as a soluble disordered polypeptide chain where it efficiently adopts a cross-β-strand polymer conformation that is the hallmark of all amyloids [43]. CsgA polypeptide contains an N-terminal 22 amino acid stretch that is required for translocation through the outer membrane secretion complex [51]. CsgA is capable of adopting the distinctive amyloid conformation under different in vitro conditions [52,53]. As shown in figure 2B, the transition of soluble disordered CsgA monomer to mature fibrils is a nucleation-dependent process and involves formation of transient intermediates [54]. Recent high resolution studies suggests that CsgA fiber formation is a one-step nucleation process wherein CsgA monomers fold and oligomerize to form a minimum fiber unit that contains all the characteristic features of mature fibers [55]. CsgA amyloid polymerization can also be accelerated or ‘seeded’ by the addition of sonicated preformed fibrils, which significantly decreases the lag phase [54]. Interestingly, E. coli CsgA exhibits seeding promiscuity and its amyloid formation can be accelerated by CsgA homologs from Citrobacter koseri and Salmonella typhimurium [56].
Due to presence of conserved amino acids, the CsgA amyloid core can be divided into five imperfect repeat regions (R1-R5) [39]. Each of the CsgA repeats contains a Ser-X5-Gln-X4-Asn-X5-Gln motif that appears conserved in all CsgA homologs from Enterobacteriaceae [54,57]. In-depth mutagenesis analysis revealed that the conserved Gln (glutamine) and Asn (asparagine) residues in the repeat regions at N-and C-terminus play an essential role in CsgB mediated curli formation in vivo and CsgA self-assembly in vitro [50]. More specifically, Gln residues at position 49 and 139 and Asn residues at 54 and 144 position were found to be indispensable for side chain interactions and curli assembly since mutating these residues renders no curli formation (Figure 3) [50]. Sequential deletion of repeat segments suggested that CsgA could make amyloids in the absence of R1-R4 (ΔR1-ΔR4) with comparable lag time, however deletion of R5 (ΔR5) increased the lag time substantially indicating the significance of R5 region in early oligomerization process during CsgA fiber assembly [58]. The specific arrangement of aspartic acid and glycine residues in the repeat regions of R2, R3 and R4 prevent spontaneous aggregation of Csg proteins inside the cell [59]. Owing the capacity for these aspartic acid residues to curtail CsgA amyloid formation, they are termed “gatekeeper residues” (Figure 3) [59].To gain insight into the functionality of individual repeat regions in curli assembly, peptides corresponding to each region were synthesized [54]. The synthetic peptides (R1 to R5) display different degrees of amyloid formation. The peptides from the R1, R3 and R5 regions were found to be highly amyloidogenic and polymerize with faster rates than peptides corresponding to R2 and R4 [54]. CsgA amyloids influence biological functions in the host through specific interactions between the R1/R5 repeat regions and human proteins such as fibronectin and tissue plasmogen activator [60,61].
Figure 3:
The details of repeat regions in CsgA amino acid sequence. The full length CsgA comprises of N-22 peptide sequence and five repeat units (R1-R5). The Gln residues at position 49 and 139 and Asn residues at position 54, 144 (all highlighted in cyan) are essential for CsgA amyloid assembly. The “gatekeeper residues” in regions R2, R3 and R4 (encircled in violet) prevent spontaneous aggregation. The conserved amino acids across repeat units are boxed together.
Inside the bacterial cell, CsgA is synthesized and secreted as an IDP [57]. The amyloid assembly of CsgA on the outer membrane is dependent on minor curli subunit CsgB which serves as a template or nucleator for the initiation of CsgA amyloidogenesis [43]. The detailed genetic, biochemical and microscopic analysis revealed that N and C- terminus of CsgA is required for CsgB templated assembly of CsgA [62].
4.2. The minor curli subunit and nucleator protein, CsgB:
CsgB, the minor curli subunit has 30% identity with CsgA at the amino acid level[63]. Under in vivo conditions, full-length CsgB is membrane associated and self-polymerize to present a template for CsgA polymerization [43]. Cells with a csgB deletion secrete CsgA, although CsgA remains unpolymerized on the cell surface[63]. CsgB is also predicted to be an IDP [49]. Purified CsgB seeds CsgA polymerization under in vitro conditions [43,64]. A truncated version of CsgB (CsgBtrunc) containing four repeats units (R1-R4) was used to understand the aggregation mechanism of CsgB under in vitro conditions. In vitro, CsgBtrunc makes amyloids similar to CsgB, although it does not complement a csgB mutant [43] in cells because CsgBtrunc does not associate with the outer membrane and is not positioned correctly to facilitate CsgA nucleation [43].
Like CsgA, CsgB also contains five imperfect repeats with a slight variation in the position of consensus sequence as compared to CsgA[43]. Sequence alignments reveal high degree of conservation in first four repeats (R1-R5) however, the fifth repeat (R5) is less conserved as compared to CsgA [43]. Biochemical and biophysical studies on different fragments of CsgB indicate that the repeat regions have redundancy and the protein still self-assemble in vitro after deletion of individual repeats [62]. An attractive model of CsgB-mediated nucleation of CsgA, suggests that CsgB rapidly forms an amyloid-like template on the cell surface that can template the folding of CsgA [64].
Curli amyloid formation can be specified to the cell surface because that is the place where the CsgA protein can interact with the CsgB nucleator. However, both CsgA and CsgB are IDPs and have some propensity to aggregate within the cell [51,65]. It is not surprising that a sophisticated network of chaperones and protein-degradation machinery is in place to keep Csg proteins from premature aggregation within the cell [34,43,66]. In the following sections, we will discuss various chaperone systems that keep the disordered Csg proteins on the right track.
5. Chaperones in E. coli: keeping the disordered Csg proteins at right place and time
5.1. Traditional molecular chaperones; Dna K, Hsp33 and Spy proteins:
Chaperones and chaperone-like proteins in cytoplasm and periplasm can prevent the inappropriate and premature aggregation of curli proteins. General chaperones such as DnaK, Spy and Hsp33 can prevent CsgA amyloid formation in vitro [66]. DnaK can transiently inhibit the aggregation of CsgA, even in the absence of DnaJ and ATP hydrolysis [66]. Similarly, the Hsp33 “holdase” can prevent CsgA amyloid formation [44]. Hsp33 is activated by oxidative stress and the oxidized form of Hsp33 is a more potent amyloid inhibitor than the reduced form [67]. The periplasmic holdase chaperone, Spy, stabilizes the monomeric soluble form of CsgA[66,68]. However, the role that these general chaperones play in keeping Csg proteins from aggregating in vivo remains largely unexplored.
5.2. A potent anti-amyloid periplasmic chaperone-like protein, CsgC:
CsgC is a periplasmic protein whose role in curli assembly is to inhibit CsgA and CsgB aggregation inside the cell [46].By preventing CsgA and CsgB from aggregating inside the cell, CsgC mitigates potential cellular toxicity [46]. CsgC is a β-sheet rich protein with a high degree of stability[69]. Strikingly, CsgC inhibits CsgA amyloid assembly at sub-stoichiometric ratios. Interestingly, CsgC can prevent amyloid formation by other bacterial and human proteins, including α-synuclein[46]. While CsgA and CsgB has 30% sequence identity, CsgA and α-synuclein share little significant sequence identity [46]. The only partially shared stretch of amino acids appears in each repeating unit of CsgA(R1-R5), and only one time in α-synuclein [46] Deletions or mutations in the shared stretch from α-synuclein renders CsgC non-responsive to α-synuclein aggregation [46]. CsgC exhibits client specificity as it is not able to prevent Aβ aggregation [46]. The mechanistic insights into the CsgC inhibitory activity highlights the role of electrostatic interactions and presence of specific amino acids for the potency of CsgC as an amyloid inhibitor [70]. Recently, a super-resolution imaging technique found that CsgC could ‘cap’ the growing CsgA fiber tip [71]. The detailed mechanism of inhibition and specificity of CsgC still remains to be understood. However, the discovery has opened avenues to explore the presence of CsgC like proteins in other kingdoms that can serve as amyloid inhibitors [61,70,72]. In sum, CsgC helps maintain a dynamic population of soluble and disordered CsgA and CsgB in the periplasm and is an elegant example of how bacteria have evolved a well-developed machinery to mitigate premature amyloid formation [73].
6. Accessory proteins: Disordered to ordered transition from inside to outside of the cell
In E. coli, controlled translocation of amyloidogenic curli subunits to the cell surface is achieved via several of the Csg (curli specific gene) proteins.
6.1. Membrane tethered, CsgF:
CsgF is a surface exposed, membrane-associated oligomeric protein [74,75]. A recent NMR study found that CsgF lacks a stable tertiary structure and contains exposed hydrophobic surfaces [76]. However, CsgF attains a stable conformation after interaction with CsgG and CsgB [76]. Cells deficient in csgF are less capable of polymerizing CsgA into amyloid [75]. Therefore, CsgF is itself an IDP that must function in concert with other Csg proteins to ensure the ordered assembly of intrinsically disordered CsgA and CsgB into curli amyloids [75]. The primary sequence of CsgF contains a high percentage of glutamine and asparagine residues, similar to CsgA and CsgB, which may act as CsgG specific export signal [51]. The C-terminus of CsgB interacts with dimeric CsgF and this interaction apparently promotes CsgB to adopt a nucleation competent conformation [76]. It is also possible that CsgF works directly on CsgA to promote amyloid formation, although this has not been directly demonstrated in cells.
6.2. The secretion pore, CsgG and CsgE:
The outer-membrane secretion channel is composed of a nonamer of the CsgG protein, which is ‘capped’ on the periplasmic side by a multimer of CsgE [51,77]. The nine subunits of CsgG form an oligomeric transport complex that gives rise to a 36-stranded β-barrel which traverses the bilayer and is connected to a cage-like vestibule in the periplasm [77]. The channel formed by the CsgG complex is 2nm wide, and is suited to export unfolded proteins [77]. The three known substrates of the CsgG secretion complex are CsgA, CsgB and CsgF, each of which is putatively disordered during secretion [51,77]. The CsgG mediated secretion of CsgA is dependent on the N-terminal 22 amino acids of soluble CsgA [51]. These residues are required for secretion and assembly of curli fibers but do not become the integral part of the fibers [51]. Structural and functional analyses of CsgG imply that it is a general secretion channel that transports protein on a diffusion based, entropy-driven transport mechanism [77]. The overexpression of CsgG results in outer-membrane permeability to antibiotics such as erythromycin [51].
CsgE acts as efficiency factor for CsgG by capping the periplasmic side of the vestibule and allowing translocation of Csg proteins to the cell surface [65]. CsgE also prevents non-specific translocation of small proteins and large molecules through CsgG pore, thus acting as a regulatory gateway to the otherwise permissible CsgG pore [65,74,78]. CsgE also inhibits CsgA polymerization in vitro suggesting that it may help keeping CsgA in soluble form in the periplasm [65,79]. Hydrogen-deuterium amide exchange revealed that CsgE exists in a dynamic ensemble of monomer and oligomers [80]. A recent NMR structure of a mutant of CsgE (W48A/F79A) highlights the detailed structural properties and localization with CsgG [74]. The NMR structure suggest that the N and C terminal of CsgE are flexible and may help in the interactions with CsgG [78]. According to the ‘head-center’ model, CsgE-CsgG interactions are driven by electrostatic forces where the flexible and negatively charged C-terminal of CsgE interacts with periplasmic domain of CsgG which is rich in positive residues [74]. The positively charged N-terminal of CsgE electrostatically interacts with soluble disordered CsgA, either as monomer or oligomeric form [74,78]. Thus, CsgE serves as a chaperone-like protein and a cap that allows an effective unidirectional transport of CsgA from periplasm to outer membrane [78].
6.3. The master regulator; CsgD:
CsgD is a key curli transcriptional regulator protein that belongs to FixJ/LuxR/UhpA transcription factor family [81]. It is the key modulator that regulates the csgBAC and csgDEFG operons required for the synthesis, secretion and assembly of curli components [82]. Numerous transcription factors and other parameters like amount of oxygen, temperature, osmolarity, starvation, high cell density plays critical role in regulating CsgD expression and thus indirectly influence csgBAC expression [83]. Transcription of CsgD is regulated by cAMP-CRP (cyclic AMP-catabolite repressor protein) complex and small RNAs [84,85]. The exact stimulus required for CsgD activation is not yet known but it is believed that the activated CsgD is phosphorylated at N-terminus by an unknown mechanism. The activation of CsgD governs the expression of curli and accessory proteins [84].
7. Curli and biofilm formation
Curli along with cellulose and extracellular DNA makes an integral part of extracellular matrix of biofilm in E. coli [34]. The extracellular matrix is a highly organized three-dimensional structure that protects bacteria from environmental stress and aids in adherence to biotic and abiotic surfaces [48,86]. Presence of biofilm renders bacteria resistant to antibiotics and host defense [87]. Furthermore, biofilm formation within the host is implicated in serious and persistent infectious diseases, including cystic fibrosis, chronic otitis media and urinary tract infection (UTI) [87]. Curli are also capable of evading host immune system and cause inflammation [88-90]. The specific and complicated interactions of curli with host environment and immune system explains the vital role of functional amyloids in host-bacterial interactions [88].
Although beneficial to bacteria, biofilms pose a great threat to humans since it is almost impossible to completely eradicate mature biofilms from biotic and abiotic surfaces. Preventing curli assembly is one of the most attractive strategies to prevent biofilm formation and subsequent damage [79]. Since curli shares similar biochemical and biophysical properties with disease-related amyloids, it is also a well suited model to design and develop general amyloid inhibitors [79,91,92].
8. Conclusion
The discovery of functional amyloids has greatly expanded our view of amyloid biology. Clearly, amyloid formation is not always deleterious from the cell’s perspective. However, there is still much to learn about amyloid formation and toxicity associated with amyloids. Many IDPs undergo liquid/solid phase transition to form hydrogels which are “solid-like amyloids” to prevent toxic effects of amyloids and imparts functional role. The most well studied functional amyloid, curli in E. coli has provided insights into how cells make amyloids efficiently at the right time and place, thus eliminates the changes of intracellular aggregation and cytotoxicity. Studies on curli not only enable us to understand the process of amyloidogenesis, but also provide a fascinating tool to discover new ways to treat amyloid-related diseases.
Highlights.
IDPs play pivotal role in biological functions.
IDPs are prone to collapse and form structured aggregates called amyloids.
Curli is a bacterial functional amyloid.
Curli is a major constituent of extracellular matrix in bacterial biofilms.
Curli biogenesis is spatio-temporally regulated to mitigate intracellular toxicity.
Acknowledgements
We thank Chapman lab members for their helpful discussions and contributory work. We are thankful to M. Goenka and S. Bhoite for their help in figure one and two, respectively. This work was supported by the National Institutes of Health R01GM118651 and R21AI137535 to MRC.
Biography

Neha Jain studied microbiology at Maharani’s college and received Bachelor’s and Master’s degree from the University of Rajasthan, Jaipur. Later she pursued PhD in Biological Sciences from the Indian Institute of Science Education and Research (IISER Mohali) with Prof. S. Mukhopadhyay. After finishing her PhD in 2013, she joined Prof. M. R. Chapman’s lab at the University of Michigan (Ann Arbor campus) for postdoctoral training. In 2018, she returned back to India. After a short stint as Assistant Professor at Ahmedabad University, she has joined the faculty of Bioscience and Bioengineering at the Indian Institute of Technology Jodhpur. Her lab is interested in understanding the interactions between human and bacterial amyloids and their role in pathophysiology of neurodegenerative diseases.

Matthew R. Chapman received his Bachelor’s degree in microbiology from the University of Nebraska at Omaha. He then moved to the Indiana University to pursue PhD with Prof. C. Kao. He joined Prof. S. J. Hultgren’s lab for postdoctoral studies where he discovered that the major proteinaceous component of the biofilm extracellular matrix belonged to a class of structurally conserved ordered aggregates called amyloid. In 2003, he started his lab at the University of Michigan, Ann Arbor. His lab has continued to work on the biogenesis and regulation of functional amyloids in bacteria.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Romero P, Obradovic Z, Kissinger C, Villafranca JE, Dunker AK, Identifying disordered regions in proteins from amino acid sequence, Proc. Int. Conf. Neural Networks, IEEE, pp. 90–95. doi: 10.1109/ICNN.1997.611643. [DOI] [Google Scholar]
- [2].Dunker AK, Brown CJ, Lawson JD, Iakoucheva A. Lilia M., Obradović§ Z, Intrinsic Disorder and Protein Function, (2002). doi: 10.1021/BI012159. [DOI] [PubMed] [Google Scholar]
- [3].Uversky VN, Dancing Protein Clouds: The strange biology and chaotic physics of intrinsically disordered proteins, J. Biol. Chem 291 (2016) 6681–6688.doi: 10.1074/jbc.R115.685859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dyson HJ, Wright PE, Intrinsically unstructured proteins and their functions, Nat. Rev. Mol. Cell Biol 6 (2005) 197–208. 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- [5].Tompa P, Structure and function of intrinsically disordered proteins, Chapman & Hall/CRC Press, 2010. [Google Scholar]
- [6].Pontius BW, Berg P, Rapid renaturation of complementary DNA strands mediated by cationic detergents: a role for high-probability binding domains in enhancing the kinetics of molecular assembly processes, Proc. Natl. Acad. Sci. U. S. A 88 (1991) 8237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dosztányi Z, Tompa P, Prediction of protein disorder, Methods Mol. Biol, 2008: pp. 103–115. doi: 10.1007/978-1-60327-058-8_6. [DOI] [PubMed] [Google Scholar]
- [8].Garner, Cannon, Romero, Obradovic, Dunker, Predicting disordered regions from amino acid sequence: Common themes despite differing structural characterization, Genome Inform. Ser. Workshop Genome Inform 9 (1998) 201–213. [PubMed] [Google Scholar]
- [9].Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK, Intrinsicdisorder and functional proteomics, Biophys. J 92 (2007) 1439–1456.doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Uversky VN, Gillespie JR, Fink AL, Why are natively unfolded proteins unstructured under physiologic conditions?, Proteins Struct. Funct. Genet 41 (2000) 415–427.doi:. [DOI] [PubMed] [Google Scholar]
- [11].Paz A, Zeev-Ben-Mordehai T, Lundqvist M, Sherman E, Mylonas E, Weiner L, Haran G, Svergun DI, Mulder FAA, Sussman JL, Silman I, Biophysical characterization of the unstructured cytoplasmic domain of the human neuronal adhesion protein neuroligin 3, Biophys. J 95 (2008) 1928–44. doi: 10.1529/biophysj.107.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL, FoldIndex(C): a simple tool to predict whether a given protein sequence is intrinsically unfolded, Bioinformatics. 21 (2005) 3435–3438.doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- [13].Chong S-H, Chatterjee P, Ham S, Computer simulations of intrinsically disordered proteins, Annu. Rev. Phys. Chem 68 (2017) 117–134. doi: 10.1146/annurev-physchem-052516-050843. [DOI] [PubMed] [Google Scholar]
- [14].Brutscher B, Felli IC, Gil-Caballero S, Hošek T, Kümmerle R, Piai A, Pierattelli R, Sólyom Z, NMR Methods for the study of instrinsically disordered proteins structure, dynamics, and interactions: General overview and practical guidelines, Adv. Exp. Med. Biol, 2015: pp. 49–122. doi: 10.1007/978-3-319-20164-1_3. [DOI] [PubMed] [Google Scholar]
- [15].Gomes G-N, Gradinaru CC, Insights into the conformations and dynamics of intrinsically disordered proteins using single-molecule fluorescence, Biochim. Biophys. Acta - Proteins Proteomics 1865 (2017) 1696–1706. doi: 10.1016/j.bbapap.2017.06.008. [DOI] [PubMed] [Google Scholar]
- [16].Uversky VN, Protein intrinsic disorder-based liquid–liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles, Adv. Colloid Interface Sci 239 (2017) 97–114. doi: 10.1016/J.CIS.2016.05.012. [DOI] [PubMed] [Google Scholar]
- [17].Alberti S, Gladfelter A, Mittag T, Leading edge primer considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates, Cell. (2019).doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McCarty J, Delaney KT, Danielsen SPO, Fredrickson GH, Shea J-E, Complete phase diagram for liquid–liquid phase separation of intrinsically disordered proteins, J. Phys. Chem. Lett 10 (2019) 1644–1652. doi: 10.1021/acs.jpclett.9b00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brangwynne CP, Phase transitions and size scaling of membrane-less organelles, J. Cell Biol 203 (2013) 875–881. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ, Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles, Mol. Cell 57 (2015) 936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Darling AL, Liu Y, Oldfield CJ, Uversky VN, Intrinsically disordered proteome of human membrane-less organelles, Proteomics. 18 (2018) 1700193.doi: 10.1002/pmic.201700193. [DOI] [PubMed] [Google Scholar]
- [22].Cuevas-Velazquez CL, Dinneny JR, Organization out of disorder: liquid-liquid phase separation in plants, Curr. Opin. Plant Biol 45 (2018) 68–74.doi: 10.1016/J.PBI.2018.05.005. [DOI] [PubMed] [Google Scholar]
- [23].Ambadipudi S, Biemat J, Riedel D, Mandelkow E, Zweckstetter M, Liquid–liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau, Nat. Commun 8 (2017) 275. doi: 10.1038/s41467-017-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bergeron-Sandoval L-P, Safaee N, Michnick SW, Mechanisms and consequences of macromolecular phase separation, Cell (2016). doi: 10.1016/j.cell.2016.05.026. [DOI] [PubMed] [Google Scholar]
- [25].Levine ZA, Larini L, LaPointe NE, Feinstein SC, Shea J-E, Regulation and aggregation of intrinsically disordered peptides., Proc. Natl. Acad. Sci. U. S. A 112 (2015)2758–63. doi: 10.1073/pnas.1418155112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kostylev MA, Tuttle MD, Lee S, Klein LE, Takahashi H, Cox TO, Gunther EC, Zilm KW, Strittmatter SM, Liquid and hydrogel phases of PrPC linked to conformation shifts and triggered by Alzheimer’s amyloid-β oligomers, Mol. Cell 72 (2018) 426–443.e12. doi: 10.1016/j.molcel.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Koo EH, Lansbury PT, Kelly JW, Amyloid diseases: Abnormal protein aggregation in neurodegeneration, Proc. Natl. Acad. Sci. U. S. A 96 (1999) 9989–90. doi: 10.1073/PNAS.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chiti F, Dobson CM, Protein misfolding, functional amyloid, and human disease, Annu. Rev. Biochem 75 (2006) 333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- [29].Fusco G, De Simone A, Arosio P, Vendruscolo M, Veglia G, Dobson CM, Structural ensembles of membrane-bound α-synuclein reveal the molecular determinants of synaptic vesicle Affinity, Sci. Rep 6 (2016) 27125. doi: 10.1038/srep27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Luk KC, Kehm V, Carroll J, Zhang B, O’Brien P, Trojanowski JQ, Lee VMY, Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice, Science (80) 338 (2012) 949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee H-J, Choi C, Lee S-J, Membrane-bound α-Synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form, J. Biol. Chem 277 (2002) 671–678. doi: 10.1074/jbc.M107045200. [DOI] [PubMed] [Google Scholar]
- [32].Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR, Diversity, biogenesis and function of microbial amyloids, Trends Microbiol. 20 (2012) 66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pham CLL, Kwan AH, Sunde M, Functional amyloid: widespread in nature, diverse in purpose, Essays Biochem. 56 (2014) 207–219. doi: 10.1042/bse0560207. [DOI] [PubMed] [Google Scholar]
- [34].Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ, Role of Escherichia coli curli operons in directing amyloid fiber formation, Science. 295 (2002) 851–5. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Raveendra BL, Siemer AB, V Puthanveettil S, Hendrickson WA, Kandel ER, McDermott AE, Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB, Nat. Struct. Mol. Biol 20 (2013) 495–501. doi: 10.1038/nsmb.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan M. “Repon”, Li L, Choi EM-L, Kannan K, Guo F, Unruh J, Slaughter B, Si K, Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory, Cell. 148 (2012) 515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- [37].Fioriti L, Myers C, Huang Y-Y, Li X, Stephan JS, Trifilieff P, Colnaghi L, Kosmidis S, Drisaldi B, Pavlopoulos E, Kandel ER, The persistence of hippocampal-based memory requires protein synthesis mediated by the Prion-like protein CPEB3, Neuron. 86 (2015) 1433–1448. doi: 10.1016/j.neuron.2015.05.021. [DOI] [PubMed] [Google Scholar]
- [38].Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KPR, Simon R, Schubert D, Eisenberg D, Rivier J, Sawchenko P, Vale W, Riek R, Functional amyloids as natural storage of peptide hormones in pituitary secretory granules, Science (80). 325 (2009) 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Evans ML, Chapman MR, Curli biogenesis: Order out of disorder, Biochim. Biophys. Acta - Mol. Cell Res 1843 (2014) 1551–1558. doi: 10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Deshmukh M, Evans ML, Chapman MR, Amyloid by design: Intrinsic regulation of microbial amyloid assembly, J. Mol. Biol 430 (2018) 3631–3641. doi: 10.1016/j.jmb.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Carballo-Pacheco M, Ismail AE, Strodel B, Oligomer Formation of Toxic and Functional amyloid peptides studied with atomistic simulations, J. Phys. Chem. B 119 (2015) 9696–9705. doi: 10.1021/acs.jpcb.5b04822. [DOI] [PubMed] [Google Scholar]
- [42].McGlinchey RP, Lee JC, Why study functional amyloids? Lessons from the repeat domain of Pmel17, J. Mol. Biol 430 (2018) 3696–3706. doi: 10.1016/j.jmb.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hammer ND, Schmidt JC, Chapman MR, The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization, Proc. Natl. Acad. Sci 104 (2007) 12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Evans ML, Schmidt JC, Ilbert M, Doyle SM, Quan S, Bardwell JCA, Jakob U, Wickner S, Chapman MR, cob chaperones DnaK E, Hsp33 and Spy inhibit bacterial functional amyloid assembly, Prion. 5 (2011) 323–334. doi: 10.4161/pri.18555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Landreh M, Rising A, Presto J, Jörnvall H, Johansson J, Specific chaperones and regulatory domains in control of amyloid formation., J. Biol. Chem 290 (2015) 26430–6. doi: 10.1074/jbc.R115.653097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Evans ML, Chorell E, Taylor JD, Åden J, Götheson A, Li F, Koch M, Sefer L, Matthews SJ, Wittung-Stafshede P, Almqvist F, Chapman MR, The bacterial curli system possesses a potent and selective inhibitor of amyloid formation., Mol. Cell 57 (2015) 445–55. doi: 10.1016/j.molcel.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mannini B, Chiti F, Chaperones as suppressors of protein misfolded oligomer toxicity, Front. Mol. Neurosci 10 (2017) 98. doi: 10.3389/fnmol.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, Heuser J, Chapman MR, Hadjifrangiskou M, Henderson JP, Hultgren SJ, Escherichia coli biofilms have an organized and complex extracellular matrix structure, MBio. 4 (2013) e00645–13. doi: 10.1128/mBio.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].DeBenedictis EP, Ma D, Keten S, Structural predictions for curli amyloid fibril subunits CsgA and CsgB, RSC Adv. 7 (2017) 48102–48112. doi: 10.1039/C7RA08030A. [DOI] [Google Scholar]
- [50].Wang X, Chapman MR, Sequence Determinants of bacterial amyloid formation, J. Mol. Biol 380 (2008) 570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Robinson LS, Ashman EM, Hultgren SJ, Chapman MR, Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein, Mol. Microbiol 59 (2006) 870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Collinson S, Parker JM, Hodges R, Kay W, Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae, J. Mol. Biol 290 (1999) 741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- [53].Dueholm MS, Nielsen SB, Hein KL, Nissen P, Chapman M, Christiansen G, Nielsen PH, Otzen DE, Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation, Biochemistry. 50 (2011) 8281–90. doi: 10.1021/bi200967c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang X, Smith DR, Jones JW, Chapman MR, In Vitropolymerization of a functional Escherichia coli amyloid protein, J. Biol. Chem 282 (2006) 3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sleutel M, Van den Broeck I, Van Gerven N, Feuillie C, Jonckheere W, Valotteau C, Dufrêne YF, Remaut H, Nucleation and growth of a bacterial functional amyloid at single-fiber resolution, Nat. Chem. Biol 13 (2017) 902–908. doi: 10.1038/nchembio.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhou Y, Smith D, Leong BJ, Brännström K, Almqvist F, Chapman MR, Promiscuous Cross-seeding between bacterial amyloids promotes interspecies biofilms, J. Biol. Chem 287 (2012) 35092–35103. doi: 10.1074/jbc.M112.383737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wang X, Chapman MR, Curli provide the template for understanding controlled amyloid propagation, Prion. 2 (2) 57–60. doi: 10.4161/pri.2.2.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang X, Hammer ND, Chapman MR, The molecular basis of functional bacterial amyloid polymerization and nucleation, J. Biol. Chem 283 (2008) 21530–21539. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wang X, Zhou Y, Ren J-J, Hammer ND, Chapman MR, Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis, Proc. Natl. Acad. Sci 107 (2010) 163–168. doi: 10.1073/pnas.0908714107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhou Y, Smith DR, Hufnagel DA, Chapman MR, Experimental manipulation of the microbial functional amyloid called curli, Methods Mol. Biol 2013: pp. 53–75. doi: 10.1007/978-1-62703-245-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Swasthi HM, Bhasne K, Mahapatra S, Mukhopadhyay S, Human fibrinogen inhibits amyloid assembly of biofilm-forming CsgA, Biochemistry. 57 (2018) 6270–6273. doi: 10.1021/acs.biochem.8b00841. [DOI] [PubMed] [Google Scholar]
- [62].Hammer ND, McGuffie BA, Zhou Y, Badtke MP, Reinke AA, Brännström K, Gestwicki JE, Olofsson A, Almqvist F, Chapman MR, The C-Terminal repeating units of CsgB direct bacterial functional amyloid nucleation, J. Mol. Biol 422 (2012) 376–389. doi: 10.1016/j.jmb.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hammar M, Bian Z, Normark S, Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli, Proc. Natl. Acad. Sci. U. S. A 93 (1996) 6562–6. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shu Q, Crick SL, Pinkner JS, Ford B, Hultgren SJ, Frieden C, The A E. coli CsgB nucleator of curli assembles to β-sheet oligomers that alter the CsgA fibrillization mechanism, Proc. Natl. Acad. Sci 109 (2012) 6502–6507. doi: 10.1073/pnas.1204161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR, CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation, Mol. Microbiol 81 (2011)486–499. doi: 10.1111/j.1365-2958.2011.07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Evans ML, Schmidt JC, Ilbert M, Doyle SM, Quan S, Bardwell JCA, Jakob U, Wickner S, Chapman MR, coli chaperones DnaK E, Hsp33 and Spy inhibit bacterial functional amyloid assembly, Prion. 5 (2011) 323–34. doi: 10.4161/pri.18555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jakob U, Muse W, Eser M, Bardwell JC, Chaperone activity with a redox switch, Cell. 96 (1999) 341–52. doi: 10.1016/S0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- [68].Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, Jakob U, Xu Z, Cygler M, Bardwell JCA, Genetic selection designed to stabilize proteins uncovers a chaperone called Spy, Nat. Struct. Mol. Biol 18 (2011) 262–269. doi: 10.1038/nsmb.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Taylor JD, Zhou Y, Salgado PS, Patwardhan A, McGuffie M, Pape T, Grabe G, Ashman E, Constable SC, Simpson PJ, Lee W, Cota E, Chapman MR, Matthews SJ, Atomic resolution insights into curli fiber biogenesis, Structure 19 (2011) 1307–16. doi: 10.1016/j.str.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Taylor JD, Hawthorne WJ, Lo J, Dear A, Jain N, Meisl G, Andreasen M, Fletcher C, Koch M, Darvill N, Scull N, Escalera-Maurer A, Sefer L, Wenman R, Lambert S, Jean J, Xu Y, Turner B, Kazarian SG, Chapman MR, Bubeck D, De Simone A, Knowles TPJ, Matthews SJ, Electrostatically-guided inhibition of curli amyloid nucleation by the CsgC-like family of chaperones, Sci. Rep, (2016). doi: 10.1038/srep24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Sleutel M, Van den Broeck I, Van Gerven N, Feuilbe C, Jonckheere W, Valotteau C, Dufrêne YF, Remaut H, Nucleation and growth of a bacterial functional amyloid at single-fiber resolution., Nat. Chem. Biol 13 (2017) 902–908. doi: 10.1038/nchembio.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jain N, Ådén J, Nagamatsu K, Evans ML, Li X, McMichael B, Ivanova MI, Almqvist F, Buxbaum JN, Chapman MR, Inhibition of curli assembly and Escherichia coli biofilm formation by the human systemic amyloid precursor transthyretin., Proc. Natl. Acad. Sci. U. S. A 114 (2017) 12184–12189. doi: 10.1073/pnas.1708805114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Otzen DE, Assembling good amyloid: Some structures at last, Structure. 19 (2011) 1207–1209. doi: 10.1016/J.STR.2011.08.005. [DOI] [PubMed] [Google Scholar]
- [74].Klein RD, Shu Q, Cusumano ZT, Nagamatsu K, Gualberto NC, Lynch AJL, Wu C, Wang W, Jain N, Pinkner JS, Amarasinghe GK, Hultgren SJ, Frieden C, Chapman MR, Structure-function analysis of the curli accessory protein CsgE defines surfaces essential for coordinating amyloid fiber formation, MBio. 9 (2018). doi: 10.1128/mBio.01349-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nenninger AA, Robinson LS, Hultgren SJ, Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF, Proc. Natl. Acad. Sci 106 (2009) 900–905. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Schubeis T, Spehr J, Viereck J, Köpping L, Nagaraj M, Ahmed M, Ritter C, Structural and functional characterization of the Curli adaptor protein CsgF, FEBS Lett. 592 (2018) 1020–1029. doi: 10.1002/1873-3468.13002. [DOI] [PubMed] [Google Scholar]
- [77].Goyal P, Krasteva PV, Van Gerven N, Gubellini F, Van den Broeck I, Troupiotis-Tsaïlaki A, Jonckheere W, Péhau-Arnaudet G, Pinkner JS, Chapman MR, Hultgren SJ, Howorka S, Fronzes R, Remaut H, Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG, Nature. 516 (2014) 250–253. doi: 10.1038/nature13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shu Q, Krezel AM, Cusumano ZT, Pinkner JS, Klein R, Hultgren SJ, Frieden C, Solution NMR structure of CsgE: Structural insights into a chaperone and regulator protein important for functional amyloid formation, Proc. Natl. Acad. Sci 113 (2016) 7130–7135. doi: 10.1073/pnas.1607222113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Andersson EK, Bengtsson C, Evans ML, Chorell E, Sellstedt M, Lindgren AEG, Hufnagel DA, Bhattacharya M, Tessier PM, Wittung-Stafshede P, Almqvist F, Chapman MR, Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones, Chem. Biol 20 (2013) 1245–1254. doi: 10.1016/J.CHEMBIOL.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wang H, Shu Q, Rempel DL, Frieden C, Gross ML, Continuous and Pulsed hydrogen-Deuterium exchange and Mass Spectrometry characterize CsgE oligomerization, Biochemistry. 54 (2015) 6475–6481. doi: 10.1021/acs.biochem.5b00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Amqvist A, Olsén A, Pfeifer J, Russell DG, Nonnark S, The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101, Mol. Microbiol 6 (2006) 2443–2452. doi: 10.1111/j.1365-2958.1992.tb01420.x. [DOI] [PubMed] [Google Scholar]
- [82].Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW, Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae, J. Bacteriol 178 (1996) 662–7. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gerstel U, Romling U, Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium, Environ. Microbiol 3 (2001) 638–48. doi : 10.1046/j.1462-2920.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- [84].Hufnagel DA, Evans ML, Greene SE, Pinkner JS, Hultgren SJ, Chapman MR, The catabolite repressor protein-Cyclic AMP complex regulates csgD and biofilm formation in uropathogenic Escherichia coli, J. Bacteriol 198 (2016) 3329–3334. doi: 10.1128/JB.00652-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mika F, Hengge R, Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli, RNA Biol. 11 (2014)494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hufnagel DA, Depas WH, Chapman MR, The biology of the Escherichia coli extracellular matrix, Microbiol. Spectr 3 (2015). doi: 10.1128/microbiolspec.MB-0014-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hufnagel DA, Tükel Ç, Chapman MR, Disease to dirt: The biology of microbial amyloids, PLoS Pathog. 9 (2013) e1003740. doi: 10.1371/journal.ppat.1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Tursi SA, Tükel Ç, Curli-Containing Enteric Biofilms Inside and Out: Matrix Composition, Immune recognition, and disease implications, Microbiol. Mol. Biol. Rev 82 (2018). doi: 10.1128/MMBR.00028-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Spaulding CN, Dodson KW, Chapman MR, Hultgren SJ, Fueling the fire with fibers: Bacterial amyloids promote inflammatory disorders., Cell Host Microbe. 18 (2015) 1–2. doi: 10.1016/j.chom.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kai-Larsen Y, Lüthje P, Chromek M, Peters V, Wang X, Holm Å, Kádas L, Hedlund K-O, Johansson J, Chapman MR, Jacobson SH, Römling U, Agerberth B, Brauner A, Uropathogenic Escherichia coli modulates Immune responses and Its curli fimbriae interact with the antimicrobial peptide LL-37, PLoS Pathog. 6 (2010) e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Andersson EK, Chapman M, Small Molecule Disruption of B. subtilis biofilms by targeting the amyloid matrix, Chem. Biol 20 (2013) 5–7. doi: 10.1016/j.chembiol.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Horvath I, Weise CF, Andersson EK, Chorell E, Sellstedt M, Bengtsson C, Olofsson A, Hultgren SJ, Chapman M, Wolf-Watz M, Almqvist F, Wittung-Stafshede P, Mechanisms of protein oligomerization: Inhibitor of functional amyloids templates α-Synuclein fibrillation, J. Am. Chem. Soc 134 (2012) 3439–3444. doi: 10.1021/ja209829m. [DOI] [PMC free article] [PubMed] [Google Scholar]