Abstract
Background:
Genotype-phenotype correlation is a statistical relationship that measures correlation between the presence of a physical trait with a group of similar mutations but is dependent upon reliable phenotyping. It can provide information regarding disease pathogenesis, future disease progression, severity or activity. Such indicators would be valuable in Hidradenitis Suppurativa.
Aims and Methods:
This study aimed to assess the inter-rater reliability of hidradenitis suppurativa clinical phenotypes and perform exploratory genotype-phenotype correlation in cases of hidradenitis suppurativa with identified sequence variants. Linkage disequilibrium between variants was assessed. Genotype-Phenotype correlations were explored using Spearman correlation coefficients. Inter-rater reliability was calculated using Cohen’s kappa. Correlation between phenotype classifications was assessed using χ2 statistic.
Results:
43 sequence variants with clinical information were identified. Clinical phenotypes were classified as LC2 (n=29, 67.4%), Scarring Folliculitis (n=18, 41.8%), atypical (n=38, 88.3%) and nodular (n=26, 60.5%). LC1 phenotype was associated with Regular (χ2=41.289, p<0.0001) and Typical (χ2=29.013, p<0.0001) phenotypes. Cohen’s kappa was highest for Van der Zee (0.815), followed by Martorell (0.813), Naasan (0.774) and Canoui (0.435) classifications. High linkage disequilibrium was seen between variants of Han Chinese pedigrees. No significant genotype-phenotype correlations were identified.
Conclusions:
These findings may be influenced by selection, publication bias and the assumption that HS is a monogenic disorder. The poor inter-rater reliability of existing phenotype measures suggests limited utility of existing measures. Further investigations into the correlation of clinical phenotypes with inflammatory biomarkers may aid in prognostic efforts for this disease.
Background:
Genotype-phenotype correlation is a statistical relationship that measures correlation between the presence of a physical trait with a given mutation or group of similar mutations1. It can provide important information regarding the pathogenesis of a disease and provide information regarding predictions for the future progression, severity or activity of a disease2. However, disease phenotypes may be modulated by additional genetic effects (termed epistasis1), epigenetic and non-genetic (environmental) factors1. Single genetic variants can also give rise to multiple (pleiotropic1) effects in different body systems. A complete understanding of the relationship between genotype and phenotype is difficult, even in simple mendelian disorders.
Hidradenitis Suppurativa is a chronic inflammatory disease characterized by painful cysts and nodules developing into discharging sinus tracts with hypertrophic scarring3. It has significant morbidity and impact upon patients’ quality of life4. Up to one third of HS patients have some degree of familial inheritance3 and genetic sequence variants have been identified in both inherited and sporadic cases of HS3. Inherited forms of HS are considered monogenic in origin3. For patients who present with limited disease, the risk of progression of their HS and development of discharging sinuses and hypertrophic scarring is currently unknown5,6. There are no clinical indicators or serum biomarkers available to help guide more aggressive treatment for those patients at risk of developing severe disease. The identification of such indicators through genotype-phenotype correlation would be valuable for patients and clinicians alike and has been highlighted as an area of need in HS research5,6.
The current HS etiological paradigm involves gamma secretase sequence variants causing alterations in Notch signaling, leading to keratinocyte hyperplasia in the follicular infundibulum, resultant follicular rupture and subsequent inflammation7,8. However, alternative hypotheses have been proposed involving mediators other than notch precipitating inflammation3 in the setting of sensitized follicular keratinocytes9.
Multiple proposed phenotype classifications exist for HS10-13. (Table 1) Most are based upon clinical distribution of lesions and lesion types (i.e. comedones, nodules, presence of scarring or tunnels), one classification is based purely upon a latent class analysis10. All classification schema take into account age, gender and comorbidities of patients, although it is well documented that there is significant overlap between types and no single descriptor is definitive for classification14,15. Valuable genotype-phenotype correlation only exists in the presence of reliable phenotypic descriptors, and the poor inter-rater reliability (IRR) of some classification systems brings into question the utility of phenotypes in HS15.
Table 1:
Various Phenotype Classification Systems for Hidradenitis Suppurativa
| Canoui-Poutrienne et al 2013 | |||||
|
Axillary-Mammary (LC1) Armpit or Breast, Hypertrophic Scars, More likely women, less likely smokers, less likely to have family history |
Follicular (LC2) Armpit or Breast Ears, Chest or other areas, Hypertrophic Scars, Comedones, Epidermal Cysts, Papules and Folliculitis, Pilonidal Sinus, Family History of HS, Severe Acne, higher proportion of men and smokers |
Gluteal (LC3) Gluteal Area, Papules and Folliculitis, Family History of HS Higher proportion of smokers, less severe disease, longer duration |
|||
| Van Der Zee and Jemec 2015 | |||||
|
Regular Most common type, typical deep-seated nodules, abscesses, hypertrophic scarring in typical areas (axillae, groin, perineum, buttock, inframammary folds), chronic and recurrent. |
Scarring Folliculitis Pustules, cysts, depressed cribriform scarring and double ended comedones. Tunnels and fistulae unusual. Frequently overweight and smokers. |
Frictional Furuncles Overweight, multiple deep nodules on sites predisposed to friction such as abdomen, thighs and buttocks, tunnels and fistulae unusual. |
Congolobata Cyst formation on back and face. Strong family history, typically men and not overweight. |
Syndromic PASH Syndrome: (Pyoderma Gangrenosum, Acne and Suppurativa Hidradenitis) PAPASH Syndrome: Pyogenic Arthritis, Pyoderma Gangrenosum, Acne and Suppurativa Hidradenitis) |
Ectopic Involving the face |
| Naasan and Affleck 2015 | |||||
|
Typical Involvement of typical sites (axillae, groin, perineum, buttock, inframammary folds) |
Atypical Involvement of atypical sites (face, retroauricular, neck, distal limbs) |
||||
| Martorell Calatayaud et al 2015 | |||||
|
Follicular (Pattern A) Follicular Lesions on a background of comedones with occasional nodules or abscesses. More common in women, less commonly abscess formation. |
Nodular (Pattern B) Predominately nodules and abscesses in the absence of comedones. More frequently in men, more severe disease with more frequent fistulae and scarring |
||||
Assessment of the relationship between identified sequence variants in HS, and phenotypic characteristics may identify certain mutation types, or clinical characteristics that can predict progression of disease, or guide early therapeutic intervention. However, the current classification schema of HS phenotypes require validation through assessment of IRR.
Aims:
This study aims to:
Assess genotype-phenotype correlation in HS.
Assess the IRR of HS clinical phenotypes
Methods:
As this study did not involve the use of human subjects, formal IRB approval was waived. Published sequence variants in hidradenitis suppurativa were identified up until September 1st 2018 using previously published systematic review methodology3. All identified variants were assessed for pathogenicity using the American College of Medical Genetics and Genomics (ACMG)16 Criteria as previously described3. All variants with ACMG ratings of ‘benign’ or ‘likely benign’ were removed from further consideration.
All clinical data including age, onset of disease, lesion types, (hypertrophic scars, epidermal cysts, comedones, papules, folliculitis), sites of lesions, Hurley staging, family history of HS, gender, comorbidities (pilonidal sinus, acne, obesity, diabetes, smoking status) were collated. Where clinical data was not available, authors were approached to provide clinical data if available. Where clinical photographs were available these were also collated. All collation of data was undertaken by an independent physician (JWF) not involved in the phenotype classification.
Three independent experts (JEH, MSW, PG) were asked to phenotypically classify each case based upon the collated data. All experts were blinded to the sequence variant and any functional data regarding each case. Phenotype classification was undertaken using the four published phenotype classifications in the literature10-13 (Table 1). After independent classification, a round table discussion produced a consensus classification for each identified case. Discussion was mediated by an independent facilitator (JWF) and discussion continued until consensus was reached for each case.
Inter-rater reliability of each phenotype classification was calculated using Cohen’s kappa17 statistic with 95% confidence intervals. Percentage agreement was also calculated. Exploratory correlation analysis (using Spearman correlation coefficients) was also undertaken to examine correlation between phenotype classifications.
Exploratory genotype-phenotype correlation was evaluated using Spearman correlation coefficient. Genotypic covariates included affected gene (NCSTN, PSENEN, PSEN1, PSTPIP1), mutation type (Missense, Truncating, Splice Site Mutation, Promoter Region Change), site of protein alteration (extracellular, intracellular, transmembrane domain.) and the effect upon downstream notch signaling (increased, decreased or no change) as per information from published reports. Linkage disequilibrium between identified variants was assessed using LDLink18. Identified instances where 2 sequence variants were present in the same individual were excluded in order to simplify the exploratory genotype-phenotype analysis. For covariates with binary outcomes (such as classification systems with only two phenotypes) Chi sqaured (χ2) Statistic was used for evaluation. Where the sample size was 10 or less, Fisher’s exact test was used. Significance was defined as two-sided alpha <0.05.
Results:
Identified Sequence Variants:
A total of 65 sequence variants were identified across 20 separate genes. 43 variants (Table 2) were with associated clinical data and 22 variants had no clinical data available. Of the 43 variants identified, 32 variants involved NCSTN, 9 variants involved PSENEN, 3 involved PSEN1 and 5 involved PSTPIP1. One case reported heterozygosity for two sequence variants (NCSTN c.582+1delG and PSEN2.Thr421Met)3 and the clinical data for this case was excluded from analysis.16 other associated genes were also reported. (Supplementary Table 1). Of the 43 variants with clinical data, 13 sequence variants involved missense mutations, 22 involved truncating variants, 5 involved splice site mutations and the remaining 3 involved promoter regions (Table 2). ACGM classification identified 21 sequence variants as likely pathogenic and 22 variants of uncertain significance (Table 2).
Table 2:
Identified Sequence Variants and Phenotype Data
| Gene | Sequence Variant | N | Mutation Type | ACGM Classification | Phenotype Classification | Reference | |||
|---|---|---|---|---|---|---|---|---|---|
| Canoui et al | Jemec et al | Naasan et al | Martorell-Calatayud | ||||||
| Nicastrin | NCSTN c.223G>A | 1 | M | Likely Pathogenic | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 |
| NCSTN c.210_211delAG | 1 | T | Likely Pathogenic | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| NCTN c.218delC | 1 | T | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Nodular | 26 | |
| NCSTN c.344_351del | 1 | T | Likely Pathogenic | LC2 | Scarring Folliculitis | Atypical | Nodular | 3 | |
| NCSTN c.349C>T | 1 | T | Uncertain Significance | LC3 | Congolobata | Atypical | Nodular | 3 | |
| NCSTN c.477 C>A | 1 | T | Likely Pathogenic | LC2 | Frictional Furuncles | Atypical | Nodular | 3 | |
| NCSTN c.487delC | 1 | T | Likely Pathogenic | LC2 | Congolobata | Atypical | Nodular | 3 | |
| NCSTN c.497C>A | 1 | T | Likely Pathogenic | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| NCSTN c.553G>A | 1 | M | Likely Pathogenic | LC1 | Regular | Typical | Nodular | 3 | |
| NCSTN c.582+1delG | 1 | SS | Likely Pathogenic | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| NCSTN c.632C>G | 1 | M | Uncertain Significance | LC3 | Congolobata | Atypical | Nodular | 3 | |
| NCSTN c.647A>C | 1 | M | Uncertain Significance | LC3 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| NCSTN c.887A>G | 1 | M | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| NCSTN c.944C>T | 1 | M | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| NCSTN c.978delG | 1 | T | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| NCSTN c.996+7G>A | 1 | SS | Uncertain Significance | LC1 | Regular | Typical | Nodular | 3 | |
| NCSTN c.1101+10A>G | 1 | SS | Uncertain Significance | LC3 | Congolobata | Atypical | Nodular | 3 | |
| NCSTN c.1101+1G>A | 1 | SS | Uncertain Significance | LC3 | Scarring Folliculitis | Atypical | Nodular | 3 | |
| NCSTN c.1258C>T | 1 | T | Likely Pathogenic | LC2 | Congolobata | Atypical | Follicular | 3 | |
| NCSTN c.1300C>T | 1 | T | Likely Pathogenic | LC2 | Syndromic | Atypical | Nodular | 3 | |
| NCSTN c1635C>G | 1 | T | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Nodular | 3 | |
| NCSTN c.1695T>G | 1 | T | Likely Pathogenic | LC2 | Congolobata | Atypical | Follicular | 3 | |
| NCSTN c.1752delG | 1 | T | Likely Pathogenic | LC2 | Congolobata | Atypical | Nodular | 3 | |
| NCSTN c.1768A>G | 1 | T | Likely Pathogenic | LC2 | Congolobata | Atypical | Nodular | 3 | |
| NCSTN c.1702C>T | 1 | T | Likely Pathogenic | LC1 | Regular | Typical | Nodular | 3 | |
| Presenilin Enhancer 2 | PSENEN c.43_56del14 | 1 | T | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Follicular | 27 |
| PSENEN c.62-1G>C | 1 | T | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Follicular | 28 | |
| PSENEN c.66delG | 1 | T | Likely Pathogenic | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| PSENEN c.66_67insG | 1 | T | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| PSENEN c.168T>G | 1 | T | Uncertain Significance | LC1 | Regular | Typical | Nodular | 22 | |
| PSENEN c.279delC | 1 | T | Likely Pathogenic | LC2 | Congolobata | Atypical | Nodular | 3 | |
| Presenilin 1 | PSEN1 c.725delC | 1 | T | Likely Pathogenic | LC2 | Congolobata | Atypical | Nodular | 3 |
| PSEN1c.837+16G>T | 1 | SS | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| PSEN1 c.953A>G | 1 | M | Uncertain Significance | LC2 | Scarring Folliculitis | Atypical | Follicular | 3 | |
| Protein Serine-Threonine Phosphatase Interacting Protein 1 | PSTPIP1 CCTG5/8 | 1 | P | Uncertain Significance | LC2 | Syndromic | Atypical | Nodular | 3 |
| PSTPIP1 c.831G>T | 1 | M | Likely Pathogenic | LC2 | Syndromic | Atypical | Nodular | 3 | |
| PSTPIP1c.1213C>T | 1 | M | Likely Pathogenic | LC2 | Syndromic | Atypical | Nodular | 3 | |
| PSTPIP1homozygous (CCTG)5 | 1 | P | Likely Pathogenic | LC3 | Syndromic | Atypical | Nodular | 29 | |
| PSTPIP1 CCTG5/ CCTG8 | 1 | P | Likely Pathogenic | LC2 | Syndromic | Atypical | Nodular | 3 | |
| Fibroblast Growth Factor Receptor 1 | FGFR1 | 1 | M | Uncertain Significance | LC1 | Regular | Typical | Nodular | 30 |
| Fibroblast Growth Factor Receptor 2 | FGFR2 c. G492C p.K164N Exon 5 | 1 | M | Uncertain Significance | LC3 | Congolobata | Atypical | Follicular | 31 |
| Protein O- Fucosyl-transferase | POFUT1 Ex4 c.430-1G>A | 1 | M | Uncertain Significance | LC1 | Regular | Typical | Nodular | 30 |
| Pyrin | MEFV | 1 | M | Uncertain Significance | LC1 | Regular | Atypical | Nodular | 19 |
Please note: No clinical information was available for the following sequence variants: NCSTN c.1799delTG NCSTN c.1551+1G>A NCSTN c1381 el C NCSTN c.1352+1G>A NCSTN c.1180– 5C>G NCSTN c.1125+1G>A NCSTN c.687insCC, SULT1B1, SULT1E1, ELOVL7, NOD2, Keratin 5, Keratin 6a, Connexin 26, IL12RB1, TNF Haplotypes, CYP1A1, ATP2A2, Keratin 17, M= Missense, T= Truncating SS= Splice Site Mutation
Clinical Phenotypes:
The phenotype classifications were collated (Supplementary Table 1) and the final consensus phenotype classifications are presented in Table 2. The majority of clinical phenotypes were classified as LC2 using the Canoui et al10 classification (n=29, 67.4%), Scarring Folliculitis using the van der Zee12 classification (n=18 41.8%), atypical using the Naasan13 classification (n=38, 88.3%) and nodular using the Martorell-Calatayud11 classification (n=26, 60.5%). 7 cases were reported as LC3 type (16.3%), 11 cases classified as congolobata type (25.6%), 6 cases classified as syndromic type (13.9%), and 17 cases were classified as follicular type (39.5%).
All cases classified as LC1 (n=7, 16.3%) were also classified as Regular and Typical. There were no Typical cases which were also classified LC2, LC3 or other non-Regular phenotypes. Only one case was classified as frictional furuncle type (n=1, 2.3%) and no cases were classified as ectopic type.
Correlation between phenotype classifications:
Significant associations were seen between the different phenotype classifications. Cases assigned the LC1 phenotype were associated with allocation of the Regular phenotype (χ2=41.289, p<0.0001) and the Typical phenotype (χ2=29.013, p<0.0001).
Cases assigned the LC2 phenotype were significantly associated with allocation of the Follicular phenotype (χ2=9.169, p<0.01). Allocation of the follicular phenotype was significantly associated with the scarring follicular phenotype (χ2=25.753,p<0.0001). Naasan’s atypical phenotype was also significantly associated with the Martorell follicular phenotype (χ2=6.664, p<0.01, Kappa=0.192).
Inter-rater reliability measurements of phenotypes:
The results of inter-rater reliability measurements are presented in Table 3. The independent experts agreed on Canoui phenotype classification in 28/43 (65.1%) cases, van der Zee phenotype classification in 36/43 (83.7%) cases, Naasan phenotype classification in 41/43 (95.3%) cases, and Martorell phenotype classification in 38/43 (88.3%) cases. Cohen’s kappa were highest for Van der Zee (κ=0.815), followed by Martorell (κ=0.813), Naasan (κ=0.774) and Canoui (κ=0.435).
Table 3:
Inter rater agreement and reliability measurements for phenotype classifications in Hidradenitis Suppurativa
| Phenotype Classification | Proportion Agreement | % Agreement | Cohen’s Kappa | 95% CI |
|---|---|---|---|---|
| Canoui et al | 28/43 | 65.1 | 0.435 | 0.39-0.48 |
| Van der Zee et al | 36/43 | 83.7 | 0.815 | 0.80-0.83 |
| Naasan et al | 41/43 | 95.3 | 0.774 | 0.76-0.79 |
| Martorell-Catalayud et al | 38/43 | 88.3 | 0.813 | 0.80-0.82 |
Linkage Disequilibrium:
The results of linkage disequilibrium analysis was limited by the rarity of variants with only 12 correlations presenting with valid R2 and D’ results. (Supplementary Figure 1). R2 ranged between 0.001 and 0.172 due to the rarity of variants, and D’ ranged from 0.257 to 0.903. The highest D’ (>0.6) were seen between NCSTN variants described in Han Chinese pedigrees with moderate D’ in those described in French pedigrees.
Genotype-Phenotype Correlations:
A weak but statistically significant inverse association was seen between the presence of NCSTN sequence variant and the LC1 phenotype (R=−0.281 p=0.038) No other significant correlations were found between Canoui phenotype classifications and the associated gene.
Significant association was seen between PSTPIP1 sequence variants and classification as syndromic (R=0.83, p=0.001). No significant association were identified between gene affected and the Van der Zee or Naasan phenotype classifications.
Regarding the type of sequence variant, the presence of a variant in the promoter region of a gene was significantly associated with the Syndromic Phenotype (G2=29.449 p=0.003). No other significant associations were seen in the Canoui, Naasan or Martorell phenotype classifications.
Regarding the site of protein alteration, no significant correlations were seen between the site of protein alteration and any of the phenotype classifications. Correlation between the presence or absence of downstream Notch signaling alteration did not correlate with phenotype classifications.
Discussion:
Of the four distinct phenotype classification schemes in HS, commentary regarding IRR has only be made regarding Canoui et al15. Our results confirm previously published results showing poor IRR for the Canoui et al Phenotype classification system for individual phenotyping15. Van Straalen notes that the Canoui-Poutrine classification is useful at a population level but inadequate for individual patient care. The remaining three classification systems11-13 show similar acceptable levels of IRR. The strong correlations between the different phenotype classification systems show that significant redundancy exists between classifications.
The near 100% concordance of the LC1-Regular-Typical Phenotype confirms this as an essential component of phenotype classification. An LC2-Atypical-Follicular-Scarring Follicular phenotype shows strong concordance but there is significant inter-rater disagreement between LC2/LC3 phenotypes and Follicular/Congolobata phenotypes. Although the van der Zee classification system returned the highest Cohen’s kappa (κ=0.815); there were no cases rated as ‘ectopic’ and only one case rated ‘frictional furuncle’ phenotype hence conclusions regarding the validity of these phenotypes cannot be made. The cases identified also suffer from selection bias (being published cases only) and these results require validation in a large cohort of inherited and spontaneous cases of HS representative of the wider population.
We present a revised phenotype classification system (Figure 1) which maximizes IRR but also enables modification to accommodate the results of future research. The inclusion of a ‘Typical’ subtype is essential, however given the clinical heterogeneity of disease, further sub-classifications under the umbrella of ‘Atypical’ disease require further validation. Evidence contributing to the validity and reliability of such a binary approach include the high IRR (κ=0.88) of cases when comparing our results to the classification of overlapping cases by Ingram and Piguet14. Syndromic disease has the potential to become a third classification, however further epidemiological evidence and clinical consensus is required to define the symptomatology sufficient for a syndromic phenotype diagnosis. It remains unclear whether a syndromic phenotype should remain restricted to PASH and PAPASH syndromes or whether other autoimmune, inherited and autoinflammatory syndromes (Familial Mediterranean Fever19, Crohn’s Disease20, Dowling Degos Disease21) should also be included under this heading.
Figure 1:
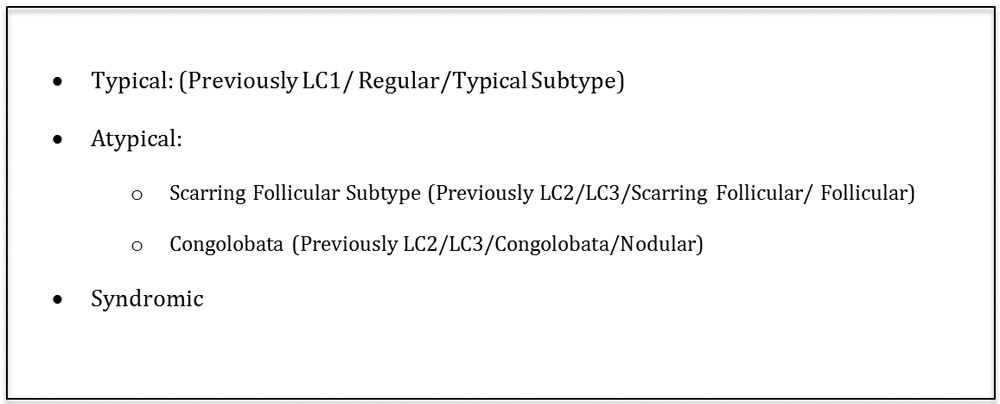
Proposed Revision to Phenotype Classification
LD was identified in 12 pairs of variants; however, the data was limited by the rarity of variants. The highest levels of LD were seen in NCSTN variants in Han Chinese pedigrees which may suggest either a founder effect or genetic admixture22. No significant correlations were identified between involved gene, type of mutation, protein alteration or impact on notch signaling with described phenotypes. The lack of correlations may be due to inadequate power of this study (contributed to by only one individual being described per variant), despite using all publicly available genetic variants and corresponding clinical data. Some contention exists regarding the pathogenicity of identified variants and some previously reported variants have been corrected as being non-pathogenic polymorphisms- particularly variants resulting in haploinsufficiency23. However, analysis of only variants classified by ACGM criteria as ‘likely pathogenic’ (Table 2) did not reveal any significant genotype-phenotype correlations by Spearman correlation coefficients (P>0.05). Given the functional data implicating gamma secretase variants in the pathogenesis of HS24, explanations and possibilities include: that the identified variants (mostly involving gamma secretase) are only one (major) contribution in a polygenic mode of inheritance; and/or that epigenetic and non-genetic factors have a strong influence on disease onset and activity. Such hypotheses could only be examined by genome wide association studies (GWAS) across multiple populations.
In the absence of valid genotype-phenotype correlation, immunophenotyping against clinical phenotype may give insight into the functional workings of inflammation in HS and would be the next step to compare valid phenotype classifications against. Examples of possible immune-phenotypes would include a stronger IL-1β signal in cases with a predominance of comedones (atypical or scarring folliculitis cases) due to evidence that IL-1β is involved in the development of comedones25. This would also help elucidate if any distinct immunological signatures can be identified related to specific clinical phenotypes which can be used as potential biomarkers for disease prognosis and patient care.
Limitations:
This study was limited by the information supplied by the authors of published articles and additional clinical information provided by the authors. Whilst the phenotype ratings were made based upon clinical descriptions and photographs, it is possible that classifications may have changed given live examination of the patient. The small sample size (43 cases) and the retrospective nature of the study also limit the power and external validity of results. However, as our results correlate with those in other independent studies14 this lends credence to the validity and reliability of the ‘Typical’/’Atypical’ duality in cases of inherited HS. Validating these results in a prospective study with a larger cohort would be informative, however the systematic nature of case collection provides us with the largest known cohort of pathogenic sequence variants in HS known to date. Not all clinical phenotypes were represented in this study. This may be a form of ascertainment bias if certain phenotypes are more strongly represented (or underrepresented) in inherited forms of HS. Hence the external validity of this study is limited to individuals with known pathogenic sequence variants in HS. Given the limited genotype information provided in published cases, no statistical analysis for pleiotropic effects was undertaken. Future studies should consider the role of principal factor analysis in order to take into account pleiotropic effects of genetic variants such as those identified in GWAS.
Conclusion
Genotype- phenotype correlation is HS is vital from the viewpoint of establishing prognostic indicators for disease severity and progression. We have identified that there is significant overlap in phenotype classification in HS. Whilst the Canoui phenotype classification may be valid for population statistics, it suffers from poor IRR and should not be used for individual phenotype classification. Within the limitations of current data, no significant genotype-phenotype correlations have been identified, However, the suggestion of high LD between certain variants in Han Chinese pedigrees suggestive of a founder or admixture effect is worthy of further investigation. The results of our study suggest revision to the existing phenotype classification for inherited forms of HS and larger prospective studies with GWAS are required to further our understanding of genotype-phenotype correlation in this disease.
Supplementary Material
Supplementary Figure 1: Linkage Disequilibrium Matrix for sequence variants in Hidradenitis Suppurativa. Associated R2 and D’ Tabulation as calculated by LD Link18 Highlighted cells identify correlations with valid R2 and D’ data.
Supplementary Table 1: Individual Phenotype Classifications Ratings
Key Points:
What is Already Known About This Topic?
Genotype-phenotype correlation can provide information regarding disease pathogenesis and predictions for future disease progression, severity or activity. The identification of such indicators in Hidradenitis Suppurativa would be valuable for patients and clinicians alike given the lack of biomarkers or clinical predictors of disease.
What Does This Study Add?
65 sequence variants across 20 separate genes were identified. There was no significant correlation between phenotype classification in four separate classification schema and gene, mutation type, or impact upon Notch signaling. The utility of current phenotype measurements are limited. The lack of genotype-phenotype correlation in HS is suggestive that the underlying assumption of inherited HS as a monogenic disorder may need revision.
Acknowledgments
Funding:
Supported in part by grant # UL1 TR001866 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program.
Footnotes
Conflicts of Interest:
The authors have no conflicts of interest to declare
References
- 1.Orgogozo V, Morizot B, Martin A. The differential view of genotype–phenotype relationships. Frontiers in Genetics 2015;6, 179 10.3389/fgene.2015.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weerakkody RA, Vandrovcova J, Kanonidou C, Mueller M, Gampawar P, Ibrahim Y, et al. Targeted next-generation sequencing makes new molecular diagnoses and expands genotype–phenotype relationship in Ehlers–Danlos syndrome Genetics in Medicine 2016;18; 1119–1127 [DOI] [PubMed] [Google Scholar]
- 3.Frew JW, Vekic DA, Woods J Cains GD A systematic review and critical evaluation of reported pathogenic sequence variants in hidradenitis suppurativa. Br J Dermatol 2017;177: 987–998 [DOI] [PubMed] [Google Scholar]
- 4.Kirby JS Sisic M Tan J Exploring Coping Strategies for Patients with Hidradenitis Suppurativa JAMA Dermatol 2016;152:1166–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elman SA, Merola JF, Armstrong AW, Duffin KC, Latella J, Garg A, Gottlieb AB The International Dermatology Outcome Measures (IDEOM) Initiative: A Review and Update. J Drugs Dermatol. 2017;16(2):119–124. [PubMed] [Google Scholar]
- 6.Hoffman LK, Mondana HG, Garg A, Hamzavi IH, Afsaneh A, Lowes MA. (2017). Major gaps in understanding and treatment of hidradenitis suppurativa. Seminars in Cutaneous Medicine and Surgery. 36 86–92. 10.12788/j.sder.2017.024. [DOI] [PubMed] [Google Scholar]
- 7.Negus D, Ahn C & Huang W An update on the pathogenesis of hidradenitis suppurativa: implications for therapy, Expert Review of Clinical Immunology, 2018;14: 275–283, DOI: 10.1080/1744666X.2018.1449647 [DOI] [PubMed] [Google Scholar]
- 8.Xu H, He Y, Hui Y, Xiao X, Zhang X, Li C, Wang B, NCSTN mutations in hidradenitis suppurativa/acne inversa do not influence cytokine production by peripheral blood mononuclear cells Br J Dermatol 2017;176:270–280 [DOI] [PubMed] [Google Scholar]
- 9.Gauntner TD Hormonal Stem Cell and Notch signaling as possible mechanisms of disease in Hidradenitis Suppurativa: A systems-level Transcriptomic Analysis. Br J Dermatol 2018; doi: 10.1111/bjd.17093 [DOI] [PubMed] [Google Scholar]
- 10.Canoui-Poitrine F, Le Thuaut A, Revuz JE, Vialette C et al. Identification of three hidradenitis suppurativa phenotypes: latent class analysis of a cross sectional study J Invest Dermatol 2013;133:1506–1511 [DOI] [PubMed] [Google Scholar]
- 11.Martorell-Calatayud A, Sanz-Motilva V, Alfaro-Rubio A, et al. Phenotypic heterogeneity in hidradenitis suppurativa J Am Acad Derm 2015;72(5):AB150 [Google Scholar]
- 12.Van der zee HH, Jemec GB New Insights into the diagnosis of hidradenitis suppurativa: Clinical presentations and phenotypes J Am Acad Dermatol 2015;73:S23–26 [DOI] [PubMed] [Google Scholar]
- 13.Naasan H, Affleck A A typical Hidradenitis Suppurativa Clin Exp Dermatol 2015;40:891–893 [DOI] [PubMed] [Google Scholar]
- 14.Ingram JR, Piguet V Phenotypic Heterogeneity in Hidradenitis Suppurativa (Acne Inversa): Classification is an Essential Step Toward Personalised Therapy J Invest Dermatol 2013;133:1453–1456 [DOI] [PubMed] [Google Scholar]
- 15.Van Straalen KR, Verhagen T, Horvath B, Ardon C, Vossen ARJV, Driessen R, Boer J, ALV Rondags, EP Prens, Zee van der HH Poor Inter-Rater Reliability of Hidradenitis Suppurativa Phenotypes J Am Acad Dermatol 2018;79(3): 577–578 [DOI] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bal S et al. Standards and Guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee M, Capazzoli M, McSweeney L, Sinha D Beyond Kappa: A Review of Interrater Agreement Measures Canadian Journal of Statistics 1999;27(1): 3–23 [Google Scholar]
- 18.Machiela MJ, Chanock SJ. LDassoc: an online tool for interactively exploring genome-wide association study results and prioritizing variants for functional investigation. Bioinformatics. 2018;34:887–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vural S, Gundogdu M, Kundakci N Ruzicka T Familial Mediterranean fever patients with hidradenitis suppurativa Int J Dermatol 2017;5:660–663 [DOI] [PubMed] [Google Scholar]
- 20.Van der Zee HH, Horvath B Jemec GB, Prens EP The Association between Hidradenitis Suppurativa and Crohn’s Disease: in Search of the Missing Pathogenic Link J Invest Dermatol 206;136(9):1747–1748 [DOI] [PubMed] [Google Scholar]
- 21.Pavlovsky M, Sarig O, Eskin-Schwartz M, Malchin N, Bochner R, Mohamad J, Gat A et al. A phenotype combining hidradenitis suppurativa with Dowling Degos Disease caused by a founder mutation in PSENEN BR J Dermatol 2018;178(2):502–508 [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Lu D, Chung YJ, Xu S Genetic Structure, divergence and admixture of Han Chinese, Japanese and Korean populations Hereditas 2018;155:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pink AE, Dafou D, Desai N, Holmes O, Hobbs B, Smith CH, Mortimer P, Simpson MA, Trembath RC, Barker JN Hidradenitis Suppurativa : Haploinsufficiency of gamma-secretase components does not affect gamma-secretase enzyme activity in vitro Br J Dermatol 2016;175(3):632–635 [DOI] [PubMed] [Google Scholar]
- 24.O’Sullivan Coyne G, Woodring TS, Lee CR, Chen AP, Kong HH Hidradenitis Suppurativa-Like Lesions Associated with Pharmacological Inhibition of Gamma-Secretase J Invest Dermatol 2018;138(4):979–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Contassot E, French LE New Insights into Acne Pathogenesis: Propionibacterium Acnes Activated the Inflammasome J Invest Dermatol 2014;134(2):310–313 [DOI] [PubMed] [Google Scholar]
- 26.Wu C, Yang J, Zhang S, Li J, Hongzhong J, Zhang X, A novel NCSTN mutation in a Chonese family with Acne Inversa Mol Genet Genom 2018; Mol Genet Genomics 2018. July 20. doi: 10.1007/s00438-018-1475-9 [DOI] [PubMed] [Google Scholar]
- 27.Kan T, Takahagi H, Shindo H, Tanaka A, Kawai M, Hide MA Unique Clinical Phenotype of a patient bearing a newly identified deletion mutation in the PSENEN gene along with the pathogenic serum desmoglein-1 antibody Clin Exp Dermatol; 2018;43:319–335 [DOI] [PubMed] [Google Scholar]
- 28.Ralser DJ, Basmanav FB, Tafazzli A, Wititsuwannakul J, Delker S, Dander S, Thiele H, Wolf S, et al. Mutations in Gamma Secretase subunit encoding PSENEN underlie Dowling-Degos disease associated with acne inversa. J Clin Invest 2017;127(4):1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonbol H, Duchatelet S, Misinkyte S, Bonsang B, Hovanian A, Misery L PASh Syndrome (pyoderma gangrenosum, acne and hidradenitis suppurativa); a disease with genetic heterogeneity Br J Dermatol 2018;178:ppe17–ppe18 [DOI] [PubMed] [Google Scholar]
- 30.Ge Qian, Liu T, Zhou C, Zhang Y Naevus Comedonicus Syndrome Complicated by Hidradenitis Suppurativa-like lesions responding to Acitretin Treatment. Acta Dermatol Venerol 2015;95(8):992–993 [DOI] [PubMed] [Google Scholar]
- 31.Higgins R, Pink A, Hunger R, Yawalkar N, Navarini AA Generalized Comedones, Acne and Hidradenitis Suppurativa in a Patient with a FGFR2 Misense Mutation. Fronteirs Med 2017;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzales-Villanueva I, Gutierrez M, Hispan P, Betlloch I, Pascual JC Novel POFUT1 mutation associated with hidradenitis suppurativa-dowling-degos disease firm up a role for Notch signaling in the pathogenesis of this disorder Br J Dermatol 2018;178:984–990 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Linkage Disequilibrium Matrix for sequence variants in Hidradenitis Suppurativa. Associated R2 and D’ Tabulation as calculated by LD Link18 Highlighted cells identify correlations with valid R2 and D’ data.
Supplementary Table 1: Individual Phenotype Classifications Ratings


