Abstract
Recent advances in genomics and imaging technologies have increased our ability to interrogate the 3D conformation of chromosomes and to better understand principles of organization and dynamics, as well as how their alteration can lead to disease. In this review we describe how these technologies have shed new light into the role of the 3D organization of the genome in defining cellular states in aging and ageassociated diseases. We compare the genomic organization in cellular senescence and cancer, discuss the role of the lamina in maintaining the structural and functional integrity of the genome, and we highlight the recent findings on how this organization breaks down in disease states.
Introduction
DNA displays a remarkable degree of compaction in living cells. If we stretched the human DNA into a linear string, it would measure roughly 2 meters. Within the cell nucleus, it is confined to a space measuring approximately 6 micrometers in diameter, a compaction of over 10,000-fold [1]. Remarkably, this degree of compaction is achieved while maintaining a complex organization that lets DNA be duplicated during mitosis, cell-type dependent sets of genes be accessible to the transcriptional machinery, and large portions of the genome be transcriptionally silenced.
Investigation of the organization of chromosomes within the nucleus has a long history and can be dated back to the pioneering microscopy studies that first revealed the presence of different states of chromatin, distinguished by their degree of staining: heterochromatin and euchromatin [2]. We now have clear evidence that the three dimensional organization of the genome plays a key role in many fundamental cellular processes including cell cycle progression [3–5], regulation of gene expression, and cell differentiation [6].
In this review, we describe the recent advances in genomic techniques to probe the 3-dimensional structure of the genome, its role in controlling gene expression, and the changes that are associated with disease and aging.
Measuring Chromosome 3-dimensional Organization and Implications in Gene Regulation
The primary genomic methodology used to probe the 3-dimensional organization of chromosomes is Chromosome Confirmation Capture (3C), a collection of technologies based on proximity ligation assays. Hi-C is a very powerful extension of 3C, and it offers the advantage of measuring long-range interactions between different loci genome-wide [7]. The typical workflow of a Hi-C assay comprises crosslinking chromatin, digesting the DNA with a restriction enzyme, and incorporating a biotinylated nucleotide into the restriction fragment ends. Next, the restriction fragments are ligated. At this step two fragments which are in proximity 3-dimensionally are joined together by the ligase enzyme, even though they may be kilobases away in an unfolded genome. The biotinylated ligation products are pulled down with streptavidin coated beads and are processed for sequencing. The sequencing data, along with the direct relationship between ligation frequency and distance in 3D, allows for the 3-dimensional organization of chromatin to be inferred computationally [8]. Typically, the event where two restriction fragments are ligated to one another is referred to as an interaction, and the genome wide interaction profiles generated by Hi-C are used to identify different features of chromatin organization.
Analysis of Hi-C interaction profiles is conducted at different levels of resolution, and at the megabase scale two separate compartments, named A and B, are notable. A compartments tend to correspond to open, gene rich, euchromatic regions, whereas B compartments correspond to closed, gene depleted, heterochromatic regions [7]. At the next level of resolution are Topologically Associated Domains (TADs), which can be thought of as insulated localities such that two loci within a TAD are more likely to be ligated together than loci which are in separate TADs [7].
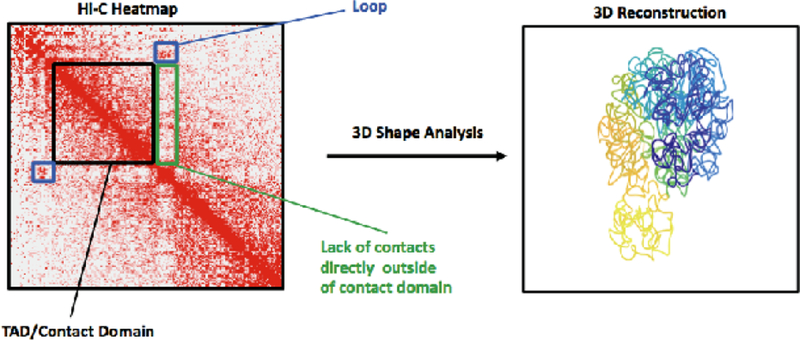
Recent advances in 3C technologies have dramatically increased the resolution of Hi-C data and have revealed additional organizational features. The advent of in-Situ Hi-C, a variant where ligation occurs within an intact nuclear structure, has shown that the A and B compartments can be further divided into six subcompartments, two within A and four within B, each demonstrating a distinct interaction profile [9]. This suggests that chromatin can be classified beyond the euchromatic and heterochromatic states which were originally described by early microscopy studies. In a similar fashion, in-Situ high-coverage Hi-C has also revealed that TADs are often composed of several smaller self-interacting regions, referred to as contact domains. Hence, folding of the genome appears to occur in a hierarchical manner, with smaller interacting regions organized into larger structures [9]. TADs and contact domains are best visualized as heatmaps, as shown in Fig. 1. A Hi-C heatmap is a visual representation of a contact matrix derived from Hi-C data; regions of the genome which interact more frequently in 3-dimensional space are shown with a darker color. Because the number of contacts between two regions can be associated to their physical distance in 3D, it is possible to infer the structure of chromosomes via 3D modelling and shape analysis [10–12].
Figure 1.
Example of a Hi-C contact map for a chromosome subregion (left). High intensity indicates the corresponding pair of genomic loci display many contacts between them. Typical features are indicated with colored boxes: a Topologically associated domain (TAD) (black), a loop anchor (blue), contact depleted region between TADs (green). Contact matrices can be used to infer the 3D structure of chromosomes (right).
The organization and functional role of TADs and contact domains is an active area of investigation. An open question is whether and how they contribute to gene regulation. Some studies have described TADs as true structures which exist inside the nucleus of a cell [13], while others have interpreted TADs to be nothing more than patterns in Hi-C data that emerge as an average property across millions of cells, hence having little to no phenotypic effect [14]. Very high resolution in-situ Hi-C datasets have demonstrated that smaller TADs tend to contain recognition sequences for the CCCTC-binding factor (CTCF) protein [9]. In what has become known as the “looping extrusion model”, the DNA in between these recognition sequences is passed through a CTCF cohesion protein complex. Once the CTCF binding sites reach this complex, the extrusion is stopped, and the result is a chromatin loop [15]. These structures have directly implicated Hi-C features in the regulation of gene expression.
Chromatin loops can facilitate the fine tuning of gene expression [9]. For example, in MECP2 knockout mice the homeobox genes DLX5 and DLX6 were overexpressed when compared to wild type mice [16]. MECP2 is implicated in the neurodevelopmental disorder Rett Syndrome, and it was shown to target chromatin modifying complexes responsible for gene silencing to the DLX5 and DLX6 loci. These genes are needed for proper development, and through a combination of ChIP and 3C based assays it was demonstrated that wild type mice possessed a chromatin loop responsible for repressing the DLX5 and DLX6 loci. However, this loop was absent in the MECP2 knockout cohort.
Chromatin organization varies between cell types, and these differences are noticeable when examining chromatin loops and the frequently interacting regions (FIREs) inside of them [17]. A large study comparing Hi-C data from different human tissues and cell types found that many chromatin loops and FIREs are tissue and cell type specific. For instance, in GM12878 cells there is a FIRE which encompasses CD70, a gene known to be expressed in B cell lymphomas [17]. However, only 100 kb away is a FIRE which is only found in brain tissue. This region contains ROBO1, a gene involved in axon regulation [17]. These types of analyses have demonstrated that the genome is capable of being differentially organized depending on the tissue of origin.
Moreover, haplotyped Hi-C matrices from GM12878 cell lines display distinct interaction profiles which correlate with gene expression. For example, H19, a gene which transcribes a long noncoding RNA responsible for downregulating cell proliferation, is only expressed in the maternal chromosome. Consistently, Rao et al. found that the chromatin loop encompassing this locus was present exclusively in the maternal chromosome [9].
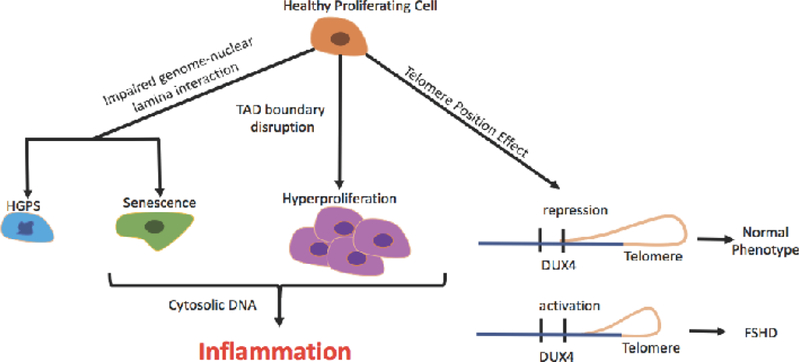
Unveiling the 3-dimensional architecture of the genome contributes to yet another layer of gene regulation, and understanding this dynamic relationship is essential for uncovering epigenetic mechanisms driving disease. For the remainder of this review we will focus on the implications of this organization on disease states, such as aging, cancer, and inflammation (Fig. 2).
Figure 2.
Schematic of some of the known 3D perturbations which can elicit different disease states. Impaired genome nuclear lamina interaction is associated with cellular senescence as well as causal to laminopathies such as Hutchinson-Gilford progeria syndrome (left). TAD disruption can lead to aberrant gene expression, contributing to hyperproliferation and cancer (center). Progressive telomere shortening can regulate gene expression over long distances, a process called Telomere position effect (TPE), and can lead to pathologies such as Facioscapulohumeral muscular dystrophy (FSHD) through the activation of the DUX4 locus.
3D Epigenomic Changes in Aging and Age Associated Diseases
Aging is a complex biological process with many contributing factors, and it alone is one of the greatest risks for many chronic diseases [18]. One of the hallmarks of aging at the cellular level is cellular senescence [19], a state of irreversible growth arrest which evolved as a mechanism to protect against cancer. The cumulative non-repaired DNA damage which each cell in an organism endures throughout its life leads to the accumulation of senescent cells [20], as well as an increase in their inflammatory products, a collection of proinflammatory signaling molecules and cytokines which are collectively referred to as the Senescence Associated Secretory Phenotype (SASP) [21]. Attenuating SASP in mice results into a marked increase in both healthspan and lifespan. For example, eliminating SASP at its source by selectively inducing the apoptosis in senescent cells extends the lifespan of mice [22,23]. These studies have helped set the stage for implicating cellular senescence in aging and disease, and, as a consequence, there is now great interest in understanding the mechanisms which drive the senescent and proinflammatory state [21].
Senescent cells are characterized by extensive epigenetic changes, including changes to the chromosome organization [24]. Hi-C was performed on fibroblasts that were passaged to replicative exhaustion [25]. In this form of senescence, referred to as replicative senescence, the shortening of telomeres which occurs during cell division leads to constitutive activation of the DNA damage response pathway, causing the cell to permanently exit the cell cycle [26]. Hi-C revealed that replicatively senescent cells have dramatically different nuclear architectures when compared to proliferating and quiescent cells [25]. One of the most prominent changes included a variety of TADs switching between active A and repressive B compartments. This switching correlated with changes in gene expression, where the genes within a TAD that switched from A to B compartments showed a decrease in expression, and vice versa [27].
Studies have also shown that Oncogene Induced Senescence (OIS) is characterized by extensive changes in nuclear architecture. OIS occurs when an activated oncogene drives a hypoerproliferative state that leads to high levels of DNA damage and subsequent exit from the cell cycle. Chandra et al. used Hi-C on WI-38 human lung fibroblasts undergoing OIS to demonstrate that regions of the genome which are typically near the nuclear periphery dissociate and localize to a more centralized area. This discovery helped to shed light on the formation of senescence-associated heterochromatic foci (SAHF), regions with high amounts of heterochromatin [28].
Hi-C interaction profiles also showed a higher number of short range contacts when compared to proliferating controls. This was thought to be because of the decrease in chromosome volume that is occurs during cellular senescence. However, it was found that some regions of the genome, such as the Lamina Associated Domains (LADs), became more accessible [27]. Typically, the LADs are heterochromatic regions which attach to the nuclear envelope via the structural proteins Lamin A/C and Lamin B [29]. In replicative senescence these regions become detached due to a decrease in Lamin B1 proteins, resulting in LADs becoming more euchromatic [30,31]. This observation is especially interesting since structural alterations in LADs are known to drive other age-related pathologies such as Gilford’s Hutchinson Progeria Syndrome [32,33].
The 3-dimensional nuclear architecture is also strongly perturbed in cancer. The uncontrolled hyperproliferation that characterizes cancerous cells often leads to somatic copy number alterations (SCNAs). These mutagenic events are defined by duplications of large portions of the genome, and these dramatic changes in DNA sequence are accompanied by modifications in nuclear organization. Recently, a computational analysis of SCNAs and Hi-C data revealed that the 3-dimesnional organization of a nucleus determines the SCNA events that happen in cancer [34].
Cell hyperproliferation is linked to many other epigenetic changes. Similar to what is seen in cellular senescence, the LADs of cancer cells detach from the nuclear lamina and become more open [35]. This type of alteration can lead to expression of the many retrotransposable elements which are within LADs, and the subsequent duplication of these repetitive sequences can further perturb the organization of the chromatin [35]. Moreover, it has been found that cancer cells have unusually high mutation rates found at CTCF sites. It is expected that these mutagenic events contribute to the altered genomic architecture and gene regulation which characterizes cancer [36].
The 3D Epigenome as a Driver of Disease
The previously mentioned Gilford Hutchinson Progeria Syndrome provides evidence for nuclear architecture having a casual role in disease. Progeria is characterized by an accelerated aging phenotype caused by a mutation in the LMN A gene [32]. Wildtype LMN A expresses Lamin A, but those with Progeria express Progerin instead. This defunct translational product prevents LADs from attaching to the nuclear envelope, causing a perturbed 3-dimensional architecture, as well as premature aging phenotypes and frailty [32,37].
Perturbations to the 3-dimensional chromatin architecture at telomere sequences can also drive disease states. As previously mentioned, continual cell division leads to telomere attrition, and this has been found to affect gene expression via a phenomenon known as the Telomere Position Effect (TPE). In young cells telomeres are able to form loops with loci megabases away, and as telomeres shorten the structure of these loops are altered [38]. TPE contributes to Facioscapulohumeral Muscular Dystrophy (FSHD), a late onset disease thought to be caused by deregulation of the DUX4 gene. DUX4 is less than 100 kilobases from the end of chromosome 4, and it was found that cells with shortened telomeres upregulate DUX4 due to their inability to repress this locus [39].
Alterations to the 3-dimensional chromatin architecture can also drive hyperproliferation. Mutating the CTCF domain that delimits the TAD boundary encompassing PDGFRA was found to give cells a proliferative advantage by increasing its expression [40]. Moreover, it was found that SCNAs in Colorectal Cancer Cell lines create a novel TAD around the IGF2 locus, leading to overexpression of the IGF2 gene due to an aberrant promoter enhancer interaction [40]. These findings suggest that the 3-dimensional organization of the genome could potentially be used as a prognostic tool. For instance, there are candidate genes whose position is highly variable between cancerous and normal tissues. However, much research remains to be done to determine if gene position can be reliably used as a biomarker for disease [41].
Perhaps the most dramatic alteration to nuclear architecture occurs when parts of the genome are completely expelled from the nucleus. This type of nuclear breakdown has been described in cancer and more recently in cellular senescence. Significantly, a growing body of evidence has shown the resulting cytosolic DNA to initiate various inflammatory pathways. The next section of this review will focus on the breakdown of the 3-dimensional architecture and how it can lead to inflammation and tissue dysfunction.
Cytosolic DNA
Recent studies have uncovered a unifying link between various cancers and cellular senescence: aberrant cytoplasmic DNA [42–45] (Fig. 3). A largely unanswered question is whether there is a connection between the 3D genome and this unique species of DNA. Indeed, for both cancer and senescence, nuclear dysmorphisms have been observed in the form of laminopathies, nuclear envelope blebbing, genomic aneuploidy, and in the display of nuclear genome in the cytoplasm [46–48]. Selected from all of these malfunctions, much is still unidentified about cancer-associated cytosolic DNA (CACD), and senescence-associated cytosolic DNA (SACD) in the context of disease etiology and progression. We will review what is known to date about these aberrant genomic species of cytoplasmic DNA, the cell intrinsic and extrinsic consequences of its spatial location, and its contribution to pathology.
Figure 3.

Nuclear dysmorphism and cytosolic DNA in cancer and senescent cells are precursors to inflammation. Nuclei from cancer and senescent cells share common aberrant nuclear structural properties such as nuclear envelope distensions (blebs) and altered lamin expression. In the cancer nucleus, nuclear blebbing and micronuclei are formed due to genotoxic stresses, lamin A/C (red) or lamin B (blue) dysregulation, and excessive force of perinuclear structural proteins (neon blue). In senescent cells the 3D genome changes represented by dissociation of chromatin (grey) from regions of the nuclear lamina. Lamin B1 (blue) is targeted for degradation with its associated DNA (grey) and is extruded into the cytosol via LC3II (yellow). Detection of aberrant DNA species in the cytosol of cancer and senescent cells by cGAS-STING, propagates a pro-inflammatory type I interferon response.
For over one hundred years, cancers have been observed and characterized to be pathologies sharing the hallmark of genomic instability. The accelerated rate at which cancer cells proliferate provides an ideal niche for events of altered gene regulation, ill-repaired DNA double stranded breaks (DSB), aneuploidy, and chromosome mis-segregation[49]. Nuclear structural proteins play an essential role in chromosome morphology and function, and due to altered proteostasis present in cancers, maintenance of nucleus shape is often severely compromised [50]. Lamins are nuclear envelope structural proteins that form the backbone of nucleus morphology. Dysregulation of lamin proteins have been implicated in multiple forms of cancers, as these proteins are directly or indirectly involved in the regulation of gene expression, DNA repair, and apoptosis [51]. Importantly, lamins are also essential for providing the nucleus with a scaffold in which extranuclear structural proteins such as actin and tubulin are coupled [52]. Due to altered expression of either or both lamin A/C or lamin B in cancers, they have been associated to the acute release of nuclear genetic material to the cytosol. We find one possible mechanism of DNA exit through the movements of metastatic cells, where the nuclear envelope is morphed by mechanotransduction in and out of tight spaces [53]. Because of the dramatic changes in lamin structure and organization, the shape of the nucleus is compromised, often forming outward distensions leading to nuclear envelope ruptures and release of genetic material into the cytoplasm [54]. Another similar mechanism has been hypothesized where extrusion of DNA from the nucleus to the cytoplasm is mediated by “pinching” of sites on the nuclear envelope that are lamin-depleted. In this case, the mechanical forces produced by perinuclear structural proteins (tubulin, actin) exceeds the anchoring and physical strength capabilities of the nuclear lamina. The pinching of the nuclear envelope eventually reaches a critical point causing nuclear envelope rupture events and subsequent leakage of chromatin into the cytoplasm [55]. A slightly divergent example of DNA exit is directly related to the nuclear envelope protein lamin B2, which plays an essential role in chromosome stability of the interphase and mitotic nucleus. Lamin B2 facilitates nuclear homeostasis by providing the necessary anchoring sites for normally repressed regions of the genome that are bound to the nuclear envelope [56]. Thus, lamin B2 directly contributes to the maintenance of nuclear content topology, and isolation of active and repressed domains of the genome. Its dysregulation has been demonstrated to be a key component in the breakdown of DNA conformation. Lamin B2 depletion was attributed to chromosome missegregation leading to genomic aneuploidy and finally formation of micronuclei, in contrast to the overexpression of lamin B2 being protective against micronucleating events. This protective role of lamin B2 was independent to the number of double stranded breaks within the nucleus [49].
Recent work in human tumor cells has probed the structure of cancer-associated cytoplasmic DNA (CACD). Interestingly, CACDs display remarkable molecular conformation heterogeneity as double stranded DNA, single stranded DNA, as well as DNA:RNA hybrids [57]. These various species of CACD was present at different total percentages as a function of tumor cell type, suggesting there are diverse mechanisms at play in the nucleus that promote and precede the exit of CACD. Although these potential mechanisms have not been fully interrogated, it is hypothesized that excreted DNA are enriched for sites that confer a high probability of DNA breaks, primarily caused by stalled or collapsed replication forks. In most cases, replication forks are impeded by commonly transcribed genomic regions, common fragility sites, DNA-binding proteins, repetitive sequences, or DNA:RNA hybrid R-loops [57,58]. At the onset of tumorigenesis there is an increased proliferative potential of the affected cell and, in theory, this will exacerbate replication fork stalling or collapse to a critical point where a cell fate decision must be determined—the fate choice to remain in a cancerous state, to induce apoptosis, or enter cellular senescence.
Though the process of aberrant DNA exit from the nucleus has been partially investigated in some cancers, due to overlapping fate choices a cell may undertake, it is convenient to speculate on observance of cytoplasmic DNA outside the scope of cancers. In that light, it becomes quickly evident that there are striking similarities between the physiology of cancers and cellular senescence [59]. Just as it has been observed in cancer cells, senescent cells are riddled with altered proteostasis, genomic instabilities, and a compromised nuclear envelope. Many groups have reported on genomic dysfunction of repetitive elements and the appearance of nuclear distensions (nuclear blebbing)in senescent cells [48,60,61]. These features preceded the observation of senescence-associated cytosolic DNA (SACD), which has been recently described by multiple groups. Though the similarities between cancer and senescence cannot be denied, the unique features in DNA conformation that define the senescent state and the genesis of disease are not yet well understood.
As mentioned in a previous section, the irreversible cell cycle arrested state fulfills essential roles in embryonic development, tissue repair and remodeling, and in tumor suppression [62]. In contrast, due to the increase in number of senescent cells retained in the body over time, they have been implicated in disease onset and in aging [63]. Senescent cells experiences dramatic nuclear morphology and genomic changes through their lifespan. These global shifts away from homeostasis are correlated with dramatic intranuclear alterations including the variable dissociation of chromatin from the nuclear envelope, and formation of senescence associated heterochromatic foci, providing additional and ample evidence that maintenance of components that dictate DNA conformation is necessary for normal function of the cell [25,64]. Beyond the observations of nuclear blebbing and nuclear envelope rupturing events in senescent cells, several groups have also reported on the appearance of cytoplasmic DNA as a result of the accumulation of DNA DSBs [49,65,66]. Dou et al. proposed a mechanism of nuclear DNA expulsion from the nucleus that showed SACD is selected based on its association with the nuclear envelope structural protein, lamin B1. Because nuclear lamins are designated for degradation in moments of cellular stress, it was hypothesized that senescent cells actively degrade lamin B1 through an autophagy-mediated process. To accomplish this, it was shown that the autophagic factor, microtubule-associated proteins 1A/1B light chain 3B II (LC3II) escorts lamin B1 with its coupled chromatin to the outside of the nucleus into autophagosomes. Autophagosome-containing SACD subsequently fuse to lysosomes, a process that typically leads to the degradation of the contained DNA. Decreasing expression of LC3II was correlated with decreased observation of SACD, directly connecting the autophagic pathway to SACD mediation and degradation [29].
Once outside of the nucleus, what are the potential consequences of these micronuclei or naked DNA being detected in the cytoplasm? Is the formation of these cytoplasmic DNA species remediation to genomic dysfunction, or are they the consequences of the dysfunction? These cytoplasmic extensions of the genome warrant further investigation to better understand their mechanism of exit from the nucleus, and the potential consequences of their presence in the cytoplasm. Answering these questions has been the subject of recent debate, with several studies converging on a unifying cell autonomous reaction to cytosolic DNA. Multiple groups have reported that the foreign body of DNA potentially acts as a Trojan Horse in the cytoplasm–involved in initiating deleterious immune responses within and outside of the affected cell [43,65]. Two of the most significant findings are that the formation of cytosolic DNA in senescence and in cancer have been linked to increased cell metastasis potential, and that cytoplasmic DNA results in cell intrinsic and extrinsic pro inflammatory responses [43,49,65]. How can this be? Anomalous DNA in the cytoplasm is sensed by the cytosolic DNA-sensing factor, cyclic GMP-AMP synthase (cGAS). Upon cGAS binding to the aberrant DNA, the stimulator of interferon genes (STING) initiates a signaling cascade that triggers the expression of type I interferons as well as other canonical pro-inflammatory factors of the innate immune system. The result is that the affected cell responds in the same manner as if it were infected by a virus, promoting the release of factors and cytokines with the intent of warning neighboring cells, including cells of the immune system. Because of the strong overlap of cytoplasmic DNA detection and immune response between cancer and senescence, this may mean that the widespread effects of cytoplasmic DNA are not dependent on cellular state or disease context. The implications of this self-propagating response are significant, with potential to pervasively effect deleterious changes on an organismal level.
Acknowledgments
This work was supported by the NIH Common Fund Program, grant U01CA200147, as a Transformative Collaborative Project Award (TCPA) to TCPA-2017-NERETTI and NIH/NIA grant 1R01AG050582-01A1 to NN, and by an Initiative to Maximize Student Development (IMSD) grant from Brown University to JH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Belmont AS, Mitotic chromosome scaffold structure: new approaches to an old controversy, Proc Natl Acad Sci U S A 99(25) (2002) 15855–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].H. E, “Das Heterochromatin der Moose: Bornträger.” Planta (1928).
- [3].Nagano T, Lubling Y, Varnai C, Dudley C, Leung W, Baran Y, Mendelson Cohen N, Wingett S, Fraser P, Tanay A, Cell-cycle dynamics of chromosomal organization at single-cell resolution, Nature 547(7661) (2017) 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J, Organization of the mitotic chromosome, Science 342(6161) (2013) 948–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, Mirny LA, Dekker J, A pathway for mitotic chromosome formation, Science 359(6376) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gorkin DU, Leung D, Ren B, The 3D genome in transcriptional regulation and pluripotency, Cell Stem Cell 14(6) (2014) 762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J, Comprehensive mapping of long-range interactions reveals folding principles of the human genome, Science 326(5950) (2009) 289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ay F, Noble WS, Analysis methods for studying the 3D architecture of the genome, Genome Biol 16 (2015) 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL, A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping, Cell 159(7) (2014) 1665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lesne A, Riposo J, Roger P, Cournac A, Mozziconacci J, 3D genome reconstruction from chromosomal contacts, Nat Methods 11(11) (2014) 1141–3. [DOI] [PubMed] [Google Scholar]
- [11].Varoquaux N, Ay F, Noble WS, Vert JP, A statistical approach for inferring the 3D structure of the genome, Bioinformatics 30(12) (2014) i26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rieber L, Mahony S, miniMDS: 3D structural inference from high-resolution Hi-C data, Bioinformatics 33(14) (2017) i261–i266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pombo A, Dillon N, Three-dimensional genome architecture: players and mechanisms, Nat Rev Mol Cell Biol 16(4) (2015) 245–57. [DOI] [PubMed] [Google Scholar]
- [14].Soler-Oliva ME, Guerrero-Martinez JA, Bachetti V, Reyes JC, Analysis of the relationship between coexpression domains and chromatin 3D organization, PLoS Comput Biol 13(9) (2017) e1005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, Geeting KP, Gnirke A, Melnikov A, McKenna D, Stamenova EK, Lander ES, Aiden EL, Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes, Proc Natl Acad Sci U S A 112(47) (2015) E6456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T, Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome, Nat Genet 37(1) (2005) 31–40. [DOI] [PubMed] [Google Scholar]
- [17].Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, Ren B, A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome, Cell Rep 17(8) (2016) 2042–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Niccoli T, Partridge L, Ageing as a risk factor for disease, Curr Biol 22(17) (2012) R741–52. [DOI] [PubMed] [Google Scholar]
- [19].Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G, The hallmarks of aging, Cell 153(6) (2013) 1194–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM, Cellular senescence in aging primates, Science 311(5765) (2006) 1257. [DOI] [PubMed] [Google Scholar]
- [21].Coppe JP, Desprez PY, Krtolica A, Campisi J, The senescence-associated secretory phenotype: the dark side of tumor suppression, Annu Rev Pathol 5 (2010) 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM, Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders, Nature 479(7372) (2011) 232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhu Y, Tchkonia T, Fuhrmann-Stroissnigg H, Dai HM, Ling YY, Stout MB, Pirtskhalava T, Giorgadze N, Johnson KO, Giles CB, Wren JD, Niedernhofer LJ, Robbins PD, Kirkland JL, Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors, Aging Cell 15(3) (2016) 428–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM, Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements, Aging Cell 12(2) (2013) 247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Criscione SW, Teo YV, Neretti N, The Chromatin Landscape of Cellular Senescence, Trends Genet 32(11) (2016) 751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cristofalo VJ, Lorenzini A, Allen RG, Torres C, Tresini M, Replicative senescence: a critical review, Mech Ageing Dev 125(10–11) (2004) 827–48. [DOI] [PubMed] [Google Scholar]
- [27].Criscione SW, De Cecco M, Siranosian B, Zhang Y, Kreiling JA, Sedivy JM, Neretti N, Reorganization of chromosome architecture in replicative cellular senescence, Sci Adv 2(2) (2016) e1500882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chandra T, Ewels PA, Schoenfelder S, Furlan-Magaril M, Wingett SW, Kirschner K, Thuret JY, Andrews S, Fraser P, Reik W, Global reorganization of the nuclear landscape in senescent cells, Cell Rep 10(4) (2015) 471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, Catanzaro JM, Ricketts MD, Lamark T, Adam SA, Marmorstein R, Zong WX, Johansen T, Goldman RD, Adams PD, Berger SL, Autophagy mediates degradation of nuclear lamina, Nature 527(7576) (2015) 105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chen H, Zheng X, Zheng Y, Age-associated loss of lamin-B leads to systemic inflammation and gut hyperplasia, Cell 159(4) (2014) 829–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bank EM, Gruenbaum Y, The nuclear lamina and heterochromatin: a complex relationship, Biochem Soc Trans 39(6) (2011) 1705–9. [DOI] [PubMed] [Google Scholar]
- [32].Arancio W, Pizzolanti G, Genovese SI, Pitrone M, Giordano C, Epigenetic involvement in Hutchinson-Gilford progeria syndrome: a mini-review, Gerontology 60(3) (2014) 197–203. [DOI] [PubMed] [Google Scholar]
- [33].Cao K, Capell BC, Erdos MR, Djabali K, Collins FS, A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells, Proc Natl Acad Sci U S A 104(12) (2007) 4949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fudenberg G, Getz G, Meyerson M, Mirny LA, High order chromatin architecture shapes the landscape of chromosomal alterations in cancer, Nat Biotechnol 29(12) (2011) 1109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Madakashira BP, Sadler KC, DNA Methylation, Nuclear Organization, and Cancer, Front Genet 8 (2017) 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kaiser VB, Semple CA, When TADs go bad: chromatin structure and nuclear organisation in human disease, F1000Res 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Del Campo L, Hamczyk MR, Andres V, Martinez-Gonzalez J, Rodriguez C, A. en nombre del Grupo de trabajo de Biologia Vascular de la Sociedad Espanola de, Mechanisms of vascular aging: What can we learn from Hutchinson-Gilford progeria syndrome?, Clin Investig Arterioscler (2018). [DOI] [PubMed]
- [38].Robin JD, Ludlow AT, Batten K, Magdinier F, Stadler G, Wagner KR, Shay JW, Wright WE, Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances, Genes Dev 28(22) (2014) 2464–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Stadler G, Rahimov F, King OD, Chen JC, Robin JD, Wagner KR, Shay JW, Emerson CP Jr., Wright WE, Telomere position effect regulates DUX4 in human facioscapulohumeral muscular dystrophy, Nat Struct Mol Biol 20(6) (2013) 671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suva ML, Bernstein BE, Insulator dysfunction and oncogene activation in IDH mutant gliomas, Nature 529(7584) (2016) 110–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Meaburn KJ, Spatial Genome Organization and Its Emerging Role as a Potential Diagnosis Tool, Front Genet 7 (2016) 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bakhoum SF, Kabeche L, Compton DA, Powell SN, Bastians H, Mitotic DNA Damage Response: At the Crossroads of Structural and Numerical Cancer Chromosome Instabilities, Trends Cancer 3(3) (2017) 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Erdal E, Haider S, Rehwinkel J, Harris AL, McHugh PJ, A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1, Genes Dev 31(4) (2017) 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, Wu H, Berger SL, Adams PD, Lysosome-mediated processing of chromatin in senescence, J Cell Biol 202(1) (2013) 129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, Takasugi M, Watanabe S, Kanemaki MT, Obuse C, Hara E, Exosomes maintain cellular homeostasis by excreting harmful DNA from cells, Nat Commun 8 (2017) 15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Andriani GA, Almeida VP, Faggioli F, Mauro M, Tsai WL, Santambrogio L, Maslov A, Gadina M, Campisi J, Vijg J, Montagna C, Whole Chromosome Instability induces senescence and promotes SASP, Sci Rep 6 (2016) 35218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].McDuffie NG, Nuclear blebs in human leukaemic cells, Nature 214(5095) (1967) 1341–2. [DOI] [PubMed] [Google Scholar]
- [48].Swanson EC, Manning B, Zhang H, Lawrence JB, Higher-order unfolding of satellite heterochromatin is a consistent and early event in cell senescence, J Cell Biol 203(6) (2013) 929–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, Duran M, Pauli C, Shaw C, Chadalavada K, Rajasekhar VK, Genovese G, Venkatesan S, Birkbak NJ, McGranahan N, Lundquist M, LaPlant Q, Healey JH, Elemento O, Chung CH, Lee NY, Imielenski M, Nanjangud G, Pe’er D, Cleveland DW, Powell SN, Lammerding J, Swanton C, Cantley LC, Chromosomal instability drives metastasis through a cytosolic DNA response, Nature 553(7689) (2018) 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Konety BR, Getzenberg RH, Nuclear structural proteins as biomarkers of cancer, J Cell Biochem Suppl 32–33 (1999) 183–91. [DOI] [PubMed] [Google Scholar]
- [51].Sakthivel KM, Sehgal P, A Novel Role of Lamins from Genetic Disease to Cancer Biomarkers, Oncol Rev 10(2) (2016) 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].De Magistris P, Antonin W, The Dynamic Nature of the Nuclear Envelope, Curr Biol 28(8) (2018) R487–R497. [DOI] [PubMed] [Google Scholar]
- [53].Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J, Nuclear envelope rupture and repair during cancer cell migration, Science 352(6283) (2016) 353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lim S, Quinton RJ, Ganem NJ, Nuclear envelope rupture drives genome instability in cancer, Mol Biol Cell 27(21) (2016) 3210–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lammerding J, Wolf K, Nuclear envelope rupture: Actin fibers are putting the squeeze on the nucleus, J Cell Biol 215(1) (2016) 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Reddy KL, Zullo JM, Bertolino E, Singh H, Transcriptional repression mediated by repositioning of genes to the nuclear lamina, Nature 452(7184) (2008) 243–7. [DOI] [PubMed] [Google Scholar]
- [57].Shen YJ, Le Bert N, Chitre AA, Koo CX, Nga XH, Ho SS, Khatoo M, Tan NY, Ishii KJ, Gasser S, Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells, Cell Rep 11(3) (2015) 460–73. [DOI] [PubMed] [Google Scholar]
- [58].Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR, Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein, Genes Dev 17(24) (2003) 3017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hernandez-Segura A, Nehme J, Demaria M, Hallmarks of Cellular Senescence, Trends Cell Biol (2018). [DOI] [PubMed]
- [60].Barascu A, Le Chalony C, Pennarun G, Genet D, Zaarour N, Bertrand P, Oxydative stress alters nuclear shape through lamins dysregulation: a route to senescence, Nucleus 3(5) (2012) 411–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD, The role of nuclear lamin B1 in cell proliferation and senescence, Genes Dev 25(24) (2011) 2579–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tominaga K, The emerging role of senescent cells in tissue homeostasis and pathophysiology, Pathobiol Aging Age Relat Dis 5 (2015) 27743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].van Deursen JM, The role of senescent cells in ageing, Nature 509(7501) (2014) 439–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW, Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence, Cell 113(6) (2003) 703–16. [DOI] [PubMed] [Google Scholar]
- [65].Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, Simithy J, Lan Y, Lin Y, Zhou Z, Capell BC, Xu C, Xu M, Kieckhaefer JE, Jiang T, Shoshkes-Carmel M, Tanim K, Barber GN, Seykora JT, Millar SE, Kaestner KH, Garcia BA, Adams PD, Berger SL, Cytoplasmic chromatin triggers inflammation in senescence and cancer, Nature 550(7676) (2017) 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yang H, Wang H, Ren J, Chen Q, Chen ZJ, cGAS is essential for cellular senescence, Proc Natl Acad Sci U S A 114(23) (2017) E4612–E4620. [DOI] [PMC free article] [PubMed] [Google Scholar]