Abstract
Background: We evaluated the strategies of resources partitioning among species, dietary overlap and niche breadth in an assemblage of carnivores integrated by top predators (Puma concolor and Panthera onca) and mesopredators(Leoparduspardalis,Leoparduswiedii,Puma yagouaroundi,Nasuanarica andUrocyoncinereoargenteus). The aim of this study was to investigate the mechanisms explaining the coexistence among species at a temperate zone in central Mexico.
Results: We collected 259 scats of carnivores and identified 45 food items. The analysis showed the common consumptionof mammals in the assemblage, and the correspondence analysis showed three guilds: 1) top predators associated with the use of medium-sized mammals and the exclusive consumption of large mammals, 2) carnivore mesopredators associated with the consumption of small mammals and birds and 3) omnivore mesopredators associated with the consumption of arthropods and plants. The dietary overlap analysis indicated a low overlap between guilds and a high overlap within them. Top predators were specialist foragers, whilst the carnivore mesopredators showed generalist consumption.
Conclusions: The coexistence in this carnivore assemblage seems to be related to body size, morphology and prey segregation because such characteristics suggest the presence of three guilds. We observed high dietary overlap within guilds and resource partitioning betweenguilds.
Keywords: Coexistence, Community, Niche breadth, Niche overlap, Trophic interactions
BACKGROUND
Theevaluation of feeding relationships between predators, prey and their environment is used to identifyfactors structuringcommunities (Carvalho and Gomes 2004). To reach this aim, it is necessary to analyse trophic interac- tions between species that use similar resources, namely guilds (Root 1967; Jaksic 1981). The guild approach allows to explain the association among species, the ecological structure and community dynamics (Carvalho and Gomes 2004; Zapata et al. 2007). Ecological interactions and the mechanisms involved in resource partitioning and species coexistenceare studied in relation to food, space or time segregation(MacArthur and Levins 1967; Schoener 1974). The knowledge of trophic structure in carnivore commu- nities improves their management andconservation, particularlyin scarcely studied communities such as tem- perate zones (Ray and Sunquist 2001; Guerrero etal. 2002; Carvalho and Gomes 2004).
The predation theory predicts a convergence of trophic niches when resources are abundant (Schoener 1982),such that the dietary overlap might suggest high abundance of resources in the environment and moder- atecompetition (Silva-Pereira et al. 2011). Because carni- vore communities are structured by intraguild predation and competition, all members play important ecological roles in the ecosystem and show different strategies to co- exist (Bianchi et al. 2011). Likewise, the presence of gener- alist predators in the ecosystem induces the exploitation of a wide range of prey by other carnivores (Pianka 1973). Thus, the opportunistic ability of some species represents an advantage in the use of resources and favours coexist- ence on the guild (Jaksic et al. 1996; Guerrero et al. 2002).
Weconsidered that the body size of predators influencesthe community structure and resource sharing between species (Simberloff and Dayan 1991;Hayward etal. 2006). We defined the top predators as species occu- pyingthe apex trophic position in a community (large- bodied) and mesopredators as species occupying trophic positions below them (medium-bodied, Ritchie and Johnson2009). We investigated the feeding strategies that may explain the coexistence among members of a carni- vore assemblage at a temperate zone of central Mexico in relationto morphological similarity. As the body size of predators is thought to shape community structure, we expectedhigh dietary overlap between Pumaconcolor and Panthera onca(toppredators), between Leopardusparda- lis, Leopardus wiedii and Puma yagouaroundi (carnivore mesopredators) and between Nasua narica and Urocyon cinereoargenteus (omnivoremesopredators).
We obtained data regarding diet composition,niche overlapand niche breadth to identify resource partitioning strategies. The study was conducted through the analysis of field-collected scats. The most effective identification of scats has been performed using molecular techniquessuch as bile acids identification using thin-layer chromatog- raphy(TLC) or DNA extraction (Ernest et al. 2000; Cazón andSühring 1999). TLC has been suggested as an effective means of identification of carnivore scats, since the pro- filesof faecal bile acids have been shown to be species- specific.Species often display a unique bile acid profile, so TLCuses specific differences in bile acid profiles to distinguishbetween species (Major et al. 1980; Fernández et al. 1997; Taber et al. 1997; Ray and Sunquist 2001). This isan inexpensive and non-invasive technique that has beeneffective with carnivore species, although some stud- ies have described a number of limitations when applied in omnivores due to the presence of vegetal pigments (Cazónand Sühring 1999). Our study is the first to de- scribe the diet of small felids (L.pardalis,L.wiedii andP. yagouaroundi)using TLC, which improves species assign- ment and allows us to analyse the associationstrategies amongguilds.
METHODS
Study area
SierraNanchititla Natural Park (SNNP) is located in cen- tral Mexico, between extreme coordinates 100° 36′ 49′′- 100° 16′ 03′′ west longitude and 18° 45′ 13′′-19° 04′ 22′′ northlatitude. Altitude ranges from 410 to 2,080 m a.s.l. andcovers 663.87 km2 (Figure 1). The vegetation types areoak forest (30%), induced grasslands (30%), deciduous tropical forest (18%), pine-oak forest (17%) and cultiva- tions (4%, Monroy-Vilchis et al. 2008). Overall, 53 species ofmammals have been recorded, belonging to 6 orders, including17 families and 38 genera; of these, 14 species arecarnivores, 11 are endemic and 9 are endangered or consideredrare (Monroy-Vilchis et al. 2011).
Fig. 1.
Figure 1 Geographic position of Sierra Nanchititla Natural Park (SNNP) in central Mexico.
Scat identification
FromSeptember 2009 to March 2012, carnivore scats were collected over different paths distributed through- outthe SNNP. The scats were identified considering the diameter, associated traces (tracks or scrapes) and ex- tractionof bile acid using TLC (Salame-Méndez et al. 2012). We collected two known-origin reference scats from individual P.concolor (threeindividuals) and P. onca(threeindividuals) and one known-origin reference scat from individual L.pardalis (sixindividuals), L.wiedii (eightindividuals) and P.yagouaroundi (sixindividuals). Thereference bile acid profiles were obtained through the following methods. We used a gram approximately of each scat collected and preserved it in ethanol; the solution was mixedand 1 μlof the supernatants was used to extract bile acids. We used two chromatography techniques to separateall bile acids by polarity, each one with a different mobile phase: 1) chloroform:methanol:acetic acid (80:12:0.5 v/v)and 2) chloroform:ethanol:acetic acid (80:45:3 v/v).
Samples were run against a known mixture of eight bile acids (cholic acid, chenodeoxycholic acid, deoxy- cholic acid, lithocholic acid, taurodeoxycholicacid, taurocholicacid, glycolic acid and dehydrocholic acid) and cholesterol, from Sigma-Aldrich® (St. Louis, MO, USA). The bile acid spots of the mixtures were identified and RF values were calculated as the ratio of the dis- tance moved by the solute (i.e. bile acid) and the dis- tance moved by the solvent front along the TLC plate. Finally, each bile acid profile obtained from field- collected scats was compared against reference bileacid profilesextracted from known-origin reference scats.
Diet composition
Thescat analysis was based on the standard method- ology reported by Monroy-Vilchis et al. (2009a) and Gómez-Ortiz et al. (2011). Food items were identified considering remains in scats (hair, bones, claws, teeth, seeds, feathers etc.). Mammalian identifications were carried out through hair identification (Monroy-Vilchis and Rubio-Rodríguez 2003). Birds, reptiles, arthropods andfruits were identified by comparing them with speci- mencollections.
Thecontribution of each food item was presented as follows: the frequency of occurrence (FO), defined as the percentage of presences of a given food item in the total number of scats, and the percentage of occurrence (PO), obtained as the number of occurrences of a given food itemdivided by the total food items identified.
Theresource partitioning among species was evalu- ated using an ordination procedure that permits the ar- rangement of similar species (Ray and Sunquist 2001). Weapplied a correspondence analysis using the frequency of occurrence of each food item. A matrix was constructed by grouping food items; mammals (commonly consumed in the assemblage) were separated into three groups ac- cordingto body size: large mammals (>6.13 kg, maximum biomass ingested by puma/day, Monroy-Vilchis et al. 2013), medium-sized mammals (>1 and <6.13 kg) and small mammals (<1 kg), birds, reptiles, arthropods and plants.
The estimates of niche were calculated through fre- quency of occurrence; niche breadth was calculated using Levins' index (B′), which ranges from 0 (specialist foraging)to 1 (generalist foraging). Dietary overlap was measuredusing Pianka's index; this ranges from 0 (total separation)to 1 (total overlap). Pairwise niche overlap values were calculated in the software EcoSim 7.0 (Gotelli and Entsminger 2005). We compared the ob- served overlap in the diet of this assemblage with null models of dietary overlap generated using the niche overlap module in this software. To determine this prob- ability, 10,000 interactions were randomly generated with a level of significance of 0.05. The RA3 algorithm was used because it preserves the specialisation ofeach species, but allows for the potential use of other re- sources(Winemiller and Pianka 1990). We assumed that allfood items were equally available to all species, due to the lack of data regarding their availability.
RESULTS
Intotal, 263 carnivore scats were identified. The largest number of scats was from U. cinereoargenteus (90), whilst the rest were from P. concolor (54), P.yagouar- oundi (42), L. pardalis (21), N. narica (20), L. wiedii (16) andP.onca (16).All scats, excluding four belonging to Canislatrans (1)and Procyonlotor (3),were used in the dietary analysis.
We found 45 food items in a total of 259 carnivore scatsexamined. We identified 51.10% of food items be- longingto mammals, 24.40% to plants, 12% to arthropods, 8.10% to birds and 4.40% to reptiles. One mammal, one birdand two plants were not identified.
Trophic structure of carnivore assemblage
Itwas found that all members of this carnivore assem- blage consumed mammals, with differences in the body size and species consumed, although these were more frequentin diet of felids (>50%). Particularly, felids showed a key association with Dasypus novemcinctus. Overall, reptilesand birds were rarely consumed, and the occa- sional consumption of lagomorphs was evident in this assemblage.
The correspondence analysis generated two axes that explainapproximately 90% of the total variation in diet. Axis I (68.27% of variance) showed a clear separation of top predators (P.oncaand P.concolor),characterised by the consumption of large- and medium-sized mammals. Axis II (19.81%) clearly separates mesopredators into twoguilds: 1) carnivore mesopredators (P.yagouaroundi, L.wiedii andL.pardalis),associated with the consump- tion of small mammals and birds, and 2) omnivore mesopredators (N. narica and U. cinereoargenteus), asso- ciatedwith the consumption of arthropods and plants (fruits,Table 1 and Figure 2).
Table 1 Diet of carnivore assemblage analysed through percentage of occurrence and frequency of occurrence, central Mexico.
| Species | Puma concolor | Panthera onca | Leopardus wiedii | Leopardus pardalis | Puma yagouaroundi | Nasua narica | Urocyon cinereoargenteus | ||||||||
| (n=54) | (n=16) | (n = 16) | (n = 21) | (n = 42) | (n = 20) | (n = 90) | |||||||||
| PO | FO | PO | FO | PO | FO | PO | FO | PO | FO | PO | FO | PO | FO | ||
| Mammals | |||||||||||||||
| Baiomys musculus | - | - | - | - | - | - | 7.41 | 9.52 | - | - | - | - | - | - | |
| Bassariscus astutus | - | - | - | - | 6.67 | 6.25 | - | - | 1.78 | 2.38 | - | - | - | - | |
| Capra hircus | 3.33 | 3.7 | 13.04 | 18.75 | - | - | - | - | - | - | - | - | - | - | |
| Conepatus leuconotus | - | - | 4.35 | 6.25 | - | - | - | - | - | - | - | - | - | - | |
| Dasypus novemcinctus | 38.33 | 42.59 | 39.13 | 56.25 | 6.67 | 6.25 | 7.41 | 9.52 | 5.36 | 7.14 | - | - | - | - | |
| Didelphis virginiana | 1.67 | 1.85 | - | - | 6.67 | 6.25 | 3.7 | 4.76 | - | - | - | - | - | - | |
| Geomyidae | - | - | - | - | - | - | - | - | 1.79 | 2.38 | - | - | - | - | |
| Liomys irroratus | 1.67 | 1.85 | - | - | 33.33 | 31.25 | 22.22 | 28.57 | 16.07 | 21.43 | 11.11 | 20 | 8.47 | 16.67 | |
| Nasua narica | 25 | 27.78 | 13.04 | 18.75 | - | - | - | - | - | - | - | - | - | - | |
| Odocoileus virginianus | 5 | 5.56 | 4.35 | 6.25 | - | - | - | - | 5.36 | 7.14 | - | - | - | - | |
| Peromyscus maniculatus | - | - | - | - | - | - | - | - | 1.79 | 2.38 | - | - | 0.56 | 1.11 | |
| Peromyscus melanotis | - | - | - | - | - | - | - | - | 1.79 | 2.38 | - | - | - | - | |
| Peromyscus aztecus | - | - | - | - | - | - | - | - | 3.57 | 4.76 | - | - | 0.56 | 1.11 | |
| Peromyscus melanophrys | - | - | - | - | - | - | - | - | 3.57 | 4.76 | 2.78 | 5 | 0.56 | 1.11 | |
| Peromyscus sp. | - | - | - | - | - | - | 7.41 | 9.52 | - | - | - | - | 1.69 | 3.33 | |
| Procyon lotor | 1.67 | 1.85 | 4.35 | 6.25 | - | - | - | - | 5.36 | 7.14 | 2.78 | 5 | - | - | |
| Sciurus aureogaster | - | - | - | - | 13.33 | 12.5 | - | - | 1.79 | 2.38 | - | - | 0.56 | 1.11 | |
| Sylvilagus cunicularius | 6.67 | 7.41 | - | - | 6.67 | 6.25 | 3.7 | 4.76 | 1.79 | 2.38 | - | - | 1.13 | 2.22 | |
| Sylvilagus floridanus | 6.67 | 7.41 | 13.04 | 18.75 | - | - | 3.7 | 4.76 | 8.93 | 11.9 | 2.78 | 5 | 1.69 | 3.33 | |
| Sylvilagus spp. | 3.33 | 3.7 | - | - | 6.67 | 6.25 | 3.7 | 4.76 | 12.5 | 16.67 | - | - | 1.13 | 2.22 | |
| Spermophilus variegatus | - | - | - | - | - | - | - | - | 1.79 | 2.38 | - | - | 1.13 | 2.22 | |
| Reitrodonthomys sp. | - | - | - | - | - | - | - | - | 1.79 | 2.38 | - | - | - | - | |
| Unidentified mammal | 5 | 5.56 | - | - | - | - | - | - | 1.79 | 2.38 | - | - | - | - | |
| Reptiles | |||||||||||||||
| Ctenosaura pectinata | 1.67 | 1.85 | 4.35 | 6.25 | - | - | - | - | - | - | - | - | 0.56 | 1.11 | |
| Sceloporus sp. | - | - | - | - | - | - | - | - | - | - | - | - | 0.56 | 1.11 | |
| Arthropods | |||||||||||||||
| Coleoptera | - | - | - | - | - | - | - | - | 3.57 | 4.76 | 13.89 | 25 | 5.65 | 11.11 | |
| Crustacea | - | - | - | - | - | - | - | - | - | - | - | - | 0.56 | 1.11 | |
| Formicidae | - | - | - | - | - | - | 3.7 | 4.76 | 1.79 | 2.38 | 11.11 | 20 | 3.39 | 6.67 | |
| Hemiptera | - | - | - | - | - | - | - | - | 1.79 | 2.38 | - | - | - | - | |
| Orthoptera | - | - | - | - | 13.33 | 12.5 | 18.52 | 23.81 | 3.57 | 4.76 | 36.11 | 65 | 24.86 | 48.89 | |
| Plants | |||||||||||||||
| Byrsonima crassifolia | - | - | - | - | - | - | 3.7 | 4.76 | 1.79 | 2.38 | 2.78 | 5 | 1.13 | 2.22 | |
| Capsicum sp. | - | - | - | - | - | - | - | - | - | - | 2.78 | 5 | - | - | |
| Crescienta sp. | - | - | - | - | - | - | - | - | - | - | - | - | 5.08 | 10 | |
| Ficus sp. | - | - | - | - | - | - | 3.7 | 4.76 | - | - | 2.78 | 5 | 5.08 | 10 | |
| Lysiloma sp. | - | - | - | - | - | - | - | - | - | - | - | - | 1.13 | 2.22 | |
| Myrmeleon sp. | - | - | - | - | 6.67 | 6.25 | - | - | - | - | - | - | - | - | |
| Physalis sp. | - | - | - | - | - | - | 3.7 | 4.76 | - | - | - | - | 0.56 | 1.11 | |
| Psidium sp. | - | - | - | - | - | - | 3.7 | 4.76 | 1.79 | 3.7 | 11.11 | 20 | 23.73 | 46.67 | |
| Zea mays | - | - | - | - | - | - | - | - | - | - | - | - | 3.39 | 6.67 | |
| Unidentified plant 1 (seeds) | - | - | - | - | - | - | - | - | - | - | - | - | 1.69 | 3.33 | |
| Unidentified plant 2 (shells) | - | - | - | - | - | - | - | - | - | - | - | - | 1.13 | 2.22 | |
| - | - | - | - | - | - | - | - | 1.79 | \ 2.38 | - | - | - | - | ||
| Birds | |||||||||||||||
| Buteogallus antharacincus | - | - | - | - | - | - | 3.7 | 4.76 | - | - | - | - | - | - | |
| Seiurus sp. | - | - | - | - | - | - | - | - | 1.79 | 2.38 | - | - | - | - | |
| Strigidae | - | - | - | - | - | - | - | - | 7.14 | 9.52 | - | - | 3.95 | 7.78 | |
| Unidentified bird | - | - | 4.35 | 6.25 | - | - | - | - | - | - | - | - | - | - | |
n, scats number.
Fig. 2.
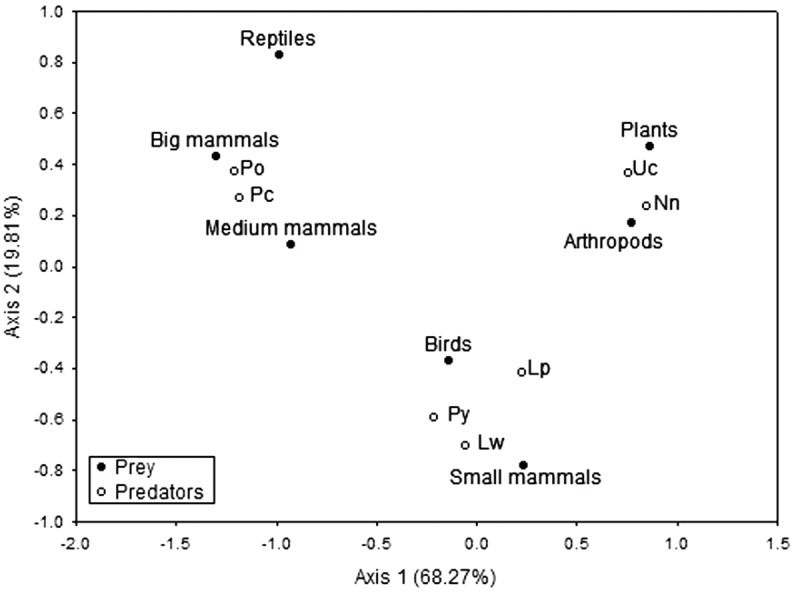
Figure 2 Trophic guild structure of carnivore assemblage from SNNP. Predators: Po, Panthera onca; Pc, Puma concolor; Lw, Leopardus wiedii; Lp, Leopardus pardalis; Py, Puma yagouaroundi; Nn, Nasua narica; and Uc, Urocyon cinereoargenteus. Food item: big mammals (>6.13 kg), medium mammals (1 kg < 6.13 kg), small mammals (<1 kg), arthropods, birds, plants and reptiles.
The dietary niche breadth calculated in thisassem- blagedescribes top predators (P.concolor andP.onca) as a specialist group and carnivore mesopredators(L. wiedii, L. pardalis and P. yagouaroundi) as a generalist group. Finally, omnivore mesopredators (N. naricaand U. cinereoargenteus) had an intermediate niche breadth betweenboth guilds. Specifically, P.concolor showedthe most specialist foraging strategies and L.wiedii themost generalist (Table 2).
Table 2 Dietary breadth and overlap niche in the diet of a carnivore assemblage in central Mexico.
| Overlap | Pumaconcolor | Pantheraonca | Leoparduswiedii | Leoparduspardalis | Pumayagouaroundi | Nasuanarica | Urocyoncinereoargenteus |
| P.concolor | 1 | 0.99 | 0.56 | 0.45 | 0.62 | 0.05 | 0.09 |
| P.onca | 1 | 0.53 | 0.43 | 0.61 | 0.04 | 0.09 | |
| L.wiedii | 1 | 0.94 | 0.96 | 0.47 | 0.42 | ||
| L.pardalis | 1 | 0.92 | 0.67 | 0.69 | |||
| P.yagoaroundi | 1 | 0.39 | 0.41 | ||||
| N.narica | 1 | 0.86 | |||||
| U.cinereoargenteus | 1 | ||||||
| Breadth | 0.1 | 0.24 | 0.86 | 0.71 | 0.54 | 0.42 | 0.42 |
Themean dietary overlap was 0.54, indicating an inter- mediate overlap in the assemblage. Comparison with null modelsindicated that the observed overlap was sig- nificantlygreater than the mean simulated overlap (0.37, p=0.002), which means that it was not random. The high- est overlap (0.86 to 0.99) occurred within generated guilds. In contrast, the lowest overlap was among guilds, particu- larlywhen top predators and omnivore mesopredators werecompared (0.05 to 0.09), which is consistent with the correspondenceanalysis results (Table 2).
DISCUSSION
Trophic structure of carnivore assemblage
Speciesin a community tend to organise according to body size, which leads them to consume prey with spe- cific characteristics. Therefore, body size differences of predators may influence their ability to hunt large prey, favouringtop predators, and restricts the mesopredators to small prey (Konecny 1989; Hayward et al. 2006; Bianchi etal. 2011; Silva-Pereira et al. 2011). Our results suggest that the organisation of species in this assemblage should be explained by body size, because this parameter sepa- rates top predators from mesopredators, favours the dif- ferential use of feeding resources and potentially reduces competition among guilds (Davies et al. 2007). Top preda- tors (P. onca and P. concolor) were associated with the consumptionof medium and large mammals and were similar to reports from other sites of sympatry of these species (Núñez et al. 2000; Scognamillo et al. 2003; Harmsen et al. 2009).
Thesimilarity in body size between species increases the likelihood to hunt the same prey, but our study showsthat the coexistence of mesopredators can also be explained by ecological foraging segregation. This is sup- ported by the fact that opportunism and plasticity in feeding of mesopredators reduces convergence in the useof similar resources (Fedriani et al. 1999; Guerrero et al. 2002; Carvalho and Gomes 2004). The diet of car- nivore mesopredators was no different to other records where small prey were consumed (Guerrero et al. 2002; Wang 2002; Abreu et al. 2008; Bianchi et al. 2011). Moreover, omnivore mesopredators have been charac- terised by the consumption of fruits and arthropods (Valenzuela 1998; Fedriani et al. 1999; Guerrero et al. 2002).
Dietary breadth and overlap
Nichebreadth has been used as a parameter to reference thedominance of species, so that the dominant species chooses times or sites in which their main prey can be found.Instead, subordinate species are confined to times orspaces in which the dominant species is absent, be- coming a generalist forager in both niche dimensions. Resourcesegregation can also be modelled by evolution- ary forces when the dominant species are sympatric leadingto a reduction in their niche (Caro and Stoner 2003; Hayward and Slotow 2009). The niche breadth patternsin the assemblage we studied support the coex- istenceof predators and the community structure, placingtop predators as specialist, carnivore mesopreda- tors as generalist and omnivore mesopredators with intermediate breadth.
Thehigh dietary overlap between species with similar morphology and diet are reported in most of the studies where these species are sympatric (Núñez et al. 2000; Guerreroet al. 2002; Scognamillo et al. 2003; Silva- Pereira et al. 2011; Gómez-Ortiz and Monroy-Vilchis 2013), because throughout their distribution, there is a similar prey base. In SNNP, competition between top predatorsshould be taken with caution due to the low abundance of jaguar in this site and different activity patterns (Monroy-Vilchis et al. 2009b; Soria-Díaz et al. 2010); also, there are insufficient data to corroborate competition, so it is necessary to carefully evaluate re- source availability and the requirements of each preda- tor. The absence or low density of a top predator lets competitive release of subordinate predators and theex- pansionof their niches by changing predation patterns (Moreno et al. 2006). In this case, such release is not ob- served, possibly due to the low diversity of large prey (Odocoileus virginianus)and the existence of a high di- versityof small and medium prey (Monroy-Vilchis et al. 2011).
In carnivore mesopredators, there is a highoverlap thatcan be supported by an adequate availability of small rodents. In SNNP, rodents are represented by a high species richness (Muridae: 13spp., Heteromidae: 1 sp., Monroy-Vilchis et al. 2011), which may favour the coexist- ence, as reported in Brazil (Silva-Pereira et al. 2011). Omnivore mesopredators also have a high overlap;how- ever, N.narica consumedmore orthopterans than fruits (Psidiumsp.)in contrast to U.cinereoargenteus.
The occasional consumption of birds, reptiles and somedomestic species has been reported in the diet of mesopredators (Emmons 1987; Wang 2002; Abreu etal. 2008).Moreover, our data suggest an occasional con- sumption on domestic goat (Capra hircus) and corn (Zea mays). In SNNR, human-wildlife conflicts have been recorded and individuals of P. concolor have been killed(Zarco-González et al. 2012), as well as the unjus- tified hunting of mesopredators (P. yagouaroundi, N. narica and U. cinereoargenteus) due to their supposed relation with predation on poultry and corn crops (Romero-Balderas et al. 2006).
Additional data on resource availability could provide moreaccurate answers regarding competition as a struc- turingmechanism in the assemblage we studied. In a previous study of prey selection in SNNP, we found that P.concolor selectsits prey (Gómez-Ortiz and Monroy- Vilchis2013). It is therefore necessary to focus future studies on assessing the availability of resources and identifyingthe role of key species, since the management and conservation of biodiversity must be maintained for the persistence of carnivores and those species that have a key role, such as D.novemcinctus inthis study.
CONCLUSIONS
The analysis allowed us to identify the main feeding strategiesof a carnivore assemblage in central Mexico. The coexistence in this carnivore assemblage appearsto berelated to body size, morphology and prey segrega- tion, because such characteristics suggest the presence ofthree guilds. We observed high dietary overlap within guilds and resource partitioning between guilds.
Acknowledgments
Acknowledgements
We thank the Mexicans and the ‘Consejo Nacional de Ciencia y Tecnología’ for the economic support for this study through project funding (105254) and scholarship (255868). We also thank ‘Programa de Mejoramiento del Profesorado’ for funding the project 103.5/10/0942. We thank all students and rangers of Sierra Nanchititla Biological Station (Universidad Autónoma del Estado de México) for their support in the field and the ‘Comisión Estatal de Parques Naturales y de la Fauna’ for providing us with carnivore scats for identification by bile acids. We express thanks to A. Salame-Méndez and H. Domínguez-Vega for their support in this study.
Footnotes
Authors’ contributions: OM-V and YG-O conceived and designed the surveys and obtained the data. YG-O, OM-V and GDM-M analysed and interpreted the data. YG-O, OM-V and GDM-M wrote the manuscript. All authors read and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
References
- Abreu K C, Moro-Rios F, Silva-Pereira J E, Miranda Jmd, Jablonski Efj, Passos F C. Feeding habits of ocelot (Leopardus pardalis) in Southern Brazil. Mamm Biol. 73:407–411. [Google Scholar]
- Bianchi R C, Rosa A F, Gatti A, Mendez S L. Diet of margay, Leopardus wiedii, and jaguarondi, Puma yagouarondi, (Carnivora: Felidae) in Atlantic Rainforest. Brazil. Zoologia. 28:127–132. [Google Scholar]
- Caro T M, Stoner C J. The potential for interspecific competition among African carnivores. Biol Conserv. 110:67–75. [Google Scholar]
- Carvalho J C, Gomes P. Feeding resources partitioning among four sympatric carnivores in the Peneda-Gerés National Park (Portugal) J Zool. 263:275–283. [Google Scholar]
- Cazón A V, Sühring S. A technique for extraction and thin layer chromatography visualization of fecal acids applied to Neotropical felid cats. Rev Biol Trop. 47:245–249. [PubMed] [Google Scholar]
- Davies T J, Meiri S, Barraclough T G, Gittleman J L. Species co-existence and character divergence across carnivores. Ecol Lett. 10:146–152. doi: 10.1111/j.1461-0248.2006.01005.x. [DOI] [PubMed] [Google Scholar]
- Emmons L H. Comparative feeding ecology of felids in a tropical rainforest. Behav Ecol Sociobiol. 20:271–283. [Google Scholar]
- Ernest H, Penedo M, May B, Sywanen S, Boyce W. Molecular tracking of mountain lions in the Yosemite Valley region in California: genetic analysis using microsatellites and fecal DNA. Molecular Ecology. 9:433–441. doi: 10.1046/j.1365-294x.2000.00890.x. [DOI] [PubMed] [Google Scholar]
- Fedriani J M, Palomares F, Delibes M. Niche relations among three sympatric Mediterranean carnivores. Oecologia. 121:138–148. doi: 10.1007/s004420050915. [DOI] [PubMed] [Google Scholar]
- Fernández G J, Corley J C, Capurro A F. Identification of cougar and jaguar feces through bile acid chromatography. J Wildl Manage. 61:506–510. [Google Scholar]
- Gómez-Ortiz Y, Monroy-Vilchis O. Feeding ecology of puma Puma concolor in Mexican montane forests with comments about jaguar Panthera onca. Wildl Biol. 19:179–187. [Google Scholar]
- Gómez-Ortiz Y, Monroy-Vilchis O, Fajardo V, Mendoza G D, Urios V. Is food quality important for carnivores? The case of Puma concolor. Anim Biol. 61:277–288. [Google Scholar]
- Gotelli N J, Entsminger G L. Burlington): Acquired Intelligence Inc. & Kesey-Bear; 2005. 7 [Google Scholar]
- Guerrero S, Badii M H, Zalapa S S, Flores A E. Dieta y nicho de alimentación del coyote, zorra gris, mapache y jaguarundi en un bosque tropical caducifolio de la costa sur del estado de Jalisco. Acta Zool Mex. 86:119–137. [Google Scholar]
- Harmsen B J, Foster R J, Silver S C, Ostro Let, Doncaster P. Spatial and temporal interactions of sympatric jaguars (Panthera onca) and (Puma concolor) in a Neotropical forest. J Mammal. 90:612–620. [Google Scholar]
- Hayward M W, Slotow R. Temporal partitioning of activity in large African carnivores: tests of multiple hypotheses. S Afr J Wildl Res. 39:109–125. [Google Scholar]
- Hayward M W, Henschel P, O'brien J, Hofmeyer M, Balme G, Kerley Gih. Prey preferences of the leopard (Panthera pardus) J Zool. 270:298–313. [Google Scholar]
- Jaksic F M. Abuse and misuse of the term "guild" in ecological studies. Oikos. 37:397–400. [Google Scholar]
- Jaksic F M, Feinsinger P, Jiménez J E. Ecological redundancy and long-term dynamics of vertebrates predators in semiarid Chile. Conserv Biol. 10:252–262. [Google Scholar]
- Konecny M J. Movement patterns and food habitats of four sympatric carnivore species in Belize, Central America. Sadhill Crane Press. pp. 243–264.
- Macarthur R H, Levins R. The limiting similarity, convergence, and divergence of coexisting species. Am Natur. 101:377–385. [Google Scholar]
- Major M, Johnson M K, Davis W S, Kellog T F. Identifying scats by recovery of bile acids. J Wildl Manage. 44:290–293. [Google Scholar]
- Monroy-Vilchis O, Rubio-Rodríguez R. Guía de identificación de mamíferos terrestres del Estado de México, a través del pelo de guardia. México): 2003. [Google Scholar]
- Monroy-Vilchis O, Zarco-González M, Rodríguez-Soto C, Suárez P, Urios V. Uso tradicional de vertebrados silvestres en la Sierra Nanchititla. México. Interciencia. 33:308–313. [Google Scholar]
- Monroy-Vilchis O, Gómez Y, Janczur M, Urios V. Food niche of Puma concolor in Central Mexico. Wildl Biol. 15:97–105. [Google Scholar]
- Monroy-Vilchis O, Rodríguez-Soto C, Zarco-González M, Urios V. Cougar and jaguar habitat use and activity patterns in central Mexico. Anim Biol. 59:145–157. [Google Scholar]
- Monroy-Vilchis O, Zarco-González M M, Ramírez-Pulido J, Aguilera-Reyes U. Diversidad de mamíferos de la Reserva Natural Sierra Nanchititla. México. Rev Mex Biodiv. 82:237–248. [Google Scholar]
- Monroy-Vilchis O, Gómez-Ortiz Y, Urios V. Expulsion rate of Puma concolor (Carnivora: Felidae) in captivity. Mamm Stud. 38:299–302. [Google Scholar]
- Moreno R S, Kays R W, Samudio R. Competitive release in diets of ocelot (Leopardus pardalis) and puma (Puma concolor) after jaguar (Panthera onca) decline. J Mammal. 87:808–816. [Google Scholar]
- Núñez R, Miller B, Lindzey F. Food habits of jaguars and pumas in Jalisco, Mexico. J Zool. 252:373–379. [Google Scholar]
- Pianka E R. The structure of lizard communities. Annu Rev Ecol Syst. 4:53–74. [Google Scholar]
- Ray J C, Sunquist M E. Trophic relations in a community of African rainforest carnivores. Oecologia. 127:395–408. doi: 10.1007/s004420000604. [DOI] [PubMed] [Google Scholar]
- Ritchie E G, Johnson C N. Predator interactions, mesopredator release and biodiversity conservation. Ecol Lett. 12:982–998. doi: 10.1111/j.1461-0248.2009.01347.x. [DOI] [PubMed] [Google Scholar]
- Romero-Balderas K G, Naranjo E J, Morales H H, Nigh R B. Daños ocasionados por vertebrados silvestres al cultivo de maíz en la selva lacandona. Interciencia. 31:276–283. [Google Scholar]
- Root R B. The niche exploitation pattern of the blue-gray gnatcatcher. Ecol Monogr. 37:317–350. [Google Scholar]
- Salame-Méndez A, Andrade-Herrera M, Zamora-Torres L, Serrano H, Sotomendoza S, Castro-Campillo A, Ramírez-Pulido J, Haro-Castellanos J. Método optimizado para evaluar ácidos biliares de muestras fecales secas o preservadas en etanol como herramienta para identificar carnívoros silvestres. Acta Zool Mex. 28:305–320. [Google Scholar]
- Schoener T W. Resource partitioning in ecological communities. Science. 185:27–39. doi: 10.1126/science.185.4145.27. [DOI] [PubMed] [Google Scholar]
- Schoener T W. The controversy over interspecific competition. Am Sci. 70:27–39. [Google Scholar]
- Scognamillo D, Maxit I, Sunquist M, Polisar J. Coexistence of jaguar (Panthera onca) and puma (Puma concolor) in a mosaic landscape in the Venezuelan llanos. J Zool. 259:269–279. [Google Scholar]
- Silva-Pereira J E, Moro-Rios R F, Bilski D R, Passos F C. Diets of three sympatric Neotropical small cats: food niche overlap and interspecies differences in prey consumption. Mamm Biol. 76:308–312. [Google Scholar]
- Simberloff D, Dayan T. The guild concept and the structure of ecological communities. Ann Rev Ecolog Syst. 22:115–143. [Google Scholar]
- Soria-Díaz L, Monroy-Vilchis O, Rodríguez-Soto C, Zarco-González M M, Urios V. Variation of abundance and density of Puma concolor in zones of high and low concentration of camera traps in central Mexico. Anim Biol. 60:361–371. [Google Scholar]
- Taber A, Novaro A, Neris N, Colman F. The food habits of sympatric jaguar and puma in the Paraguayan Chaco. Biotropica. 29:204–213. [Google Scholar]
- Valenzuela D. Natural history of the white-nosed coati, Nasua narica, in a tropical dry forest of western México. RMM. 3:26–44. [Google Scholar]
- Wang E. Diets of ocelots (Leopardus pardalis), margays (L. wiedii), and oncillas (L. tigrinus) in the Atlantic rainforest in southeast Brazil. Stud Neotrop Fauna E. 37:207–212. [Google Scholar]
- Winemiller K O, Pianka E R. Organization in natural assemblages of desert lizards and tropical fishes. Ecol Monogr. 60:27–55. [Google Scholar]
- Zapata S C, Travaini A, Ferreras P, Delibes M. Analysis of trophic structure of two carnivore assemblages by means of guild identification. Eur J Wildl Res. 53:276–286. [Google Scholar]
- Zarco-González M M, Monroy-Vilchis O, Rodríguez-Soto C, Urios V. Spatial factors and management associated with livestock predations by Puma concolor in Central Mexico. Hum Ecol. 40:631–638. [Google Scholar]