Abstract
Long non-coding RNAs (lncRNAs) have been known to partake in the development and the immune escape of hepatocellular carcinoma (HCC). The initial microarray analysis of GSE115018 expression profile revealed differentially expressed lncRNA fetal-lethal non-coding developmental regulatory RNA (FENDRR) in HCC. Therefore, this study’s main purpose was to explore the mechanism of tumor suppressor lncRNA FENDRR in regulating the immune escape of HCC cells. Notably, it was further validated through this study that lncRNA FENDRR competitively bound to microRNA-423-5p (miR-423-5p), and miR-423-5p specifically targeted growth arrest and DNA-damage-inducible beta protein (GADD45B). The effects that lncRNA FENDRR and miR-423-5p have on the cell proliferation and apoptosis, the immune capacity of regulatory T cells (Tregs), and the tumorigenicity of HCC cells were examined through overexpressing or the knocking down of lncRNA FENDRR and miR-423-5p both in vitro and in vivo. Subsequently, lncRNA FENDRR and GADD45B were revealed to have poor expressions in HCC. Meanwhile, miR-423-5p was highly expressed in HCC. Importantly, overexpressed lncRNA FENDRR and downregulated miR-423-5p diminished cell proliferation and tumorigenicity, and promoted apoptosis in HCC cells, thus regulating the immune escape of HCC mediated by Tregs. Taken conjointly, lncRNA FENDRR inhibited the Treg-mediated immune escape of HCC cells by upregulating GADD45B by sponging miR-423-5p.
Keywords: long non-coding FENDRR, microRNA-423-5p, GADD45B, hepatocellular carcinoma, immune escape
Introduction
Hepatocellular carcinoma (HCC) is recognized as the sixth most common cancer and is ranked as the third major cause of cancer-correlated death.1 The development of HCC commonly occurs in people who have chronic liver disease, such as cirrhosis of the liver. In addition, it is affected by risk factors including hepatitis B virus (HBV) and hepatitis C virus (HCV) in serum, diabetes mellitus (DM), albumin expression, age at sustained virologic response (SVR), alcohol, and smoking.2, 3, 4 Radiology and biopsy represent two methods that are commonly used for diagnosing HCC.5 Currently, many studies have reported novel treatments for HCC. To elaborate, examples of novel treatments would be the antiviral treatment and immunotherapy.6, 7 Tumor immune escape is noted to be a phenomenon in which tumor cells proliferate and metastasize by evading recognition and attack of the immune system by different mechanisms. This represents a significant strategy for tumor survival and development.8 More importantly, the occurrence of programmed death-1 (PD-1)-targeted therapy has established the values of this pathway in constraining anticancer T cell immunity in numerous cancers and extending overall survival.9 Interestingly, long non-coding RNAs (lncRNAs) have been emerging as the new targets for pharmacological intervention in HCC.10
lncRNAs are expressed as the non-coding transcripts ranging from 200 nt to 100 kb in length, and they function as novel factors in the cancer. They play the roles of acting both as oncogenic and tumor-suppressive factors.11 lncRNA fetal-lethal non-coding developmental regulatory RNA (FENDRR), a gene that consists of seven exons, is transcribed from transcription factor-coding gene Foxf1 and located 1,250 bp upstream of the 50-end of Foxf1.12 Furthermore, a previous study has demonstrated that lncRNA FENDRR was considered to be one of the favorable diagnostic lncRNA biomarkers for HCC.13 In addition, a recent study proposed that lncRNA FENDRR methylated Glypican-3 (GPC3) promoted and downregulated GPC3 expression. Therefore, it suppresses HCC cell proliferation, migration, and invasion.14 However, this current study discovered that lncRNA FENDRR mediated the immune escape process of HCC, and evaluated the underlying mechanism. It is intriguing to find that microRNA-423-5p (miR-423-5p) has the potential to bind to lncRNA FENDRR and target growth arrest and DNA-damage-inducible beta (GADD45B). microRNAs (miRNAs), which are short non-coding RNAs, have been noted to play crucial roles in various malignancies, including HCC.15 Notably, miR-423-5p acted as an important predictor to sorafenib in HCC patients and regulated the autophagy in HCC cells.16 Additionally, GADD45B gene plays a fundamental role in regulating different physiological events.17 It has been reported that GADD45B was involved in sorafenib-induced cell apoptosis in HCC.18 Therefore, it is safe to hypothesize that lncRNA FENDRR has an impact on the immune escape of HCC cells through the interaction with miR-423-5p and GADD45B. The main objective of this study is to provide clinical insights for future treatments of HCC.
Results
Overexpression of lncRNA FENDRR Suppresses the Proliferation and Immune Escape of HCC Cells
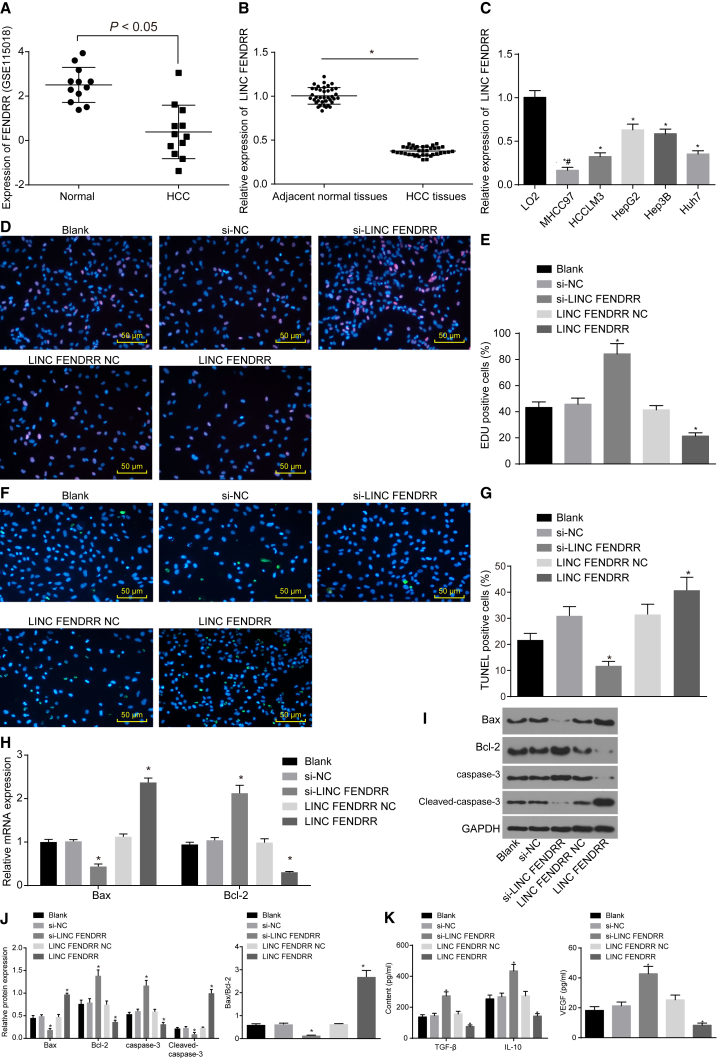
Initially, microarray-based analysis was conducted. Through differential analysis for normal samples and HCC samples in the GSE115018 microarray, it was discovered that lncRNA FENDRR had low expressions in HCC (Figure 1A). Then, the results of qRT-PCR (Figures 1B and 1C) indicated that the expression of lncRNA FENDRR was significantly lower in HCC tissues and cell lines (MHCC97, HCCLM3, HepG2, Hep3B, and Huh7) than that in adjacent normal tissues and normal hepatic cells (L02), with the largest fold changes (FCs) observed in the MHCC97 cell line, which was thus selected for the following experiment. By using 5-ethynyl-2′-deoxyuridine (EdU) and terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) assays, it was revealed in this study that cells treated with si-NC and lncRNA FENDRR NC displayed no significant difference in proliferation and apoptosis compared with cells without treatment (all p > 0.05). Cells transfected with si-lncRNA FENDRR exhibited increased proliferation and decreased apoptosis rate, and cells transduced with lncRNA FENDRR demonstrated an opposite trend (all p < 0.05; Figures 1D–1G). According to the results of qRT-PCR and western blot analysis (Figures 1H–1J), there was no significant difference in the expression of Bax, Bcl-2, cleaved-caspase-3, and caspase-3 among cells without treatment and cells treated with si-NC and lncRNA FENDRR NC (all p > 0.05). In comparison with the cells that were exposed to no treatment, the expression of Bax and cleaved-caspase-3 was significantly reduced, and that of Bcl-2 and caspase-3 was significantly elevated in cells transduced with si-lncRNA FENDRR. However, an opposite trend was found in cells transfected with lncRNA FENDRR (all p < 0.05). Furthermore, ELISA was conducted, and it presented the results that the expression of transforming growth factor-β (TGF-β), interleukin-10 (IL-10), and vascular endothelial growth factor (VEGF) had no coherent difference among cells without treatment and cells treated with si-NC and lncRNA FENDRR NC (all p > 0.05). However, the expression of TGF-β, IL-10, and VEGF was remarkably increased in cells transduced with si-lncRNA FENDRR but significantly decreased in cells transduced with lncRNA FENDRR compared with cells without treatment (all p < 0.05; Figure 1K). Taking the results from above into account, it was demonstrated that overexpressed lncRNA FENDRR has the potential to inhibit the proliferation and secretion of immune-related factors, but also promote the apoptosis of HCC cells.
Figure 1.
Upregulated lncRNA FENDRR Represses the Proliferation and Immune Escape of HCC Cells
(A) The expression of lncRNA FENDRR in the HCC-related dataset GSE115018. (B) The expression of lncRNA FENDRR in HCC tissues and adjacent normal tissues (n = 38). *p < 0.05 compared with adjacent normal tissues. (C) The expression of lncRNA FENDRR in HCC cell lines and normal hepatic cell L02. *p < 0.05 compared with normal hepatic cell L02; #p < 0.05 compared with HCC cell lines HCCLM3, HepG2, Hep3B, and Huh7. The MHCC97 cell line was treated with si-lncRNA FENDRR or lncRNA FENDRR with si-NC or lncRNA FENDRR NC as controls. (D) Cell proliferation after different treatments examined by EdU assay (×200). (E) EdU-positive cells after different treatments. (F) Cell apoptosis examined by TUNEL assay (×200). (G) TUNEL-positive cells after different treatments. (H) The relative expression of mRNA of apoptosis-related factors in cells with different treatments determined by qRT-PCR. (I) The protein bands of apoptosis-related factors in cells with different treatments. (J) The protein expression of apoptosis-related factors in cells with different treatments. (K) The expression of TGF-β, IL-10, and VEGF in cells with different treatments examined by ELISA. *p < 0.05 compared with cells without treatment. In the EdU assay, the cells stained in red were considered positive, and the results were measurement data. In the TUNEL assay, cells stained in green were considered positive, and the results were measurement data. In qRT-PCR and western blot analysis, the results were measurement data and were expressed as mean ± SD. The data between two groups were analyzed by paired t test, and data among multiple groups were analyzed by one-way ANOVA, followed by Tukey’s post hoc test. Experiments were repeated three times. EdU, 5-ethynyl-2′-deoxyuridine; HCC, hepatocellular carcinoma; IL-10, interleukin-10; lncRNA FENDRR, long non-coding RNA fetal-lethal non-coding developmental regulatory RNA; TGF-β, transforming growth factor-β; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling; VEGF, vascular endothelial growth factor.
Inhibition of miR-423-5p Suppresses the Proliferation and Secretion of Immune-Related Factors, and Promotes the Apoptosis of HCC Cells by Targeting GADD45B
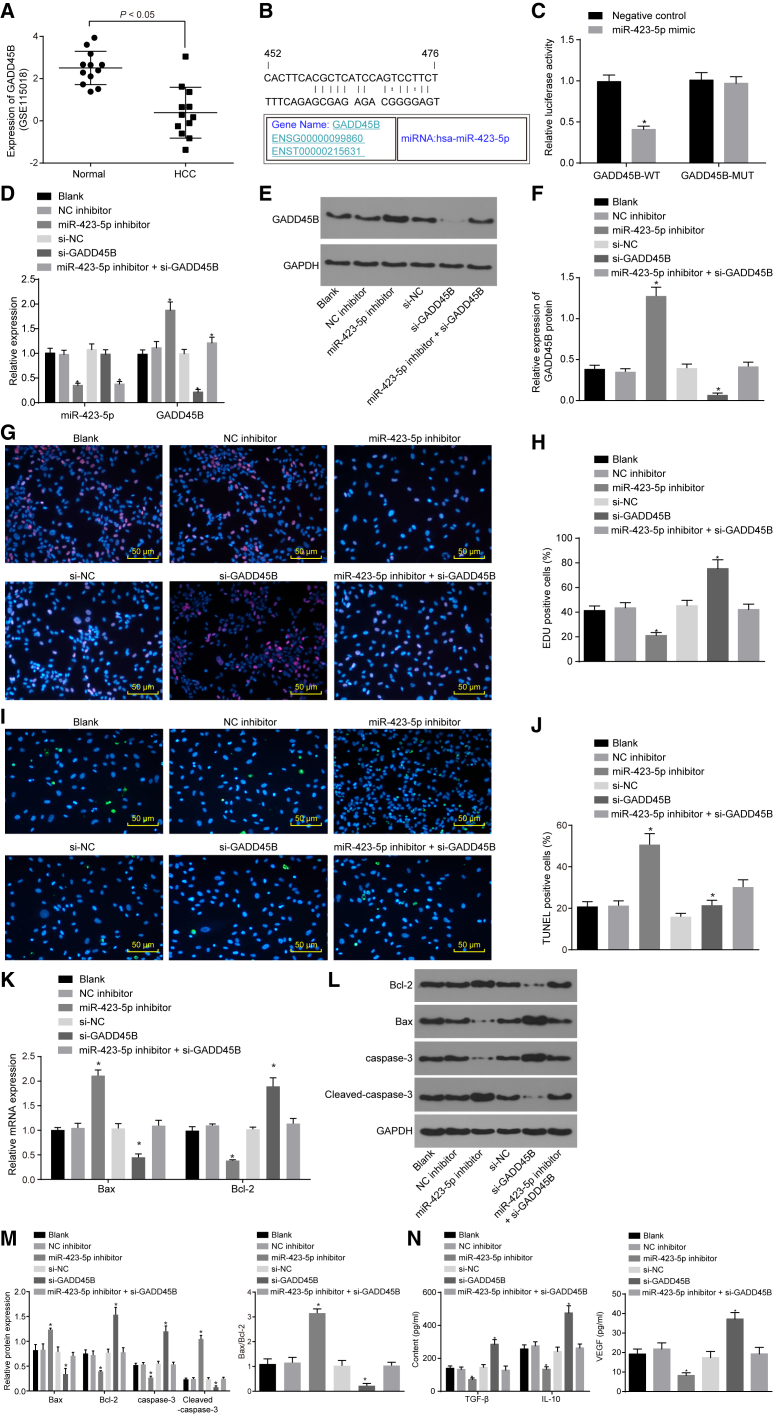
It was previously reported that GADD45B, as tumor-inhibiting factor, was downregulated in HCC,19 and loss of GADD45B increased the number of regulatory T cells (Tregs).20 Although differential analysis for the normal samples and HCC samples in the GSE115018 expression profile revealed that GADD45B was poorly expressed in HCC (Figure 2A). Moreover, the prediction by online tool exhibited a binding site between miR-423-5p and GADD45B (Figure 2B). The results of dual-luciferase reporter gene assay showed that, compared with the NC group, the luciferase activity was significantly decreased in the miR-423-5p mimic group in GADD45B-WT (wild-type) plasmid (p < 0.05), and no significant difference in the luciferase activity of the GADD45B-MUT (mutant type) plasmid was found (p > 0.05), demonstrating a direct interaction between miR-423-5p and GADD45B (Figure 2C). Next, qRT-PCR and western blot analysis were carried out in order to examine the expression of miR-423-5p and GADD45B, and the results (Figures 2D–2F) presented that, compared with cells without treatment, no significant difference in the expression of miR-423-5p and GADD45B was observed in cells treated with NC inhibitor and si-NC (both p > 0.05), and cells treated with both miR-423-5p inhibitor and si-GADD45B exhibited decreased miR-423-5p expression, without obvious change in GADD45B expression (p > 0.05). Cells treated with miR-423-5p inhibitor revealed lower miR-423-5p expression but higher GADD45B expression, and cells transfected with si-GADD45B exhibited reduced GADD45B expression (p < 0.05), without significant difference in miR-423-5p expression (p > 0.05). Moreover, EdU assay (Figures 2G and 2H), TUNEL assay (Figures 2I and 2J), qRT-PCR (Figure 2K), western blot analysis (Figures 2L and 2M), and ELISA (Figure 2N) were conducted successively. The results revealed that no significant difference was found in cell proliferation, apoptosis, and the expression of Bax, Bcl-2, cleaved-caspase-3, caspase-3, TGF-β, IL-10, and VEGF among cells without treatment and cells treated with NC inhibitor, si-NC, and both miR-423-5p inhibitor and si-GADD45B (all p > 0.05). However, decreased proliferation and expression of Bcl-2, caspase-3, TGF-β, IL-10, and VEGF, accompanied by increased apoptosis and expression of Bax and cleaved-caspase-3, were discovered in cells treated with miR-423-5p inhibitor, but an opposite trend was detected in the cells transfected with si-GADD45B, in contrast with cells without treatment (all p > 0.05). Therefore, it could be drawn from the results that downregulation of miR-423-5p repressed the proliferation and secretion of immune-related factors, and enhanced the apoptosis of HCC cells by targeting GADD45B.
Figure 2.
Downregulation of miR-423-5p Inhibits the Proliferation and Immune Escape, and Elevates the Apoptosis of HCC Cells by Upregulating GADD45B
(A) The expression of GADD45B in the HCC-related dataset GSE115018. (B) The binding site between miR-423-5p and GADD45B. (C) Dual-luciferase reporter gene assay for confirmation of the targeting relationship between miR-423-5p and GADD45B. *p < 0.05 compared with the NC group. The MHCC97 cell line was treated with miR-423-5p inhibitor and/or si-GADD45B. (D) The miR-423-5p expression and mRNA expression of GADD45B in cells with different treatments examined by qRT-PCR. (E) The protein expression of GADD45B and GAPDH in cells with different treatments detected by western blot analysis. (F) The statistical analysis of (E). (G) Cell proliferation in cells with different treatments examined by EdU assay (×200). (H) EdU-positive cells after different treatments. (I) Cell apoptosis examined by TUNEL assay (×200). (J) TUNEL-positive cells after different treatments. (K) The mRNA expression of apoptosis-related factors in cells with different treatments detected by qRT-PCR. (L) The protein expression of apoptosis-related factors in cells with different treatments. (M) The statistical analysis of (L). (N) The expression of TGF-β, IL-10, and VEGF in cells with different treatments examined by ELISA. *p < 0.05 compared with cells without treatment. The results of luciferase activity detection and qRT-PCR were measurement data and expressed as mean ± SD. In the EdU assay, the cells stained in red were considered positive, and the results were measurement data. In the TUNEL assay, cells stained in green were considered positive, and the results were measurement data. In qRT-PCR and western blot analysis, the results were measurement data and were expressed as mean ± SD. Data between two groups were analyzed by paired t test and among multiple groups were analyzed by one-way ANOVA, followed by Tukey’s post hoc test. Experiments were repeated three times. EdU, 5-ethynyl-2′-deoxyuridine; GADD45B, growth arrest and DNA-damage-inducible beta; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCC, hepatocellular carcinoma; IL-10, interleukin-10; miR-423-5p, microRNA-423; NC, negative control; TGF-β, transforming growth factor-β; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling; VEGF, vascular endothelial growth factor.
lncRNA FENDRR Regulates GADD45B by Competitively Binding to miR-423-5p
Through differential analysis between the normal samples and HCC samples, it was found that lncRNA FENDRR was positively correlated with GADD45B (Figure 3A). A former study has demonstrated that lncRNA FENDRR was poorly expressed in gastric cancer and related to unfavorable prognosis.21 Also, lncRNA FENDRR regulated the expression of VEGFA through competitively binding to miR-126.22 Therefore, it was speculated that lncRNA FENDRR regulated the expression of GADD45B by competitively binding to miRNA. In order to explore the interactions among lncRNA FENDRR, miR-423-5p, and GADD45B, initially the starBase website (http://starbase.sysu.edu.cn/index.php) was utilized to predict the miRNAs bound to both lncRNA FENDRR and GADD45B. Next, these miRNAs were intersected using an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/), which revealed that miR-495-3p, miR-423-5p, and miR-421 could bind to both lncRNA FENDRR and GADD45B (Figure 3B). Subsequently, the expression of miR-495-3p, miR-423-5p, and miR-421 in HCC tissues and adjacent normal tissues was examined. In comparison with the adjacent normal tissues, the expression of miR-495-3p, miR-423-5p, and miR-421 was significantly increased in HCC tissues (all p < 0.05), with miR-423-5p showing the largest FC (p < 0.05; Figure 3C). Given that a previous study has demonstrated that miR-423 could elevate the growth of HCC cells,23 we selected miR-423-5p for our study subject and hypothesized that lncRNA FENDRR could suppress the Treg-mediated immune escape of HCC cells through regulating GADD45B by sponging miR-423-5p.
Figure 3.
lncRNA FENDRR Regulates the Expression of GADD45B through Sponging miR-423-5p
(A) The correlation analysis between lncRNA FENDRR and GADD45B in the GSE115018 dataset. (B) The miRNAs bound to both lncRNA FENDRR and GADD45B predicted by starBase. (C) The expression of miR-495-3p, miR-423-5p, and miR-421 in HCC tissues and adjacent normal tissues (n = 38). *p < 0.05 compared with adjacent normal tissues. (D) The binding site between lncRNA FENDRR and miR-423-5p. (E) Dual-luciferase reporter gene assay for confirmation of the targeting relationship between lncRNA FENDRR and miR-423-5p. *p < 0.05 compared with the NC group. (F) The binding ability between lncRNA FENDRR and miR-423-5p analyzed by RNA pull-down assay. (G) The binding ability between lncRNA FENDRR and Ago2 detected by RIP assay. *p < 0.05 compared with IgG. (H) The location of lncRNA FENDRR examined by FISH assay (×400). The MHCC97 cell line was treated with si-lncRNA FENDRR or lncRNA FENDRR with si-NC or lncRNA FENDRR NC as controls. (I) The expression of miR-423-5p and GADD45B in cells with different treatments examined by qRT-PCR. (J) The protein expression of GADD45B and GAPDH in cells with different treatments detected by western blot analysis. (K) The statistical analysis of (J). *p < 0.05 compared with cells without treatment. The results of luciferase activity detection, qRT-PCR, and western blot analysis were measurement data and were expressed as mean ± SD. Data in (C) were analyzed by paired t test. Data between two groups were analyzed by unpaired t test and among multiple groups were analyzed by one-way ANOVA, followed by Tukey’s post hoc test. Experiments were repeated three times. FISH, fluorescence in situ hybridization; GADD45B, growth arrest and DNA-damage-inducible beta; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCC, hepatocellular carcinoma; IgG, immunoglobulin G; lncRNA FENDRR, long non-coding RNA fetal-lethal non-coding developmental regulatory RNA; miR-423-5p, microRNA-423; NC, negative control.
The online prediction tool confirmed that a binding site existed between miR-423-5p and lncRNA FENDRR (Figure 3D). Through dual-luciferase reporter gene assay, it was asserted that, in contrast with the NC group, the luciferase activity of lncRNA FENDRR-WT was significantly decreased in the miR-423-5p mimic group (p < 0.05), and no significant difference was found in the lncRNA FENDRR-MUT plasmid (p > 0.05), suggesting the presence of a binding site between miR-423-5p and lncRNA FENDRR (Figure 3E). After conducting RNA immunoprecipitation (RIP) and RNA pull-down assays, the results (Figures 3F and 3G) revealed that lncRNA FENDRR enrichment was higher in the WT-bio-miR-423-5p group than that in the MUT-miR-423-5p and Bio-NC groups (p < 0.05). RIP further confirmed that lncRNA FENDRR could interact with miR-423-5p. Additionally, fluorescence in situ hybridization (FISH) assay showed that lncRNA FENDRR was mainly located in the cytoplasm (Figure 3H).
Furthermore, to analyze and verify the relationship between lncRNA FENDRR and miR-423-5p, the ectopic expression and depletion experiments were performed in the cell line. As depicted in Figures 3I–3K, no significant difference in expression of lncRNA FENDRR was found between cells without treatment and cells treated with si-NC and lncRNA FENDRR (all p > 0.05). In contrast with cells without treatment, the GADD45B expression was lower in cells transfected with si-lncRNA FENDRR, and an opposite trend was observed in cells transfected with lncRNA FENDRR (all p < 0.05).
All of the results above suggested that lncRNA FENDRR might upregulate GADD45B by competitively binding to miR-423-5p.
Overexpressed lncRNA FENDRR and Downregulated miR-423-5p Inhibit the Secretion of Immune-Related Factors in HCC Cells by Suppressing the Immune-Suppressive Capacity of Tregs
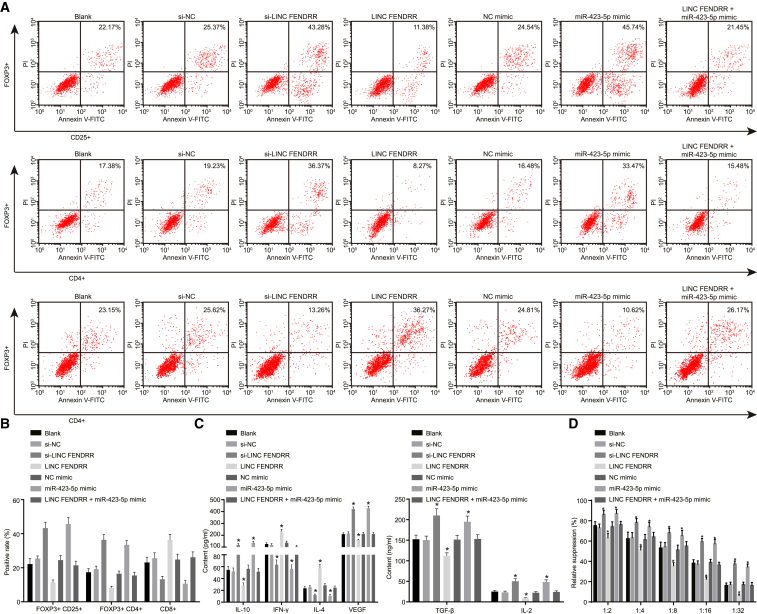
After co-culture of HCC cell lines and Tregs, the positive rate of forkhead box P3+ (FOXP3+) CD4+ Tregs, FOXP3+ CD25+ Tregs, and CD8+ Tregs was detected using flow cytometry. Compared with cells without treatment, there was no prominent difference of positive rate of FOXP3+ CD4+ Tregs, FOXP3+ CD25+ Tregs, and CD8+ Tregs after treatment of NC, NC mimic, and both lncRNA FENDRR and miR-423-5p mimic (all p > 0.05). The positive rate of FOXP3+ CD4+ Tregs and FOXP3+ CD25+ Tregs was decreased after treatment of lncRNA FENDRR, accompanied by an increased rate of CD8+ Tregs. Conversely, the treatment of si-lncRNA FENDRR and miR-423-5p mimic led to the opposite trends (all p < 0.05; Figures 4A and 4B). Next, the results of ELISA (Figure 4C) revealed no obvious difference of the levels of TGF-β, VEGF, IL-10, interferon (IFN)-γ, IL-2, and IL-4 among cells without treatment and cells treated with NC, NC mimic, and both lncRNA FENDRR and miR-423-5p mimic (all p > 0.05). In contrast with the cells that were exposed to no treatment, cells transfected with lncRNA FENDRR exhibited lower levels of TGF-β, VEGF, IL-2, and IL-10 and higher levels of IFN-γ and IL-4 (all p < 0.05), and cells treated with si-lncRNA FENDRR and miR-423-5p mimic displayed an opposite result (all p < 0.05). Furthermore, suppression assay was carried out to detect the immune-suppressive capacity of Tregs and eventually presented the finding that there was no remarkable change in immune-suppressive capacity of Tregs after treatment of NC, NC mimic, and both lncRNA FENDRR and miR-423-5p mimic compared with cells without treatment (all p > 0.05). The immune-suppressive capacity of Tregs was significantly inhibited after treatment of lncRNA FENDRR and enhanced after treatment of si-lncRNA FENDRR and miR-423-5p mimic (all p < 0.05; Figure 4D). Therefore, it was safe to conclude that overexpressed lncRNA FENDRR and downregulated miR-423-5p could suppress immune-suppressive capacity of Tregs, thus inhibiting the Treg-mediated immune escape of HCC.
Figure 4.
Upregulation of lncRNA FENDRR or Downregulation of miR-423-5p Inhibits the Treg-Mediated Immune Escape of HCC Cells
The MHCC97 cell line was treated with si-lncRNA FENDRR, lncRNA FENDRR, and/or miR-423-5p mimic and co-cultured with Tregs. (A) The FOXP3+CD4+, FOXP3+CD25+, and CD8+ Tregs examined by flow cytometry after different treatments. (B) The statistical analysis of (A). (C) The levels of IL-10, IFN-γ, TGF-β, VEGF, IL-2, and IL-4 after different treatments detected by ELISA. (D) The immune-suppressive capacity of Tregs after different treatments. *p < 0.05 compared with cells without treatment. The results of positive staining examined by flow cytometry and the results of ELISA were measurement data and expressed as mean ± SD. Data among multiple groups were analyzed by one-way ANOVA, followed by Tukey’s post hoc test. Experiments were repeated three times. HCC, hepatocellular carcinoma; IFN-γ, interferon-γ; IL-4, interleukin-4; IL-10, interleukin-10; lncRNA FENDRR, long non-coding RNA fetal-lethal non-coding developmental regulatory RNA; miR-423-5p, microRNA-423; Treg, regulatory T cell.
Overexpression of lncRNA FENDRR and Downregulation of miR-423-5p Inhibit the Tumorigenicity and Secretion of Immune-Related Factors of HCC Cells In Vivo
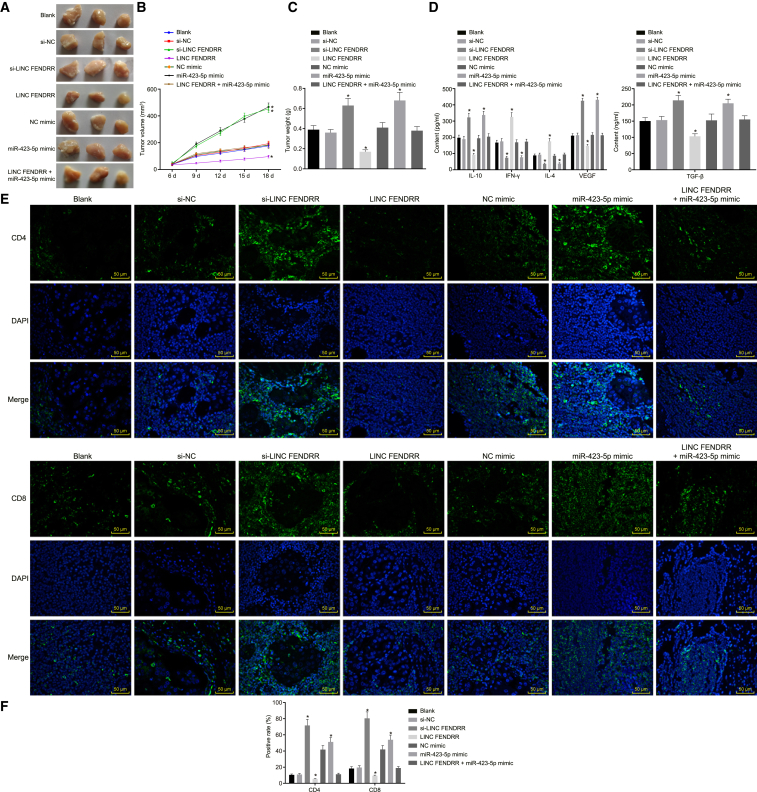
In order to examine the effects of lncRNA FENDRR and miR-423-5p on the tumorigenicity in vivo, we have established the tumor xenograft and orthotopic HCC nude mouse models. After measuring the volume and weight of transplanted tumors, no significant difference existed between the volume and weight of tumors and the expression of TGF-β, VEGF, IL-10, IFN-γ, and IL-4 of nude mice after injection with cells without treatment and cells treated with NC, NC mimic, and both lncRNA FENDRR and miR-423-5p mimic (all p > 0.05). The volume and weight of tumors were significantly inhibited in nude mice after injection with cells treated with lncRNA FENDRR, which were increased in nude mice after injection with cells treated with si-lncRNA FENDRR and miR-423-5p mimic (all p < 0.05; Figures 5A–5C). Following this, we conducted ELISA and immunofluorescence assays, and we found that there were no obvious changes in the expression of TGF-β, VEGF, IL-10, IFN-γ, and IL-4 and the positive rates of CD4 and CD8 proteins among tumor tissues without treatment and those treated with NC, NC mimic, and both lncRNA FENDRR and miR-423-5p mimic (all p > 0.05). In contrast with tumor tissues without treatment, the tumor tissues treated with lncRNA FENDRR exhibited reduced expression of TGF-β, VEGF, IL-10, and positive rates of CD4 and CD8 proteins but increased expression of IFN-γ and IL-4 (all p < 0.05), and tumor tissues treated with si-lncRNA FENDRR and miR-423-5p mimic revealed an opposite result (all p < 0.05; Figures 5E and 5F). From the findings above, we concluded that overexpressed lncRNA FENDRR and downregulation of miR-423-5p could suppress the tumorigenicity and secretion of immune-related factors of HCC cells in vivo.
Figure 5.
Overexpressed lncRNA FENDRR or Downregulation of miR-423-5p Inhibits the Tumorigenicity and Immune Escape of HCC Cells In Vivo
Nude mice were injected with the MHCC97 cell line treated with si-lncRNA FENDRR, lncRNA FENDRR, and/or miR-423-5p mimic. (A) The subcutaneous transplanted tumor of nude mice after injection with different treated cells. (B) The volume of tumor in nude mice. (C) The weight of tumor in nude mice. (D) The expression of TGF-β, VEGF, IL-10, IFN-γ, and IL-4 examined by ELISA. (E) The immunofluorescence staining of CD4 and CD8 proteins (×200). (F) The statistical analysis of (E). *p < 0.05 compared with cells without treatment. Data among multiple groups were analyzed by one-way ANOVA, and data at different time points were compared using repeated-measurement ANOVA, followed by Tukey’s post hoc test. HCC, hepatocellular carcinoma; IFN-γ, interferon-γ; IL-4, interleukin-4; IL-10, interleukin-10; lncRNA FENDRR, long non-coding RNA fatal-lethal non-coding developmental regulatory RNA; miR-423-5p, microRNA-423.
Discussion
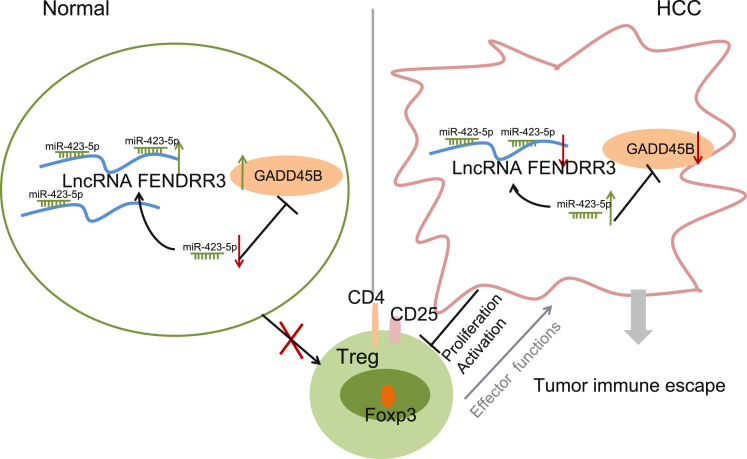
In spite of the advances in treating HCC, the existing treatment options are still inefficient for all of the patients, which is attributed to a lack of understanding of the physiology of HCC overall,24, 25 underscoring the need for more effective therapy. It is reported that lncRNAs function as crucial factors in the development of HCC by regulating the biological events including cell growth and tumorigenesis.10 In addition, immune escape continues to represent the stumbling obstacle in effectively treating cancers,26 which may be regulated by lncRNAs. Additionally, miR-423 has been reported to participate in the development of HCC by promoting cell growth and mediating G1/S transition through p21Cip1/Waf1 in HCC.23 Thus, we studied the effect of lncRNA FENDRR on the immune escape of HCC cells through interaction with miR-423-5p. Eventually, the findings from the present study demonstrated that overexpression of lncRNA FENDRR suppressed the Treg-mediated immune escape of HCC cells by miR-423-5p-mediated upregulation of GADD45B (Figure 6).
Figure 6.
The Molecular Mechanism Involved in lncRNA FENDRR in Regulating the Progression of HCC
lncRNA FENDRR was downregulated but miR-423-5p was upregulated in HCC. Overexpressed lncRNA FENDRR and downregulation of miR-423-5p artificially inhibited the Treg-mediated immune escape of HCC by upregulating GADD45B. GADD45B, growth arrest and DNA-damage-inducible beta; HCC, hepatocellular carcinoma; lncRNA FENDRR, long non-coding RNA fetal-lethal non-coding developmental regulatory RNA; miR-423-5p, microRNA-423; Treg, regulatory T cell.
At first, the results obtained in this study revealed that lncRNA FENDRR was poorly expressed, and miR-423-5p was upregulated in HCC, which resulted in unfavorable outcomes for patients. Moreover, another study displayed that lncRNA FENDRR exhibited low expression in HCC and acted as an important factor in HCC development.13 It has been reported that the expression of miR-423-5p in serum was increased in HCC after sorafenib treatment,16 and upregulation of miR-423-5p in HCC was negatively related to the recurrence-free survival.27 In the present study, we uncovered that lncRNA FENDRR competitively bound to miR-423-5p and verified the targeting relationship between miR-423-5p and GADD45B. Consistently, previous research revealed that lncRNA FENDRR could sponge miR-18a-5p in prostate cancer.28 Furthermore, lncRNA FENDRR elevated the apoptosis of human brain microvascular endothelial cells (HBMECs) by upregulation of VEGFA through competitively binding miR-126.22 Therefore, the findings above were consistent with the results of the present study. This supports the conclusion that highly expressed lncRNA FENDRR and poorly expressed miR-423-5p were involved in the progression of HCC, and miR-423-5p was the intermediary in the regulation between lncRNA FENDRR and GADD45B.
On one hand, we found that overexpressed lncRNA FENDRR and downregulation of miR-423-5p inhibited cell proliferation and tumorigenicity, but enhanced apoptosis of HCC cells as evidenced by lowered levels of TGF-β, IL-10, and VEGF. IL-10-stimulated tumor rejections rely on the granzymes in tumor-resident CD8+ T cells and the release of major histocompatibility complex molecules, indicating the unique values of IL-10 in simultaneously inducing antitumor immunity and impairing tumor-related inflammation.29 VEGF could block the function of T cells and elevate the recruitment of Tregs.30 To elaborate, TGF-β is recognized to exert a suppressive role in immunity and host immunosurveillance by directly disarming T and cell effector functions.31 Similarly, overexpressed lncRNA FENDRR repressed the cell proliferation and tumorigenicity, and elevated apoptosis of breast cancer cells.32 In addition, the inhibitory effect of downregulation of miR-423-5p on cell proliferation of glioblastomas has been previously noted.33 On the other hand, we showed that overexpressed lncRNA FENDRR reduced the immune-suppressive capacity of Tregs through upregulation of GADD45B. It has been understood that Treg-mediated immunosuppression promoted immune escape in tumors.34 In addition, loss of GADD45B was demonstrated to lead to more Tregs.20 Therefore, these evidences coincide with this study and could prove the suppressive role overexpressed lncRNA FENDRR plays in proliferation, tumorigenicity, and immune escape of HCC cells through increasing GADD45B through competitively binding to miR-423-5p.
In conclusion, the fundamental findings of the present study suggested the inhibitory effect of lncRNA FENDRR on the progression of HCC. This study has revealed that overexpression of lncRNA FENDRR inhibited the Treg-mediated immune escape of HCC cells by upregulating GADD45B through sponging miR-423-5p. This study has the potential to offer novel insights for immunotherapy for HCC. Nevertheless, the promotive roles of exosomal miR-423-5p in gastric cancer progression35 and exosomal PD-L1 in immunosuppression36 have been underscored recently. The focus of the future study may place emphasis on the exosomal miRNA-mediated inhibition. To conclude, the critical role of lncRNA FENDRR in this study provided a possible strategy for a promising approach to HCC treatment.
Materials and Methods
Ethics Statement
The study was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-Sen University. The written informed consents were obtained from all participants. All experiments that incorporated animals were carried out in accordance with the principles and procedures of Guide for the Care and Use of Laboratory Animals by the NIH.
Microarray-Based Analysis
HCC-related microarray dataset GSE115018 was acquired from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Then, differential analysis was conducted using R language with thresholds of |log FC| > 2 and a p value <0.05 set as the conditions that are necessary to be met for the differentially expressed genes (DEGs).
Study Subjects
A total of 38 patients with HCC (aged 53.97 ± 4.65 years) within the time period from July 2016 to April 2018 in the Third Affiliated Hospital of Sun Yat-Sen University were enrolled in this study. These patients consisted of 25 males and 13 females; 6 cases were known to be in stage T1, 17 cases in stage T2, 13 cases in stage T3, and 2 cases in stage T4. All patients were not exposed to any radiotherapy or chemotherapy prior to the surgery. The histopathological types of HCC were evaluated and judged in accordance with the World Health Organization (WHO) classification. HCC tissues and adjacent normal liver tissues 5 cm away from the tumor (confirmed by histopathology) were collected and then preserved in liquid nitrogen within 10 min after sampling. Subsequently, they were utilized for following analysis.
Cell Culture
Five common HCC cell lines, MHCC97, HCCLM3, HepG2, Hep3B, and Huh7, and human normal hepatic cell L02 that were attained from the Cell Bank of Chinese Academy of Sciences (http://www.cellbank.org.cn) (Shanghai, China) were involved in this study. After recovery, HCC cell lines MHCC97, HCCLM3, HepG2, Hep3B, and Huh7 were cultured with RPMI 1640 medium (GIBCO, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin. Normal hepatic cell L02 was cultured with DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin at a temperature of 37°C consisting of 5% CO2. Upon reaching 90% confluence, cells were sub-cultured, followed by qRT-PCR in order to determine the cell line with the lowest expression of lncRNA FENDRR for the subsequent experiment.
qRT-PCR
Total RNA was extracted with the use of TRIzol (15596026; Invitrogen, Carlsbad, CA, USA); then RNA concentration and purity were determined using a NanoDrop ND-1000 spectrophotometer. According to the instructions of the PrimeScript RT reagent Kit (RR047A; Takara, Kyoto, Japan) and TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA), RNA was reversely transcribed into cDNA. Following this part of the procedure, the primers were designed and synthetized by Shanghai Sangon Biotechnology (Shanghai, China) (Table 1). qRT-PCR was carried out in accordance with the instructions of the SYBR Premix Ex Taq II kit (Takara Biotechnology, Dalian, Liaoning, China) using the ABI 7500 PCR instrument (Applied Biosystems, Carlsbad, CA, USA). The relative expression of the targeted genes, with U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as housekeeping genes, was calculated based on the 2−ΔΔCt method.37 The experiment was repeated three times.
Table 1.
Primer Sequences for qRT-PCR
| Genes | Primer Sequences (5′–3′) |
|---|---|
| lncRNA FENDRR | F: TCTTGTCTTTGTAATCAGGCAG |
| R: GGAGGTATTTCAGTTCTGTCGT | |
| miR-423-5p | F: GCCTGAGGGGCAGAGAGC |
| R: CCACGTGTCGTGGAGTC | |
| miR-495-3p | F: ACACTCCAGCTGGGAAACAAA CATGGTGCA |
| R: TGGTGTCGTGGAGTCG | |
| miR-421 | F: TATGGTTGTTCTGCTCTCTGTGTC |
| R: CTCACTCACATCAACAGACATTAATT | |
| GADD45B | F: TACGAGTCGGCCAAGTTGATG |
| R: GGATGAGCGTGAAGTGGATTT | |
| Bax | F: CCCGAGAGGTCTTTTTCCGAG |
| R: CCAGCCCATGATGGTTCTGAT | |
| Bcl-2 | F: GGTGGGGTCATGTGTGTGG |
| R: CGGTTCAGGTACTCAGTCATCC | |
| U6 | F: CGGTTCAGGTACTCAGTCATCC |
| R: AACGCTTCACGAATTTGCGT | |
| GAPDH | F: GGAGCGAGATCCCTCCAAAAT |
| R: GGCTGTTGTCATACTTCTCATGG |
Bax, Bcl-2-associated X protein; Bcl-2, B cell lymphoma 2; F, forward; GADD45B, growth arrest and DNA-damage-inducible beta; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; lncRNA FENDRR, long non-coding RNA fetal-lethal non-coding developmental regulatory RNA; miR, microRNA; R, reverse.
Western Blot Analysis
The tissue or cell lines in each group were collected and added with radioimmunoprecipitation assay (RIPA) lysis buffer (P0013B; Beyotime Institute of Biotechnology, Shanghai, China) containing PMSF and phosphatase inhibitor. The proteins were isolated by SDS-PAGE and transferred onto a nitrocellulose (NC) membrane. It was then blocked for 1.5 h with 5% skimmed milk powder, which was prepared by Tris-buffered saline with Tween (TBST). Next, the membrane was incubated overnight with the primary antibody, rabbit anti-human antibodies to GADD45B (ab205252, 1:1,000), Bax (ab32503, 1:1,000), Bcl-2 (ab32124, 1:1,000), caspase-3 (ab13847, 1:500), cleaved caspase-3 (ab32042, 1:500), and GAPDH (ab9485, 1:2,500) at a temperature of 4°C. The following day, the membrane was washed with TBST and incubated with the secondary antibody, horseradish peroxidase (HRP)-labeled goat anti-rabbit immunoglobulin G (IgG; ab205718, 1:2,000–1: 50,000), at room temperature for 2 h. All antibodies were purchased from Abcam (Cambridge, MA, USA). Then, the membrane was developed in the enhanced chemiluminescence (ECL) and photographed with the use of SmartView Pro 2000 (UVCI-2100; Major Science, Saratoga, CA, USA). The Quantity One software was employed in the experiment in order to quantify the corresponding protein bands.38
FISH
Cells were seeded into 24-well plates at the density of 6 × 104 cells/well until cell confluence reached 60%–70%. After being fixed with 4% paraformaldehyde, the cells in each well were added with 1 mL precooling permeate solution at 4°C for about 5 min. After this part of the procedure, the cells were blocked with 20 μL prehybridization solution at a temperature of 37°C for about 30 min. It was then added with Stellaris RNA FISH probe hybridization solution (Biosearch Technologies, Hoddesdon, UK), which contains a lncRNA FENDRR probe to hybridize at 37°C overnight, avoiding any exposure to light. Subsequently, cells were washed with washing buffer I, II, and III, respectively, at 42°C, averting exposure to light, and nuclei were stained by DAPI staining solution for 10 min, washed with PBS, and mounted. Lastly, cells were examined with the use of a fluorescence microscope (Olympus Optical, Tokyo, Japan).
Dual-Luciferase Reporter Gene Assay
The target genes of miR-423-5p were predicted by the biological prediction software. Moreover, the dual-luciferase reporter gene assay was applied to confirm the fact that lncRNA FENDRR and GADD45B represented direct targets of miR-423-5p. The sequences of WT and MUT in mRNA 3′ UTR of lncRNA FENDRR and GADD45B were artificially synthetized.
Furthermore, the pmiR-RB-REPORT lncRNA FENDRR-3′ UTR and pmiR-RB-REPORT-GADD45B-3′ UTR plasmids (Guangzhou RiboBio, Guangzhou, Guangdong, China) were treated using restriction enzymes, and the synthetic target gene fragments WT and MUT were inserted into the pmiR-RB-REPORT vector (Guangzhou RiboBio, Guangzhou, Guangdong, China) with miR-423-5p mimic, respectively. Then, luciferase reporter plasmids WT and MUT were employed for the purpose of subsequent detection. After 48 h, the cells were collected, lysed, and centrifuged for 3–5 min with the supernatant obtained. The luciferase detection kit (RG005; Shanghai Beyotime Biotechnology, Shanghai, China) was implemented in the experiment to dissolve Renilla luciferase detection buffer solution and firefly luciferase detection agent, respectively. Following this, the relative luciferase activity was calculated and measured by the relative light unit (RLU) ratio of firefly luciferase to Renilla luciferase.39
RIP
The binding of lncRNA FENDRR with Ago2 protein was detected with the use of a RIP kit (Millipore, Billerica, MA, USA). The MHCC97 cells were lysed with RIPA lysis buffer (P0013B; Beyotime Biotechnology, Shanghai, China) for 5 min in an ice bath. After centrifugation, the collected lysate (100 μL) was then incubated with RIP buffer containing A + G magnetic beads conjugated with human anti-Ago2 antibody (ab186733, 1:50; Abcam). To elaborate, IgG (ab109489, 1:100; Abcam) was incorporated in the experiment as a negative control (NC). Samples and input were treated by protease K to extract RNA for subsequent PCR detection.
RNA Pull-Down
MHCC97 cells were transfected with 50 nM biotin-labeled WT-bio-miR-423-5p and MUT-bio-miR-423-5p (Wuhan Genecreate Biological Engineering, Wuhan, Hubei, China). After about 48 h, cells were collected, washed with PBS, and incubated in a lysis buffer (Ambion, Austin, TX, USA) for 10 min. Biotinylated FENDRR was incubated overnight with streptavidin magnetic beads at a temperature of 4°C. Furthermore, the beads were rinsed two times with pre-cooled lysis buffer, three times with low-salt buffer, and one time with high-salt buffer. The bound RNA was purified by the TRIzol method, and lncRNA FENDRR enrichment was determined by qRT-PCR.40
Cell Treatment
The monocytes were isolated from the blood of HCC patients. The CD4+CD25+ T cells were separated from monocytes and purified with over 95% purity determined by flow cytometry. Next, MHCC97 cells in the logarithmic phase were selected for the process of transfection with the following plasmids and small RNA: si-NC plasmid, si-lncRNA FENDRR plasmid, NC plasmid, lncRNA FENDRR plasmid, NC inhibitor sequence, miR-423-5p inhibitor sequence, GADD45B plasmid, and si-GADD45B plasmid. Consequently, CD4+CD25+ T cells and MHCC97 cells were co-cultured and transfected with si-NC plasmid, lncRNA FENDRR plasmid, si-lncRNA FENDRR plasmid, NC mimic, and miR-423-5p mimic. All of the plasmids above were obtained from Dharmacon (Lafayette, CO, USA). Next, MHCC97 cells were inoculated in six-well plates at the density of 3 × 105 cells/well and cultured using a Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA, USA) when cell confluence reached 50%–80%. Subsequently, 4 μg target plasmid and 10 μL Lipofectamine 2000 were diluted independently with 250 μL serum-free Opti-MEM (GIBCO Company, Grand Island, NY, USA). The two mixtures were mixed and added into the culture well for incubation at 37°C in a 5% CO2 incubator. After approximately 6 h, cells were cultured in complete culture medium and were then collected after 48 h of incubation.
EdU Assay
Cells in each group were seeded in a 96-well plate at a density of 1.6 × 105 cells/well for 48 h. The cells in each well were added with 50 mM EdU (Cell-Light EdU Apollo 488 In Vitro Imaging Kit; RiboBio, Guangzhou, Guangdong, China) and cultured at 37°C for 4 h. Cells were subsequently fixed with 4% formaldehyde solution for 15 min and treated with 0.5% Triton X-100 for permeabilization. After PBS washing, cells in each well were incubated with 100 mL Apollo mixture at room temperature for about 30 min and stained with 100 mL Hoechst33342 staining solution for 30 min. The cells were finally observed and photographed under a fluorescence microscope (Olympus Optical, Tokyo, Japan). The Image-Pro Plus (IPP) 6.0 software (Media Cybernetics, Bethesda, MD, USA) was implemented in order to measure the number of EdU-positive cells (erythrocytes). The EdU incorporation rate was expressed as the ratio of EdU-positive cells to total cells (blue cells). The experiment was repeated three times to determine the mean value.
TUNEL Assay
After dewaxing and dehydration, the cell slides were treated with protease K (Boster Biological Technology, Wuhan, Hubei, China) at room temperature for 20 min, incubated with TUNEL reaction mixture at 37°C for 60 min in accordance with the instructions of the kit (Boster Biological Technology, Wuhan, Hubei, China), and washed with PBS. Then, the cell slides were observed with the use of a fluorescence microscope (BX-60; Olympus Optical, Tokyo, Japan) using a filter at excitation wavelength of 450–500 nm and emission wavelength of 515–565 nm. The NC slide glass was incubated with label solution (terminal-free transferase) instead of TUNEL reaction solution. The positive control slide glass was incubated with bovine pancreas DNase I (Boster Biological Technology, Wuhan, Hubei, China). Lastly, IPP 6.0 software was used to count the number of apoptosis cells.
ELISA
The culture supernatant of cells in each group was utilized for detecting the expression of TGF-β, IL-10, VEGF, interferon-γ (IFN-γ), and IL-4 according to the instructions of TGF-β, IL-10, VEGF, IFN-γ, and IL-4 ELISA kits (RapidBio, West Hills, CA, USA). The diluted samples were added into the reaction wells of the ELISA plate (100 μL per well), with NC and positive control set with duplicated wells detection. Thus, samples in each well were added with 100 μL enzyme conjugate and reacted at 37°C for 30 min. Cells were added with 100 μL HRP substrate solution for developing at 37°C for 10–20 min. Once significant color change in the positive control or slight color change in the NC appeared, 50 μL termination solution was added to each well to prevent the reaction from continuing any further. The optical density (OD) value of each well was measured at 450 nm using the microplate reader (SpectraMAX M5; Molecular Devices Corporation, Sunnyvale, CA, USA) within 20 min.41
Flow Cytometry
In accordance to the instructions of the Tregs staining kit (eBioscience, San Diego, CA, USA), the process of flow cytometry was carried out. Then, the surface molecules were stained by fluorescein isothiocyanate (FITC)-CD4 antibody and allophycocyanin (APC)-conjugated CD25 antibody at 4°C for 15 min. Next, cells were washed by flow cytometry staining buffer, added with permeation working solution, and stained with phycoerythrin (PE)-conjugated FOXP3 antibody at 4°C for 30 min. Finally, cells were washed, resuspended by flow cytometry staining buffer, and analyzed with a Gallios flow cytometer (Beckman Coulter, Chaska, MN, USA). WinMDI 2.8 software (The Scripps Institute, San Diego, CA, USA) was employed to evaluate the ratio of positive cells of Tregs.42
Suppression Assay
The in vitro-suppressive capacity of expanded Tregs was assessed with a carboxyfluorescein succinimidyl ester (CFSE) inhibition assay as previously mentioned.43 In brief, peripheral blood mononuclear cells (PBMNCs) were purified, labeled with CFSE (Invitrogen, Carlsbad, CA, USA), and stimulated with anti-CD3 monoclonal antibody (mAb)-coated beads ± cultured tTregs (1:2–32 tTregs/PBMNCs).
In Vivo Assay
In the study, a total of 56 male nude mice (BALB/c, aged 5 weeks, weighing 15–25 g) were acquired from Shanghai Laboratory Animal Center of Chinese Academy of Sciences (Shanghai, China). For the establishment of a subcutaneous xenograft HCC model, the nude mice were separated evenly into seven groups (n = 8) and then subcutaneously injected at the left axilla with MHCC97 cells (1 × 107 cells). When the tumor volume reached 100 mm3 after about 10 days, the nude mice were injected with cells after treatment of si-NC, lncRNA FENDRR, si-lncRNA FENDRR, NC mimic, miR-423-5p mimic, and both lncRNA FENDRR and miR-423-5p mimic, respectively. This whole process of intratumoral injection was carried out every 2 days for 2 weeks. The tumor volume was calculated every week with the use of the formula: V (volume) = W (width)2 × L (length)/2. On the 35th day, the nude mice were euthanized by using CO2, with tumor weight measured. The siRNA was packed with liposome before intratumoral injection. According to the instructions (Altogen Biosystems, Las Vegas, NV, USA), this study used cationic liposomes-based in vivo transfer reagent for the purpose of preparing the si-RNA transfer reagent complex.44
For the orthotopic HCC model, a single tumor nodule could be analyzed in the liver after 6 days of an injection of 2 × 106 treated MHCC97 cells into the left liver lobe of nude mice. The total observation duration lasted 70 days, and the nude mice were exposed to anesthesia and then euthanized if moribund.45
Immunofluorescence Assay
The immunofluorescence analysis for orthotopic HCC tissues was performed in a way reported in a previous study.46 The orthotopic HCC tumor tissues were fixed in 10% formalin for 48 h, embedded in paraffin before sectioning, and then blocked with protein block serum-free solution. A suspension of LX2 cells (1 × 106 cells/mL) was fixed with ice-cold acetone on ice for 15 min and then blocked with 5% (w/v) BSA. Subsequently, cells were incubated with the primary antibody, rabbit anti-human antibodies to CD4 (ab133616, 1:100) and CD8 (ab4055, 1:100) at 4°C overnight, and the secondary antibody, fluorescence-labeled goat anti-rabbit antibody to IgG (ab205718, 1:2,000–1:50,000), at room temperature for 1 h. Subsequently, the cells were stained with propidium iodide (PI; eBioscience, San Diego, CA, USA), washed, mounted, and visualized by fluorescence microscopy (Olympus IX51; Olympus Optical, Tokyo, Japan).
Statistical Analysis
All data were processed by SPSS 22.0 statistical software (IBM, Armonk, NY, USA). Measurement data were expressed as mean ± SD. All data were calculated using a normal distribution test and homogeneity of variance test. Data conformed to normal distribution and homogeneity of variance were compared by paired t test within groups, by unpaired t test between two groups, and by one-way ANOVA or repeated-measurement ANOVA among multiple groups, followed by Tukey’s post hoc test. If the p value was <0.05, it was considered to be statistically significant.
Author Contributions
H.L., C.Q., J.Z., and X.Y. designed the study. H.T. collated the data, carried out data analyses, and produced the initial draft of the manuscript. Z.Y., H.Z., and X.F. contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper.
Contributor Information
Xiaomeng Yi, Email: yixiaom@mail.sysu.edu.cn.
Hui Tang, Email: chenux@aliyun.com.
Jianwen Zhang, Email: zhjianw2@mail.sysu.edu.cn.
References
- 1.Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Chaturvedi V.K., Singh A., Dubey S.K., Hetta H.F., John J., Singh M.P. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb. Pathog. 2019;128:184–194. doi: 10.1016/j.micpath.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Hedenstierna M., Nangarhari A., Weiland O., Aleman S. Diabetes and Cirrhosis Are Risk Factors for Hepatocellular Carcinoma After Successful Treatment of Chronic Hepatitis C. Clin. Infect. Dis. 2016;63:723–729. doi: 10.1093/cid/ciw362. [DOI] [PubMed] [Google Scholar]
- 4.Dragani T.A. Risk of HCC: genetic heterogeneity and complex genetics. J. Hepatol. 2010;52:252–257. doi: 10.1016/j.jhep.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Sherman M. Hepatocellular carcinoma: epidemiology, surveillance, and diagnosis. Semin. Liver Dis. 2010;30:3–16. doi: 10.1055/s-0030-1247128. [DOI] [PubMed] [Google Scholar]
- 6.Colombo M., Iavarone M. Role of antiviral treatment for HCC prevention. Best Pract. Res. Clin. Gastroenterol. 2014;28:771–781. doi: 10.1016/j.bpg.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Greten T.F., Wang X.W., Korangy F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut. 2015;64:842–848. doi: 10.1136/gutjnl-2014-307990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang X., Wang J., Deng X., Xiong F., Ge J., Xiang B., Wu X., Ma J., Zhou M., Li X. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer. 2019;18:10. doi: 10.1186/s12943-018-0928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J.M., Chen D.S. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure) Ann. Oncol. 2016;27:1492–1504. doi: 10.1093/annonc/mdw217. [DOI] [PubMed] [Google Scholar]
- 10.Zou H., Shao C.X., Zhou Q.Y., Zhu G.Q., Shi K.Q., Braddock M., Huang D.S., Zheng M.H. The role of lncRNAs in hepatocellular carcinoma: opportunities as novel targets for pharmacological intervention. Expert Rev. Gastroenterol. Hepatol. 2016;10:331–340. doi: 10.1586/17474124.2016.1116382. [DOI] [PubMed] [Google Scholar]
- 11.Qi P., Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod. Pathol. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 12.Grote P., Wittler L., Hendrix D., Koch F., Währisch S., Beisaw A., Macura K., Bläss G., Kellis M., Werber M., Herrmann B.G. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mou Y., Wang D., Xing R., Nie H., Mou Y., Zhang Y., Zhou X. Identification of long noncoding RNAs biomarkers in patients with hepatitis B virus-associated hepatocellular carcinoma. Cancer Biomark. 2018;23:95–106. doi: 10.3233/CBM-181424. [DOI] [PubMed] [Google Scholar]
- 14.Wang B., Xian J., Zang J., Xiao L., Li Y., Sha M., Shen M. Long non-coding RNA FENDRR inhibits proliferation and invasion of hepatocellular carcinoma by down-regulating glypican-3 expression. Biochem. Biophys. Res. Commun. 2019;509:143–147. doi: 10.1016/j.bbrc.2018.12.091. [DOI] [PubMed] [Google Scholar]
- 15.Sadri Nahand J., Bokharaei-Salim F., Salmaninejad A., Nesaei A., Mohajeri F., Moshtzan A., Tabibzadeh A., Karimzadeh M., Moghoofei M., Marjani A. microRNAs: Key players in virus-associated hepatocellular carcinoma. J. Cell. Physiol. 2019;234:12188–12225. doi: 10.1002/jcp.27956. [DOI] [PubMed] [Google Scholar]
- 16.Stiuso P., Potenza N., Lombardi A., Ferrandino I., Monaco A., Zappavigna S., Vanacore D., Mosca N., Castiello F., Porto S. MicroRNA-423-5p Promotes Autophagy in Cancer Cells and Is Increased in Serum From Hepatocarcinoma Patients Treated With Sorafenib. Mol. Ther. Nucleic Acids. 2015;4:e233. doi: 10.1038/mtna.2015.8. [DOI] [PubMed] [Google Scholar]
- 17.Cretu A., Sha X., Tront J., Hoffman B., Liebermann D.A. Stress sensor Gadd45 genes as therapeutic targets in cancer. Cancer Ther. 2009;7(A):268–276. [PMC free article] [PubMed] [Google Scholar]
- 18.Ou D.L., Shen Y.C., Yu S.L., Chen K.F., Yeh P.Y., Fan H.H., Feng W.C., Wang C.T., Lin L.I., Hsu C., Cheng A.L. Induction of DNA damage-inducible gene GADD45beta contributes to sorafenib-induced apoptosis in hepatocellular carcinoma cells. Cancer Res. 2010;70:9309–9318. doi: 10.1158/0008-5472.CAN-10-1033. [DOI] [PubMed] [Google Scholar]
- 19.Qiu W., David D., Zhou B., Chu P.G., Zhang B., Wu M., Xiao J., Han T., Zhu Z., Wang T. Down-regulation of growth arrest DNA damage-inducible gene 45beta expression is associated with human hepatocellular carcinoma. Am. J. Pathol. 2003;162:1961–1974. doi: 10.1016/s0002-9440(10)64329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y., Boyle D.L., Hammaker D., Edgar M., Franzoso G., Firestein G.S. Suppression of collagen-induced arthritis in growth arrest and DNA damage-inducible protein 45β-deficient mice. Arthritis Rheum. 2011;63:2949–2955. doi: 10.1002/art.30497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu T.P., Huang M.D., Xia R., Liu X.X., Sun M., Yin L., Chen W.M., Han L., Zhang E.B., Kong R. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J. Hematol. Oncol. 2014;7:63. doi: 10.1186/s13045-014-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong B., Zhou B., Sun Z., Huang S., Han L., Nie H., Chen G., Liu S., Zhang Y., Bao N. LncRNA-FENDRR mediates VEGFA to promote the apoptosis of brain microvascular endothelial cells via regulating miR-126 in mice with hypertensive intracerebral hemorrhage. Microcirculation. 2018;25:e12499. doi: 10.1111/micc.12499. [DOI] [PubMed] [Google Scholar]
- 23.Lin J., Huang S., Wu S., Ding J., Zhao Y., Liang L., Tian Q., Zha R., Zhan R., He X. MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis. 2011;32:1641–1647. doi: 10.1093/carcin/bgr199. [DOI] [PubMed] [Google Scholar]
- 24.Wörns M.A., Galle P.R. Future perspectives in hepatocellular carcinoma. Dig. Liver Dis. 2010;42(Suppl 3):S302–S309. doi: 10.1016/S1590-8658(10)60521-X. [DOI] [PubMed] [Google Scholar]
- 25.Marquardt J.U., Galle P.R., Teufel A. Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): an emerging field for advanced technologies. J. Hepatol. 2012;56:267–275. doi: 10.1016/j.jhep.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Vinay D.S., Ryan E.P., Pawelec G., Talib W.H., Stagg J., Elkord E., Lichtor T., Decker W.K., Whelan R.L., Kumara H.M.C.S. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 2015;35(Suppl):S185–S198. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Wu L.M., Ji J.S., Yang Z., Xing C.Y., Pan T.T., Xie H.Y., Zhang F., Zhuang L., Zhou L., Zheng S.S. Oncogenic role of microRNA-423-5p in hepatocellular carcinoma. HBPD INT. 2015;14:613–618. doi: 10.1016/s1499-3872(15)60038-8. [DOI] [PubMed] [Google Scholar]
- 28.Zhang G., Han G., Zhang X., Yu Q., Li Z., Li Z., Li J. Long non-coding RNA FENDRR reduces prostate cancer malignancy by competitively binding miR-18a-5p with RUNX1. Biomarkers. 2018;23:435–445. doi: 10.1080/1354750X.2018.1443509. [DOI] [PubMed] [Google Scholar]
- 29.Oft M. IL-10: master switch from tumor-promoting inflammation to antitumor immunity. Cancer Immunol. Res. 2014;2:194–199. doi: 10.1158/2326-6066.CIR-13-0214. [DOI] [PubMed] [Google Scholar]
- 30.Yang J., Yan J., Liu B. Targeting VEGF/VEGFR to Modulate Antitumor Immunity. Front. Immunol. 2018;9:978. doi: 10.3389/fimmu.2018.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gigante M., Gesualdo L., Ranieri E. TGF-beta: a master switch in tumor immunity. Curr. Pharm. Des. 2012;18:4126–4134. doi: 10.2174/138161212802430378. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Zhang W., Liu P., Xu Y., Tang L., Chen W., Guan X. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. OncoTargets Ther. 2018;11:1403–1412. doi: 10.2147/OTT.S149511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S., Zeng A., Hu Q., Yan W., Liu Y., You Y. miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro-oncol. 2017;19:55–65. doi: 10.1093/neuonc/now129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J., Ding T., Pan W., Zhu L.Y., Li L., Zheng L. Increased intratumoral regulatory T cells are related to intratumoral macrophages and poor prognosis in hepatocellular carcinoma patients. Int. J. Cancer. 2009;125:1640–1648. doi: 10.1002/ijc.24556. [DOI] [PubMed] [Google Scholar]
- 35.Yang H., Fu H., Wang B., Zhang X., Mao J., Li X., Wang M., Sun Z., Qian H., Xu W. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol. Carcinog. 2018;57:1223–1236. doi: 10.1002/mc.22838. [DOI] [PubMed] [Google Scholar]
- 36.Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., Yu Z., Yang J., Wang B., Sun H. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arocho A., Chen B., Ladanyi M., Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn. Mol. Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- 38.Alexopoulou A.N., Leao M., Caballero O.L., Da Silva L., Reid L., Lakhani S.R., Simpson A.J., Marshall J.F., Neville A.M., Jat P.S. Dissecting the transcriptional networks underlying breast cancer: NR4A1 reduces the migration of normal and breast cancer cell lines. Breast Cancer Res. 2010;12:R51. doi: 10.1186/bcr2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collin S.P. Topographic organization of the ganglion cell layer and intraocular vascularization in the retinae of two reef teleosts. Vision Res. 1989;29:765–775. doi: 10.1016/0042-6989(89)90089-8. [DOI] [PubMed] [Google Scholar]
- 40.Luan W., Zhou Z., Ni X., Xia Y., Wang J., Yan Y., Xu B. Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J. Cancer Res. Clin. Oncol. 2018;144:531–542. doi: 10.1007/s00432-018-2582-z. [DOI] [PubMed] [Google Scholar]
- 41.Engvall E., Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 42.Hou P.F., Zhu L.J., Chen X.Y., Qiu Z.Q. Age-related changes in CD4+CD25+FOXP3+ regulatory T cells and their relationship with lung cancer. PLoS ONE. 2017;12:e0173048. doi: 10.1371/journal.pone.0173048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y., Gao J., Zhang S., Gu J., Lu H., Xia Y., Zhu Q., Qian X., Zhang F., Zhang C. miR-142-3p regulates autophagy by targeting ATG16L1 in thymic-derived regulatory T cell (tTreg) Cell Death Dis. 2018;9:290. doi: 10.1038/s41419-018-0298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 45.Xiao J., Xing F., Liu Y., Lv Y., Wang X., Ling M.T., Gao H., Ouyang S., Yang M., Zhu J. Garlic-derived compound S-allylmercaptocysteine inhibits hepatocarcinogenesis through targeting LRP6/Wnt pathway. Acta Pharm. Sin. B. 2018;8:575–586. doi: 10.1016/j.apsb.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y., Xi D., Ding W., Wang F., Zhou H., Ning Q. Soluble FGL2, a novel effector molecule of activated hepatic stellate cells, regulates T-cell function in cirrhotic patients with hepatocellular carcinoma. Hepatol. Int. 2014;8:567–575. doi: 10.1007/s12072-014-9568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]