Abstract
Endosymbioses between bacteria and eukaryotes are enormously important in ecology and evolution, and as such are intensely studied. Despite this, the range of investigated hosts is narrow in the context of the whole eukaryotic tree of life: most of the information pertains to animal hosts, while most of the diversity is found in unicellular protists. A prominent case study is the ciliate Euplotes, which has repeatedly taken up the bacterium Polynucleobacter from the environment, triggering its transformation into obligate endosymbiont. This multiple origin makes the relationship an excellent model to understand recent symbioses, but Euplotes may host bacteria other than Polynucleobacter, and a more detailed knowledge of these additional interactions is needed in order to correctly interpret the system. Here, we present the first systematic survey of Euplotes endosymbionts, adopting a classical as well as a metagenomic approach, and review the state of knowledge. The emerging picture is indeed quite complex, with some Euplotes harbouring rich, stable prokaryotic communities not unlike those of multicellular animals. We provide insights into the distribution, evolution and diversity of these symbionts (including the establishment of six novel bacterial taxa), and outline differences and similarities with the most well-understood group of eukaryotic hosts: insects.
Keywords: Devosia, Euplotes, Francisellaceae, Holosporales, prokaryote–eukaryote symbioses, Rickettsiales
1. Background
Endosymbiosis is defined as a highly interconnected relationship between two organisms of different species, one of which (the endosymbiont) lives inside the other (the host), and is a widespread and important phenomenon deeply affecting ecology and evolution [1,2]. Symbiotic events were involved in several milestones of the history of life, including the origin of mitochondria and plastids [3], the ability of animals to digest plant material [4] and the building of coral reefs [5]. Because of their ubiquity and importance, bacteria–eukaryote symbioses are the subject of a vast literature. However, nearly all model systems are focused on a single type of host: insects [4,6,7]. Studies on insect symbioses over several decades, especially mutualisms in hosts restricted to nutritionally poor foods (plant sap, vertebrate blood, wood), have provided important insights into the endosymbiosis process. Nevertheless, to understand universal rules, we need to expand the range of investigations to a variety of hosts and ecological contexts, and identify suitable systems among all eukaryotic lineages (e.g. [8,9]).
One such model that has been developing over recent years is Euplotes, a speciose genus of unicellular ciliates found in many aquatic environments [10]. All Euplotes species in the ‘clade B’ group [11] harbour endocytoplasmic bacteria that are both obligate (they cannot survive outside their host) and essential (the host survival and reproduction depend on them) [12–14]. The most common of these bacteria belong to the genus Polynucleobacter [15], and are coopted from an abundant free-living pool in the water column [16]. Extant symbiotic Polynucleobacter arose multiple times independently and relatively recently [17], evolving from very similar ancestors and experiencing similar selective pressures. They therefore represent the end products of a natural evolutionary experiment rerunning the evolutionary transition from free-living to obligate endosymbiont. Access to free-living strains closely related to each symbiotic lineage has allowed us to address questions that cannot be answered in systems where symbiosis originated only once, in the distant past.
But endosymbioses between Euplotes and prokaryotes are by no means limited to Polynucleobacter. A minority of populations in clade B depend on different bacteria, members of the genera Devosia [18] and ‘Candidatus Protistobacter’ [14,19]. Either of them may be a remnant of the original symbiotic event, indeed co-evolving with its host but being replaced in most cases by Polynucleobacter. In addition to these essential symbionts, many Euplotes also harbour ‘accessory’ bacteria that are probably not required for host survival since they are not present in all strains of their host species, always co-occur with known essential symbionts and usually belong to groups of intracellular parasites [12,20–24]. Finally, bacterial symbioses in species of Euplotes outside clade B are considerably less studied, but have been occasionally reported [25–27].
Here, we provide the first detailed survey of the diversity of bacteria harboured by Euplotes. We examined a large number of Euplotes strains, integrating the standard ‘Full-Cycle rRNA Approach’ [28] that involves characterization of the 16S rRNA gene and validation with fluorescent in situ hybridization (FISH), with metagenomic screening to enhance the completeness of the survey. Further developing Euplotes as a useful model for the study of endosymbiosis requires an understanding of all the components of the system. This is the most comprehensive attempt to date to achieve this goal. We report and discuss previously unknown symbiotic taxa (25 bacterial strains including six new species and three new genera), their host distribution and their features of interest in order to understand the intricacies of these multi-partner relationships.
2. Methods
(a). Overview and source of Euplotes
Novel data are provided for 17 Euplotes monoclonal strains (14) or populations (3), each coming from a different sampling site and representing in total eight morphospecies. Two freshwater strains were never previously characterized: Eae4 and Eae6, both assigned to Euplotes aediculatus based on their 18S rRNA gene sequences. Both were collected in Tuscany (Italy) and maintained as reported elsewhere (e.g. [17]). Other Euplotes were previously identified at the species level (see electronic supplementary material, table S1 for a complete list), often together with some of their symbionts (see electronic supplementary material, table S2). We describe additional bacteria harboured by these Euplotes strains and populations, using FISH experiments to validate 16S rRNA gene sequences of putative symbionts obtained by PCR amplification and Sanger sequencing [28] and later integrated with metagenomic screening. In most instances, live stocks were available. In three cases (Euplotes eurystomus EM, Euplotes octocarinatus FL(12)-VI and Euplotes woodruffi POH1), metagenomic screening could be conducted on old DNA extractions, but no live cell was available for FISH. Only one strain, Euplotes enigma MaS2, died before either DNA of sufficient quality for metagenomics or fixed cells for FISH could be collected; the detection of bacterial symbionts in this strain is based on 16S rRNA gene amplification, cloning and Sanger sequencing.
(b). Molecular methods
DNA extractions, Illumina library preparations and MiSeq sequencing were performed as reported previously [17] for 13 of the Euplotes strains and populations (Eae1–6, Eda1, Eoc1/2, Ewo1, POH1, Fsp1.4, Na2 and LIV5). Archived extracted DNA from other Euplotes was obtained as described in the corresponding reference papers (electronic supplementary material, table S1). Accessory symbionts in population EMP and strain MaS2 were characterized through alphaproteobacterial-specific PCR amplification of the 16S rRNA gene and cloning as described in [14], with the exception of Caedimonas in EMP, whose 16S rRNA gene sequence was amplified with primers 16S_F35Caedcar [29] and 1492R (modified from [30]) and sequenced directly.
(c). Metagenomic screening
Raw metagenomic reads were trimmed as reported previously [17] and screened for 16S rRNA gene sequences with PhyloFlash v. 3.3b1 [31]. Full-length 16S and 18S rRNA gene sequences were then extracted from the targeted PhyloFlash assembly. A total metagenome assembly was also carried out in SPAdes v. 3.12.0 using default settings [32]. The resulting assembly graph was inspected using Bandage [33] and the assembly was checked by BlobTools [34] to confirm multiple closely related symbionts present in a single host. Only fully assembled 16S rRNA were considered. Sequences from putative symbionts (e.g. those belonging to groups of exclusively intracellular bacteria or related to previously described protist symbionts) had usually far higher coverage than those from common environmental contaminants living in the cultures.
(d). Oligonucleotide probe design and fluorescence in situ hybridization protocol
Species-specific probes were designed for the newly described symbiont species and for Francisella adeliensis. The probe-design tool from the ARB software package was used [35], based on the SILVA 128 database [36]. The specificity of each new probe was also tested in silico on the Ribosomal Database Project [37]; their sequences are reported in electronic supplementary material, table S2. Fluorescently labelled oligonucleotides were synthesized by Eurofins Genomics (Ebersberg, Germany). FISH was performed according to [38], using two probes of different specificity and emission wavelength in each experiment, adding DAPI to visualize the ciliate nucleus and employing negative controls with no probes to test for autofluorescence. Hybridized ciliates were observed with a Zeiss Axioplan epifluorescence microscope equipped with a Nikon Digital Sight DS-U1 camera and pictures were captured by ACT-2U software. At least 20 cells per host strain were observed in each experiment. Most FISH were performed or repeated at least a year after DNA was obtained for metagenomics libraries, and hence attest bacterial populations stable at this temporal scale.
(e). Phylogenetic inference
16S rRNA sequences were aligned with the linsi algorithm in MAFFT [39]. Character matrices were trimmed at both ends to remove columns with more than 50% missing data. Maximum-likelihood trees were inferred using IQ-TREE v. 1.6.6 [40], using the best-fitting model according to the Bayesian information criterion.
3. Results
(a). Symbionts of Euplotes aediculatus (clade B)
16S rRNA sequences from Eae2, Eae3 and Eae5 libraries did not suggest the presence of putative symbionts beyond the previously reported Polynucleobacter (Betaproteobacteria, Burkholderiales) [17], and the cytoplasmic signal from eubacterial and Polynucleobacter-specific fluorescent probes confirmed this. The same results were obtained for the newly analysed strain Eae4.
In strains Eae1 and Eae6, harbouring the essential symbionts Polynucleobacter [17] and ‘Ca. Protistobacter’, respectively, the metagenomic screening additionally detected ‘Ca. Nebulobacter yamunensis' (Gammaproteobacteria, Thiotrichales) and ‘Ca. Cyrtobacter zanobii’ (Alphaproteobacteria, Rickettsiales). Species-specific oligonucleotide probes confirmed the presence of both bacteria in the cytoplasm of all inspected cells (electronic supplementary material, figure S1A–C). 16S rRNA sequences, bacterial shape and size, and abundance of symbionts matched those in the original descriptions [21,22].
(b). Symbionts of Euplotes daidaleos (clade B)
Strain Eda1 harbours Polynucleobacter [17]. Additionally, a 16S rRNA gene sequence affiliated to ‘Ca. Finniella’ (Alphaproteobacteria, Holosporales) was detected in the metagenome. The sequence shares 95.4% identity with that of ‘Ca. Finniella lucida’ from the cercozoan Orciraptor [41]. The species-specific oligonucleotide probe Fin_1025 was designed for FISH experiments, confirming the presence of this bacterium in the cytoplasm of all inspected Eda1 cells. Relatively short (about 1.7 µm) rod-like bacteria were visible in some hosts, while in others, a second, extremely elongated (up to more than 25 µm) form was present. The two morphotypes occasionally occurred in the same host cell (figure 1a).
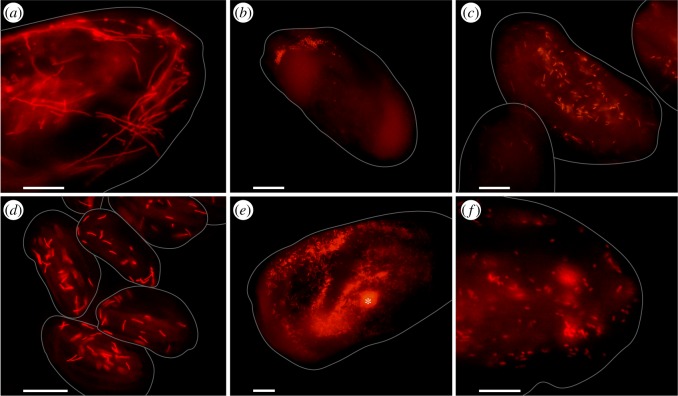
Figure 1.
FISHs with species-specific oligonucleotide probes for the six novel endosymbiotic taxa. (a) ‘Ca. Finniella dimorpha’ in E. daidaleos Eda1. (b) ‘Ca. Fujishimia apicalis' in E. octocarinatus Eoc1/2. (c) ‘Ca. Anadelfobacter sociabilis’ in E. octocarinatus Eoc1/2. (d) ‘Ca. Euplotella sexta’ in E. octocarinatus Eoc1/2. (e) ‘Ca. Bandiella numerosa’ in E. woodruffi Ewo1 (the asterisk marks autofluorescence signal from an undigested alga). (f) ‘Ca. Parafinniella ignota’ in Euplotes sp. EMP. Grey outlines represent Euplotes cells and were drawn based on the corresponding bright field pictures. Bars represent 10 µm. (Online version in colour.)
(c). Symbionts of Euplotes eurystomus (clade B)
Strain EM, now extinct, was originally described as a host of ‘Ca. Protistobacter’ [14]. The metagenomic screening on archived DNA revealed 16S rRNA gene sequences from ‘Ca. Protistobacter’ as well as four additional putative symbionts: (i) ‘Ca. Megaira polyxenophila’, a common symbiont found in many protists [42] (Alphaproteobacteria, Rickettsiales); (ii) an uncultured bacterium belonging to the family ‘Ca. Midichloriaceae’ (Alphaproteobacteria, Rickettsiales; 94.3–95.2% identity with representatives of the genus ‘Ca. Cyrtobacter’); (iii) a bacterium sharing high sequence identity (99.7%) with the ‘Ca. Finniella’ accessory symbiont of E. daidaleos Eda1; and (iv) a bacterium affiliated to ‘Ca. Endonucleariobacter rarus' (Gammaproteobacteria), an endosymbiont of the opisthokont Nuclearia [43] (96.5–97.3% identity). Live cells were not available for FISH experiments, but since all described Rickettsiales and Holosporales live intracellularly, it is safe to assume that at least three of the four mentioned bacteria are indeed endosymbiotic.
(d). Symbionts of Euplotes octocarinatus (clade B)
The monoclonal strain Eoc1 harboured Polynucleobacter [17]. Data presented here were obtained from the population Eoc1/2 that strain belonged to. At least five putative alphaproteobacterial accessory symbionts were predicted by the metagenomic screening: (i) a bacterium closely related to Holospora-like infectious symbionts (82.4% 16S rRNA identity to ‘Ca. Hafkinia simulans’, accession: MH319377); (ii) a second Holosporales symbiont resembling the ‘Ca. Finniella’ already mentioned for E. daidaleos Eda1 and E. eurystomus EM (99.0 –99.1% sequence identity); (iii) ‘Ca. Megaira polyxenophila’; and two representatives of the family ‘Ca. Midichloriaceae’, one (iv) affiliated to the genus ‘Ca. Anadelfobacter’ (95.4% identity to ‘Ca. Anadelfobacter veles', accession: FN552695), and the other (v) only distantly related to described bacteria (best BLAST hit: uncultured bacterium T47, 91.8% identity, accession: KU524857). The presence of ‘Ca. Megaira’ in the cytoplasm of Euplotes was confirmed using the oligonucleotide probe MegPol436 [23] (electronic supplementary material, figure S1D). Species-specific probes Fuji_838, Ana2_436 and EocBan_828 were designed and tested for the Holospora-related bacterium (figure 1b), ‘Ca. Anadelfobacter’ (figure 1c; electronic supplementary material, figure S1E) and the divergent ‘Ca. Midichloriaceae’ bacterium (figure 1d), respectively. They gave positive signals in all inspected host cells, except for Fuji_838. This probe matched small coccoid bacteria preferentially distributed at the anterior end of the cell, and sometimes entirely absent. Probe Fin_1025, validated on the ‘Ca. Finniella’ of Eda1, did not work on population Eoc1/2, despite several attempts at various formamide concentrations. It is possible that in this case, the symbiont was lost in the time between DNA extraction and FISH experiments.
The extinct strain FL(12)-VI was reported to harbour ‘Ca. Megaira polyxenophila’ [23] as well as the essential symbiont ‘Ca. Protistobacter’ [14]. Our metagenomic screening additionally found the same ‘Ca. Anadelfobacter’ (99.9% 16S rRNA gene sequence identity) described in the conspecific Eoc1/2.
(e). Symbionts of Euplotes woodruffi (clade B)
Strain Ewo1 harbours Polynucleobacter [17]. In the metagenomic screening, two accessory alphaproteobacteria were also found: ‘Ca. Megaira venefica’, originally described in Paramecium [42], and a bacterium associated with ‘Ca. Bandiella’, belonging to ‘Ca. Midichloriaceae’ and previously also observed in E. woodruffi [44] (95.8% 16S rRNA gene identity, accession: LN864514). FISH probes MegVene_95 [42] and the newly designed BanNum_173 confirmed the localization of the bacteria in the cytoplasm of all host cells, in very high number in the case of ‘Ca. Bandiella’ (figure 1e). A very similar ‘Ca. Bandiella’ (99.8% sequence identity), but no other accessory symbiont, emerged from the metagenomic screening of the extinct E. woodruffi strain POH1, which harboured ‘Ca. Protistobacter’ as its essential symbiont.
(f). Symbionts of Euplotes sp. (clade B)
Population EMP could not be unambiguously assigned to any known Euplotes morphospecies, but it is deeply nested within clade B and harbours Polynucleobacter [14]. Three accessory symbionts could be characterized by PCR amplification, cloning and FISH experiments: (i) ‘Ca. Megaira polyxenophila’ (electronic supplementary material, figure S1F); (ii) Caedimonas (formerly Caedibacter) varicaedens (Alphaproteobacteria, Holosporales), a ‘killer-symbiont’ of Paramecium [29,45] never detected before in Euplotes (electronic supplementary material, figure S1G); and (iii) a bacterium in the family Paracaedibacteraceae, like ‘Ca. Finniella’, but not closely related to any described symbiont (85.4% identity with ‘Ca. Finniella lucida’, accession: KT343635). The species-specific Paraf_838 probe was designed and tested for the Paracaedibacteraceae bacterium, targeting numerous small cytoplasmic bacteria (figure 1f).
(g). Symbionts of Euplotes platystoma (clade B)
Euplotes platystoma (some strains of which were previously misclassified as Euplotes harpa [46]) is more distantly related to all other clade B Euplotes species, and it is often sampled in low salinity rather than freshwater environments. Strain Fsp1.4 harbours Polynucleobacter [13], while strain Na2 is unique in clade B for harbouring a member of the genus Devosia (Alphaproteobacteria, Rhizobiales), ‘Ca. Devosia symbiotica’, as the essential symbiont [18]. Metagenomic screenings on these strains did not detect any additional 16S rRNA gene sequence that is likely to belong to accessory symbionts.
(h). Symbionts in marine Euplotes species of clade A
Strain LIV5 of Euplotes magnicirratus, like all previously screened strains of this species, depends on ‘Ca. Devosia euplotis' for reproduction and long-term survival [25]. Our metagenomic screening also recovered F. adeliensis (Gammaproteobacteria, Thiotrichales), described as a symbiont of Euplotes petzi [27], which belongs to the distantly related clade E. The probe Franci_199 confirmed the presence of the bacterium in the cytoplasm of LIV5 cells, although in relatively low amount (electronic supplementary material, figure S1H).
The single strain of Euplotes enigma we had access to did not survive long enough to perform a thorough investigation of its symbionts. Through PCR amplification and cloning, however, a partial 16S rRNA gene sequence similar to those of symbiotic Devosia in other Euplotes (96.5% identity with ‘Ca. Devosia euplotis’ and 97.1% identity with ‘Ca. Devosia symbiotica’) was obtained.
(i). Phylogenetic analysis
All symbiotic Polynucleobacter strains, including the newly described symbiont of E. aediculatus Eae4, fall within the PnecC clade, that originally coincided with the species Polynucleobacter necessarius [47] (figure 2a). Their relationship with free-living strains cannot be resolved using the 16S rRNA gene. The new ‘Ca. Protistobacter’ is the first reported in E. aediculatus, and clusters within the genus. The sister group status of Polynucleobacter and ‘Ca. Protistobacter’ within the family Burkholderiaceae is not strongly supported.
Figure 2.
16S rRNA-based phylogenetic affiliations of bacterial endosymbionts of Euplotes. (a) Phylogenetic tree of family Burkholderiaceae (Betaproteobacteria), including symbiotic Polynucleobacter forming a polyphyletic group in the otherwise free-living clade ‘PnecC’, and the exclusively symbiotic genus ‘Ca. Protistobacter’. (b) Phylogenetic tree of orders Rickettsiales and Holosporales (Alphaproteobacteria), entirely composed of intracellular bacteria harboured by diverse hosts. (c) Phylogenetic tree of the closely related families Francisellaceae and Fastidiosibacteraceae, including obligate and opportunistic endosymbionts as well as free-living bacteria. (d) Phylogenetic tree of Devosia (Alphaproteobacteria) and closely related genera. Euplotes endosymbionts are highlighted, and colour-coded according to their host species. Numbers in square brackets represent the number of sequences in collapsed nodes. Standard bootstrap supports, when at or above 70%, are provided close to the corresponding node. Bars stand for an estimated sequence divergence of 0.1.
Alphaproteobacterial symbionts belonging to Rickettsiales and Holosporales cluster within established families of obligate intracellular symbionts, in various relationships with existing genera (figure 2b). Three of the new strains are particularly long-branching and not reliably associated with described bacteria: one of the two ‘Ca. Midichloriaceae’ bacteria in Eoc1/2, the Holosporaceae bacterium in the same host and the Paracaedibacteraceae bacterium in Euplotes sp. EMP.
In Gammaproteobacteria, most Euplotes symbionts cluster in the related families Francisellaceae and Fastidiosibacteraceae (figure 2c). Finally, the partial sequence of Devosia obtained from E. enigma MaS2 belongs to a clade of symbionts together with the two previously described species found in Euplotes, although bootstrap support for the clade is low, as is the case for all subgenus relationships in Devosia (figure 2d).
4. Discussion
(a). Establishment of novel bacterial taxa
Defining bacterial ‘species’ is notoriously tricky, and ‘genus’ is an even more artificial concept. Due to the universal use of nucleotide sequences as standard data, most discrimination is based on nucleotide identity thresholds. When establishing new taxa, we applied a 94.5% 16S rRNA gene sequence identity threshold for genera and a 98.7% threshold for species [48], while also taking into account the support for taxa monophyly. Polynucleobacter is a slightly more complex case: symbiotic Polynucleobacter lineages are scattered in the clade that once corresponded to P. necessarius, but that has since been split into several species [47], all extremely similar at the 16S rRNA gene sequence level but differing considerably in gene content [49]. Symbiotic Polynucleobacter are here classified only as Polynucleobacter sp.
Of the 25 newly detected symbiotic strains, 11 belong to already established species: ‘Ca. Protistobacter heckmanni’ (1), ‘Ca. Megaira polyxenophila’ (3), ‘Ca. Megaira venefica’ (1), ‘Ca. Cyrtobacter zanobii’ (2), Caedimonas varicaedens (1), Francisella adeliensis (1) and ‘Ca. Nebulobacter yamunensis' (2). Of these, ‘Ca. Megaira venefica’ and Caedimonas varicaedens were never previously reported in Euplotes. The strain of ‘Ca. Megaira polyxenophila’ in E. eurystomus EM actually shares only 96.9% sequence identity with its conspecifics, but this is probably due to the low quality of this metagenomic sequence, and the phylogenetic analysis confirms its placement within this species.
Seven symbiotic strains were assigned to three new species in existing genera: ‘Ca. Anadelfobacter sociabilis' sp. nov. (in E. octocarinatus Eoc1/2 and FL(12)-VI), ‘Ca. Bandiella numerosa’ sp. nov. (in E. woodruffi Ewo1 and POH1) and ‘Ca. Finniella dimorpha’ sp. nov. (in E. daidaleos Eda1, E. eurystomus EM and E. octocarinatus Eoc1/2, the latter being the most divergent). Three strains warranted the establishment of as many novel genera. This was the case for ‘Ca. Euplotella sexta’ gen. nov., sp. nov., a ‘Ca. Midichloriaceae’ symbiont in E. octocarinatus Eoc1/2; ‘Ca. Fujishimia apicalis' gen. nov., sp. nov., the coccoid Holosporaceae bacterium infecting some cells of the same host population; and ‘Ca. Parafinniella ignota’ gen. nov., sp. nov., the Paracaedibacteraceae bacterium harboured by Euplotes sp. EMP. Formal descriptions of the new taxa are provided in electronic supplementary material, text S1.
Finally, three of the characterized putative symbionts belong to undescribed taxa that cannot be formally established in the absence of a successful FISH with a specific probe. The uncultured ‘Ca. Midichloriaceae’ in E. eurystomus EM is probably a new species of ‘Ca. Cyrtobacter’ according to sequence identity and phylogenetic position. Similarly, the gammaproteobacterial endosymbiont in the same host differs enough from ‘Ca. Endonucleariobacter rarus' to be considered a different species of the same genus. We have little information on the Devosia harboured by E. enigma, which is a very close relative of Euplotes symbionts ‘Ca. Devosia euplotis’ and ‘Ca. Devosia symbiotica’.
(b). Taxonomy, distribution and biology of the bacterial endosymbionts of Euplotes
A detailed synopsis of all known bacterial symbionts of Euplotes, with an in-depth review of the literature, can be found in the electronic supplementary material, text S2. Their distribution pattern is shown in figure 3.
Figure 3.
Synopsis of all Euplotes strains and populations screened for the presence of bacterial endosymbionts with molecular techniques. On the left, a simplified phylogeny of the Euplotes species investigated is presented. On columns, symbionts are organized first by their characterization as ‘essential’ or ‘accessory’, and then by taxonomy. Asterisks mark bacterial species found in hosts other than Euplotes. The ‘absence’ status is employed for negative FISH or negative metagenomic screening results. Black dots represent presences inferred by the recovery of 16S rRNA gene sequences (through Sanger or high-throughput sequencing) but not confirmed by FISH. (Online version in colour.)
Characterizations of bacterial endosymbionts in Euplotes are not uncommon, but have until now been mostly anecdotal, with descriptions of individual taxa selected from larger prokaryotic communities, additionally biased by the narrowness of the employed screening methods and the situational interest of the researchers. In order for Euplotes to become a robust model system, more information on the identity and distribution of its intracellular bacteria is needed. We have here attempted to provide a comprehensive picture by including metagenomic data mining and by investigating old, partially characterized Euplotes strains alongside new ones.
Some features emerge as generalized. But for a single report [50], all known symbionts of Euplotes have been observed in the cytoplasm, either free or enclosed in a host-derived membrane, whereas other ciliates may have conspicuous ectosymbionts [51,52] or harbour bacteria in their nuclear apparatus [24,29]. One explanation for the rarity of bacteria in the Euplotes macronucleus might be its relatively small diameter, although impressive Holospora infections can take place in the tiny micronuclei of certain Paramecium [53]. Alternatively, the complex ‘replication band’ structures in Euplotes and related ciliates, responsible for the duplication of DNA before cell division, might render the nucleus inhospitable. Another general feature is that no known symbiont of Euplotes is motile or possesses flagella, although more ultrastructural studies are needed to confirm this.
At least 15 genera and 20 species of bacteria have now confirmed representatives in Euplotes. However, they belong to relatively few large lineages. All Euplotes symbionts are Proteobacteria, and the vast majority is confined to the family Burkholderiaceae in Betaproteobacteria and the specialized intracellular orders Rickettsiales and Holosporales in Alphaproteobacteria.
In contrast with their limited phylogenetic affiliations, the accessory endosymbionts show an extensive range of distribution and co-distribution patterns. A single Euplotes can harbour from zero to six prokaryotic species stably coexisting in its cytoplasm (over several years in laboratory cultures). Most Rickettsiales and Holosporales, as well as Francisella, are found in different host species, but not in all strains or populations of those species. The essential symbionts are notably different: either Polynucleobacter or ‘Ca. Protistobacter’ are always present in clade B Euplotes species (with the single exception of E. platystoma Na2, harbouring ‘Ca. Devosia symbiotica’ instead), and ‘Ca. Devosia euplotis' is always present in the marine E. magnicirratus. No strong correlation with host taxonomy can be inferred for other bacteria. In clade B, in particular, accessory alpha- and gammaproteobacteria do not match the presence of either Polynucleobacter or ‘Ca. Protistobacter’, suggesting little, if any, taxon-specific interaction with these betaproteobacteria. Euplotes harbouring Devosia have not been intensely investigated yet, but they seem to be less rich in accessory symbionts. Finally, no clear pattern of co-occurrence among different accessory symbionts emerges, with an intriguing exception: ‘Ca. Cyrtobacter zanobii’ and ‘Ca. Nebulobacter yamunensis' from E. aediculatus are always detected together. Should this observation stand the test of time, it would definitely be interesting to look at their genomes for signs of metabolic integrations as reported in co-occurring symbionts of insects (e.g. [54]).
Phylogenetic analyses can provide many indirect insights on the biology of these bacteria. It was through phylogenomics that the multiple establishments of symbiosis in Polynucleobacter were proven [17]. Strains of ‘Ca. Megaira’, ‘Ca. Bandiella’ and Francisella in Euplotes are scattered in clades including symbionts of diverse hosts, sometimes from unlike environments. This provides strong evidence for horizontal transmission of these bacteria, by no means confined to ciliates. Details of the ecology of infectious bacteria in aquatic environments are largely unknown, and it would be important to assess if ciliates and other protists play a role in their spread, as arthropods do in terrestrial environments [55]. Horizontal transmission in culture has been observed only for ‘Ca. Bandiella woodruffi’, but it did not lead to long-term establishment in secondarily infected Euplotes [44].
It is tempting to conclude that at least the infectious taxa are probably parasitic. There is, however, no evidence for any harmful effect on the Euplotes hosts. The prevalence of most of these bacteria is close to 100% in isolated host strains, and the symbionts are usually present in high numbers (roughly correlating with the size of the bacteria) in each host cell, a footprint of well-adapted parasites or commensals. It cannot be excluded that some might even be beneficial to their hosts. Polynucleobacter, ‘Ca. Protistobacter’ and Devosia certainly are (for reasons still unclear [56]), and yet cannot be described as mutualists in the absence of long-term benefits for the bacterium.
(c). Comparison with insect symbioses: are Euplotes endosymbioses suitable model systems?
Protists are hugely diverse and far less known than metazoans and plants, which makes them intriguing as well as challenging model systems that require specific expertise. Euplotes is becoming the most deeply and widely sampled protist when it comes to symbiotic interactions with bacteria. This window into the diversity and evolutionary history of Euplotes symbionts allows us to draw preliminary comparisons to insect symbioses, that have been studied with molecular methods for three decades [57] and note a few interesting similarities and differences. This is made particularly relevant by the prominent position held by ciliates, among protists, as model organisms for several fundamental processes shared with metazoans [58], despite their extreme divergence in the evolutionary history of eukaryotes.
First, the narrow taxonomic diversity of Euplotes endosymbionts is strikingly mirrored by insect symbioses where clades such as Wolbachia, Rickettsia (both Rickettsiales), Sodalis, Arsenophonus (both Gammaproteobacteria in the family Enterobacteriaceae) and ‘Candidatus Cardinium’ (Bacteroidetes) are extremely common symbionts due to their ability to infect eukaryotic cells and spread horizontally among species [59]. Within groups that are common symbionts of all eukaryotes, such as Rickettsiales, the total diversity of protist symbionts is much higher, probably reflecting the evolutionary time for these symbioses to originate and diversify in protists, the bacterivorous nature of the hosts, and their lack of complex immune systems.
Second, the most evolutionarily successful bacteria associated with arthropods and nematodes are reproductive manipulators that shift the sex ratios of their hosts to increase their chance of maternal transmission, including Wolbachia, Rickettsia and ‘Ca. Cardinium’. No such manipulation is needed in single-celled eukaryotes, but we predict that some of the ciliate symbionts are probably just parasites that are good at (i) staying in both daughter cells after the host divides, (ii) avoiding host defence against bacteria and (iii) spreading horizontally by infectious stages (e.g. spores) or when their original host is eaten by a different protist. On the other hand, accessory mutualists in insects were shown to have a diverse array of functions, particularly nutritional and defensive [60]. Whether some of the numerous accessory symbionts in Euplotes confer protection from pathogens or provide nutrients to the host or co-symbionts remains to be elucidated, although we predict that nutritional symbioses will not be very common in bacterivorous organisms.
Third, this study shows that up to six different symbionts can co-occur in the cytoplasm of a single Euplotes species. Of course, this is not easily comparable with much larger, multicellular animals that often house different bacterial symbionts in distinct bacteriocyte cells, and yet less than 10 different species of intracellular symbionts are known from the most-understood insects such as whiteflies from the Bemisia tabaci species complex or pea aphids [60]. In the case of whiteflies, five accessory symbionts (‘Candidatus Hamiltonella’, Arsenophonus, ‘Ca. Cardinium’, Wolbachia and Rickettsia) can even co-occur with an essential ‘Candidatus Portiera’ symbiont in the same host cell [61] and either compete or cooperate in diverse metabolic interactions [62]. Unlike in insects, it is difficult to sample the same protist species from multiple geographical locations, so drawing conclusions about prevalence and abundance across populations is premature. Nevertheless, some of the ciliate symbionts appear to be generalists infecting various protists and some appear to be species-specific, again drawing parallels with insect symbioses [60].
Our view of eukaryotic symbioses is biased by our model systems that currently do not even come close to representing the possible range of eukaryotic host diversity. Due to the long history of research, increasing amount of data, and ease of laboratory culture of both the host and free-living relatives of some of the symbionts, we view Euplotes symbioses as a valuable model for understanding symbioses in single-celled eukaryotes and identify generalized features of bacteria–eukaryote symbioses.
Supplementary Material
Acknowledgements
The authors wish to thank Stanley Prescott, Alessandro Ristori, Marta Stancampiano and Charissa Wall for help with PCR and FISH experiments. We acknowledge Simone Gabrielli for his help with artwork preparation.
Data accessibility
18S and 16S rRNA gene sequences are deposited in the GenBank/EMBL/ENA database (accession numbers: LR588889-90, FR873728-30 and LR585330-53).
Competing interests
We declare we have no competing interests.
Funding
This work was supported by a grant (RGPIN-2014-03994) from the Natural Sciences and Engineering Research Council of Canada to P.J.K. and by funding from the University of Pisa (565-60%2017, 565-60%2018) and the Italian Ministry of University and Research (565-FFABR 2017) to C.V. F.H. was supported by an EMBO fellowship (ALTF 1260-2016).
References
- 1.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Müller DB, Vogel C, Bai Y, Vorholt JA. 2016. The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50, 211–234. ( 10.1146/annurev-genet-120215-034952) [DOI] [PubMed] [Google Scholar]
- 3.Dyall SD, Brown MT, Johnson PJ. 2004. Ancient invasions: from endosymbionts to organelles. Science 304, 253–257. ( 10.1126/science.1094884) [DOI] [PubMed] [Google Scholar]
- 4.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. New York, NY: Interscience. [Google Scholar]
- 5.Baker AC. 2003. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689. ( 10.1146/annurev.ecolsys.34.011802.132417) [DOI] [Google Scholar]
- 6.Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189. ( 10.1146/annurev.micro.59.030804.121041) [DOI] [PubMed] [Google Scholar]
- 7.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10, 13–26. ( 10.1038/nrmicro2670) [DOI] [PubMed] [Google Scholar]
- 8.Spribille T, et al. 2016. Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science 353, 488–492. ( 10.1126/science.aaf8287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowack ECM, Weber APM. 2018. Genomics-informed insights into endosymbiotc organelle evolution in photosynthetic eukaryotes. Annu. Rev. Plant Biol. 69, 51–84. ( 10.1146/annurev-arplant-042817-040209) [DOI] [PubMed] [Google Scholar]
- 10.Boscaro V, Syberg-Olsen MJ, Irwin NAT, del Campo J, Keeling PJ. 2019. What can environmental sequences tell us about the distribution of low-rank taxa? The case of Euplotes (Ciliophora, Spirotrichea), including a description of Euplotes enigma sp. nov. J. Eukaryot. Microbiol. 66, 281–293. ( 10.1111/jeu.12669) [DOI] [PubMed] [Google Scholar]
- 11.Syberg-Olsen MJ, Irwin NAT, Vannini C, Erra F, Di Giuseppe G, Boscaro V, Keeling PJ. 2016. Biogeography and character evolution of the ciliate genus Euplotes (Spirotrichea, Euplotia), with description of Euplotes curdsi sp. nov. PLoS ONE 11, e0165442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heckmann K, Ten Hagen R, Görtz H-D. 1983. Freshwater Euplotes species with a 9 type 1 cirrus pattern depend upon endosymbionts. J. Protozool. 30, 284–289. ( 10.1111/j.1550-7408.1983.tb02917.x) [DOI] [Google Scholar]
- 13.Vannini C, Petroni G, Verni F, Rosati G. 2005. Polynucleobacter bacteria in the brackish-water species Euplotes harpa (Ciliata Hypotrichia). J. Eukaryot. Microbiol. 52, 116–122. ( 10.1111/j.1550-7408.2005.04-3319.x) [DOI] [PubMed] [Google Scholar]
- 14.Vannini C, Ferrantini F, Ristori A, Verni F, Petroni G. 2012. Betaproteobacterial symbionts of the ciliate Euplotes: origin and tangled evolutionary path of an obligate microbial association. Environ. Microbiol. 14, 2553–2563. ( 10.1111/j.1462-2920.2012.02760.x) [DOI] [PubMed] [Google Scholar]
- 15.Heckmann K, Schmidt HJ. 1987. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus. Int. J. Syst. Bacteriol. 37, 456–457. ( 10.1099/00207713-37-4-456) [DOI] [Google Scholar]
- 16.Jezberová J, Jezbera J, Brandt U, Lindström ES, Langenheder S, Hahn MW. 2010. Ubiquity of Polynucleobacter necessarius ssp. asymbioticus in lentic freshwater habitats of a heterogeneous 2000 km2 area. Environ. Microbiol. 12, 658–669. ( 10.1111/j.1462-2920.2009.02106.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boscaro V, Kolisko M, Felletti M, Vannini C, Lynn DH, Keeling PJ. 2017. Parallel genome reduction in symbionts descended from closely related free-living bacteria. Nat. Ecol. Evol. 1, 1160–1167. ( 10.1038/s41559-017-0237-0) [DOI] [PubMed] [Google Scholar]
- 18.Boscaro V, Fokin SI, Petroni G, Verni F, Keeling PJ, Vannini C. 2018. Symbiont replacement between bacteria of different classes reveals additional layers of complexity in the evolution of symbiosis in the ciliate Euplotes. Protist 169, 43–52. ( 10.1016/j.protis.2017.12.003) [DOI] [PubMed] [Google Scholar]
- 19.Vannini C, Ferrantini F, Verni F, Petroni G. 2013. A new obligate bacterial symbiont colonizing the ciliate Euplotes in brackish and freshwater: ‘Candidatus Protistobacter heckmanni’. Aquat. Microb. Ecol. 70, 233–243. ( 10.3354/ame01657) [DOI] [Google Scholar]
- 20.Vannini C, Ferrantini F, Schleifer K-H, Ludwig W, Verni F, Petroni G. 2010. ‘Candidatus Anadelfobacter veles' and ‘Candidatus Cyrtobacter comes,’ two new Rickettsiales species hosted by the protist ciliate Euplotes harpa (Ciliophora, Spirotrichea). Appl. Environ. Microbiol. 76, 4047–4054. ( 10.1128/AEM.03105-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boscaro V, Vannini C, Fokin SI, Verni F, Petroni G. 2012. Characterization of ‘Candidatus Nebulobacter yamunensis' from the cytoplasm of Euplotes aediculatus (Ciliophora, Spirotrichea) and emended description of the family Francisellaceae. Syst. Appl. Microbiol. 35, 432–440. ( 10.1016/j.syapm.2012.07.003) [DOI] [PubMed] [Google Scholar]
- 22.Boscaro V, Petroni G, Ristori A, Verni F, Vannini C. 2013. ‘Candidatus Defluviella procrastinata’ and ‘Candidatus Cyrtobacter zanobii’, two novel ciliate endosymbionts belonging to the ‘Midichloria clade’. Microb. Ecol. 65, 302–310. ( 10.1007/s00248-012-0170-3) [DOI] [PubMed] [Google Scholar]
- 23.Schrallhammer M, Ferrantini F, Vannini C, Galati S, Schweikert M, Görtz H-D, Verni F, Petroni G. 2013. ‘Candidatus Megaira polyxenophila’ gen. nov., sp. nov.: considerations on evolutionary history, host range and shift of early divergent rickettsiae. PLoS ONE 8, e72581 ( 10.1371/journal.pone.0072581) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vannini C, et al. 2014. Flagellar movement in two bacteria of the family Rickettsiaceae: a re-evaluation of motility in an evolutionary perspective. PLoS ONE 2, e87718 ( 10.1371/journal.pone.0087718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vannini C, Rosati G, Verni F, Petroni G. 2004. Identification of the bacterial endosymbionts of the marine ciliate Euplotes magnicirratus (Ciliophora, Hypotrichia) and proposal of ‘Candidatus Devosia euplotis'. Int. J. Syst. Evol. Microbiol. 54, 1151–1156. ( 10.1099/ijs.0.02759-0) [DOI] [PubMed] [Google Scholar]
- 26.Schrallhammer M, Schweikert M, Vallesi A, Verni F, Petroni G. 2011. Detection of a novel subspecies of Francisella noatunensis as endosymbiont of the ciliate Euplotes raikovi. Microb. Ecol. 61, 455–464. ( 10.1007/s00248-010-9772-9) [DOI] [PubMed] [Google Scholar]
- 27.Vallesi A, et al. 2019. A new species of the γ-proteobacterium Francisella, F. adeliensis sp. nov., endocytobiont in an Antarctic marine ciliate and potential evolutionary forerunner of pathogenic species. Microb. Ecol. 77, 587–596. ( 10.1007/s00248-018-1256-3) [DOI] [PubMed] [Google Scholar]
- 28.Amann R, Springer N, Ludwig W, Görtz H-D, Schleifer K-H. 1991. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature 351, 161–164. ( 10.1038/351161a0) [DOI] [PubMed] [Google Scholar]
- 29.Schrallhammer M, Castelli M, Petroni G. 2018. Phylogenetic relationships among endosymbiotic R-body producer: bacteria providing their host the killer trait. Syst. Appl. Microbiol. 41, 213–220. ( 10.1016/j.syapm.2018.01.005) [DOI] [PubMed] [Google Scholar]
- 30.Lane DJ. 1991. 16S/23S rRNA sequencing. In Nucleic acid techniques in bacterial systematics (eds E Stackebrandt, M Goodfellow), pp. 115–175 New York, NY: Wiley. [Google Scholar]
- 31.Gruber-Vodicka HR, Seah BKB, Pruesse E. 2019. phyloFlash—Rapid SSU rRNA profiling and targeted assembly from metagenomes. bioRxiv ( 10.1101/521922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. ( 10.1089/cmb.2012.0021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. ( 10.1093/bioinformatics/btv383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laetsch DR, Blaxter ML. 2017. BlobTools: interrogation of genome assemblies. F1000Research 6, 1287 ( 10.12688/f1000research.12232.1) [DOI] [Google Scholar]
- 35.Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acid. Res. 32, 1363–1371. ( 10.1093/nar/gkh293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO.. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. ( 10.1093/nar/gks1219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145. ( 10.1093/nar/gkn879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15, 593–600. ( 10.1016/S0723-2020(11)80121-9) [DOI] [Google Scholar]
- 39.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hess S, Suthaus A, Melkonian M. 2016. ‘Candidatus Finniella’ (Rickettsiales, Alphaproteobacteria), novel endosymbionts of viridiraptorid amoeboflagellates (Cercozoa, Rhizaria). Appl. Environ. Microbiol. 82, 659–670. ( 10.1128/AEM.02680-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanzoni O, Sabaneyeva E, Modeo L, Castelli M, Lebedeva N, Verni F, Schrallhammer M, Potekhin A, Petroni G. 2019. Diversity and environmental distribution of the cosmopolitan endosymbiont ‘Candidatus Megaira’. Sci. Rep. 9, 1179 ( 10.1038/s41598-018-37629-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dirren S, Salcher MM, Blom JF, Schweikert M, Posch T. 2014. Ménage-à-trois: the amoeba Nuclearia sp. from Lake Zurich with its ecto- and endosymbiotic bacteria. Protist 165, 745–758. ( 10.1016/j.protis.2014.08.004) [DOI] [PubMed] [Google Scholar]
- 44.Senra MVX, Dias RJP, Castelli M, Silva-Neto ID, Verni F, Soares CAG, Petroni G. 2016. A house for two—double bacterial infection in Euplotes woodruffi Sq1 (Ciliophora, Euplotia) sampled in Southeastern Brazil. Microb. Ecol. 71, 505–517. ( 10.1007/s00248-015-0668-6) [DOI] [PubMed] [Google Scholar]
- 45.Pond FR, Gibson I, Lalucat J, Quackenbush RL. 1989. R-body-producing bacteria. Microbiol. Rev. 53, 25–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lian C, Luo X, Fan X, Huang J, Yu Y, Bourland W, Song W. 2018. Morphological and molecular redefinition of Euplotes platystoma Dragesco & Dragesco-Kerneis, 1986 and Aspidisca lynceus (Müller, 1773) Ehrenberg, 1830, with reconsideration of a ‘well-known’ Euplotes ciliate, Euplotes harpa Stein, 1859 (Ciliophora, Euplotida). J. Eukaryot. Microbiol. 65, 531–543. ( 10.1111/jeu.12499) [DOI] [PubMed] [Google Scholar]
- 47.Hahn MW, Schmidt J, Pitt A, Taipale SJ, Lang E. 2016. Reclassification of four Polynucleobacter necessarius strains as representatives of Polynucleobacter asymbioticus comb. nov., Polynucleobacter duraquae sp. nov., Polynucleobacter yangtzensis sp. nov. and Polynucleobacter sinensis sp. nov., and emended description of Polynucleobacter necessarius. Int. J. Syst. Evol. Microbiol. 66, 2883–2892. ( 10.1099/ijsem.0.001073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarza P, et al. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. ( 10.1038/nrmicro3330) [DOI] [PubMed] [Google Scholar]
- 49.Hahn MW, Jezberová J, Koll U, Saueressig-Beck T, Schmidt J. 2016. Complete ecological isolation and cryptic diversity in Polynucleobacter bacteria not resolved by 16S rRNA gene sequences. ISME J. 10, 1642–1655. ( 10.1038/ismej.2015.237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosati G, Verni F. 1975. Macronuclear symbionts in Euplotes crassus. Boll. Zool. 42, 231–232. ( 10.1080/11250007509431435) [DOI] [Google Scholar]
- 51.Bright M, Espada-Hinojosa S, Lagkouvardos I, Volland J-M. 2014. The giant ciliate Zoothamnium niveum and its thiotrophic epibiont Candidatus Thiobios zoothamnicoli: a model system to study interspecies cooperation. Front. Microbiol. 5, 145 ( 10.3389/fmicb.2014.00145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seah BKB, Schwaha T, Volland J-M, Huettel B, Dubilier N, Gruber-Vodicka HR. 2017. Specificity in diversity: single origin of a widespread ciliate-bacteria symbiosis. Proc. R. Soc. B 284, 20170764 ( 10.1098/rspb.2017.0764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Görtz H-D, Fujishima M. 1983. Conjugation and meiosis of Paramecium caudatum infected with the micronucleus-specific bacterium Holospora elegans. Eur. J. Cell Biol. 32, 86–91. [PubMed] [Google Scholar]
- 54.McCutcheon JP, von Dohlen CD.. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21, 1366–1372. ( 10.1016/j.cub.2011.06.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Husnik F. 2018. Host–symbiont–pathogen interactions in blood-feeding parasites: nutrition, immune cross-talk and gene exchange. Parasitology 145, 1294–1303. ( 10.1017/S0031182018000574) [DOI] [PubMed] [Google Scholar]
- 56.Boscaro V, et al. 2013. Polynucleobacter necessarius, a model for genome reduction in both free-living and symbiotic bacteria. Proc. Natl Acad. Sci. USA 110, 18 590–18 595. ( 10.1073/pnas.1316687110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Unterman BM, Baumann P, McLean DL. 1989. Pea aphid symbiont relationships established by analysis of 16S rRNAs. J. Bacteriol. 171, 2970–2974. ( 10.1128/jb.171.6.2970-2974.1989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruehle MD, Orias E, Pearson CG. 2016. Tetrahymena as a unicellular model eukaryote: genetic and genomic tools. Genetics 203, 649–665. ( 10.1534/genetics.114.169748) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190. ( 10.1146/annurev.genet.41.110306.130119) [DOI] [PubMed] [Google Scholar]
- 60.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55, 247–266. ( 10.1146/annurev-ento-112408-085305) [DOI] [PubMed] [Google Scholar]
- 61.Gottlieb Y, Ghanim M, Gueguen G, Kontsedalov S, Vavre F, Fleury F, Zchori-Fein E. 2008. Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J. 22, 2591–2599. ( 10.1096/fj.07-101162) [DOI] [PubMed] [Google Scholar]
- 62.Opatovski I, et al. 2018. Modeling trophic dependencies and exchanges among insects' bacterial symbionts in a host-simulated environment. BMC Genomics 19, 402 ( 10.1186/s12864-018-4786-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
18S and 16S rRNA gene sequences are deposited in the GenBank/EMBL/ENA database (accession numbers: LR588889-90, FR873728-30 and LR585330-53).