Abstract
BACKGROUND: Gene expression can be posttranscriptionally regulated by a complex network of proteins. N1-methyladenosine (m1A) is a newly validated RNA modification. However, little is known about both its influence and biogenesis in tumor development. METHODS: This study analyzed TCGA data of patients with five kinds of gastrointestinal (GI) cancers. Using data from cBioPortal, molecular features of the nine known m1A-related enzymes in GI cancers were investigated. Using a variety of bioinformatics approach, the impact of m1A regulators on its downstream signaling pathway was studied. To further confirm this regulation, the effect of m1A writer ALKBH3 knockdown was studied using RNA-seq data from published database. RESULTS: Dysregulation and multiple types of genetic alteration of putative m1A-related enzymes in tumor samples were observed. The ErbB and mTOR pathways with ErbB2, mTOR, and AKT1S1 hub genes were identified as being regulated by m1A-related enzymes. The expression of both ErbB2 and AKT1S1 was decreased after m1A writer ALKBH3 knockdown. Furthermore, Gene Ontology analysis revealed that m1A downstream genes were associated with cell proliferation, and the results showed that m1A genes are reliably linked to mTOR. CONCLUSION: This study demonstrated for the first time the dysregulation of m1A regulators in GI cancer and its signaling pathways and will contribute to the understanding of RNA modification in cancer.
Introduction
Researchers in recent years have begun to explore the crucial role of reversible RNA modifications in regulating gene expression [1], [2], [3], [4]. This is an understudied area in contrast to numerous studies conducted on epigenetic regulations of DNA and histones [5]. Posttranscriptional modification of RNA forms an emerging layer of genetic expression regulation and analogous in its potential role in posttranslational modification of proteins level [6]. Recent high-throughput sequencing approaches came up with more than 100 types of RNA modifications [7], [8].
N1-methyladenosine (m1A) is a crucial posttranscriptional modification in RNA which was first documented more than five decades ago [9], [10]. Adding a methyl group at the N1 position of adenosine forms m1A [11], [12]. Methyl group of m1A is located at the Watson-Crick base pairing interface to disrupt base pairing, and the positive charge carried by m1A affects local RNA structure or protein-RNA interaction [13]. Meanwhile, m1A is also found to have a strong enrichment effect on translation in 5′UTR [14]. Previous studies pointed to the presence of m1A in tRNA, rRNA, mRNA, and mitochondrial (mt) transcripts [15].
The unique physicochemical properties of m1A play a crucial role in maintaining the correct structure and function of these noncoding RNAs (ncRNAs) [16]. The “writer” (TRMT10C, Trmt61B, TRMT6/61A), “reader” (YTHDF1, YTHDF2, YTHDF3, and YTHDC1), and “eraser” (ALKBH1, ALKBH3) proteins of the m1A of mRNAs and ncRNAs are important gene regulators at the posttranscriptional level [10], [17], [18], [19]. Trmt61B and TRMT6/61A catalyze m1A at position 58 of human cell mt and cyt tRNA. Alternatively, TRMT10C catalyzes it at position 9 [18], [19], [20]. ALKBH1 and ALKBH3 catalyze demethylation of m1A in single-stranded (ss) DNA and RNA [21], [22], [23]. YTH domain-containing proteins, such as YTHDF1, YTHDF2, YTHDF3, and YTHDC1, directly bind to m1A-bearing RNA as readers [10].
Gastrointestinal (GI) cancer is a group of most common tumors referring to esophageal, gastric, liver, gallbladder, pancreatic, colon, and rectum cancers [24], [25]. Genetic mutations have been document to play an important role in GI cancer formation [26], [27]. Aberrant changes in DNA methylations have also been reported in GI cancers [28]. However, RNA modifications have been considered relatively static and stable epigenetic marks [3]. Recently, there has been an increase in interest in RNA methylation, such as N6-methyladenosine (m6A), 5-methyl cytidine (m5C; also known as 5mC), and m1A [29]. Studies have revealed that m6A RNA methylation provides an effective direction for early diagnosis and treatment of pancreatic cancer and hepatocellular carcinoma [30]. It was found that hTrm6p/hTrm61p m1A transmethylase forms m1A and promotes urinary bladder cancer [31], [32]. Targeted therapy is increasingly employed in tumor-related cancers [33], [34]. An early diagnosis of most GI cancers is difficult, and there is a risk of advanced diagnosis. This clinical challenge emphasizes the importance of early detection and/or prediction of GI cancers; however, there are few reports on m1A in GI cancers [17].
To study the prognostic and diagnostic role of m1A methylation as a robust molecular marker in GI cancers, this study capitalized on nine m1A regulators in GI cancers through The Cancer Genome Atlas (TCGA) database. The method is based on genetic alteration and dysregulated function to speculate the m1A modification status in the tumor. Gene ontology (GO) and pathways enrichment, as well as protein-protein interaction (PPI) network of these differential expressed genes, were performed. Furthermore, the next-generation RNA sequencing was used to investigate the hub gene expression pattern and mutation under the condition of ALKBH3 knockdown to provide an insight into m1A modification and GI tumor mechanism. These observations suggest the potential of m1A in regulating human GI cancers.
Results
m1A Regulator Expression in GI Cancers
In order to illustrate m1A regulator gene expression in GI cancers, tissue samples obtained from The Genotype-Tissue Expression (GTEx) and TCGA database were processed (Figure 1A). The researchers downloaded and normalized gene expression levels (TPM) of RNA-seq data from 3433 samples, including 1537 from TCGA and 1896 from GTEx project. Samples with low alignment rates and samples not used in the final GTEx study were discarded. The m1A regulators like writer (“TRMT6,””TRMT61A,””TRMT10C”), eraser (“ALKBH1,””ALKBH3”), and reader (“YTHDF1-3,” “YTHDC1”) were dysregulated in five types of GI cancers. Expression levels in tumor samples were significantly higher than normal, except the TRMT61A in esophageal carcinoma (ESCA) and colorectal adenocarcinoma (COADREAD).
Figure 1.
Expression of m1A regulators in GI Cancer patients. (A) Normalized expression across tissue and cancer types for nine known m1A-related genes. The normalized expression of RNA sequence (TPM) detected from the tumor (TCGA) and normal (GTEX) database, which contain 1537 complete tumor patients and 1896 normal samples. Orange and turquoise boxes represented tumor and normal samples, respectively. *P < .05, **P < .01, ***P < .001, by two-tailed t test. (B) PCA of the normalized expression data in five subtypes of GI cancers. Two-dimensional plots have principal components calculated by performing PCA of the gene expression values of ESCA, STAD, COADREAD, LIHC, and PAAD samples from TCGA. (C) Survival differences of GI cancer patients with high and low m1A-related genes expression. The overexpression of m1A-related genes in LIHC was significantly associated with poor overall survival. High expression of ALKBH1 in STAD and COADREAD was also connected with poor prognosis. Statistical analysis was carried out by Kaplan-Meier analysis with best separation. (D) m1A-related genes expression levels by neoplasm stage. Analyzed using one-way ANOVA.
In liver hepatocellular carcinoma (LIHC), the m1A regulator gene expression levels were significantly higher than normal (Figure 1A). An unsupervised principal component analysis (PCA) was performed in the TCGA with sample from the same study (Figure 1B). The TPCA showed an obvious separation of LIHC and other four cancer types based on genetic expression profile presented in m1A regulator genes. Thus, known m1A regulated genes are differentially expressed in LIHC compared with other four tumors, suggesting that m1A activates distinct biological function in LIHC. Kaplan-Meier analysis showed higher m1A regulator gene expression which is associated with poor prognosis in LIHC (Figure 1C). Overexpression of ALKBH1 is negatively connected with overall survival in STAD, COADREAD, and LIHC; in contrast, lower expression of ALKBH3 in ESCA and COADREAD was mostly associated with worse overall survival.
After filtering incomplete clinical data from TCGA based on GISTIC annotation [35], total GI cancer patients with different cluster of regulator genes were categorized as altered and unaltered groups. The cancer history, neoplasm stage, and race information of patients are described in (Table 1). The tumor neoplasm stage is the most used risk classification. The association between m1A regulator genes and GI neoplasm stage (Figure 1D) was later assessed. Patients with higher neoplasm stage (G1-G3) had higher expression of TRMT6, TRMT61A, and TRMT10C as well as ALKBH3 and YTHDF2.
Table 1.
Different m1A Regulator Gene Types and Clinical-Pathological Characteristics of GI Cancer Patients in the TCGA Database
| Writer |
Eraser |
Reader |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes | No | χ2 | P | Yes | No | χ2 | P | Yes | No | χ2 | P | ||
| Cancer history | Tumor | 77 | 77 | 2.16 | .153 | 72 | 82 | 5.217 | .022 | 92 | 62 | 0.803 | .37 |
| Normal | 308 | 400 | 261 | 447 | 395 | 313 | |||||||
| Neoplasm grade | G1 | 62 | 51 |
9.737 |
.021 | 50 | 63 | 3.938 | .268 | 64 | 49 | 7.573 | .056 |
| G2 | 201 | 290 | 179 | 312 | 261 | 230 | |||||||
| G3 | 199 | 263 | 165 | 297 | 269 | 193 | |||||||
| G4 | 9 | 5 | 7 | 7 | 12 | 2 | |||||||
| Race | White | 435 | 542 | 3.015 | .221 | 371 | 606 | 4.189 | .123 | 538 | 439 | 6.939 | .031 |
| Asian | 138 | 172 | 112 | 198 | 188 | 122 | |||||||
| African | 53 | 46 | 47 | 52 | 66 | 33 | |||||||
m1A Regulator Gene Alteration in GI Cancers
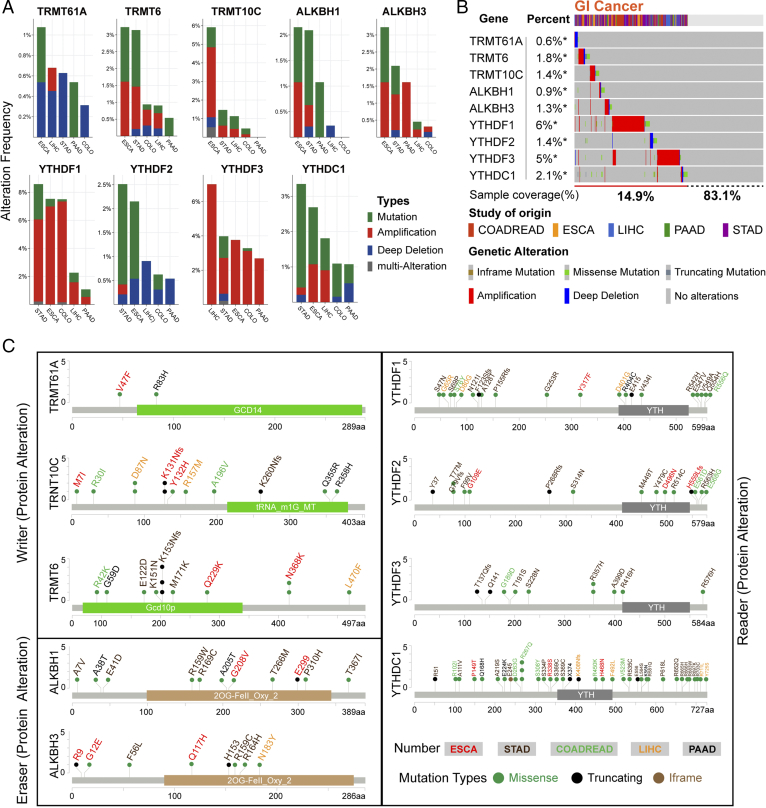
Somatic mutations are associated with prognosis and chemotherapy sensitivity [36]. The cBio Cancer Genomics Portal (cBioPortal) database was accessed to analyze the nine regulator genes of five different cancer statuses collected from TCGA database. The results showed reclassification of genomic alteration frequency of amplification, mutation, deletion, and multiple alteration (Figure 2A). The mutation and copy-number amplification were obvious in GI cancers; meanwhile, TRMT6 genetic alteration in ESCA and STAD was more than 3%. TRMT10C alteration in ESCA was close to 6%. In LIHC, the reader YTHDF3 copy-number amplification was more than 6%.
Figure 2.
The frequency and type of m1A regulator alteration in cancer (cBioPortal). (A) The alteration frequency of m1A regulators in various cancer studies. The total alteration frequency was according to decreasing sizes. The alteration type included mutations (green), amplification (red), deep deletions (blue), and multiple alterations (gray). (B) Summarizing the detailed alteration. The mutation profile and putative copy-number alterations (CNAs) of m1A regulators in TCGA (Provisional) cohort. Missense mutations (green), truncating mutations (dark gray), inframe mutations (brown), amplification (red), deep deletions (blue), and no alterations (gray). In this graphical summary, individual cases are represented as columns. (C) m1A regulator mutations in GI cancer studies. This graphic shows the Pfam protein domains and the position of m1A regulators mutations in various cancers. The colored structure represents the common domain of the protein.
Despite the m1A regulator gene alteration generally observed in GI cancers, there is no regular routine. Approximately 15.9% GI cancer patients had different kinds of m1A regulators alteration, including mutation, copy-number amplification, or deep deletion (Figure 2B). TRMT6 and TRMT10C have alteration frequency of 1.8% and 1.4%, respectively. Moreover, the reader YTHDF1 (6% of all sample) and YTHDF3 (5% of all sample) alterations had the most activities in GI cancer patients. Meanwhile, TRMT61A was rarely altered in GI cancers with a lowest alteration frequency at only 0.6% and mainly processed with copy-number deep deletion. Figure 2C summarizes details of all mutations including Missense, Iframe, and Truncating mutations in GI cancers. In 2OG-Fell domain of ALKBH1, RC169C missense mutation reached level 3 (the number of patients with the same mutation site) in the Catalogue of Somatic Mutations in Cancer database, suggesting that it as an important region for its function.
Signaling Pathway Regulated by m1A Modification
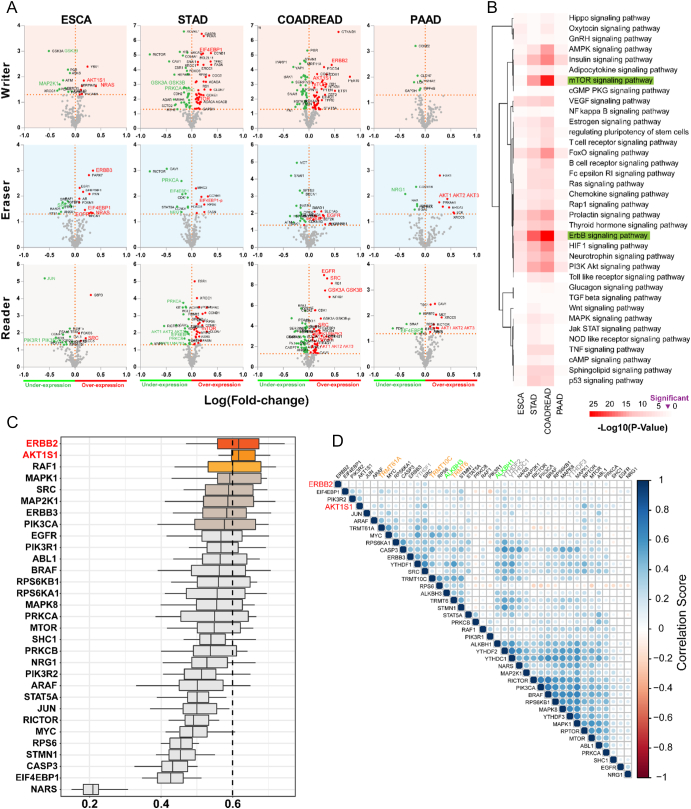
We later applied a correlational analysis of protein expression to validate associated pathway activation of m1A. The protein expression changes (both unphosphorylated and phosphorylated proteins) along with m1A regulator gene alteration in GI cancers were analyzed through cBioPortal with the Reverse Phase Protein Arrays (RPPA) (Figure 3A). The main altered proteins (with expression and over expression) (P < .05) are shown in different colors. The significantly changed proteins in different kinds of tumors from RPPA database were screened. The protein coding genes were later calculated using Functional Annotation Tool online [Database for Annotation Visualization and Integrated Discovery (DAVID), https://david.ncifcrf.gov/]. These protein-coding genes were discovered to be mainly involved in 36 kinds of signaling pathways (Figure 3B), especially in mTOR (the mammalian target of rapamycin) and ErbB (Her2) signaling pathways.
Figure 3.
A prediction of potential targets of m1A regulators. (A) Protein enrichment in GI cancers. Protein enrichment in four types of GI cancers was studied within the cluster of writer, eraser, or reader alteration. Reliability of over-/underexpressed protein is displayed as initial conditions or phosphorylation with Student’s t test significance (P < .05). The distance of expression level change was based on log fold-change (mean in altered/mean in unaltered). (B) Heatmap of pathway-related protein alterations. In GI cancers, protein coding genes of m1A regulators alterations were investigated based on KEGG pathway database. Red emphasized, mTOR and ErbB signaling pathways are most likely involved in dysregulated expression of m1A regulators. It is arranged in order of the log (P value). (C) Summary of functional similarities of mTOR and ErbB signaling pathway interactome in alteration with m1A regulators. The distributions of functional similarities are summarized as boxplots. The lines in the boxes indicate the mean of the functional similarity. Proteins with a higher average functional similarity (cutoff >0.6) are defined as party proteins; the dashed line represents the cut-off value. (D) Relationship of m1A regulator genes with mTOR and ErbB expression. Correlogram shows direct cross-correlation of major m1A regulator genes with signaling proteins in the total GI cancer patients. The blue represents the positive correlation, while red represents vice versa.*P < .05.
Later, the crucial protein-coding genes in mTOR and ErbB signaling pathways were filtered to affirm the most important factors. Functional similarities were further calculated by geometric mean of semantic similarities in biological processes (BPs), molecular functions (MFs), and cellular components (CCs), which is measured through the GOSemSim package by taking the GO topological structure into account in a more precise and unbiased manner [37]. The distribution of functional similarities is shown in Figure 3C. Average of similarities score was used to rank proteins in the m1A altered-regulator interactome. A cutoff value of 0.6 was chosen, and ErbB2 and AKT1S1 showed the highest similarity in all the protein members. Furthermore, direct comparison of m1A regulator mRNA levels and selected signaling proteins was performed (Figure 3D). It showed a lower correlation between ErbB2 and writer (TRMT61A, TRMT6, TRMT10C) (r < 0.1) than ErbB2 and reader (YTHDF1-2) (r > 0.15). DNA methylation levels of individual CpGs in the promoters of the selected genes were also studied. As shown in Supplementary Figure S1, only PIK3R1, STAT5A, SHC1, PRKCB, and NRG1 were found to be frequently influenced by DNA methylation in GI cancers, suggesting that RNA modification was likely involved in regulating this gene expression.
Figure S1.
DNA methylation signatures identify different cancer types and normal samples. Unsupervised hierarchical clustering and heatmap of TCGA cohort including five types of GI cancer patients and normal samples based on CpG islands using Human Methylation 450 BeadChip assay. The color key indicates methylation level; hypermethylated sample shows an increased value, and it turns red. The black annotation represents the tumor, and green represents the normal cells.
Genomic View of m1A Writer ALKBH3 Knockdown
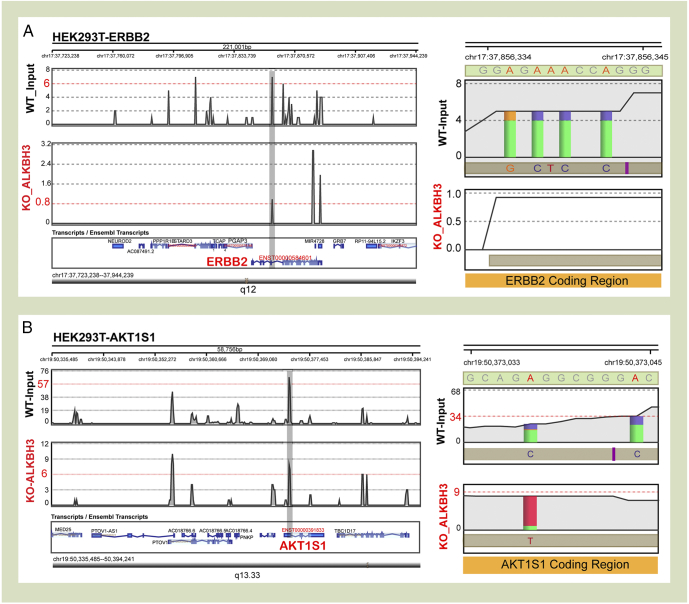
The high-throughput sequence analysis shows that knockdown of the m1A ALKBH3 demethylase in HEK293T cells resulted in downregulating the ErbB2 read density peak and the misincorporated adenines disappeared in ErbB2 coding region (Figure 4A). Similarly, the AKT1S1 read density peak also dropped when knockdown of ALKBH3 was performed, and misincorporation in adenine sites was also decreased (Figure 4B).
Figure 4.
Representative views of mTOR and ErbB consensus sequence in ALKBH3-knockdown HEK293T cell. (A) Examples of coverage from WT_input (black) and ALKBH3-knockdown (red) experiments across selected genes in HEK293T cell. Coverage plot along the density of code region footprints was compared between two groups. The gray shade displays a representative change. (B) Misincorporation and coverage plots for putative adenine sites were identified in this study. The graphical representation of ALKBH3, the misincorporation rate (A/G, A/C), and overall coverage (gray shade) in sequence window neighboring the adenine sites were decreased in coding regions. The RNA-seq data were taken from a published database.
Function and Network Analysis
The correlation between m1A regulators and two major involved signaling pathways was calculated, and a total of 31 m1A regulator downstream candidate genes were processed via GO functional analysis. The results of BP (Figure 5A) was obtained, so the genes were enriched in several bioprocesses, including intracellular signal transduction, positive regulation of gene expression, and positive regulation of cell migration and apoptotic process. The GO BP analysis indicated that the functions of downstream genes involved in GI cancers are associated with cell proliferation. A total of 31 downstream candidate genes and 9 m1A regulator genes network containing 217 nodes and 4512 edges were obtained from STRING online database and Cytoscape software (Figure 5B). The m1A regulators were further proved to interact with ErbB pathway and mTOR pathway in correlation network. The PPI network showed the detailed protein interaction. It was found the m1A methylase genes are reliably linked to mTOR and indirectly to other signaling genes, among which the ErbB and mTOR signaling pathways probably are the key connection to m1A regulators’ function.
Figure 5.
Functional analyses showed that functions of m1A modification candidate genes are closely related to the progression of GI tumors. (A) Significantly enriched GO annotation of m1A regulators alterations related pathway. BP analysis refers to the ratio of genes enriched in entries. The color of the circle represents the P value, and the size of the circle refers to the number of genes enriched in the same entry. (B) Based on STRING and Cytoscape database, the correlation of m1A regulators and downstream candidate genes is demonstrated by PPI network analysis.
Discussion
To date, more than a hundred types of posttranscriptional modifications have been reported [11]. Cellular RNA modification has emerged to be an important regulator of gene expression, and dynamic modification represents a novel layer of genetic information [16]. The data obtained highlight the importance of m1A methylase, demethylase, and specific protein dysregulation in GI cancer patients. It also used TCGA high-throughput data analysis to investigate the function of dysregulated expression in the pathogenesis of GI cancers.
In mammals, nine members of m1A regulators have been identified catalyzed by writers (“TRMT6,” “TRMT61A,” “TRMT10C”) [38], [39], reversed by erasers (“ALKBH1,” “ALKBH3”) [40], [41], and specially recognized by readers (“YTHDF1-3,” “YTHDC1”) with YTH domain [10], all of which are associated with RNA metabolism. Tumor cells often acquired genetic and epigenetic alterations that always contribute to oncogene dysregulation and widespread changes in gene expression [42]. Up to now, only ∼2570 m1A modification sites have been validated in humans [43], and little is known about the relationship between m1A modification and GI cancers.
Hence, thousands of TCGA samples were employed to study the expression pattern; it was suggested that the dysregulated expression level of m1A-related genes may also be linked to the GI cancer tumorigenesis. It has been previously shown that the overexpression of ALKBH1 is significantly associated with poor prognosis and metastasis in gastric cancer [44]. Meanwhile, high expression of ALKBH3 is also positively correlated with advanced tumor stage in pancreatic cancer [45]. Thus, dysregulation of m1A-related genes may closely be linked to GI cancer progress. Using PCA of RNA-seq data, m1A gene expression pattern in GI cancers was found to be similar among the five types of tumor, and the distinctive expression quantification in LIHC indicated that study-specific biases still accounted for RNA-seq expression levels within each tissue type. Notably, results of KM plotter in overall survival showed that higher expression level of m1A-related genes in LIHC is significantly associated with poor prognosis. Moreover, high expression level of ALKBH1 in STAD and COADREAD was also negatively associated with overall survival.
Using publicly available clinical data from TCGA, the study investigated the clinical feature of the m1A regulator gene alterations in GI cancer patients. It was found that m1A regulator expression level was positively associated with the tumor malignance; however, the poorly differentiated tumors presented a significantly higher expression level of TRMT6 and TRMT10C. In addition, the G4 stage did not show a consistent result which might be due to the limitation of the G4 stage available data. Thus, it is speculated that TRMT6 and TRMT10C are involved in the pathogenesis of GI cancer.
Previous studies have shown TRMT10C missense mutation can influence the mitochondrial ribonuclease P protein 1 (MRPP1) decrease in mitochondria, causing the mitochondrial disease by affecting the protein stability and mt-tRNA processing [46]. Thus, the dysregulation of m1A regulator not only influences the m1A function, but it can also contribute to other disease. Analysis of data from cBioPortal [47] revealed that m1A regulators possessed a high-frequency alteration percentage in GI cancer patients, and more than 14% samples were tested for different types of genetic alterations (mutation, copy-number amplification, copy-number deep deletion). Genetic mutations usually cause phenotypic changes, which are closely related to carcinogenesis and aging [34]. The genomic translocation is a key factor in chimeric transcripts expression in tumor [48]. Therefore, analysis of m1A regulator alteration is important in understanding the role of m1A in GI tumorigenesis and for planning treatment.
In studying the oncogenic mechanism of m6A RNA methylation, it was found that when METTL14 or METTL3 mutation is reduced, AKT signaling is activated and endometrial cancer cells’ proliferation and tumorigenicity are developed [49]. In order to investigate this biological function, data were obtained from RPPA database. First, the protein enrichment in each type of GI cancers demonstrated that m1A regulator alterations influenced the multiproteins function in tumor. Results showed that ErbB and PI3K/AKT/mTOR signaling pathways are associated with m1A methylation. In selecting the protein potentially significant in the major signaling pathway, we applied further bioinformatics analysis by ranking through integration of protein semantic similarity. ErbB2 and AKT1S1 are top-ranked proteins potentially playing central roles in ErbB and mTOR interactome in m1A regulators. AKT1S1, mTOR, Raptor, and DEPTOR have been previously demonstrated as part of mTORC1 complex [50]. Thus, results suggest that ErbB2 and AKT1S1 may be connected to m1A regulators. Based on the correlation analysis, m1A regulators have been found to be positively correlated with two signaling genes.
Similar to DNA and histones, RNA can also be chemically modified [3]. RNA modification has emerged as a major regulator to control of genetic information [16]. The m1A is a rare internal modification and presented at very low stoichiometry [6]. Promoter of DNA methylation may represent a key epigenetic mechanism in regulating gene expression [51]. Thus, it was important to explore in-depth whether DNA methylation influences signaling activation. The results showed that several genes, such as SHC1, STAT5, PRKCB, NRG1, and PIK3R1, in GI cancer patients were affected by DNA methylation. Meanwhile, other signaling members, such as ErbB2, mTOR, and AKT1S1, were not affected by DNA methylation. The HEK293T cell ALKBH3-knockdown experiment RNA-seq data from the NCBI Gene Expression Omnibus (GEO) database were also collected. Unsurprisingly, both ErbB2 and AKT1S1 expression density peaks were downregulated after the ALKBH3 knockdown. However, at the transcriptomic level, the m1A can induce the modification site mutation in rRNA [52]. As ErbB2 is also connected to the mitogen-activated protein kinase pathway and in order to find new oncogenic markers [53], [54], [55], the mRNA underlying mechanism required further investigation.
In the final phase, the study utilized the enrichment proteins and m1A regulators to construct PPI network based on Cytoscape and String database. Several studies have reported that aberrant PPIs were the basis of cancer occurrence and progression [56], [57]. The correlation network revealed that m1A regulators were mainly associated with mTOR and ErbB signaling pathways and affected multiple human biological functions.
Materials and Methods
Data Processing
A total of 1696 GI cancer samples that were included in the final whitelist for TCGA (http://cancergenome.nih.gov) with clinical data and 1874 normal samples from GTEX (https://gtexportal.org/) were analyzed. The GSE73941 RNA-seq data were obtained from the GEO database. The TCGA data were first normalized and log2 transformed; GTEX data were normalized to TCGA. The somatic mutation data (amplification, deep deletion, and missense mutations) of GI cancers were downloaded from TCGA through cBioPortal and GISTIC [35], [47]. Proteomic data were also taken from the TCGA database as normalized RPPA data through the cBioPortal. The value in the methylation profile represents the methylation degree calculated by Illumina Infinium Human Methylation 450K arrays through the DiseaseMeth version 2.0 [58].
Pathway-Level Analysis of Conditional Gene Selection
Proteomic data were collected by RPPA based on TCGA in GI cancers. The RPPA quality control and methodology have been explained previously [59]. Through the online Functional Annotation Tool online (DAVID, https://david.ncifcrf.gov/), selected genes of each type of cancer were used to predict statistical confident P value score of significant pathways in KEGG database. Functional similarity defined as the geometric mean of their semantic similarities in BP, MF, and CC aspect of GO is designed for measuring the strength of the relationship between each protein and its partners by considering their function and location. Two pathway members in BP, MF and CC, were measured through the GOSemSim package [37], applying the Wang method, which is both accurate and unbiased. This takes the GO topological structure into account [60]. A cutoff value of 0.6 was chosen.
RNA-seq Data Reads Mapping
The high-throughput sequence data were transformed to FASTQ file and handled with StrandsNGS (www.strand-ngs.com). The raw data were first aligned and then filtered by Title quantity and Duplicates with default parameters. Reference transcriptome was prepared based on the Refseq annotation of human (hg19) downloaded from UCSC database. For each peak/site, the genomic location, misincorporation column, and read counts were displayed in the newly lightweight Browse.
Association of m1A Regulators and Genes
m1A related genes were used to perform human GO enrichment analysis. DAVID (https://david.ncifcrf.gov) provides a set of complete web-based annotation tool to understand the biological significance of m1A regulators related genes. The STRING (https://string-db.org/) database provides PPI information, including direct (physical) and indirect (functional) associations [61]. Pathway from KEGG and the extend network was constructed for m1A regulators and related protein coding genes signatures by the Cytoscape 3.5.1 [62].
Quantification and Statistical Analysis
Using R language (https://www.r-project.org/), analysis of PCA, correlation coefficient, and tumor and normal comparison were mainly performed based on several special publicly available packages. The SPSS 21.0 statistical software program was used for Pearson’s chi-squared test statistical analysis, t test in discrepancy of two-group comparison, one-way ANOVA to compare multiple group, and Pearson analysis to calculate the correlation between two classes. Overall survival analysis was carried out as Kaplan-Meier cure with P value calculated using the log-rank test. P values less than .05 were considered statistically significant.
Conclusions
This study depicted for the first time the dysregulation of m1A regulators and its association with clinicopathological parameters in GI cancer using bioinformatics method and underlined the importance of this newly validated RNA modification mechanism. Results suggested that m1A regulators most probably modulate ErbB2 and mTOR pathways. The impact of m1A regulators on the key pathway genes still needs to be further validated.
The following are the supplementary data related to this article.
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant nos. 81503093, 81602166, and 81672444) and the Joint Funds of the Southwest Medical University & Luzhou (2016LZXNYD-T01, 2017LZXNYD-Z05, and 2017LZXNYD-J09).
Conflict of Interests
Authors have no conflict of interests.
Author Contributions
Methodology, Q. Zh.; software, Y. Z., Q. Zh.; validation, P. J. K, Y. Z., and Z. Z.; formal analysis, J. Sh., Q. Zh.; investigation, H. Zh., X. L, L. Zh., L. W., Q. W.; resources, M. L., X. W., L. L., L. W., Q. W.; data curation, M. L., Y. W., J. L.; writing – original draft preparation, Y. Zh., Q. Zh., P. J. K.; writing – review & editing, P. J. K., J. L., Zh. X.; visualization, J. Y., T. Y.; supervision, Zh. X.; project administration, C. H. C., Zh. X.; funding acquisition, Zh. X.
Contributor Information
Jing Li, Email: jing.li9@hotmail.com.
Zhangang Xiao, Email: xzg555898@hotmail.com.
References
- 1.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008 Jun;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 2.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007 Apr;8(4):307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 3.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010 Dec;6(12):863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 4.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014 May;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 5.Cao Q, Mani R-S, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR, Kim JH, Brenner JC, Jing X, Cao X. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell. 2011 Aug;20(2):187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz S. m(1)A within cytoplasmic mRNAs at single nucleotide resolution: a reconciled transcriptome-wide map. RNA (New York, NY). 2018 Nov;24(11):1427–36. [DOI] [PMC free article] [PubMed]
- 7.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018 Jan;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013 Dec;155(6):1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn DB. The occurrence of 1-methyladenine in ribonucleic acid. Biochim Biophys Acta. 1961 Jan;46:198–200. doi: 10.1016/0006-3002(61)90668-0. [DOI] [PubMed] [Google Scholar]
- 10.Dai X, Wang T, Gonzalez G, Wang Y. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal Chem. 2018 Jun;90(11):6380–6384. doi: 10.1021/acs.analchem.8b01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013 Jan;41(Database issue):D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong X, Li X, Yi C. N(1)-methyladenosine methylome in messenger RNA and non-coding RNA. Curr Opin Chem Biol. 2018 Aug;45:179–186. doi: 10.1016/j.cbpa.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016 May;12(5):311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 14.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016 Feb;530(7591):441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Jia G. Reversible RNA modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genomics Proteomics Bioinformatics. 2018 Jun;16(3):155–161. doi: 10.1016/j.gpb.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017 Jun;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, Lu Z, Zheng Z, Dai Q, Wang H. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res 2018 Dec. [DOI] [PMC free article] [PubMed]
- 18.Chujo T, Suzuki T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA (New York, NY). 2012 Dec;18(12):2269–76. [DOI] [PMC free article] [PubMed]
- 19.Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017 Nov;551(7679):251–255. doi: 10.1038/nature24456. [DOI] [PubMed] [Google Scholar]
- 20.Vilardo E, Rossmanith W. Molecular insights into HSD10 disease: impact of SDR5C1 mutations on the human mitochondrial RNase P complex. Nucleic Acids Res. 2015 May;43(10):5112–5119. doi: 10.1093/nar/gkv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci U S A. 2002 Dec;99(26):16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003 Feb;421(6925):859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 23.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002 Sep;419(6903):174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 24.Vedeld HM, Goel A, Lind GE. Epigenetic biomarkers in gastrointestinal cancers: The current state and clinical perspectives. Semin Cancer Biol. 2018 Aug;51:36–49. doi: 10.1016/j.semcancer.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015 Mar;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 26.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010 Oct;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 27.Stoffel EM. Screening in GI cancers: the role of genetics. J Clin Oncol. 2015 Jun;33(16):1721–1728. doi: 10.1200/JCO.2014.60.6764. [DOI] [PubMed] [Google Scholar]
- 28.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983 Jan;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 29.Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA modifications: what have we learned and where are we headed? Nat Rev Genet. 2016 Jun;17(6):365–372. doi: 10.1038/nrg.2016.47. [DOI] [PubMed] [Google Scholar]
- 30.Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019 Feb;112:108613. [DOI] [PubMed]
- 31.Shi L, Yang X-M, Tang D-D, Liu G, Yuan P, Yang Y, Chang L-S, Zhang L-R, Song D-K. Expression and significance of m1A transmethylase, hTrm6p/hTrm61p and its related gene hTrm6/hTrm61 in bladder urothelial carcinoma. Am J Cancer Res. 2015;5(7):2169–2179. [PMC free article] [PubMed] [Google Scholar]
- 32.Barraud P, Golinelli-Pimpaneau B, Atmanene C, Sanglier S, Van Dorsselaer A, Droogmans L, Dardel F, Tisne C. Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J Mol Biol. 2008 Mar;377(2):535–550. doi: 10.1016/j.jmb.2008.01.041. [DOI] [PubMed] [Google Scholar]
- 33.Birkeland AC, Ludwig ML, Spector ME, Brenner JC. The potential for tumor suppressor gene therapy in head and neck cancer. Discov Med. 2016 Jan;21(113):41–47. [PMC free article] [PubMed] [Google Scholar]
- 34.Jabbarzadeh Kaboli P, Leong MP-Y, Ismail P, Ling K-H. Antitumor effects of berberine against EGFR, ERK1/2, P38 and AKT in MDA-MB231 and MCF-7 breast cancer cells using molecular modelling and in vitro study. Pharmacological reports [Internet]. 2018 Aug; Available from: https://linkinghub.elsevier.com/retrieve/pii/S173411401730676X [DOI] [PubMed]
- 35.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome biology. 2011;12(4):R41. [DOI] [PMC free article] [PubMed]
- 36.Liu Y, Yasukawa M, Chen K, Hu L, Broaddus RR, Ding L, Mardis ER, Spellman P, Levine DA, Mills GB. Association of somatic mutations of ADAMTS genes with chemotherapy sensitivity and survival in high-grade serous ovarian carcinoma. JAMA Oncol. 2015 Jul;1(4):486–494. doi: 10.1001/jamaoncol.2015.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics (Oxford, England). 2010 Apr;26(7):976–8. [DOI] [PubMed]
- 38.Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA (New York, NY). 2005 Aug;11(8):1281–90. [DOI] [PMC free article] [PubMed]
- 39.Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase—extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012 Dec;40(22):11583–11593. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016 Dec;167(7):1897. doi: 10.1016/j.cell.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 41.Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K, Tsujikawa K. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017 Feb;7:42271. doi: 10.1038/srep42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osmanbeyoglu HU, Pelossof R, Bromberg JF, Leslie CS. Linking signaling pathways to transcriptional programs in breast cancer. Genome Res. 2014 Nov;24(11):1869–1880. doi: 10.1101/gr.173039.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xuan J-J, Sun W-J, Lin P-H, Zhou K-R, Liu S, Zheng L-L, Qu L-H, Yang J-H. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018 Jan;46(D1):D327–D334. doi: 10.1093/nar/gkx934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Dig Dis Sci 2019 Jan; [DOI] [PMC free article] [PubMed]
- 45.Yamato I, Sho M, Shimada K, Hotta K, Ueda Y, Yasuda S, Shigi N, Konishi N, Tsujikawa K, Nakajima Y. PCA-1/ALKBH3 contributes to pancreatic cancer by supporting apoptotic resistance and angiogenesis. Cancer Res. 2012 Sep;72(18):4829–4839. doi: 10.1158/0008-5472.CAN-12-0328. [DOI] [PubMed] [Google Scholar]
- 46.Metodiev MD, Thompson K, Alston CL, Morris AAM, He L, Assouline Z, Rio M, Bahi-Buisson N, Pyle A, Griffin H, Siira S, Filipovska A, Munnich A, Chinnery PF, McFarland R, Rotig A, Taylor RW. Recessive mutations in TRMT10C cause defects in mitochondrial RNA processing and multiple respiratory chain deficiencies. Vol. 99, American Journal of Human Genetics. United States; 2016. p. 246. [DOI] [PMC free article] [PubMed]
- 47.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013 Apr;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenner JC, Chinnaiyan AM. Translocations in epithelial cancers. Biochim Biophys Acta. 2009 Dec;1796(2):201–215. doi: 10.1016/j.bbcan.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, Eckert MA, Harada BT, Liu S-M, Lu Z, Yu K, Tienda SM, Chryplewicz A, Zhu AC, Yang Y. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018 Sep;20(9):1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inoki K, Corradetti MN, Guan K-L. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005 Jan;37(1):19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 51.Aznar MA, Labiano S, Diaz-Lagares A, Molina C, Garasa S, Azpilikueta A, Etxeberria I, Sanchez-Paulete AR, Korman AJ, Esteller M. CD137 (4-1BB) costimulation modifies DNA methylation in CD8(+) T cell-relevant genes. Cancer Immunol Res. 2018 Jan;6(1):69–78. doi: 10.1158/2326-6066.CIR-17-0159. [DOI] [PubMed] [Google Scholar]
- 52.Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, Mao Y, Lv J, Yi D, Chen X-W, Wang C, Qian S-B, Yi C. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Molecular cell. 2017 Dec;68(5):993–1005.e9. [DOI] [PMC free article] [PubMed]
- 53.Verhaegen M, Bauer JA. Martin de la Vega C, Wang G, Wolter KG, Brenner JC, Nikolovska-Coleska Z, Bengtson A, Nair R, Elder JT, Van Brocklin M, Carey TE, Bradford CR, Wang S, Soengas MS. A novel BH3 mimetic reveals a mitogen-activated protein kinase-dependent mechanism of melanoma cell death controlled by p53 and reactive oxygen species. Cancer Res. 2006 Dec;66(23):11348–11359. doi: 10.1158/0008-5472.CAN-06-1748. [DOI] [PubMed] [Google Scholar]
- 54.Jabbarzadeh Kaboli, Parham; Ismail, Patimah; Ling K-H. Molecular modeling, dynamics simulations, and binding efficiency of berberine derivatives: a new group of RAF inhibitors for cancer treatment. PLOS ONE. 2018;in press. [DOI] [PMC free article] [PubMed]
- 55.Jabbarzadeh Kaboli P, Rahmat A, Ismail P, Ling KH. Targets and mechanisms of berberine, a natural drug with potential to treat cancer with special focus on breast cancer. European Journal of Pharmacology [Internet] 2014;740:584–595. doi: 10.1016/j.ejphar.2014.06.025. Available from: [DOI] [PubMed] [Google Scholar]
- 56.Vishnubalaji R, Hamam R, Abdulla M-H, Mohammed MAV, Kassem M, Al-Obeed O, Aldahmash A, Alajez NM. Genome-wide mRNA and miRNA expression profiling reveal multiple regulatory networks in colorectal cancer. Cell Death Dis. 2015 Jan;6 doi: 10.1038/cddis.2014.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Z, Duan H, Li H. Identification of gene expression pattern related to breast cancer survival using integrated TCGA datasets and genomic tools. Biomed Res Int. 2015;2015:878546. doi: 10.1155/2015/878546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong Y, Wei Y, Gu Y, Zhang S, Lyu J, Zhang B, Chen C, Zhu J, Wang Y, Liu H. DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res. 2017 Jan;45(D1):D888–D895. doi: 10.1093/nar/gkw1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Zhao W, Akbani R, Liu W, Ju Z, Ling S, Vellano CP, Roebuck P, Yu Q, Eterovic AK. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell. 2017 Feb;31(2):225–239. doi: 10.1016/j.ccell.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang JZ, Du Z, Payattakool R, Yu PS, Chen C-F. A new method to measure the semantic similarity of GO terms. Bioinformatics (Oxford, England). 2007 May;23(10):1274–81. [DOI] [PubMed]
- 61.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015 Jan;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]