Abstract
Increasing incidence of multidrug-resistant bacteria presents an imminent risk to global health. Polymyxins are ‘last-resort’ antibiotics against Gram-negative ‘superbugs’; however, nephrotoxicity remains a key impediment in their clinical use. Molecular mechanisms underlying this nephrotoxicity remain poorly defined. Here we examined the pathways which led to polymyxin B-induced cell death in vitro and in vivo. Human proximal tubular cells were treated with polymyxin B (12.5–100 μM) for up to 24 h and showed significant increase in micronuclei frequency, as well as abnormal mitotic events (over 40% in treated cells, p<0.05). Time-course studies were performed using a mouse nephrotoxicity model (cumulative 72 mg/kg). Kidneys were collected over 48 h and investigated for histopathology and DNA damage. Notable increases in γH2AX foci (indicative of double stranded breaks) were observed in both cell culture (up to ~44% cells with 5+ foci at 24 h, p<0.05) and mice treated with polymyxin B (up to ~25%, p<0.05). Consistent with these results, in vitro assays showed high binding affinity of polymyxin B to DNA. Together, our results indicate that polymyxin B nephrotoxicity is associated with DNA damage, leading to chromosome missegregation and genome instability. This novel mechanistic information may lead to new strategies to overcome the nephrotoxicity of these important last-line antibiotics.
Keywords: Polymyxins, cellular toxicity, nephrotoxicity, DNA damage, cell biology
Introduction
With increasing incidence of antibiotic-resistant ‘superbugs’ globally and an estimated 10 million deaths per year by 2050, the world is in danger of reverting to the pre-antibiotic era (Boucher et al. 2013; Nathan and Cars 2014). Given the paucity of new antibiotics, last-line antibiotics for patients infected with multidrug-resistant (MDR) pathogens have received renewed attention (Boucher et al. 2009; Jabes 2011; Livermore 2004). Polymyxins are ‘last-line’ lipopeptide antibiotics used against Gram-negative ‘superbugs’, namely A. baumannii, K. pneumoniae and P. aeruginosa which are on the 2017 WHO Priority 1 (Critical) Pathogens List deemed to pose the greatest threat to human health (Falagas and Bliziotis 2007; Tacconelli and Magrini 2017). Polymyxins were first used clinically in the late 1950s, but their use was discontinued in the 1970s because of nephro- and neurotoxicity (Katz 1963; Koch-Weser et al. 1970; Li et al. 2006; Price and I. 1970; Tallgren et al. 1965; Velkov et al. 2013). Despite renewed use of polymyxins due to MDR pathogens since the 1990s, clinic usage remains limited because of nephrotoxicity, which can occur in up to ~60% of patients in a dose-dependent manner (Kubin et al. 2012; Pogue et al. 2011). This has led to sub-optimal dosage regimens and increased likelihood of resistance to these ‘last-resort’ antibiotics (Landman et al. 2008; Roberts et al. 2015).
Polymyxins have never been subject to contemporary drug development and regulatory procedures (Li et al. 2006; Lim et al. 2010). Little was known about their pharmacokinetics/pharmacodynamics and mode of action until recently (Nation et al. 2017; Sandri et al. 2013; Velkov et al. 2014). Animal and clinical studies have shown that polymyxins are extensively reabsorbed by kidney tubular cells (Li et al. 2003; Sandri et al. 2013; Zavascki et al. 2008).. The mechanism of toxicity is likely to initiate, at least partially, through receptor-mediated uptake by membrane-bound transporters such as megalin, PEPT2 and OCTN1, after which polymyxins accumulate intracellularly and ultimately cause cell death (Lu et al. 2016; Ma et al. 2006; Suzuki et al. 2013).
Recent in vitro and in vivo studies have shown at least three major apoptosis pathways – mitochondrial, endoplasmic reticulum and death receptor – play key roles in polymyxin-induced nephrotoxicity (Dai et al. 2014a; Dai et al. 2014b, Azad et al. 2015). It has been reported that antioxidants such as melatonin and lycopene display protective effects against the toxicity caused by polymyxins (Dai et al. 2014b, Yousef et al. 2011). In addition, the autophagy pathway is activated as a protective response to the damage by polymyxins, with high levels of autophagosomes and LC3 gene expression post-treatment with colistin (Zhang et al. 2015) Though increased research efforts have moved forward to shed light on polymyxin toxicity, there still remains much unknown regarding the molecular mechanisms, especially with short-term effects of polymyxin-treatment in cell culture. Given the structure and amphipathic nature of polymyxins, we were interested in investigating its effect on mammalian DNA and cell division.
In this study, we employed molecular techniques to investigate the effect of polymyxin B on DNA damage, repair and cell death. We discovered that polymyxin B induces DNA damage, leading to chromosome missegregation and genome instability in vitro and in vivo. Our data provide important novel insight into polymyxin nephrotoxicity.
Results
Cellular viability
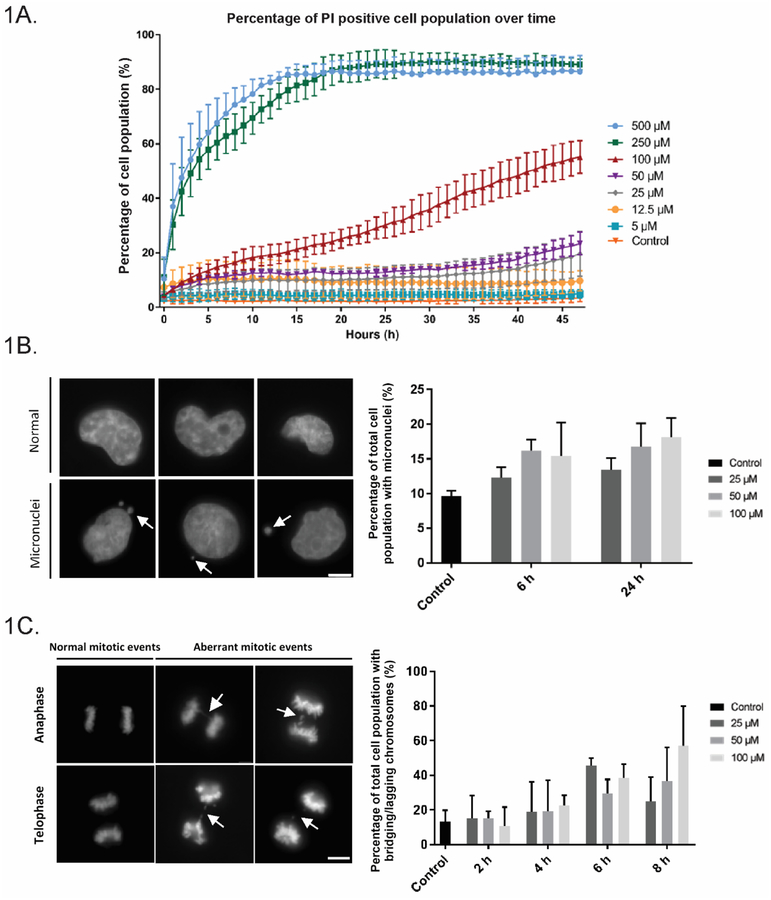
Polymyxin B toxicity has been investigated in cell lines including rat proximal tubular (NRK-52E), rat adrenal medulla (PC-12), porcine kidney epithelial (LLC-PK1) and human proximal tubular (HK-2) cells, all of which demonstrated toxicity (Azad et al. 2013; Keirstead et al. 2014; Mingeot-Leclercq et al. 2012; Zhang et al. 2016a). We were most interested in the effect of polymyxin B on HK-2 cells given its clinical use. Polymyxin B concentrations over 100 μM (250 and 500 μM) resulted in over 70% cell death early on after 12 h, with cell death plateauing out after around 20 h, while concentrations of 100 μM and 50 μM resulted in around 50% and 20% cell death at 40 h, respectively. Lower concentrations of 25 μM and below only showed marginal cell death, even after treatment over 48 h (Fig. 1A). Interestingly, following an initial peak in cell death, death of cell populations treated at 25 and 50 μM appeared to plateau before a slow increase in propidium iodide (PI) uptake again from ~34 h onwards; however, this is not observed at lower concentrations of 5 and 12.5 μM (Fig. 1A). Therefore, polymyxin B concentrations of 25, 50 and 100 μM were used in most subsequent HK-2 experiments.
Fig 1.
Polymyxin B (PMB) treatment in human proximal tubular (HK-2) cells causes chromosome missegregation and genome instability. (A) Dose-dependent toxicity of PMB. Cells were imaged over 48 h and the population viability was assessed with PI staining. PMB at 5 μM ( ),12.5 μM (
),12.5 μM ( ), 25 μM (
), 25 μM ( ), 50 μM (
), 50 μM ( ), 100 μM (
), 100 μM ( ),250 μM (
),250 μM ( ), 500 μM (
), 500 μM ( ), and control (
), and control ( ); and data are presented as mean ± SD, n = 300 in triplicate. (B) PMB-treated HK-2 cells showed significant increases in micronuclei. HK-2 cells were treated with PMB (25–100 μM) for 6 or 24 h, stained with DAPI and quantified (n=200, in triplicate). Representative images of HK-2 cells. Scale bar = 5 microns. (C) Analysis of chromosome segregation events in PMB-treated HK-2 cells. HK-2 cells were treated with PMB (25–100 μM) for 2–8 h and stained with DAPI and quantified for abnormal mitotic events. A higher incidence of abnormal mitotic events (bridging and lagging chromosome) was observed at shorter time-points after PMB treatment (n=30, in triplicate).
); and data are presented as mean ± SD, n = 300 in triplicate. (B) PMB-treated HK-2 cells showed significant increases in micronuclei. HK-2 cells were treated with PMB (25–100 μM) for 6 or 24 h, stained with DAPI and quantified (n=200, in triplicate). Representative images of HK-2 cells. Scale bar = 5 microns. (C) Analysis of chromosome segregation events in PMB-treated HK-2 cells. HK-2 cells were treated with PMB (25–100 μM) for 2–8 h and stained with DAPI and quantified for abnormal mitotic events. A higher incidence of abnormal mitotic events (bridging and lagging chromosome) was observed at shorter time-points after PMB treatment (n=30, in triplicate).
Mitotic aberrations in polymyxin B-treated HK-2 cells
As polymyxin B interacts with DNA ((Kong et al. 2011; Wang et al. 2015) and later section), we hypothesized that polymyxin B treatment led to genome instability. We first measured the appearance of micronuclei, a hallmark of genome instability (Fenech 2002). HK-2 cells treated with 50 and 100 μM at 6 and 24 h showed significant increase in micronuclei (16.2%, 16.8% for 6 h; 15.4%, 18.1% for 24 h, respectively), compared to the untreated control (9.6%, p<0.05) (Fig. 1B). No significant changes were observed at 25 μM relative to the control (12.3% and 13.4% at 6 and 24 h respectively, p<0.05) (Fig. 1A). As micronuclei are generally a downstream phenomenon of mitotic errors, we then assessed the effect of polymyxin B treatment on mitotic defects (Fenech et al. 2011; Hudson et al. 2016). Results at short time-intervals of 2–8 h show time-dependent increases in ana/telophase defects of chromosome missegregation (chromosomal bridging and lagging chromosomes) were evident. These events increased from 15.0% to 24.9% after 2–8 h in cells treated with 25 μM polymyxin B, to more pronounced increases from 15.0% to 36.5% (50 μM) over 2–8 h, and from 10.4% to 56.9% at the highest concentration examined (100 μM) at 2–8 h (Fig. 1C). Missegregation was particularly noticeable at 6 and 8 h, where significant increases relative to controls were seen across 50 and 100 μM (27.1 and 39.5% at 6 h; 36.5 and 56.9% at 8 h respectively, relative to 17.8% in the control group, p<0.05).
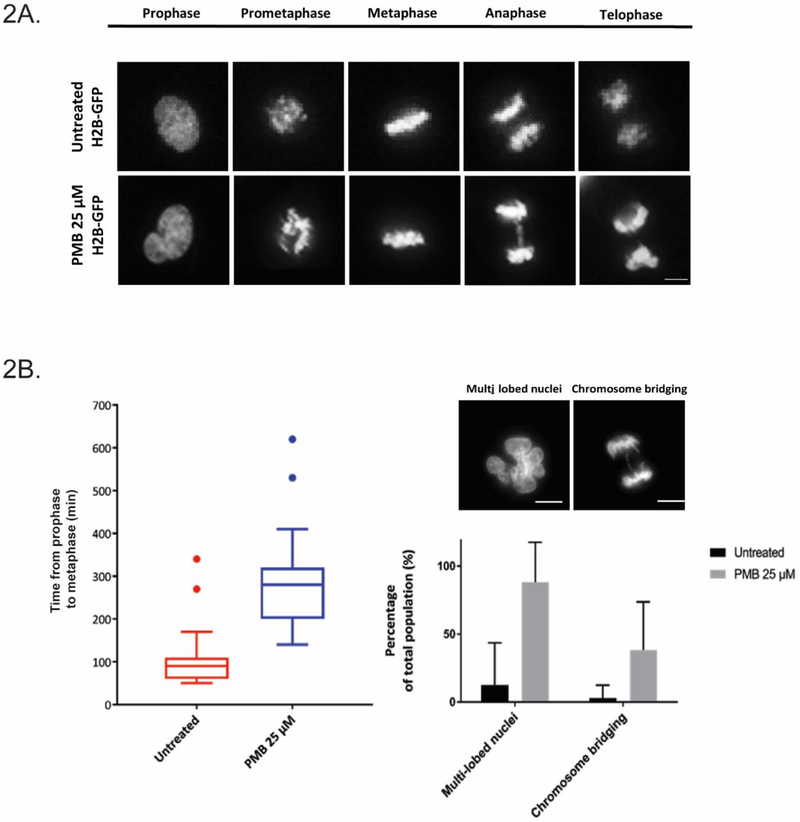
To understand this in real time, live-cell imaging with H2B-GFP HK-2 cells incubated with 25 μM polymyxin B was performed (Fig. 2A). The data correlated well with fixed cell studies, with lagging chromosomes evident (40.6% of the total population irregularities compared to 3.0% in control, p<0.05) (Fig. 2B). We also observed appearance of multi-lobed nuclei in HK-2 cells which increased from 12.5% in the control to 90.0% over 24 h. The time taken for a mitotic cell to progress from prophase to anaphase also increased from an average of 78.5 min to 113.6 min in cells exposed to polymyxin B, suggesting mitotic delay from drug treatment.
Fig. 2.
PMB-induced mitotic errors are evident in the live-imaging experiments with HK-2 cells. (A) Representative images of live-imaging of H2B-GFP cells. Cells were imaged over 24 h with FITC and BF channels. (B) Observed prolonged transition time between prophase and metaphase, appearance of multi-lobular nuclei and chromosome bridging over 24 h in PMB-treated cells. All data are presented as mean ± SD. Scale bar = 5 microns.
DNA damage and G2 checkpoint in polymyxin B treated HK-2 cells
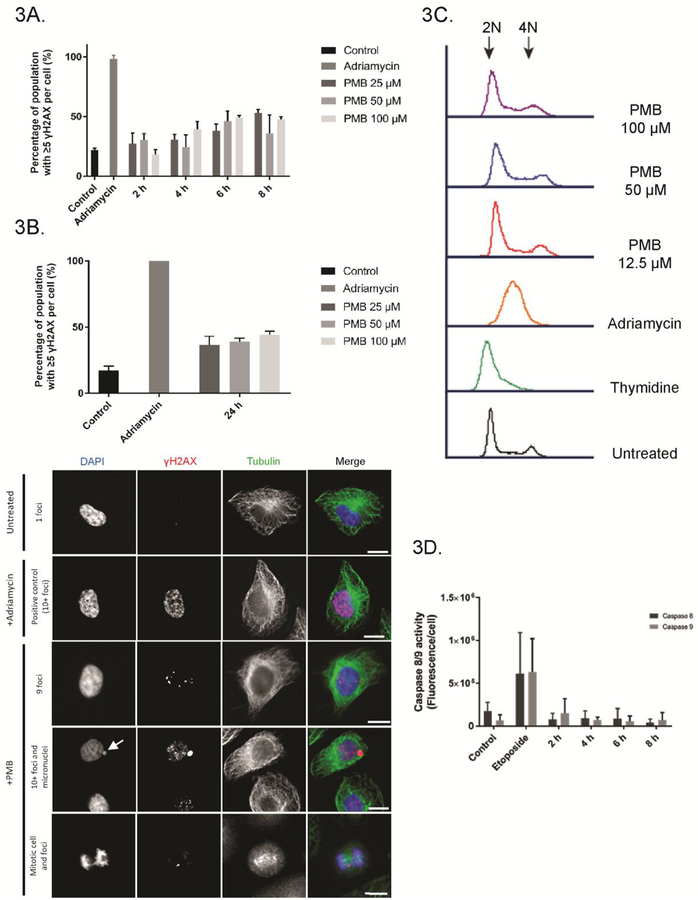
A likely cause of the mitotic defects is upstream DNA damage, which causes errors in DNA replication and can lead to mitotic and genomic defects if left unresolved(Liu et al. 2014; Nielsen and Hickson 2016; Payne and Hickson 2009). Therefore, it was important to examine if and when DNA damage occurred and whether it was an early effect of polymyxin B treatment. Double stranded breakages (DSB) can occur as a result of direct DNA damage, or as an intermediate when the repair process deals with stalled or converging replication forks and other intermediates (van Gent et al. 2001). Cells were stained for γH2AX, which recognises DSBs and is used to assess DNA damage (Kuo and Yang 2008). Time-dependent increases of γH2AX foci (5+ per cell) were observed in HK-2 cells from 2–8 h of polymyxin B treatment at 25, 50 and 100 μM, with notable differences arising at 4 h post-incubation with 100 μM polymyxin B (39.4% v 22.0% in untreated control, p<0.05) (Fig. 3A). At 6 and 8 h, the difference became more evident, with significant increases observed at concentrations of 25, 50 and 100 μM (38.2%, 46.1% and 49.2%, respectively at 6 h; 53.1%, 35.9% and 47.6%, respectively at 8 h, relative to untreated control at 22.0%, p<0.05). At 24 h, cells also showed significant differences in foci occurrence at higher concentrations (Fig. 3B). Flow cytometry analysis of the HK-2 cells treated with 12.5, 50 and 100 μM polymyxin B showed no obvious perturbations in the cell cycle (Fig. 3C), while the thymidine control showed a G1/S-arrested population and Adriamycin showed G2/M accumulation as expected.
Fig. 3.
γH2AX foci notably increased in cell culture after PMB-treatment. (A) Shorter time-course experiment of PMB-treated cells (2, 4, 6, 8 h at 25, 50 and 100 μM) stained for γH2AX showed notable increases in foci at 6 h and afterwards (n=200 in triplicate). (B) γH2AX foci also appeared elevated in cell culture at 24 h. Treated cells were stained with anti-γH2AX antibodies and quantified using fluorescence microscopy. Representative images of HK-2 cells with foci. Scale bar = 10 microns. (C) The cell cycle remained unperturbed by PMB treatment at 24 h. All data are presented as mean ± SD. (D) Activated caspase 8/9 staining in HK-2 cells treated with PMB (25 μM) over 8 h. Adriamycin (300 nM) incubated for 24 h was used for all positive controls across these experiments.
Lack of caspase pathway activation in HK-2 cells treated with 25 μM polymyxin B
Previously, cell death by polymyxins have been attributed to activation of caspase pathways (Azad et al. 2015; Dai et al. 2014b). To test whether apoptosis was occurring before mitotic defects and DNA damage, we conducted caspase 8/9 assays to detect early apoptosis. Results show no significant difference between the amounts of activated caspase 8/9 of control and polymyxin B-treated cells (25 μM), whereas etoposide-treated positive control showed over 5-fold increase (Fig. 3D). This suggests that the DNA damage induced by polymyxin B at 25 μM was an early effect independent of an apoptotic response.
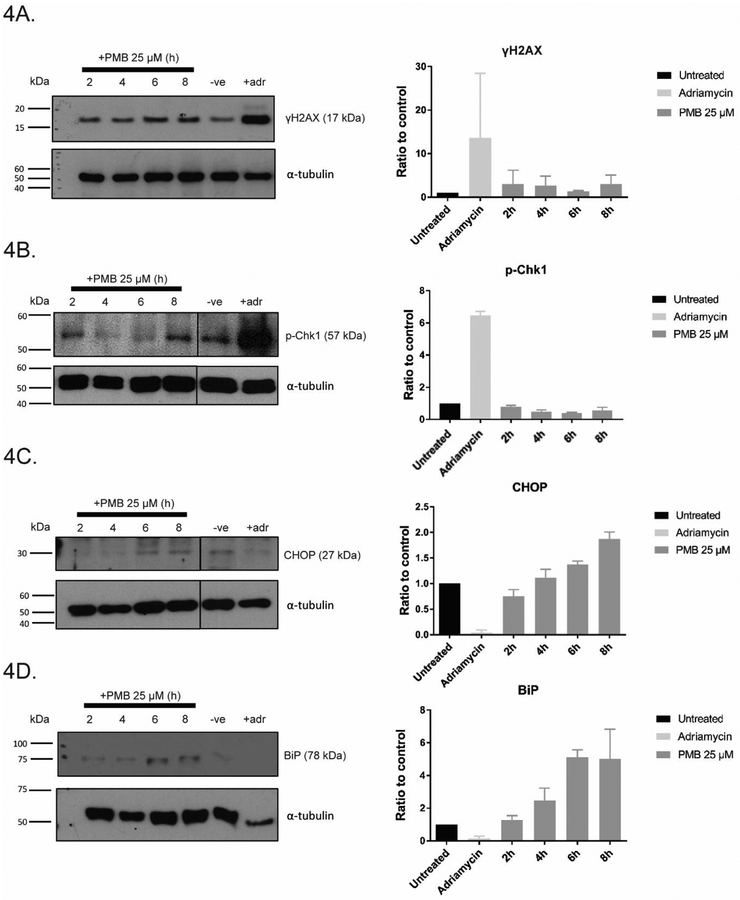
To test whether the G2 checkpoint was activated after polymyxin B treatment, immunoblotting was conducted to detect γH2AX and phospho-Chk1 (pChk1). Consistent with the immunofluorescence data, γH2AX signal increased in a time-dependent manner (×1.2 at 2 h to ×1.6 at 8 h, relative to control) (Fig. 4A). Interestingly, pChk1 signal gradually decreased, with the lowest at ×0.4 relative to control, suggesting that the G2 checkpoint was gradually suppressed in prolonged polymyxin B treatment (Fig. 4B). Both markers of ER stress, BiP and CHOP, increased in cells exposed to polymyxin B over time (Figs. 4C and D) (Dai et al. 2014a).
Fig. 4.
γH2AX and other markers of stress and DNA damage were elevated in HK-2 cells after PMB treatment. HK-2 cells were either untreated (-ve) or treated for 2, 4, 6, 8 h at 25 μM PMB and analyzed by western blotting using antibodies for (A) γH2AX, (B) p-Chk1, (C) CHOP, and (D) BiP. All data are presented as mean ± SD in triplicate.
DNA-binding affinity
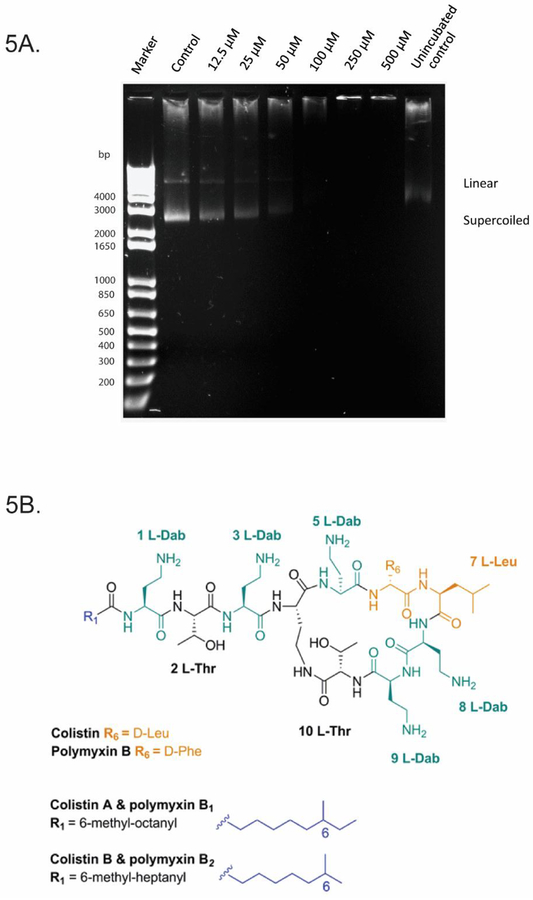
Multiple studies have suggested that polymyxin B binds to DNA (Kong et al. 2011; Wang et al. 2015). Given its amphipathic structure, it is thought that polymyxin B binds DNA non-covalently via electrostatic attraction, groove-binding, intercalation or a combination of these (Kong et al. 2011; Wang et al. 2015). Kong et al. previously utilised polymyxin B as a probe to determine the DNA concentrations of calf thymus, salmon and herring sperm (Kong et al. 2011). No studies to date have investigated polymyxin binding B to plasmid DNA. In our experiments, migration of the super-coiled and linear forms of plasmid DNA was inhibited by polymyxin B at concentrations of ≥25 μM, demonstrating affinity of polymyxin B to plasmid DNA (Fig. 5A). A notable difference was evident at 50 μM, where almost all DNA remained in the well; higher concentrations (100, 250 and 500 μM) also showed DNA remaining in the respective wells. In vitro binding of polymyxin B to DNA is consistent with our finding that polymyxin B treatment causes DNA damage and suggests this could be an early defect. The data is consistent with the previous reported modelling of polymyxin B, which showed that negatively charged grooves on the DNA helix were bound by positively charged Dab (diaminobutyrate) residues on the polymyxin molecule (Fig. 5B). The presence of five Dab residues on the molecule may also potentially result in crosslinking of different DNA molecules.
Fig 5.
PMB had high affinity for DNA and inhibited electrophoretic migration of plasmid at higher concentrations. (A) 60 ng of pBlueScript II DNA incubated with PMB (12.5–500 μM) in a topoisomerase I buffer for 2 h at 37 °C. Samples were resolved by electrophoresis. (B) Chemical structure of PMB with potential DNA binding sites (including the Dab residues) highlighted.
Polymyxin-induced nephrotoxicity in mice
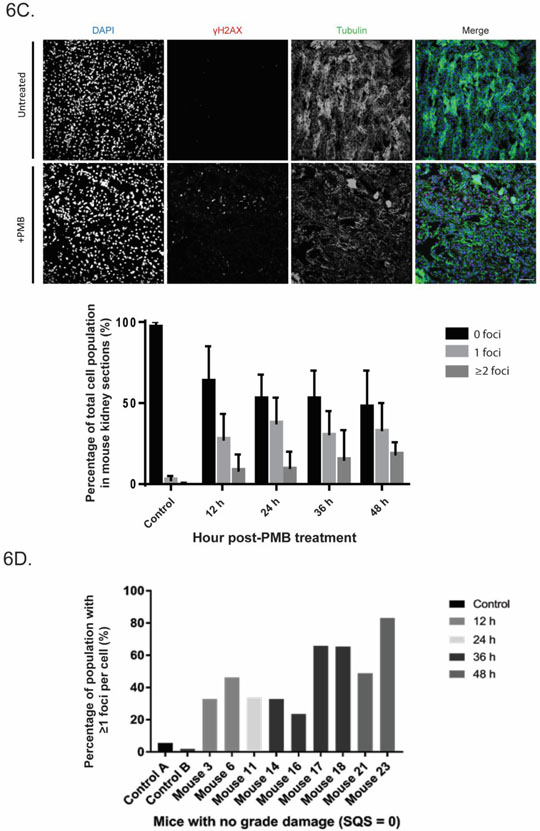
We next investigated the nephrotoxic effect of polymyxin B in animals (Fig. 6). Nephrotoxicity was confirmed and graded from histopathological hematoxylin and eosin (H&E) staining of the left kidney of each mouse. Here we used 72 mg/kg/day after considering animal scaling and clinically relevant dosage regimens (Nair and Jacob 2016). All treated mice showed increased kidney damage relative to both untreated and saline controls. Histopathological results varied between mice at each time-point with damage ranging from no grade to grade 2 (SQS from 0.1 to 2) - from mild tubular damage with tubular dilution and casts, to cortical necrosis of tubules and glomeruli, sometimes with papillary necrosis (Fig. 6A). Overall, kidney damage in mice appeared to peak at 24 h post-treatment with polymyxin B (72 mg/kg), with 5 of the 6 mice showing high percentile damage scoring. This was followed by a general overall decrease in damage across time (Fig. 6B).
Fig. 6.
PMB-induced nephrotoxicity in mice showed histological damage and DNA damage and repair. (A) Representative histopathological changes in mouse kidneys after PMB treatment, showing identifiers used to assess damage including tubular necrosis and degeneration, tubular cases and necrotic tubules. (B) Graphed damage of time-course mouse model of polymyxin-induced nephrotoxicity showed different degrees of damage (n=2000 cells for each mouse, n=6 for each time-point). (C) Time-course of PMB-induced nephrotoxicity in mice showed mean increase in foci to cell percentile of total population as time post-treatment increased, with representative examples of undamaged and damaged mouse kidney sections showing γH2AX foci staining. All data are presented as mean ± SD. (D) Foci percentage of individual mice which showed no obvious histological damage (n=8).
We then investigated γH2AX occurrence in mice. Increases in γH2AX foci in cells (≥1) were observed in sections as early as 12 h post-treatment of polymyxin B; and there was little evidence to suggest recovery over time despite cessation of drug administration (Fig. 6C). Interestingly, of the 8 individual mice that showed no significant histopathological damage (no grade damage, SQS 0.1), characteristic DSB γH2AX foci still appeared in a time-dependent manner (Fig. 6D). Taken together, the animal and cell culture data showed that an unwanted side effect of polymyxin B is DNA damage.
Discussion
Nephrotoxicity has long been a standing issue with polymyxin use in patients(Landman et al. 2008; Lopez-Novoa et al. 2011). Recent studies have shed light on the mechanistic pathways of polymyxin-induced nephrotoxicity (Azad et al. 2015; Dai et al. 2014a; Dai et al. 2014b). However, there remains a lack of understanding of early stage pathways activated by polymyxins. Together, our data and others demonstrate polymyxin B affinity for DNA (Fig. 5A) (Kong et al. 2011; Wang et al. 2015). A previous study using fluorescent polymyxin probes showed that polymyxin primarily localizes in the ER and mitochondria in cultured rat renal tubular cells, with only modest signals in the nucleus (Yun et al. 2015) This discrepancy is likely due to the recognition epitope of the antibody, reported to specifically recognise the conformational hydrophobic entities (fatty acyl tail and position 6/7 hydrophobic residues) and cyclic backbone (Velkov et al. 2016). A polymyxin molecule where these components are hidden or blocked from binding to DNA (or other proteins) would not be bound by the antibody, whereas free polymyxin B would be (Fig. 5B). Regardless, the effects of polymyxin B interference with the cell division process is apparent through the complementary experiments conducted in this study.
The cell cycle is monitored by specific checkpoints (Harashima et al. 2013). In G2/M phase, the G2 checkpoint is activated upon intrinsic/extrinsic insult to the replicated DNA, and the cell cycle is halted until DNA damage is repaired (Mankouri et al. 2013). In cells with deficiency and/or perturbation of this checkpoint, the cycle will proceed regardless, leading to chromosome missegregation and eventual micronuclei formation (Kastan and Bartek 2004; Zhang et al. 2016b). In our studies with polymyxin B-treated cells, significant increases in micronuclei and aberrant mitotic events were evident (Fig. 1C). Additionally, there was a notable increase in multi-lobed nuclei in our live cell studies, another effect of genome instability from segregation defects (Fig. 2B) (Vaz et al. 2013). Similarly, transition time from prophase to metaphase was substantially increased by ~2-fold in cells treated with 25 μM polymyxin B, suggesting difficulties in progression to chromosome segregation (Fig. 2B). Regardless, the cell cycle appeared to continue unperturbed in these cells (Fig. 3C). This can be explained by either: (1) following polymyxin B treatment, the cell cycle was halted and DNA damage repaired, or (2) cell cycle was transiently halted, but continued without repairs due to checkpoint perturbations caused directly/indirectly by polymyxin B. We favour the second possibility. FACS data lacked indication of cell cycle arrest in polymyxin B-treated cells, suggesting continued progress to mitosis (Fig. 3C). Furthermore, time-course experiments showed increase in γH2AX, suggesting persistent DNA damage in polymyxin B-treated cells; whereas pChk1, a hallmark for an activated G2 checkpoint showed gradual decreases from 2 to 6 h, implying that polymyxin B might play a role in perturbing this checkpoint (Figs. 4A and B) (Liu et al. 2000). As such, these cells with unresolved DNA damage are a likely source of the abnormal mitotic events and micronuclei formation following polymyxin B treatment, and escaped the checkpoint to proceed to mitosis (Figs. 1B and C).
Accumulation of oxidative stress markers BiP and CHOP was also observed (Fig. 4C and D), consistent with previous reports (Dai et al. 2014a). Polymyxins are known to induce oxidative stress through disruption of ER and mitochondria pathways (Azad et al. 2015; Dai et al. 2014a). CHOP, a proapoptotic factor, activates downstream effector caspases including caspase 9 after prolonged ER stress (Shiraishi et al. 2006). In our study, elevation of CHOP signal was evident across the short time-course, despite absence of activated caspase. While unclear whether this is a primary or secondary effect of drug treatment, mitotic defects and DSB appeared at a similar time, suggesting multiple, concurrent pathways of cellular toxicity. An indicator of DNA damage, γH2AX, also appeared significantly elevated around this time-point across all evaluated concentrations (Kuo and Yang 2008).
Lagging chromosomes arise when a chromosome mass falls behind during anaphase, leading to micronuclei formation (Liu et al. 2014). Chromosomal bridges are stretches of condensed chromatin connected during anaphase, often due to DNA entanglements unresolved prior to segregation (van Gent et al. 2001). This can lead to uneven distribution of DNA in the two daughter cells, formation of micronuclei (Figure 1B) or even multi-lobular nuclei as seen in live cell imaging with polymyxin B-treated cells (Fenech 2002). At shorter time-points up to 8 h, these chromosomal aberrations were observed in mitotic cells post-treatment across all concentrations tested (25–100 μM polymyxin B) (Fig. 1C). Interestingly, Operetta data with HK-2 at lower concentrations (25 and 50 μM) also showed more gradual toxicity effect (Fig. 1A), and this may be due to the accumulation of errors as the cells underwent mitosis.
A similar effect is also evident in the mouse model of polymyxin toxicity with clinically relevant doses (cumulative dose 72 mg/kg) (Fig. 6). Perhaps the most interesting observation arising from this study is the gradual overall increase in the average number of cells showing ≥1 γH2AX foci, as the time post-treatment continues (from 12× at post-12 h treatment relative to the control, to 16× at post-48 h treatment). This is observed despite mice demonstrating modest recovery at 36–48 h post-treatment histopathologically, and corroborates the cell culture results. Of particular interest are the sub-group of kidneys which showed no grade damage (SQS 0.1) (Fig. 6D). Despite very low levels of observable damage, individual kidneys were affected on a molecular level, shown by the general post-treatment time-dependent increase in appearance of γH2AX foci per cell. This further supports the aforementioned findings of potential accumulation of molecular errors over time, and suggests the possibility of continued latent damage following treatment, despite mice appearing healthy by standard assessment (SQS scoring).
Our data cannot rule out the possibility that other targets could elicit a similar response indirectly, and further proteomics studies are warranted. Overall, we have demonstrated that polymyxin B causes DNA damage and suggest that polymyxin nephrotoxicity may have more long-term effects than originally envisioned, in particular from delayed emergence of damage after cessation of treatment. Translated to the clinical setting, this may warrant closer patient monitoring as well as extended follow-up studies and vigilance.
Conclusions
In this study, we discovered that polymyxin B causes DNA damage and leads to genome instability. These results pinpoint a defect that needs to be investigated and further characterized to counter the toxic effects of polymyxins in the clinic. Our findings may provide new impetuses for targeted alterations in the polymyxin structure that might remove the DNA damage side effect. Polymyxins remain effective against Gram-negative ‘superbugs’ and any improvement in their efficacy and safety will take pressure off an already strained system in the need of new antibiotics.
Materials and methods
Chemicals
Polymyxin B (#81334, 6,500 IU/mg), thymidine (both Sigma-Aldrich, NSW, Australia) and Adriamycin (LC, Laboratories, Massachusetts, US) were sterilized by 0.22-μm syringe filters (Millex-GV; Millipore) and solubilized accordingly.
Cell culture
Immortalized human proximal tubular cells (HK-2; ATCC, Manassas, VA, USA), were grown in keratinocyte medium supplemented with pituitary growth hormone (0.05 mg/mL) and human recombinant epidermal growth factor (5 ng/mL) (Invitrogen, Victoria, Australia). HK-2 cells were grown in supplemented KSFM at 37°C in a humidified atmosphere of 5% CO2 until 60–70% confluent.
Plasmid band shift assay
The assay described by Dunstan et al. (Dunstan et al. 2002) was used to test the ability of polymyxin B to bind DNA. In brief, a 20-μL reaction containing 60 ng pBlueScript II SK(+) plasmid DNA (Stratagene), polymyxin B (12.5–500 μM), or ddH2O (control) was incubated at 37 °C for 2 h in topoisomerase I buffer (NEB). Samples were resolved by electrophoresis on1% agarose gel and migration of the plasmids were visualized on a G:BOX Chemi XT4 gel doc system.
Operetta
HK-2 cells were seeded in triplicate into 96-well plates at increasing concentrations at 0, 5, 12.5, 25, 50, 100, 200 and 500 μM and were imaged over 48 h in the presence of PI (1 μg/mL) with the Operetta High-Content Imaging System (PerkinElmer). Percentage cell viability across time was analysed and graphed.
Caspase 8/9 assay
CaspGLOW red active caspase-8 and −9 assay kits were employed according to the manufacturer’s instructions (CaspGLOW red active caspase-8 and −9 assay kit; BioVision, USA). HK-2 cells were treated with 25 μM polymyxin B for 2, 4, 6 and 8 h, or etoposide (100 μM) and caffeine (300 μM), incubated for 24 h as a positive apoptosis control. For each analysis, Images were acquired with a DeltaVision widefield deconvolution microscope, 40× oil objective at Ex/Em: 561/566–685 nm (Applied Precision). ImageJ was employed to quantify background-subtracted average integrated intensity per cell by counting. 50 cells per sample were randomly selected from 5+ fields.
Micronuclei and abnormal mitotic events
HK-2 cells were incubated with polymyxin B (12.5, 50 and 100 μM) for 6 or 24 h and stained with DAPI (10 μg/mL) for assessment of micronuclei. 500 cells in triplicate were counted. Cells were also incubated with polymyxin B (25, 50 and 100 μM) at shorter intervals (2, 4, 6 and 8 h) and stained with DAPI for quantification of mitotic aberrations - chromosome bridging and lagging chromosomes. For each time-point and concentration, 30 random mitotic cells were analysed in triplicate.
Flow cytometry
FACS analysis was performed as described by Zhang et al. (Zhang et al. 2016c). In brief, cells were treated with polymyxin B (12.5 and 100 μM), thymidine (3 mM) or adriamycin (300 nM) for 24 h. Cells were harvested and fixed with 80% ethanol at 4 °C overnight, washed with 0.1% Triton X-100, treated with RNase (1 mg/mL), and stained with PI (50 μg/mL) at 37°C for 2 h. Cells were analysed using FACSCalibur (Becton Dickinson).
Immunofluorescence staining
HK-2 cells incubated with polymyxin B (25, 50 and 100 μM) were fixed in 4% PFA, permeabilized with 0.3% Triton X-100 for 10 min, blocked with 5% BSA-PBS for 1 h and incubated with rabbit anti-γH2AX antibody (1:2000) overnight at 4°C. Samples were incubated with secondary anti-rabbit AlexaFluor594 (1:1000) for 2 h at 37°C, and stained with DAPI (10 μg/mL). γH2AX foci were quantified with Ex/Em of 590/617 nm. Adriamycin-treated (300 nM) cells were used as a positive control. 300 cells per sample were quantified in triplicate.
Western blotting
Cell pellets were incubated in a protease/phosphatase inhibitor cocktail (Roche) on ice for 30 min, ultrasonicated and centrifuged at 120,000 × g for 10 min at 4 °C. Protein concentration was determined using Bradford’s assay (Bio-rad), with standard curve of 0.025 – 2 mg/mL BSA. For each well, 40 μg protein was loaded. The following primary antibodies were employed: rabbit polyclonals against p-Chk1, BiP, CHOP (all 1:1000, Cell Signalling), γH2AX (1:2000, Abcam), and mouse polyclonals against α-tubulin (1:2000, Sigma) and β-tubulin (1:1000, Sigma). Secondary HRP antibodies were goat anti-rabbit IgG or rabbit anti-mouse (1:5000–1:10000, Cell Signalling). Protein band intensity was measured and normalized to either α or β-tubulin and analysed using ImageJ (imagej.nih.gov/ij).
Live time-course of H2B-GFP labelled HK-2 cells
HK-2 cells were transiently transfected with CellLight 2.0 Histone-2B-GFP reagent according to manufacturer’s instructions and incubated for at least 18 h before commencement of live-imaging. Live-imaging of cells incubated with 25 μM of polymyxin B was conducted over 26 h on a DeltaVision wide-field deconvolution microscope (× 40 oil objective) (Applied Precision) equipped with an environment control chamber containing 5% CO2 at 37°C. Images were taken every 10 min of 10 fields of view, with(Ex/Em 488/510 nm). Cells were analysed for 3 mitotic errors: time from prophase to anaphase, multi-lobed nuclei and chromosome bridging.
Mouse model of polymyxin-induced nephrotoxicity
Animal studies were approved by the Monash Institute of Pharmaceutical Sciences Animal Ethics Committee and performed in accordance with the Australian National Health and Medical Research Council Guidelines for the Care and Use of Animals for Scientific Purposes. Female Swiss mice (6–8 weeks, 20–25 g, Monash Animal Research Platform, Victoria, Australia) were used. Mice had free access to food and water at all times. Studies were conducted using our validated mouse nephrotoxicity model (Roberts et al. 2015). Mice were separated into polymyxin B (n = 24) and control (n = 8) groups. In the experimental group, mice were administered polymyxin B (12 mg/kg) subcutaneously every 2 h over day one (accumulated dose 72 mg/kg), euthanized via isoflurane overdose at 12, 24, 36 and 48 h post-treatment (n = 6 each time point) and kidneys harvested. In control group, polymyxin B was replaced with saline (n = 2 per time point). The right kidney of each mouse was flash frozen and sectioned for immunofluorescence, the left kidney was fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned and stained with H&E. Histopathological scoring was conducted and tubular damage evaluated using SQS score (Yousef et al. 2011). Three grades were employed: grade 1, mild acute tubular damage with tubular dilation, prominent nuclei, and several pale tubular casts; grade 2, severe acute tubular damage with necrosis of tubular epithelial cells and numerous tubular casts; and grade 3, acute cortical necrosis/infarction of tubules and glomeruli with or without papillary necrosis. Staining of sectioned kidneys followed a similar procedure as that of cell culture, with frozen sections fixed with 4% PFA and permeabilized.. Foci quantification was performed blind using ImageJ and macros script with 2000 cells per kidney section.
Statistical analysis
All experiments were conducted with a minimum of three replicates, and results are presented as mean ± standard deviation (SD) unless specified otherwise. Statistical significance denotes Tukey’s test with p<0.05 with one-way analysis of variance (ANOVA) using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA).
Acknowledgements
J.L. and T.V. are supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI111965). Part of this work was supported by National Health and Medical Research Council (Australia) project grant GNT1127209 (D.H.) and by the Victorian Government’s Operational Infrastructure Support Program. J.L. is an NHMRC Senior Research Fellow and T.V. is an NHMRC Career Development Industrial Fellow. This study utilised Australian Phenomics Network Histopathology and Organ Pathology Service, University of Melbourne for analysis of mouse kidneys.
Footnotes
Disclaimer
All authors declare no competing interests. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
References
- Azad MA, Atker J, Rogers KL, Nation RL, Velkov T, Li J (2015) Major pathways of polymyxin-induced apoptosis in rat kidney proximal tubular cells. Antimicrob Agents Chemother 59:2136–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad MA et al. (2013) Polymyxin B induces apoptosis in kidney proximal tubular cells. Antimicrob Agents Chemother 57:4329–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher HW et al. (2013) 10 x ‘20 Progress--development of new drugs active against Gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher HW et al. (2009) Bad bugs, no drug: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12 [DOI] [PubMed] [Google Scholar]
- Dai C, Li J, Tang S, Li J, Xiao X (2014a) Colistin-induced nephroxicity in mice involves the mitochondria, death receptor, and endoplasmic reticulum pathways. Antimicrob Agents Chemother 58:4075–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C et al. (2014b) Lycopene attenuates colistin-induced nephrotoxicity in mice via activating the Nrf2/HO-1 pathway. Antimicrob Agents Chemother 59:579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan HM et al. (2002) Cell-based assays for identification of novel double-strand break-inducing agents. J Natl Cancer Inst 94:88–94 [DOI] [PubMed] [Google Scholar]
- Falagas ME, Bliziotis IA (2007) Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? Int J Antimicrob Agents 29:630–636 [DOI] [PubMed] [Google Scholar]
- Fenech M (2002) Chromosomal biomarkers of genomic instability relevant to cancer. Drug Discov Today 7:1128–1137 [DOI] [PubMed] [Google Scholar]
- Fenech M et al. (2011) Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26:125–132 [DOI] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Schnittger A (2013) Cell cycle control across the eukaryotic kingdom. Trends Cell Biol 23:345–356 [DOI] [PubMed] [Google Scholar]
- Hudson DF, Amor DJ, Boys A, Butler K, Williams L, Zhang T, Kalitsis P (2016) Loss of RM12 increases genome instability and causes a bloom-like syndrome PLoS Genetics 12:e1006483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabes D (2011) The antibiotic R&D pipeline: an update. Curr Opin Microbiol 14:564–569 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 435:316–323 [DOI] [PubMed] [Google Scholar]
- Katz R (1963) Renal and possibly hepatic toxicity from coly-mycin. Report of a case. Med Ann Dist Columbia 32:408–413 [PubMed] [Google Scholar]
- Keirstead ND et al. (2014) Early prediction of polymyxin-induced nephrotoxicity with next generation urinary kidney injury biomarkers. Toxicol Sci 137:278–291 [DOI] [PubMed] [Google Scholar]
- Koch-Weser J, Sidel VW, Federman EB, Kanarek P, Finer DC, Eaton AE (1970) Adverse effects of sodium colistimethate. Manifestations and specific reaction rates during 317 courses of therapy. Ann Intern Med 72:857–868 [DOI] [PubMed] [Google Scholar]
- Kong L, Liu Z, Hu X, Liu S (2011) Interaction of polymyxin with ds-DNA, and determination of DNA or polymyxin B via resonance Rayleigh scattering and resonance non-linear scattering spectra. Microchim Acta 173:207–213 [Google Scholar]
- Kubin CJ, Ellman TM, Phadke V, Haynes LJ, Calfee DP, Yin MT (2012) Incidence and predictors of acute kidney injury associated with intravenous polymyxin B therapy. J Infect 65:80–87 [DOI] [PubMed] [Google Scholar]
- Kuo LJ, Yang L (2008) γ-H2AX - A novel biomarker for DNA double-strand breaks. In vivo 22:305–309 [PubMed] [Google Scholar]
- Landman D, Georgescu C, Martin DA, Quale J (2008) Polymyxins revisited. Clin Microbiol Rev 21:449–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Milne RW, Nation RL, Turnidge JD, Smeaton TC, Coulthard K (2003) Use of high-performance liquid chromatography to study the pharmacokinetics of colistin sulfate in rats following intravenous administration. Antimicrob Agents Chemother 47:1766–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayne CR, Paterson DL (2006) Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601 [DOI] [PubMed] [Google Scholar]
- Lim LM et al. (2010) Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy 30:1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q et al. (2000) Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes & Dev 14:1448–1459 [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nielsen CF, Yao Q, Hickson ID (2014) The origins and processing of ultra fine anaphase DNA bridges. Curr Opin Genet Dev 26:1–5 [DOI] [PubMed] [Google Scholar]
- Livermore DM (2004) The need for new antibiotics. Clin Microbiol Infect 10:1–9 [DOI] [PubMed] [Google Scholar]
- Lopez-Novoa JM, Quiros Y, Vincente L, Morales AI, Lopez-Hernandez FJ (2011) New insights into the mechanism of aminoglycoside nephrotoxicity: an integrative point of view. Kidney Int 1:33–45 [DOI] [PubMed] [Google Scholar]
- Lu X et al. (2016) Human oligopeptide transporter 2 (PEPT2) mediates cellular uptake of polymyxins. J Antimicrob Chemother 71:403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z et al. (2009) Renal disposition of colistin in the isolated perfused rat kidney. Antimicrob Agents Chemother 53:2857–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Huttner D, Hickson ID (2013) How unfinished business from S-phase affects mitosis and beyond EMBO J 32:2661–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingeot-Leclercq MP, Tulkens PM, Denamur S, Vaara T, Vaara M (2012) Novel polymyxin derivatives are less cytotoxic than polymyxin B to renal proximal tubular cells. Peptides 35:248–252 [DOI] [PubMed] [Google Scholar]
- Nair AB, Jacob S (2016) A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm 7:27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Cars O (2014) Antibiotic resistance - problems, progress and prospects. N Engl J Med 371:1761–1763 [DOI] [PubMed] [Google Scholar]
- Nation RL et al. (2017) Dosing guidance for intravenous colistin in critically ill patients. Clin Infect Dis 64:565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CF, Hickson ID (2016) PICH promotes mitotic chromosome segregation: identification of a novel role in rDNA disjunction. Cell Cycle 15:2704–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne M, Hickson ID (2009) Genomic instability and cancer: lessons from analysis of Bloom’s syndrome. Biochem Soc Trans 37:553–559 [DOI] [PubMed] [Google Scholar]
- Pogue JM et al. (2011) Incidence of and risk factors for colistin-associated nephrotoxicity in a large academic health system. Clin Infect Dis 53:879–884 [DOI] [PubMed] [Google Scholar]
- Price DJE I. GD (1970) Effects of large doses of colistin sulphomethate sodium on renal function. Br Med J 28:525–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KD et al. (2015) Antimicrobial activity and toxicity of the major lipopeptide components of polymyxin B and colistin: last-line anibotics against multidrug-resistant gram-negative bacteria. ACS Infect Dis 1:568–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri AM et al. (2013) Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57:524–531 [DOI] [PubMed] [Google Scholar]
- Shiraishi H, Okamoto H, Yoshimura A, Yoshida H (2006) ER stress-induced apoptosis and caspase-12 activation occurs downstream of mitochondrial apoptosis involving Apaf-1. J Cell Sci 1:3958–3966 [DOI] [PubMed] [Google Scholar]
- Suzuki T et al. (2013) Megalin contributes to kidney accumulation and nephrotoxicity of colistin. Antimicrob Agents Chemother 57:6319–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E, Magrini N (2017) Global priority list of antibiotic-resistant bacteria to guide research, discovery and development of new antibiotics. World Health Organization. [Google Scholar]
- Tallgren LG, Liewendahl K, Kuhlback B (1965) The therapeutic success and nephrotoxicity of colistin in acute and chronic nephropathies with impaired renal function. Acta Medica Scandinavica 177:717–728 [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R (2001) Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2:196–206 [DOI] [PubMed] [Google Scholar]
- Vaz B, Halder S, Ramadan K (2013) Role of p97/VCP (Cdc48) in genome stability. Front Genet 4:doi: 10.3389/fgene.2013.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov T, Roberts KD, Nation RL, Thompson PE, Li J (2013) Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future microbiol 8:711–724 doi: 10.2217/fmb.13.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov T, Roberts KD, Nation RL, Wang J, Thompson PE, Li J (2014) Teaching ‘old’ polymyxins new tricks: new-generation lipopeptides targeting Gram-negative ‘superbugs’. ACS Chem Biol 16:1172–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov T et al. (2016) A novel chemical biology approach for mapping of polymyxin lipopeptide antibody binding epitopes. ACS Infect Dis 2:341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu S, Liang W, Li D, Yang J, He Y (2015) Detection of DNA utilizing a fluorescent reversible change of a biosensor based on the electron transfer from quantum dots to polymyxin B sulfate. J Colloid Interface Sci 448:257–264 [DOI] [PubMed] [Google Scholar]
- Yousef JM, Chen G, Hill PA, Nation RL, Li J (2011) Melatonin attenuates colistin-induced nephrotoxicity in rats. Antimicrob Agents Chemother 55:4044–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B et al. (2015) Cellular uptake and localization of polymyxins in renal tubular cells using rationally designed fluorescent probes. Antimicrob Agents Chemother 59:7489–7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavascki AP et al. (2008) Pharmacokinetics of intravenous polymyxin B in critically ill patients. Clin Infect Dis 47:1298–1304 [DOI] [PubMed] [Google Scholar]
- Zhang L et al. (2015) Autophagy regulates colistin-induced apoptosis in PC-12 cells. Antimicrob Agents Chemother 59:2189–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L et al. (2016a) p53 mediates colistin-induced autophagy and apoptosis in PC-12 cells. Antimicrob Agents Chemother 60:5294–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Paulson JR, Bakhrebah M, Kim JH, Nowell CJ, Kalitsis P, Hudson DF (2016b) Condensin I and II behaviour in interphase nuclei and cells undergoing premature chromosome condensation. Chromosome Res 24:243–269 [DOI] [PubMed] [Google Scholar]
- Zhang T, Si-Hoe SL, Hudson DF, Surana U (2016c) Condensin recruitment to chromatin is inhibited by Chk2 kinase in response to DNA damage. Cell Cycle 15:3454–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]