Abstract
Aim:
Dadih samples from two different origins (Kamang and Gadut in West Sumatra) manufactured with different methods (back-slopping or spontaneous fermentation) were evaluated for the diversity of lactic acid bacteria (LAB).
Materials and Methods:
Four dadih samples manufactured with two different fermentation methods were obtained from Kamang and Gadut regions. Both genotypic and phenotypic characteristic (16S rRNA partial gene sequence analysis and carbohydrate fermentation profile) were used to analyze the diversity of dadih LAB population.
Results:
This study showed that LAB count in back-slopping fermented dadih was one log cycle higher than spontaneous fermented dadih. LAB isolates from the two regions were divided into three genera, namely Lactococcus, Lactobacillus, and Pediococcus. Sequencing results showed that 41.6% (five isolates) were identified as Lactococcus lactis ssp. lactis, 25% (three isolates) were identified as Lactobacillus plantarum ssp. plantarum, 16.6% (two isolates) were identified as L. lactis ssp. cremoris, and 8.3% (one isolate each) were identified as Pediococcus pentosaceus and Lactobacillus pentosus.
Conclusion:
Five species were determined in back-slopping fermented dadih, i.e., L. lactis ssp. lactis, L. lactis ssp. cremoris, L. plantarum ssp. plantarum, L. pentosus, and P. pentosaceus. On the other hand, spontaneous fermented dadih only contained three different species, namely L. lactis ssp. lactis, L. lactis ssp. cremoris, and L. plantarum ssp. plantarum. This research showed that back-slopping fermentation offers greater abundance and diversity compared to spontaneous fermentation in dadih.
Keywords: back-slopping, dadih, lactic acid bacteria, spontaneous fermentation
Introduction
Dadih is naturally fermented buffalo milk from West Sumatra and has become an integral part of Minangkabau diet. The dadih has “yogurt-like” consistency, i.e. soft texture, whitish cream color, sour, and pleasant taste. Dadih surface is smooth and shiny, clean without air bubbles in the middle of the product [1]. The fermentation process occurs spontaneously; bamboo tubes are filled with fresh buffalo milk and each tube is covered with banana leaves that are then incubated at room temperature (28-30°C) for approximately 24-48 h [2]. Nevertheless, there are still some artisans that apply back-slopping fermentation methods during dadih production. Back-slopping method is carried out by adding a small amount of fermented products into fresh ingredient and then is left to be fermented in room temperature. Finally, a stable microbial community is formed after a few cycles [3].
Various types of indigenous microflora dominated by lactic acid bacteria (LAB) in spontaneous fermentation cannot be fully controlled or correctly predicted, even though this process has been practiced for many years [4]. Microorganism might be derived from milk or the environment, i.e. bamboo tubes, banana leaves, or artisans. This traditional processing of dadih is obtained from generation to generation and is accompanied by a low quality of sanitation and hygiene during the process. The biggest risks for natural milk fermentation are the low hygiene conditions during processing, the high initial contamination, and the absence of a heating stage before the process [5]. These problems can be overcome gradually by applying the back-slopping method so that the uniformity and safety of the dadih can be correctly predicted. Various types of LAB are isolated from dadih and play a role in spontaneous fermentation, specifically Leuconostoc paramesenteroides, Lactobacillus casei ssp. rhamnosus, L. casei ssp. casei, Lactococcus lactis ssp. lactis, Lactobacillus brevis, Lactobacillus plantarum, Lactobacillus paracasei, Lactobacillus fermentum, Pediococcus pentosaceus, L. rhamnosus, and Enterococcus faecium [6-9].
Spontaneous and back-slopping fermentation methods are still performed during dadih production in Gadut and Kamang regions, West Sumatra. No study has been conducted on comparing these two fermentation methods that are applied during dadih production. We assume that LAB population is affected by the different fermentation methods. The aim of this study was, therefore, to highlight the diversity of LAB isolated from dadih in Gadut and Kamang, West Sumatra.
Materials and Methods
Ethical approval
No live animals were used in the present study. Samples were purchased from herdsmen from Kamang and Gadut region in West Sumatra.
Study area
Kamang is located in Agam District; meanwhile, Gadut is located in Limapuluhkota District. Their distance from Bukittinggi city is 16 km and 42.4 km, respectively.
Sampling
Four dadih samples were purchased from two regions in West Sumatra, namely Kamang and Gadut. Dadih sample from Kamang was processed with back-slopping fermentation by adding approximately 50 g (1 tablespoon) of previous dadih into 1600 ml of fresh milk and then placed into bamboo tube. Fermentation process occurred for 48 h at room temperature. Spontaneous fermentations were applied in dadih from Gadut. Fresh buffalo milk was inserted into the bamboo tube and let it fermented for 48 h at room temperature. pH of dadih sample was measured using Hanna HI8424 microcomputer pH meter and then 5 g were aseptically removed in a sterile tube and transferred to the laboratory under cool condition (4°C) for further analyses.
Isolation
About 1 g of each dadih sample was mixed with 45 ml of sterile NaCl 0.85%. An appropriate dilution (107-108) was made and inoculated on de Man-Rogosa-Sharpe (MRS) Agar (Merck) + 0.5% CaCO3 medium by double layer technique and incubated at 37°C for 48 h. The number of LAB colonies was expressed as log unit colony count (log cfu) per gram sample. Ten colonies with the surrounding clear zone were randomly selected in each plate. Non-motile rod and coccus, Gram-positive and catalase-negative isolates were plated on fresh MRS plates to obtain a pure culture. LAB isolates were stored in 10% (v/v) of sterile glycerol at −20°C [10].
Molecular identification
The genome DNA of the selected LAB isolates was extracted using Presto™ Mini gDNA Bacteria Kit (Geneaid). The DNA pellets were suspended in 50 µL TE buffer (Tris-EDTA) and stored at −20°C. LAB 16S rRNA gene was amplified using a universal primer 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) [11]. A total volume of polymerase chain reaction (PCR) of 50 ml consisted of 25 µL MyTaq HS Red Mix (Geneaid), 2 µl of each primer (10 pmol), and distilled water to achieve the final volume. Amplification was performed in Takara PCR Thermal Cycler SimpliAmp with PCR conditions as follows: Pre-heating at 94°C for 1.5 min, denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and elongation at a 72°C for 1.5 min, this cycle was repeated at 30 cycles and finally performed at 72°C for 5 min. PCR products were electrophoresed in 1% agarose gel. PCR products were sent to the 1st BASE for sequencing 16S rDNA gene. The sequencing results were subjected into National Center for Biotechnology Information (NCBI) GenBank (www.ncbi.nlm.nih.gov) and basic local alignment search tool (BLAST) was adopted to search GenBank database for sequence homology analysis to determine the genera of LAB. Species with similarity more than 97% was considered as the same. Phylogenetic tree of LAB was constructed with MEGA 10 software, (Proprietary Freeware, Pennsylvania State University), and neighbor-joining methods were performed to test confidence with bootstrap data set of 1000 times [12].
Fermentation profile
Identification of LAB isolates was carried out through observation of carbohydrate fermentation patterns using APIÒ 50 CHL kit (bioMérieux, France). The fermentation profile of isolates was determined using APILAB Plus software version 3.3.3. from bioMérieux [12].
Results
Isolation
Fifty-three presumptive LAB colonies with clear zone around the colony were randomly selected. The morphological test results showed 31 isolates that were categorized as non-motile rod and coccus, Gram-positive bacteria, and negative to catalase (Table-1). Based on antibacterial and proteolytic activity (data not shown), 12 LAB isolates were selected for further analysis.
Table-1.
Number of isolates, pH, total LAB, and antimicrobial properties against Escherichia coli, Staphylococcus aureus, Salmonella Typhi, and proteolytic activity.
| Variables | Gadut | Kamang |
|---|---|---|
| Number of isolates | 12 | 19 |
| pH | 4.57 | 4.52 |
| Total LAB (log cfu/g) | 8.56 | 9.04 |
| Isolates with antimicrobial properties | 9 | 13 |
| Isolates with proteolytic activity | 5 | 7 |
LAB=Lactic acid bacteria
Molecular identification
Twelve isolates were determined and compared with the partial 16S rRNA gene sequence. The DNA size of all isolates was approximately 1000-1500 bp (Figure-1). To confirm the species, these sequences were determined and compared with related bacteria using BLAST program at NCBI. LAB isolates from the two regions were divided into three genera, namely Lactococcus, Lactobacillus, and Pediococcus. Sequencing results showed that 41.6% (5 isolates) were identified as L. lactis ssp. lactis, 25% (three isolates) were identified as L. plantarum ssp. plantarum, 16.6% (two isolates) were identified as L. lactis ssp. cremoris, and 8.3% (one isolate each) were identified as P. pentosaceus and Lactobacillus pentosus (Table-2).
Figure-1.
Polymerase chain reactions product lactic acid bacteria (LAB) isolates in 1% agarose gel. Line 1 and 14 (Ladder DNA 1 kb); Line 2-13 (LAB isolate); DK=Dadih from Kamang, DG=Dadih from Gadut.
Table-2.
Identification of 12 LAB isolate by 16S rRNA sequence.
| Isolate code | Origin | Species | % ID in NCBI | Accession number |
|---|---|---|---|---|
| DK13 | Kamang | Lactobacillus pentosus | 99 | CP032757.1 |
| DG21 | Gadut | Lactobacillus plantarum ssp. plantarum | 99 | KY762263.1 |
| DK4 | Kamang | Lactococcus lactis ssp. lactis | 97 | KF879153.1 |
| DG30 | Gadut | Lactococcus lactis ssp. cremoris | 98 | MF098152.1 |
| DG17 | Gadut | Lactobacillus plantarum ssp. plantarum | 98 | CP031771.1 |
| DG15 | Gadut | Lactococcus lactis ssp. cremoris | 99 | MF098152.1 |
| DK10 | Kamang | Pediococcus pentosaceus | 99 | AB494722.1 |
| DK12 | Kamang | Lactococcus lactis ssp. lactis | 98 | KJ095659.1 |
| DG7 | Gadut | Lactococcus lactis ssp. lactis | 98 | KF148962.1 |
| DK28 | Kamang | Lactococcus lactis ssp. lactis | 98 | KF879153.1 |
| DK5 | Kamang | Lactococcus lactis ssp. cremoris | 98 | KM485587.1 |
| DK2 | Kamang | Lactobacillus plantarum ssp. plantarum | 98 | KY762263.1 |
DK=Dadih from Kamang, DG=Dadih from Gadut, LAB=Lactic acid bacteria, NCBI=National Center for Biotechnology Information
Diversity and phylogenetic tree
LAB in dadih from Kamang (DK) origin had greater diversity than that originated from Gadut (Figure-2). It consisted of three genera, namely Lactococcus, Pediococcus, and Lactobacillus, while that in dadih from Gadut was only dominated by two genera, i.e. Lactococcus and Lactobacillus.
Figure-2.
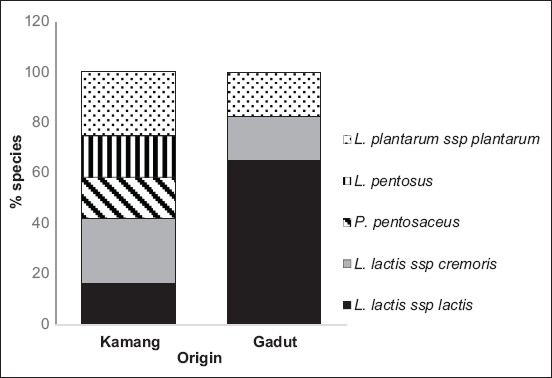
Greater diversity in lactic acid bacteria Dadih from Kamang origin than Gadut origin.
The neighbor-joining method and p-distance parameter model were applied to construct the phylogenetic tree among the aligned 16S rRNA sequences of all LAB isolates and reference strains (Figure-3). In the study, LAB isolates phylogenetic tree consisted of two clusters based on their genetic type. The first cluster belonged to Lactococcus genus, which was composed of one species, namely L. lactis. The second cluster belonged to Lactobacillus and Pediococcus genera. Lactobacillus genus consisted of two species, i.e. L. plantarum ssp. plantarum and L. pentosus, while Pediococcus genus was identified as P. pentosaceus.
Figure-3.
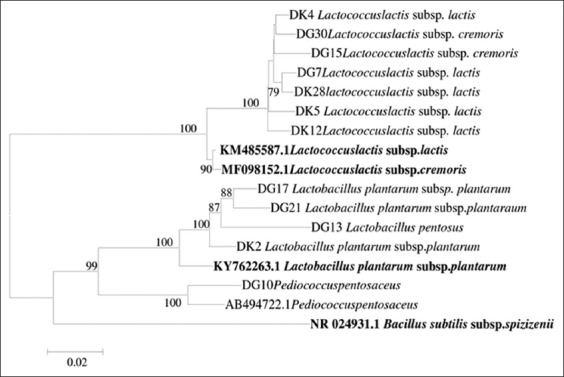
Neighbor-joining tree of LAB dadih from Kamang and Gadut origin showing phylogenetic relationship between two genera cocci and bacilli. Bacillus subtilis ssp spizizenii as an out group.
Fermentation profile
Fermentation profile of LAB isolates was carried out based on the ability to ferment various carbohydrates using API 50 CHL Kit test (Table-3). API 50 HCL kit test confirmed an accuracy rate above 99% apart from DK13 (96.8%) and DK10 (97%) isolates, although these results are still relatively high. The fermentation profile supports the results of genotypic identification based on 16S rRNA partial sequences gene and matches with their biochemical characteristics.
Table-3.
Fermentation profile of LAB isolates from Kamang and Gadut in API 50 CHL kit test.
| Isolate code* | Accuration level (%) | Identification |
|---|---|---|
| DK13 | 96.8 very good identification | Lactobacillus pentosus |
| DG21 | 99.9 excellent identification | Lactobacillus plantarum ssp. plantarum |
| DK4 | 99.8 very good identification | Lactococcus lactis ssp. lactis |
| DG30 | 99.9 excellent identification | Lactococcus lactis ssp. cremoris |
| DG17 | 99.9 excellent identification | Lactobacillus plantarum ssp. plantarum |
| DG15 | 99 very good identification | Lactococcus lactis ssp. cremoris |
| DK10 | 97 very good identification | Pediococcus pentosaceus |
| DK12 | 99.9 excellent identification | Lactococcus lactis ssp. lactis |
| DG7 | 99.9 excellent identification | Lactococcus lactis ssp. lactis |
| DK28 | 99.8 very good identification | Lactococcus lactis ssp. lactis |
| DK5 | 99.8 very good identification | Lactococcus lactis ssp. cremoris |
| DK2 | 99.9 excellent identification | Lactobacillus plantarum ssp. plantarum |
DK=Dadih from Kamang, DG=Dadih from Gadut, LAB=Lactic acid bacteria
Discussion
Dadih fermentation methods in the West Sumatra are generally classified as spontaneous. The product quality and safety are associated with the diversity and the population of microorganisms contained in the buffalo milk. Quality limitations from spontaneous fermentation caused a shift to back-slopping fermentation, which has been proven to accelerate the fermentation rate and increase safety, sensory quality, and product nutrition content [13-15]. Dadih from Gadut region was produced by spontaneous fermentation. On the other hand, specifically in Kamang, dadih is processed by back-slopping fermentation. The difference between these methods could dramatically affect the diversity of LAB as a dominant bacterial agent for dadih fermentation.
The results of the study (Table-1) showed that pH of the product ranged between 4.52 and 4.57. LAB relative abundance in dadih fermented with back-slopping method was 1 log cycle higher than that produced by spontaneous fermentation. Similar results were also shown in Kivunde and Ogi product that are fermented foods made from cassava and corn [13,16]. This high bacterial population was possibly an effect of back-slopping method application. A small amount of product from previous fermentation that was added in the buffalo milk not only caused this increase but also accelerated the fermentation time.
The results of the genotype analysis of the 12 LAB isolates showed that they were grouped into three genera, namely Lactococcus (seven isolates), Lactobacillus (four isolates), and Pediococcus (one isolate). Seven Lactococcus isolates were identified as L. lactis ssp. lactis and L. lactis ssp. cremoris; four Lactobacillus isolates were identified as L. pentosus and L. plantarum ssp. plantarum; and there was only one isolate identified as P. pentosaceus. All isolates had the same homology ≥97% with those in the reference sequence GenBank database at NCBI (Table-2). All species isolated from two processing methods have also been isolated from various other fermented milk products such as traditional fermented dairy foods from Mongolia [17,18], China [19], Africa [20], Algeria [21], Russia [22,23] Tibet [24-26], India [27], and Iran [28].
Greater diversity of LAB species was found in dadih fermented by back-slopping method. Five LAB stains had been isolated, specifically L. lactis ssp. lactis, L. lactis ssp. cremoris, L. pentosus, L. plantarum ssp. plantarum, and P. pentosaceus. Meanwhile, at spontaneous fermented dadih, only three species were found, namely L. lactis ssp. lactis, L. lactis ssp. cremoris, and L. plantarum ssp. plantarum (Figure-2). Most of the identified LAB were classified in the LAB homofermentative group, apart from L. plantarum ssp. plantarum which belongs to facultative heterofermentative group. Besides LAB originating from spontaneous fermentation, there was also found heterofermentative group in back-slopping fermentation [29]. The diversity of LAB population in back-slopping fermentation is greater than spontaneous fermentation due to the addition of some products that trigger the fermentation process [15]. In the current study, the present of L. lactis, Lb. plantarum, and L. pentosus in dadih was assumed originated from fresh buffalo milk. In a study by Sharma et al. [30] and Rizqiati et al. [31], on isolation of autochthonous LAB in buffalo milk, L. lactis, L. plantarum, L. pentosus, and L. brevis were isolated and characterized. Meanwhile, P. pentosaceus suggest originated from the stable LAB community from previous dadih. The previous study indicated that P. pentosaceus is the most typical isolated LAB from dadih [7].
L. lactis contains the majority of the isolated species in both fermentation methods. L. lactis ssp. lactis and L. lactis ssp. cremoris are important group of LAB during milk fermentation and serve as starter cultures; they produce lactic acid from lactose, hydrolyze casein and also play a significant role in citric acid fermentation and flavor formation (especially L. lactis ssp. cremoris) [32-34]. When the pH value is reduced, the growth of specific microflora is triggered, namely Lactobacillus spp. and Pediococcus spp. that are both acid tolerance. The existence of these species in the final product is predominantly caused by the presence of other species who grown first and provide a suitable environment for their growth [35]. During growth, the microbes can interact and influence each other’s growth and metabolism by antagonism, metabiosis, or cell-to-cell communication [36,37].
This succession phenomenon is a process that takes place synergistically during dadih fermentation. This same phenomenon also occurs in Kivuguto, a traditional fermented milk from East Africa. LAB succession in African fermented milk mostly initiates when Lactococcus and Leuconostoc genera grown at the beginning and in the middle of fermentation (0-8 h). Isoelectric pH (4.6) will be reached by L. lactis after 8 h of fermentation, while Leuconostoc spp. need more time, approximately 14 h to reach the same pH value [38]. The acceleration of Lactobacilli genus will dominate the substrate until 24th h fermentation. Acid tolerance is the property that enables that Lactobacilli to survive in the final product [39].
According to 16S rRNA gene sequence, a phylogenetic tree was constructed to describe a genetic relationship between isolates. The result (Figure-3) showed that there is no clear grouping in LAB isolates found in two fermentation methods. However, every identified group shared the same ancestor. As shown in Figure-3, Lactobacillus and Pediococcus genera were stand in same cluster, distinct from Lactococcus genus. Genus Pediococcus is an integral, not peripheral, part of the genus Lactobacillus [40] since the 16S rRNA gene sequence of genus Pediococcus was reported to fall within Lb. casei branch of LAB [41]. The phenotypic properties (fermentation profile in Table-3) support the genotypic characteristic of all isolates.
The diversity of LAB in the samples manufactured by the two examined methods was influenced by many factors. More extensive in-depth research is necessary to evaluate the contributing factor that affects the diversity of LAB in dadih. The different methods, recipes, raw material, environmental temperature, and location difference, may also cause some variation in strain [17]. Furthermore, anthropogenic variables (e.g., competition among sellers and organoleptic preferences) are important in shaping microbial community structure [3].
Conclusion
The different methods in dadih fermentation significantly affected LAB diversity. The back-slopping fermentation method provided greater diversity of LAB than spontaneous fermentation in dadih (two different species had been isolated, namely L. pentosus and P. pentosaceus). Understanding the role of complex, bacterial population and diversity could be useful to adjust the fermentation methods from spontaneous to back-slopping in dadih manufacturing.
Authors’ Contributions
MBS supervised the research. CUW and EAS carried out the analyses. MBS, DWL, IW, and EAS supervised laboratory work. CUW wrote the manuscript with the help of MBS, DWL, and IW. All authors read and approved the final manuscript.
Acknowledgments
This research was supported by Beasiswa Unggulan Dosen Indonesia, Dalam Negeri, Indonesia scholarship from LPDP (Lembaga Pengelola Dana Pendidikan), Ministry of Finance, Republic of Indonesia.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Surono I.S. Traditional Indonesian dairy foods. Asia Pac. J. Clin. Nutr. 2015;24(1):S26–S30. doi: 10.6133/apjcn.2015.24.s1.05. [DOI] [PubMed] [Google Scholar]
- 2.Sunaryanto R, Marwoto B. Isolasi, identifikasi, dan karakterisasi bakteri asam laktat dari dadih susu kerbau. J. Sains Teknol. Indones. 2012;14(3):228–233. [Google Scholar]
- 3.Schoustra S.E, Kasase C, Toarta C, Kassen R, Poulain A.J. Microbial community structure of three traditional Zambian fermented products:Mabisi, chibwantu and munkoyo. PLoS One. 2013;8(5):1–12. doi: 10.1371/journal.pone.0063948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Widyastuti Y.R, Febrisiantosa A. The role of lactic acid bacteria in milk fermentation. Food Nutr. Sci. 2014;5(4):435–442. [Google Scholar]
- 5.Inatsu Y, Bari M.D.L. Food safety challenges associated with traditional fermented food. In: Ray R.C, Montent D, editors. Microorganism and Fermentation of Traditional Foods. New York (US): CRC Press; 2014. [Google Scholar]
- 6.Surono I.S. In vitro probiotic properties of indigenous dadih lactic acid bacteria. Asian Australas. J. Anim. Sci. 2003;16(5):726–731. [Google Scholar]
- 7.Mustopa A.Z, Fatimah F. Diversity of lactic acid bacteria isolated from Indonesian traditional fermented foods. Microbiol. Indones. 2014;8(2):48–57. [Google Scholar]
- 8.Syukur S, Rijal F, Jamsari A, Purwati E. Isolation and molecular characterization of lactic acid bacteria by using 16s rRNA from fermented buffalo milk (Dadih) in Sijunjung, West Sumatera. Res. J. Pharm. Biol. Chem. Sci. 2014;5(6):871–876. [Google Scholar]
- 9.Nuraida L. A review:Health promoting lactic acid bacteria in traditional Indonesian fermented foods. Food Sci. Hum. Wellness. 2015;4(2):47–55. [Google Scholar]
- 10.Dzhakibaeva T.G, Kebekbaeva K.M. Comparison storage methods of lactic acid bacteria. J. Int. Sci. Publ. Agric. Food. 2010;2(2014):316–321. [Google Scholar]
- 11.Pang H, Tan Z, Qin G, Wang Y, Li Z, Jin Q, Cai Y. Phenotypic and phylogenetic analysis of lactic acid bacteria isolated from forage crops and grasses in the Tibetan plateau. J. Microbiol. 2012;50(1):63–71. doi: 10.1007/s12275-012-1284-5. [DOI] [PubMed] [Google Scholar]
- 12.Jannah S.N, Dinoto A, Wiryawan K.G, Rusmana I. Molecular diversity pattern of intestinal lactic acid bacteria in Cemani chicken, Indonesian native chicken, as revealed by terminal restriction fragment length polymorphisms. Malays. J. Microbiol. 2016;12(1):102–111. [Google Scholar]
- 13.Teniola O.D, Holzapfel W.H, Odunfa S.A. Comparative assessment of fermentation techniques useful in the processing of ogi. World J. Microbiol. Biotechnol. 2005;21(1):39–43. [Google Scholar]
- 14.Kabak B, Dobson A.D.W. An introduction to the traditional fermented foods and beverages of Turkey. Crit. Rev. Food Sci. Nutr. 2011;51(3):248–260. doi: 10.1080/10408390903569640. [DOI] [PubMed] [Google Scholar]
- 15.Shrivastava N, Ananthanarayan L. Use of the backslopping method for accelerated and nutritionally enriched idli fermentation. J. Sci. Food Agric. 2014;95(10):2081–2087. doi: 10.1002/jsfa.6923. [DOI] [PubMed] [Google Scholar]
- 16.Kimaryo V.M, Massawe G.A, Olasupo N.A, Holzapfel W.H. The use of starter culture in the fermentation of cassava for the production of ''kivunde'', a traditional Tanzanian food product. Int. J. Food Microbiol. 2000;56(2-3):179–190. doi: 10.1016/s0168-1605(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Wang W.H, Menghe B.L.G, Jiri M.T, Wang H.M, Liu W.J, Bao Q.H, Lu Q, Zhang J.C, Wang F, Xu H.Y, Sun T.S, Zhang H.P. Diversity of lactic acid bacteria associated with traditional fermented dairy products in Mongolia. J. Dairy Sci. 2011;94(7):3229–3241. doi: 10.3168/jds.2010-3727. [DOI] [PubMed] [Google Scholar]
- 18.Gao M.L, Hou H.M, Teng X.X, Zhu Y.L, Hao H.S, Zhang G.L. Microbial diversity in raw milk and traditional fermented dairy products (Hurood cheese and Jueke) from Inner Mongolia, China. Genet. Mol. Res. 2017;16(1):1–13. doi: 10.4238/gmr16019451. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W.Y, Yun Y.Y, Sun T.S, Menghe B, Zhang H.P. Isolation and identification of dominant microorganisms involved in naturally fermented goat milk in Haixi region of Qinghai, China. Ann. Microbiol. 2008;58(2):213–217. [Google Scholar]
- 20.Jans C, Bugnard J, Njage P.M.K, Lacroix C, Meile L. Lactic acid bacteria diversity of African raw and fermented camel milk products reveals a highly competitive, potentially health-threatening predominant microflora. LWT Food Sci. Technol. 2012;47(2):371–379. [Google Scholar]
- 21.Mechai A, Debabza M, Kirane D. Screening of technological and probiotic properties of lactic acid bacteria isolated from Algerian traditional fermented milk products. Int. Food Res. J. 2014;21(6):2451–2457. [Google Scholar]
- 22.Abed T.A. Evaluation of methods for the extraction and purification of DNA of cultured Lactobacillus colony isolated from dairy products. Int. J. Appl. Microbiol. 2013;1(2013):20–25. [Google Scholar]
- 23.Yu J, Wang H.M, Zha M.S, Qing Y.T, Bai N, Ren Y, Xi X.X, Liu W.J, Menghe L.G, Zhang H.P. Molecular identification and quantification of lactic acid bacteria in traditional fermented dairy foods of Russia. J. Dairy Sci. 2015;98(8):5143–5154. doi: 10.3168/jds.2015-9460. [DOI] [PubMed] [Google Scholar]
- 24.Yu J, Sun Z, Liu W, Zhang J, Sun T, Bao Q, Zhang H. Rapid identification of lactic acid bacteria isolated from home-made fermented milk in Tibet. J. Gen. Appl. Microbiol. 2009;55(3):181–190. doi: 10.2323/jgam.55.181. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Xi X, Sudu Q, Kwok L, Guo Z, Huo Q, Menhe B, Sun T, Zhang H. High-throughput sequencing reveals microbial community diversity of Tibetan naturally fermented yak milk. Ann. Microbiol. 2015;65(3):1741–1751. [Google Scholar]
- 26.Ren L, Suo H. Molecular identification of lactic acid bacteria isolated from the traditional fermented yak yogurt in Western Sichuan region. Adv. Comput. Sci. Res. 2017;76(2017):1248–1256. [Google Scholar]
- 27.Shangpliang H.N.J, Rai R, Keisam S, Jeyaram K, Tamang J.P. Bacterial community in naturally fermented milk products of Arunachal Pradesh and Sikkim of India analyzed by high-throughput amplicon sequencing. Sci. Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-19524-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehsani A, Hashemi M, Afshari A, Aminzare M. Probiotic white cheese production using coculture with Lactobacillus species isolated from traditional cheeses. Vet. World. 2018;11(5):726–730. doi: 10.14202/vetworld.2018.726-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Vuyst L, Neysens P. The sourdough microflora:Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2005;16(1-3):43–56. [Google Scholar]
- 30.Sharma R, Sanodiya B.S, Thakur G.S, Tjaiswal P, Pal S, Sharma A, Bisen P.S. Characterization of lactic acid bacteria from raw milk samples of cow, goat, sheep, camel and buffalo with special elucidation to lactic acid production. Br. Microbiol. Res. J. 2013;3(34):743–752. [Google Scholar]
- 31.Rizqiati H, Sumantri C, Noor R.R, Damayanthi E, Rianti E.I. Isolation and identification of indigenous lactic acid bacteria from North Sumatra river buffalo milk. Indones. J. Anim. Vet. Sci. 2015;20(2):9–16. [Google Scholar]
- 32.Hutkins R.W. Microbiology and Technology of Fermented Foods. US: IFT Press, Blackwell Publishing; 2006. [Google Scholar]
- 33.Samaržija D, Antunac N, Havranek J.L. Taxonomy, physiology and growth of Lactococcus lactis A review. Mljekarstvo. 2001;51(1):35–48. [Google Scholar]
- 34.Song A.A.L, In L.L.A, Lim S.H.E, Rahim R.A. A review on Lactococcus lactis:From food to factory. Microb. Cell Fact. 2017;16(1):1–15. doi: 10.1186/s12934-017-0669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lien M. Growth of Different Coliform Bacteria in Milk and Their Effect on the Growth and Metabolism of Lactic Acid Bacteria [Thesis] Norway (NO): Agricultural University of Norway; 2002. [Google Scholar]
- 36.Gram L, Ravn L, Rasch M, Bartholin J, Christensen A.B, Givskov M. Food spoilage-interactions between food spoilage bacteria. Int. J. Food Microbiol. 2002;78(1-2):79–97. doi: 10.1016/s0168-1605(02)00233-7. [DOI] [PubMed] [Google Scholar]
- 37.Ivey M, Massel M, Phister T.G. Microbial interactions in food fermentations. Annu. Rev. Food Sci. Technol. 2013;4(1):141–162. doi: 10.1146/annurev-food-022811-101219. [DOI] [PubMed] [Google Scholar]
- 38.Karenzi E, Mashaku A, Nshimiyimana A.M, Munyanganizi B, Thonart P. Kivuguto traditional fermented milk and the dairy industry in Rwanda. A review. Biotechnol. Agron. Soc. Environ. 2013;17(2):383–391. [Google Scholar]
- 39.Van Beek S, Priest F.G. Evolution of the lactic acid bacterial community during malt whisky fermentation:A polyphasic study. Appl. Environ. Microbiol. 2002;68(1):297–305. doi: 10.1128/AEM.68.1.297-305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng J, Ruan L, Sun M, Gänzle M. A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl. Environ. Microbiol. 2015;81(20):7233–7243. doi: 10.1128/AEM.02116-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobson C.M, Deneer H, Lee S, Hemmingsen S, Glaze S, Ziola B. Phylogenetic analysis of the genus Pediococcus including Pediococcus claussenii spp. a novel lactic acid bacterium isolated from beer. Int. J. Syst. Evol. Microbiol. 2002;52(6):2003–2010. doi: 10.1099/00207713-52-6-2003. [DOI] [PubMed] [Google Scholar]


