Abstract
There have been limited options for people who suffer from fibroblast growth factor receptor (FGFR) signaling disorders. In this study, we developed RNA aptamers specific for FGFR3 as potential therapeutic agents. Using a structured aptamer library, we performed ten rounds of SELEX (systematic evolution of ligands by exponential enrichment) against mouse FGFR3c protein. Using an engineered BaF3 cell line, one aptamer clone from round 6 of the selection inhibited FGF-dependent cell growth with a concentration at which 50% of growth is observed (IC50) of ∼260 nM and bound both mouse and human FGFR3 but not FGFR1 or FGFR2. This inhibitor of FGFR3 signaling (iR3), when dimerized using a template-driven approach, resulted in a functional activator of FGFR3 (aR3). We validated the activity and specificity of iR3 and aR3 on engineered BaF3 cell lines, mouse and human FGFR protein, and primary cultures of neuroepithelial precursor cells.
Keywords: aptamer, FGFR3, drug
Introduction
The fibroblast growth factors (FGFs) are a family of 19 protein ligands that have diverse roles during embryonic development and adulthood in mammals.1, 2, 3 Not surprisingly, aberrant FGF signaling is involved in a multitude of disease states, including cancer and developmental disorders.3 Of the five known FGF receptor (FGFR) family members, FGFR1–FGFR4 are classified as type I transmembrane receptor tyrosine kinases.1, 2 Most if not all of the FGFs bind to each FGFR with varying affinities.4 Extracellular binding of two FGF molecules to two receptor monomers with heparin as a cofactor results in stabilization of an FGFR dimer conformation, which activates the intracellular tyrosine kinase domain.5 Initiation of intracellular phosphorylation cascades, particularly through the phosphatidylinositol 3-kinase (PI3K), protein kinase B (AKT), or mitogen associated protein kinase (MAPK) pathways, results in changes in gene transcription and cellular processes.6, 7, 8, 9
FGFR3 has been implicated in a number of different physiological and pathological processes. For example, FGFR3 is a negative regulator of bone growth. Gain-of-function (GOF) mutations of FGFR3 result in bone growth defects such as craniosynostosis, achondroplasia (dwarfism), and thanotophoric dysplasia, a lethal form of dwarfism.10, 11 FGFR3 GOF mutations have also been identified in multiple types of cancer, including bladder, myeloma, lung, cervical, prostate, testicular, and head and neck cancer.12, 13, 14 Loss-of-function (LOF) mutations of FGFR3 have also been documented. For example, lacrimo-auriculo-dento-digital (LADD) syndrome results in a mutation that decreases FGFR3 signaling, leading to pathogenesis.14
To date, there have not been any effective molecular therapies to treat these and other conditions related to dysregulated FGFR3 signaling. The vast majority of molecular therapeutic agents that have been developed for the FGF signaling pathway have been multitarget kinase inhibitors.1, 15 For example, the small molecules TKI258 (dovitinib) and BIBF (the main metabolite of nintedanib, a small molecule tyrosine kinase inhibitor) 1120 inhibit signaling not only through the FGFRs but also platelet-derived growth factor (PDGFR) and the VEGFR25.16, 17 Anti-FGFR antibodies, on the other hand, may provide specificity but have not yet been proven effective in clinical trials.18, 19 For example, preclinical studies of an anti-FGFR1 antibody were halted because its intraperitoneal injection resulted in rodents and monkeys becoming anorexic, possibly because of FGFR inhibition in the hypothalamus.18 The human antibody D11 blocks signaling specifically through FGFR3 but has not yet demonstrated efficacy in clinical trials.19 On the other hand, there have been no reports of an FGFR3 agonist, except for natural ligands, which have promiscuous activity toward FGFRs.20, 21
To develop agonists and antagonists that have high affinity and specificity for FGFR3, we turned to aptamers, RNA ligands that can be selected using systematic evolution of ligands by exponential enrichment (SELEX). Aptamers have been raised against a range of targets and antigens, including cell surface receptors, viral capsids, metals, and even entire cells.22, 23 Compared with their antibody counterparts, aptamers are smaller (250 kDa versus 25 kDa), which enables better tissue penetration.22 Aptamers are also less likely to cause an immunogenic response and are stable at a wide range of pH levels and temperatures.22, 24 Moreover, dimerizing aptamers that act as ligands has been shown to confer agonist activity; e.g., the Ox40, VEGFR2, and CD28 aptamers.25, 26, 27
Here we selected aptamers based on their ability to bind specifically to FGFR3. The selection yielded a number of aptamers that bound FGFR3 with nanomolar affinity; some of these molecules inhibited receptor activity. In addition, we found that dimerization of our best antagonistic molecule, inhibitor of FGFR3 (iR3), converted it into an agonist for FGFR3. We confirmed our findings on recombinant protein, cell lines, and primary cell cultures of neuroepithelial progenitor cells.
Results
Selection of Aptamers for FGFR3
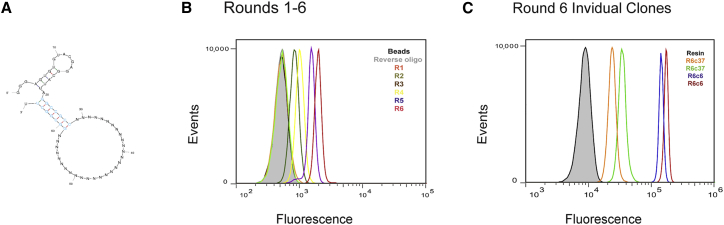
We sought to identify nuclease-resistant aptamers that (1) specifically bound FGFR3 and not FGFR1 or FGFR2 (FGFR4 was not examined because it is not expressed in the nervous system, our primary tissue of interest); (2) inhibited or activated FGFR3; and (3) could be readily minimized after selection. To enable aptamer minimization, we utilized a library designed to include a stem region consisting of 6 G-C pairs and a non-canonical G-U wobble pair (Figure 1A, turquoise). This would, in theory, allow the randomized region consisting of 30 nt to contact the target while excluding the forward and reverse regions, which were used for amplification, from partaking in binding events and, subsequently, enable omission of the constant regions during chemical synthesis. To obtain nuclease-resistant aptamers, a variant of T7 RNA polymerase was employed (Y639F) that incorporated 2′-fluoro cytidine triphosphate (CTP) and 2′-fluoro uridine triphosphate (UTP) during transcription.28, 29
Figure 1.
Selection of Aptamers that Bind FGFR3
(A) Schematized structure of the aptamers in the library, with an engineered stem loop where the stem allows protrusion of the randomized sequence away from the forward and reverse sequences. (B) Aptamers from the first 6 rounds of selection were incubated with a labeled reverse primer and screened for binding against mouse FGFR3 protein by flow cytometry. (C) Aptamer clones from round 6 were annealed to a labeled reverse primer and screened against mouse and human FGFR3c for binding via flow cytometry; R6c6 on mouse FGFR3c, R6c6 on human FGFR3c, R6c37 on mouse FGFR3c, and R6c37 on human FGFR3c. See the details of selection in Materials and Methods and Table S1.
Ten iterative rounds of selection were performed with the structured library, in which the first six rounds were on an Fc fusion of the extracellular domain of FGFR3 and the last four rounds on FGFR3-expressing Neuro-2A cells. To eliminate non-specific binding on protein G resin and on the Fc tag, a negative selection step was incorporated, starting from the second round of selection. To increase the stringency of aptamer binding, the amount of protein was dropped by 90% (5 μg → 0.5 μg) during the third round of selection (see Table S1 for a summary of the selection parameters).
To monitor the progress of the selection, a fluorescently labeled reverse primer was hybridized to the 3′ end of the RNA pools and screened via flow cytometry against FGFR3c protein immobilized on resin. A significant shift in fluorescence between the initial library was observed as early as round 3 and improved steadily through round 6 (Figure 1B). Following the sixth round of selection, we performed an additional 4 rounds of selection using FGFR3-expressing cells (Table S1). As shown in Figure S1A, these subsequent rounds displayed only modest, if any, improvement over the round 6 library.
To identify individual aptamers, we cloned and sequenced the pools from rounds 6 and 10. A total of 66 RNAs were cloned and sequenced, 31 from round 6 and 35 from round 10. Individual aptamers were then screened for binding to the extracellular FGFR3-Fc fusion using a fluorescently labeled reverse primer annealed to the 3′ end of the aptamers, followed by flow cytometry. Thirty-three aptamers generated from rounds 6 and 10 of the selection were tested, and all of these bound not only mouse FGFR3c but also human FGFR3c with varying degrees (see example aptamers in Figure 1C and Figure S1B from rounds 6 and 10, respectively).
Functional Screen of Aptamers Using Engineered BaF3 Cells
To determine whether individual aptamers could inhibit or activate FGFR3, we performed a screen on cells whose proliferation depends on FGFR signaling. Cultured BaF3 cells are ideal for assaying FGFR function because they do not express their own FGFRs and can instead be transduced with constructs that selectively express any FGFR to study their activity.20, 21 Because BaF3 cells transfected with the full-length receptor do not survive and proliferate in response to FGF2 (Figure S1D), BaF3 cells were used that express a chimeric receptor, a fusion between the mouse extracellular domain and transmembrane domain of FGFR3c and the intracellular domain of FGFR1c, which confers more potent kinase activity capable of sustaining cell growth (BaF3-R3c:R1c).21 BaF3-R3c:R1c cell proliferation was stimulated by a range of FGF2 concentrations, with cell growth plateauing at 0.4–0.5 nmol/L at 48 or 72 h for seeding densities of 5,000 and 10,000 cells (Figure S1E).
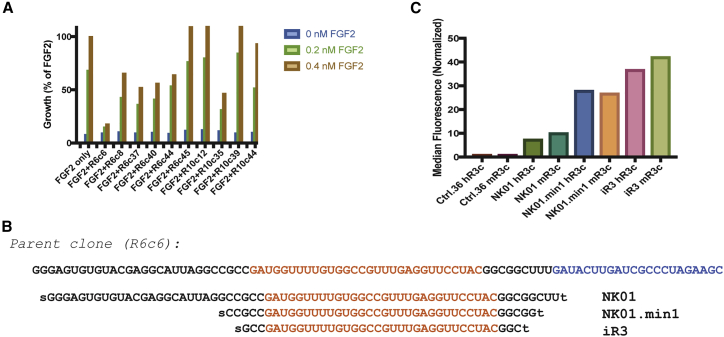
BaF3-R3c:R1c cells were incubated with aptamers at a concentration of 1 μmol/L for 1 h at 37°C prior to challenging the cells with 0.2 nmol/L and 0.4 nmol/L FGF2 for 72 h. An 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was then performed to test for cell viability and density. We observed that one of the aptamer clones from round 6 (round 6, clone 6 [R6c6]) inhibited FGF2-mediated growth of the BaF3-R3c:R1c cells (Figure 2A). Because this molecule robustly inhibited cell growth, we characterized it further.
Figure 2.
Screening Aptamer Clones from Round 6 and Round 10 from Selection and Identifying a Potent Inhibitor, iR3, for FGFR3
(A) Examples of aptamers from round 6 and round 10 of selection competing with 0.2 and 0.4 nM FGF2 on BaF3-R3c:R1c cells. Values are shown as the percentage of growth compared with 0.4 nM FGF2. Round 6 clone 6 (R6c6) robustly inhibited cell growth at a concentration of 1 μmol/L compared with other aptamers from either round 6 or round 10. (B) Full-length parent R6c6 with the reverse primer shown in blue. NK01 is the parent clone without the reverse primer, and the two minimized sequences are NK01.min1 and iR3 with shortened stems. The t denotes an inverted dT placed at the 3′ end during synthesis, whereas s denotes a thiol placed at the 5′ end. (C) Binding of NK01, NK01.min1, and iR3 to mouse and human FGFR3c at a concentration of 100 nmol/L. iR3 retains its ability to bind the target upon minimization from the parent clone.
Characterization of iR3, an Inhibitor of FGFR3 Signaling
To minimize the length of R6c6 to its smallest functional size, we performed a sequence analysis using the mfold application. In our analysis, we excluded the 3′ end of the aptamer where the reverse primer annealed (Figure 2B, blue). The predicted secondary structure of the inhibitor suggested that the molecule adopted two primary folds that (1) did not utilize the primer binding region (Figure S2A, arrowheads) and (2) shared a long stem that could be truncated during synthesis, as initially anticipated during development of the library. We chemically synthesized different variants from the parent molecule R6c6. Of the minimized sequences synthesized, three aptamers, NK01 (NK stands for the initials of the first author), NK01.min1, and iR3, retained binding capacity, as assessed by flow cytometry (Figure 2C). During synthesis, an additional inverted deoxy-thymidine (dT) was added for protection against exonucleases, and all molecules were modified with a 5′ thiol for dye conjugation (denoted by “t” and “s” for the inverted dT and thiol, respectively). NK01 was 67 nt, excluding the inverted t, consisting of the forward primer-binding region and the 30-nt putative target-binding sequence (Figure 2B, red) but excluding the reverse primer sequence. NK01.min1, a 40-mer, and iR3, a 36-mer, were derived from NK01 and lack the forward region but retain a shortened stem consisting of 6 G-C pairs (NK01.min1) or 4 G-C pairs (iR3). The synthesized aptamers were conjugated to DyLight650 and tested for binding to mouse and human FGFR3 protein (Figure 2C). At a concentration of 100 nmol/L, all three constructs bound FGFR3 compared with a non-targeting negative aptamer, ctrl.36. The predicted secondary structures of iR3 illustrate that this smallest variant of R6c6 retains the two primary folds of the parent molecule (Figure S2B).
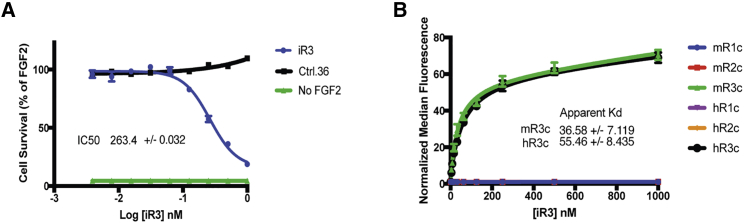
We assessed the inhibitor activity of iR3 as well as the other R6c6 truncates using BaF3-R3c:R1c cells. iR3 inhibited FGF2-induced cell growth (a measure of proliferation and survival) similarly as NK01 and NK01.min1 when titrated in the BaF3-R3c:R1c cell assay (Figure 3A; Figure S2C). Compared with the non-targeting control, ctrl.36, iR3 inhibited growth of BaF3 cells by ∼95% at 1 μmol/L when competing with 0.4 nmol/L FGF2. The calculated a concentration at which 50% of growth is observed (IC50) of iR3 is ∼260 nM (Figure 3A). The apparent Kd of iR3 on both mouse and human FGFR3c protein was approximately 36.5 and 55.5 nM, respectively (Figure 3B). The apparent Kd on BaF3-R3c:R1c cells was similar (data not shown). Importantly, iR3 specifically bound the mouse and human FGFR3c, showing no detectable affinity for mouse or human FGFR1c and FGFR2c at a range of concentrations (Figure 3B). The specificity of iR3 for FGFR3 isoforms was tested on BaF3 cells transfected with a full-length mouse FGFR3b construct. When incubated with a concentration of 200 nmol/L, iR3 also bound to FGFR3b-expressing BaF3 cells, as indicated by a shift on flow cytometry (Figures S3A–S3C). In addition to binding mouse FGFR3b, iR3 also bound human FGFR3b with an apparent Kd of ∼62 and ∼48 nmol/L on mouse and human FGFR3b, respectively (Figure S3D).
Figure 3.
iR3 Inhibits Growth of BaF3-R3c:R1c Cells and Is Specific for FGFR3
(A) iR3 inhibits growth of BaF3-R3c:R1c cells after 72 h with a calculated IC50 of 263.4 ± 0.032 nmol/L. Cells are treated with 0.4 nM FGF2, except for the no-FGF2 control. (B) iR3 binds only mouse or human FGFR3c protein but not mouse or human FGFR1c or FGFR2c. The calculated apparent Kd of iR3 on mouse and human FGFR3c are 36.6 ± 7.1 nmol/L and 55.5 ± 8.4 nmol/L, respectively. Data were acquired from two independent experiments.
To assess whether iR3 competes with FGF2 for binding to FGFR3, beads bearing immobilized FGFR3 were incubated with increasing concentrations of FGF2 (from 0–100 nM) for 30 min, followed by an additional 30-min incubation with DyLight650-labeled iR3 at 20 nM. At higher FGF2 concentrations, iR3 was prevented from binding to both mouse and human FGFR3, as revealed by a stark reduction in bead fluorescence (Figure S4). These findings are consistent with overlapping binding sites for iR3 and FGF2 on FGFR3.
Construction and Testing of Bivalent Aptamers
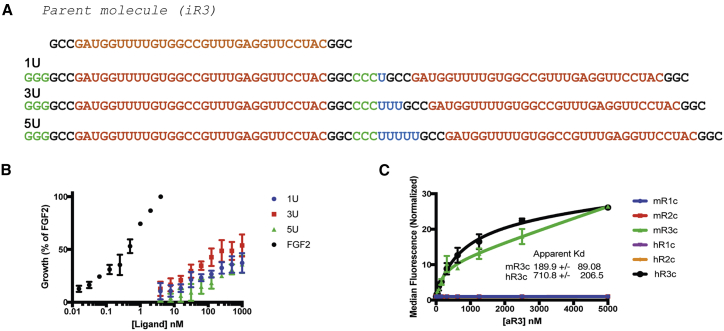
Because FGFs bind as dimers to the receptors,1, 5, 30 we constructed a dimeric version of our aptamers to mimic FGF dimers. Using a template-driven approach, a T7 forward primer was placed in front of iR3 for transcription. Three additional G-C pairs were installed, flanking the first monomer of iR3 to make the template suitable for transcription. The second monomer was linked to the first monomer with a series of 2′ F-uridine linkers (1xU, 3xU, or 5xU), and the 3′ end was extended for the reverse primer region (Figure 4A, only the final transcript product is shown; the reverse primer and T7 promoter regions are omitted; predicted secondary structure in Figure S5B). These constructs were hybridized to a reverse primer labeled with DyLight650 and assayed for binding to FGFR3 at a concentration of 100 nmol/L, which showed variable but significant binding comparable with the parent monomer (Figure S5A).
Figure 4.
aR3 Stimulates Growth of BaF3-R3c:R1c Cells
(A) Bivalent aptamer transcript sequences constructed to test for agonist activity toward BaF3-R3c:R1c cells. The bivalent aptamers were developed from a modified iR3, the parent molecule of the bivalent aptamers. Different uridine (U) linkers are highlighted in blue, the core randomized sequence in red, and the additional G-C pair in the first monomer in green. (B) Stimulation of BaF3-R3c:R1c cells with the different constructs compared with 0.4 nM FGF2. (C) Calculation of the apparent Kd of aR3 on mouse and human FGFR3c yields a value of ∼189.9 ± 89.1 μmol/L and 710.8 ± 206.5 μmol/L, respectively. All data were acquired from two or more independent experiments.
We assayed the iR3 dimers for FGF-like activity on BaF3-R3c:R1c cells. As shown in Figure 4B, addition of the dimers to BaF3-R3c:R1c cells resulted in a modest stimulation of cell growth, indicating that these constructs were functioning as agonists, not antagonists. Although not statistically significantly, a linker length of three 2′ F-uridines resulted in the greatest level of stimulation compared with the single or quintuple uridine linker (Figure 4B; Figure S5C). We call this construct aR3 (activator of FGFR3). aR3 has an apparent Kd of approximately 190 nM on mouse FGFR3c and 710 nM on human FGFR3c.
Assaying iR3 and aR3 on Cells Expressing Physiological Levels of FGFR3
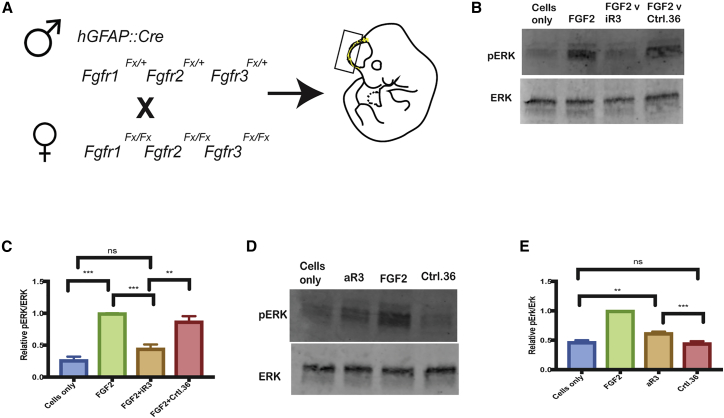
To validate the function of iR3 and aR3 in a more physiologically relevant context that has natural expression levels of FGFR3 and to confirm the specificity of the aptamers toward FGFR3 and not FGFR1 or FGFR2, we used embryonic cortical cells that depend on FGF signaling to activate the extracellular-signal regulated kinase (ERK) pathway in culture and that can be rendered null for one or more of the three expressed FGFR genes: Fgfr1, Fgfr2, and Fgfr3.31, 32 Mice carrying floxed Fgfr1, Fgfr2, and Fgfr3 alleles were crossed to a hGFAP::Cre line that expresses Cre starting at embryonic day 13 throughout dorsal cortical precursor cells,31, 33 yielding embryos with cortical cells that have deleted one or both alleles of Fgfr1, Fgfr2, and Fgfr3 alone or in combination (Figure 5A, boxed region on the embryo).
Figure 5.
Validation of iR3 and aR3 in a Primary Cell Culture Model
(A) Cross used to generate embryos to test aptamers on cells with or without FGFR1, FGFR2, and FGFR3. The dorsal neocortex tissue (boxed region) on embryonic day 14.5 was dissected and dissociated to obtain a single-cell suspension of neocortical precursor cells. (B) Western blot analysis using cells from hGFAP:Cre;Fgfr1Fx/Fx;Fgfr2Fx/Fx;Fgfr3Fx/+ embryos treated with FGF2 alone or FGF2 and iR3 (or the control Ctrl.36 aptamer) at a concentration of 1 μmol/L. (C) Quantitation of western blots. There was a significant reduction in pERK when cortical cells were incubated with iR3 (n = 3 embryos/condition; FG2+Ctrl.36 versus FGF2+iR3, p = 0.0046; FGF2+iR3 versus FGF2, p = 0.0008). (D) Western blot of cells as in (C) but without FGF2. (E) aR3 demonstrated a modest but significant increase in pERK. N = 7 embryos; aR3 versus cells only, p = 0.0027; aR3 versus FGF2, p = 0.001; aR3 versus ctrl.36, p = 0.0007.
For iR3, cortical cells were incubated with the aptamer at a concentration of 1 μmol/L for 1 h before challenge with FGF2 at a concentration of 0.4 nmol/L. The cells were lysed, and a western blot was performed to detect the levels of phospho-ERK (pERK), an intracellular mediator of FGF signaling. When FGFR1 or FGFR2 were left undeleted (i.e., hGFAP::Cre-negative hGFAP::Cre+;Fgfr1Fx/+;Fgfr2Fx/Fx;Fgfr3Fx/+ or Fx or hGFAP::Cre;Fgfr1Fx/Fx;Fgfr2Fx/+;Fgfr3Fx/+ or Fx embryos), pERK levels were not decreased in response to FGF2 (Figures S6A–S6F), indicating that FGF2 stimulation through FGFR1 and FGFR2 was not inhibited by iR3. However, when FGFR1 and FGFR2 were deleted and FGFR3 was left intact (hGFAP::Cre;Fgfr1Fx/Fx;Fgfr2Fx/Fx;Fgfr3Fx/+), a substantial reduction in pERK levels was observed, indicating that iR3 specifically inhibits signaling through FGFR3 (Figures 5B and 5C). When tested on BaF3 cells, iR3’s inhibition of the FGF2-induced increase in pERK levels was not significantly different between 5-, 10-, 20-, and 30-min incubation times (Figure S7). Although pERK was consistently induced by FGF2 and inhibited by iR3, basal levels of pAKT were not induced in BaF3 cells by FGF2 and, therefore, not surprisingly, this basal level was not inhibited by iR3 (Figure S8A).
For aR3, the aptamer was annealed to a labeled reverse primer and incubated for 30 min with the cortical cells in the absence of FGF2. The cells were then lysed, and western blot analysis revealed a significant increase in pERK compared with the controls (Figures 5D and 5E). These results indicate that aR3 can function as an FGFR3 agonist in a physiologically relevant context. Neither iR3 nor aR3 in these assays, performed on BaF3 cells and an astrocyte cell line (mouse cortical astrocytes immortalized with human telomerase reverse transcriptase [hTERT]), respectively, affected the levels of FGFR protein (using an anti-FGFR3 antibody raised against the N terminus of FGFR3 to allow detection of chimeric R3:R1 protein in BaF3 cells; Figure S8B). Note that the levels of FGFR3:R1 protein in BaF3 cells are lower than the levels of FGFR3 in astrocytes, indicating that BaF3 cells express levels of FGFR that are physiological (Figure S9).
Discussion
Here we developed an aptamer, iR3, that behaves as an inhibitor against FGFR3 in its monomeric form and as an agonist when dimerized (aR3). Using a structured library, aptamers could be readily minimized after their selection. Upon minimization, iR3 displayed enhanced binding for FGFR3, as determined by flow cytometry (Figure 2C). To our knowledge, iR3 is the first aptamer that specifically inhibits signaling through FGFR3 and that does not interact with FGFR1 or FGFR2. Although the high specificity of iR3 for FGFR3 suggests that it would not interact with FGFR4, this remains a formal untested possibility. iR3 also binds both mouse and human FGFR3, which demonstrates its potential not only as a tool to detect FGFR3b or FGFR3c but also as a therapeutic for FGFR3-related disorders. Although we did not show binding data for FGFR1b or FGFR2b isoforms, such binding is highly unlikely because iR3 does not bind FGFR1c or FGFR2c, and the IIIb versus the IIIc isoforms share a high degree of homology (>95%).34
Although we performed ten rounds of selection, the last four of which were performed on cell lines that express FGFR3, the molecule we pursued was raised in round 6. Our analysis of the round 10 pool did not yield any strong inhibitors or agonists even though individual aptamer clones from this round displayed binding of FGFR3 protein by flow cytometry (Figure S1B). This was surprising because we retrieved sequences that were taken up by cells following a cell internalization protocol (Materials and Methods). This could be due to several reasons. For instance, the aptamers selected did not bind with sufficient affinity to produce a response by the receptor, or, when they did bind with sufficient affinity, they bound without prohibiting FGF2 from binding to the receptor.
The stimulation of BaF3-R3c:R1c cells with aR3 over a course of 3 days is a result of aR3 interacting with the extracellular FGFR3 domain that triggers signaling through the intracellular FGFR1 domain. Because of the chimeric nature of the receptor, the potency of aR3 to stimulate FGFR3 was not conclusive in this assay. However, by using a primary cell culture model of FGF signaling in which only FGFR1, FGFR2, or FGFR3 were expressed in a physiological context, the efficacy and specificity of both iR3 and aR3 were demonstrated. These results suggest that these molecules function with physiologically relevant levels of natural receptors.
Interestingly, aR3, as a dimeric version of iR3, had a weaker affinity for FGFR3 than iR3 toward both the mouse and human protein (Figure 4C). This was somewhat unexpected because dimerizing often enhances binding through avidity effects.35 However, because this assay was performed using recombinant protein on beads, it may not reflect the true affinity of this dimeric molecule for its homodimeric target on the cell surface.
Other aptamers have also demonstrated efficacy as agonists when presented to their targets as dimers, including the Ox40 receptor aptamer, the VEGFR2 aptamer, AptDivalent, and the CD28 receptor aptamers CD28Apt2 and CD28Apt7. For example, the Ox40 aptamer does not function as an inhibitor as a monomer but stimulates the Ox40 receptor as a dimer when assembled on a DNA scaffold.25 Similarly, the dimeric anti-VEGFR2 aptamer AptDivalent has been shown to stimulate VEGFR2 through the AKT pathway.26 Like Ox40, the CD28 aptamer CD28Apt7 does not have a function when binding to the CD28 receptor as a monomer but does activate the CD28 receptor as a dimer,27 whereas the CD28 aptamer CD28Apt2 functions as an inhibitor as a monomer and functions as an agonist as a dimer, not unlike aR3. aR3, however, is produced from transcription off a DNA template with a triplicate uridine linker, whereas CD28Apt2 and CD28Apt7 have the greatest stimulatory effect as dimers without any linkers.27 This is most likely due to the precise docking of aR3 on FGFR3, which is determined by an exact distance between two FGF monomers that is simulated upon having three uridines between aR3.
To date, there have been limited interventional therapeutic agents targeting FGFRs and FGFR3 specifically. With the aptamers described here, it may be feasible to target FGFR3-related cancers with iR3, including lung and bladder cancers, whereas aR3 can be used as a preventative measure for disorders such as LADD syndrome.14, 19
Materials and Methods
Cell Lines and Conditions for Growth
BaF3 cells, both untransfected and expressing the chimeric FGFR3c extracellular domain and FGFR1c intracellular domain, were a generous gift from Dr. David Ornitz (Washington University, St. Louis, MO, USA). BaF3 cells were grown in RPMI-1940 medium supplemented with 10% newborn bovine calf serum (BCS), 10% WEHI-3 cell conditioned medium, 2% L-glutamate, and 1% penicillin/streptomycin/β-mercaptoethanol (P/S/Bme). NIH 3T3 and Neuro2A cells were grown in DMEM, 10% fetal bovine serum (FBS), and 1% P/S.
Electroporation of FGFR3b in BaF3 Cells
Electroporation of a plasmid carrying the Fgfr3b cDNA in BaF3 cells was performed as described previously.21 Briefly, the MIRB-FGFR3b plasmid (provided by the laboratory of Dr. Ornitz) was linearized with ClaI, and 20 μg was electroporated in approximately 106 untransfected BaF3 cells. The cells were selected in medium containing 600 μg/mL G418 and 10% WEHI-3 conditioned medium for 10 days. Transfection was confirmed with iR3 binding studies because BaF3 cells transfected with full-length FGFR3b are not induced to proliferate in response to FGFs.
Identification of Nuclease-Stabilized Anti-FGFR3 Aptamers
See Table S1 for an outline of the specific conditions for the ten rounds of selection. The library for the selection had the sequence 5′-GGGAGTGTGTACGAGGCATTAGGCCGCC-Nx30-GGCGGCTTTGATACTTGATCGCCCTAGAAGC-3′. The library was designed to display a secondary stem structure that exposed a random region consisting of 30 nt (Nx30) (Figure 1A). During construction, these 30 nt had an equal probability of being an A, T, C, or G. The 3′ end of the library was blocked with the reverse primer binding sequence 5′-GCTTCTAGGGCGATCAAGTATCA-3′. The single-stranded DNA was reverse transcribed with Klenow and then directly transcribed to produce the RNA. The library was transcribed with the transcriptase Y639F enzyme with 2′-fluoro pyrimidines. Ten rounds of selection were completed in total. The first six rounds were performed on the extracellular domain of mouse FGFR3c fused to the Fc domain of human immunoglobulin G1 (IgG1). The final four rounds were performed on Neuro2A cells expressing FGFR3. The first round of selection was performed with 5 μg of FGFR3 protein and 1 nmol of the input RNA library, which consisted of roughly 3 copies of the library of 1015 unique sequences. Prior to incubation with the target protein for round 1 and all subsequent rounds, the library was resuspended in Dulbecco’s phosphate buffered saline (DPBS) with a 1.5× molar excess of reverse primer to block the 3′ end of the library and MgCl2 at a final concentration of 0.1 mM. The pool was then thermally equilibrated for 3 min at 70°C and cooled for 15 min at room temperature. In addition, BSA (1%) was added as a blocking agent. 5 μg FGFR3 protein was added to Dynabeads protein G resin in 30 μL for 30 min, mixing occasionally. The RNA pool was then added to the protein-bead mixture and incubated at 37°C for 30 min. After incubation, the pool was washed three times with Hank’s balanced salt solution (HBSS), and the RNA molecules bound to FGFR3 were eluted with 7 M urea. The pool was then ethanol precipitated and resuspended in 20 μL of water. For the first round, the entire 20 μL was used for reverse transcription, and in subsequent rounds, only half (10 μL) of the round was processed. The reverse-transcribed product was PCR amplified, and transcription was carried out with a T7 RNA polymerase that bears two mutations: P266L and Y639F.
In the second round of selection, a negative round of selection was implemented to eliminate species that bound the linker region of IgG1 of FGFR3 or that bound the protein G resin. In addition, the concentration of the input RNA library was dropped by 90% to 0.1 nmol. After 30 min at 37°C, the negative resin was pulled down, the supernatant was then incubated with the positive resin, and the protocol was carried out as in round 1. To increase the stringency of the selection, in round 3, 10% of the protein (0.5 μg) was used. Rounds 4, 5, and 6 proceeded similarly as round 3.
For the remaining 4 rounds of selection, positive selection continued with cells that expressed FGFR3, Neuro2A, and the negative selection step was with NIH 3T3 cells for rounds 8–10 only. 100,000 cells were seeded overnight at 37°C in a 24-well plate. The next day, the medium was removed and DMEM with 10% salmon sperm DNA (ssDNA) was added at a volume of 270 μL. The round 6 pool was prepared with a 1.5× molar excess of reverse primer and incubated with the cells for 1 h. The medium with the round 6 pool was then removed, and the cells were washed three times with HBSS + 0.1 NaN3. Thereafter, the cells were raised off the dish with 10 mM EDTA, collected with 1 mL HBSS + 0.1% NaN3, placed in a 1.5-mL centrifuge tube, and spun at 300 × g for 5 min. The cells were resuspended in 100 μL of HBSS + 0.1% NaN3 and 5 μL of Riboshredder nuclease cocktail for 15 min at room temperature to eliminate RNA species that had not been internalized. Cells were washed three times with 1 mL HBSS + 0.1% NaN3 and subsequently lysed with Trizol. RNA was retrieved using a phase lock centrifuge tube and ethanol precipitated. The RNA was reverse transcribed, and the selection then proceeded as described. Rounds 8, 9, and 10 were performed identically to round 7, except that a negative selection with NIH 3T3 cells was performed prior to incubation of the library with Neuro2A cells. Briefly, 100,000 NIH 3T3 cells were seeded overnight at 37°C. The next day, the medium was removed, and the RNA pool from the previous round was prepared and incubated with the cells for 30 min. The RNA was then extracted from the cells as described above and incubated with Neuro2A cells.
Chemical Synthesis of RNA Aptamers and Dye Labeling
The minimized aptamers without the reverse primer region were synthesized on an Expedite 8909 DNA synthesizer (Applied Biosystems, Carlsbad, CA, USA). Using standard solid-phase synthesis, aptamers were produced with 2′-fluoro-deoxycytidine and 2′-fluoro-deoxyuridine phosphoramidites on 1,000-Å controlled pore glass (CPG) with an inverted dT. A 5′ thiol modification was added after the initial synthesis using thiol-modified C6 S-S phosphoramidites. The aptamers were then cleaved and deprotected by standard methods using trifluoroacetic acid and a 1:1 of 30% ammonium hydroxide:40% aqueous methylamine. The sequence for the synthesized iR3 is 5′-s GCCGATGGTTTTGTGGCCGTTTGAGGTTCCTACGGCt-3′. The sequence of the control non-targeting aptamer, ctrl.36, is 5′-sGGCGTAGTGATTATGAATCGTGTGCTAATACACGCCt-3′. s represents the thiol at the 5′ end, whereas t represents the inverted thymidine. All synthesized sequences retained the dimethoxytrityl group to purify the sequences after deprotection via reverse-phase high-performance liquid chromatography (HPLC). A 10 × 50-mm Xbridge C18 column was used with a linear gradient of acetonitrile in 0.1 mol/L triethylammonium acetate (TEAA) at pH 7.0.
The aptamers were subsequently labeled with DyLight650 for cytometry. 10 nmol of thiolated aptamer was reduced with tricarboxyethylphosphine at a concentration of 10mmol/L in a total volume of 50 μL of 0.1M TEAA. The reduced aptamer was desalted using a Biospin-6 size exclusion column in 50 μL of PBS. 50 nmol of DyLight650-poly(ethylene glycol) (PEG)-maleimide was resuspended in 5 μL dimethylformamide, which was then reacted with ∼10 nmol of reduced aptamer overnight at 4°C. The labeled aptamer was ethanol precipitated and spun through a Biospin-6 size exclusion column to discard excess dye. The extent of labeling was determined by absorption at 260 nm and 650 nm for the aptamer and dye, respectively, and followed by reverse-phase HPLC to confirm the absence of the dye.
Competition Experiments with iR3 and FGF2
Fc-tagged mouse and human FGFR3 were immobilized on 2.8-μm protein G beads (Dynabeads). After washing to remove unbound protein, the beads were incubated with increasing concentrations of FGF2 from 0–100 nM for 30 min in HBSS containing 1% BSA. DyLight650-labeled iR3 was then added to the wells at a final concentration of 20 nM and allowed to incubate for an additional 30 min. The beads were subsequently washed with HBSS and 1% BSA and analyzed by flow cytometry.
Assembly and Transcription of Bivalent Aptamers
To construct bivalent aptamers, iR3 dimer DNA templates were assembled with a T7 promoter placed upstream of the first monomer with varying lengths of thymidine between the two monomers and a reverse primer region. A triple G-C pair was installed flanking the first monomer. For example, the bivalent aptamer DNA template consisting of one thymidine was 5′-TCTTAATACGACTCACTATAGGGGCCGATGGTTTTGTGGCCGTTTGAGGTTCCTACGGCCCCTGCCGATGGTTTTGTGGCCGTTTGAGGTTCCTACGGCTGATACTTGATCGCCCTAGAAGC-3′. The T spacer is shown in italics. The T7 promoter and the reverse primer region are underlined, the iR3 sequence is in bold, and the single T is the linker. The 3′ end was blocked with the reverse primer 5′-GCTTCTAGGGCGATCAAGTATCA-3′. After transcription, complete synthesis and the lengths of the oligos and hybridization of the reverse primer to the reverse end were verified with non-denaturing PAGE.
Before assays on protein or cells, the bivalent aptamers were mixed with a reverse primer in PBS, heated to 95°C, and equilibrated in a thermal cycler back to room temperature for optimal folding and hybridization.
Cell Growth Assay
For the inhibition assay, 5,000 BaF3 cells were washed in medium (RPMI, 10% newborn bovine calf serum [NBCS], 1% P/S/Bme, 2% L-glutamate [L-glut], and 1 μg/mL heparin) and plated in wash medium in a 96-well plate. The cells were then incubated with the aptamer, which was first heated at 70°C for 3 min and allowed to equilibrate at room temperature for 15 min. When the aptamer required a reverse primer to block the 3′ end, the aptamer was also mixed with a 1.5× molar excess of labeled reverse primer. The cells were then given FGF2 at a final concentration of 0.4 nmol/L. For control stimulation of BaF3 cells, increasing concentrations of 0–4 nM FGF1 or FGF2 were incubated with the cells. 48 or 72 h later, an MTT assay was performed as described elsewhere.36 Briefly, 15 μL of tetrazole was added to each well and placed in the incubator at 37°C for 4 h. 100 μL solubilization mix was subsequently added to the wells and incubated for 5–10 min. An absorbance value at 570 nm was then recorded on a plate reader.
Testing the Binding of Aptamers by Flow Cytometry
Aptamers were initially prepared in DPBS at appropriate concentrations. If aptamers were transcribed with the reverse primer at the 3′ end, then the complementary labeled reverse primer was hybridized at a 1.5× molar excess. If the aptamer was chemically synthesized and minimized, then the aptamer was heated and cooled and plated at appropriate concentrations. The aptamers were prepared either as individual clones or as rounds. Either Dynabeads protein G beads or Ni-NTA beads were washed twice with wash buffer (HBSS and 0.05% Tween 20). 0.240 μg of Fc-tagged protein can theoretically be loaded on 1 μL of protein G resin and 1.65 μg of His-tagged protein on Ni-NTA beads. To load the protein G and Ni-NTA magnetic beads to completion, 1 μg per 1 μL of Fc-tagged protein was incubated with 2 μg per 1 μL of Ni-NTA magnetic beads. The FGFRs used in the study were mouse FGFR1c-Fc, human FGFR1c-His, mouse FGFR2c-Fc, human FGFR2c-His, mouse FGFR3c-Fc, human FGFR3c-Fc, mouse FGFR3b-Fc, and human FGFR3b-Fc. Protein was incubated with the appropriate bead resin for 30 min at room temperature. Human IgG was used as a negative control. The protein-bead mixture was pulled down with a magnet, washed twice in HBSS to remove excess protein, and resuspended in 1.5 mL fluorescence-activated cell sorting (FACS) buffer, which was supplemented with 1 mg/mL ssDNA. The beads were plated in a 96-well plate and incubated with aptamer either at a final concentration of 100 nmol/L or as a dilution series from 1 μmol/L to 0 μmol/L. The samples were then incubated for 1 h at 37°C. Following incubation, the samples were thoroughly washed three times with FACS buffer and run through a SONY cytometer.
Aptamer binding was also tested on cells. For BaF3 suspension cells, parental and engineered cell lines, 100,000 cells were seeded with 1 mg/mL ssDNA. Aptamers were prepared as stated above and incubated with the cells at appropriate concentrations for 1 h at 37°C. Following incubation, cells were rigorously washed three times with PBS and resuspended in FACS buffer. The samples were then run through the cytometer.
Determination of the Apparent Kd of Aptamers
Half serial dilutions of aptamers of interest were prepared at final concentrations of 5 μmol/L to 0 μmol/L (for aR3) or 1 μmol/L to 0 μmol/L in DPBS. The aptamers were incubated for 1 h at 37°C, with receptors immobilized on the appropriate bead resin (protein G or Ni-NTA) in FACS buffer. The resin was pulled down using a magnet, washed three times, and subsequently run through the flow cytometer.
Testing Aptamer Efficacy on Embryonic Cortical Cells
The hGFAPCre, Fgfr1flox, Fgfr2flox, and Fgfr3flox mice have been described previously.10, 33, 37, 38, 39 All experiments described here were approved by and meet the strictest standards of the Institutional Animal Care and Use Committee (IACUC) of the Albert Einstein College of Medicine. The females used in the cross to generate Fgfr mutants were maintained as Fgfr1flox/flox;Fgfr2flox/flox;Fgfr3flox/flox by mating triple homozygous males and females. The stud males used to generate mutant embryos were either hGFAPCre;Fgfr1flox/+;Fgfr2flox/+;Fgfr3flox/+ or hGFAPCre;Fgfr1flox/flox;Fgfr2flox/flox;Fgfr3flox/+. To generate Fgfr mutants, Fgfr1flox/flox;Fgfr2flox/flox;Fgfr3flox/flox females were mated with the stud males overnight. At noon on the day when a plug was observed was considered embryonic day 0.5. On embryonic day 14.5 (E14.5), the mother was euthanized by CO2 inhalation followed by cervical dislocation, and the embryos were harvested. The dorsal cortex was collected from both hemispheres. For each embryo, the cortical tissue was placed individually in 48-well plates in DMEM supplemented with 1% P/S, 2% L-glut, and 5 μg/mL heparin. The tissue was triturated and passed through a 30-μm mesh filter to get rid of clumps. The cells were then equally divided among four 1.5-mL centrifuge tubes in dissection medium. One tube was left untreated (cells only control), and the remaining three tubes received either FGF2 for 5 min, aR3, or a negative control aptamer.
To test for inhibition, the aptamer was prepared at a 1 μmol/L final concentration in PBS. iR3 and the non-targeting aptamer ctrl.36 were incubated for 1 h at 37°C prior to challenge with FGF2. FGF2 was then added to the samples for 5 min at 37°C prior to centrifugation for 5 min at 10,000 × g. The supernatant was evacuated from the tubes, and the cells were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with a cocktail consisting of phosphoprotease and protease inhibitors in 50 μl. The samples were placed at −80°C while genotyping of the individual embryos was completed. 5 μg of protein determined by bicinchoninic acid (BCA) quantitation was run on a 4%–20% gradient protein gel for western blot analysis. Antibodies used were anti-phospho-P42/P44 (pERK1/2) (polyclonal; Cell Signaling Technology, 9001, 1:1,000) and anti-P42/P44 (polyclonal; Cell Signaling Technology, 9002,1:1,000).
Author Contributions
N.K. performed the experiments. N.K., M.L., and J.M.H. designed and interpreted the experiments and wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by NIH MH070596 and MH083804. Astrocyte cells were a kind gift from David Spray.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.06.020.
Contributor Information
Nachiket Kamatkar, Email: nachiket.kamatkar@phd.einstein.yu.edu.
Matthew Levy, Email: matthew.levy@einstein.yu.edu.
Jean M. Hébert, Email: jean.hebert@einstein.yu.edu.
Supplemental Information
References
- 1.Beenken A., Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh N., Ornitz D.M. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J. Biochem. 2011;149:121–130. doi: 10.1093/jb/mvq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hébert J.M. FGFs: Neurodevelopment’s Jack-of-all-Trades - How Do They Do it? Front. Neurosci. 2011;5:133. doi: 10.3389/fnins.2011.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ornitz D.M. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. BioEssays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 5.Plotnikov A.N., Schlessinger J., Hubbard S.R., Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 6.Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008;99:1319–1325. doi: 10.1111/j.1349-7006.2008.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotoh N., Laks S., Nakashima M., Lax I., Schlessinger J. FRS2 family docking proteins with overlapping roles in activation of MAP kinase have distinct spatial-temporal patterns of expression of their transcripts. FEBS Lett. 2004;564:14–18. doi: 10.1016/S0014-5793(04)00287-X. [DOI] [PubMed] [Google Scholar]
- 8.Nandi S., Gutin G., Blackwood C.A., Kamatkar N.G., Lee K.W., Fishell G., Wang F., Goldfarb M., Hébert J.M. FGF-Dependent, Context-Driven Role for FRS Adapters in the Early Telencephalon. J. Neurosci. 2017;37:5690–5698. doi: 10.1523/JNEUROSCI.2931-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goetz R., Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013;14:166–180. doi: 10.1038/nrm3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng C., Wynshaw-Boris A., Zhou F., Kuo A., Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 11.Horton W.A., Hall J.G., Hecht J.T. Achondroplasia. Lancet. 2007;370:162–172. doi: 10.1016/S0140-6736(07)61090-3. [DOI] [PubMed] [Google Scholar]
- 12.Kalff A., Spencer A. The t(4;14) translocation and FGFR3 overexpression in multiple myeloma: prognostic implications and current clinical strategies. Blood Cancer J. 2012;2:e89. doi: 10.1038/bcj.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chae Y.K., Ranganath K., Hammerman P.S., Vaklavas C., Mohindra N., Kalyan A., Matsangou M., Costa R., Carneiro B., Villaflor V.M. Inhibition of the fibroblast growth factor receptor (FGFR) pathway: the current landscape and barriers to clinical application. Oncotarget. 2017;8:16052–16074. doi: 10.18632/oncotarget.14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talebi F., Ghanbari Mardasi F., Mohammadi Asl J., Bavarsad A.H., Tizno S. Identification of a novel missence mutation in FGFR3 gene in an Iranian family with LADD syndrome by Next-Generation Sequencing. Int. J. Pediatr. Otorhinolaryngol. 2017;97:192–196. doi: 10.1016/j.ijporl.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Gavine P.R., Mooney L., Kilgour E., Thomas A.P., Al-Kadhimi K., Beck S., Rooney C., Coleman T., Baker D., Mellor M.J. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 16.Konecny G.E., Finkler N., Garcia A.A., Lorusso D., Lee P.S., Rocconi R.P., Fong P.C., Squires M., Mishra K., Upalawanna A. Second-line dovitinib (TKI258) in patients with FGFR2-mutated or FGFR2-non-mutated advanced or metastatic endometrial cancer: a non-randomised, open-label, two-group, two-stage, phase 2 study. Lancet Oncol. 2015;16:686–694. doi: 10.1016/S1470-2045(15)70159-2. [DOI] [PubMed] [Google Scholar]
- 17.Rolfo C., Raez L.E., Bronte G., Santos E.S., Papadimitriou K., Buffoni L., van Meerbeeck J.P., Russo A. BIBF 1120/ nintedanib : a new triple angiokinase inhibitor-directed therapy in patients with non-small cell lung cancer. Expert Opin. Investig. Drugs. 2013;22:1081–1088. doi: 10.1517/13543784.2013.812630. [DOI] [PubMed] [Google Scholar]
- 18.Sun H.D., Malabunga M., Tonra J.R., DiRenzo R., Carrick F.E., Zheng H., Berthoud H.R., McGuinness O.P., Shen J., Bohlen P. Monoclonal antibody antagonists of hypothalamic FGFR1 cause potent but reversible hypophagia and weight loss in rodents and monkeys. Am. J. Physiol. Endocrinol. Metab. 2007;292:E964–E976. doi: 10.1152/ajpendo.00089.2006. [DOI] [PubMed] [Google Scholar]
- 19.Yin Y., Ren X., Smith C., Guo Q., Malabunga M., Guernah I., Zhang Y., Shen J., Sun H., Chehab N. Inhibition of fibroblast growth factor receptor 3-dependent lung adenocarcinoma with a human monoclonal antibody. Dis. Model. Mech. 2016;9:563–571. doi: 10.1242/dmm.024760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Ibrahimi O.A., Olsen S.K., Umemori H., Mohammadi M., Ornitz D.M. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ornitz D.M., Xu J., Colvin J.S., McEwen D.G., MacArthur C.A., Coulier F., Gao G., Goldfarb M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 22.Maier K.E., Levy M. From selection hits to clinical leads: progress in aptamer discovery. Mol. Ther. Methods Clin. Dev. 2016;5:16014. doi: 10.1038/mtm.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoltenburg R., Reinemann C., Strehlitz B. SELEX--a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Lao Y.H., Phua K.K., Leong K.W. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano. 2015;9:2235–2254. doi: 10.1021/nn507494p. [DOI] [PubMed] [Google Scholar]
- 25.Dollins C.M., Nair S., Boczkowski D., Lee J., Layzer J.M., Gilboa E., Sullenger B.A. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 2008;15:675–682. doi: 10.1016/j.chembiol.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramaswamy V., Monsalve A., Sautina L., Segal M.S., Dobson J., Allen J.B. DNA Aptamer Assembly as a Vascular Endothelial Growth Factor Receptor Agonist. Nucleic Acid Ther. 2015;25:227–234. doi: 10.1089/nat.2014.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastor F., Soldevilla M.M., Villanueva H., Kolonias D., Inoges S., de Cerio A.L., Kandzia R., Klimyuk V., Gleba Y., Gilboa E., Bendandi M. CD28 aptamers as powerful immune response modulators. Mol. Ther. Nucleic Acids. 2013;2:e98. doi: 10.1038/mtna.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sousa R., Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padilla R., Sousa R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 2002;30 doi: 10.1093/nar/gnf138. e138–e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason I. Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nat. Rev. Neurosci. 2007;8:583–596. doi: 10.1038/nrn2189. [DOI] [PubMed] [Google Scholar]
- 31.Kang W., Wong L.C., Shi S.H., Hébert J.M. The transition from radial glial to intermediate progenitor cell is inhibited by FGF signaling during corticogenesis. J. Neurosci. 2009;29:14571–14580. doi: 10.1523/JNEUROSCI.3844-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutin G., Fernandes M., Palazzolo L., Paek H., Yu K., Ornitz D.M., McConnell S.K., Hébert J.M. FGF signalling generates ventral telencephalic cells independently of SHH. Development. 2006;133:2937–2946. doi: 10.1242/dev.02465. [DOI] [PubMed] [Google Scholar]
- 33.Zhuo L., Theis M., Alvarez-maya I., Brenner M., Willecke K. Vol. 94. 2001. pp. 85–94. (hGFAP-cre Transgenic Mice for Manipulation of Glial and Neuronal Function In Vivo). [DOI] [PubMed] [Google Scholar]
- 34.Smith L.B., Bannasch D.L., Young A.E., Grossman D.I., Belanger J.M., Oberbauer A.M. Canine fibroblast growth factor receptor 3 sequence is conserved across dogs of divergent skeletal size. BMC Genet. 2008;9:67. doi: 10.1186/1471-2156-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasegawa H., Taira K.-I., Sode K., Ikebukuro K. Improvement of Aptamer Affinity by Dimerization. Sensors (Basel) 2008;8:1090–1098. doi: 10.3390/s8021090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miagkov A.V., Kovalenko D.V., Brown C.E., Didsbury J.R., Cogswell J.P., Stimpson S.A., Baldwin A.S., Makarov S.S. NF-κB activation provides the potential link between inflammation and hyperplasia in the arthritic joint. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13859–13864. doi: 10.1073/pnas.95.23.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hébert J.M., Lin M., Partanen J., Rossant J., McConnell S.K. FGF signaling through FGFR1 is required for olfactory bulb morphogenesis. Development. 2003;130:1101–1111. doi: 10.1242/dev.00334. [DOI] [PubMed] [Google Scholar]
- 38.Malatesta P., Hack M.A., Hartfuss E., Kettenmann H., Klinkert W., Kirchhoff F., Götz M. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 39.Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E.N., Towler D.A., Ornitz D.M. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.