Abstract
Background:Inthis study, the genetic substructure and morphology of the species Neusticomysmonticolus was evaluated. A nuclear marker and mitochondrial maker were used to examine phylogeographic structure and to estimategenetic distances. Two statistical measurement analyses were applied to morphological data.
Results: These data recovered two morphologically distinct phylogeographic groups corresponding to populations on the eastern and western slopes of the Andes. Further, these eastern and western Andean slope populations of N. monticolus are 8.5 % divergent using sequence data from cytochrome-b (0.8 % divergent in the interphotoreceptor retinoid-binding protein gene).
Conclusions:Populationscurrently assigned to N.monticolus constitutea species complex. The name N.monticolus is here restricted to western Andean slope populations. Populations on the eastern slope of the Andes are assigned to a new species, to which the authors assign the name Neusticomys vossi sp.nov.
Keywords: Andes, Cricetidae; Ecuador; Ichthyomyini; Muroidea; Neusticomys monticolus; Neusticomys vossi sp. nov
BACKGROUND
Sigmodontine rodents constitute a diverse group of New World rodents; extant diversity encompasses ca. 400 species arranged in 86 genera (D’Elía and Pardiñas2015). Traditionally, sigmodontine taxa have been ar- ranged into tribes (Reig 1980; Musser and Carleton 2005; D’Elía et al. 2007), one of which is the tribe Ichthyomyini comprising five genera and 17 species ran- ging from Bolivia, central Brazil, and the Guianas to southern Mexico (Jenkins and Barnett 1997; Musser and Carleton 2005). Ichthyomyines are small to medium- sized semiaquatic animalivorous rodents (Voss 1988), constituting a remarkably distinct group within the sigmodontine radiation. Several morphological features differentiate them from other sigmodontines and as such have been interpreted as supporting the monophyly of the tribe (Voss 1988). Recent phylogenetic analyses, based on nuclear DNA sequences which include repre- sentatives of only two ichthyomyine genera (Rheomys and Neusticomys), have questioned the monophyly of the tribe (Martínez et al. 2012; Parada et al. 2013; Salazar‐Bravo et al. 2013). However, a recent inspection of the sequence of Neusticomys (EU649036) indicates that it may be a chimeric sequence that includes a fragment retrieved from an oryzomyine. This would therefore weaken the argument against a monophyletic Ichthyomyini.
The ichthyomyine genus that deviates the least from the general sigmodontine body plan is Neusticomys(Voss 1988); for example, on its hind feet, it lacks thestiff hairs (presumably an adaptation for swimming) found in other members of the tribe. This genus was nominated by Anthony (1921) to contain the Andean species Neusticomys monticolus. Almost 7 decades later, Voss (1988) subsumed the predominantly lowland Dapt- omys Anthony (1929) under Neusticomys. Recently, Percequillo et al. (2005) described another lowland spe- cies of Neusticomys from central Brazil increasing the known diversity of the genus to six species. Of these, N. monticolus is the most widely distributed species, being endemic to the Andes Mountains of Colombia and Ecuador, at elevations between 1800 and 3750 m (Lee et al. 2006a, 2006b; Musser and Carleton 2005). The lifestyle of thespecies (living near fast-moving streams) has made it difficult to collect. Individuals either are caught in pitfall traps set next to streams, or Sherman traps set in the stream, and they do not seem to be attracted to typical baits, rather those that are caught seem to have been attracted due to curiosity or by insects gathered around typical rodent bait (Personal observation of TEL). These characteristics result in few museum specimens of N. monticolusavailablefor study, with still fewer available tis- sue samples. In 1988, there were ~50 known specimens of N. monticolus (Voss 1988), none of which had associated frozen tissue that we could identify. An additionalsix specimens were collected during field work conducted in Ecuador during 2003, 2005, 2007, and 2008 (Lee et al. 2008; M. Pinto personal communication).
The only taxonomic assessment of N. monticolus todate is that presented by Voss (1988), and no published taxo- nomic oriented study based on genetic data has focused on this species or any other Neusticomys. Here weattempt to characterize the genetic variation of N. monticolus by examining the nucleotide sequence of the mitochondrial cytochrome-b gene (Cytb) and nuclear IRBP gene (Rbp3) of individuals from populations on both sides of the Andes. Given the already noticed morphological differ- ences among the phylogeographic units here uncovered, we describe a new species ofNeusticomys.
METHODS
Sampling
Four field trips were conducted to survey the mammalian fauna of Ecuador. Animals were collected following methods approved by the American Society of Mammalo- gists Animal Care and Use Committee (Sikes 2011). Two specimens of Neusticomys were collected on the western sideof the Andes near the type locality of N.monticolus in 2003 as well as an additional two from a different locality in 2008. One specimen each was collected in 2005 and in 2007 at two different localities in the eastern Andes.These specimens are housed at various natural history collec- tions (Appendix). Further, the tissue was obtained for another member of Ichthyomyini (Rheomys raptor), and sequences were obtained from GenBank for eight additional members of the subfamily Sigmodontinae (one representative of seven tribes within Oryzomalia, one member of Sigmodontini) and four non-sigmodontine members of the family Cricetidae(Appendix).
Morphological analysis
The following measurements (Table 1, defined in Voss 1988) were examined for 41 specimens of Neusticomys (including four specimens not presented in Voss (1988); Appendix): TL—total length, HBL—head body length, LT—length of tail, HF.C—length of hindfoot collected, E.C—length of ear collected, CIL—condylo-incisive length, LD—length of diastema, LM—length of maxillary molars, LIF—length of the incisive foramina, BIT—- breadth of the incisor tips, BIF—breadth of the incisive foramina, BPB—breadth of the palatal bridge, LN—length of nasals, BN—breadth of nasals, LIB—least interorbital breadth, ZB—zygomatic breadth, BB—breadth of brain- case, BZP—breadth of the zygomatic plate, BM1—breadth of M1, HI—height of incisor, DI—depth of incisor, and BOC—breadth of occipital condyle. The combined meas- urement data were first examined with principle compo- nents analysis (PCA) in the R programming package (R Development Core Team 2009) to determine if distinct groupswere identified. Defined groups were then analyzed with linear discriminant analysis (LDA) using the R pro- gramming package (R Development Core Team 2009). LDA was used to graphically explain differences between phylogeographic groups using multiple variables.
Table1 Meanmeasurements and ranges (in mm) for distinct sets of specimens of Neusticomys;specimen QCAZ 7830 is the holotype of N.vossi sp.nov.
| N.monticolus ♀ | N.monticolus ♂ | N.monticolus Antiquia | N.vossi sp.nov. ♀ | N.vossi sp.nov. ♂ | QCAZ7830 | |
| TL | 216.56(196–287) | 226.44(187–313) | 206.8(191–217) | 199.5(188–211) | 205.57(193–221) | 211 |
| HBL | 115.67(95–195) | 122.88(96–211) | 106.8(100–112) | 102.82(97–110.99) | 105.17(101–108) | 110.99 |
| LT | 100.89(90–109) | 103.56(87–114) | 100(91–105) | 101.25(96–108) | 99(92–108) | 108 |
| HFC | 25.56(25–26) | 26.25(24–36) | 24.8(24–26) | 26(25–27) | 25.43(24–27) | 27 |
| EC | 10(6–12) | 10.25(8–14) | 11(11–11) | 10.25(8–13) | 9.86(9–11) | 13 |
| CIL | 24.36(23.1–25) | 24.96(24.2–25.8) | 23.75(22.7–24.9) | 23.7(23.2–25.01) | 24.13(23.4–24.9) | 25.01 |
| LD | 6.01(5.5–6.2) | 6.09(5.4–6.6) | 5.76(5.5–6.1) | 5.66(5.34–6.1) | 6.04(5.7–6.4) | 6.1 |
| LM | 4.16(4–4.91) | 4.07(3.9–4.4) | 4.06(3.9–4.1) | 4.12(4–4.2) | 4.09(4–4.3) | 4.17 |
| LIF | 4.74(4.3–5.9) | 4.69(4.4–5.06) | 4.46(4.2–4.7) | 4.39(4.11–4.7) | 4.86(4.7–5.2) | 4.45 |
| BIT | 1.44(1.3–1.75) | 1.44(1.3–1.6) | 1.42(1.4–1.5) | 1.28(1.1–1.47) | 1.27(1.2–1.4) | 1.47 |
| BIF | 2.11(1.9–2.4) | 2.09(1.9–2.3) | 2.16(2.1–2.2) | 1.99(1.97–2) | 2.15(2–2.3) | 1.99 |
| BPB | 2.93(2.7–3.1) | 2.96(2.7–3.4) | 2.88(2.5–3.3) | 2.87(2.7–3.14) | 2.85(2.6–3.1) | 3.14 |
| LN | 9.65(8.6–11.24) | 9.48(8.5–10.2) | 9.74(9.2–10.7) | 9.1(8.81–9.5) | 9.46(8.9–10.1) | 8.89 |
| BN | 2.89(2.6–3.2) | 2.99(2.7–3.2) | 2.94(2.7–3.1) | 2.37(2.27–2.52) | 2.7(2.5–3) | 2.52 |
| LIB | 4.81(4.6–5) | 4.85(4.6–5.2) | 4.84(4.6–5.1) | 4.59(4.5–4.67) | 4.61(4.4–4.9) | 4.57 |
| ZB | 12.83(12.2–13.3) | 13.16(12.3–13.4) | 12.1(12.1–12.1) | 12.03(11.73–12.49) | 12.54(11.9–13) | 12.49 |
| BB | 12.13(11.8–12.3) | 12.16(11.7–12.5) | 11.38(11.2–11.5) | 11.73(11.52–11.91) | 12.13(11.9–12.5) | 11.91 |
| BZP | 1.31(1–2.06) | 1.14(1–1.3) | 1.02(0.9–1.1) | 1.17(1.1–1.25) | 1.13(1.1–1.3) | 1.25 |
| BM1 | 1.45(1.4–1.57) | 1.44(1.3–1.5) | 1.44(1.3–1.6) | 1.22(1.01–1.4) | 1.38(1.3–1.5) | 1.16 |
| HI | 4.3(2.36–4.9) | 4.68(4.1–5.2) | 4.36(4–4.7) | 3.77(3.03–4.3) | 4.49(4.2–5) | 3.03 |
| DI | 1.33(1–1.5) | 1.42(1.3–1.5) | 1.42(1.4–1.5) | 1.21(1.2–1.23) | 1.3(1.2–1.4) | 1.23 |
| BOC | 7.09(6.7–7.39) | 7.04(6.6–7.5) | 6.75(6.6–7) | 6.65(6.47–6.8) | 6.85(6.8–7) | 6.63 |
Measurementsacronyms and definitions are found in Voss (1988)
Molecular methods
Genomic DNA was isolated from approximately 0.1 g of either the liver or muscle, using a phenol extraction method (Longmire et al. 1997). The polymerase chain reaction (PCR) was used to amplify the complete 1143 bp of Cytb for the seven specimens and up to 1266 bp of Rbp3. Reaction concentrations (25 μl volume) included ≤300 ng genomic DNA, 0.07 mM dNTPs, 2.86mM MgCl, 5 μl10× buffer, 1.25 U Taq (Go Taq, Promega, Madison, Wisconsin), and 0.286 μMof primers L14115 and H15288 (Cytb; Martin et al. 2000)or A1 and B2 then A1-F and E2-B2 (Rbp3; Stanhope et al. 1992; Weksler 2003). Thermal profiles for PCR in- cluded an initial denaturation step at 95 °C (2 min), 30 to 40 cycles with denaturation at 95 °C (45 s), annealing at 48 °C-Cytb or 54 °C-Rbp3 (1 min), extension at 72 °C (1 min 30 s), and a final extension cycle of 72 °C (8 min). Amplicons were purified using the QIAquick PCR purification kit (Qiagen, Inc., Valencia, California) and then sequenced using ABI Prism Big Dye Termin- ator v3.1 ready reaction mix (Applied Biosystems, Foster City, California) and a 3100-Avant automated sequencer (Applied Biosystems, Foster City, California). Cytb primers L14115 and H1528 and the internal primers O400R (Hanson and Bradley 2008), F1 (Whiting et al. 2003), O700H (Hanson and Bradley 2008), and 700 L (Peppers and Bradley 2000) and Rbp3 primers A1 andB2 and the internal primers F and E2, 395R (Hanson et al. 2010), and C and D2 (Jansa and Voss 2000; Weksler 2003) were used for cycle sequencing at 95 °C (30 s) denaturing, 50 °C (20 s) annealing, and 60 °C (4 min) extension. Fol- lowing 30 to 40 cycles, reactions were purified and precipitated in isopropanol. Sequencher 4.1.4 (Gene Codes, Ann Arbor, Michigan) was used to proof se- quences. DNA sequences were deposited in GenBank, and accession numbers are listed in Appendix.
Genetic analyses
ClustalW, MUSCLE, and manual approaches were used in MEGA4 (Tamura et al. 2007) to align nucleotide se- quences and gave identical alignments. As no additional ichthyomyine Cytb or Rbp3 sequence were available in GenBank(the one sequence of Rbp3available for Neustic- omys has been identified as chimeric and was regenerated in this paper), newly obtained sequences of Neusticomys and Rheomys were integrated into a matrix withsequences gathered from eight Sigmodontinae tribes (one represen- tative of seven tribes within Oryzomalia, one member of Sigmodontini) and four non-Sigmodontinae tribes which were used to form the out-group. When available, we in- cluded full-length sequences gathered from a specimen of the type species of the type genus of each tribe. The matri- ces were analyzed using a Bayesian approach (Rannala and Yang 1996) in MrBayes version 3.1.2 (Ronquist and Huelsenbeck 2003). MrModeltest (Nylander 2004) was used to estimate the most appropriate model of sequence evolution (GTR + I + G). Bayesian analysis was performed with sequences partitioned by codon using site-specific gamma distribution allowing for a proportion of invariable sites. Runs, consisting of four Markov chains, were allowed to proceed for ten million generations and were sampled every 1000 generations. The first 1000 trees were discarded as “burnin” based on stabilization of likelihood scores; the remaining trees were used to compute a 50 % majority rule consensus tree and obtain posterior prob- ability (PP) estimates for each clade. Additionally, Cytb pairwise genetic distances were calculated between the re- covered phylogroups using MEGA4 (Tamura et al. 2007) and the Kimura two-parameter model (Kimura 1980).
RESULTS
Morphological data
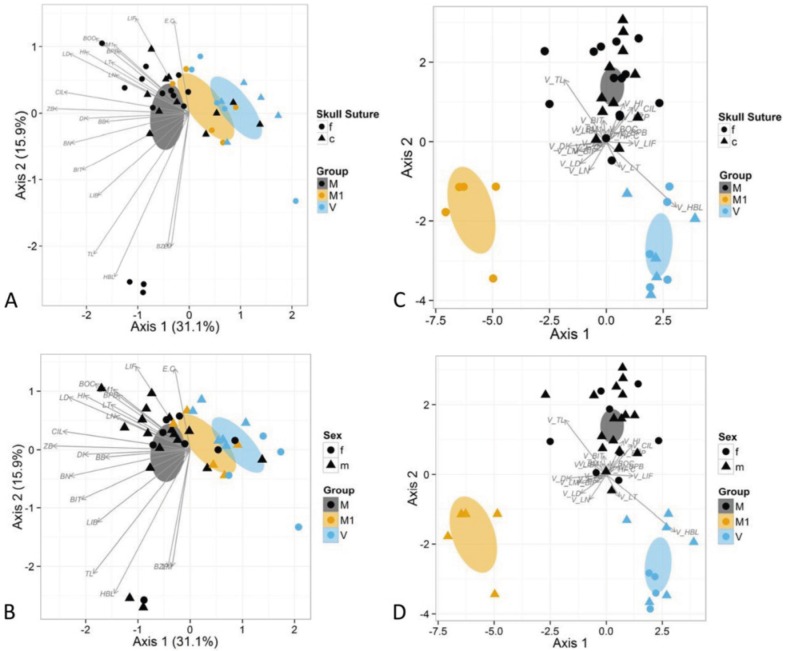
Resultsof the PCA of morphometric data (Fig. 1a, b) are congruent with the genetic data in identifying two puta- tively allopatric groups in Ecuador, one (V) formed by specimens collected on the eastern slopes of the Andes and the other (M) formed by specimens collected on the western slopes of the Andes. In addition, a second west- ern group, distributed north near Antioquia, Colombia, was also recovered (there is currently no genetic data available for this group). The eastern and western groups marginally overlap. This overlap decreased when age groups and sexes were examined separately (Fig. 1a, b, respectively). The two western subgroups overlap more than the eastern and western groups but still are well separated. When the three groups were examined using LDA (Fig. 1c, d), the separation was complete in both age and sex analyses (Fig. 1c, d).
Fig. 1.
Fig. 1 Graphical representation of the PCA analyses (a, b) and LDA analysis (c, d). Vectors labeled as in Table 1. a, c Samples identified by locality and age. Eastern samples (N. vossi sp. nov.: V) are light gray, western samples (N. monticolus: M) are dark gray, specimens from Antioquia (N. monticolus: M1) are mid-gray. Adults (fused craniosutures) are circles, and subadults (closed craniosutures) are triangles. b, d Samples identified by locality and sex. Location symbols same as above, females—circles, males—triangles
Genetic data
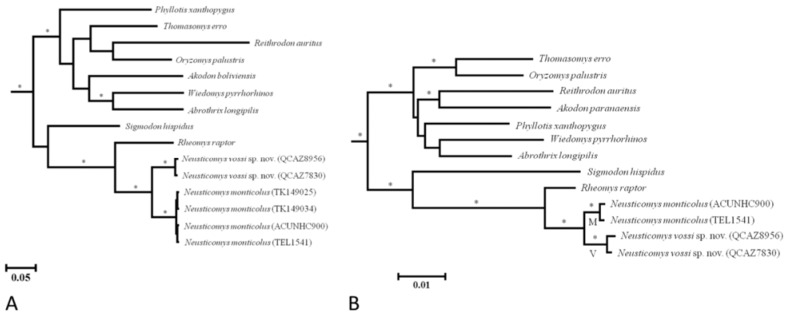
Both phylogenetic analyses of mitochondrial and nuclear DNA sequences (Fig. 2) depict a strongly monophyletic Sigmodontinae (clade I; PP = 1.00). Importantly, the tribe Ichthyomyini also is recovered as monophyletic (clade II; PP = 1.00); Rheomys appears sister to Neusticomys. Within Neusticomys, two distinct groups are indicated, each highly supported (PP = 1.00). The groups appear to be allopatric; clade V is distributed east of the Andes and clade M west of the Andes (Fig. 3). Average genetic distances among the eastern and western groups were 8.5% (Cytb)and 0.8 % (Rbp3);a much higher value than that observed within each group (eastern: 0.4 and 0.0 %; western: 0.1 and 0.1 %, respectively).
Fig. 2.
Fig. 2 Phylogenetic tree of sigmodontine rodents based on Cytb (a) and Rbp3 (b) DNA sequences. Bayesian posterior probability values greater than 0.95 are represented by asterisks placed above the branch
Fig. 3.
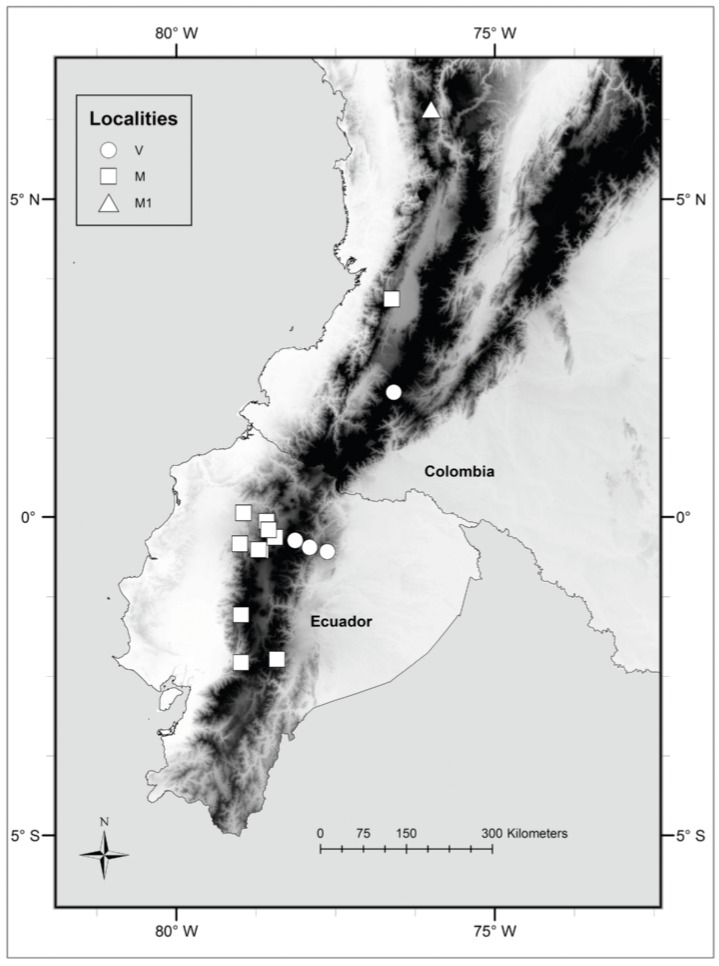
ig. 3 Map of recording localities of specimens of Neusticomys analyzed in the present study. Eastern samples (N. vossi sp. nov.: V) are circles, western samples (N. monticolus: M) are squares, and specimens from Antioquia (N. monticolus: M1) are triangles
Taxonomic implications
Despite the small sample size examined, we consider that when taken together, available data provide suffi- cient evidence to justify recognition of an additional spe- cies of Neusticomys. As no name is available, we name and describe it below.
Neusticomys vossi sp. nov.
Voss’ fish-eating rat urn:lsid:zoobank.org:act:FB716D66- 3D57-40A0-9E48-0E8A5A9B402E
Holotype—QCAZ 7830, an adult lactating female col- lected by T. E. Lee (personal catalog number TEL1846) in August 2005 with a Sherman trap placed in a forested mountain stream near a small (1 m tall) waterfall (Fig. 4). The specimen is preserved as the skin, skull and skel- eton, and frozen tissue and deposited at El Museo de Zoología, Pontificia Universidad Católica del Ecuador, Quito, Ecuador.
Fig. 4.

Fig. 4View of the type locality of Neusticomys vossi sp. nov
Type locality—12 km by road northwest of Cosanga (0° 31′ 70″ S, 77° 52′ 99″ W), Napo Province, Ecuador (1900m).
Diagnosis—As stated by Percequillo et al. (2005), spe- cies of Neusticomys are difficult to diagnose using pre- sumptive autapomorphies; rather, unique combinations of character states are operationally useful for species recognition. Neusticomys vossi sp. nov. is a species of Ichthyomini, that can be distinguished from other ichthyomyine species by its smaller size, reduced hallux, single cusped last molar, large interparietal bone, and an apparent less advanced aquatic specialization, and can be distinguished from other congeners (except N. monti- colus) by having dull grayish rather than brownish pelage and by occipital condyles not projecting posteriorly be- yond rest of occiput. N. vossi sp. nov. can be distin- guished from N. monticolus by its smaller overall sizeand by its narrower incisors, braincase, occipital con- dyles, and rostrum as well as shorter condylo-incisive length, length of diastema, length of maxillary molars, zygomatic breadth, breadth of braincase, and breadth of occipital condyles.
Holotypemeasurements—Headbody length (103 mm), tail length (108 mm), hindfoot (27 mm), ear (13 mm), breadthof nasals (2.52 mm), length of nasals (8.89 mm), depth of incisor (1.23 mm), length of incisors (3.03 mm), breadth of M1 (1.16 mm), and the length of the incisive foramina (4.45 mm).
Description—Dull grayish black pelage (Fig. 5). Ventral pelage color often slightly lighter than dorsal pelage. Mystacial vibrissae and oral margins usually silver. Tail uniformly colored with whitish hairs along ventral sur- face and occasionally a white tip. Some individuals may have white midpectoral blazes and irregular whitish dorsal spotting. Manus and pes covered with whitish silver hairs turning to dark at the wrist, ankles, and metapodials.
Fig. 5.

Fig. 5 Skin of the holotype of Neusticomys vossi sp. nov. (QCAZ 7830)
Rostrum slender with threadlike zygomatic arch (Fig. 6). Infraorbital foramina very large. Palatal foramina very large and medially broad, reaching the anterior lobe of large palate, well surpassing the posterior end of the toothrow. Square mesopterygoid fossa lacking a median palatine process. Braincase broad, flat, and smooth. Cor- onoid process long and narrow. No ridges on frontals or parietals. Small depression at base of nasals, plane of na- sals continuous with frontals. Large interparietal bone. Occipital condyles not projecting posteriorly beyond rest of occiput, and not exposed dorsally. Well-developed gnathic process. Peglike process on maxillary roots of zygomata external to molars. Upper incisors ungrooved and orthodont. Parallel upper molar rows. Molar cusps nearly opposite. High crowned molars with deep flexes almost touching in the middle line of molars. M1 much larger than M2 and M3. M1 with three similar-sized lobes M2 with paracone-protocone pair much wider than the metacone-hypocone pair, M3 reduced.
Fig. 6.
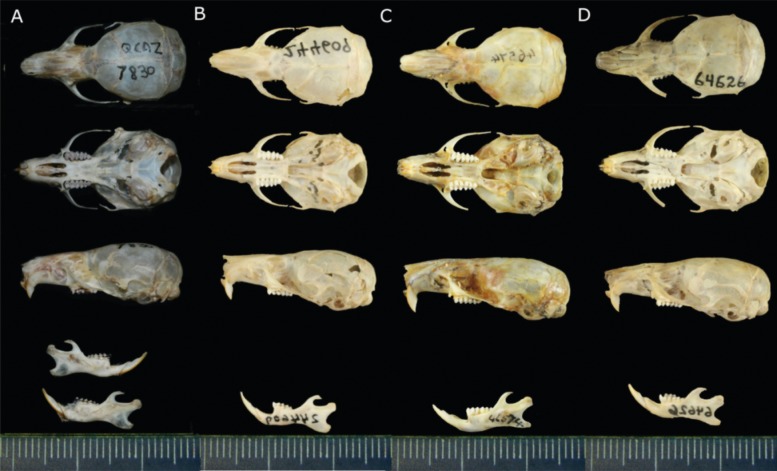
Fig. 6 Skulls and jaw for Neusticomys vossi sp. nov. (a QCAZ 7830 and b AMNH 244609) and N. monticolus (c AMNH 46574 and d AMNH 64626). a, c are the respective type specimens. All skulls are from adult females with closed cranial sutures except for c which is a juvenile male with open sutures
Comparisons—Voss (1988) noted that specimens from Papallacta in the eastern Andes, referred here to N. vossi sp. nov., average slightly smaller than N. monticolus from Guarumal in the western Andes; the same is true for the specimens, including the holotype of N. vossi sp. nov., collected by us. The breadth of the occipital condyles ap- pears nearly diagnostic between the two groups in Voss (1988) study. BOC in N. vossi sp. nov. samples is ≤6.9mm with females being smaller than males. For the most part, N. monticolus shows a BOC of ≥6.9 mm with females being smaller than males.
Similar to Neusticomys venezuelae and Neusticomys mussoi, Voss’ fish-eating rat is differentiated from Neus- ticomys ferreirai, Neusticomys oyapocki, andNeusticomys peruviensis in that the posterior edge of theinferior zygomatic root lies above the anterecone of M1. N. vossi sp. nov. differs from N. oyapocki and N. ferreirai in hav- ing three upper and lower molars.
Distribution—Known from four sites on the eastern slopes of the Andes in northern Ecuador and southern Colombia ranging from 1° 58′ N, 76° 35′ W in the north to 0° 33′ N, 77° 36′ W and from 1900 to 3750 m.
Etymology—Neusticomys vossi sp. nov. is named to honor Dr. Robert S. Voss of the American Museum of Natural History. Rob Voss is the author of a large series of key contributions towards the understanding of South American mammals; in particular, he authored a now classical monographon ichthyomyine rodents. As such, he was the first to recognize the morphological differences between the new species described here and N.monticolus. Aspart of an ichthyomyine review, Voss (1988) wrote the following as a commenton N. monticolus, “The seriesfrom Guarumal to- gether with those from nearby Las Machinas and from the Rio Pita closely resemble the Volcan Pichincha specimens, but the Papallacta series ex- hibits some metric differences.”
Natural history—One N. vossi sp. nov. was recorded at Papallacta with two large embryos on the 15th of May (Voss 1988). The type collected in August is a lactating female. The type was taken by a small waterfall about 1 m tall, with traps exposed to the spray of water from the fall, which is congruent with the description by Tate (1931) of Neusticomys habitat. The stream was rocky and about 1 m across with fast rapids (Fig. 4). This specimen represents a low eleva- tion record in Ecuador at 1900 m for the eastern Andes (Lee et al. 2006a). Oreoryzomys balneator and Thomasomys erro were collected in the same trap line or in nearby forests, as the type specimen (Lee et al. 2006a). Vegetation along the stream consisted of plants with large waxy leaves. Plants of the families Araceae, Arecaceae, Cecropiaceae, Chloranthaceae, Cyatheaceae, Cyclanthaceae, Flacourtiaceae, Lauraceae, Lobeliaceae, Melastomataceae, Meliaceae, Moraceae, Piperaceae, and Poaceae were found along the stream bank (Lee et al. 2006a).
DISCUSSION
Samples from eastern and western localities were se- quenced to examine phylogeographic structure of N. monticolus. Two highly differentiated and potentially allopatric groups were recovered; these groups are con- gruent with the morphological forms previously identi- fied by Voss (1988). Externally, the two groups do not appear to present differences besides pelage color in a few individuals but show differences in certain cranial characteristics as well as an overall smaller size for the eastern animals (Voss 1988). However, Voss (1988) con- sidered these differences insufficient to consider both forms as distinct species.
Genetic and genealogical results combined with mor- phological differences presented herein suggest that pop- ulations currently allocated to N. monticolus east and west of the Andes represent two distinct biological en- tities. Mean Cytb genetic distance between the two Neusticomys haplogroups (8.5 %) is in the order of mean genetic distances shown between species in other groups of sigmodontine rodents (e.g., Abrothrix ca. 5 to 10 % [Feijoo et al. 2010; D’Elía et al. in press]; species of the Akodon boliviensis species group: 2.8–7.7 % [Jayat et al. 2010]; Eligmodontia: 4.6–11.4 % [Mares et al. 2008]; Juli- omys:ca. 12 % [Pardiñas et al. 2008]; Melanomys:4.5–7.6 % [Hanson and Bradley 2008]; Nectomys: 7.36 % [Hanson and Bradley 2008]; Oligoryzomys: 4.45–15 % [Hanson et al. 2011; Palma et al. 2010; Richter et al. 2010; Rogers et al. 2009]; Oryzomys: 4.5–12.1 % [Hanson et al. 2010]; Oxymycterus: 2.5–9.6 % [Jayat et al. 2008]); Rhipidomys: 4.1–12.4 % [Costa et al. 2011]; Scapteromys: ca. 4.5 % [D'Elía and Pardiñas 2004]; Sigmodon: 8.5–20.8 % [Henson and Bradley 2009]). Although the pecies used for comparison are not the closest possible relatives to Neusticomys, all belong to other sigmodon- tine tribes and provide a diverse array of reference points. Further, the Rbp3 genetic distance between the two Neusticomys groups (0.8 %) is significant when com- pared to the 1.5 % difference between them and Rhe- omys (the only other Ichthyomini examined genetically). For a frame of reference, the Rbp3 genetic distance be- tween Neusticomys and other members of the Sigmo- dontinae is only 6–8 % (compared to 15–20 % in Cytb). Eastern and western Andean clades of Neusticomys pre- viously assigned to N. monticolus showed enough gen- etic differentiation—despite the low sample size—to warrant further examination using other source of evi- dence (e.g., karyological, morphological, ecological stud- ies; see Baker and Bradley 2006; Bradley and Baker 2001). In this particular situation, the additional investi- gation was mostly conducted some 21 years (Voss 1988) prior to the discovery of the highly differentiated hap- logroups, as well as complemented here.
In addition to the new species described above, the morphometric data further support a unique group near Antiquia, Colombia, which Voss (2015) identified in his account of N. monticolus saying, “The few available spec- imens from the western Andes (Cordillera Occidental) of Colombia exhibit morphological differences from Ecua- dorean material and may represent a distinct species.” We identify this subgroup as one requiring further molecular examination.
CONCLUSIONS
Ichthyomyini is the least studied tribe of sigmodontine rodents; studies on these mice are rare in the literature, limiting in the current century to the reporting of new geographic records (e.g., Leite et al. 2007; Miranda et al. 2012; Nunes 2002; Pacheco and Ugarte-Núñez 2011), the presentation of new data in reports of mammalian surveys (e.g., Lee et al. 2006a, 2006b, 2008; Voss et al. 2001), synthesis of available knowledge for one species (Packer and Lee 2007), and the description of a new spe- cies (e.g., Percequillo et al. 2005). As such, aside from Voss (1988) study, no comprehensive phylogenetic hy- pothesis is available for the tribe. Therefore, currently, it is not feasible to advance either a robust biogeographic hypothesis accounting for ichthyomyine diversification or one centered on Neusticomys. Similarly, given the scarcity of ichthyomyines in natural history collections and the difficulty of collecting them in the field (but see Pacheco and Ugarte-Núñez 2011), the task of perform- ing an exhaustive phylogeographic study of any ichthyo- myine species may be difficult to accomplish.
The present study is the first study of ichthyomyine alphataxonomy since Percequillo et al. (2005) described N. ferrerai and the first taxonomic study focused on members of the tribe Ichthyomyini using sequence data. Given the results of our morphological assessment (and that of Voss 1988), it would be informative to perform an analysis of DNA sequences from specimens collected near Antioquia, Colombia.
Resumen
En este estudio se evaluó la estructura genética y morfo- lógica de la variación de la especie Neusticomys montico- lus. Distancias genéticas y estructura filogeográfica fueron estimadas en base a secuencias de un gen nucleary otro mitocondrial. Los datos morfológicos fueron ana- lizados estadísticamente. Los datos indican la existencia de dos grupos filogeográficos que difieren morfológica- mente y corresponden a poblaciones de las vertientes este y oeste de los Andes. Estos grupos difieren en pro- medio en 8.5 % en el gen del citocromo b (0.8 % en el gen de la proteína de unión interfotorreceptor retinoide). Las poblaciones actualmente asignadas a N. monticolus constituyen un complejo de especies. El nombre N. mon- ticolus es aquí restringido a las poblaciones de la ver- tiente occidental de los Andes. Las poblaciones de las estribaciones orientales de los Andes son asignadas a una nueva especies, a la cual es dado el nombre Neustic- omys vossi sp.nov.
Acknowledgements
An Abilene Christian University Math/Science Research Council grant funded the trip to Cosanga Ecuador. M. Pinto, D. Wilson, and K. Helgen graciously loaned tissue from their field work. M. Pinto and A. Camacho provided pictures of the type and reference individuals. M. Mauldin assisted with the map edits. R. Voss allowed access to the original data sheets for his 1988 manuscript. Abilene Christian Natural History Collections, El Museo de Zoología, Pontificia Universidad Católica del Ecuador, The Museum of Texas Tech Natural Science Research Laboratory, and the University of Kansas Museum of Natural History for facilitating the tissue loans. Ecuadorian specimens were collected under the following research permits by the Ecuadorian Ministry of Environment: 012-IC-FAU-DNBAPVS/N (July 15th, 2005); 020-IC-FAU-DNBAPVS/MA (July 20th, 2006); 002-IC-FAU/FLO-DRFN-P/ MA (July 30th, 2007); and 02–12 IC-FAU-FLO-DPAC/MA (May 18th, 2012).
Appendix
Specimens examined—Specimens are arranged by spe- cies and locality. Specific identification numbers (mu- seum or collector numbers) and GenBank accession numbers (Cytb/Rbp3; for specimens included in molecular analysis) are listed in parentheses, respectively. Museum acronyms are as follows: Abilene Christian University Natural History Collections (ACUNHC); El Museo de Zoología, Pontificia Universidad Católica del Ecuador (QCAZ);Departamento de Microbiología, Universidad del Valle (HTC); Field Museum of Natural History (FMNH); Museum of Southwestern Biology (MSB); Museum of Vertebrate Zoology (MVZ); The Museum of Texas Tech University (TTU, TK—tissue collection); Thomas E. Lee (TEL; vouchers available at ACUNHC or QCAZ); University of Kansas Museum of Natural History (KU); and University of Michigan Museum of Zoology (UMMZ)
Akodon boliviensis—BOLIVIA: Tarija; 4.5 km E of Iscayachi (MSB68571, Cytb [GenBank:KC841367]).
Akodon paranaensis—PARAGUAY (TTU108159, Rbp3 [EU649035]).
Abrothrix longipilis—ARGENTINA: Río Negro, La Veranda (MVZ1554494, Rbp3 [AY163557]). CHILE: Araucania; Fundo Hermanos Garcia (MSB205660, Cytb [GenBank:GU564083]).
Arvicola terrestri—SWITZERLAND (MVZ155884, [Gen- Bank:AY275106/AY277407]).
Cricetus cricetus—AUSTRIA: Niederosterreich; 1 km NE Gutramsdorf (MVZ155880, [GenBank:AY275109/ AY277410]).
Neusticomys monticolus—COLOMBIA: Antioquia, Santa Bárbara (FMNH71218; FMNH71220; FMNH71221; FMNH71222; FMNH71223). COLOMBIA: Valle, Pichindé (HTC1372; HTC2180; HTC2237; HTC3365). ECUADOR: Chimborazo, Pauchi (AMNH66848). ECUADOR: Coto- paxi, Reserva Otonga (TK149025, [GenBank:KF359517]; TK149034, [GenBank:KF359518]). ECUADOR: Pichincha, Guarumal (UMMZ126298; UMMZ126299; UMMZ126298; UMMZ126299; UMMZ155789; UMMZ155790; UMMZ 155791; UMMZ155793; UMMZ155794). ECUADOR: Pichincha, Las Machinas (AMNH64634; AMNH64636; AMNH64637;AMNH64638; AMNH64639). ECUADOR: Pinchincha, Rio Pita (AMNH64627; AMNH64628; AMNH64629;AMNH64631; AMNH64632). ECUADOR: Pinchincha, San Ignacio (AMNH64625; AMNH64626). ECUADOR: Pichincha, Tandayapa Valley (QCAZ6446, [GenBank:KF359515/KR105606]; ACUNHC900/QCAZ 6531, [GenBank:KF359516/KR105605]).
Neusticomys vossi sp. nov.—COLOMBIA: Huila, San Antonio (FMNH71224; FMNH71225). ECUADOR: Bolivar, Sinche (AMNH62920). ECUADOR: Napo; 11 km SE of Baeza (QCAZ7830, [GenBank:KF359513/ KR105608]). ECUADOR:Napo, Papallacta (AMNH244608; AMNH244609; UMMZ126297; UMMZ155604; UMMZ 155605; UMMZ155606). ECUADOR: Napo; Volcan Sumaco (QCAZ8956, [GenBank:KF359514/KR105607]).
Oryzomys palustris—USA: Texas; Galveston, Virginia Point (TTU82920, [GenBank:DQ185382/EU273431]).
Phyllotis xanthopygus—ARGENTINA: Río Negro, Pilcanieyeu (MVZ182703, Rbp3 [GenBank:AY163632]). CHILE: Tarapaca; Parinacota; Arica Cytb (FMNH133830, [GenBank:U86831]).
Reithrodon auritus—ARGENTINA: Rio Negro, Las Victorias (MVZ182704, [GenBank:EU579474/AY163634]).
Rheomys raptor—COSTA RICA: Puntarenas; Monteverde Cloud Forest Reserve, Quebrada Cuecha (KU159017, [GenBank:KF359512/AY163635]).
Sigmodon hispidus—USA: Texas; Cameron, Brownsville (TK32481, Cytb [GenBank:AF425199]). USA: Kansas; Ellis, Hays (OK5840, Rbp3 [EU635707]).
Thomasomys erro—ECUADOR: Napo; 12 km N Cosanga (ACUNHC1137/QCAZ7649, [GenBank:EU57 9476/EU649057]).
Wiedomys pyrrhorhinos—BRAZIL: Minas Gerais; Ponte do Colatino (MVZ197566, Cytb [GenBank:EU579477). BRAZIL: Bahia, Jaborandi (CRB1839, Rbp3 [AY163644]).
Footnotes
Authors’ contributions: JDH supervised the molecular work, drafted the manuscript, and processed the measurements. GD helped draft the manuscript, and proofed and modified the species description. SBA performed the molecular work and helped drafting the manuscript. SBC performed the statistical analyses. TEL and SB collected the specimens and measurements and provided guidance on the natural history of the genus. All authors read and approved the final version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
References
- Anthony H E. Preliminary report on Ecuadorean mammals. 1:1–6. [Google Scholar]
- Anthony H E. Two genera of rodent from South America. Am Mus Novit. 383:1–6. [Google Scholar]
- Baker R J, Bradley R D. Speciation in mammals and the genetic species concept. J Mammal. 87:643–662. doi: 10.1644/06-MAMM-F-038R2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley R D, Baker R J. A test of the genetic species concept: cytochrome-b sequences and mammals. J Mammal. 82:960–973. doi: 10.1644/06-MAMM-F-038R2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Bma, Geise L, Pereira L G, Costa L P. Phylogeography of Rhipidomys (Rodentia: Cricetidae: Sigmodontinae) and description of two new species from southeastern Brazil. J Mamm. 249:945–962. [Google Scholar]
- 'elía D, Pardiñas G. Subfamily Sigmodontinae Wagner, 1843. Rodents. The University of Chicago Press. 2:63–70. [Google Scholar]
- 'elía D, Pardiñas G, Teta Ufj, Patton P. Definition and diagnosis of a new tribe of sigmodontine rodents (Cricetidae: Sigmodontinae), and a revised classification of the subfamily. Gayana. 71:187–194. [Google Scholar]
- 'elía D, Teta G, Upham P, Pardiñas N S, Patterson Ufj. Description of a new soft-haired mouse, genus Abrothrix (Sigmodontinae), from the temperate Valdivian rainforest. J Mamm; [Google Scholar]
- 'elía D, Pardiñas G. Systematics of argentinean, Paraguayan, and uruguayan swamp rats of the genus Scapteromys (Rodentia, Cricetidae, Sigmodontinae) J Mammal. 85:897–910. [Google Scholar]
- R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria): 2009. [Google Scholar]
- Feijoo M, 'elía D, Pardiñas G, Lessa Ufj. Systematics of the southern Patgonian-Gueguian endemic Abrothrix lanosus (Rodentia: Soigmodontinae): phylogenetic position, karyotypic and morphological data. Mamm Biol. 75:122–137. [Google Scholar]
- Hanson J D, Bradley R D. Molecular diversity within Melanomys caliginosus (Rodentia: Oryzomyini): evidence for multiple species. Occas Papers Mus Texas Tech Univ. 275:1–11. [PMC free article] [PubMed] [Google Scholar]
- Hanson J D, Indorf J L, Swier V J, Bradley R D. Molecular diversity within the Oryzomys palustris complex: evidence for multiple species. J Mammal. 91:336–347. [Google Scholar]
- Hanson J D, Utrera A, Fulhorst C F. The delicate pygmy rice rat (Oligoryzomys delicatus) is the principal host of Maporal virus (family Bunyaviridae, genus Hantavirus) Vector Borne Zoonotic Dis. 11:691–696. doi: 10.1089/vbz.2010.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson D D, Bradley R D. Molecular systematics of the genus Sigmodon: results from mitochondrial and nuclear gene sequences. Can J Zool. 87:211–220. doi: 10.1139/z09-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansa S A, Voss R S. Phylogenetic studies on didelphid marsupials I. Introduction and preliminary results from nuclear IRBP gene sequences. J Mamm Evol. 7:43–77. [Google Scholar]
- Jayat J P, 'elía D, Pardiñas G, Miotti Ufj, Ortiz M D. A new species of the genus Oxymycterus (Mammalia: Rodentia: Cricetidae) from the vanishing Yungas of Argentina. Zootaxa. 1911:31–51. [Google Scholar]
- Jayat J P, Ortiz P E, Pardiñas Ufj, Salazar-Bravo J, 'elía D. The Akodon boliviensis species group (Rodentia: Cricetidate: Sigmodontinae) in Argentina: species limits and distribution, with the description of a new entity. Zootaxa. 2409:1–61. [Google Scholar]
- Jenkins P D, Barnett A A. A new species of water mouse, of the genus Chibchanomys (Rodentia, Muridae, Sigmodontinae) from Ecuador. Bull Nat His Mus Lond (Zool) 63:123–128. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:11–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Lee T E, Alvarado-Serrano D, Platt R N, Goodwiler G G. Report on a mammal survey of the Cosanga River drainage. Occas Papers Mus Texas Tech Univ. 260:1–10. [Google Scholar]
- Lee T E, Packer J B, Alvarado-Serrano D. Results of a mammal survey of the Tandayapa Valley. Occas Papers Mus Texas Tech Univ. 290:1–14. [Google Scholar]
- Lee T E, Burneo S F, Marchán M R, Roussos S A, Vizcarra-Váscomez Sebastian. The mammals of the temperate forests of Volcán Sumaco. Occas Papers Mus Texas Tech Univ. 276:1–10. [Google Scholar]
- Leite R N, Silva Da, Gardner Mnf. New records of Neusticomys oyapocki (Rodentia, Sigmodontinae) from a human-dominated forest landscape in northeastern Brazilian Amazonia. Mastozool Neotrop. 14:257–261. [Google Scholar]
- Longmire J L, Maltbie M, Baker R J. Use of lysis buffer in DNA isolation and its implications for museum collections. Occas Papers Mus Texas Tech Univ. 163:1–4. [Google Scholar]
- Mares M A, Braun J K, Coyner B S, Van Den Bussche R A. Phylogenetic and biogeographic relationships of gerbil mice Eligmodontia (Rodentia, Cricetidae) in South America, with a description of a new species. Zootaxa. 1753:1–33. [Google Scholar]
- Martin Y, Gerlach G, Schlotter C, Meyer A. Molecular phylogeny of European muroid rodents based on complete cytochrome-b sequences. Mol Phylogenet Evol. 16:37–47. doi: 10.1006/mpev.1999.0760. [DOI] [PubMed] [Google Scholar]
- Martínez J J, Ferro L I, Mollerach M I, Barquez R M. The phylogenetic relationships of the Andean swamp rat genus Neotomys (Rodentia, Cricetidae, Sigmodontinae) based on mitochondrial and nuclear markers. Acta Theriol. 57:277–287. [Google Scholar]
- Miranda C L, Rogério R V, Semedo Tbf, Flores T A. New records and geographic distribution extension of Neusticomys ferreirai and N. oyapocki (Rodentia, Sigmodontinae) Mammalia. 76:335–339. [Google Scholar]
- Musser G G, Carleton M D. Family Cricetidae. Johns Hopkins University Press. 2:894–1522. [Google Scholar]
- Nunes A. First record of Neusticomys oyapocki (Muridae, Sigmodontinae) from the Brazilian Amazon. Mammalia. 66:445–447. [Google Scholar]
- Nylander Jaa. Mr.Modeltestv2. Program distributed by the author. Evolutionary Biology Centre; 2004. [Google Scholar]
- Pacheco V, Ugarte-Núñez J. New records of Stolzmann's fish-eating rat Ichthyomys stolzmanni (Cricetidae, Sigmodontinae) in Peru: a rare species becoming a nuisance. Mamm Biol. 76:657–661. [Google Scholar]
- Packer J B, Lee T E. Neusticomys monticola. Mammal Species. 805:1–3. [Google Scholar]
- Palma R E, Rodríguez-Serrano E, Rivera-Milla E, Hernandez C E, Salazar-Bravo J, Carma M I, Belmar-Lucero S, Gutierrez-Tapia P, Zeballos H, Yates T L. Phylogenetic relationships of the pygmy rice rats of the genus Oligoryzomys Bangs, 1900 (Rodentia: Sigmodontinae) Zool J Linn Soc. 160:551–566. [Google Scholar]
- Parada A, Pardiñas U F, Salazar-Bravo J, 'elía D, Palma G. Dating an impressive Neotropical radiation: molecular time estimates for the Sigmodontinae (Rodentia) provide insights into its historical biogeography. Mol Phyl Evol. 66:960–968. doi: 10.1016/j.ympev.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Pardiñas U F, Teta P, 'elia D, Galliari G. Rediscovery of Juliomys pictipes (Rodentia: Cricetidae) in Argentina: emended diagnosis, geographic distribution, and insights on genetic structure. Zootaxa. 1758:29–44. [Google Scholar]
- Peppers L L, Bradley R D. Molecular systematics of the genus Sigmodon. J Mammal. 81:332–343. [Google Scholar]
- Percequillo A R, Carmignotto A P, Silva M J. A new species of Neusticomys (Ichthyomyini, Sigmodontinae) from central Brazilian Amazonia. J Mammal. 86:873–880. [Google Scholar]
- Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol. 43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- Reig O A. A new fossil genus of South American cricetid rodents allied to Wiedomys, with an assessment of the Sigmodontinae. J Zool. 192:257–281. [Google Scholar]
- Richter M H, Hanson J D, Cajimat M N, Milazzo M L, Fulhorst C F. Geographical range of Rio Mamore virus (family Bunyaviridae, genus Hantavirus) in association with the small-eared pygmy rice rat (Oligoryzomys microtis) Vector Borne Zoonotic Dis. 10:613–620. doi: 10.1089/vbz.2009.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D S, Arenas E A, González-Cózatl F X, Hardy D K, Hanson J D, Lewis-Rogers N. Molecular phylogenetics of Oligoryzomys fulvescens based on cytochrome b gene sequences, with comments on the evolution of the genus Oligoryzomys. Unam): 2009. [Google Scholar]
- Ronquist F, Huelsenbeck J P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Salazar-Bravo J, Pardiñas U F, 'elía D. A phylogenetic appraisal of Sigmodontinae (Rodentia, Cricetidae) with emphasis on phyllotine genera: systematics and biogeography. Zool Scripta. 42:250–261. [Google Scholar]
- Sikes R S, Gannon W L. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. American Society of Mammalogists. 92:235–253. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhope M J, Czelusniak J, Si J S, Nickerson J, Goodman M. A molecular perspective on Mammalian evolution from the gene encoding interphotoreceptor retinoid binding protein with convincing evidence for bat monophyly. Mol Phylogenet Evol. 1:148–160. doi: 10.1016/1055-7903(92)90026-d. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tate Ghh. Random observations on habits of South American mammals. J Mammal. 12:248–256. [Google Scholar]
- Voss R S. Systematics and ecology of ichthyomyine rodents (Muroidea): patterns of morphological evolution in a small adaptive radiation. Bull Am Mus Nat His. 188:259–493. [Google Scholar]
- Voss R S. Tribe Ichthyomyini Vorontsov. Rodents. The University of Chicago Press. 2:279–291. [Google Scholar]
- Voss R S, Lunde D P, Simmons N B. The mammals of Paracou, French Guiana: a neotropical lowland rainforest fauna part 2. Nonvolant species. Bull. Am. Mus. Nat. His. pp. 3–236.
- Weksler M. Phylogeny of Neotropical Oryzomyine rodents (Muridae: Sigmodontinae) based on the nuclear IRBP exon. Mol Phylogenet Evol. 29:331–349. doi: 10.1016/s1055-7903(03)00132-5. [DOI] [PubMed] [Google Scholar]
- Whiting A S, Bauer A M, Sites J W. Phylogenetic relationships and limb loss in sub-Saharan African scincines lizards (Squamata: Scincidae) Mol Phylogenet Evol. 29:582–598. doi: 10.1016/s1055-7903(03)00142-8. [DOI] [PubMed] [Google Scholar]