Abstract
The widely accepted dogma of intrauterine sterility and initial colonization of the newborn during birth has been blurred by recent observations of microbial presence in meconium, placenta, and amniotic fluid. Given the importance of a maternal-derived in utero infant seeding, it is crucial to exclude potential environmental or procedural contaminations and to assess fetal colonization before parturition. To this end, we analyzed sterilely collected intestinal tissues, placenta, and amniotic fluid from rodent fetuses and tissues from autoptic human fetuses. Total bacterial DNA was extracted from collected samples and analyzed by Next Generation Sequencing (NGS) techniques using hypervariable 16S ribosomal RNA (rRNA) regions (V3-V4). Colonizing microbes were visualized in situ, using labeled probes targeting 16S ribosomal DNA by fluorescent in situ hybridization. The NGS analysis showed the presence of pioneer microbes in both rat and human intestines as well as in rodent placentas and amniotic fluids. Microbial communities showed fetus- and dam-dependent clustering, confirming the high interindividual variability of commensal microbiota even in the antenatal period. Fluorescent in situ hybridization analysis confirmed the microbes’ presence in the lumen of the developing gut. These findings suggest a possible antenatal colonization of the developing mammalian gut.
Keywords: mammalian gut, embryonic development, microbiota, 16S rRNA gene sequencing
Introduction
Fetus, amniotic fluid, and chorioamnion tissue have long been considered sterile until birth or rupture of the amniotic sac. However, recent evidence shows that the intragestational sac environment harbors a diversity of microorganisms even in physiological pregnancies,1–3 contradicting the long-standing dogma of “womb sterility”.4 Indeed, the cultivable genera Enterococcus, Streptococcus, Staphylococcus, and Propionibacterium have been isolated from umbilical cord blood of healthy neonates born by cesarean delivery.5 Within the abundant literature on human placenta microbiome composition,3,6,7 a recent study combining 16S ribosomal DNA-based and whole-genome shotgun metagenomic analyses showed the presence of a unique placental microbiota, strongly resembling the maternal oral bacteria, with the dominance of Firmicutes, Tenericutes, Proteobacteria, Bacteroidetes, and Fusobacteria phyla.3 In addition, amniotic fluid and placenta were found to have similar microbial communities, consistent across individuals.6 Lactic acid bacteria and enteric bacteria have been reported in meconium collected after birth,8 and a certain degree of similarity has been demonstrated between meconium and amniotic fluid,6 probably related to liquid swallowing by the fetus during pregnancy.
All these data suggest that humans might come into contact with bacteria before birth, and, depending on the time of gestation and the type of bacteria that first seed the fetus, this antenatal colonization might have important physiological and clinical consequences. Indeed, microbes, either true pioneer or transient species, could expose the developing fetus to a diverse array of antigens9 that educate the fetal immune system toward tolerance and participate in the full development of the gut-associated lymphoid tissue.10
At the same time, toxins and viable microorganisms, through active and passive transport from maternal circulation to the placenta, could gain direct entry into fetal circulation, eliciting infective and/or inflammatory processes.11
Despite these recent advances in the field, a conclusive analysis of antenatal microbial colonization has not been reported,12 leaving a gap in the knowledge of this important developmental process. The present study is aimed at ascertaining antenatal microbial colonization of mammalian fetal intestinal tissues. To address this issue, a rodent animal model was used to allow sterile experimental conditions. This was compared to human gut samples from fetal autopsies that were studied using a 16S rRNA amplicon-based NGS approach, validated by in situ detection.
Results
Microbial Species Are Identifiable in Rodent Fetuses in Utero
We collected, under sterile conditions, intestine, placenta, and amniotic fluid from 5 rat fetuses: 3 fetuses (numbered 1-3) from 1 dam (dam A) and 2 (numbered 4-5) from the other (dam B). The tissues were analyzed by Next Generation Sequencing (NGS).
An average of 259 465 reads were obtained per sample, giving a total of 4 670 364 reads overall. Paired-end reads generated from the original DNA fragments using Illumina MiSeq NGS were merged and quality-filtered producing a total of 1 560 296 sequence tags from the gut samples and 982 017 and 900 070 from placentas and amniotic fluids, respectively.
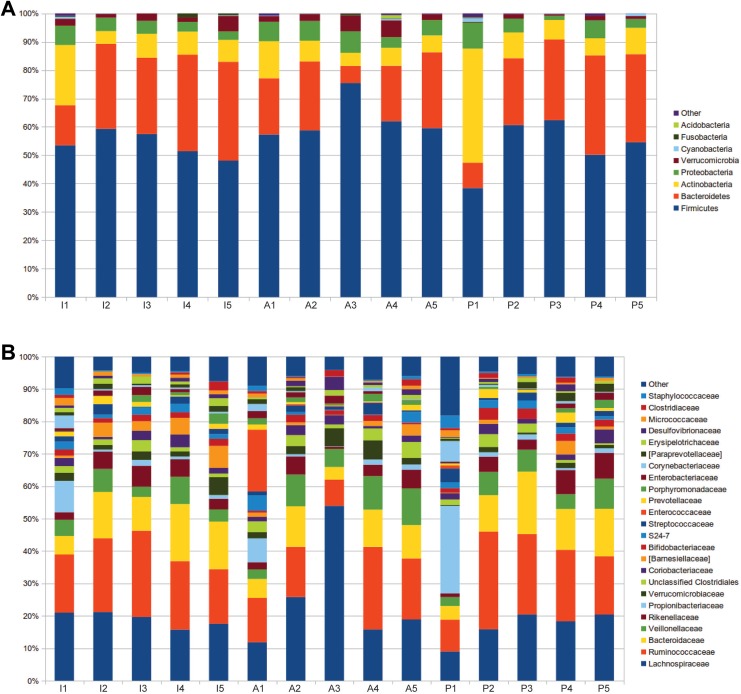
Nine different bacterial phyla were identified in rat fetal samples following negative control subtraction. The most represented phyla (Figure 1A), using a cutoff applied of a relative abundance greater than 1% in at least one experimental group, were Firmicutes (mean relative abundance [SD], 57.0 [8.6]), Bacteroidetes (23.7 [8.7]), Actinobacteria (10.3 [8.4]), Proteobacteria (5.0 [2.1]), and Verrucomicrobia (2.8 [1.9]).
Figure 1.
Pioneer microbiota in the developing rodent gut bar charts representing the relative abundance of 5 fetal intestines (I1-I5), amniotic fluids (A1-A5), and placentas (P1-P5). The figure shows relative abundance of bacterial (A) phyla and (B) families.
The most abundant families (Figure 1B) were Ruminococcaceae (20.9 [7.6]), Lachnospiraceae (20.5 [9.3]), Bacteroidaceae (11.4 [4.4]), Veillonellaceae (5.9 [3]), Rikenellaceae (4.2 [2.3]), and Propionibacteriaceae (3.5 [6.3]).
Microbial Community Is Characteristic of Fetuses and Dams
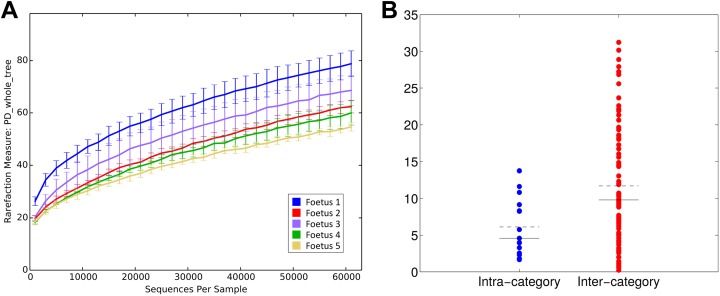
In order to understand the main determinants constituting the microbial diversity, we evaluated the differences among the samples on both richness and composition. Tissues (ie, intestine, placenta, and amniotic fluid), dams (ie, A and B), and fetuses were considered for microbiota profiling. The analysis of samples biodiversity (α-diversity) showed clustering according to dam and fetus rather than analyzed tissue. Faith’s phylogenetic diversity (Figure 2) measured based on distances and observed species metrics showed a significant separation dependent on fetus (Mann-Whitney U test: P = .016 and .019, respectively). Both the metrics showed a separation dependent also on dam (permutation-based test: P = .004 and .036, respectively). Interestingly, no significant separation was observed based on tissue type (P > .05), independent of the metric used to compare distributions.
Figure 2.
Microbial biodiversity (α-diversity) is fetus-specific (A) α-diversity rarefaction curves according to faith’s phylogenetic distance (“PD whole Tree”). X-axis reports the number of sequences per sample, whereas Y-axis shows the value of the metric. Samples are grouped based on fetus number. (B) Distribution of distances between α-diversity PD whole tree values; Distances are labeled as “intra-” or “inter-category” according to fetus number. Dashed black line represents the mean of the distances, whereas the solid black line represents the median.
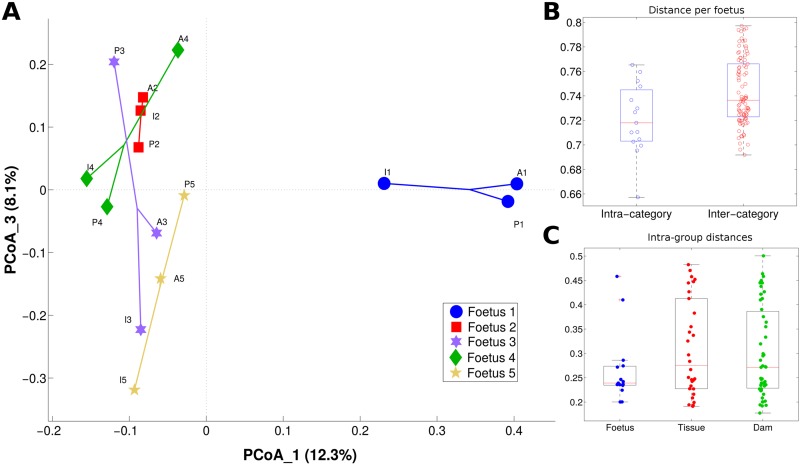
To evaluate whether different samples were characterized by distinct microbiota composition profiles (β-diversity, Figure 3A), the distribution of UniFrac distances was assessed. Both unweighted, which gives equal importance to rare and common taxa, and weighted UniFrac distances, which gives a higher importance to the most abundant bacteria, were used.
Figure 3.
Microbiota composition (β-diversity) is fetus-specific (A) principal coordinates analysis (PCoA) of the unweighted UniFrac distances; PCoA components 1 and 3 are reported. Samples are connected together on the basis of fetus number. (B) Boxplots of intra- and intercategory unweighted UniFrac distances among samples; categories are based on the fetus number. (C) Boxplots of intracategory weighted UniFrac distances among samples; samples are grouped according to fetus, tissue, or dam.
As with the α-diversity, the β-diversity analyses clustered according to fetus (adonis test: P = .009 and .006 on unweighted and weighted UniFrac distances, respectively) and to dam (adonis test P = .015 on weighted UniFrac distances).
Distributions of unweighted UniFrac distances (Figure 3B) on fetus were statistically different (P = .01), whereas weighted UniFrac distances were not (P = .09), indicating that significant differences are present in subdominant components of the microbiota. In detail, fetus 1 was characterized by a high relative abundance of Propionibacteriaceae (14.3% compared to an average of 1.1% in other fetuses) and Corynebacteriaceae (4.1% compared to an average of 0.4% in other fetuses). Fetus 3 showed an increased abundance of Erysipelotrichaceae (1.7% compared to an average of 0.9% in other fetuses), and fetus 5 was enriched in Porphyromonadaceae (average relative abundance: 2.4% compared to 1.0% in other fetuses; Supplemental Figures 1 and 2A).
Analysis of observed species metric (P = .036) and Faith’s phylogenetic distances (P = .004) allowed for a significant separation among dams, with dam B presenting with a lower biodiversity (Supplemental Figure 3). Analysis of distances based on sample origin (ie, fetus, tissue, and dam) showed a trend indicating similarity in microbial profiles among fetuses (Figure 3C).
The microbiota signature for each tissue appeared less distinct, with only some hints of a lower presence of Verrucomicrobiaceae in placenta tissue (relative abundance: 1.1% compared to 2.7% and 3.7% in amniotic fluid and intestine, respectively) and a trend of higher relative abundance of Barnesiellaceae in the intestine (4.0% compared to 1.4% and 1.5% in placenta and amniotic liquid, respectively; Supplemental Figures 2C and 4). Few differences emerged in the comparison between dams A and B (Supplemental Figures 2B and 5).
Bacteria Are Visualized in the Gut During Rodent Fetal Development
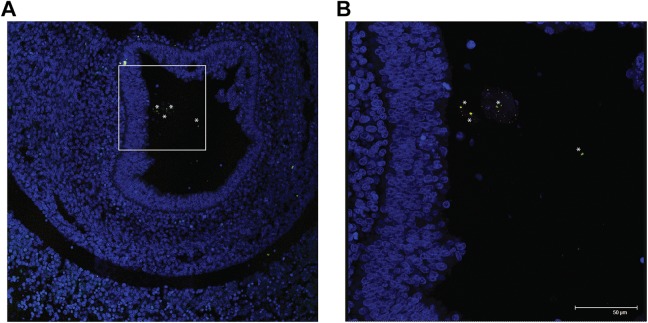
To assess bacteria distribution, in situ analysis was performed on whole sectioned fetuses. Fluorescent detection revealed the presence of bacteria in the gut lumen of developing rat fetuses (Figure 4A and B). In particular, eubacteria (green fluorescence) could be visualized on the different analyzed sections, confirming that bacteria colonize the rodent intestine before birth.
Figure 4.
Eubacteria in the developing rodent gut lumen confocal microscopy images showing (A) eubacteria (in green) in the lumen of a 16-day post coitum rat fetus; (B) at a higher magnification (inset is represented as a white box in A) note the typical bacterial morphology and it is possible to identify few bacteroides spp. (yellow). In blue is DAPI (4′,6-diamidino-2-phenylindole) (nuclei) and 50-μm scale bar is reported in B. Asterisks indicate bacterial cells.
Probe for Staphyloccaceae did not give positive fluorescent signal in any of the tissue sections analyzed.
Bacteria Are Present in the Gut During Human Fetal Development
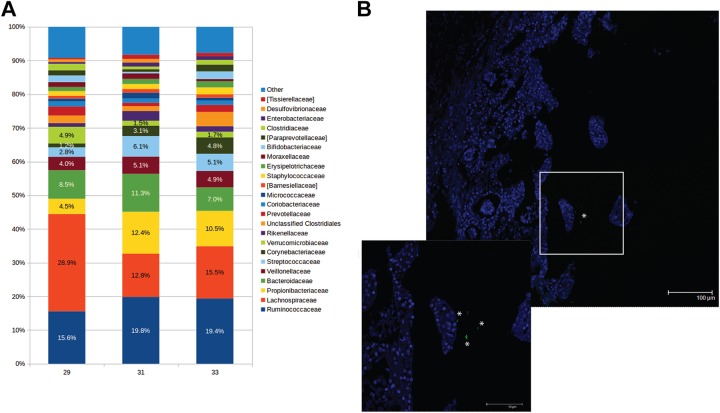
Paraffin-embedded intestinal tissues from 3 third-trimester (gestational age 29, 31, and 33 weeks) human fetuses were screened for the presence of fetal microbiota.
In all analyzed samples, bacteria were observed. The most represented phyla (Figure 5A) were Firmicutes (57.3 [4.5]), Bacteroidetes (17.4 [1.2]), Actinobacteria (16.8 [5.4]), Proteobacteria (4.9 [2.2]), and Verrucomicrobia (2.7 [1.7]). At family level, the most abundant taxa were Lachnospiraceae (19.0 [8.6]), Ruminococcaceae (18.2 [2.4]), Propionibacteriaceae (9.1 [4.1]), Bacteroidaceae (8.9 [2.2]), Streptococcaceae (4.7 [1.7]), and Veillonellaceae (4.7 [0.6]).
Figure 5.
Eubacteria in the developing human gut (A) bar charts representing the family relative abundance at family level of 3 fetal human intestines (29, 31, and 33 weeks of gestation, respectively). (B) Representative confocal microscopy images of in situ hybridization showing the presence of eubacteria (in green-, inset showing higher magnification) in the lumen human fetuses. In blue is DAPI (nuclei) and 100 μm and 50 μm (inset) scale bars are reported. Asterisks indicate bacterial cells.
The presence of bacteria indicated by the data obtained by NGS analysis was further validated by visualizing eubacteria in the lumen of the developing gut by fluorescent in situ hybridization (Figure 5B).
Discussion
The presence of a meconium microbiota supports the existence of maternal microbial transmission in utero.8 In this study, we show that bacteria are present in anatomical fetal mammalian gut sections. Reported relative abundance in meconium6,8,13 indicate a consistent presence of Proteobacteria, whereas in this study, both in human and in rat developing gut, Firmicutes were found to be more represented. These changes in microbiota may be a consequence of physiological changes that occur during birth. The present study does not suffer for the rodent (not the human) sampling of the known different oxygen levels given the immediate freezing of dissected tissues.
Supporting the important role of the microbiota community in the developing mammalian gut, the phyla composition found in analyzed samples (Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Verrucomicrobia) closely resembles those reported in healthy adult human gut.14,15 It is important to note that, compared to adult and childhood tissues,16,17 the developing gut is enriched in Actinobacteria and depleted in Bacteroidetes. Actinobacteria abundance has been shown to progressively increase in infant feces during lactation and to then decrease when solid food is introduced in the diet. The opposite has been reported for Bacteoidetes.18–20 Hence, the difference between fetal and adult gut composition is in line with infant fecal microbiota, suggesting that solid food might be responsible for the switch. It could also suggest that the presence of a maternally provided reservoir of bifidobacteria, which with human milk oligosaccharides are known to be fundamental for the development of a balanced infant microbiota and a fully functional gastrointestinal tract.18,21–23
Another difference found in our study compared to bacteria composition reported in meconium collected after birth6,8 is that Staphylococcaceae and Streptococcaceae appear to be less abundant during uterine life. It is known that Staphylococci are characteristic of higher respiratory tract and skin microbiota; hence, it is conceivable that colonization occurs during birth or from the first days of life through contact with maternal tissues. It has also recently been shown that both genera are abundant in colostrum and maternal milk,24 indicating a possible dual colonizing path.
Analysis of 16S rRNA amplicon data showed that microbiota composition is fetus- and dam specific rather than tissue specific. Indeed, bacterial families found in amniotic fluid and placenta overlap with those found in the corresponding fetus. Importantly, the specificity seems to be independent of growing environment (ie, uterine tissues), but it seems to relate to micro niches (ie, embryonic implant). This is true not only for the microbial community but also for other developmental determinants, such as genetic, metabolic, biochemical, or epigenetic components, known to be specific to the single developing organism and not always shared among all siblings. This is in addition to the possible confounding factors of gender, time, site of implantation, and so on.
Given that bacteria colonize mammalian gut during intrauterine life, the fundamental question remains as to the source and the path of this in utero seeding or exposure. Recent findings3 have described how, analyzing a significant number of placentas collected after birth, the placental microbiota shares more similarities with that of the oral composition compared to vaginal, skin, and/or gut communities. Rather than identifying the origin of placental microbiota, these data support the exclusion of a passive dispersion through excreting organs. In the literature, a possible microbiota colonization in utero has been often hypothesized as a consequence of pathogens known to be able to reach the developing fetus. Our data suggest an alternative mechanism, where pathogens may pass the maternal barrier as a consequence of the necessary permissiveness to commensal bacteria,4 instead of resulting from infectious events (reviewed in Doran et al25).
Clearly, considering the accumulating evidence for a strict relationship between microbiome and health status, in all studied settings (age, gender, ethnicity, mtDNA SNP, haplogroup, etc)26–31 and the importance of maintaining or replenishing the microbial community in pathological conditions,29,32,33 the present article reports data contributing to the open field of investigation relating to the management of healthy pregnancies.34 Future studies will be devoted to overcome the limitations of the present research, that is, prospective collection of fetal human samples, increase number of rodent fetuses, and implementing culture techniques for growing commensal microorganisms.
Materials and Methods
Animals and Housing
CD albino rats (Sprague-Dawley) were maintained in standard conditions (light 6 am to 6 pm, T = 22°C ± 2°C, humidity = 55% ± 5%) with tap water and food (Mucedola standard diet) ad libitum. Virgin females were caged overnight with males of proven fertility. The day of positive vaginal smear was considered as day 0. The pregnant rats were housed individually. Animals were euthanized by carbon dioxide inhalation, and bilaterally pregnant uterine samples were collected in the morning of gestational day 16. Amniotic fluid, placenta, and fetal intestines were dissected, collected in sterile conditions, and stored at −80°C until use. For dissection, uteruses were placed in sterile saline solution under laminar flow cabinet, and all procedures were conducted in the hood using UV-sterilized equipment within a sterile field created by a Bunsen burner, until samples were placed in sterile tubes. For in situ hybridization, whole fetuses were collected in paraformaldehyde 4% (vol/vol) and kept at 4°C in rotation for 4 days. Samples were then washed twice in phosphate saline solution, rehydrated through a graded series of alcohols, and paraffin embedded.
All animal procedures were conducted in accordance with the ethical guidelines approved by the University of Milan in compliance with national (Dlgs 26/2014) and international laws and policies (EEC Council Directive 86/609).
Human Samples
Human samples were included based on stillborn nonmacerated fetuses and gut tissue availability. The 3 fetuses were not malformed and with normal karyotype. Pregnancies were reported as uneventful, including infection and inflammation disease or premature rupture of the membranes, until intrauterine fetal death. Autopsy was performed following international protocols.35
Four micrometer thick tissue sections from formalin-fixed, paraffin-embedded tissue samples were cut and processed for in situ hybridization by deparaffinization in xylene and rehydration through a graded series of alcohols. For NGS analysis, tissue sections, cut under laminar flow cabinet using sterile blades and placed in sterile tubes, were washed twice with 1 mL of Histo-Clear (Sigma Aldrich, Milan, Italy) for 15 minutes with rotation at 56°C or until diaphanization. Tissue was recovered by centrifugation, washed in ethanol, and dried for DNA extraction.
The study was exempt from institutional review board approval because, following Italian Data Protection Act 9/2013, and autopsy material sampled for diagnostic purposes can be used for research as long as patient privacy is ensured. This law is in line with European Commission recommendation n. Rec(2006)4.
Next Generation Sequencing Analysis
Total bacterial DNA extraction was performed using the QIAamp DNA Microbiome Kit (QIAGEN, Hilden, Germany), following manufacturer’s instructions. Particular attention was paid to avoid environmental contamination of collected samples and cross-contamination between samples. Samples were individually processed for DNA extraction under laminar flow cabinet, following UV sterilization. Empty tubes, processed in parallel during tissue recovery and DNA extraction, were used as negative controls. A number of corrective measures have been applied. In particular, UV irradiation of surfaces and instruments using disposable equipment or autoclave-based sterilization. Negative controls were run in parallel and processed for detecting possible contaminating microorganisms. 16s rRNA sequencing results did not show commonly reported environmental and reagent contaminants.36
16S rRNA gene amplicon libraries were performed with a 2-step barcoding approach according to Illumina 16S Metagenomic Sequencing Library Preparation (Illumina, San Diego, California), which amplifies 2 hypervariable regions (ie, V3 and V4) of the 16S rRNA bacterial gene. Library concentration and exact product size were measured using a KAPA Library Quantification Kit (Kapa Biosystems, Woburn, Massachusetts) and Agilent 2100 Bioanalyzer System (Agilent, Santa Clara, California), respectively. Agilent analysis for evaluating the correct predicted size of amplicons showed no bands in negative controls, extracted, and processed in parallel with samples (Supplemental Figure 6). Prior to sequence, libraries were pooled using Amicon Ultra 0.5 mL Centrifugal Filters (Merck Millipore Ltd, Tullagreen, Carrigtwohill Co, Cork, Ireland).
The resulting library was loaded on a MiSeq 500 cycle-v2 cartridge to obtain a paired-end 2 × 250 bp sequencing. Demultiplexed FASTQ files were generated by Illumina MiSeq Reporter, and 2.5 Gbases were obtained.
Raw sequence data determined in this study are available at NCBI Short Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under Accession numbers PRJNA379373 and PRJNA379370.
Fluorescence In Situ Hybridization Analysis
Paraffin-embedded tissue specimens (4-μm thick) were deparaffinized by sequential steps in xylene. Then samples were rehydrated in 95% ethanol, 90% ethanol, and finally deionized water. The slides were air-dried prior to hybridization.
Fluorescence in situ hybridization probe sequences for Eubacteria (EUB 338-I, 5′-GCTGCCTCCCGTAGGAGT-3′; EUB 338-III, 5′-GCTGCCACCCGTAGGTGT-3′), encompassing all bacterial species in Bacteria domain (labeled with fluorescein isothiocyanate [FITC]), Bacteroides (BAC303, 5′-TCCTCCATATCTCTGCGC-3′, Cy3), and Lachnospiraceae (LACHNO, 5′-TTCCCATCTTTCTTGCTGGC-3′, Cy5) were obtained from probeBase website.37 Negative control probe (complementary to EUB 338-I probe, NON-EUB, 5′-ACTCCTACGGGAGGCAGC-3′) was also hybridized to evaluate nonspecific binding. Staphylococcaceae probe (STAPHY, 5′-TCCTCCATATCTCTGCGC-3′, Cy3) sequence was designed as described by Gey et al38 and used to assess possible environmental contaminants in sampling. Probes were purchased from Integrated DNA Technologies (San Jose, California). Hybridization was carried out using standard methods.38,39 Briefly, sections were deparaffinized and rehydrated in serial solutions. Following section air-drying, specific oligonucleotide probes were hybridized using conditions optimized for each probe for stringent hybridizations: BAC303 at 48°C and 10% formamide; STAPHY, and LACHNO at 48°C and 30% formamide; EUB 338-I, and EUB 338-III, at 48°C and 10% or 30% formamide according to the paired probes. DAPI (4′,6-diamidino-2-phenylindole) counterstaining was applied to assess prokaryotic and eukaryotic nuclear morphology.
In this set of experiments, a number of controls were used: sections hybridized with STAPHY probe that resulted negative, to exclude common contaminants; sections hybridized with NON-EUB probe, as negative control; and artificially contaminated sections hybridized with STAPHY probe, that resulted positive, as technical control.
Images of probe-labeled sections were acquired using a confocal laser scanning microscope (TCS SP2, Leica, Wetzlar, Germany). Microorganisms were checked for position, size, and morphology. Confocal images were acquired by series and sequential scan mode. Photomultiplier tube detectors were adjusted to minimize the bleed-through of fluorescent emissions and to optimize signal/noise ratio, in particular versus tissue autofluorescence.
Data Analysis
Sequencing reads were processed, filtered, and analyzed following similar procedures described in Borghi et al.40 Briefly, read pairs were merged together by PandaSeq software41 discarding fragments of length <300 bases or >900 bases as well as nonoverlapping sequences. Then, fragments were quality-filtered, clustered into operational taxonomic units, and taxonomically classified against the 13.8 release of the Greengenes bacterial 16S rRNA database (http://greengenes.lbl.gov) using the QIIME suite (version 1.8.042).
Biodiversity (α-diversity) was evaluated by permutation-based t tests, whereas “adonis” of the R package “vegan” was used for bacterial composition (β-diversity). In addition, due to the reduced number of samples per category, we devised an alternative strategy for comparing the distributions of distances within and between each experimental group for both α-diversity and β-diversity evaluations. Each sample was assigned to an experimental group according to one of the associated labels (ie, tissue type and dam or fetus number); then, a distance between each sample and all the others was calculated. This allowed distinguishing distances between samples belonging to the same (“intracategory” distance) or to a different (“intercategory” distance) experimental group. This strategy was applied for evaluating the absolute difference for α-diversity indexes (ie, chao1, Shannon index, observed species, and Faith’s phylogenetic distance) and the weighted or unweighted UniFrac distances43 (β-diversity). A Mann-Whitney U test was applied for comparing the distributions of “intra-” and “intercategory” distances. Details of statistical methods are provided as Supplemental information.
Supplemental Material
Supplementary_Information for Antenatal Microbial Colonization of Mammalian Gut by Elisa Borghi, Valentina Massa, Marco Severgnini, Grazia Fazio, Laura Avagliano, Elena Menegola, Gaetano Pietro Bulfamante, Giulia Morace and Francesca Borgo in Reproductive Sciences
Acknowledgments
We thank Raffaella Adami for her assistance in confocal laser microscopy techniques, Francesca Di Renzo and Silvia Rigamonti for technical assistance. We are grateful to Ms Dawn Savery and Dr Jon Wilson for commenting on the manuscript.
Authors’ Note: E.B., V.M., and F.B. conceived and designed the study; E.M. and V.M. performed rodent housing and sampling; L.A. and G.B. performed human tissue sampling; E.B., F.B., and G.F. performed experiments; M.S. performed bioinformatics analysis; M.S., E.B., and F.B. analyzed the data; V.M. and E.B. wrote the article; G.M. supervised the project. All authors made comments on the manuscript. E.B. and V.M. equally contributed to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was done at the Università degli Studi di Milano, Milan, Italy. This study was funded by a Research Grant 2016 of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) to F.B., and by Università degli Studi di Milano.
ORCID iD: Elisa Borghi, PhD  https://orcid.org/0000-0002-1893-0455
https://orcid.org/0000-0002-1893-0455
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Brugman S, Perdijk O, van Neerven RJJ, Savelkoul HFJ. Mucosal immune development in early life: setting the stage. Arch Immunol Ther Exp. 2015;63(4):251–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ardissone AN, de la Cruz DM, Davis-Richardson AG, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One. 2014;9(3):e90784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 2013;11(8):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiménez E, Fernández L, Marín ML, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol. 2005;51(4):270–274. [DOI] [PubMed] [Google Scholar]
- 6. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6:23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prince AL, Ma J, Kannan PS, et al. The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am J Obstet Gynecol. 2016;214(5):627.e1–627.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiménez E, Marín ML, Martín R, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159(3):187–193. [DOI] [PubMed] [Google Scholar]
- 9. Avagliano L, Marconi AM, Candiani M, Barbera A, Bulfamante G. Thrombosis of the umbilical vessels revisited. An observational study of 317 consecutive autopsies at a single institution. Hum Pathol. 2010;41(7):971–979. [DOI] [PubMed] [Google Scholar]
- 10. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fardini Y, Chung P, Dumm R, Joshi N, Han YW. Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun. 2010;78(4):1789–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the ‘sterile womb’ and ‘in utero colonization’ hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23(3):314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huttenhower C; Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing: commentary. Nature. 2010;11:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Greenhalgh K, Meyer KM, Aagaard KM, Wilmes P. The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ Microbiol. 2016;18(7):2103–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Riva A, Borgo F, Lassandro C, et al. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ Microbiol. 2016;19(1):1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turroni F, Ribbera A, Foroni E, van Sinderen D, Ventura M. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek. 2008;94(1):35–50. [DOI] [PubMed] [Google Scholar]
- 19. Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. 2012;69(1):52–60. [DOI] [PubMed] [Google Scholar]
- 20. Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(suppl 1):4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Asakuma S, Hatakeyama E, Urashima T, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286(40):34583–34592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang M, Li M, Wu S, et al. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr. 2015;60(6):825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rasmussen SO, Martin L, Østergaard MV, et al. Human milk oligosaccharide effects on intestinal function and inflammation after preterm birth in pigs. J Nutr Biochem. 2017;40:141–154. [DOI] [PubMed] [Google Scholar]
- 24. Drago L, Toscano M, De Grandi R, Grossi E, Padovani EM, Peroni DG. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. 2016;11(4):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Doran KS, Banerjee A, Disson O, Lecuit M. Concepts and mechanisms: crossing host barriers. Cold Spring Harb Perspect Med. 2013;3(7):a010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hullar MA, Fu BC. Diet, the gut microbiome, and epigenetics. Cancer J. 2014;20(3):170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strati F, Di Paola M, Stefanini I, et al. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front Microbiol. 2016;7:1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aagaard K, J Petrosino J, Keitel W, et al. The human microbiome project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013;27(3):1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma J, Coarfa C, Qin X, et al. mtDNA haplogroup and single nucleotide polymorphisms structure human microbiome communities. BMC Genomics. 2014;15:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peterson CT, Sharma V, Elmén L, Peterson SN. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol. 2015;179(3):363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends Mol Med. 2015;21(2):109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mecacci F, Serena C, Avagliano L, et al. Stillbirths at term: case control study of risk factors, growth status and placental histology. PLoS One. 2016;11(12):e0166514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Greuter D, Loy A, Horn M, Rattei T. ProbeBase-an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Res. 2016;44(D1):D586–D589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gey A, Werckenthin C, Poppert S, Straubinger RK. Identification of pathogens in mastitis milk samples with fluorescent in situ hybridization. J Vet Diagn Invest. 2013;25(3):386–394. [DOI] [PubMed] [Google Scholar]
- 39. Berry D, Schwab C, Milinovich G, et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012;6(11):2091–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Borghi E, Borgo F, Severgnini M, Savini MN, Casiraghi MC, Vignoli A. Rett syndrome: a focus on gut microbiota. Int J Mol Sci. 2017;18(2):E344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics. 2012;13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Information for Antenatal Microbial Colonization of Mammalian Gut by Elisa Borghi, Valentina Massa, Marco Severgnini, Grazia Fazio, Laura Avagliano, Elena Menegola, Gaetano Pietro Bulfamante, Giulia Morace and Francesca Borgo in Reproductive Sciences