Abstract
Background
Dihydrofolate reductase (DHFR) is an important target for antimetabolite class of antimicrobials because it participates in purine synthesis. 2-mercaptobenzimidazole (2MBI) has similar structural features as purine nucleotides. Given that benzimidazole and similar heteroaromatics have been broadly examined for their anticancer potential, so, we hereby report the design, synthesis and biological studies (i.e. antimicrobial and anticancer studies) of 2MBI derivatives.
Methodology
The antimicrobial activity of synthesized 2MBI derivatives were evaluated against Gram positive and Gram negative bacterial species as well as fungal species by tube dilution technique whereas their anticancer activity was assessed against human colorectal carcinoma cell line (HCT116) by Sulforhodamine B (SRB) assay. They were also structurally characterized by IR, NMR, MS and elemental analyses.
Results, discussion and conclusion
The antimicrobial activity findings revealed that compound N1 (MICbs,st,ca = 1.27, 2.54, 1.27 µM), N8 (MICec= 1.43 µM), N22 (MICkp,an= 2.60 µM), N23 and N25 (MICsa= 2.65 µM) exhibited significant antimicrobial effects against tested strains, i.e. Gram-positive, Gram-negative (bacterial) and fungal strains. The anticancer screening results demonstrated that compounds N9, N18 (IC50 = 5.85, 4.53 µM) were the most potent compounds against cancer cell line (HCT116) even more than 5-FU, the standard drug (IC50 = 9.99 µM). 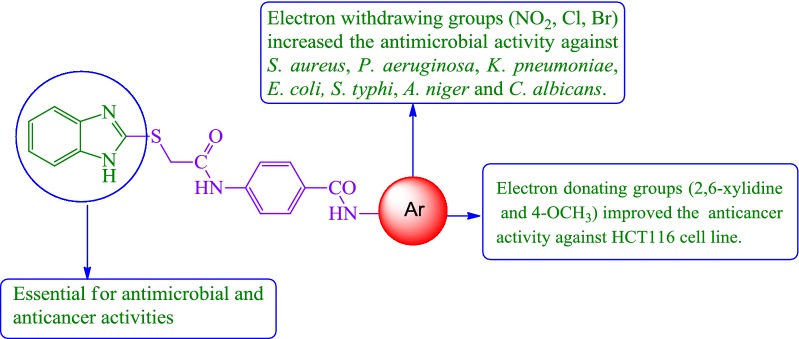
Keywords: p-Amino benzoic acid, Antibacterial, Anticancer, Antifungal, 2-Mercaptobenzimidazole derivatives
Background
The global statistics of infectious diseases have reached a worrying level following the rise of multi-drug resistant Gram-positive and Gram-negative microorganisms. Advanced treatments against infections which rely on a long multidrug schedule are often impeded by patient noncompliance and the emergence of multidrug resistant pathogens. The issue of resistance could be potentially overcome by exploring new effective agents. In this regard, rational drug design has been proven to be much beneficial as the biochemical basis of mechanisms of intrinsic and acquired resistance is largely known [1].
In terms of cancer, presently used chemotherapeutic agents restrain the growth of tumour through suppression of DNA replication and transcription. Chemotherapy is, however, often compromised by development of multidrug resistance, endurance of cancer cells in anaerobic environment, involvement of ribonucleotide reductase (RNR), topoisomerase I (Topo I) and topoisomerase II (Topo II) in neoplasm growth. Nevertheless, the attempt of discovering new curative anticancer agents in last decade has led to targets of specific molecular modifications in tumour cells. The new approach now focuses mainly on the development of small biologically active molecules containing significant activity without toxicity related to the usual chemotherapy [2].
Substituted benzimidazole compounds, with excellent antimicrobial activity against bacteria and ability to permeate mammalian cell membranes as well as maintain the activity with low to no associated toxicity have made these lead compounds attractive for further research work in the field of medicine [3]. The search for new active, antitumour drugs with lower toxicity against normal cells and tissues, however, remains as one of the most significant problems of modern antitumour chemotherapy [4].
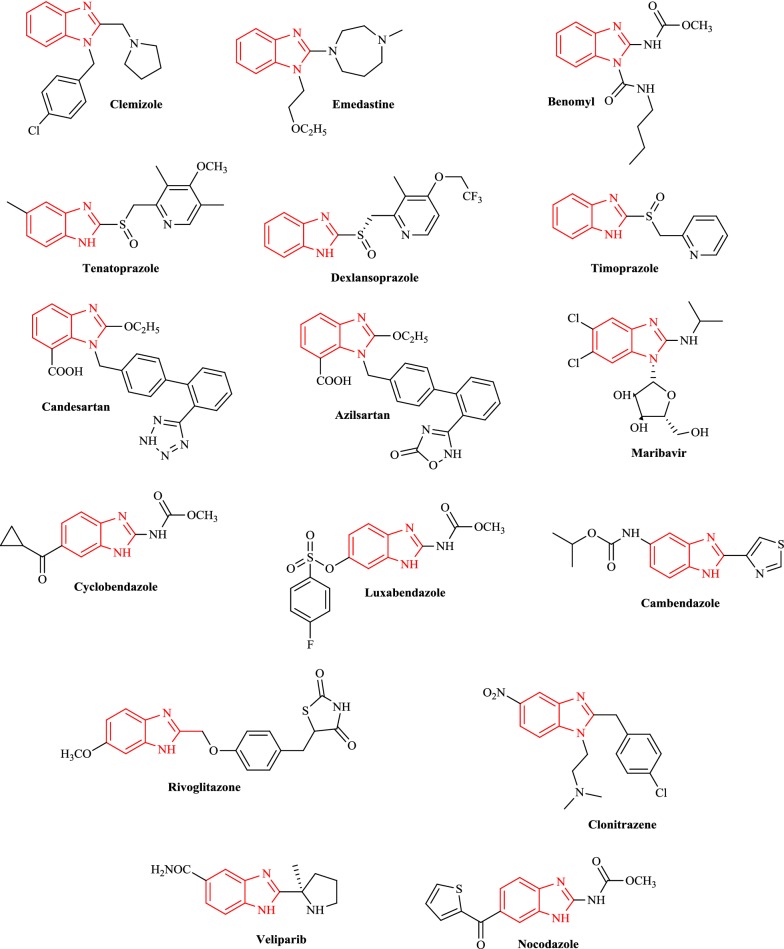
In modern drug discovery and medicinal chemistry, benzimidazole nucleus has been proven to be a very significant pharmacophore with a wide variety of activities [5] including antihistaminic (H1-receptor antagonists, e.g. clemizole and emedastine) [6], antifungal (systemic fungicide, e.g. benomyl) [7], antiulcer (proton pump inhibitors (PPIs), e.g. tenatoprazole, dexlansoprazole and timoprazole) [8], antihypertensive (angiotensin II receptor blockers, e.g. candesartan and azilsartan) [9], antiviral (oral ribosyl benzimidazole, e.g. maribavir) [10], antiparasitic (specifically anthelmintics, e.g. cyclobendazole, luxabendazole and cambendazole) [11], antidiabetic (PPAR gamma agonist, e.g. rivoglitazone) [12], analgesic (opioid analgesic, e.g. clonitrazene) [13] and anticancer (antimitotic agent, e.g. nocodazole, PARP inhibitor, e.g. veliparib) [14] (Fig. 1).
Fig. 1.
Marketed formulations having benzimidazole moiety
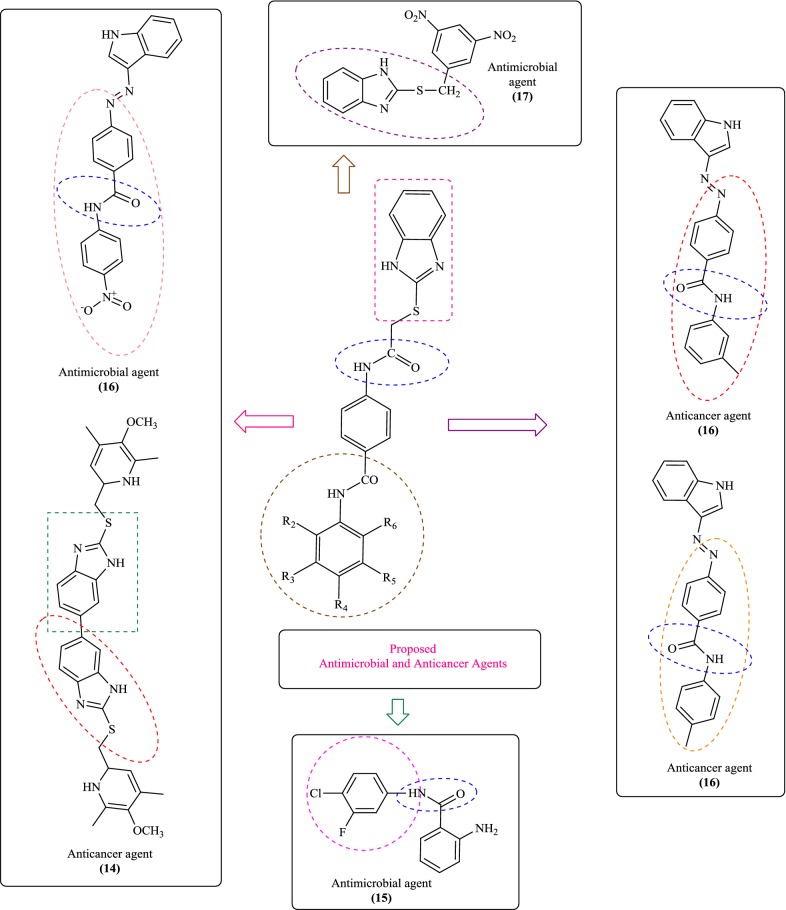
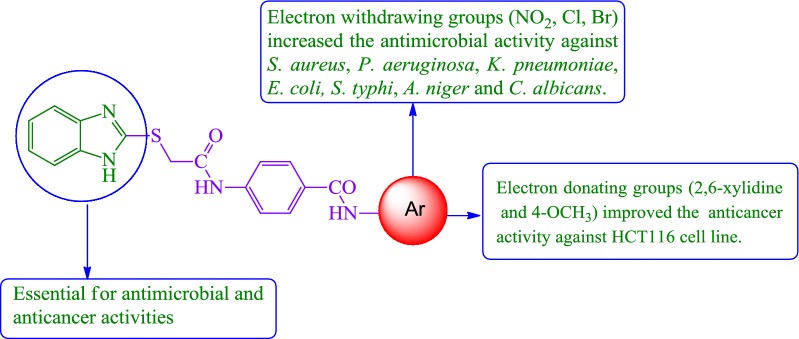
Figure 2 shows some antimicrobial and anticancer activities of benzimidazole derivatives which were reviewed based on a literature survey [15–19]. As part of our continuing effort in search for new therapeutic agents, we had synthesized a number of pharmacologically important heterocyclic compounds (N1–N26). In order to study the impact of electron-donating and accepting groups within the moieties, an attempt has been made to combine 2MBI and p-amino benzoic acid moieties as hybrid and substitute them with different substituted amines. In this regard, a library of substituted benzimidazole was designed, synthesized, spectroscopically characterized and evaluated for potential antimicrobial and anticancer effects in vitro.
Fig. 2.
Design of benzimidazole analogues for antimicrobial and anticancer activity based on literature study
Results and discussion
Chemistry
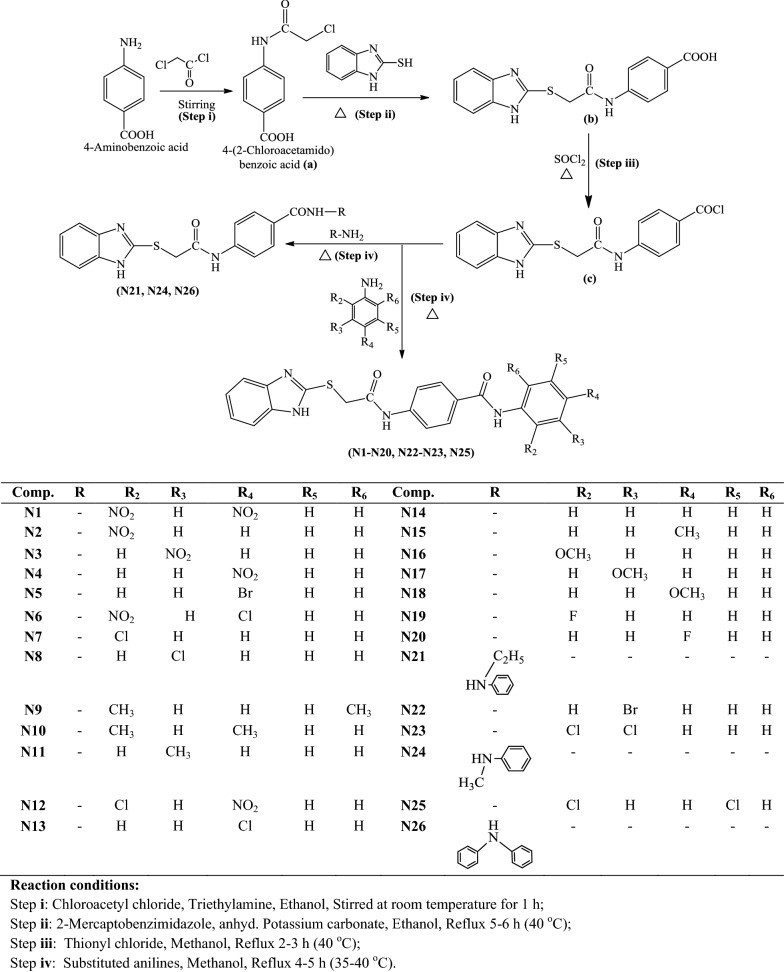
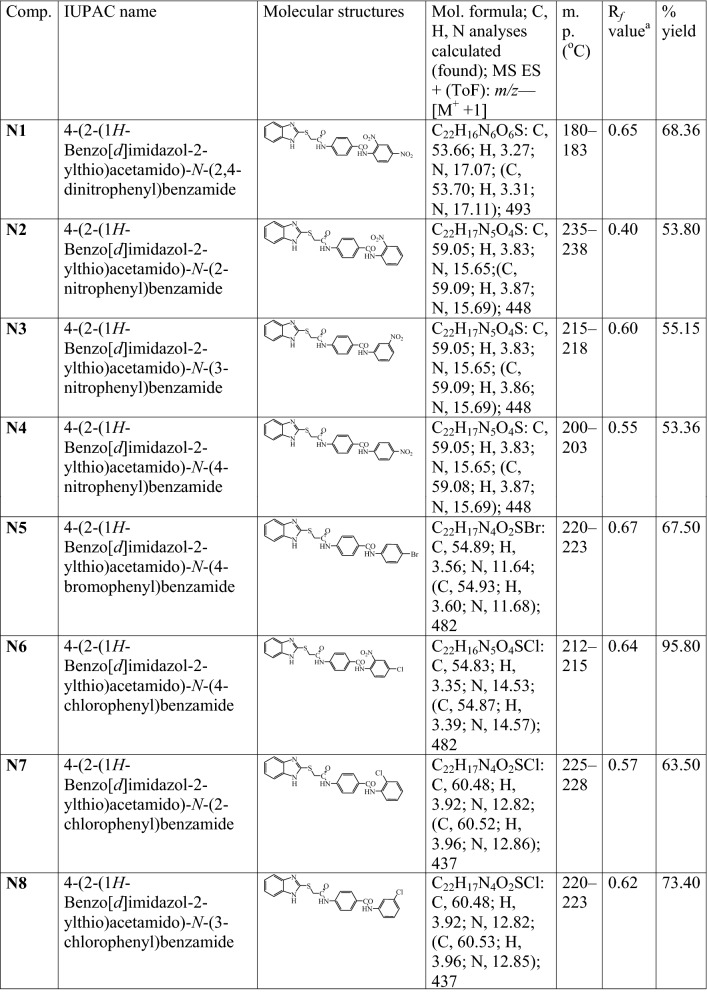
A series of 2MBI benzamides has been synthesized as discussed in Scheme 1. The 4-(2-chloro acetamido) benzoic acid (a) was prepared by the reaction of chloroacetyl chloride with p-amino benzoic acid, which on further reaction with 2-mercaptobenzimidazole yielded 4-(2-(1H-benzo[d]imidazol-2-ylthio)acetamido) benzoic acid (b). The reaction of b with thionyl chloride resulted in the formation of 4-(2-(1H-benzo[d]imidazol-2-ylthio)acetamido)benzoyl chloride (c). The reaction of above synthesized benzoyl chloride (c) with different substituted anilines in methanolic/ethanolic solvent yielded the title scaffolds (N1–N26). The physicochemical properties, elemental analyses and mass spectral studies of title compounds (N1–N26) are presented in Table 1. The molecular structures of synthesized compounds were determined by FT-IR, 1H/13C-NMR (Table 2). The halogenated compounds, N5 and N22 (Ar–Br) showed peak around 633–616 cm−1, N19 and N20 (Ar-F) at 1117–1116 cm−1 and N6, N7, N8, N12, N13, N23 and N25 (Ar–Cl) around 771–738 cm−1. The existence of Ar–NO2 group (N1, N2, N3, N4, N6 and N12) was indicated by the appearance of stretching at 1549–1513 cm−1. The spectral data of aryl alkyl ether group (C–O–C, Ar–OCH3) in compounds N16, N17 and N18 was in between 2837 and 2831 cm−1. The band at 2948–2860 cm−1 in the spectral data of N9, N10, N11 and N15 depicted the presence of Ar–CH3. The IR stretching around 714–689 cm−1 (C–S), 1692–1635 cm−1 (–CONH–), 3110–3018 cm−1, ~ 1600 cm−1 (C–H and C=C, Ar) and 1361–1253 cm−1 (C=N) found in all synthesized 2MBI derivatives. In 1H-NMR spectra, the existence of multiplet signals between 6.51 and 9.10 δ ppm indicated the presence of aromatic proton of synthesized derivatives (N1–N26). Due to presence of Ar-CH3, compounds N9, N10, N11 and N15 showed singlet at 2.23–2.53 δ ppm and because of Ar–OCH3, N16, N17 and N18 showed singlet at range of 3.73–3.74 δ ppm. The synthesized molecules exhibited singlet at 7.93–8.01 δ ppm, 2.55–4.38 δ ppm and 4.36–4.53 δ ppm due to the existence of –CONH, –CH2 groups and –NH of imidazole ring. The findings of elemental analyses of synthesized 2MBI derivatives were recorded within theoretical results of ± 0.4%. Conclusively, the 13C-NMR spectra of synthesized benzamides were in DMSO-d6 and their molecular structures were in accordance with the spectral signals. Mass spectra of the synthesized derivatives reflected the characteristic molecular ion peaks.
Scheme 1.
Synthesis of 4-(2-(1H-benzo[d]imidazol-2-ylthio)acetamido)-N-(substitutedphenyl) benzamide derivatives
Table 1.
The physicochemical properties of synthesized 2-mercaptobenzimidazole derivatives
aTLC mobile phase-Ethyl acetate
Table 2.
Spectral interpretation of synthesized derivatives
| Comp. | IR (KBr cm−1) | 13C-NMR (DMSO-d6, δ ppm) | 1H-NMR (DMSO-d6, δ ppm) |
|---|---|---|---|
| N1 | [3104 (C–H str.), 1602 (C=C str.) pn(phenyl nucleus)], 1626 (–CONH str.), 1333 (N=CH str. (30 amine)), 1256 (C–N str. (20 amine)), 714 (C–S str., CH2–S), 2838 (C–H str., –CH2–), 1516 (C–NO2 str., C6H5NO2) | 39.02, 113.68, 118.35, 122.48, 123.21, 128.54, 129.10, 137.44, 137.45, 142.67, 149.72, 166.25, 166.78 | 7.25–8.77 (m, 11H, Ar–H), 4.42 (s, 1H, NH of imidazole), 7.93 (s, 2H, (CONH)2) |
| N2 | [3058 (C–H str.), 1599 (C=C str.) pn, 1684 (–CONH str.), 1332 (N=CH str.), 1293 (C–N str.), 716 (C–S str., CH2–S), 2931 (C–H str., –CH2–), 1513 (C–NO2 str., C6H5NO2) | 3.6.41, 115.43, 118.44, 119.13, 122.59, 125.47, 130.25, 130.43, 135.62, 137.42, 142.72, 149.80, 166.28, 166.83 | 7.25–7.66 (m, 12H, Ar–H), 4.45 (s, 1H, NH of imidazole), 7.96 (s, 2H, (CONH)2) |
| N3 | [3109 (C–H str.), 1599 (C=C str.) pn], 1666 (–CONH str.), 1331 (N=CH str.), 1299 (C–N str.), 713 (C–S str., CH2–S), 2833 (C–H str., –CH2–), 1540 (C–NO2 str., C6H5NO2) | 36.42, 113.65, 118.42, 120.05, 122.48, 125.46, 129.83, 130.43, 137.43, 137.63, 142.74, 149.80, 166.32, 166.83 | 7.24–7.97 (m, 12H, Ar–H), 4.45 (s, 1H, NH of imidazole), 7.97 (s, 2H, (CONH)2) |
| N4 | [3110 (C–H str.), 1600 (C=C str.) pn], 1681 (–CONH str.), 1301 (N=CH str.), 1280 (C–N str.), 712 (C–S str., CH2–S), 2934 (C–H str., –CH2–), 1542 (C–NO2 str., C6H5NO2) | 113.63, 118.43, 118.55, 122.62, 125.47, 126.31, 130.43, 137.34, 142.73, 149.81, 166.27, 166.82; | 7.26–7.97 (m, 12H, Ar–H), 4.47 (s, 1H, NH of imidazole), 7.97 (s, 2H, (CONH)2) |
| N5 | 3107 (C–H str.), 1598 (C=C str.)pn], 1669 (–CONH str.), 1330 (N=CH str.), 1313 (C–N str.), 711 (C–S str., CH2–S), 2944 (C–H str., –CH2–), 633 (C–Br str., C6H5Br) | 39.02, 113.79, 118.38, 121.81, 125.42, 130.79, 149.72, 166.58, 166.84 | 7.19–7.97 (m, 12H, Ar–H), 4.38 (s, 1H, NH of imidazole), 4.34 (s, 1H, CH2), 7.97 (s, 2H, (CONH)2) |
| N6 | [3101 (C–H str.), 1599 (C=C str.) pn], 1635 (–CONH str.), 1338 (N=CH str.), 1293 (C–N str.), 693 (C–S str., CH2–S), 2833 (C–H str., –CH2–),762 (C–Cl str., C6H5Cl), 1549 (C–NO2 str., C6H5NO2) | 36.47, 113.56, 121.13, 122.84, 123.98, 125.41, 125.47, 129.95, 130.41, 130.43, 135.31, 136.81, 142.65, 142.70, 149.84, 166.15, 166.80 | 7.27–7.95 (m, 11H, Ar–H), 4.47 (s, 1H, NH of imidazole), 7.95 (s, 2H, (CONH)2) |
| N7 | [3109 (C–H str.), 1599 (C=C str.) pn], 1686 (–CONH str.), 1318 (N=CH str.), 1293 (C–N str.), 694 (C–S str., CH2–S), 2870 (C–H str., –CH2–),750 (C–Cl str., C6H5Cl) | 36.41, 115.59, 118.41, 122.56, 125.45, 125.56, 127.60, 128.92, 130.43, 137.29, 137.49, 142.73, 149.80, 166.27, 166.80 | 7.23–7.97 (m, 12H, Ar–H), 4.42 (s, 1H, NH of imidazole), 7.93 (s, 2H, (CONH)2) |
| N8 | [3106 (C–H str.), 1598 (C=C str.) pn, 1683 (–CONH str.), 1331 (N=CH str.), 1293 (C–N str.), 694 (C–S str., CH2–S), 2868 (C–H str., –CH2–), 750 (C–Cl str., C6H5Cl) | 36.38, 113.67, 118.44, 122.42, 125.46, 130.44, 135.62, 137.68, 142.73, 149.76, 166.38, 166.86; | 7.19–7.94 (m, 12H, Ar–H), 4.41 (s, 1H, NH of imidazole), 7.94 (s, 2H, (CONH)2) |
| N9 | [3018 (C–H str.), 1598 (C=C str.) pn], 1668 (–CONH str.), 1360 (N=CH str.), 1281 (C–N str.), 713 (C–S str., CH2–S), 2915 (C–H str., –CH2–), 2948 (C–H str., CH3) | 39.01, 118.37, 121.64, 125.41, 130.46, 142.81, 149.71, 166.65, 166.85; | 7.16–7.96 (m, 11H, Ar–H), 4.36 (s, 1H, NH of imidazole), 7.97 (s, 2H, (CONH)2), 2.53 (s, 6H, (–CH3)2) |
| N10 | [3055 (C–H str.), 1599 (C=C str.)pn], 1667 (–CONH str.), 1331 (N=CH str.), 1298 (C–N str.), 714 (C–S str., CH2–S), 2934 (C–H str., –CH2–), 2888 (C–H str., CH3) | 17.63, 20.37, 38.99, 113.78, 118.39, 121.82, 124.03, 125.42, 126.43, 130.46, 130.76, 138.78, 138.84, 142.79, 149.72, 166.59, 166.85 | 7.18–7.96 (m, 11H, Ar–H), 4.39 (s, 1H, NH of imidazole), 4.30 (s, 2H, CH2), 7.98 (s, 2H, (CONH)2), 2.23 (s, 6H, (–CH3)2) |
| N11 | [3054 (C–H str.), 1597 (C=C str.) pn], 1667 (–CONH str.), 1360 (N=CH str.), 1311 (C–N str.), 713 (C–S str., CH2–S), 2834 (C–H str., –CH2–), 2877 (C–H str., CH3) | 21.12, 39.01, 116.25, 118.45, 121.73, 124.18, 125.41, 128.59, 130.39, 130.46, 137.99, 138.72, 138.80, 138.92, 142.80, 149.73, 166.62, 166.85 | 7.17–7.96 (m, 12H, Ar–H), 4.39 (s, 1H, NH of imidazole), 4.33 (s, 2H, CH2), 7.97 (s, 2H, (CONH)2), 2.28 (s, 3H, –CH3) |
| N12 | [3047 (C–H str.), 1604 (C=C str.) pn], 1650 (–CONH str.), 1318 (N=CH str.), 1253 (C–N str.), 692 (C–S str., CH2–S), 2905 (C–H str., –CH2–), 744 (C–Cl str., C6H5Cl), 1546 (C–NO2 str., C6H5NO2) | 36.54, 115.49, 118.44, 123.23, 124.53, 125.50, 125.55, 130.40, 135.87, 142.68, 149.88, 166.02, 166.81 | 7.31–8.11 (m, 11H, Ar–H), 4.53 (s, 1H, NH of imidazole), 2.55 (s, 2H, CH2), 7.98 (s, 2H, (CONH)2) |
| N13 | 3051 (C–H str.), 1598 (C=C str.) pn], 1631 (–CONH str.), 1331 (N=CH str.), 1315 (C–N str.), 694 (C–S str., CH2–S), 2951 (C–H str., –CH2–), 749 (C–Cl str., C6H5Cl) | 38.96, 113.67, 118.42, 122.39, 125.45, 130.44, 137.72, 142.74, 149.77, 166.37, 166.84 | 7.31–7.94 (m, 12H, Ar–H), 4.53 (s, 1H, NH of imidazole), 7.97 (s, 2H, (CONH)2) |
| N14 | [3059 (C–H str.), 1599 (C=C str.) pn, 1683 (–CONH str.), 1333 (N=CH str.), 1315 (C–N str.), 692 (C–S str., CH2–S), 2921 (C–H str., –CH2–) | 39.02, 113.67, 118.40, 122.31, 122.38, 123.52, 125.43, 128.77, 130.43, 137.85, 138.74, 142.75, 149.77, 166.79, 166.82; | 7.07–7.93 (m, 13H, Ar–H), 4.41 (s, 1H, NH of imidazole), 4.38 (s, 2H, CH2), 7.94 (s, 2H, (CONH)2) |
| N15 | [3053 (C–H str.), 1599 (C=C str.) pn], 1667 (–CONH str.), 1361 (N=CH str.), 1310 (C–N str.), 714 (C–S str., CH2–S), 2934 (C–H str., –CH2–), 2860 (C–H str., CH3) | 36.26, 118.34, 119.02, 121.52, 125.37, 129.14, 130.45, 132.39, 136.32, 142.81, 149.68, 165.83, 166.82 | 7.11–7.92 (m, 12H, Ar–H), 4.34 (s, 1H, NH of imidazole), 4.28 (s, 2H, CH2), 7.94 (s, 2H, (CONH)2), 2.51 (s, 3H, –CH3) |
| N16 | [3055 (C–H str.), 1599 (C=C str.) pn], 1683 (–CONH str.), 1332 (N=CH str.), 1315 (C–N str.), 694 (C–S str., CH2–S), 2941 (C–H str., –CH2–), 1250 (C–O–C str., phenyl ether), 2837 (C–H str., O-CH3) | 38.87, 113.64, 118.46, 122.51, 125.48, 130.44, 137.50, 142.72, 149.77, 166.36, 166.88 | 7.26–7.82 (m, 12H, Ar–H), 4.49 (s, 1H, NH of imidazole), 4.38 (s, 2H, CH2), 8.00 (s, 2H, (CONH)2), 3.73 (s, 3H, –OCH3) |
| N17 | [3107 (C–H str.), 1599 (C=C str.) pn], 1685 (–CONH str.), 1332 (N=CH str.), 1316 (C–N str.), 694 (C–S str., CH2–S), 2917 (C–H str., –CH2–), 1250 (C–O–C str., phenyl ether), 2837 (C–H str., O-CH3) | 38.93, 113.67, 118.43, 122.38, 125.45, 130.44, 137.75, 142.74, 149.77, 166.39, 166.86 | 7.24–7.98 (m, 12H, Ar–H), 4.45 (s, 1H, NH of imidazole), 4.40 (s, 2H, CH2), 7.98 (s, 2H, (CONH)2), 3.74 (s, 3H, –OCH3) |
| N18 | [3054 (C–H str.), 1600 (C=C str.) pn], 1679 (–CONH str.), 1334 (N=CH str.), 1314 (C–N str.), 691 (C–S str., CH2–S), 2933 (C–H str., –CH2–), 1248 (C–O–C str., phenyl ether), 2831 (C–H str., O-CH3) | 38.95, 55.08, 113.89, 118.40, 120.66, 121.67, 125.43, 130.48, 131.92, 139.06, 149.72, 155.38, 165.61, 166.88; | 6.92–7.99 (m, 12H, Ar–H), 4.40 (s, 1H, NH of imidazole), 4.33 (s, 2H, CH2), 7.99 (s, 2H, N(CONH)2), 3.74 (s, 3H, –OCH3) |
| N19 | [3106 (C–H str.), 1597 (C=C str.) pn], 1667 (–CONH str.), 1361 (N=CH str.), 1313 (C–N str.), 692 (C–S str., CH2–S), 2934 (C–H str., –CH2–), 1117 (C–F str., C6H5F) | 38.93, 113.64, 118.44, 122.52, 125.47, 130.44, 137.46, 142.74, 149.79, 166.34, 166.86 | 7.26–7.99 (m, 12H, Ar–H), 4.49 (s, 1H, NH of imidazole), 8.00 (s, 2H, (CONH)2) |
| N20 | [3066 (C–H str.), 1598 (C=C str.) pn], 1667 (–CONH str.), 1361 (N=CH str.), 1310 (C–N str.), 713 (C–S str., CH2–S), 2937 (C–H str., –CH2–), 1116 (C–F str., C6H5F) | 39.03, 118.36, 121.63, 125.39, 130.46, 142.80, 149.70, 166.64, 166.83 | 7.15–7.94 (m, 12H, Ar–H), 4.36 (s, 1H, NH of imidazole), 4.31 (s, 2H, CH2), 7.95 (s, 1H, CONH) |
| N21 | [3103 (C–H str.), 1597 (C=C str.) pn], 1666 (–CONH str.), 1360 (N=CH str.), 1310 (C–N str.), 711 (C–S str., CH2–S), 2935 (C–H str., –CH2–), 2911 (C–H str., CH3) | 39.03, 118.36, 121.63, 125.40, 130.46, 142.80, 149.70, 166.64, 166.83 | 7.16–7.96 (m, 13H, Ar–H), 4.36 (s, 1H, NH of imidazole), 7.96 (s, 1H, CONH) |
| N22 | [3104 (C–H str.), 1599 (C=C str.) pn], 1684 (–CONH str.), 1331 (N=CH str.), 1316 (C–N str.), 695 (C–S str., CH2–S), 28,668 (C–H str., –CH2–), 616 (C–Br str., C6H5Br) | 39.03, 113.68, 118.40, 122.36, 125.43, 130.43, 137.78, 142.75, 149.78, 166.36, 166.81 | 7.23–7.94 (m, 12H, Ar–H), 4.41 (s, 1H, NH of imidazole), 7.94 (s, 2H, (CONH)2) |
| N23 | [3107 (C–H str.), 1602 (C=C str.) pn], 1650 (–CONH str.), 1350 (N=CH str.), 1316 (C–N str.), 691 (C–S str., CH2–S), 2840 (C–H str., –CH2–), 738 (C–Cl str., C6H5Cl) | 39.02, 114.73, 118.53, 123.26, 125.50, 127.92, 130.41, 131.50, 136.04, 146.61, 166.02, 166.81 | 6.80–7.94 (m, 11H, Ar–H), 4.53 (s, 1H, NH of imidazole), 7.95 (s, 2H, (CONH)2) |
| N24 | [3108 (C–H str.), 1599 (C=C str.) pn], 1685 (–CONH str.), 1332 (N=CH str.), 1316 (C–N str.), 694 (C–S str., CH2–S), 2916 (C–H str., –CH2–), 2874 (C–H str., CH3) | 38.98, 113.61, 118.43, 122.64, 125.46, 130.42, 137.23, 142.72, 149.80, 166.27, 166.83 | 7.26–7.95 (m, 13H, Ar–H), 4.46 (s, 1H, NH of imidazole), 7.96 (s, 1H, CONH) |
| N25 | [3109 (C–H str.), 1604 (C=C str.) pn], 1651 (–CONH str.), 1351 (N=CH str.), 1318 (C–N str.), 691 (C–S str., CH2–S), 2837 (C–H str., –CH2–), 771 (C–Cl str., C6H5Cl) | 36.52, 115.62, 118.45, 123.13, 125.49, 130.18, 130.42, 131.91, 136.21, 136.27, 142.70, 149.87, 166.07, 166.82; | 1H-NMR: 6.88–7.95 (m, 11H, Ar–H), 4.52 (s, 1H, NH of imidazole), 7.96 (s, 2H, (CONH)2) |
| N26 | [3107 (C–H str.), 1600 (C=C str.) pn, 1650 (–CONH str.), 1351 (N=CH str.), 1316 (C–N str.), 689 (C–S str., CH2–S), 2839 (C–H str., –CH2–), 1650 (phenyl conjugation) | 36.55, 116.67, 118.41, 119.57, 123.21, 125.48, 125.51, 129.07, 130.43, 136.15, 142.72, 143.38, 149.90, 166.05, 166.83 | 7.10–7.56 (m, 17H, Ar–H), 4.53 (s, 1H, NH of imidazole), 7.96 (s, 1H, CONH) |
Antimicrobial screening results
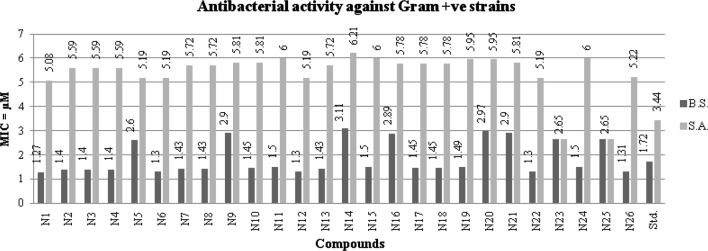
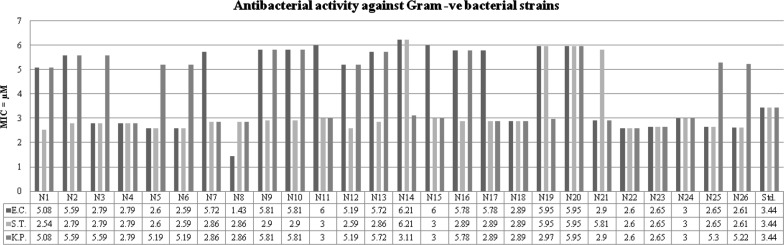
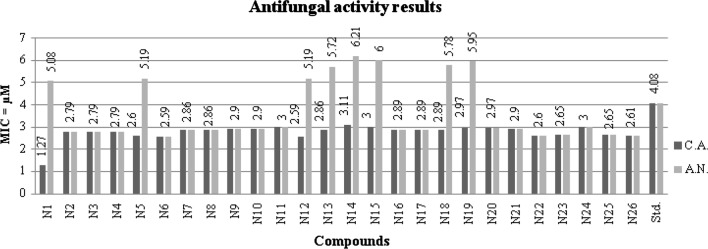
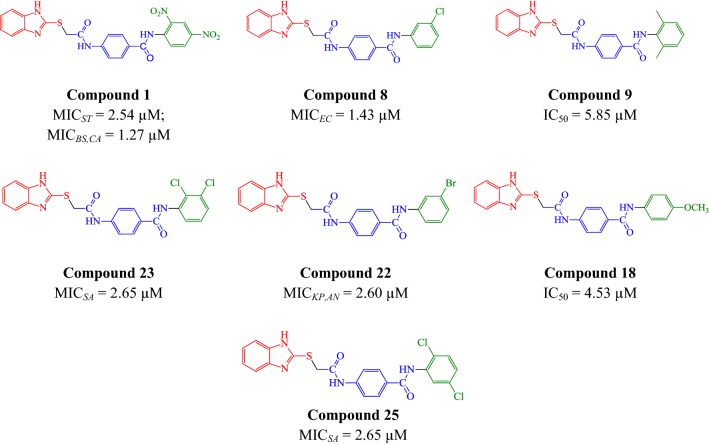
The results of synthesized 2MBI benzamides (N1–N26) are presented in Table 3, Figs. 3, 4 and 5. The synthesized compounds were screened for their antimicrobial potential using cefadroxil (antibacterial) and fluconazole (antifungal) as standard drugs. Among the synthesized compounds, 2, 4-dinitro substituted benzamide, i.e. compound N1 (MICbs,st,ca = 1.27, 2.54, 1.27 µM) exhibited significantly potent antimicrobial effect against B. subtilis, S. typhi and C. albicans, respectively. The meta substituted (–Cl and –Br) derivatives, i.e. compounds N8 (MICec = 1.43 µM) and N22 (MICkp,an = 2.60 µM) displayed promising activity against E. coli, K. pneumoniae and A. niger, respectively. Furthermore, the 2, 3 and 2,5-dichloro substituted ones, i.e. compounds N23 and N25 (MICsa = 2.65µM) were found to be more potent against S. aureus. From the antimicrobial screening results, it was found that all the scaffolds had excellent activity compared with the reference drugs.
Table 3.
Antimicrobial and anticancer screening results of synthesized compounds
| Compounds | MIC (minimum inhibitory concentration = µM) | IC50 (μM) | ||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial strains | Fungal strains | Cancer cell line (HCT116) | ||||||
| Gram +ve | Gram −ve | |||||||
| MICbs | MICsa | MICec | MICst | MICkp | MICca | MICan | ||
| N1 | 1.27 | 5.08 | 5.08 | 2.54 | 5.08 | 1.27 | 5.08 | > 20.31 |
| N2 | 1.40 | 5.59 | 5.59 | 2.79 | 5.59 | 2.79 | 2.79 | > 22.35 |
| N3 | 1.40 | 5.59 | 2.79 | 2.79 | 5.59 | 2.79 | 2.79 | > 22.35 |
| N4 | 1.40 | 5.59 | 2.79 | 2.79 | 2.79 | 2.79 | 2.79 | > 22.35 |
| N5 | 2.60 | 5.19 | 2.60 | 2.60 | 5.19 | 2.60 | 5.19 | > 20.77 |
| N6 | 1.30 | 5.19 | 2.59 | 2.59 | 5.19 | 2.59 | 2.59 | > 20.75 |
| N7 | 1.43 | 5.72 | 5.72 | 2.86 | 2.86 | 2.86 | 2.86 | > 22.89 |
| N8 | 1.43 | 5.72 | 1.43 | 2.86 | 2.86 | 2.86 | 2.86 | 13.73 |
| N9 | 2.90 | 5.81 | 5.81 | 2.90 | 5.81 | 2.90 | 2.90 | 5.85 |
| N10 | 1.45 | 5.81 | 5.81 | 2.90 | 5.81 | 2.90 | 2.90 | > 23.23 |
| N11 | 1.50 | 6.00 | 6.00 | 3.00 | 3.00 | 3.00 | 3.00 | > 24.01 |
| N12 | 1.30 | 5.19 | 5.19 | 2.59 | 5.19 | 2.59 | 5.19 | > 20.75 |
| N13 | 1.43 | 5.72 | 5.72 | 2.86 | 5.72 | 2.86 | 5.72 | > 22.89 |
| N14 | 3.11 | 6.21 | 6.21 | 6.21 | 3.11 | 3.11 | 6.21 | 12.42 |
| N15 | 1.50 | 6.00 | 6.00 | 3.00 | 3.00 | 3.00 | 6.00 | 19.21 |
| N16 | 2.89 | 5.78 | 5.78 | 2.89 | 5.78 | 2.89 | 2.89 | > 23.12 |
| N17 | 1.45 | 5.78 | 5.78 | 2.89 | 2.89 | 2.89 | 2.89 | > 23.12 |
| N18 | 1.45 | 5.78 | 2.89 | 2.89 | 2.89 | 2.89 | 5.78 | 4.53 |
| N19 | 1.49 | 5.95 | 5.95 | 5.95 | 2.97 | 2.97 | 5.95 | > 23.78 |
| N20 | 2.97 | 5.95 | 5.95 | 5.95 | 5.95 | 2.97 | 2.97 | 14.27 |
| N21 | 2.90 | 5.81 | 2.90 | 5.81 | 2.90 | 2.90 | 2.90 | > 23.23 |
| N22 | 1.30 | 5.19 | 2.60 | 2.60 | 2.60 | 2.60 | 2.60 | > 13.86 |
| N23 | 2.65 | 2.65 | 2.65 | 2.65 | 2.65 | 2.65 | 2.65 | > 21.22 |
| N24 | 1.50 | 6.00 | 3.00 | 3.00 | 3.00 | 3.00 | 3.00 | 20.41 |
| N25 | 2.65 | 2.65 | 2.65 | 2.65 | 5.30 | 2.65 | 2.65 | > 21.22 |
| N26 | 1.31 | 5.22 | 2.61 | 2.61 | 5.22 | 2.61 | 2.61 | > 20.90 |
| Broth control | NG | NG | NG | NG | NG | NG | NG | – |
| Std. | 1.72a | 3.44a | 3.44a | 3.44a | 3.44a | 4.08b | 4.08b | 9.99c |
Bacillus subtilis MTCC 441-bs, Staphylococcus aureus MTCC 3160-sa, Escherichia coli MTCC 443-ec, Salmonella typhi MTCC 3231-st, Klebsiella pneumoniae MTCC 9024-kp, Candida albicans MTCC 227-ca and Aspergillus niger MTCC 281-an
DMSO dimethyl sulfoxide, NG no growth
Std. drugs: a Cefadroxil; b Fluconazole; c 5-FU (5-Fluorouracil)
Fig. 3.
Antibacterial screening results against Gram positive species
Fig. 4.
Antibacterial screening results against Gram negative species
Fig. 5.
Antifungal screening results against fungal species
Anticancer screening results
The anticancer activity of synthesized analogues (N1–N26) was evaluated against human colorectal cancer cell line (HCT116) by SRB assay. Table 3 shows the antiproliferative outcome of the synthesized analogues and 5-FU, the standard drug against HCT116. The anticancer screening results indicated that 2,6-xylidine and para-methoxy substituted scaffolds, i.e. compounds N9 (IC50 = 5.85 µM) and N18 (IC50 = 4.53 µM) to exhibit the greatest anticancer activity among the synthesized scaffolds. The anticancer effect was even more potent than 5-FU (IC50 = 9.99 µM).
Toxicity study
For the selectivity index calculation of the most active compounds (N9 and N18), these compounds were tested against normal human embryonic kidney cell line (HEK-293). Compounds were dissolved into 0.1% DMSO solution. The compounds were diluted in concentration (2 μM, 4 μM, 6 μM, 8 μM and 10 μM). The cells were incubated with these compounds for 24 h and more than almost 100% of HEK-293 cells were viable at IC50 for growth inhibition of each studied compound. Results showed the significant viability difference between the test compounds treated and control cells (at zero concentration) after 24 h with (P < 0.01). The 50% of the cells were viable at the lethal dose (LD50) 8.43 μM and 8.22 μM of the active compounds, N9 and N18, respectively. As we know that higher the LD50 value than the IC50 higher will be the selectivity that implied that the compounds may have better safety of the each of two compounds since the IC50 is much lower the LD50 (Table 4).
Table 4.
Lethal dose and selectivity index calculation of most active compounds (N9 and N18)
| Compound | Lethal dose (LD50) | IC50 | Selectivity index (LD50/IC50) |
|---|---|---|---|
| N9 | 8.43 | 5.85 | 1.44 |
| N18 | 8.22 | 4.53 | 1.81 |
MTT assay
Human embryonic kidney (HEK-293) cells were maintained in Dulbecco’s modified Eagle’s medium (10% heat-inactivated FBS). Antibiotics penicillin and streptomycin were added and were placed at 37 °C in a 5% CO2 incubator for colorimetric based assay using MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) compounds N9 and N18 were seeded with five thousand HEK-293 cells (viability 98%) into 96-well plate for 24 h. Wells were added with MTT 5 mg/mL after 24 h incubation for 4 h [20]. Absorbance at 580 nm was recorded using Synergy/HTX MultiScan reader (BioTek) and lethal dose LD50 was calculated and for selectivity index (SI) was calculated.
Structure–activity relationship studies
Electronic discussion
The substitution pattern of the 2MBI benzamide derivatives was carefully selected to confer different electronic environment to the molecules. Thus, electron donating groups to aromatic ring, such as methyl and methoxy and electron withdrawing groups from aromatic ring, such as halogens, nitro were chosen as substituents on the molecular structure of the target compounds [20]. Most of 2MBI benzamides derivatives were found to possess moderate antimicrobial activity except those compounds which has electron withdrawing groups. Suggestions are made that the negative inductive effect plays a significant role in medicinal field. This means that compounds with high electron density gave poor antimicrobial activity which makes the diffusion of compounds more difficult throw the body of the bacteria cell while presence of electron withdrawing groups will cause a decrease of electronic density in 2MBI benzamides compared with electron donating groups, thereby facilitating entry of the 2MBI benzamides into the cell. This is likely to increase the antimicrobial potency. The increase in antimicrobial activity is due to faster diffusion of the 2MBI benzamides with electron withdrawing groups because the reducing the total electron density on 2MBI benzamide compounds make the diffusion faster through the bacteria cell [21].
Because of their significant medicinal importance, the synthesis of substituted benzimidazoles has become a focus of synthetic organic chemistry. The antiproliferative effect and mechanism of induction of apoptosis by various bioactive heterocyclic compounds and the impact of electron releasing and withdrawing groups on the induction of apoptosis is supported by the studies of Gowda et al. [22]. Structure–activity relationship studies of the synthesized compounds based on electronic discussion, antimicrobial and anticancer screening results, the following SAR (Fig. 6) can be assumed:
-
i.
Substitution of aromatic ring with 2,6-dimethyl substituent (compound N9, IC50 = 5.85 µM) considerably improved the anticancer activity of benzamide derivatives whereas substitution with 2,4-dimethyl group (compound N10, IC50 = 23.23 µM) had no such effect. The para-OCH3 substituted aromatic ring (compound N18, IC50 = 4.53 µM) significantly improved the anticancer activity of benzamide derivatives while the ortho and meta-OCH3 substituents (compound N16–N17, IC50 = 23.12 µM) does not improved the anticancer activity of synthesized compounds.
-
ii.
The substitution of aromatic amine with di-NO2 group (compound N1) had significant effect on B. subtilis, S. typhi and fungal strain C. albicans, compared to compounds with single –NO2 substitution (N2, N3 and N4). The meta-Cl and -Br substituted aromatic amines (compounds N8, N22) resulted in improved antimicrobial activity against E. coli, K. pneumoniae and fungus A. niger, respectively. The 2,3 and 2,5-dichloro substituted aromatic amines (compounds N23, N25) noticeably improved the antibacterial activity against S. aureus.
Fig. 6.
Structural requirements for the antimicrobial and anticancer activities of synthesized benzimidazole analogues
Thus, it very well may be stated that nitro, chloro and bromo substituents bearing derivatives are the most suitable scaffolds for accomplishing the best antimicrobial range. The role of electron withdrawing group in improving antimicrobial activities is supported by the studies of Kumar et al. [19].
Experimental section
The reactants and reagents for the synthesis were got from Hi-media Laboratory, Loba Chemie and CDH Pvt. Ltd. Microbial type cell cultures (MTCC) for biological study were procured from Institute of Microbial Technology and Gene bank, Chandigarh. Reaction steps forward was monitored by thin layer chromatography (TLC) using ethyl acetate as mobile phase. The synthetic scheme was drawn via Chem Draw 8.03. Determination of melting point was done using labtech melting point equipment. IR spectrum was recorded on Bruker 12060280, spectrometer via KBr in the range of 4000–400 cm−1. 1H/13C-NMR spectra recorded at 600 and 150 MHz, respectively on Bruker Avance III 600 NMR spectrometer. For Mass spectra, Waters Micromass Q-ToF Micro instrument was used. Elemental analyses of 2MBI derivatives were performed on Perkin-Elmer 2400 C, H and N analyzer.
Procedure to synthetic Scheme 1
Step i: Synthesis of 4-(2-chloroacetamido) benzoic acid (a)
p-Aminobenzoic acid (0.01 mol) and triethylamine (0.01 mol) were stirred properly in ethanol to get a clear solution and cooled it for half an hour. Now the solution was added with chloroacetylchloride (0.01 mol) and stirred for 1 h and the precipitated compound a, was strained via filtering, desiccated and recrystallized with ethanol [1].
Step ii: Synthesis of 4-(2-(1H-benzo[d]imidazol-2-ylthio)acetamido)benzoic acid (b)
Amixture of 4-(2-chloroacetamido) benzoic acid (a, 0.01 mol), 2-mercapto benzimidazole (0.01 mol) and potassium carbonate (0.01 mol) in ethanol (50 ml) was refluxed for 5–6 h and then cooled at room temperature followed by evaporation to dryness. The resultant residue was washed with water and recrystallized from ethanol [23].
Step iii: Synthesis of 4-(2-(1H-benzo[d]imidazol-2-ylthio)acetamido)benzoyl chloride (c)
Thionyl chloride (0.3 mol) was added to 4-(2-(1H-benzo[d]imidazol-2-ylthio) acetamido) benzoic acid (b, 0.25 mol) and the mixture were refluxed for 2–3 h . The excess of thionyl chloride was removed by distillation [17].
Step iv: Synthesis of final (N1-N26) 2MBI derivatives
The reaction mixture of 4-(2-(1H-benzo[d]imidazol-2-ylthio)acetamido)benzoyl chloride (c, 0.01 mol) and substituted aniline (0.01 mol) in suitable solvent was refluxed for appropriate time and thin layer chromatography was used to monitor the reaction. After completion of reaction, it was poured into ice cold water and the resultant precipitate was filtered, desiccated and recrystallized using ethanol [24].
Biological study
Antimicrobial evaluation (in vitro)
The antimicrobial potential of new synthesized compounds (N1–N26) was validated against Gram-positive, Gram-negative bacteria using cefadroxil and fungal strains with fluconazole, by serial dilution method. Dilutions were set up in nutrient broth I.P. for bacterial (incubated at 37 ± 1 °C for 24 h) and Sabouraud dextrose broth I.P. for fungal species (25 ± 1 °C for 7 days for A. niger) and (37 ± 1 °C for 48 h for C. albicans) [25, 26].
Anticancer evaluation (in vitro)
The antiproliferative activity (expressed as IC50) was assessed against HCT116 using SRB B assay. This assay was based on the ability of SRB dye to bind electrostatically and its pH-dependence on protein basic amino acid residues of trichloroacetic acid-fixed cells [27]. Briefly, HCT116 was seeded at 2500 cell/well (96 well plates) and allowed to attach overnight before being exposed to 2MBI compounds (stock solution was suspended in DMSO) for 72 h and subjected to SRB assay. DMSO less than 1% did not kill the cells and the concentration of DMSO in each compound was 0.1%. Treated cells were fixed with 10% cold trichloroacetic acid and stained in 0.4% SRB. Unincorporated dye was rinsed off with 1% acetic acid and plates were left to air-dry at room temperature overnight. The air-dried plates were placed on a plate shaker and bound SRB was solubilised in 10 mM Tris base solution. Absorbance was measured by a computer-interfaced 96-well plate spectrophotometer at 570 nm.
MTT assay
Human embryonic kidney (HEK-293) cells were maintained in Dulbecco’s modified Eagle’s medium (10% heat-inactivated FBS). Antibiotics penicillin and streptomycin were added and were placed at 37 °C in a 5% CO2 incubator for colorimetric based assay using MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) compounds N9 and N18 were seeded with five thousand HEK-293 cells (viability 98%) into 96-well plate for 24 h. Wells were added with MTT 5 mg/mL after 24 h incubation for 4 h [28]. Absorbance at 580 nm was recorded using Synergy/HTX MultiScan reader (BioTek) and lethal dose LD50 was calculated and for selectivity index (SI) was calculated.
Conclusion
The present work involves the synthesis of new substituted benzamides linked to p-amino benzoic acid and 2-mercaptobenzimidazole which possess antibacterial, antifungal and antiproliferative activities. The synthesized compounds were evaluated in vitro against seven representative microorganisms along with the anticancer activity against carcinoma cell line (HCT116). Antimicrobial results demonstrated that the presence of nitro and halo groups in aromatic ring enhanced the antimicrobial potential. Anticancer results revealed that 4-(2-(1H-benzo[d]imidazol-2-ylthio)acetamido)-N-(2,6-dimethyl phenyl)benzamide (N9, IC50 = 5.85 µM) and 4-(2-(1H-benzo[d]imidazol-2-ylthio)acetamido)-N-(4methoxyphenyl) benzamide (N18, IC50 = 4.53 µM) were the most potent anticancer agents even higher than the standard drug 5-FU (IC50 = 9.99 µM). These compounds were more selective towards cancer cells rather than macrophages. Further the toxicity study revealed the better selectivity index against the HEK-293 cell lines at the respective IC50 concentration. Study suggested that compound may be safer as anticancer after required experimental evaluation. The molecular structures of active compounds are mentioned in Fig. 7.
Fig. 7.
Molecular structures of the most active compounds
Authors’ contributions
BN and ST have designed synthesized and carried out the antimicrobial activity and SML, KR, VM and SAAS have carried out the spectral analysis, interpretation and anticancer evaluation of synthesized compounds. All authors read and approved the final manuscript.
Acknowledgements
The authors are thankful to Head, Department of Pharmaceutical Sciences, Maharshi Dayanand University, Rohtak, for providing necessary facilities to carry out this research work.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Funding
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sumit Tahlan, Email: sumittahlan1989@gmail.com.
Kalavathy Ramasamy, Email: kalav922@gmail.com.
Siong Meng Lim, Email: stvlsm@gmail.com.
Syed Adnan Ali Shah, Email: benzene301@yahoo.com.
Vasudevan Mani, Email: vasumpharmacol@gmail.com.
Balasubramanian Narasimhan, Phone: +91 9416649342, Email: naru2000us@yahoo.com.
References
- 1.Turan-Zitouni G, Ozdemir A, Kaplancikli ZA, Cevikbas A, Gurbuz B, Gurer US. Studies on some new pyrazolo[3,4-c]pyridine derivatives as antimicrobial agents. Turk J Pharm Sci. 2009;6(2):63–72. [Google Scholar]
- 2.Sharma A, Luxami V, Paul K. Synthesis, single crystal and antitumor activities of benzimidazole–quinazoline hybrids. Bioorg Med Chem Lett. 2013;23:3288–3294. doi: 10.1016/j.bmcl.2013.03.107. [DOI] [PubMed] [Google Scholar]
- 3.Kumar K, Awasthi D, Lee S-Y, Cummings JE, Knudson SE, Slayden RA, Ojima I. Benzimidazole-based antibacterial agents against Francisella tularensis. Bioorg Med Chem. 2013;21:3318–3326. doi: 10.1016/j.bmc.2013.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodionov AN, Zherebker KY, Snegur LV, Korlyukov AA, Arhipov DE, Peregudov AS, Ilyin MM, Jr, Ilyin MM, Nikitin OM, Morozova NB, Simene AA. Synthesis, structure and enantiomeric resolution of ferrocenylalkyl mercaptoazoles. Antitumor activity in vivo. J Organomet Chem. 2015;783:83–91. doi: 10.1016/j.jorganchem.2015.01.031. [DOI] [Google Scholar]
- 5.Hranjec M, Starcevic K, Pavelic SK, Lucin P, Pavelic K, Zamola GK. Synthesis, spectroscopic characterization and antiproliferative evaluation in vitro of novel Schiff bases related to benzimidazoles. Eur J Med Chem. 2011;46:2274–2279. doi: 10.1016/j.ejmech.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Lavrador-Erb K, Ravula SB, Yu J, Zamani-Kord S, Moree WJ, Petroski RE, Wen J, Malany S, Hoare SRJ, Madan A, Crowe PD, Beaton G. The discovery and structure-activity relationships of 2-(piperidin-3-yl)-1H-benzimidazoles as selective, CNS penetrating H1-antihistamines for insomnia. Bioorg Med Chem Lett. 2010;20:2916–2919. doi: 10.1016/j.bmcl.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Ansari KF, Lal C. Synthesis and evaluation of some new benzimidazole derivatives as potential antimicrobial agents. Eur J Med Chem. 2009;44:2294–2299. doi: 10.1016/j.ejmech.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 8.Kubo K, Oda K, Kaneko T, Satoh H, Nohara A. Synthesis of 2-[[(4-fluoroalkoxy-2-pyridyl)methyl]sulfinyl]-1H-benzimidazoles as antiulcer agents. Chem Pharm Bull. 1990;38(10):2853–2858. doi: 10.1248/cpb.38.2853. [DOI] [PubMed] [Google Scholar]
- 9.Bai R, Wei Z, Liu J, Xie W, Yao H, Wua X, Jiang J, Wang Q, Xu J. Synthesis and biological evaluation of 4′-[(benzimidazole-1-yl)methyl]biphenyl-2-sulfonamide derivatives as dual angiotensin II/endothelin A receptor antagonists. Bioorg Med Chem. 2012;20:4661–4667. doi: 10.1016/j.bmc.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J, Xie J, Luo X. Synthesis and antiviral activity against Coxsackie virus B3 of some novel benzimidazole derivatives. Bioorg Med Chem Lett. 2005;15:267–269. doi: 10.1016/j.bmcl.2004.10.087. [DOI] [PubMed] [Google Scholar]
- 11.Andrzejewska M, Yepez-Mulia L, Tapia A, Cedillo-Rivera R, Laudy AE, Starosciak BJ, Kazimierczuk Z. Synthesis, and antiprotozoal and antibacterial activities of S-substituted 4,6-dibromo- and 4,6-dichloro-2-mercaptobenzimidazoles. Eur J Pharm Sci. 2004;21:323–329. doi: 10.1016/j.ejps.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 12.Vinodkumar R, Vaidya SD, Kumar BVS, Bhise UN, Bhirud SB, Mashelka UC. Synthesis, anti-bacterial, anti-asthmatic and anti-diabetic activities of novel N-substituted-2-(4-phenylethynyl-phenyl)-1H-benzimidazoles and N-substituted 2[4-(4,4-dimethyl-thiochroman-6-yl-ethynyl)-phenyl)-1Hbenzimidazoles. Euro J Med Chem. 2008;43:986–995. doi: 10.1016/j.ejmech.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Achar KCS, Hosamani KM, Seetharamareddy HR. In-vivo analgesic and anti-inflammatory activities of newly synthesized benzimidazole derivatives. Eur J Med Chem. 2010;45:2048–2054. doi: 10.1016/j.ejmech.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 14.Paul K, Sharma A, Luxami V. Synthesis and in vitro antitumor evaluation of primary amine substituted quinazoline linked benzimidazole. Bioorg Med Chem Lett. 2013;24:624–629. doi: 10.1016/j.bmcl.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Yang YH, Cheng MS, Wang QH, Nie H, Liao N, Wang J, Chen H. Design, synthesis and antitumor evaluation of novel symmetrical bis-benzimidazoles. Eur J Med Chem. 2009;44:1808–1812. doi: 10.1016/j.ejmech.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Mahiwal K, Kumar P, Narasimhan B. Synthesis, antimicrobial evaluation, ot-QSAR and mt-QSAR studies of 2-amino benzoic acid derivatives. Med Chem Res. 2012;21:293–307. doi: 10.1007/s00044-010-9537-5. [DOI] [Google Scholar]
- 17.Kumar H, Kumar P, Narasimhan B, Ramasamy K, Mani V, Mishra RK, Majeed ABA. Synthesis, in vitro antimicrobial, antiproliferative and QSAR studies of N-(substituted phenyl)-2/4-(1H-indol-3-ylazo)-benzamides. Med Chem Res. 2013;22:1957–1971. doi: 10.1007/s00044-012-0181-0. [DOI] [Google Scholar]
- 18.Klimesova V, Koci J, Pour M, Stachel J, Waisser K, Jarmila K. Synthesis and preliminary evaluation of benzimidazole derivatives as antimicrobial agents. Eur J Med Chem. 2002;37:409–418. doi: 10.1016/S0223-5234(02)01342-9. [DOI] [PubMed] [Google Scholar]
- 19.Kumar P, Narasimhan B, Sharma D. Substituted benzoic acid benzylidene/furan-2-yl-methylene hydrazides: synthesis, antimicrobial evaluation and QSAR analysis. ARKIVOC. 2008;13:159–178. [Google Scholar]
- 20.Ozkay Y, Tunali Y, Isikdag Karaca H. Antimicrobial activity and a SAR study of some novel benzimidazole derivatives bearing hydrazone moiety. Eur J Med Chem. 2010;45:3293–3298. doi: 10.1016/j.ejmech.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Hania MM. Synthesis and antibacterial activity of some transition metal complexes of oxime, semicarbazone and phenylhydrazone. E-J Chem. 2009;6(S1):S508–S514. doi: 10.1155/2009/204714. [DOI] [Google Scholar]
- 22.Gowda NRT, Kavitha CV, Chiruvella KK, Joy O, Rangappa KS, Raghavan SC. Synthesis and biological evaluation of novel 1-(4-methoxyphenethyl)-1H-benzimidazole-5-carboxylic acid derivatives and their precursors as antileukemic agents. Bioorg Med Chem Lett. 2009;19:4594–4600. doi: 10.1016/j.bmcl.2009.06.103. [DOI] [PubMed] [Google Scholar]
- 23.Turan-Zitouni G, Kaplancikli ZA, Ozdemir A, Revial G, Guven K. Synthesis and antimicrobial activity of some 2-(benzo[d]oxazol/benzo[d]imidazol-2-ylthio)-N-(9H-fluoren-9-yl)acetamide derivatives. Phosphorus Sulfur Silicon. 2007;182:639–646. doi: 10.1080/10426500601047016. [DOI] [Google Scholar]
- 24.Kumar S, Lim SM, Ramasamy K, Vasudevan M, Shah SAA, Narasimhan B. Bis-pyrimidine acetamides: design, synthesis and biological evaluation. Chem Cent J. 2017;11:80. doi: 10.1186/s13065-017-0312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cappuccino JG, Sherman N. In microbiology-a laboratory manual. 4. California: Addison Wesley Longman Inc; 1999. p. 263. [Google Scholar]
- 26.Pharmacopoeia of India . Controller of publication. New Delhi: Ministry of Health Department, Govt. of India; 2007. p. 37. [Google Scholar]
- 27.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 28.Selvaraj V, Bodapati S, Murray E, Rice KM, Winston N, Shokuhfar T, Zhao Y, Blough E. Cytotoxicity and genotoxicity caused by yttrium oxide nanoparticles in HEK293 cells. Int J Nanomed. 2014;9:1379–1391. doi: 10.2147/IJN.S52625. [DOI] [PMC free article] [PubMed] [Google Scholar]