Abstract
Background
Urease are responsible for several pathogenic states in human as well as in animals and its inhibition is utmost urgent. Clinically used drugs are associated with many side effects; recently several researches have shown that flavonoids have good urease inhibition properties. Morin, a natural flavonoid has been investigated for urease inhibition studies which includes designing of library of morin analogues and their in-silico evaluation with the help of Schrodinger’s maestro package of molecular docking software against crystallographic complex of plant enzyme Jack bean urease (PDB ID: 3LA4) followed by synthesis and in vitro evaluation.
Results
Best thirteen derivatives of morin were selected on the basis of their interaction energy and dock score for synthesis and further investigated for in-vitro antioxidant, urease inhibitory and Anti-H. Pylori activity. In-vitro results revealed that a large number of synthesized compounds were found to possess excellent antioxidant and urease Inhibition properties.
Conclusions
Among the synthesized compounds, N-(2-chlorophenyl)-N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)thiourea (M2b) and N-(4-bromophenyl)-N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)thiourea (M2i) were found to be most potent urease inhibitor and antioxidant with IC50 value 10.74 ± 0.018, 11.12 ± 0.033 and 7.37 ± 0.024, 7.73 ± 0.015and 7.795 ± 0.003 µM. Derivative M2i exhibited good anti-H. pylori activity having MIC = 500 μg/ml and zone of inhibition 15.00 ± 0.00 mm as compared to standard AHA having MIC = 1000 μg/ml and zone of inhibition 9.00 ± 0.50 mm determined against H. Pylori bacterium (ATCC 43504, DSM 4867) by well diffusion technique. Furthermore, molecular docking study explained the binding pattern of synthesized ligand within active cavity of jack bean protein and drug similarity was explained by ADME studies by quikprop module of molecular docking software.
Electronic supplementary material
The online version of this article (10.1186/s13065-019-0562-2) contains supplementary material, which is available to authorized users.
Keywords: Morin, Urease inhibition, Natural phenolic compounds, Antioxidant
Introduction
Urease (urea amidohydrolase; E.C. 3.5.1.5) is a nickel containing metalloenzyme brings catalytic hydrolysis of urea and leads to the formation of ammonia and carbamate which instinctively disintegrates, at normal functioning pH, to give another ammonia molecule and bicarbonate [1]. It’s presence in soil was first reported by Rotini [2]. It increases rate of biochemical dissociation of urea by 1014 times [3]. Urease plays role of key enzyme for global nitrogen cycle and supplies nitrogen to plants for seed germination and for growth [4]. However, high urease action is responsible for release of unusually a lot of ammonia into climate which may prompt natural and monetary issues [1, 4]. Ureases have been found among plants, microscopic organisms like bacteria, algae, fungi and invertebrates [12]. Catalytic mechanism of plant and microbial originated urease is similar although they possesses structural differences, probably as they have similar pattern of amino acids and Ni+2 ions at active center which indicates its emergence from a common ancestry [4–6].
Urease are responsible for several pathogenic states in both human as well as in animals such as urinary and GIT infections, gastric cancer, stone formation in kidney, pyelonephritis, encrustation in catheter, ammonia encephalopathy, hepatic coma [7–11]. Urease is also a virulence factor found in pathogenic bacteria H. Pylori and is one of the main cause for its spreading in gastric environment by catalyzing the urea present there. Released ammonia thereby causes elevation in pH and makes a comfortable environment for pathogen H. Pylori to survive and spread colonies. The presence of excess of urease cause breakdown of fertilizer urea and ammonia released in high concentration in the climate and in addition plants get damaged due to toxicity of ammonia and elevation in soil pH, consequently posturing noteworthy ecological and financial troubles [10].
Morin (3, 5, 7, 2′, 4′-pentahydroxyflavone), a bioflavonoid found in fruits, vegetables and various medicinal herbs such as figs, mulberry, Osage orange (Maclura pomifera), sweet chestnut (Castanea sativa, Family-Fagaceae) guava (Psidium guajana L.) leaves, Brazilian wood (Chlorophora tinctoria), almond (Prunus dulcis, Family-Rosaceae), onion, seaweeds, apple, Chinese and Asian medicinal herb as well as in several beverages [12–18] was initially isolated from the Moraceae family [12]. It has been documented that morin is enriched with wide range of biological activities like anti-inflammatory activity [13], antiallergen [15], hepatoprotective [19], antihypertensive, antioxidant, nephroprotective, antidiabetic, anticlastogenic activities, cytoprotective effects, xanthine oxidase–hypoxanthine activity decreases the frequency of septic shock [14] and various types of cancers like liver cancer, breast cancer, colon cancer, oral tumor and have been used as an herbal medicine from long time [10–21].
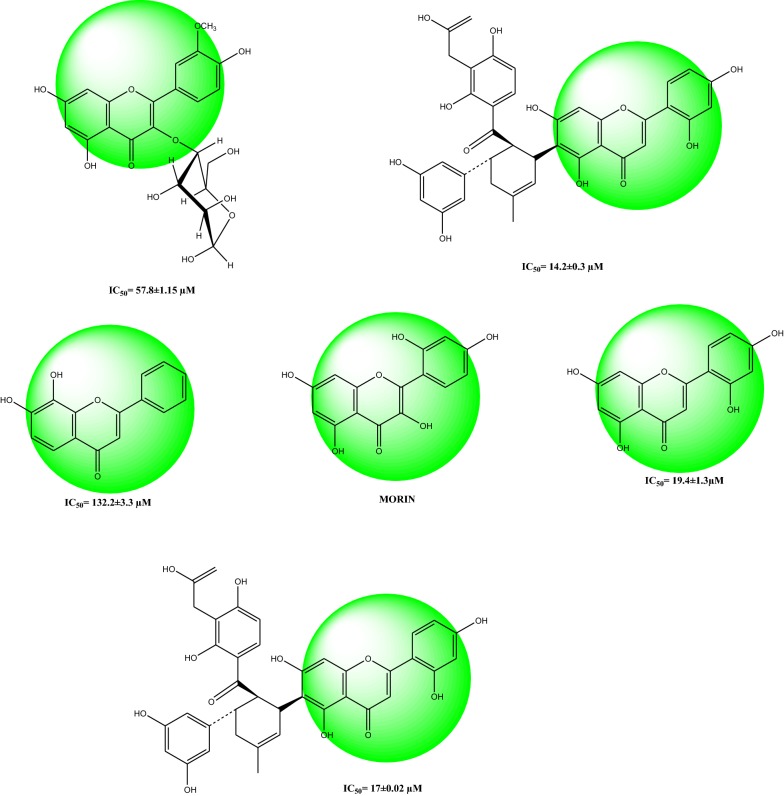
Drugs used clinically for the treatment of ailments caused by urease producing bacteria includes bismuth complex, phosphoramidates, imidazole derivatives and hydroxamic acids are associated with numerous side effects like teratogenic effects shown by hydroxamic acid and rapid disintegration of phosphoramidates at low pH. Pharmaceutical industry is in continuous search for better safe and effective drug for treating such disease. Naturally occurring flavonoids have shown urease inhibitory as well as anti H. Pylori activity as reported by several researchers. Awllia Jalaluddin et al. [22] recently evaluated nine flavonoids for inhibition of jack bean urease and five of them were found to be good in activity having IC50 values 14.2 ± 0.3 to 132.9 ± 3.3 μM shown in Fig. 1 [23]. Similarity in pharmacophore of morin with these flavonoids i.e. 2-phenyl-4H-chromen-4-one and pharmacological enrichment of morin motivated the researchers to design more potent derivatives of it by using docking studies and then evaluation was done for antioxidant, urease inhibition and anti-H. Pylori activity by in-vitro studies.
Fig. 1.
Reported flavonoids having same pharmacophore with morin possessing urease inhibitory activity. flavonoids
Results and discussion
Chemistry
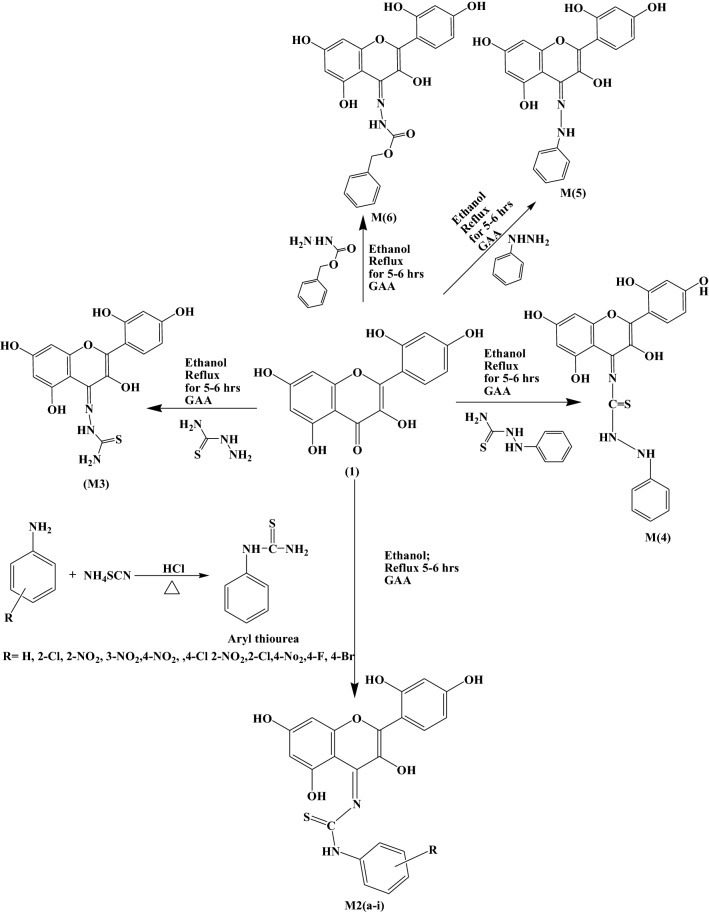
In the current study, morin derivatives (M2a–i, M3–6) were synthesized by following the procedure illustrated in Scheme 1 [24–26]. Synthetic scheme involve two-step process involving formation of substituted aryl thiourea by reaction of substituted anilines with ammonium thiocyanate in presence of hydrochloric acid. Morin derivatives were synthesized in second step by reaction with equimolar concentration of arylthiourea(M2a-i)/thiosemicarbazide(M3)/phenylthiosemicarbazide(M4)/phenyhydrazine(M5)/benzylcarbazate(M6) in ethanol with glacial acetic acid as catalyst. Monitoring of reaction was done by thin layer chromatography and completion of reaction was confirmed by single spot in TLC under UV lamp. Evaluation of structure of novel derivatives was confirmed by spectroscopic methods such as IR, 1H NMR, 13CNMR, elemental analysis. Derivatisation was confirmed by downward shift of peak at 1680 (C=O)str to 1615–1625 (C=N)str. Appearance of a peak at 1539–1571 of N–C=S str was observed in compound M2a–M2i which further confirmed the formation of derivatives. Whereas in compounds M3, M4, M5 and M6 additional peak at 1033–1078 for N–Nstr was also observed which confirmed formation of M3–M6 compounds. 1HNMR signals were interpreted by their value of chemical shift for particular protons of synthesized derivatives, coupling constant and multiplicities of signals. For instance in compound M2a appearance of singlet at δ 12.68, 10.68, 9.7, 9.68 was noticed for OH, OH, NH and OH groups respectively and 7.53–6.30 for aromatic protons, wheras in 13C NMR signals at δ 187 (C–S) and 157 (C–N) confirmed formation of compound. Further and final confirmation process involves analyzing their mass spectrum for determination of molecular weight in which Q-ToF Micro instrument was used as ion source. Maximum number of the derivatives showed peak at M+ (molecular ion peak), (M++1), (M++2) in positive chemical ionization and (M+1), (M+2), M+ during negative chemical ionization mode. Elemental analysis of diosmin derivative was carried out in CHNS analyzer where C, H and N in percent were found within acceptable range.
Scheme 1.
Synthetic scheme for the synthesis of morin derivatives
Biological activity
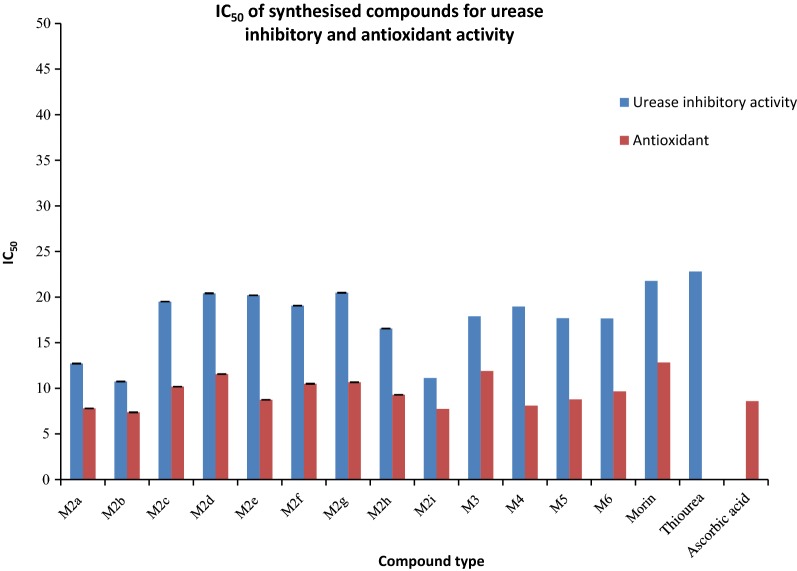
The novel synthesized Morin derivatives were examined for in- vitro urease inhibitory action by determining the ammonia concentration released during the reaction by indophenol technique as described by Weatherburn [27] as well as for antioxidant nature [28, 29] by determining the ability of compounds to donate hydrogen and electrons using DPPH method. All the synthesized compounds were found to be potent inhibitors of jack bean urease and showed good antioxidant potential with IC50 value ranging between 10.74–20.48 µM and 7.37–11.9 µM respectively. Among them compounds M2b, M2i and M2a displayed the excellent urease inhibition with IC50 value 10.74 ± 0.018, 11.12 ± 0.033 and 12.71 ± 0.027 µM even two folds more active than standard drug thiourea and also displayed good antioxidant behavior with IC50 value 7.37 ± 0.024, 7.73 ± 0.015and 7.795 ± 0.003 µM against DPPH using ascorbic acid as standard as shown in Tables 1 and 2 and Figs. 2, 3 and 4.
Table 1.
In-vitro urease inhibition activity and IC50 of synthesized morin derivatives
| Compound | IC50(µM)a | Compound | IC50(µM)a |
|---|---|---|---|
| M2a | 12.71 ± 0.027 | M2i | 11.12 ± 0.033 |
| M2b | 10.74 ± 0.018 | M3 | 17.9 ± 0.007 |
| M2c | 19.5 ± 0.005 | M4 | 18.96 ± 0.011 |
| M2d | 20.41 ± 0.031 | M5 | 17.68 ± 0.08 |
| M2e | 20.2 ± 0.01 | M6 | 17.66 ± 0.02 |
| M2f | 19.06 ± 0.015 | Morin | 21.77 ± 0.016 |
| M2g | 20.48 ± 0.026 | Thiourea | 22.80 ± 0.011 |
| M2h | 16.55 ± 0.011 |
aValues related for the evaluated compound absorption which provide 50% inhibition of Urease inhibition action, and are the mean SEM; statistical significance: p < 0.05 against the equivalent IC50 values achieved against urease, as identified through ANOVA/Dunnett’s test
Table 2.
In-vitro DPPH radical scavenging activities and IC50 of synthesized morin derivatives (antioxidant activity)
| Compound | IC50 (µM)a | Compound | IC50 (µM)a |
|---|---|---|---|
| M2a | 7.795 ± 0.003 | M2i | 7.73 ± 0.015 |
| M2b | 7.37 ± 0.024 | M3 | 11.9 ± 0.031 |
| M2c | 10.18 ± 0.006 | M4 | 8.099 ± 0.018 |
| M2d | 11.56 ± 0.013 | M5 | 8.78 ± 0.008 |
| M2e | 8.744 ± 0.008 | M6 | 9.65 ± 0.022 |
| M2f | 10.50 ± 0.036 | Morin | 12.83 ± 0.028 |
| M2g | 10.66 ± 0.019 | Ascorbic acid | 8.59 ± 0.004 |
| M2h | 9.290 ± 0.007 |
aValues related for the evaluated compound absorption which provide 50% inhibition of Urease inhibition action, and are the mean SEM; statistical significance: p < 0.05 against the equivalent IC50 values achieved against urease, as identified through ANOVA/Dunnett’s test
Fig. 2.
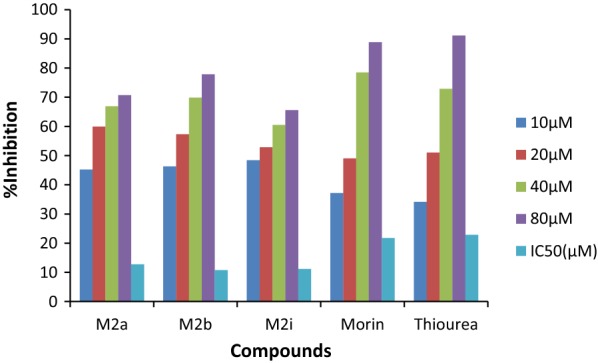
In-vitro urease inhibitory activity of most potent compounds
Fig. 3.
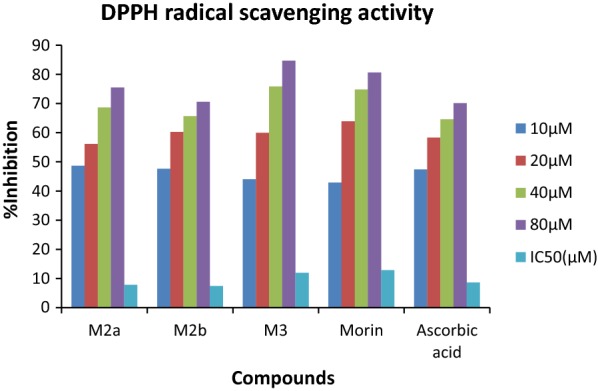
DPPH radical scavenging activity of most potent compounds
Fig. 4.
IC50 value synthesized compounds for urease inhibition and antioxidant activity
Compounds M2b and M2i which were found potent in urease inhibition and antioxidant activity as well as in terms of docking score were tested further against antibacterial efficiency against Helicobacter pylori ATCC 43504, DSM 4867 using AHA as a standard and DMSO as control. MIC50 values were calculated for compounds and the results revealed that M2i displayed its potency with good zone of inhibition i.e. 15.00 ± 0.00 mm as compared to standard 9.00 ± 0.50 mm and MIC50 value was found to be comparable to Standard i.e. 500 µg/ml. It could be concluded that compound M2i indicated excellent antibacterial action against H. Pylori, antioxidant and urease inhibitory activity.
Enzyme kinetics
Study of inhibitory effect of morin derivatives on jack bean urease was performed to check the inhibitory potential, kinetics studies and mechanism of inhibition in phosphate buffer and 1 mM EDTA at pH 8.2. Lineweaver–Burk plots (1/absorbance versus 1/urea) was constructed from kinetic data to determine the mechanism of enzyme inhibition by varying the concentration of substrate urea in the presence of different concentrations of most potent compound M2i. Inhibition constant (Ki) was determined as the intersection on x-axis of the plot of 1/Vmax and varying concentration of inhibitor obtained from Lineweaver-Burke plot and all experiments were performed in triplicate. Inhibition was found to be non-competitive as Km was constant but Vmax was changed. Binding confirmations from molecular simulation studies also confirm the mode of inhibition as shown in Fig. 5.
Fig. 5.
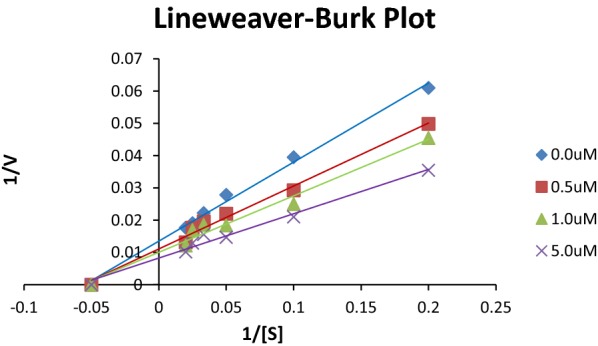
Lineweaver-burke plot for compound M2i
Structure activity relationship
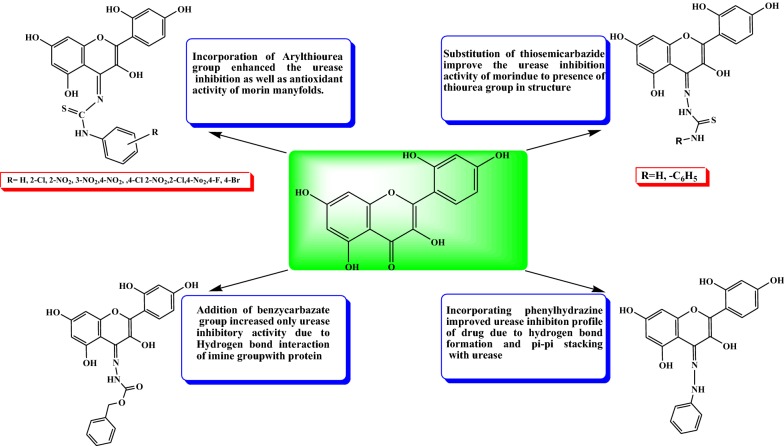
Studying results from antioxidant nature and urease inhibition activities of newly synthesized morin derivatives, structure activity relationship can be derived (Fig. 6).
Incorporation of substituted arylthiourea group enhanced the urease inhibition as well as antioxidant behavior of morin manyfolds such as compounds (M2a–M2i) having IC50 value for urease inhibition from 10.74 ± 0.018 to 20.48 ± 0.026 µM.
Attachment of bromo substituted arylthiourea increased the effectiveness of molecule (M2i) in inhibiting growth of pathogenic bacteria H. Pylori with MIC value 500 µg/ml and zone of inhibition 15.00 ± 0.00 mm as compared to standard acetohydroxamic acid having MIC 500 µg/ml and zone of inhibition 9.00 ± 0.50 mm.
Substitution of thiosemicarbazide and phenylthiosemicarbazide groups in morin leads to improvement of urease inhibition activity to greater extent due to presence of thiourea groups in the structure.
Phenylhydrazine and benzylcarbazate addition improved of activity of morin due to stronger interaction of compounds with enzyme through H-bond formation by imine group and pi–pi stacking interactions.
Presence of hydroxyl groups improved the antioxidant activity.
Fig. 6.
Structure activity relationship of synthesized derivative of morin
Molecular docking study
Newly designed ligands were studied for molecular simulation studies with the help of Schrodinger’s maestro package of molecular docking software [30–34]. Molecules were docked into Jack bean urease crystallographic complex (PDB ID: 3LA4) by Induced fit docking (IFD) method. The predicted binding pattern revealed that synthesized ligand bind within catalytic cavity firmly via. hydrogen bond formation, pi–pi stacking and hydrophobic interaction. Position and alignment of particular substituents on molecules was found to be responsible for perfect binding of ligand with enzyme. In the binding model of the most active compound (M2b) eight hydrogen bonds were noticed between residues Lys 709, Glu 718, Try 32, Val 744, Pro 717, Ash 730, Ser 421 with hydroxyl and NH groups of synthesized ligands. Hydrophobic interactions among ligand and residues Ala 16, Leu 13, Leu 839, Pro 717, Phe 712, Tyr 32, Val 744, Met 746, Val 36, Ala37 was noticed. The second most potent compound (M2i) was found to form six hydrogen bonds among Pro 717, Ser 421, Glu 742, Try 32, Ash 730 Lys 709 with hydroxyl and NH groups, a salt bridge formation was also observed between oxygen and Lys 716 residue. Ligand was found to interact with Phe 712, Pro 717, Tyr 32, Met 746, Val 744, Pro 743, Val 36, Ala 37, Leu 13, Ala 16, Leu 839 residues hydrophobically. Similarly six hydrogen bond formations was observed in inhibitor of third rank (M2a) between Glu 718, Ash 730, Lys 716, Val 744, Met 746 residues and hydroxyl, thiol and NH group of ligand. Docking studies revealed that hydrogen bond interactions fix the ligands firmly and tightly in the active site. In-silico studies and in-vitro results comply with each other, according to the which, compounds M2b, M2i and M2a were found to be most potent ligands. Molecular docking parameters were shown in Table 3 and interaction pattern of ligands within the pocket were described in Table 4.
Table 3.
Docking parameters of the designed derivatives
| Compound | Docking score | Binding energy | Glide hbond | Glide ecoul | Glide evdw |
|---|---|---|---|---|---|
| M2a | − 9.225 | − 61.834 | − 2.212 | − 15.894 | − 45.94 |
| M2b | − 10.977 | − 59.062 | − 2.364 | − 13.986 | − 45.076 |
| M2c | − 7.94 | − 52.778 | − 1.989 | − 4.563 | − 48.214 |
| M2d | − 7.117 | − 48.622 | − 1.073 | − 4.498 | − 44.124 |
| M2e | − 8.306 | − 52.965 | − 1.907 | − 4.693 | − 48.002 |
| M2f | − 7.634 | − 52.788 | − 1.34 | − 5.445 | − 47.343 |
| M2g | − 7.202 | − 55.996 | − 0.83 | − 7.193 | − 48.803 |
| M2h | − 8.576 | − 48.949 | − 1.65 | − 4.621 | − 44.329 |
| M2i | − 10.273 | − 45.27 | − 2.955 | − 11.623 | − 33.6447 |
| M3 | − 8.266 | − 47.885 | − 1.293 | − 12.371 | − 35.514 |
| M4 | − 8.12 | − 54.757 | − 1.81 | − 10.458 | − 44.298 |
| M5 | − 8.306 | − 49.449 | − 1.69 | − 7.449 | − 42 |
| M6 | − 8.536 | − 59.562 | − 1.424 | − 12.371 | − 47.323 |
| Thiourea | − 3.459 | − 21.156 | − 1.484 | − 8.152 | − 13.004 |
| AHA | − 3.049 | − 17.454 | − 1.311 | − 8.523 | − 8.936 |
Table 4.
Protein-ligand interaction details of synthesized derivatives
| Ligands | Interaction diagram | Interaction description |
|---|---|---|
| M2a |
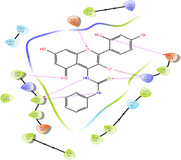
|
Hydrogen bond interaction: Glu 718, Ash 730, Lys 716, Val 744, Met 746 Hydrophobic interaction: Phe 712, Pro 717, Try 32, Val 36, Ala 37, Ala 16, Leu 13, Pro 743, Val 744, Met 746, Leu 839 |
| M2b |
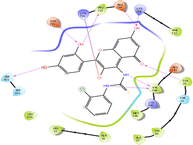
|
Hydrogen bond interaction: Lys 709, Glu 718, Tyr 32, Val 744, Pro 717, Ash 730, Ser 421 Hydrophobic interaction: Pro 717, Phe 712, Tyr 32, Val 744, Val 36, Met 746, Ala 37, Ala 16, Leu 13, Leu 839 |
| M2c |
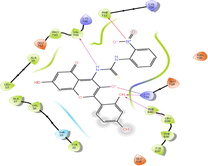
|
Hydrogen bond interaction: Phe 712, Val 744, Lys 716, Leu 839 Hydrophobic interaction: Phe 712, Phe 840, Leu 839, Phe 838, Tyr 837, Val 744, Pro 743, Ala 16, Leu 13, Ala 37, Val, 36, Tyr 32 |
| M2d |
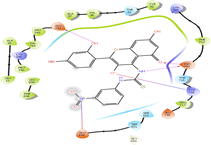
|
Hydrogen bond interaction: Ash 730, Glu 742 Hydrophobic interaction: Phe 712, Pro 717, Tyr 32, Val 36, Ala 37, Pro 743, Val 744, Met 746, Leu 839, Phe 838, Leu 13, Ala 16 |
| M2e |
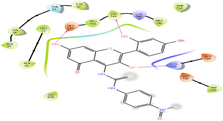
|
Hydrogen bond interaction: Val 744, Glu 742 Hydrophobic interaction: Trp 728, Phe 712, Met 746, Val 744, Pro 743, Leu 839, Tyr 32, Leu 13, Ala 16, Val 36, Ala 37 |
| M2f |
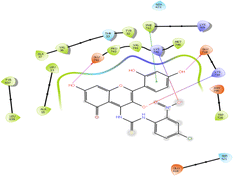
|
Hydrogen bond interaction: Glu 718, Glu 742 Hydrophobic interaction: Trp 728, Met 746, Phe 712, Val 744, Tyr 32, Pro 743, Val 36, Ala 37, Tyr 837, Leu 839 π–π stacking: Phe 712 |
| M2g |
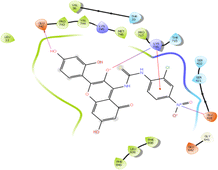
|
Hydrogen bond interaction: Glu 742 Hydrophobic interaction: Pro 717, Phe 838, Leu 839, Phe 840, Met 746, Val 744, Val 36, Pro 743, Leu 13 π–cation interaction: Lys 716 |
| M2h |
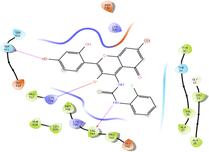
|
Hydrogen bond interaction: Ser 421, Val 744 Hydrophobic interaction: Tyr 32, Leu 13, Val 36, Ala 37, Ala 16, Pro 717, Met 746, Val 744, Pro 743, Phe 838, Leu 839 |
| M2i |
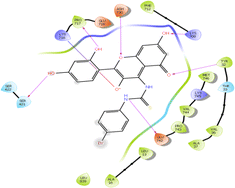
|
Hydrogen bond interaction: Pro 717, Ser 421, Glu 742, Tyr 32, Ash 730, Lys 709 Hydrophobic interaction: Phe 712, Tyr 32, Met 746, Val 36, Val 744, Pro 743, Ala 37, Leu 13, Pro 717 |
| M3 |
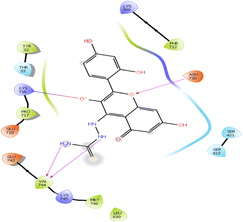
|
Hydrogen bond interaction: Ash 730, Val 744 Hydrophobic interaction: Phe 712, Tyr 32, Pro 717, Val 744, Met 746, Leu 839 |
| M4 |
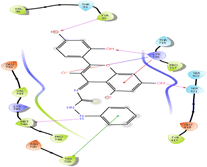
|
Hydrogen bond interaction: Tyr 32, Lys 716, Ser 421, Met 746 Hydrophobic interaction: Pro 717, Tyr 417, Tyr 32, Val 36, Val 744, Met 746, Pro 748, Trp 728 π–π stacking: Trp 728 π–cation interaction: Lys 716 |
| M5 |
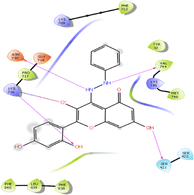
|
Hydrogen bond interaction: Val 744, Ash 730, Lys 716, Ser 421 Hydrophobic interaction: Phe 712, Tyr 32, Val 744, Met 746, Pro 717, Phe 838, Leu 839, Phe 840 |
| M6 |
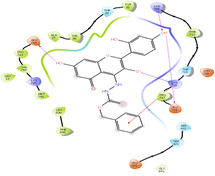
|
Hydrogen bond interaction: Lys 709, Glu 718, Glu 742 Hydrophobic interaction: Phe 712, Pro 717, Tyr 32, Val 36, Ala 37, Pro 743, Val 744, Met 746, Leu 839, Phe 839, Leu 13 π–cation interaction: Pro 717 |
Hydrogen bond formations were considered as most important for perfect fitting of ligand within the enzyme. Each one of molecule demonstrated good docking score from − 7.117 to − 10.977 as compared to − 3.459 and − 3.049 of standard thiourea and Acetohydroxamic acid as well as excellent binding energy ranges from − 45.27 to − 61.834 kJ/mol as compared to − 21.156 kJ/mol and − 17.454 kJ/mol of standard thiourea and Acetohydroxamic acid. Docking studies concluded that designed ligands have excellent binding capability as compared to parent and standard compounds; a correlation has been set up in docking score and IC50 values of synthesized ligands having R2 value 0.7085 has been shown in Fig. 7 signfied that molecule having greater the docking score have lesser value of IC50 for urease inhibition.
Fig. 7.
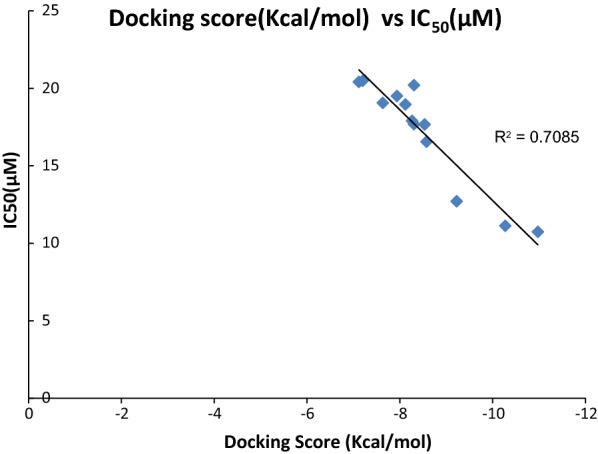
Correlation among docking score and IC50 value of synthesized compounds for urease inhibition studies
Admet studies
In drug discovering the ADME profile of drug like molecules is very important and for this purpose Schrodinger’s maestro molecular modeling package Qikprop module was utilized. The Absorption, distribution, metabolism and excretion details of the designed molecules are given in Table 5.
Table 5.
Simulation studies of derivatives by Quikprop
| Synthesised Ligands | QPlogS | QPlog HERG | QPPCaco | QPlogBB | QPlogKp | HBD | HBA | LogP o/w |
Rule of Five |
|---|---|---|---|---|---|---|---|---|---|
| M2a | − 5.333 | − 6.948 | 85.491 | − 1.979 | 9.89 | 5 | 7 | 2.629 | 1 |
| M2b | − 5.373 | − 6.405 | 96.414 | − 1.705 | 9.83 | 5 | 7 | 2.9 | 1 |
| M2c | − 5.573 | − 6.912 | 9.439 | − 3.243 | 10.254 | 5 | 8 | 1.937 | 2 |
| M2d | -5.267 | − 6.734 | 8.44 | − 3.256 | 9.974 | 5 | 8 | 1.713 | 2 |
| M2e | − 5.366 | − 6.611 | 10.739 | − 3.046 | 10.589 | 5 | 8 | 1.933 | 2 |
| M2f | − 6.14 | − 6.647 | 19.107 | − 2.648 | 10.422 | 5 | 8 | 2.661 | 3 |
| M2g | − 5.831 | − 6.414 | 13.503 | − 2.778 | 11.635 | 5 | 8 | 2.395 | 3 |
| M2h | − 4.782 | − 6.086 | 109.727 | − 1.611 | 10.606 | 5 | 7 | 2.657 | 1 |
| M2i | − 5.646 | − 6.361 | 88.46 | − 1.695 | 9.959 | 5 | 7 | 3.004 | 2 |
| M3 | − 3.201 | − 5.146 | 18.445 | − 2.331 | 11.441 | 7 | 8 | 0.073 | 1 |
| M4 | − 4.438 | − 6.8 | 54.098 | − 2.341 | 10.667 | 7 | 8.25 | 1.869 | 1 |
| M5 | − 5.597 | − 8.051 | 64.909 | − 2.614 | 11.971 | 5 | 6.5 | 2.412 | 1 |
| M6 | − 5.323 | − 7.262 | 20.721 | − 3.082 | 10.963 | 5 | 8 | 2.057 | 1 |
Blood brain barrier partition coefficient (QPlogBB), estimated IC50 value for HERG K+ channels obstruction (log HERG), permeation through skin estimation (QPlogKp), apparent Caco-2 cell permeability estimation in nm/sec (QPPCaco) and apparent MDCK cell permeability estimation in nm/sec (QPPMDCK), partition coefficient in octanol and water Log P, solubility in aqueous media log S, Lipinski’s rule of five. Results revealed that ADME parameters of each ligand within the bounds of satisfactory range without violating Lipinski’s rules.
ADMET calculations showed that synthesised ligands quietly obey rule of five without any considerable violations also many ligands possessed in range values of QPlog S, OPlogHERG, OPPCaco, QPlogBB, QPPMDCK, QPlogKp, QPlogKhsa, HBD, HBA, Log P, % HOA which made them ligands of choice for urease protein [35–39].
Experimental
Materials used
Jack bean urease and morin was purchased from Sigma Aldrich and Himedia respectively. Analytical grade reagents and solvents were utilized as a part of study and obtained locally. Progression of reaction was observed via. Thin layer chromatography and recrystallization of products was done in order to purify the compounds which were again checked for purity by thin layer chromatography (TLC) performed on plates covered with silica gel G. Measurement of melting point was done in open capillary tubes on a melting point apparatus and was uncorrected. The spectral data, IR and 1H NMR, 13CNMR were measured by standard procedures. Brucker 12060280, Germany Software: OPUS 7.2.1394 spectrophotometer in cm−1 was used for recording IR spectra of derivatives and elemental analysis was done on Perkin–Elmer 2400 C, H, N analyzer. The 1HNMR and 13CNMR spectra were recorded in DMSO-d6 on a Brucker DRX-300 FTNMR instrument.
Synthetic procedure
Morin derivatives (M2a–i, M3–6) synthesis was accomplished via. a simple and efficient way by two steps which have been outlined in Scheme 1. The synthesis of compouds M2a–M2i was started by formation of substituted aryl thiourea using different substituted anilines then derivatives of morin were prepared reacting synthesized substituted aryl thiourea/thiosemicarbazide/phenylthiosemicarbazide/phenyl hydrazine/benzylcarbazate solution with equimolar concentration of morin in ethanol in presence of acetic acid as catalyst according to previously reported procedure with slight modification [24–26]. Characterisation of all the synthesized compounds was done by IR, 1 H NMR, 13CNMR and elemental analysis and was found in full accordance with their depicted structures.
General synthetic procedure for preparation of derivatives of morin
Step 1: synthesis of substituted aryl thiourea
Substituted Aniline (0.32 mol) was taken in a 250 ml round bottom flask and, to this, conc. hydrochloric acid (32.19 mL, 0.32 mol) was added dropwise with continues stirring. 100 mL of water was added after appearance of turbidity about after 20 min. followed by addition of ammonium thiocyanate solution (29.42 g, 0.38 mol). This reaction mixture was heated untill the solution becomes turbid, after discontinuing the heating it was poured over ice cold water, and filtration of precipitates was done which were finally dried. Crude product obtained was recrystallized by ethanol.
Step 2: synthesis of schiff bases of morin
Equimolar concentration of Morin (0.01 mmol) and substituted arylthiourea/thiosemicarbazide/phenylthiosemicarbazide/benzylcarbazate (0.01 mmol) were solubilized in ethanol (50 ml). Small amount of glacial acetic acid (1–2 ml) was added to the reaction mixture followed by refluxing for 5–6 h. Reaction completion was monitored by thin layer chromatography. Reaction mixture was concentrated; precipitates formed were filtered off and dried. Recrystallization of crude product was done by ethanol and compounds (M2a–i, M3–6) were obtained.
N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)-N-phenylthiourea (M2a)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.64; Yield = 71.2%; m.p (246–248 °C); IR cm−1 3649 (O–H str), 3523 (N–Hstr), 1651 (C=Cstr), 1614 (C=N str),1539 (N–C=Sstr), 1313 (C–O str), 1172 (C–N str); 1H NMR (400 MHz, DMSO-d6) δ 12.68 (s, 1H, OH), 10.68 (s, 1H, OH), 9.74 (s, 1H, NH), 9.68 (s, 1H, OH), 9.38 (s, 2H Ar–H), 7.53–7.25 (m, 5H, Ar–H), 7.24 (d, 1H, Ar–H), 7.18–6.90 (m, 2H, Ar–H), 6.14–6.30 (m, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 187.81, 166.55, 163.81, 159.55, 157.75, 157.20, 146.11, 139.95, 135.22, 129.13, 127.42–127.13, 123.12, 123.11–122.88, 112.83, 107.64, 104.87, 102.35–102.06, 99.77, 91.34; MS ES + (ToF): m/z 436.07 [M+]; CHNS: Calc (C22H16N2O6S): C, 60.54; H, 3.70; N, 6.42; S, 7.35; Found: C, 60.57; H, 3.69; N, 6.41; S, 7.33.
N-(2-chlorophenyl)-N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)thiourea (M2b)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.66; Yield = 72.8%; m.p (202–204 °C); IR cm−1 3651 (O–H str), 3591 (N–Hstr), 1652 (C=Cstr), 1621 (C=N str), 1541 (N–C=Sstr), 1316 (C–O str), 1174 (C–N str),750 (C–Cl); 1H NMR (400 MHz, DMSO-d6) δ 13.01 (s, 1H, OH), 10.57 (s, 1H, OH), 9.67 (s, 1H, OH), 9.57 (s, 1H, NH), 9.40 (s, 2H, OH), 7.66 (dd, 1H, OH), 7.59–7.33 (m, 3H, Ar–H), 7.05 (m, 1H, Ar–H), 6.62–6.55 (m, 2H, Ar–H), 6.43–6.34 (m, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 188.57, 166.92, 163.11, 159.53, 157.93, 157.18, 142.31, 136.37, 135.13, 130.75–129.18, 128.20, 127.89–127.74, 127.28, 126.07, 124.72–124.43, 113.43, 107.63, 104.85, 101.20–101.07, 97.68, 91.51; MS ES + (ToF): m/z 470.02 [M+]; CHNS: Calc (C22H15ClN2O6S): C, 56.11; H, 3.21; N, 5.95; S, 6.81; Found: C, 56.09; H, 3.22; N, 5.94; S, 6.80.
N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)-N-(2-nitrophenyl)thiourea (M2c)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.62; Yield = 65.7%; m.p (238–240 °C); IR cm−1 3670 (O–H str), 3565 (N–Hstr), 1652 (C=Cstr), 1625 (C=N str), 1571 (N–C=Sstr), 1508 (N–O str), 1344 (C–O str), 1170 (C–N str); 1H NMR (400 MHz, DMSO-d6) δ 12.83 (s, 1H, OH), 10.44 (s, 2H, OH), 9.88 (s, 1H, OH), 9.59 (s, IH, NH), 9.42 (s, 2H, OH), 8.50 (dd, 1H, Ar–H), 8.28 (dd, 1H, Ar–H), 7.71 (td, 1H, Ar–H), 7.37–7.28 (m, 2H, Ar–H), 6.63–6.55 (m, 2H, Ar–H), 6.43–6.34 (m, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 187.91, 166.57, 163.81, 159.94, 157.93, 157.19, 145.11, 137.36, 136.04, 135.18, 132.22, 127.90–127.67, 126.54–126.25, 125.44–125.14, 118.88, 112.43, 107.62, 104.85, 102.36–102.10, 99.68, 91.73; MS ES + (ToF): m/z 481.06 [M+]; CHNS: Calc (C22H15N3O8S): C, 54.88; H, 3.14; N, 8.73; S, 6.66; Found: C, 54.85; H, 3.16; N, 8.72; S, 6.66.
N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)-N-(3-nitrophenyl)thiourea (M2d)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.63; Yield = 71.9%; m.p (240–242 °C); IR cm−1 3690 (O–H str), 3549 (N–Hstr), 1698 (C=Cstr), 1614 (C=N str),1541 (N–C=Sstr), 1507 (N–O) 1302 (C–O str), 1172 (C–N str); 1H NMR (400 MHz, DMSO-d6) δ 13.05 (s, 1H, OH), 10.48 (s, 1H, OH), 9.87 (s, 1H, OH), 9.65 (s, 1H, NH), 9.41 (s, 2H, OH), 8.54–8.48 (m, 1H, Ar–H), 7.96–7.86 (m, 2H, Ar–H), 7.55 (m, 1H, Ar–H), 7.34 (d, 1H, Ar–H), 6.60–6.52 (m, 2H, Ar–H), 6.54–6.36 (m, 2H, Ar–H); 13C NMR 13C NMR (400 MHz, CDCl3)δ = 187.75, 166.57, 163.81, 159.94, 157.93, 157.19, 148.76, 145.11, 140.08, 135.18, 131.13, 128.33, 127.88–127.79, 117.85, 116.90, 112.43, 107.62, 104.85, 102.38–102.10, 99.67, 91.73; MS ES + (ToF): m/z 481.9 [M+]; CHNS: Calc (C22H15N3O8S): C, 54.88; H, 3.14; N, 8.73; S, 6.66; Found: C, 54.86; H, 3.13; N, 8.75; S, 6.65.
N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)-N-(4-nitrophenyl)thiourea (M2e)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.65; Yield = 69.4%; m.p (240–242 °C); IR cm−1 3650 (O–H str), 3523 (N–Hstr), 1656 (C=Cstr), 1611 (C=N str), 1539 (N–C=Sstr), 1508 (N–O), 1307 (C–O str), 1173 (C–N str); 1H NMR (400 MHz, DMSO-d6) δ 13.08 (s, 1H, OH), 11.61 (s, 1H, OH), 11.14 (s, 1H, OH), 10.40 (s, 1H, OH), 9.88 (s, 1H, NH), 8.34–8.26 (m, 2H, Ar–H), 7.78–7.70 (m, 2H, Ar–H), 7.34 (d, 1H, Ar–H), 6.64–6.57 (m, 2H, Ar–H), 6.41–6.32 (m, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 187.83, 166.57, 163.81, 159.94, 157.93, 157.18, 145.11, 143.20, 135.18, 127.95–127.69, 125.73–125.44, 121.40, 112.43, 107.62, 104.85, 102.36–102.09, 99.66, 91.74; MS ES + (ToF): m/z 481.09[M+]; CHNS: Calc (C22H15N3O8S): C, 54.88; H, 3.14; N, 8.73; S, 6.66; Found: C, 54.86; H, 3.15; N, 8.72; S, 6.67.
N-(4-chloro-2-nitrophenyl)-N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)thiourea (M2f)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.69; Yield = 68.5%; m.p (228–230 °C); IR cm−1 3669 (O–H str), 3587 (N–Hstr), 1655 (C=Cstr), 1627 (C=N str),1540 (N–C=Sstr), 1506 (N–O str), 1295 (C–O str), 1174 (C–N str), 791 (C–Cl); 1H NMR (400 MHz, DMSO-d6) δ 13.15 (s, 1H, OH), 12.99 (s, 1H, NH), 11.45 (s, 1H, OH), 11.14 (s, 1H, OH), 10.44 (s, 1H, OH), 9.86 (s, 1H, OH), 8.08 (s, 1H, Ar–H), 7.79 (d, 1H, Ar–H), 7.55 (d, 1H, Ar–H), 7.29 (d, 1H, Ar–H), 6.62–6.55 (m, 2H, Ar–H), 6.41–6.32 (m, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 187.91, 166.57, 163.81, 159.94, 157.93, 157.18, 145.11, 136.04, 135.18, 133.46, 127.95–127.66, 127.51–127.20, 125.21, 112.43, 109.18–30.08; MS ES + (ToF): m/z 515.02 [M+]; CHNS: Calc (C22H14ClN3O8S): C, 51.22; H, 2.74; N, 8.15; S, 6.22; Found: C, 51.24; H, 2.75; N, 8.12; S, 6.20.
N-(2-chloro-4-nitrophenyl)-N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)thiourea (M2 g)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.61; Yield = 70.4%; m.p (226–228 °C); IR cm−1 3649 (O–H str), 3547 (N–Hstr), 1650 (C=Cstr), 1616 (C=N str),1524 (N–C=Sstr), 1502 (N–O str),1304 (C–O str), 1173 (C–N str), 814 (C–Cl str); 1H NMR (400 MHz, DMSO-d6) δ 13.19 (s, 1H, OH), 12.41 (s, 2H, NH), 11.45 (s, 1H, OH), 10.44 (s, 1H, OH), 9.88 (s, 1H, OH), 8.24 (d, 1H, OH), 8.07 (dd, 1H, Ar–H), 7.81 (d, 1H, Ar–H), 7.29 (d, 1H, Ar–H), 6.67–6.55 (m, 2H, Ar–H), 6.41–6.32 (m, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 188.48, 166.57, 163.81, 159.94, 157.93, 157.18, 145.11, 140.54, 139.78, 135.18, 127.92–127.69, 126.10–125.82, 124.63, 123.27–123.01, 122.80, 112.43, 107.62, 104.85, 102.38–102.07, 99.67, 91.74; MS ES + (ToF): m/z 515.22 [M+]; CHNS: Calc (C22H14ClN3O8S): C, 51.22; H, 2.74; N, 8.15; S, 6.22; Found: C, 51.24; H, 2.75; N, 8.12; S, 6.20.
N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)-N-(2-fluorophenyl)thiourea (M2 h)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.71; Yield = 74.2%; m.p (206–208 °C); IR cm−1 3648 (O–H str), 3529 (N–Hstr), 1651 (C=Cstr), 1614 (C=N str), 1539 (N–C=Sstr), 1313 (C–O str), 1204 (C–F str), 1172 (C–N str); 1H NMR (400 MHz, DMSO-d6) δ 13.16 (s, 1H, OH), 11.49 (s,1H, OH), 11.16 (s, 1H, OH), 10.61 (s, 1H, NH), 10.39 (s, 1H, OH), 9.80 (s, 1H, OH), 7.95 (m, 1H, Ar–H), 7.30 (d, 1H, Ar–H), 7.22–6.98 (m, 4H, Ar–H), 6.61–6.56 (m, 2H, Ar–H), 6.41–6.32 (m, 2H, Ar–H); 13C NMR(400 MHz, CDCl3)δ = 188.46, 166.57, 163.81, 159.94, 157.93, 157.18, 145.11, 135.18, 129.03–62.61; MS ES + (ToF): m/z 545.06 [M+]; CHNS: Calc (C22H15FN2O6S): C, 58.15; H, 3.33; N, 6.16; S, 7.06; Found: C, 58.12; H, 3.34; N, 6.14; S, 7.08.
N-(4-bromophenyl)-N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene)thiourea (M2i)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.74; Yield = 67.3%; m.p (222–224 °C); IR cm−1 3628 (O–H str), 3566 (N–Hstr), 1684 (C=Cstr), 1616 (C=N str),1508 (N–C=Sstr), 1369 (C–O str), 1189 (C–N str), 613 (C–Br str); 1H NMR (400 MHz, DMSO-d6) δ 13.11 (s, 1H, OH), 11.45 (s, 1H, OH), 11.34 (s, 1H, NH), 11.14 (s, 1H, OH), 10.47 (s, 1H, OH), 9.88 (s, 1H), 7.56–7.44 (m, 4H, Ar–H), 7.34 (d, 1H, Ar–H), 6.62–6.55 (m, 2H, Ar–H), 6.43–6.34 (m, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 230.03–126.59 (m), 126.59–25.04 (m); MS ES + (ToF): m/z 515.98 [M+]; CHNS: Calc (C22H15BrN2O6S): C, 51.27; H, 2.93; N, 5.44; S, 6.22; Found: C, 51.25; H, 2.90; N, 5.47;; S, 6.20.
(1Z)-1-(3,5,7-trihydroxy-2-(2,4-dihydroxyphenyl)-4H-chromen-4-ylidene)thiosemicarbazide (M3)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.68; Yield = 68.8%; m.p (204–206 °C); IR cm−1 3631 (O–H str), 3521 (N–Hstr), 1650 (C=Cstr), 1623 (C=N str), 1539 (N–C=Sstr), 1319 (C–O str), 1225 (C–N str), 1033 (N–N str); 1H NMR (400 MHz, DMSO-d6) δ 12.21 (s, 1H, OH), 11.70 (s, 1H, NH), 10.12 (s, 1H, OH), 9.95 (s, 1H, OH), 9.88 (s, 1H),9.54 (s, OH, 1H), 8.64 (s, 2H, NH2), 7.59 (t, 1H, Ar–H), 6.59–6.51 (m, 2H, Ar–H), 6.43–6.27 (m, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 180.16, 163.73, 159.94, 157.11, 155.70, 154.25, 143.84, 131.07, 130.47, 127.92–127.71, 111.89, 107.62, 102.38–102.09, 101.83, 99.06, 91.12; MS ES + (ToF): m/z 375.05 [M+]; CHNS: Calc (C16H13N3O6S): C, 51.20; H, 3.49; N, 11.19; S, 8.54; Found: C, 51.20; H, 3.49; N, 11.19; S, 8.54.
(4Z)-4-(3,5,7-trihydroxy-2-(2,4-dihydroxyphenyl)-4H-chromen-4-ylidene)-1-phenylthiosemicarbazide (M4)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.72; Yield = 73.1%; m.p (198–200 °C); IR cm−1 3616 (O–H str), 3562 (N–Hstr), 1651 (C=Cstr), 1601 (C=N str), 1521 (N–C=Sstr), 1338 (C–O str), 1186 (C–N str), 1062 (N–N str); 1H NMR (400 MHz, DMSO-d6) δ 13.21 (s, 1H, OH), 11.45 (s, 1H, OH), 10.44 (s, 1H), 9.85 (s, 1H), 9.68 (s, 1H, OH), 9.46 (s, 1H, OH), 7.34 (d, 1H, Ar–H), 7.23–7.13 (m, 2H, Ar–H), 7.07–6.99 (m, 2H, Ar–H), 6.86 (t, 1H, Ar–H), 6.62–6.55 (m, 2H, Ar–H), 6.43–6.34 (m, 2H), 5.70 (d, 1H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 182.86, 169.47, 163.81, 159.94, 157.93, 157.18, 147.22, 145.11, 135.18, 128.66, 127.95–127.69, 120.66, 114.89, 112.43, 107.62, 104.85, 102.38–102.07, 99.67, 91.74; MS ES + (ToF): m/z 451.08 [M+]; CHNS: Calc (C22H17N3O6S): C, 58.53; H, 3.80; N, 9.31; S, 7.10; Found: C, 58.50; H, 3.82; N, 9.30; S, 7.09.
(4Z)-2-(2,4-dihydroxyphenyl)-4-(2-phenylhydrazinylidene)-4H-chromene-3,5,7-triol (M5)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.75; Yield = 62.8%; m.p (210–212 °C); IR cm−1 3639 (O–H str), 3568 (N–Hstr), 1652 (C=Cstr), 1570 (C=N str), 1363 (C–O str), 1163 (C–N str), 1042 (N–N)str); 1H NMR (400 MHz, DMSO-d6) δ 12.24 (s, 1H, OH), 11.42 (s, 1H, OH), 11.10 (s, 1H, OH), 10.44 (s, 1H, NH), 9.86 (s, 1H, OH), 9.72 (s, 1H, OH), 7.50– 7.42 (m, 2H), 7.33–7.24 (m, 2H, Ar–H), 7.09–7.00 (m, 1H, Ar–H), 6.58–6.51 (m, 2H, Ar–H), 6.40–6.30 (m, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 163.73, 159.94, 157.11, 155.69, 154.25, 143.84, 143.62, 131.08, 130.65, 128.97, 127.93–127.69, 119.95, 116.25–115.92, 111.89, 107.62, 102.38–102.09, 101.86, 99.06, 91.12; MS ES + (ToF): m/z 392.10 [M+]; CHNS: Calc (C21H16N2O6): C, 64.28; H, 4.11; N, 7.14; Found: C, 64.26; H, 4.12; N, 7.13.
(4Z)-2-(2,4-dihydroxyphenyl)-4-(2-phenylhydrazinylidene)-4H-chromene-3,5,7-triol (M6)
Rf = Toluene: Chloroform: methanol (4:4:1) = 0.77; Yield = 58.6%; m.p (250–252 °C); IR cm−1 3670 (O–H str), 3520 (N–H str), 1651 (C=C str), 1618 (C=N str), 1338 (C–O str), 1170 (C–N str), 1078 (N–N str); 1H NMR (400 MHz, DMSO-d6) δ 14.19 (s, 1H, NH), 11.45 (s, 2H, OH), 11.14 (s, 1H, OH), 10.49 (s, 1H, OH), 9.88 (s, 1H, OH), 9.70 (s, 1H, OH), 7.39–7.24 (m, 6H, Ar–H), 6.57 (dd, 2H, Ar–H), 6.43–6.34 (m, 2H, Ar–H), 5.21 (d, 2H, Ar–H); 13C NMR (400 MHz, CDCl3)δ = 163.73, 159.94, 157.11, 155.70, 154.25, 154.02, 143.84, 137.12, 131.07, 130.71, 128.64–113.49, 111.89, 107.62, 102.40–102.07, 101.68, 99.06, 91.12, 66.08; MS ES + (ToF): m/z 450.11 [M+]; CHNS: Calc (C23H18N2O8): C, 61.33; H, 4.03; N, 6.22; Found: C, 61.35; H, 4.01; N, 6.24.
In-silico study protocol
In-silico studies were done to study the interaction pattern of newly designed ligands in the catalytic cavity of jack bean urease enzyme. X-ray crystal structure of Jack Bean Urease enzyme having resolution 2.05 Å was downloaded from Protein Data Bank (http://www.rcsb.org) (PDB code: 3LA4) from the Research Collaboratory for Structural bioinformatics (RCSB). Structure of protein was prepared first of all before docking studies by using pre-process method of Prepwiz module of Schrodinger’s Maestro molecular modeling. Bond orders were assigned, partial charges and hydrogen were added, side chains and loops having missing atoms were fabricated, all waters (with exception of those which were coordinated to metals) were deleted. Structure of proposed Ligands was drawn using Chemdraw Ultra 8.0 (ChemOffice Package) software which was saved as mol. file. Preparation of ligand was done prior to docking by Ligprep module of Schrodinger’s Maestro molecular modeling. Optimization of structure was done by the Gaussian 03 package. Ideal conformation of ligands was acquired by the adding or removing hydrogen bonds, computing partial charges according to the OPLS-2005 force field with 32 stereo isomers, tautomers, and other options like ionization at the physiological pH 7.2 were set as default options. Ligands each conformation was docked within radii of 20 Å into the receptor lattice at the coupling destinations of receptor structures as per their minimum potential energy. Glide/XP by default joined with Induced Fit docking conventions was connected for the forecast of energy of binding and interaction within protein and ligand at the catalytic sites of enzyme. Induced Fit Docking (IFD) methods give the springiness to ligand as well as the residues present at active site all over the docking study where it was rotated in all three angles [37–40]. Finally, the result in Glide score (GScore) scoring function was obtained as the output for each ligand given in Table 3.
Urease inhibitiory assay
Urease enzyme inhibition investigation studies for all synthesized compounds were done by the method developed by Weatherburn [27] named Indophenol against Jack Bean Urease. Briefly, incubation of solution of Jack bean protein 25 μL was done at temperature of 30 °C for a period of 15 min with 55 μL of buffers solution having 100 mM urea and 5 μL of test solution in 96-well plates. 45 μL phenol solution composing phenol 1% w/v and solution of sodium nitroprusside 0.005% w/v and alkali reagent 70 μL composed of sodium hydroxide 0.5% w/v and active sodium hypochloride solution 0.1% were mixed to every well. The measurement of increase in absorption at 625 nm was done after 50 min by help of a microplate reader (Molecular Device, USA). Readings were recorded in triplicate set in a final volume of 200 μL using thiourea as standard. Each assay was performed at pH 8.2 (0.01 M K2HPO4.3H2O, 0.01 M LiCl2 and 1 mM EDTA). Calculation of Inhibition in percentage of synthesized derivatives was done by the formula given as:
where, Acontrol is control absorbance; Asample is test absorbance
Dpph assay for antioxidant activity
DPPH method was adopted for measuring the antioxidant nature of the synthesized derivatives. According to this method synthesized compounds were allowed to react for 0.5 h. at 37 °C with stable free radical, 1, 1-diphenyl-2-picrylhydrazyl radical (DPPH). The DPPH was taken in a concentration of 300 µM. Decrease in value of absorption at 515 nm was measured after the period of incubation using a microplate reader using ascorbic acid as a standard. Molecule which can donate a proton to DPPH and cause its reduction act as an antioxidant. Following reduction deep violet coloured DPPH solution changes to yellow depending upon the nature of antioxidant compound, which brings a measurable decrease in value of absorption at 517 nm. The proton or electron donating capacity of the derivative was estimated from the fading of deep purple colored methanolic solution of 1, 1-diphenylpicrylhydrazyl (DPPH). Incubation of mixture was done at room temperature for 30 min and measurements were done at 517 against blank [28, 29]. Calculation of percentage inhibition was done by following formula:
where, Acontrol is control absorbance; Asample is test absorbance
Anti-H. pylori activity
Novel synthesized derivatives were investigated against H. pylori bacterium (ATCC 43504 DSM 4867, AHA as standard) for their antibacterial activity by using Well diffusion technique. The stock solution of compounds in DMSO (1000 μg/mL) were prepared. Cell suspension was prepared from culture grown on BHI broth. The cell suspensions of all the cultures were adjusted to 1–2 × 105cells/ml. Helicobacter pylori (100 µl) was inoculated by spread plate technique on agar plates (90 mm). Agar wells (5 mm) were made on H. pylori inoculated media and impregnated with 2500 µg of each sample and standard which were incubated @ 35 °C for 24–48 h with 5% CO2 and observed for zone of inhibition around the well. MIC was determined against Helicobacter pylori by micro broth dilution technique as per NCCLS method [41–45].
Kinetic parameters of urease activity
Kinetic parameters i.e. Km, Vmax and Michaelis constant were calculated by verifying the concentration effect of substrate on the rate of reaction of urease enzyme in presence as well as in absence of synthesized compounds. Mechanism behavior of derivatives was studied by dissolving 1% of them in dimethyl sulfoxide and inhibition mode studied by Lineweaver–Burk plots was found to be non-competitive as Km was contant but Vmax changed. Urease enzyme kinetic studies of interaction with synthesized was done GraphPad Prism 7 software.
Statistical analysis
Output of statistical analysis has been depicted in mean ± SEM whereas statistical examination of data collected experimentally was done by one-way analysis of variance (ANOVA). Considerable difference revealed by ANOVA p < 0.05 was regarded significant. Evaluation of statistical data was done by Graph Pad Prism 5.0 Version for Windows (San Diego, CA, USA).
Conclusion
In conclusion, thirteen Morin derivatives have been synthesized successfully via. a simple two step reaction and evaluated for their DPPH- free radical scavenging activity and urease inhibitory activity against jack bean urease. Among the series M2b, M2i and M2a (IC50 110.74 ± 0.018, 11.12 ± 0.033 and 12.71 ± 0.027 µM even two folds more active than standard drug thiourea and also displayed good antioxidant behavior with IC50 value 7.37 ± 0.024, 7.73 ± 0.015 and 7.795 ± 0.003 µM. Moreover these are found to be most potent by having excellent dock score 10.977, − 10.273, − 9.225 and binding energy − 59.062, − 45.27, − 61.834 kJ/mol as compared to standard drugs − 3.459, − 3.049 and − 21.156 kJ/mol and − 17.454 kJ/mol of standard thiourea and Acetohydroxamic acid. These top molecules were again further examined for anti-H. Pylori activity and M2i was found to be more potent as compared to standard drug AHA with MIC = 500 μg/ml and zone of inhibition 15.00 ± 0.00 mm as compared to standard having MIC = 1000 μg/ml and zone of inhibition 9.00 ± 0.50 mm. Derivative M2i can be the potential lead compound in future for treatment of pathologies caused by urease as well as against H. Pylori infection.
Additional file
Additional file 1. Supplementary file for spectral data.
Authors’ contributions
Both RK and AK have equal contributions. Both authors read and approved the final manuscript.
Acknowledgements
We are really thankful to Schrödinger team, especially Vinod Devraji Schrodinger, Inc. (New York, USA) for providing necessary help required to complete the computational work and Skanda life sciences Pvt. Ltd, Banglore for carrying out H. Pylori studies.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request (Additional file 1).
Funding
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ADMET
absorption distribution metabolism excretion
- MIC
minimum inhibitory concentration
- TLC
thin layer chromatography
- UV
ultra violet
- DPPH
2, 2-diphenyl-1-picrylhydrazyl
- SEM
standard error mean
- ANOVA
analysis of variance
- DMSO
dimethyl sulphoxide
- AHA
acetohydroxamic acid
- HOA
human oral absorption
- RCSB
Research Collaboratory for Structural bioinformatics
- NCCLS
National Committee for Clinical Laboratory Standards
References
- 1.Mobley HLT, Hausinger RP. Microbial ureases: significance, regulation and molecular characterization. Microbiol Rev. 1989;53:85–103. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rotini OT. Latrasformazione enzimatica dell' urea nel tereno. Annali Laboratorio Ricerca Spalllanzani. 1935;3:143–154. [Google Scholar]
- 3.Tamaddon F, Ghazi S. Urease: a highly biocompatible catalyst for switchable Biginelli reaction and synthesis of 1, 4-dihydropyridines from the in situ formed ammonia. Catal Commun. 2015;72:63–67. doi: 10.1016/j.catcom.2015.09.006. [DOI] [Google Scholar]
- 4.Mobley HLT, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001;8:505–509. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 6.Jabri E, Carr MB, Hausinger RP, Karplus PA. The crystal structure of urease from Klebsiella aerogenes. Science. 1995;268:998–1004. doi: 10.1126/science.7754395. [DOI] [PubMed] [Google Scholar]
- 7.Jang HJ, Choi MH, Shin WG, Kim KH, Chung YW, Kim KO, Kim HY. Has peptic ulcer disease changed during the past ten years in Korea? A prospective multi-center study. Dig Dis Sci. 2008;53(6):1527–1531. doi: 10.1007/s10620-007-0028-6. [DOI] [PubMed] [Google Scholar]
- 8.Mobley HL, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Rev. 1995;59(3):451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins CM, D’Orazio SE. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol Microbiol. 1993;9:907–913. doi: 10.1111/j.1365-2958.1993.tb01220.x. [DOI] [PubMed] [Google Scholar]
- 10.Sujoy B, Aparna A. Potential clinical significance of urease enzyme. Eur Sci J. 2013 doi: 10.19044/esj.2013.v9n21p%p. [DOI] [Google Scholar]
- 11.Ciurli S, Benini S, Rypniewski WR. Structural properties of the nickel ions in urease: novel insights into the catalytic and inhibition mechanisms. Coord Chem Rev. 1999;192:331–355. doi: 10.1016/S0010-8545(99)00093-4. [DOI] [Google Scholar]
- 12.Li HW, Zou TB, Jia Q. Anticancer effects of morin-7-sulphate sodium, a flavonoid derivative, in mouse melanoma cells. Biomed Pharmacother. 2016;84:909–916. doi: 10.1016/j.biopha.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Galvez J, Coelho G, Crespo ME. Intestinal anti-inflammatory activity of morin on chronic experimental colitis in the rat. Aliment Pharmacol Ther. 2001;15(12):2027–2039. doi: 10.1046/j.1365-2036.2001.01133.x. [DOI] [PubMed] [Google Scholar]
- 14.Fang SH, Hou YC, Chang WC, Hsiu SL, Chao PD, Chiang BL. Morin sulfates/glucuronides exert anti-inflammatory activity on activated macrophages and decreased the incidence of septic shock. Life Sci. 2003;74(6):743–756. doi: 10.1016/j.lfs.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 15.Kim JW, Lee JH, Hwang BY. Morin inhibits Fyn kinase in mast cells and IgE-mediated type I hypersensitivity response in vivo. Biochem Pharmacol. 2009;77(9):1506–1512. doi: 10.1016/j.bcp.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Dilshara MG, Jayasooriya RG, Lee S, Choi YH, Kim GY. Morin downregulates nitric oxide and prostaglandin E 2 production in LPS-stimulated BV2 microglial cells by suppressing NF-κB activity and activating HO-1 induction. Environ Toxicol Pharmacol. 2016;44:62–68. doi: 10.1016/j.etap.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Khazan M, Hdayati M. The role of nitric oxide in health and diseases. Scimetr. 2014;3(1):e20987. doi: 10.5812/scimetr.20987. [DOI] [Google Scholar]
- 18.Paoli P, Cirri P. Caselli A (2013) The insulin-mimetic effect of Morin: a promising molecule in diabetes treatment. Biochim Biophys Acta Gen Subj. 1830;4:3102–3111. doi: 10.1016/j.bbagen.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Lee HS, Jung KH, Hong SW. Morin protects acute liver damage by carbon tetrachloride (CCl4) in rat. Arch Pharmacal Res. 2008;31(9):1160–1165. doi: 10.1007/s12272-001-1283-5. [DOI] [PubMed] [Google Scholar]
- 20.Pal R, Chaudhary MJ, Tiwari PC, Babu S, Pant KK. Protective role of theophylline and their interaction with nitric oxide (NO) in adjuvant-induced rheumatoid arthritis in rats. Int Immunopharmacol. 2015;29(2):854–862. doi: 10.1016/j.intimp.2015.08.031. [DOI] [PubMed] [Google Scholar]
- 21.Mobley HL, Hausinger RP. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev. 1989;53(1):85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Awllia Jalaluddin JAJ, Al-Ghamdi M, Huwait E, Javaid S, Rasheed S, Iqbal Choudhary M. Flavonoids as natural inhibitors of jack bean urease Enzyme. Lett Drug Des Discov. 2016;13(3):243–249. doi: 10.2174/1570180812666150914220050. [DOI] [Google Scholar]
- 23.Aslam MAS, Mahmood SU, Shahid M, Saeed A, Iqbal J. Synthesis, biological assay in vitro and molecular docking studies of new Schiff base derivatives as potential urease inhibitors. Eur J Med Chem. 2011;46(11):5473–5479. doi: 10.1016/j.ejmech.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Ansari MM, Deshmukh SP, Khan R, Musaddiq M. Synthesis antimicrobial and anticancer evaluation of 1-Aryl-5-(o-methoxyphenyl)-2-S-benzyl isothiobiurets. Int J Med Chem. 2014 doi: 10.1155/2014/352626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mughal EU, Ayaz M, Hussain Z, Hasan A, Sadiq A, Riaz M, Malik A, Hussain S, Choudhary MI. Synthesis and antibacterial activity of substituted flavones, 4-thioflavones and 4-iminoflavones. Bioorg Med Chem. 2006;14(14):4704–4711. doi: 10.1016/j.bmc.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 26.Brodowska K, Sykuła A, Garribba E, Łodyga-Chruścińska E, Sójka M. Naringenin Schiff base: antioxidant activity, acid–base profile, and interactions with DNA. Transition Met Chem. 2016;41(2):179–189. doi: 10.1007/s11243-015-0010-7. [DOI] [Google Scholar]
- 27.Weatherburn MW. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39(8):971–974. doi: 10.1021/ac60252a045. [DOI] [Google Scholar]
- 28.Hanif M, Shoaib K, Saleem M, Hasan Rama N, Zaib S, Iqbal J. Synthesis, urease inhibition, antioxidant, antibacterial, and molecular docking studies of 1, 3, 4-oxadiazole derivatives. ISRN Pharmacol. 2012;13:2012. doi: 10.5402/2012/928901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhary MI, Begum A, Abbaskhan A, Musharraf SG, Ejaz A. Two new antioxidant phenylpropanoids from Lindelofia stylosa. Chem Biodivers. 2008;5(12):2676–2683. doi: 10.1002/cbdv.200890221. [DOI] [PubMed] [Google Scholar]
- 30.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 31.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. Enrichment factors in database screening. J Med Chem. 2004;47(7):1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 32.Glide, version 6.6, Schrödinger, LLC, New York, NY, 2015
- 33.Lee SK, Mbwambo ZH, Chung Luyengi L, Gamez EJ, Mehta RG, Kinghorn AD, Pezzuto JM. Evaluation of the antioxidant potential of natural products. Comb Chem High Throughput Screening. 1998;1(1):35–46. [PubMed] [Google Scholar]
- 34.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Exp Op Drug Disc. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godschalk F, Genheden S, Söderhjelm P, Ryde U. Comparison of MM/GBSA calculations based on explicit and implicit solvent simulations. Phys Chem Chem Phy. 2013;15:7731–7739. doi: 10.1039/c3cp00116d. [DOI] [PubMed] [Google Scholar]
- 36.Sengupta D, Verma D, Naik PK. Docking-MM-GB/SA and ADME screening of HIV-1 NNRTI inhibitor: nevirapine and its analogues. Silico Biol. 2008;8:275–289. [PubMed] [Google Scholar]
- 37.Cecchelli R, Berezowski V, Lundquist S, Culot M, Renftel M, Dehouck MP, Fenart L. Modelling of the blood-brain barrier in drug discovery and development. Nat Rev Drug Discov. 2007;6:650–661. doi: 10.1038/nrd2368. [DOI] [PubMed] [Google Scholar]
- 38.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2012;64:4–17. doi: 10.1016/j.addr.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 40.Hanif M, Saleem M, Hussain MT, Rama NH, Zaib S, Aslam MA, Jones PG, Iqbal J. Synthesis, urease inhibition, antioxidant and antibacterial studies of some 4-amino-5-aryl-3H-1, 2, 4-triazole-3-thiones and their 3, 6-disubstituted 1, 2, 4-triazolo [3, 4-b] 1, 3, 4-thiadiazole derivatives. J Braz Chem Soc. 2012;23(5):854–860. doi: 10.1590/S0103-50532012000500010. [DOI] [Google Scholar]
- 41.Xiao ZP, Shi DH, Li HQ, Zhang LN, Xu C, Zhu HL. Polyphenols based on isoflavones as inhibitors of Helicobacter pylori urease. Bioorg Med chem. 2007;15(11):3703–3710. doi: 10.1016/j.bmc.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 42.Li HQ, Xiao ZP, Yan T, Lv PC, Zhu HL. Amines and oximes derived from deoxybenzoins as Helicobacter pylori urease inhibitors. Eur J Med Chem. 2009;44(5):2246–2251. doi: 10.1016/j.ejmech.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Xiao ZP, Wang XD, Peng ZY, Huang S, Yang P, Li QS, Zhou LH, Hu XJ, Wu LJ, Zhou Y, Zhu HL. Molecular docking, kinetics study, and structure–activity relationship analysis of quercetin and its analogous as Helicobacter pylori urease inhibitors. J Agric Food Chem. 2012;60(42):10572–10577. doi: 10.1021/jf303393n. [DOI] [PubMed] [Google Scholar]
- 44.Xiao ZP, Peng ZY, Dong JJ, He J, Ouyang H, Feng YT, Lu CL, Lin WQ, Wang JX, Xiang YP, Zhu HL. Synthesis, structure–activity relationship analysis and kinetics study of reductive derivatives of flavonoids as Helicobacter pylori urease inhibitors. Eur J Med Chem. 2013;63:685–695. doi: 10.1016/j.ejmech.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 45.Numao N, Iwahori A, Hirota Y, Sasatum M, Kondo I, Onimura K, Sampe R, Yamane S, Itoh S, Katoh T, Kobayashi S. Antibacterial activity of two alkylamines integrated an indane scaffold: mimicry of a complementary unit on magainin. Biol Pharm Bull. 1997;20(7):800–804. doi: 10.1248/bpb.20.800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary file for spectral data.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request (Additional file 1).