Abstract
A guide RNA (gRNA) directs the function of a CRISPR protein effector to a target gene of choice, providing a versatile programmable platform for engineering diverse modes of synthetic regulation (edit, silence, induce, bind). However, the fact that gRNAs are constitutively active places limitations on the ability to confine gRNA activity to a desired location and time. To achieve programmable control over the scope of gRNA activity, here we apply principles from dynamic RNA nanotechnology to engineer conditional guide RNAs (cgRNAs) whose activity is dependent on the presence or absence of an RNA trigger. These cgRNAs are programmable at two levels, with the trigger-binding sequence controlling the scope of the effector activity and the target-binding sequence determining the subject of the effector activity. We demonstrate molecular mechanisms for both constitutively active cgRNAs that are conditionally inactivated by an RNA trigger (ON → OFF logic) and constitutively inactive cgRNAs that are conditionally activated by an RNA trigger (OFF → ON logic). For each mechanism, automated sequence design is performed using the reaction pathway designer within NUPACK to design an orthogonal library of three cgRNAs that respond to different RNA triggers. In E. coli expressing cgRNAs, triggers, and silencing dCas9 as the protein effector, we observe a median conditional response of ≈4-fold for an ON → OFF “terminator switch” mechanism, ≈15-fold for an ON → OFF “splinted switch” mechanism, and ≈3-fold for an OFF → ON “toehold switch” mechanism; the median crosstalk within each cgRNA/trigger library is <2%, ≈2%, and ≈20% for the three mechanisms. To test the portability of cgRNA mechanisms prototyped in bacteria to mammalian cells, as well as to test generalizability to different effector functions, we implemented the terminator switch in HEK 293T cells expressing inducing dCas9 as the protein effector, observing a median ON → OFF conditional response of ≈4-fold with median crosstalk of ≈30% for three orthogonal cgRNA/trigger pairs. By providing programmable control over both the scope and target of protein effector function, cgRNA regulators offer a promising platform for synthetic biology.
Short abstract
Conditional guide RNAs (cgRNAs) change conformation in bacterial or mammalian cells to enable conditional regulation of a target gene of choice depending on the presence or absence of an RNA trigger.
Introduction
Dynamic RNA nanotechnology holds great promise as a paradigm for introducing synthetic regulatory links into living cells and organisms. We envision small conditional RNAs (scRNAs) that, upon detection of a programmable nucleic acid input, change conformation to produce a programmable output that up-regulates or down-regulates the activity of a biological pathway. In this scenario, the input controls the scope of regulation, and the output controls the target of regulation, with the scRNA performing signal transduction to create a logical link between the two.1,2 Any pathway that recognizes RNA is a potential candidate for conditional regulation by scRNAs (e.g., RNA interference, RNase H, PKR, RIG-1); the CRISPR/Cas pathway is a particularly attractive candidate because of its functional versatility, high regulatory dynamic range, and portability between species.3−5
The repurposing of RNA-guided CRISPR effectors through development of modified guide RNAs (gRNAs) and CRISPR-associated (Cas) proteins has yielded a suite of powerful tools for biological research and synthetic biology. Precision genome editing has been achieved in a variety of organisms using gRNAs to direct the nuclease activity of Cas9 and Cas12a (Cpf1) to a target gene of choice.3,6−8 Mutation of the nuclease domains to produce a catalytically dead Cas9 (dCas9) has enabled silencing of genetic expression via inhibition of transcriptional elongation,4,9 or induction (or silencing) of genetic expression using dCas9 fusions that incorporate transcriptional regulatory domains.5 Other dCas9 fusions have mediated target-binding to enable visualization of genomic loci,10,11 epigenetic modification,12 and single-base editing at a specific genomic locus.3,13 Hence, gRNA:effector complexes combine the benefits of the rich functional vocabulary of the protein effector (edit, silence, induce, bind) and the programmability of the gRNA in targeting effector activity to a gene of choice.
Because gRNAs are constitutively active, additional measures are needed to restrict effector activity to a desired location and time. Temporal control can be achieved by small-molecule induction of gRNAs14,15 or Cas9,16 but this comes with limitations in terms of multiplexing and spatial control. Spatiotemporal control has been achieved by regulation of Cas9 via photoactivation17 or via tissue-specific promoters18,19 or microRNAs,20 which comes with the unwelcome restriction that all gRNAs are subject to the same regulatory scope. Systematic mapping of the structure and sequence properties of functional gRNAs has revealed that Cas9 activity is tolerant to significant modifications to the standard gRNA structure,21,22 facilitating introduction of auxiliary domains that enable conditional control of gRNA activity via structural changes induced by small-molecules,23−25 protein-bound RNAs,26 nucleases,27 or nuclease-recruiting DNAs.27 Alternatively, the activity of standard gRNAs has been modulated by antisense RNAs28 or by photolysis of antisense DNAs incorporating photocleavable groups.29 For generality, it is highly desirable to control the regulatory scope in a manner that is both conditional and programmable, a tantalizing prospect central to the proposed scRNA paradigm based on dynamic RNA nanotechnology.
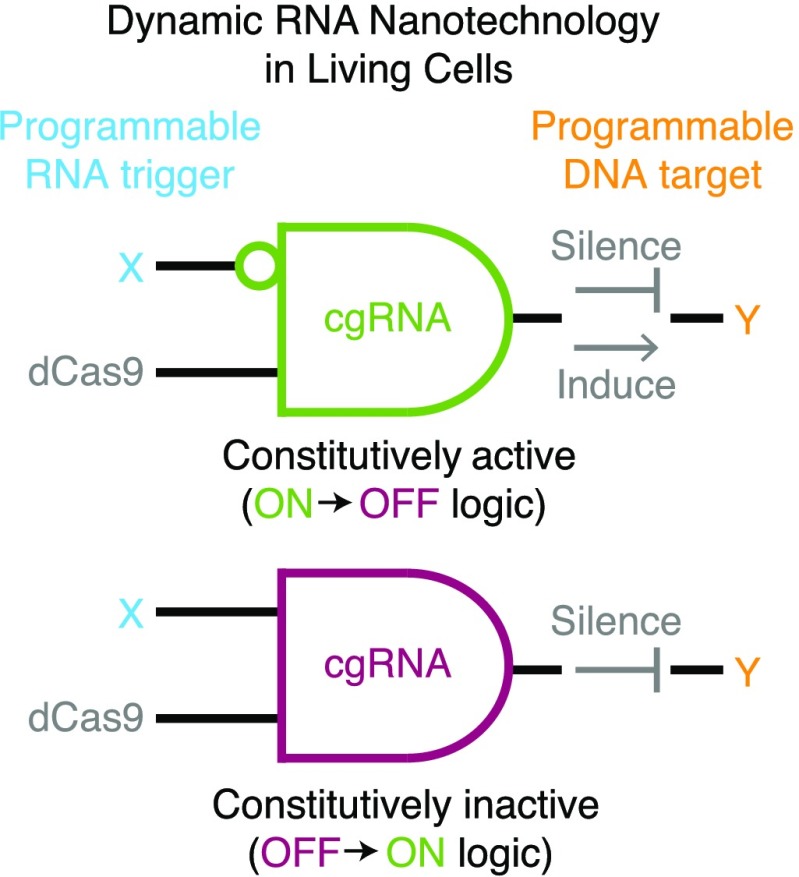
With this paradigm in mind, we set out to engineer conditional guide RNAs (cgRNAs) that change conformation in response to an RNA trigger X to conditionally direct the function of dCas9 to a target gene Y. Unlike a standard gRNA, a cgRNA is programmable at two levels, with the trigger-binding sequence controlling the scope of cgRNA activity and the target-binding sequence determining the subject of effector activity. Functionally, the cgRNA must perform sequence transduction between X and Y as well as shape transduction between active/inactive conformations. In principle, cgRNA activity can be engineered to toggle either OFF → ON (as was recently demonstrated by Siu and Chen30) or ON → OFF in response to a cognate RNA trigger X; this conditional control can be exerted over dCas9 variants that either edit, silence, induce, or bind the target Y, emphasizing the broad functional potential available via interplay between cgRNA logic and protein effector function (Figure 1a). For example, by selecting an endogenous transcript X with a desired spatiotemporal expression profile during development, the downstream regulatory effect on target Y could be restricted to a desired tissue and developmental stage within a model organism (Figure 1b). Alternatively, in a therapeutic context, X could be a disease marker and Y an independent therapeutic target, enabling selective treatment of diseased cells leaving healthy cells untouched.
Figure 1.
Programmable regulators. (a) A conditional guide RNA (cgRNA) changes conformation in response to a programmable trigger X to conditionally direct the activity of a protein effector to a programmable target Y. Top: a constitutively active cgRNA is conditionally inactivated by X (ON → OFF logic). Bottom: a constitutively inactive cgRNA is conditionally activated by X (OFF → ON logic). (b) Molecular logic of programmable regulation using a standard gRNA (“not Y”) vs programmable conditional regulation using a cgRNA (“if X then not Y”). In this conceptual illustration, the standard gRNA silences Y in all tissues, while the cgRNA silences Y only in tissues where and when X is expressed, exerting spatiotemporal control over regulation. (c) A standard guide RNA (gRNA) is constitutively active, directing the function of protein effector dCas9 to a target gene Y; different dCas9 variants implement different functions (edit, silence, induce, bind). From 5′ to 3′, a standard gRNA comprises a target-binding region, a Cas9 handle recognized by the protein effector, and a terminator region.
Results and Discussion
Constitutively Active Terminator Switch cgRNAs (ON → OFF Logic) with Silencing dCas9 in Bacteria
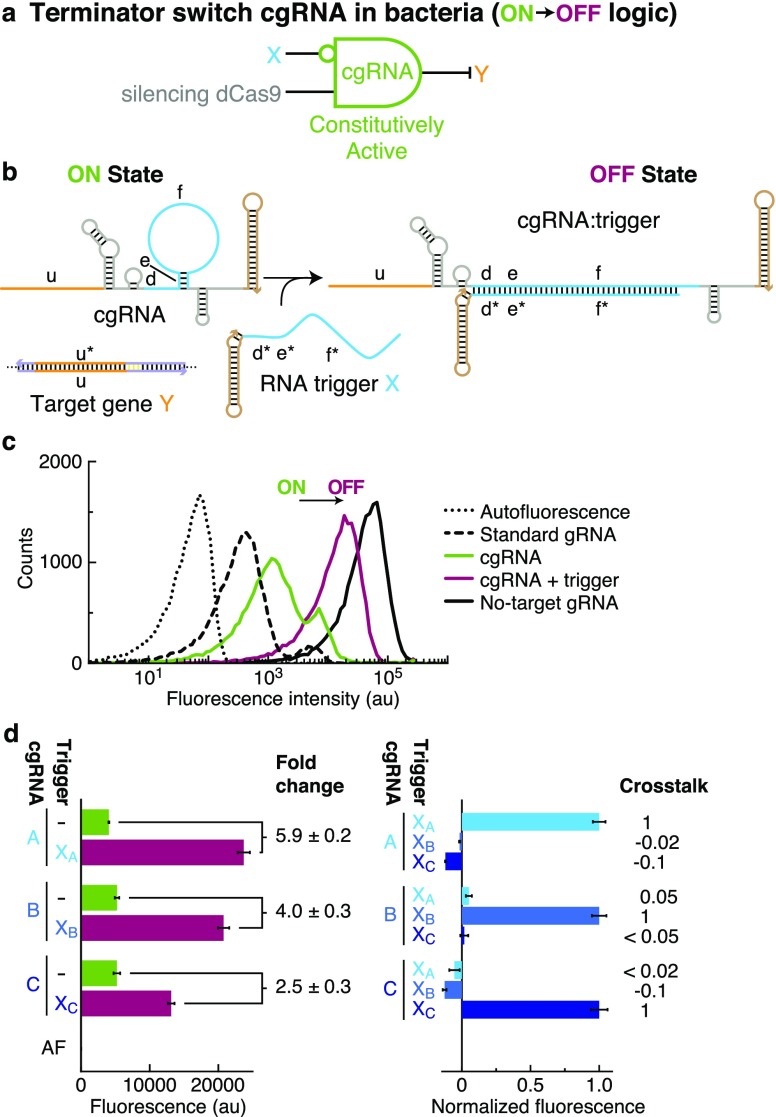
As a starting point, consider the constitutively active “terminator switch” cgRNA mechanism of Figure 2b that is conditionally inactivated by RNA trigger X (ON → OFF logic). Compared to a standard gRNA (Figure 1c), the cgRNA has a modified terminator region with an extended loop and rationally designed sequence domains “d–e–f”. Hybridization of the RNA trigger X to these modified domains is intended to form a structure incompatible with cgRNA mediation of dCas9 function. We validated the cgRNA mechanism in vivo in E. coli expressing silencing dCas94 as the protein effector and a fluorescent protein reporter (mRFP) as the target gene Y (conditional logic: “if not X then not Y”; Figure 2a). An E. coli strain expressing the cgRNA exhibits low fluorescence (ON state) while a strain expressing both the cgRNA and the cognate RNA trigger exhibit high fluorescence (OFF state), achieving a conditional ON → OFF response (Figure 2c). With the terminator switch mechanism, the sequences of the RNA trigger X and the silencing target Y are fully independent, with the cgRNA mediating allosteric regulation—the trigger down-regulates cgRNA:dCas9 function not by sequestering the target-binding region (orange in Figure 2b) but by hybridizing to the distal trigger-binding region (blue). To test programmability, we used NUPACK31,32 to design a library of three orthogonal cgRNA/trigger pairs (Figure 2d), achieving a median ≈4-fold conditional ON → OFF response to expression of the cognate trigger (left) and median crosstalk below 2% between noncognate cgRNA/trigger combinations (right). Ideally, a cgRNA would have a strong ON state with activity equivalent to a standard gRNA (ideal ON state) and a clean OFF state with minimal activity equivalent to a no-target gRNA lacking the target-binding region (ideal OFF state). For this cgRNA mechanism, there is room for improvement in both the ON and OFF states (Figure 2c and Table S13a). The bimodality of the fluorescence distributions observed for both the standard gRNA control strain and the cgRNA-only strain (Figure 2c) is a property of the assay and not of the terminator switch mechanism; the same gRNA and cgRNA sequences yield unimodal fluorescence distributions in E. coli strains created using a different plasmid layout (Figure S34a).
Figure 2.
Constitutively active terminator switch cgRNAs (ON → OFF logic) with silencing dCas9 in bacteria. (a) Conditional logic: if not X then not Y. (b) cgRNA mechanism: the constitutively active cgRNA is inactivated by hybridization of RNA trigger X. Rational sequence design of cgRNA terminator region (domains “d–e–f” comprising 6 nt linker, 4 nt stem, 30 nt loop) and complementary trigger region (domains “f*–e*–d*”). (c) Expression of RNA trigger X (40 nt unstructured + synthetic terminator hairpin) toggles the cgRNA from ON → OFF, leading to an increase in fluorescence. Single-cell fluorescence intensities via flow cytometry. Induced expression (aTc) of silencing dCas9 and constitutive expression of mRFP target gene Y and either: standard gRNA (ideal ON state), cgRNA (ON state), cgRNA + RNA trigger X (OFF state; trigger expression is IPTG-induced), no-target gRNA that lacks target-binding region (ideal OFF state). Autofluorescence (AF): cells with no mRFP. (d) Programmable conditional regulation using 3 orthogonal cgRNAs (A, B, C). Left: raw fluorescence depicting ON → OFF conditional response to cognate trigger (fold change = OFF/ON = [cognate trigger–AF]/[no trigger–AF]). Right: normalized fluorescence depicting orthogonality between noncognate cgRNA/trigger pairs (crosstalk = [noncognate trigger–no trigger]/[cognate trigger–no trigger]). Bar graphs depict mean ± estimated standard error calculated based on the mean single-cell fluorescence over 20 000 cells for each of N = 3 replicate wells (fold change and crosstalk calculated with uncertainty propagation).
Single and Double Sequence Inserts for Construction of Allosteric cgRNAs in Bacteria
Seeking to improve cgRNA performance for ON → OFF conditional logic, we undertook a systematic study of single-stranded sequence inserts into the standard gRNA structure, seeking to identify inserts that satisfied two key properties: (1) strong ON state – inserts well-tolerated by dCas9; (2) clean OFF state – cgRNA inactivated by hybridization of complementary trigger to inserted domains. We created a total of 71 E. coli strains to test designed sequence inserts for each of three lengths (15, 25, 35 nt) at each of four insert sites (5′-extension, Cas9 handle loop, terminator loop 1, terminator loop 2; Figure S39) or at pairwise combinations of insert sites. Each of these modified gRNAs represented a candidate allosteric cgRNA mechanism, as the trigger sequence X is fully independent of the target gene Y. Interestingly, all of the single and double inserts were well-tolerated by dCas9 with a strong ON state comparable to the standard gRNA, but most inserts did not mediate effective silencing when the cognate trigger was expressed (Figure S39 and Table S16). A notable exception was the modified gRNA with 35 nt inserts in both the dCas9 handle loop and one of the terminator loops, providing the basis for the “splinted switch” cgRNA mechanism presented next.
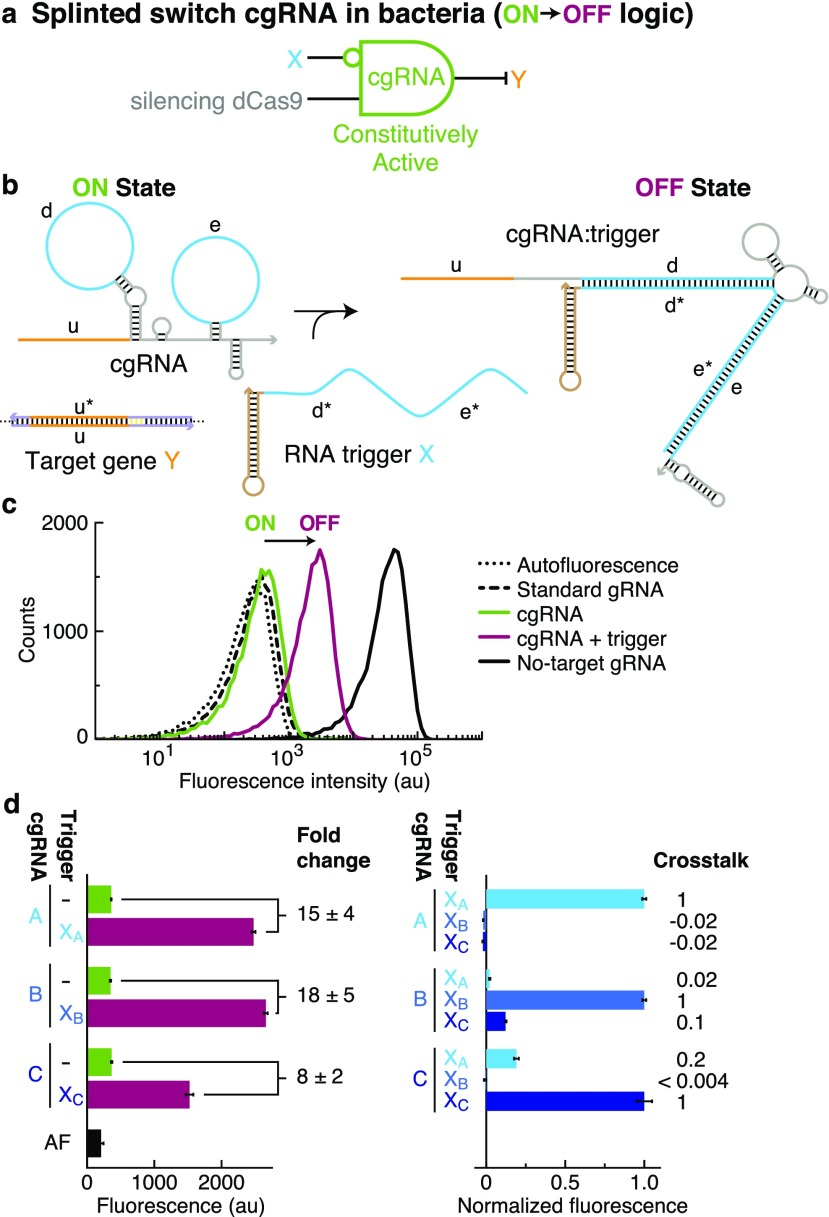
Constitutively Active Splinted Switch cgRNAs (ON → OFF Logic) with Silencing dCas9 in Bacteria
The constitutively active “splinted switch” cgRNA mechanism (Figure 3b) has extended loops in both the Cas9 handle (domain “d”) and terminator (domain “e”). Hybridization of RNA trigger X to both loops is intended to form a splint that is structurally incompatible with cgRNA mediation of dCas9 function. In E. coli expressing silencing dCas9 and a fluorescent protein reporter (sfGFP) as the target gene Y (conditional logic: “if not X then not Y”; Figure 3a), the splinted switch exhibits a conditional ON → OFF response to expression of RNA trigger X (Figure 3c). Examining a library of three orthogonal splinted switch cgRNA/trigger pairs designed using NUPACK (Figure 3d), we observe a median ≈15-fold ON → OFF conditional response to expression of the cognate trigger and median crosstalk of ≈2% between noncognate cgRNA/trigger combinations. As expected from our insert studies (Figure S39 and Table S16), splinted switch cgRNAs exhibit a strong ON state comparable to the ideal ON state of a standard gRNA, and the OFF state could still be improved relative to the ideal OFF state of a no-target gRNA lacking the target-binding region (Figure 3c and Table S13b). As with the terminator switch mechanism, splinted switch cgRNAs are allosteric regulators—the trigger down-regulates cgRNA:dCas9 function by hybridizing to extended loops (blue in Figure 3b) distal to the target-binding region (orange). The resulting full sequence independence between RNA trigger X and target gene Y provides the flexibility for X to control regulatory scope independent of the choice of Y.
Figure 3.
Constitutively active splinted switch cgRNAs (ON → OFF logic) with silencing dCas9 in bacteria. (a) Conditional logic: if not X then not Y. (b) cgRNA mechanism: the constitutively active cgRNA is inactivated by hybridization of RNA trigger X. Rational sequence design of the 35 nt Cas9 handle loop (domain “d”) and an extended 35 nt terminator hairpin loop (domain “e”). (c) Expression of RNA trigger X (70 nt unstructured + synthetic terminator hairpin) toggles the cgRNA from ON → OFF, leading to an increase in fluorescence. Single-cell fluorescence intensities via flow cytometry. Induced expression (aTc) of silencing dCas9 and constitutive expression of sfGFP target gene Y and either: standard gRNA (ideal ON state), cgRNA (ON state), cgRNA + RNA trigger X (OFF state), or no-target gRNA that lacks target-binding region (ideal OFF state). Autofluorescence (AF): cells with no sfGFP. (d) Programmable conditional regulation using 3 orthogonal cgRNAs (A, B, C). Left: raw fluorescence depicting ON → OFF conditional response to cognate trigger (fold change = OFF/ON = [cognate trigger–AF]/[no trigger–AF]). Right: normalized fluorescence depicting orthogonality between noncognate cgRNA/trigger pairs (crosstalk = [noncognate trigger–no trigger]/[cognate trigger–no trigger]). Bar graphs depict mean ± estimated standard error calculated based on the mean single-cell fluorescence over 20 000 cells for each of N = 3 replicate wells (fold change and crosstalk calculated with uncertainty propagation).
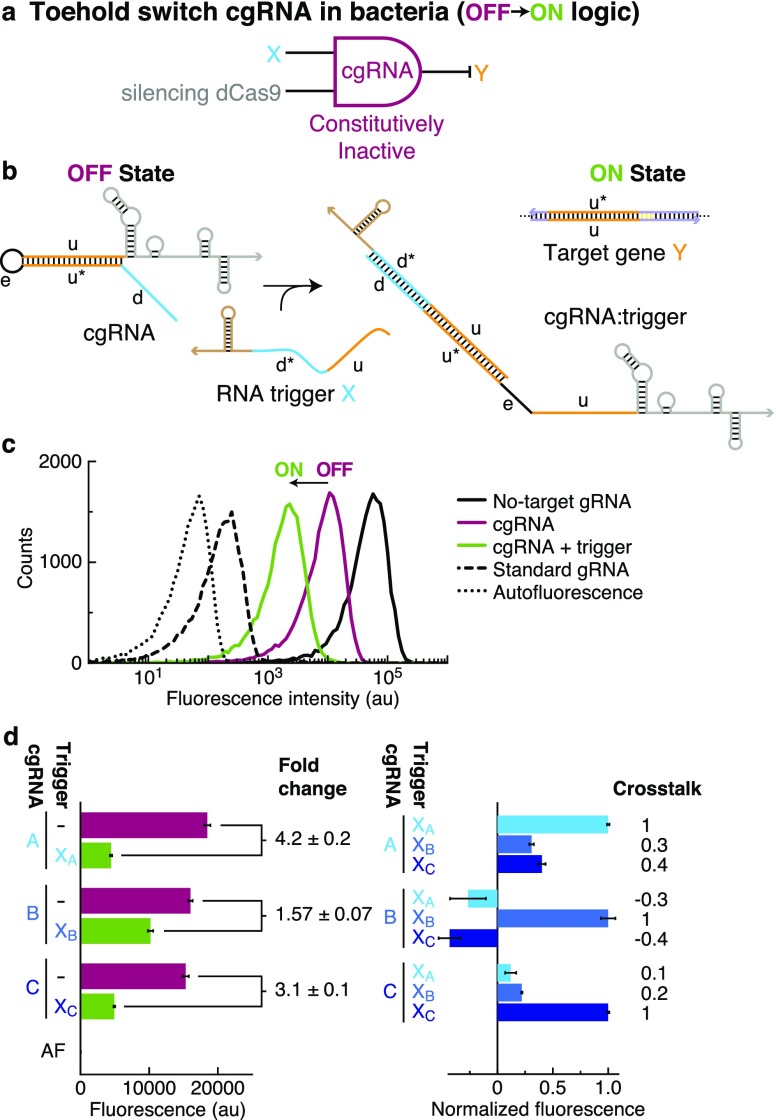
Constitutively Inactive Toehold Switch cgRNAs (OFF → ON Logic) with Silencing dCas9 in Bacteria
To reverse the conditional logic, we then tested a constitutively inactive “toehold switch” cgRNA mechanism (Figure 4b) that is conditionally activated by RNA trigger X (OFF → ON logic). The target-binding region of the cgRNA (domain “u”) is initially sequestered by a 5′ extension to inhibit recognition of target gene Y; hybridization of trigger X to this extension is intended to desequester the target-binding region and enable cgRNA direction of dCas9 function to target gene Y. In E. coli expressing silencing dCas9 and a fluorescent protein reporter (mRFP) as the target gene Y (conditional logic: “if X then not Y”; Figure 4a), the toehold switch cgRNA exhibits a conditional OFF → ON response to expression of RNA trigger X (Figure 4c). In this case, the OFF state is imperfect relative to the ideal OFF state (no-target gRNA control), and the ON state is imperfect relative to the ideal ON state (standard gRNA control) (Figure 4c and Table S13c). For a library of three orthogonal toehold switch cgRNA/trigger pairs designed using NUPACK (Figure 4d), we observe a median ≈3-fold OFF → ON conditional response to expression of the cognate trigger and median crosstalk of ≈20% between noncognate cgRNA/trigger combinations. Recently, Siu and Chen demonstrated a median ≈6.6-fold OFF → ON conditional response using toehold switch cgRNAs with subtly different structural details in the sequestration of the target-binding region.30 Unlike the terminator switch and splinted switch mechanisms for ON → OFF logic, toehold switch cgRNAs for OFF → ON logic are not allosteric, as the cgRNA initially down-regulates cgRNA:dCas9 function by sequestering the target-binding region (orange domain “u” in Figure 4b) with a portion of the trigger-binding region (orange domain “u*”). As a result, the toehold switch cgRNAs offer only partial sequence independence between the trigger X and the target gene Y (“u” is a subsequence of both X and Y). This partial sequence dependence is not necessarily limiting for synthetic biology applications where the trigger can be rationally designed and expressed exogenously but does pose a limitation in situations where X and Y are both endogenous sequences.
Figure 4.
Constitutively inactive toehold switch cgRNAs (OFF → ON logic) with silencing dCas9 in bacteria. (a) Conditional logic: if X then not Y. (b) cgRNA mechanism: the constitutively inactive cgRNA is activated by hybridization of RNA trigger X. Rational sequence design of the toehold (domain “d”; 15 nt) and loop (domain “e”; 8 nt) flanking the sequestration domain “u*” (20 nt). (c) Expression of RNA trigger X (35 nt unstructured + synthetic terminator hairpin) toggles the cgRNA from OFF → ON, leading to a decrease in fluorescence. Single-cell fluorescence intensities via flow cytometry. Induced expression (aTc) of silencing dCas9 and constitutive expression of mRFP target gene Y and either: no-target gRNA that lacks target-binding region (ideal OFF state), cgRNA (OFF state), cgRNA + RNA trigger X (ON state), or standard gRNA (ideal ON state). Autofluorescence (AF): cells with no mRFP. (d) Programmable conditional regulation using 3 orthogonal cgRNAs (A, B, C). Left: raw fluorescence depicting OFF → ON conditional response to cognate trigger (fold change = OFF/ON = [no trigger–AF]/[cognate trigger–AF]). Right: normalized fluorescence depicting orthogonality between noncognate cgRNA/trigger pairs (crosstalk = [noncognate trigger–no trigger]/[cognate trigger–no trigger]). Bar graphs depict mean ± estimated standard error calculated based on the mean single-cell fluorescence over 20 000 cells for each of N = 3 replicate wells (fold change and crosstalk calculated with uncertainty propagation).
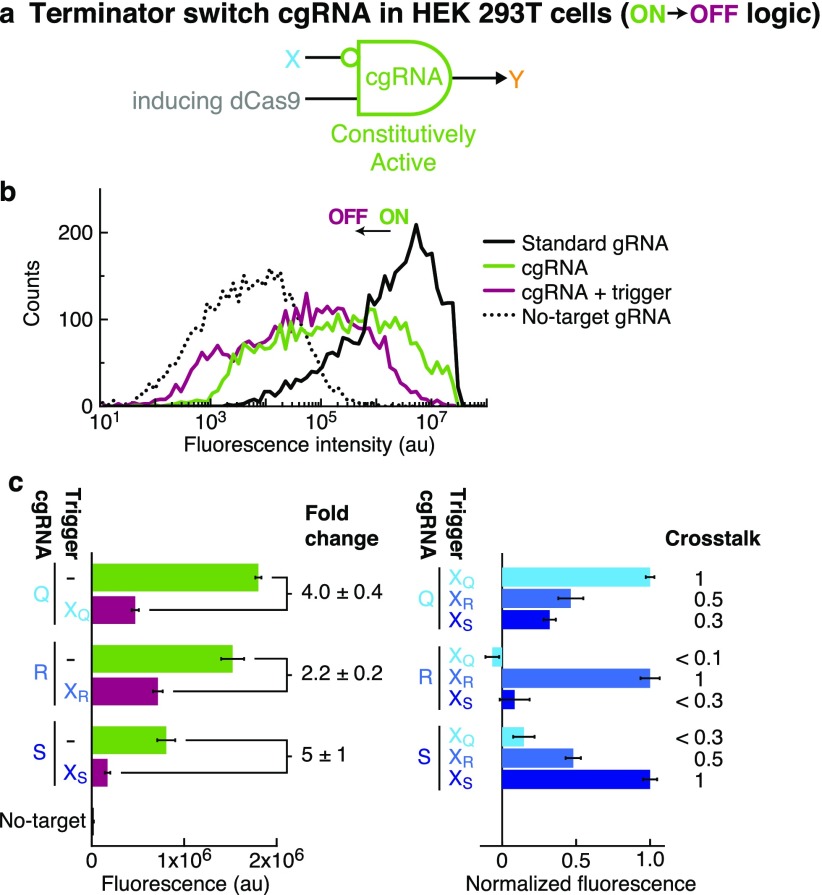
Constitutively Active Terminator Switch cgRNAs (ON → OFF Logic) with Inducing dCas9 in Mammalian Cells
To test the portability of cgRNAs prototyped in bacteria, we migrated the constitutively active terminator switch cgRNA mechanism (ON → OFF logic) to mammalian cells. Moreover, to test generalizability to different effector functions, for mammalian studies we employed inducing rather than silencing dCas9. In HEK 293T cells expressing the cgRNA, inducing dCas9-VPR as the protein effector33 (in contrast to the silencing dCas9 tested in E. coli; cf. Figure 2), and a fluorescent protein reporter (dTomato)34,35 as the target gene Y, we expect fluorescence to decrease with expression of the RNA trigger X (conditional logic: “if not X then Y”; Figure 5a), and indeed we observe this conditional ON → OFF response (Figure 5b). A library of three orthogonal terminator switch cgRNA/trigger pairs designed using NUPACK (Figure 5c) exhibits a median ≈4-fold ON → OFF conditional response to expression of the cognate trigger and median crosstalk of ≈30% between noncognate cgRNA/trigger pairs. The strength of the mean conditional response is similar to that for the bacterial terminator switch (compare the left bar graphs of Figures 2d and 5c), but the distributions for the bacterial strains are more sharply peaked and hence better separated (Figure 2c and Figures S26 and S27) compared to those for mammalian cells transiently transfected with a mixture of four plasmids (Figure 5b and Figures S32a and S33a). The replicate histograms of Figures S32a and S33a show a consistent shift to the left (lower fluorescence) at the high end of the distribution for the OFF state (cgRNA + cognate trigger) relative to the ON state (cgRNA-only or cgRNA + noncognate trigger), contributing to a measurable mean conditional response (Figure 5c) despite the large overlap in distributions. To further assess the significance of this shift, Figures S32b and S33b display the corresponding empirical cumulative distribution functions (ECDFs) with bootstrapped 95% confidence intervals.36,37 The confidence intervals are tight around the ECDFs, and the OFF state replicates (cgRNA + cognate trigger) exhibit a consistent shift to the left (lower fluorescence) at the top right corner of the ECDFs relative to the ON state replicates (cgRNA-only or cgRNA + noncognate trigger), supporting the interpretation that the shift is significant. Further improvement in the mammalian cgRNAs and/or the mammalian assay will be needed to better separate the ON and OFF state distributions.
Figure 5.
Constitutively active terminator switch cgRNAs (ON → OFF logic) with inducing dCas9 in mammalian cells. (a) Conditional logic: if not X then Y. See Figure 2b for cgRNA mechanism: the constitutively active cgRNA is inactivated by hybridization of RNA trigger X (note that the mammalian cgRNA and trigger do not include the depicted synthetic terminator hairpins). (b) Expression of RNA trigger X (40 nt + hU6 terminator) toggles the cgRNA from ON → OFF, leading to a decrease in fluorescence. Single-cell fluorescence intensities via flow cytometry. Transfection of plasmids expressing inducing dCas9-VPR, dTomato target gene Y, and either: standard gRNA (ideal ON state), cgRNA (ON state), cgRNA + RNA trigger X (OFF state), or no-target gRNA that lacks target-binding region (ideal OFF state). Background (BACK): characterized using no-target gRNA control. (c) Programmable conditional regulation using 3 orthogonal cgRNAs (Q, R, S). Left: raw fluorescence depicting ON → OFF conditional response to cognate trigger (fold change = ON/OFF = [no trigger–BACK]/[cognate trigger–BACK]). Right: normalized fluorescence depicting orthogonality between noncognate cgRNA/trigger pairs (crosstalk = [noncognate trigger–no trigger]/[cognate trigger–no trigger]). Bar graphs depict mean ± estimated standard error calculated based on the mean single-cell fluorescence over 426–7714 cells for each of N = 3 replicate wells (fold change and crosstalk calculated with uncertainty propagation).
Computational Sequence Design of Libraries of Orthogonal cgRNA/Trigger Pairs Using NUPACK
For each cgRNA mechanism (Figures 2–5), sequence design was performed using the reaction pathway designer within NUPACK.31,32 Following Wolfe et al.,32 sequence design was formulated as a multistate optimization problem using target test tubes to represent reactant and product states of cgRNA/trigger hybridization as well as to model crosstalk between orthogonal cgRNAs (Figure 6a). Each reactants tube (Step 0) and products tube (Step 1) contains a set of desired “on-target” complexes (each with a target secondary structure and target concentration), corresponding to the on-pathway hybridization products for a given step, and a set of undesired “off-target” complexes (each with a target concentration of 0 nM), corresponding to on-pathway reactants and off-pathway hybridization crosstalk for a given step. Hence, these elementary step tubes are designed for full conversion of cognate reactants into cognate products and against local hybridization crosstalk between these same reactants. To simultaneously design N orthogonal systems, elementary step tubes are specified for each system (Figure 6a; left). Furthermore, to design against off-pathway interactions between systems, a single global crosstalk tube is specified (Figure 6a; right). In the global crosstalk tube, the on-target complexes correspond to all reactive species generated during all elementary steps (m = 0, 1) for all systems (n = 1, ..., N); the off-target complexes correspond to noncognate interactions between these reactive species. Crucially, the global crosstalk tube ensemble omits the cognate products that the reactive species are intended to form (they appear as neither on-targets nor off-targets). Hence, all reactive species in the global crosstalk tube are forced to either perform no reaction (remaining as desired on-targets) or undergo a crosstalk reaction (forming undesired off-targets), providing the basis for minimization of global crosstalk during sequence optimization. Note that, for design of a library of N orthogonal cgRNA/trigger pairs, all N cgRNAs have the same on-target structure, and all N triggers have the same on-target structure; within a library, the only difference between cgRNA/trigger pairs is the designed sequence.
Figure 6.
Computational cgRNA sequence design using NUPACK.31,32 (a) Target test tubes for design of 3 orthogonal cgRNAs A, B, and C (terminator switch mechanism of Figure 2). Left: elementary step tubes. Reactants tube (Step 0): cgRNA and trigger. Products tube (Step 1): cgRNA:trigger complex. Each target test tube contains a set of desired “on-target” complexes (each with the depicted target secondary structure and a target concentration of 10 nM) corresponding to the on-pathway hybridization products for a given step and a set of undesired “off-target” complexes (all complexes of up to 2 strands, each with a target concentration of 0 nM; not depicted) corresponding to on-pathway reactants and off-pathway hybridization crosstalk for a given step. To design 3 orthogonal systems, there are two elementary step tubes for each system A, B, and C. Right: global crosstalk tube. Contains the depicted on-target complexes corresponding to reactive species generated during Steps 0 and 1 (each with the depicted target secondary structure and a target concentration of 10 nM) as well as off-target complexes (all complexes of up to 2 strands, each with a target concentration of 0 nM; not depicted) corresponding to off-pathway interactions between these reactive species. To design 3 orthogonal systems, the global crosstalk tube contains a set of on-targets and off-targets for each system A, B, and C. (b) Analysis of design quality.31,38 Left: tubes depict the target structure and predicted concentration for each on-target complex with nucleotides shaded to indicate the probability of adopting the depicted base-pairing state at equilibrium. For this design, all on-targets are predicted to form with quantitative yield at the 10 nM target concentration, but some nucleotides have unwanted base-pairing interactions (nucleotides not shaded dark red). Right: computational orthogonality study. Predicted equilibrium concentration of each cgRNA:trigger complex for the 3 orthogonal systems of Figure 2 (one cgRNA species and one RNA trigger species per tube). RNA at 37 °C in 1 M Na+.39
Sequence design is performed subject to complementarity constraints inherent to the reaction pathway (Figure 2b; domain “d” complementary to “d*”, etc.), as well as to biological sequence constraints imposed by the silencing target Y (mRFP, sfGFP, or dTomato), the protein effector (dCas9), or the synthetic terminator; see the constraint shading in Figure 6a. The sequence is optimized by reducing the ensemble defect quantifying the average fraction of incorrectly paired nucleotides over the multitube ensemble.32,40,41 Within the ensemble defect, defect weights were applied to prioritize design effort.32 Optimization of the ensemble defect implements both a positive design paradigm, explicitly designing for on-pathway elementary steps, and a negative-design paradigm, explicitly designing against off-pathway crosstalk.32
Figure 6b displays the reactants and products tubes for a completed sequence design (cgRNAs of Figure 2). For cgRNA A (left panel), on-target complexes are predicted to form with quantitative yield at the target concentrations but with some unintended base-pairing (nucleotides not shaded dark red). These structural defects within the ensemble of on-target complexes reflect the real-world challenges of designing a cgRNA that satisfies biological sequence constraints, changes conformation in response to a cognate RNA trigger, and operates orthogonally to a library of other cgRNAs. For the corresponding library of orthogonal cgRNAs (A, B, C), each cgRNA is predicted to interact appreciably only with its cognate RNA trigger (right panel).
Conceptual Opportunities for Biological Research Tools, Therapeutics, and Synthetic Biology Using Dynamic RNA Nanotechnology
To date, dynamic DNA nanotechnology in a test tube42,43 has received far more research emphasis than dynamic RNA nanotechnology in the cell,44−47 although it is the latter that has the potential to enable diverse modes of programmable conditional regulation in living organisms. The ability to rationally design cgRNAs suggests a conceptual framework for enabling biologists to exert spatiotemporal control over regulatory perturbations in living organisms using CRISPR/Cas technology. In principle, Cas activity could be restricted to a desired cell type, tissue, or organ by selecting an endogenous RNA trigger X with the desired spatial and temporal expression profile (Figure 1b). To shift conditional regulation to a different tissue or developmental stage, the cgRNA would be reprogrammed to recognize a different trigger sequence. Signal transduction with cgRNAs would also have attractive therapeutic potential, with trigger X as a programmable disease marker and target Y as an independent programmable therapeutic target, enabling selective treatment of diseased cells. Synthetic biology provides another attractive arena for use of cgRNAs. Traditional synthetic biology regulators have relied on protein:protein and protein:DNA interactions mined from existing genomes, placing limits on scalability due to crosstalk and the limited number of available regulators. cgRNA regulators offer a promising platform for scalable synthetic biology.
In working toward these applications, it remains to measure and optimize cgRNA conditional response times, which are expected to depend on a variety of factors including whether triggers can toggle the state of both free cgRNA and cgRNA in complex with Cas (possibly a mechanism-specific property) and the production and degradation rates of the participating chemical species. As a starting point for further study, induction of the trigger at different time points following dCas9 induction reveals a 1–2 h conditional response time for gene silencing mediated by a splinted switch cgRNA (Figure S40).
Comparison of cgRNAs to Other scRNAs
It is interesting to compare the present work engineering cgRNAs (a particular class of scRNAs with notable properties) to the scRNAs previously demonstrated in buffer and human cell lysate working toward the goal of conditional RNA interference (RNAi).1,2 In both cases, the scRNAs are intended to perform signal transduction between detection of a programmable RNA input and production of a biologically active programmable output. In the case of conditional RNAi, the scRNAs detect an mRNA input X and interact to produce a substrate that is processed by Dicer to produce an siRNA output targeting independent silencing target mRNA Y for destruction. Because Dicer substrates are structurally simple, comprising predominantly a duplex containing the target-binding sequence,48 signal transduction between X and Y and inactive/active states is performed by scRNAs upstream of formation of the biologically active Dicer substrate. For example, the simplest mechanism devised to date involves a dimer scRNA that conditionally generates a monomer Dicer substrate anti-Y upon detection of mRNA X.1,2 By contrast, not only are the standard gRNAs that serve as substrates for Cas9 protein effectors structurally more complex than Dicer substrates (involving multiple duplexes, loops, and tails), but Cas9 also appears to be more permissive of modifications to the standard structure, providing hooks for engineering programmable conditional regulation. As a result, it is possible to perform signal transduction between X and Y and inactive/active states (for either ON → OFF or OFF → ON logic) all within a single cgRNA (i.e., a single monomer scRNA). A benefit of this mechanistic simplicity is that monomer cgRNAs can be readily expressed, while expression of well-formed multimer scRNAs such as those developed for conditional Dicer substrate formation appears more challenging, possibly necessitating delivery with chemical reagents.
Conclusions
The present work represents only a first step toward our long-term goal of engineering programmable conditional regulators that function robustly in living organisms. Here, we describe progress on multiple fronts: (1) In E. coli expressing cgRNA regulators and RNA triggers we demonstrate mechanisms for both logical directions of conditional regulation, ON → OFF logic with constitutively active cgRNAs that are conditionally inactivated by a cognate RNA trigger and OFF → ON logic with constitutively inactive cgRNAs that are conditionally activated by a cognate RNA trigger. (2) To leverage the programmability of these dynamic regulators, we establish a computational framework for automated sequence design of libraries of orthogonal cgRNA/trigger pairs using the reaction pathway engineering tools within NUPACK. (3) To test the portability of cgRNA mechanisms prototyped in bacteria into mammalian cells, we demonstrate constitutively active cgRNAs (ON → OFF logic) in HEK 293T cells. (4) To establish that cgRNAs can exert conditional regulation over dCas9 variants with different downstream functions, we demonstrate conditional gene silencing in bacteria (if X then not Y, if not X then not Y) and conditional gene induction in mammalian cells (if not X then Y). (5) These contributions demonstrate the applicability of dynamic RNA nanotechnology for programmable conditional regulation in both bacterial and mammalian cells.
To develop cgRNAs into a versatile platform for biological research, a number of major improvements are needed. First, it is desirable to engineer improved cgRNA mechanisms that exploit the full regulatory dynamic range of standard gRNAs to achieve ≈100-fold conditional responses. Toward this end, further understanding of the structure/function relationships between cgRNAs, triggers, and Cas effectors is needed to ascertain how to robustly achieve both a strong ON state and a clean OFF state depending on the presence/absence of the cognate trigger. Second, to enable tissue-selective regulation in living organisms, it is critical that cgRNAs are able to efficiently detect a trigger that is a subsequence of a longer endogenous RNA (e.g., a subsequence of an mRNA). Detection of a subsequence of a full-length mRNA poses significant additional challenges relative to detection of a short RNA trigger,2,30 increasing the degree of difficulty in achieving a conditional response that exploits the full dynamic range. Third, in common with the terminator switch and splinted switch mechanisms studied here (but unlike the toehold switch mechanisms studied here and elsewhere30), it is important that cgRNA regulators be allosteric, so that the sequence of the target gene Y places no restriction on the sequence of the RNA trigger X, enabling independent control over the regulatory scope (using X) and the regulatory target (using Y). Significant effort and innovation are needed to achieve these goals and develop cgRNAs that operate as plug-and-play programmable conditional regulators within endogenous biological circuits in living organisms.
Methods Summary
For each mechanism, orthogonal cgRNA/trigger pairs were designed using the reaction pathway engineering tools within NUPACK (nupack.org).31,32 For bacterial studies, a control gRNA or a cgRNA/trigger plasmid was transformed into a modified E. coli MG1655 strain expressing genomically incorporated mRFP and sfGFP.4 Strains were grown overnight in EZ-RDM (Teknova) and then diluted and grown to mid log phase (≈4 h). Cell density was normalized with fresh medium containing aTc for induction of silencing dCas9 expression (and IPTG for the bacterial terminator switch experiments only). Induced cells were grown for 12 h, with end-point fluorescence measured via flow cytometry. For mammalian studies, a cgRNA expression plasmid and a trigger expression plasmid were cotransfected with a plasmid expressing an inducing dCas9-VPR fusion33 and a reporter plasmid containing a gRNA binding site upstream of a minimal CMV promoter for dTomato expression.34,35 The four plasmids were transiently transfected into HEK 293T cells with Lipofectamine 3000 and grown for 24 h, with end-point fluorescence measured via flow cytometry. Data analysis was performed on cells expressing high levels of both cgRNA and trigger fluorescent protein transfection controls. No unexpected or unusually high safety hazards were encountered.
Acknowledgments
We thank S. Qi for the gift of E. coli expressing mRFP and sfGFP, N. J. Porubsky for assistance with reaction pathway engineering using NUPACK, J. S. Bois for discussions on data analysis, A. Hou and J. Kishi for performing preliminary studies, and R. Phillips for discussions on allosteric regulation. This work was funded by the Defense Advanced Research Projects Agency (HR0011-17-2-0008; the findings are those of the authors and should not be interpreted as representing the official views or policies of the US Government), by the Caltech Center for Environmental Microbial Interactions (CEMI), by the National Institutes of Health (5T32GM112592), by the Rosen Bioengineering Center at Caltech, by the Natural Sciences and Engineering Research Council (NSERC) of Canada, by the National Science Foundation Molecular Programming Project (NSF-CCF-1317694), by a Professorial Fellowship at Balliol College (University of Oxford), and by the Eastman Visiting Professorship at the University of Oxford.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.9b00340.
Methods, sequences, plasmids, schematics, flow cytometry replicates, and additional studies (PDF)
Author Contributions
∥ M.H.H.-H. and Z.C. contributed equally. cgRNA project conceived by N.A.P. cgRNA mechanism invention by all coauthors. Exploratory mechanism studies by M.H.H.-H., Z.C., and J.H. in bacteria. Computational sequence design by M.H.H.-H. (splinted switch and toehold switch for bacteria, terminator switch for mammalian) and Z.C. (terminator switch for bacteria). Experimental design and data presentation approach developed by all coauthors. Presented cgRNA mechanisms prototyped and optimized in bacteria by Z.C. (terminator switch and toehold switch) and M.H.H.-H. (splinted switch and toehold switch), extended to mammalian cells by L.M.H. (terminator switch). Final data collected by M.H.H.-H. (bacterial) and L.M.H. (mammalian). Paper written by M.H.H.-H. and N.A.P. Supporting Information written by M.H.H.-H., L.M.H., and N.A.P. Paper edited and approved by all coauthors.
The authors declare the following competing financial interest(s): Filed patents.
Supplementary Material
References
- Hochrein L. M.; Schwarzkopf M.; Shahgholi M.; Yin P.; Pierce N. A. Conditional Dicer substrate formation via shape and sequence transduction with small conditional RNAs. J. Am. Chem. Soc. 2013, 135, 17322–17330. 10.1021/ja404676x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochrein L. M.; Ge T. J.; Schwarzkopf M.; Pierce N. A. Signal transduction in human cell lysate via dynamic RNA nanotechnology. ACS Synth. Biol. 2018, 7, 2796–2802. 10.1021/acssynbio.8b00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott G. J.; Doudna J. A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866. 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S.; Larson M. H.; Gilbert L. A.; Doudna J. A.; Weissman J. S.; Arkin A. P.; Lim W. A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A.; Larson M. H.; Morsut L.; Liu Z.; Brar G. A.; Torres S. E.; Stern-Ginossar N.; Brandman O.; Whitehead E. H.; Doudna J. A.; Lim W. A.; Weissman J. S.; Qi L. S. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L.; Ran F. A.; Cox D.; Lin S.; Barretto R.; Habib N.; Hsu P. D.; Wu X.; Jiang W.; Marraffini L. A.; Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P.; Yang L.; Esvelt K. M.; Aach J.; Guell M.; DiCarlo J. E.; Norville J. E.; Church G. M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B.; Heidenreich M.; Mohanraju P.; Fedorova I.; Kneppers J.; DeGennaro E. M.; Winblad N.; Choudhury S. R.; Abudayyeh O. O.; Gootenberg J. S.; Wu W. Y.; Scott D. A.; Severinov K.; van der Oost J.; Zhang F. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 2017, 35, 31–34. 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M. H.; Gilbert L. A.; Wang X.; Lim W. A.; Weissman J. S.; Qi L. S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180–2196. 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.; Gilbert L. A.; Cimini B. A.; Schnitzbauer J.; Zhang W.; Li G.-W.; Park J.; Blackburn E. H.; Weissman J. S.; Qi L. S.; Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013, 155, 1479–1491. 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S. C.; Tjian R.; Doudna J. A. Genomes in focus: Development and applications of CRISPR-Cas9 imaging technologies. Angew. Chem., Int. Ed. 2018, 57, 4329–4337. 10.1002/anie.201709201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton I. B.; D’Ippolito A. M.; Vockley C. M.; Thakore P. I.; Crawford G. E.; Reddy T. E.; Gersbach C. A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli N. M.; Komor A. C.; Rees H. A.; Packer M. S.; Badran A. H.; Bryson D. I.; Liu D. R. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey B. J.; Kelly G. L.; Kueh A. J.; Brennan M. S.; O’Connor L.; Milla L.; Wilcox S.; Tai L.; Strasser A.; Herold M. J. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015, 10, 1422–1432. 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Bertero A.; Pawlowski M.; Ortmann D.; Snijders K.; Yiangou L.; Cardoso de Brito M.; Brown S.; Bernard W. G.; Cooper J. D.; Giacomelli E.; Gambardella L.; Hannan N. R. F.; Iyer D.; Sampaziotis F.; Serrano F.; Zonneveld M. C. F.; Sinha S.; Kotter M.; Vallier L. Optimized inducible shRNA and CRISPR/Cas9 platforms for in vitro studies of human development using hPSCs. Development 2016, 143, 4405–4418. 10.1242/dev.138081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.; Gao D.; Zhang R.; Zeng G.; Yan H.; Lim E.; Liang F.-S. Chemically controlled epigenome editing through an inducible dCas9 system. J. Am. Chem. Soc. 2017, 139, 11337–11340. 10.1021/jacs.7b06555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y.; Otabe T.; Sato M. Emerging approaches for spatiotemporal control of targeted genome with inducible CRISPR-Cas9. Anal. Chem. 2018, 90, 429–439. 10.1021/acs.analchem.7b04757. [DOI] [PubMed] [Google Scholar]
- Shen Z.; Zhang X.; Chai Y.; Zhu Z.; Yi P.; Feng G.; Li W.; Ou G. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell 2014, 30, 625–636. 10.1016/j.devcel.2014.07.017. [DOI] [PubMed] [Google Scholar]
- Ablain J.; Durand E. M.; Yang S.; Zhou Y.; Zon L. I. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell 2015, 32, 756–764. 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosawa M.; Fujita Y.; Parr C. J. C.; Hayashi K.; Kashida S.; Hotta A.; Woltjen K.; Saito H. Cell-type-specific genome editing with a microRNA-responsive CRISPR–Cas9 switch. Nucleic Acids Res. 2017, 45, e118. 10.1093/nar/gkx309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briner A. E.; Donohoue P. D.; Gomaa A. A.; Selle K.; Slorach E. M.; Nye C. H.; Haurwitz R. E.; Beisel C. L.; May A. P.; Barrangou R. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol. Cell 2014, 56, 333–339. 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Nowak C. M.; Lawson S.; Zerez M.; Bleris L. Guide RNA engineering for versatile Cas9 functionality. Nucleic Acids Res. 2016, 44, 9555–9564. 10.1093/nar/gkw908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Zhan Y.; Chen Z.; He A.; Li J.; Wu H.; Liu L.; Zhuang C.; Lin J.; Guo X.; Zhang Q.; Huang W.; Cai Z. Directing cellular information flow via CRISPR signal conductors. Nat. Methods 2016, 13, 938–944. 10.1038/nmeth.3994. [DOI] [PubMed] [Google Scholar]
- Tang W.; Hu J. H.; Liu D. R. Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat. Commun. 2017, 8, 15939. 10.1038/ncomms15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundert K.; Lucas J. E.; Watters K. E.; Fellmann C.; Ng A. H.; Heineike B. M.; Fitzsimmons C. M.; Oakes B. L.; Qu J.; Prasad N.; Rosenberg O. S.; Savage D. F.; El-Samad H.; Doudna J. A.; Kortemme T. Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat. Commun. 2019, 10, 2127. 10.1038/s41467-019-09985-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. J.; Hoynes-O’Connor A.; Leong M. C.; Moon T. S. Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system. Nucleic Acids Res. 2016, 44, 2462–2473. 10.1093/nar/gkw056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry Q. R. V.; Lyutova R.; Fulga T. A. Rational design of inducible CRISPR guide RNAs for de novo assembly of transcriptional programs. Nat. Commun. 2017, 8, 14633. 10.1038/ncomms14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mückl A.; Schwarz-Schilling M.; Fischer K.; Simmel F. C. Filamentation and restoration of normal growth in Escherichia coli using a combined CRISPRi sgRNA/antisense RNA approach. PLoS One 2018, 13, e0198058 10.1371/journal.pone.0198058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain P. K.; Ramanan V.; Schepers A. G.; Dalvie N. S.; Panda A.; Fleming H. E.; Bhatia S. N. Development of light-activated CRISPR using guide RNAs with photocleavable protectors. Angew. Chem., Int. Ed. 2016, 55, 12440–12444. 10.1002/anie.201606123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-H.; Chen W. Riboregulated toehold-gated gRNA for programmable CRISPR–Cas9 function. Nat. Chem. Biol. 2019, 15, 217–220. 10.1038/s41589-018-0186-1. [DOI] [PubMed] [Google Scholar]
- Zadeh J. N.; Steenberg C. D.; Bois J. S.; Wolfe B. R.; Pierce M. B.; Khan A. R.; Dirks R. M.; Pierce N. A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- Wolfe B. R.; Porubsky N. J.; Zadeh J. N.; Dirks R. M.; Pierce N. A. Constrained multistate sequence design for nucleic acid reaction pathway engineering. J. Am. Chem. Soc. 2017, 139, 3134–3144. 10.1021/jacs.6b12693. [DOI] [PubMed] [Google Scholar]
- Chavez A.; Scheiman J.; Vora S.; Pruitt B. W.; Tuttle M.; P R Iyer E.; Lin S.; Kiani S.; Guzman C. D.; Wiegand D. J.; Ter-Ovanesyan D.; Braff J. L.; Davidsohn N.; Housden B. E.; Perrimon N.; Weiss R.; Aach J.; Collins J. J.; Church G. M. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P.; Aach J.; Stranges P. B.; Esvelt K. M.; Moosburner M.; Kosuri S.; Yang L.; Church G. M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L.; Perli S. D.; Fridkin A.; Perez-Pinera P.; Lu T. K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell 2014, 54, 698–710. 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman L.All of Statistics: A Concise Course in Statistical Inference; Springer: New York, 2004. [Google Scholar]
- Efron B.; Hastie T.. Computer Age Statistical Inference; Cambridge University Press: New York, 2016. [Google Scholar]
- Dirks R. M.; Bois J. S.; Schaeffer J. M.; Winfree E.; Pierce N. A. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 2007, 49, 65–88. 10.1137/060651100. [DOI] [Google Scholar]
- Serra M. J.; Turner D. H. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995, 259, 242–261. 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- Zadeh J. N.; Wolfe B. R.; Pierce N. A. Nucleic acid sequence design via efficient ensemble defect optimization. J. Comput. Chem. 2011, 32, 439–452. 10.1002/jcc.21633. [DOI] [PubMed] [Google Scholar]
- Wolfe B. R.; Pierce N. A. Sequence design for a test tube of interacting nucleic acid strands. ACS Synth. Biol. 2015, 4, 1086–1100. 10.1021/sb5002196. [DOI] [PubMed] [Google Scholar]
- Bath J.; Turberfield A. J. DNA nanomachines. Nat. Nanotechnol. 2007, 2, 275–284. 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- Zhang D. Y.; Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011, 3, 103–113. 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- Chappell J.; Watters K. E.; Takahashi M. K.; Lucks J. B. A renaissance in RNA synthetic biology: new mechanisms, applications and tools for the future. Curr. Opin. Chem. Biol. 2015, 28, 47–56. 10.1016/j.cbpa.2015.05.018. [DOI] [PubMed] [Google Scholar]
- Li J.; Green A. A.; Yan H.; Fan C. Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem. 2017, 9, 1056–1067. 10.1038/nchem.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M.; Fussenegger M. Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol. 2018, 19, 507–525. 10.1038/s41580-018-0024-z. [DOI] [PubMed] [Google Scholar]
- Jani M. S.; Veetil A. T.; Krishnan Y.. Precision immunomodulation with synthetic nucleic acid technologies. Nat. Rev. Mater. 2019, in press. 10.1038/s41578-019-0105-4 [DOI] [Google Scholar]
- Kim D. H.; Rossi J. J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007, 8, 173–184. 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.