Short abstract
Neutrophils are the most abundant immune cell of the innate immune system and participate in essential immune functions. Heterogeneity within neutrophils has been documented, but it is difficult to distinguish if these are altered activation states of a single population or separate subpopulations of neutrophils determined at the time of differentiation. Several groups have identified a subset of human neutrophils that express olfactomedin 4 (OLFM4) and increased OLFM4+ neutrophils during sepsis is correlated with worse outcome, suggesting these neutrophils or the OLFM4 they secrete may be pathogenic. We tested if mice could be used as a model to study OLFM4+ neutrophils. We found the OLFM4 expressing subset of neutrophils is conserved in mice. Depending on the strain, 7–35% of murine neutrophils express OLFM4 and expression is determined early in neutrophil differentiation. OLFM4+ neutrophils phagocytose and transmigrate with similar efficiency as OLFM4− neutrophils. Here we show that within neutrophil extracellular traps (NETs) OLFM4+ and OLFM4− neutrophils undergo NETosis and OLFM4 colocalizes. Finally, we generated an OLFM4 null mouse and show that these mice are protected from death when challenged with sepsis, providing further evidence that the OLFM4 expressing subpopulation of neutrophils, or the OLFM4 they secrete, may be pathogenic during overwhelming infection.
Keywords: Neutrophils, sepsis, olfactomedin, heterogeneity, inflammation
Introduction
Cellular barriers and innate immunity are the first line of defence against invading pathogens throughout the animal kingdom. A key component of innate immunity is a specialized cell that carries both innate immune receptors to recognize pathogens and the ability to provide a rapid response to potential danger. Early in the animal kingdom, these cells are termed coelomocytes or simply phagocytes because of their ability to consume pathogens.1 In mammals, there is increased variety of these cells including monocytes, macrophages and the primary cell of the innate immune system, the neutrophil.
Neutrophils are the most abundant white blood cell in the human blood stream and participate in both innate and adaptive immune responses.2–4 While many other white blood cell types have been divided into different subpopulations with unique roles, traditionally neutrophils have been considered a homogenous population of cells. Recently there has been increasing evidence for neutrophil heterogeneity and plasticity.5–7 However, it remains unclear if these subpopulations of neutrophils are simply activation states or stages of maturation of the general population of neutrophils or if there are actual separate subpopulations of neutrophils determined at the time of granulocyte differentiation with unique functions.
Olfactomedin 4 (OLFM4) is a glycoprotein that was identified as a target for the important myeloid transcription factor PU.1 and shown to be expressed in gastrointestinal tumour cells.8,9 In tumour cells, it was shown that OLFM4 had anti-apoptotic properties which may explain its increased expression in gastrointestinal tumour cells.10–12 Recently, OLFM4 has been shown to signal via the Frizzled receptors and to compete with Wnt ligands to decrease Wnt signalling in intestinal adenocarcinomas.13 Apart from tumours, Lgr-5 expressing stem cells that reside at the base of intestinal and colonic crypts also express OLFM4. The role for OLFM4 in these stem cells is unclear as the investigators were unable to find a stem cell-related phenotype in OLFM4 null animals.14
OLFM4 has also been shown to be expressed in a subset of neutrophils within the granules. In healthy human donors, approximately 25% of neutrophils express OLFM4.15,16 Our group and others recently identified OLFM4 as one of the most up-regulated genes, in terms of fold increase, during sepsis.17 We also showed that increased percentage of neutrophils expressing OLFM4 at the time of presentation to the intensive care unit independently associated with a poor outcome from septic shock.18 In mice, OLFM4 has been shown to be expressed in the bone marrow, prostate and gut, and furthermore, mice null for OLFM4 have been shown to be protected from death by intraperitoneal injection of bacteria, suggesting OLFM4 participates in immune responses.9,19–21 These same studies found that OLFM4 inhibited cathepsin C, leading the authors to conclude that lack of OLFM4 led to increased antimicrobial activities of cathepsin C and provided protection to the animal from bacterial challenge.22 However, when the OLFM4 null mouse was crossed onto a cathepsin C null mouse, lack of OLFM4 still provided protection from bacterial challenge, suggesting OLFM4 participated in other immune mechanisms.
All the experiments conducted on murine neutrophils have been based on the presumption that all murine neutrophils expressed OLFM4. This presumption is based on Western blot experiments and immunohistochemistry showing expression in neutrophils using the OLFM4 specific Abs that are available. This would make the mouse different from human who only express OLFM4 in a subset of neutrophils. However, here we show for the first time that mice, like humans, only express OLFM4 in a subset of neutrophils. We further characterize OLFM4 expression in murine neutrophils to establish the mouse as a tool for delineating the role of OLFM4 in sepsis and immune responses.
Materials and methods
Animal strains
C57Bl/6, 129, and BALB/c strains were obtained from Charles River, Wilmington, MA or breeding pairs from commercial vendors and then bred in-house. Mice were maintained with standard housing, food and day/night regulation. All animal experiments were approved by the Cincinnati Children’s Hospital Institutional Animals Care and Use Committee. For generation of OLFM4 null mice, we targeted the fourth exon with guide RNA’s using CRISPR cas9 method.23,24 Pups born from injected implanted embryos were screened and sequenced to identify insertions or deletions leading to frame shifts and premature stop codons.
Quantitative PCR
Neutrophils, CD11b+ Ly6g+, or other cells, all cells not defined as CD11b+ Ly6g+, were isolated by FACS cell sorting on a FACSAria II (Becton Dickinson, Franklin Lakes, NJ) or using neutrophil negative selection isolation kit from Miltenyi Biotec, Santa Barbara, CA for the LPS stimulation experiments. Cell sort purities were greater than 95% while isolation with negative selection kits were greater than 90%. Gating strategy for maturing neutrophils can be seen in Supplemental Figure 1. Isolated cells were then preserved in Trizol (Thermo Fisher, Waltham, MA) before RNA extraction. cDNA generation was done using the SuperScript III (Thermo Fisher) according to manufacturer’s protocol. TaqMan probes from Applied Biosystems were used for qPCR: OLFM4-Mm01320260, HPRT-Mm03024075, Mmp9-Mm00442991 and MPO-Mm01298424. TaqMan qPCR assays were run using the Taqman Universal Fast Mast Mix (2×) according to the manufacturer’s protocol.
Polyclonal Ab
Full-length murine OLFM4 was cloned into an eukaryotic protein expression system in frame with a tobacco echo virus (TEV) linker sequence followed by human IgG1 domain. This vector was transfected into HEK293T cells. Over the next 6 d we collected supernatant from the transfected cells. Supernatants from several experiments were combined and run over a protein G column (GE Life Sciences, Chicago, IL). Purified protein was then dialyzed into PBS and used for subsequent rabbit immunization. For immunization, we mixed purified OLFM4 protein with Complete Freund’s Adjuvant for initial subcutaneous immunization followed by Incomplete Freund’s Adjuvant for subsequent subcutaneous immunizations. The initial immunization was with 250 μg of OLFM4-IgG fusion protein, with subsequent booster immunization with approximately 100 μg of fusion or cleaved and purified OLFM4 protein alone. Following second immunization, we were able to detect high titer anti-OLFM4 Abs by flow cytometry (plasma could be diluted 2000× and still detect OLFM4 protein by flow cytometry).
Staining for flow cytometry
Standard flow cytometric protocols were used. Briefly, following bone marrow harvest and red cell lysis, cells were washed with 1 ml FACS buffer (1× PBS with 0.1% BSA and 1 mM sodium azide). Surface Fc block was done with 20 µl of conditioned media from hybridoma 24G.2 for 5 min at 4°C. Primary Abs for myeloid lineage marker CD11b and Ly6g (Tonbo biosciences, San Diego, CA) in FACS buffer were added to cells for 1 h incubation at 4°C. The cells were then washed with 1 ml FACS buffer, and were fixed with 2% PFA for 5 min at 4°C. Then the cells were permeabilized with incubation in intracellular cytokine staining (ICCS) buffer (1× PBS with 10 mM HEPES, 0.1% BSA, 0.1% saponin). Two washes were done with 1 ml ICCS. Incubation was with Rabbit anti-mouse OLFM4 Ab conjugated to Alexa Fluor 647 in ICCS buffer for 1 h at room temperature. The cells were then washed with ICCS, followed by FACS buffer, and fixed in 2% PFA at 4°C overnight. Finally, cells were washed with 1 ml FACS buffer and reconstituted in 300 µl of FACS buffer prior to flow analysis.
Neutrophil phagocytosis assay
Neutrophil phagocytosis was done using Escherichia coli Bioparticles Alexa Fluor 488 conjugate (Invitrogen, Carlsbad, CA). The Bioparticles were reconstituted according to manufacturer’s recommended protocol. Bioparticles were opsonized with mouse plasma (100 µl) by incubation at 37°C for 30 min. The particles were then washed with 1 ml of PBS and reconstituted in 500 µl of growth medium (RPMI with 10 FBS) at 4°C. Neutrophils were isolated from bone marrow following RBC lysis with ACK lysis buffer (Gibco, Gaithersburg, MD). Opsonized Bioparticles (10 million in 50 µl of growth medium) were added to the neutrophils (1 million) at 10:1 ratio. The Bioparticle neutrophil suspension mixture was incubated in 150 µl of growth medium for 2 h at 37°C. Following incubation, the cells were washed twice with 1 ml PBS. Phagocytosis process was stopped by adding 300 µl 2% PFA to cells followed by 5 min incubation at 4°C. Cells were then processed for flow cytometry.
Neutrophil migration
Neutrophils were isolated from bone marrow of BALB/c mice following RBC lysis using gradient centrifugation using a previously published protocol.25 Briefly, Histopaque 1119 (Sigma, St. Louis, MO) was overlaid with Histopaque 1077 followed by bone marrow cells overlaid on top of Histopaque1077. Cells were then centrifuged at 700 g for 30 min without brake. The interface between the two layers was collected and washed. Cells were re-suspended in RPMI with 10% FBS and placed in the upper chamber of a 3-micrometer pore Transwell (Corning, Corning, NY) plate that had been coated with 10 μg/ml of fibrinogen. The lower chamber contained 600 μl of RPMI 10% FBS with 100 nM fMLP. Cells were then incubated for 2 h at 37°C before harvesting the cells in top and bottom chambers for flow cytometric analysis.
Immunofluorescence
Mouse neutrophils were purified from bone marrow using gradient centrifugation. Purified neutrophils were resuspended in RPMI 10% FBS and then incubated on glass slides coated with poly-L-lysine for 3–4 h at 37°C. PMA (100 nM) was added to some of these for immune stimulation (for degranulation images) or 5 μm calcium ionophore A23187 (for neutrophil extracellular trap (NET) images; Sigma-Aldrich, St Louis, MO). Following incubation slides were gently washed and then fixed in 2% PFA for 1 min. Cells are washed again with PBS and ice-cold methanol is added for 1 min. Cells are again washed and then blocked with blocking buffer in ICCS for 15 min. Cells are then stained with polyclonal anti-OLFM4 for 2 h, washed with PBS-tween and then secondary anti-rabbit AF488 (Jackson Immunoresearch, West Grove, PA) was added for 1 h. Hoechst dye is added to the secondary Ab mixture to counterstain DNA. Images were collected on a Nikon A1R inverted microscope.
Animal models of inflammation and sepsis
For LPS induction of neutrophil migration into the lung, animals were anaesthetized with isoflurane. Animals were then suspended on their backs at a 70° angle. The tongue is retracted and 50 μl of saline containing 33 μg of LPS were placed in the posterior pharynx. The nose is occluded with gentle finger pressure for 10 s to allow aspiration of the LPS. For sepsis challenge survival studies, we used the cecal ligation and puncture (CLP) model, as described previously.26 Briefly, male animals were anaesthetized and sterilely prepped. The cecum was externalized through a midline incision. 1.5 cm of cecum was ligated and punctured 3 times through and through with 23-gauge needle. The cecum was returned to the peritoneum and the abdominal incision was closed. Animals were given 1 ml saline, pain medications and 25 mg/kg of imipenem every 12 h for 6 doses (antibiotics were started 6–8 h after the procedure). For flow cytometry, we used the cecal slurry model as described previously.27 Briefly, cecal contents from donor mice were combined in a suspension, filtered to remove large particulates and glycerol added prior to freezing. Subsequently, animals were given 0.6 mg/kg of cecal slurry IP injection and sacrificed at specified times for sample collection.
Statistical analysis
For statistical comparisons we used SigmaPlot software (Systat Software Inc.). Percentage of OLFM4+ neutrophils and comparing OLFM4 MFI was done using a two tailed t-test. For comparing qPCR group results, we used Mann-Whitney U-test for non-parametric and t-test for parametric comparisons. For survival studies we used Kaplan-Meier Log-Rank Survival Analysis.
Results
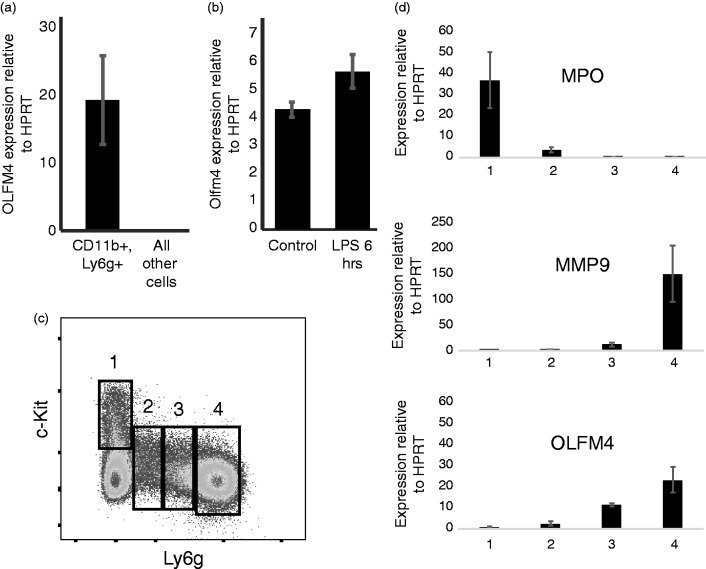
As Olfm4 has previously been shown to be expressed in the bone marrow compartment of mice,9 we sought to test if Olfm4 was expressed solely in neutrophils, similar to humans. We sorted CD11b+, Ly6g+ bone marrow neutrophils from C57Bl/6 mice and performed quantitative PCR (qPCR) comparing neutrophils to all other bone marrow cells. Olfm4 expression was almost exclusively expressed in neutrophils in the bone marrow (Figure 1a).
Figure 1.
OLFM4 is expressed solely in neutrophils. (a) qPCR of bone marrow cells showing OLFM4 is expressed exclusively in neutrophils, P = 0.008. (b) qPCR showing increased expression of OLFM4 6 h following LPS stimulation P = 0.006. (c) Histogram of bone marrow lineage depleted cells looking at c-Kit and Gr1, demonstrating maturation of neutrophils starting from 1 (immature) to 4 (mature neutrophil).30 (d) qPCR from sorted populations of maturing neutrophils showing immature gene Mpo present only in population 1 and the mature neutrophil gene Mmp9 only expressed in population 4. Olfm4 expression can be detected in population 2 and increasing transcript in 3 and 4.
Because humans show a dramatic increase in OLFM4 expression during sepsis,18,28,29 we tested the effect of bacterial LPS exposure in mice. Mice were injected intraperitoneally with LPS and then 6 h later neutrophils were harvested for qPCR and flow cytometric analysis. Similar to humans with septic shock, mice showed increased expression of Olfm4 transcript 6 h after injection with LPS (Figure 1b).
We next used the method described by Satake et al.30 to evaluate at what stage during neutrophil maturation is Olfm4 expressed. This method evaluates lineage negative cells based on expression of c-Kit and Ly6g. As neutrophils develop from myeloblasts (c-Kit+, Ly6g−; Figure 1c, box 1) to mature multi-nucleated neutrophils, they first lose c-Kit expression and then demonstrate increasing expression of Ly6g (Figure 1c, boxes 2–4). Using qPCR with gene probes specific for each stage of neutrophil development, we found that Olfm4 was first expressed during the promyelocyte and myelocyte stage of neutrophil development and increased with neutrophil maturation (Figure 1d).
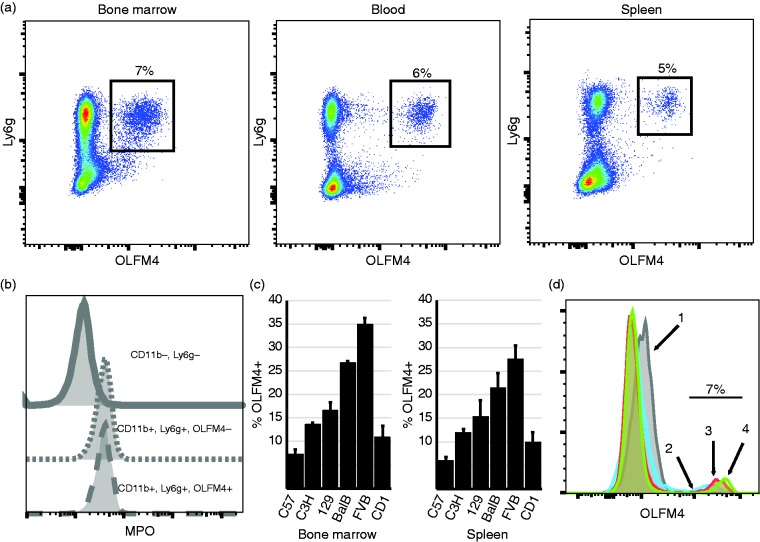
To test OLFM4 protein expression in murine neutrophils, we generated murine OLFM4 recombinant protein and immunized a rabbit to generate anti-murine OLFM4 polyclonal Abs. Because OLFM4 is a neutrophil granule protein, we used intracellular staining of permeabilized bone marrow cells. We found that OLFM4 was expressed solely in a subpopulation of CD11b+, Ly6g+ neutrophils (Figure 2a). To confirm the differential staining of neutrophils by the anti-OLFM4 rabbit polyclonal was not due to variability in permeabilization, we performed co-staining with another intracellular protein, myeloperoxidase (MPO). We found that all CD11b+ and Ly6g+ cells stained for MPO, even those that were negative for OLFM4, suggesting variability in OLFM4 staining was not due to incomplete permeabilization (Figure 2b). In C57Bl/6 mice, OLFM4 was expressed in 6–8% of neutrophils isolated from the bone marrow, spleen or peripheral blood (Figure 2a). We also tested other common strains of inbred mice, C3H, 129, BalbC, FVB and one strain of outbred mice, Cd1 and found percentage of neutrophils that express OLFM4 ranged from 8% in C57Bl/6 to 35% in FVB mice. In all cases the percentage of OLFM4 expression was similar between bone marrow, spleen and peripheral blood (Figure 2c; blood is not shown).
Figure 2.
Only a subset of murine neutrophils express OLFM4. (a) Flow cytometry dot plot showing mouse bone marrow, blood and spleen cells, gating on CD11b+ cells. OLFM4 expression can be see only in a subset of Ly6g+ cells (black box, percentage is of CD11b, Ly6g+ cells.) (b) Histograms showing MPO intracellular staining is present in both OLFM4+ and OLFM4− neutrophils but not present in non-neutrophils (CD11b−, Ly6g−). (c) Percentage of neutrophils that express OLFM4 from 6 common stains of mice. Both bone marrow and peripheral splenic CD11b, Ly6g+ neutrophils are shown. (d) Histograms overlay of the four populations of maturing neutrophils from sort windows identical to those shown in Figure 1c. Windows 2, 3, 4 are all have 7% OLFM4+ with increasing OLFM4 MFI.
To test at what stage during neutrophil maturation is OLFM4 protein expressed we used a similar approach as described above for qPCR combined with intracellular staining for OLFM4. OLFM4 protein expression matched that of mRNA expression and was detected as soon as c-Kit expression was lost and low levels of Ly6g could be detected (Figure 1c, boxes 2–4). While the OLFM4 mean fluorescence intensity (MFI) increased with neutrophil maturation, the percentage of neutrophils that express OLFM4 did not change during each stage of neutrophil maturation (Figure 2d).
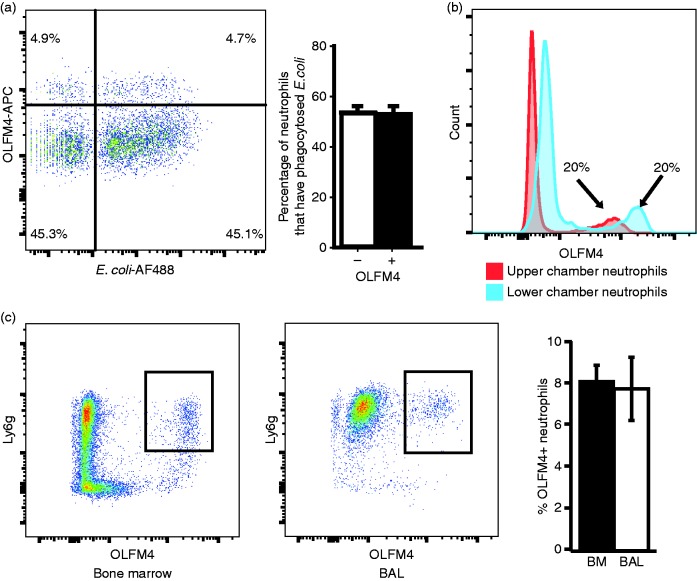
As a major function of neutrophils is to phagocytose pathogens, we tested for differences in ability to phagocytose bacteria between OLFM4+ and OLFM4− neutrophils. We incubated murine bone marrow neutrophils with fluorescently labelled bacteria and then fixed and permeabilized cells to perform OLFM4 staining. Both OLFM4+ and OLFM4− neutrophils equally phagocytosed bacteria, suggesting there is no difference in ability to phagocytose bacteria (Figure 3a).
Figure 3.
OLFM4+ and OLFM4− neutrophils phagocytose and migrate similarly. (a) Flow cytometric histogram of fixed bone marrow neutrophils following incubation with fluorescently labelled bacteria. (b) Histogram showing percentage of OLFM4+ neutrophils in the upper and lower chambers of a transwell following 3 h of incubation. Of note neutrophils in the lower chamber are exposed to higher concentration of fMLP and have higher OLFM4 MFI. (c) Dot plots showing gated CD11b+ cells from the bone marrow and BAL 24 h following treatment with intratracheal LPS.
We also tested neutrophil migration in vitro and in vivo. Neutrophil transwell migration was performed by placing bone marrow neutrophils in the upper chamber and media containing N-formylmethionine-leucyl-phenylalanine (fMLP) in the lower chamber. Neutrophils were then incubated at 37°C to allow migration toward the fMLP gradient. Flow cytometry of upper and lower chambers showed the percentage of OLFM4+ neutrophils remained the same in both chambers (Figure 3b). We did note that those neutrophils that had undergone transmigration into the lower chamber containing fMLP tended to have increased MFI for OLFM4 staining (Figure 3b). To test neutrophil migration in vivo we instilled LPS into the lungs of C57Bl/6 mice. Some 24 h after LPS instillation we performed alveolar lavage and performed flow cytometry on lavage cells and bone marrow of mice. In all cases, the percentage of OLFM4+ neutrophils was the same among the migrated neutrophils in the lung and that of the bone marrow, suggesting that OLFM4+ or OLFM4− neutrophils did not have differences in transmigration ability in vivo (Figure 3c).
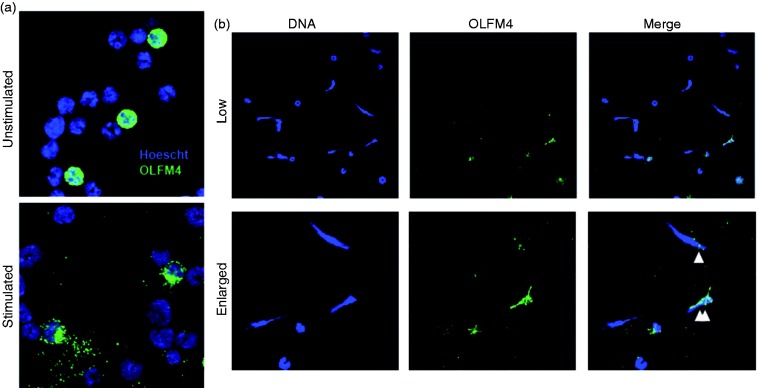
Studies of human OLFM4 found that it could be located within the webs of DNA in NETs.16 We used immunofluorescence to evaluate the morphology and location of OLFM4 before and after neutrophil stimulation to induce NET formation. OLFM4 expressing neutrophils could easily be identified among purified neutrophils, having intense cytoplasmic granular staining. Following stimulation with phorbol 12-myristate 13-acetate (PMA), neutrophils underwent activation and increased adherence and spreading onto the glass slide. OLFM4 staining following stimulation was more diffuse and localized to larger cytoplasmic granules (Figure 4a). We next used calcium ionophore to induce NET formation. Both OLFM4+ and OLFM4− neutrophils could be seen undergoing NET formation (Figure 4b). Even in OLFM4− NETs OLFM4 staining could be seen colocalizing with then DNA NET (Figure 4b).
Figure 4.
Murine OLFM4 colocalizes with neutrophil NETs. (a) Immunofluorescence of purified bone marrow neutrophils from BalbC mice using Hoescht dye to stain DNA (blue) and Af488 to stain OLFM4 (green) showing approximately 20% OLFM4+. Following stimulation with PMA, OLFM4 staining can be seen more toward the periphery of the activated neutrophil. (b) Immunofluorescence staining of low magnification (60×; top) and enlarged (bottom) magnification of stimulated neutrophils showing formation of NETs. Single white triangle designates an OLFM4 negative neutrophil that has a few small areas of OLFM4 colocalized staining. Double white triangles designate and OLFM4+ neutrophil that has undergone NETosis.
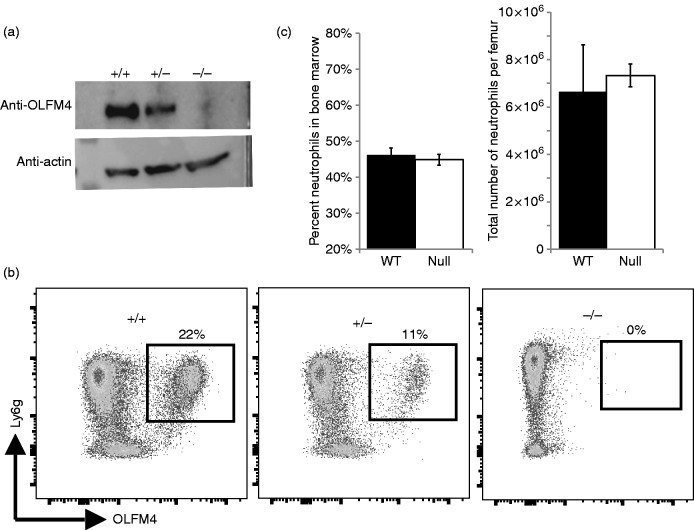
We also tested if OLFM4 might be important for neutrophil development. To do this, we used the CRISPR/Cas9 methodology to generate a null mutation within the OLFM4 mouse allele. We targeted the fourth exon as all identified splice variants contain this exon and it is upstream of the olfactomedin domain, which is the only recognizable protein domain in the OLFM4 locus. Pups were screened by sequencing and those containing frame shifts leading to premature stop codons were maintained for further analysis. Western blotting and flow cytometry both demonstrated loss of the OLFM4 protein and confirmed specificity of the polyclonal Ab we generated (Figure 5a and b). Notably, when we compared heterozygous mice with littermate homozygous null and wild type, heterozygotes always had MFI equal to wild type littermate, but the percentage of OLFM4+ cells was half of the wild type in both BALB/c and C57BL/6 (BALB/C shown in Figure 5c). These mice grew and reproduced normally, similar to that reported previously by other groups who generated OLFM4 null mice.14,20 We tested for loss of OLFM4 on neutrophil development. Both the percentage of cells in the bone marrow that were CD11b+, Ly6g+ neutrophils and the number of neutrophils per femur were unaffected by loss of OLFM4 (Figure 5c).
Figure 5.
The OLFM4 null mouse has normal neutrophil counts. (a) Western blot for OLFM4 from the bone marrow of wild type, heterozygous and OLFM4 null mouse. (b) Flow cytometric dot plots of CD11b+ bone marrow cells from wild type BALB/c, heterozygous and OLFM4 null mouse, crossed onto BALB/c background for 5 generations, showing the loss of OLFM4 in null mice and confirming specificity of the Ab. (c) Bar graphs showing there is no difference in the number of neutrophils in terms of percentage or number of neutrophils per femur when comparing OLFM4 null mice with wild type mice.
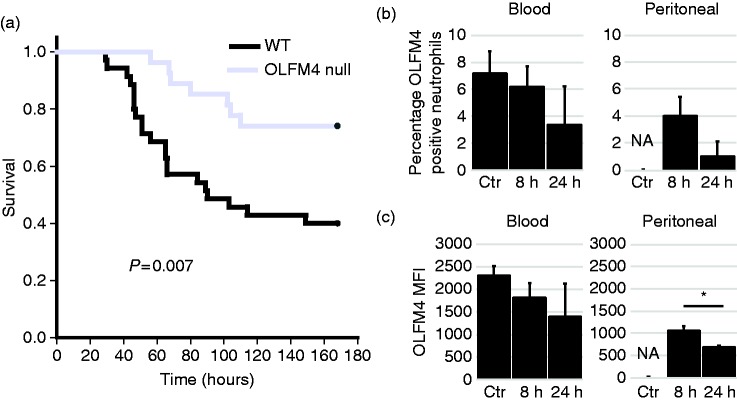
Because humans with high percentage of OLFM4+ neutrophils at the time of admission with septic shock have worse outcomes, we tested if OLFM4 deletion affected outcome of murine sepsis. We used CLP model to induce peritonitis and polymicrobial sepsis. OLFM4 null male mice were protected from death compared to wild type controls (Figure 6a). We also tested for changes in OLFM4 expression within neutrophils using cecal slurry model of polymicrobial sepsis. We found that there was a trend toward decreased percentage of OLFM4+ positive neutrophils in the blood and peritoneum (Figure 6b). There was also a trend toward decreased OLFM4 MFI within neutrophils in the blood and a clear decrease in MFI from neutrophils from the peritoneal space (Figure 6c).
Figure 6.
OLFM4 null mice are protected from septic shock. (a) Kaplan-Meier Survival curve showing OLFM4 null mice (n = 25) have less mortality following cecal ligation and puncture when compared to wild type mice (n = 29). (b) Percentage of OLFM4+ neutrophils from the blood and peritoneum from control and animals that were injected with cecal slurry 8 and 24 h previously. There is a trend toward decrease expression. Percentage of neutrophils could not be determined in control peritoneal samples because there were two few neutrophils to accurately measure. (c) Graphs showing OLFM4 MFI from the blood and peritoneal cells. In the blood there is a trend toward decreased expression and in the peritoneal there is a significant difference between the two time points (*P = 0.004).
Discussion
Neutrophil heterogeneity and plasticity have been proposed for some time, however, neutrophils have lagged far behind other leukocytes in our ability to identify unique subpopulations. This is largely because of the lack of surface markers that allow for easy identification of these subpopulations. In the rare cases where surface markers that identify unique subpopulations of neutrophils have been identified, such as CD177, it has been difficult to find functional differences between those cells that express CD177 and those that do not.31,32 Other markers such as, CXCR4 and CD62L have been shown to mark aged neutrophils, marked for clearance from the circulation.33
In humans, OLFM4 has been shown to be expressed in a subpopulation of neutrophils, around 25% in healthy controls with variation between 10–30%. Clemmensen et al. reported that in healthy volunteers, sampling the same individual over time, the percentage of OLFM4 expressing neutrophils did not change.15 Our group showed that in paediatric patients admitted to the intensive care unit with septic shock and increased percentage of OLFM4 expressing neutrophils had greater end organ injury.18 Welin et al. characterized OLFM4+ and OLFM4− neutrophil populations from humans, but was unable to identify difference in terms of phagocytosis, migration, or apoptosis. They did find OLFM4 associated with NETs and speculated on its potential role in NETosis.16 There is little data regarding control of OLFM4 expression. Chin et al. showed that in mice, there are several NF-κB transcription factor binding sights located in the promoter region and showed that these were important for expression for OLFM4 expression in cell lines.34
Because of the technical limitations in human research, we tested if the mouse might be a good model to understand the role of OLFM4 in neutrophil biology. We first confirmed that murine neutrophils expressed Olfm4 and found, like humans, in the blood and bone marrow compartment, OLFM4 was expressed exclusively in neutrophils. The expression of Olfm4 begins early in neutrophil development and it has a similar expression pattern as the neutrophil marker Ly6g. Olfm4 expression increases with neutrophil maturation and is also increased during LPS exposure in mice, similar to humans where OLFM4 was found to be highly up-regulated during septic shock.18
We developed a polyclonal Ab to conduct flow cytometry in murine cells. We used full-length murine OLFM4 expressed in 293T cells, to ensure glycosylation and eukaryotic protein folding, as an immunogen in rabbit. Following two immunizations the rabbit serum accurately identified a subpopulation of 6–8% of neutrophils in C57Bl/6 mice. This staining was only present with permeabilization of the cells, similar to human neutrophils and consistent with the subcellular localization in granules. The percentage of neutrophils in the bone marrow and peripheral blood differed between strains of mice, suggesting genetic regulators of the OLFM4 locus. Evaluating expression of OLFM4+ neutrophils during myeloid development showed that OLFM4 protein expression closely matches the mRNA expression and OLFM4 staining can be seen early in the promyelocyte and myelocyte stage. Interestingly, the percentage of cells that express OLFM4 from promyelocyte to mature neutrophil did not change, suggesting that OLFM4 expression is not simply a protein expressed during neutrophil maturation, but that from the early stages of neutrophil differentiation, there is dichotomous pathways to either express OLFM4 or not. In addition, stimulation with LPS led to increased Olfm4 transcript, but not an increase in the percentage of neutrophils expressing OLFM4 (not shown), suggesting OLFM4 is not simply an activation marker. Thus, the subpopulation of OLFM4 expressing neutrophils is conserved in mice and humans.
We tested for differences in neutrophil functions between OLFM4+ and OLFM4− neutrophils. Mixing cells with opsonized fluorescent bacteria showed no difference between the two subpopulations. We also tested migration in vitro (transwell) and in vivo (LPS lung lavage) and again found no difference between the two subpopulations. Both of these findings are similar to human OLFM4+ and OLFM4− neutrophils.16 Because others have reported that OLFM4 associates with NETs, we tested if OLFM4 associated with NETs in mice. We identified both OLFM4+ and OLFM4− neutrophils undergoing NET formation. We also found that most NETs we identified had small amounts of OLFM4 colocalizing within the DNA NET. We cannot be sure if this is simply sticky DNA binding secreted OLFM4 or if there is a functional role for OLFM4 within the NET. Thus, mice mimic human studies showing OLFM4 colocalization with neutrophil NETs and may be useful if further testing OLFM4’s role in NETosis.
We also generated an OLFM4 null mouse to test the effect on the neutrophil population. This null mouse was also helpful in confirming the specificity of our polyclonal Ab. Interestingly, comparing homozygous and heterozygous animals showed that OLFM4 MFI was unchanged. However, the percentage of OLFM4+ neutrophils was exactly half of the wild type animals. This suggests monoallelic expression from the OLFM4 locus but requires further testing. The fact that the percentage and number of neutrophils in the bone marrow was unaffected by OLFM4 deletion, suggests that OLFM4 is not essential to the development of this subpopulation of neutrophils.
We found that OLFM4 null mice are protected when challenged with the CLP model of polymicrobial sepsis. This finding is consistent with previous work showing that mice deficient of OLFM4 are protected, compared to wild type mice, from challenge with intraperitoneal injection of E. coli and Staphylococcus aureus. These studies together strongly suggest OLFM4 plays a pathogenic role in immune responses.21 OLFM4 binds and inhibits cathepsin C, a serine protease that potentiates several cellular antimicrobial functions. Two lines of evidence suggest that OLFM4 participates in immune functions independent of cathepsin C. First, crossing the OLFM4 null mouse onto cathepsin C null mice still provided protection from challenges by E. coli and S. aureus.22 Second, the bacterial challenge experiments were carried out on the C57Bl/6 background, where only 6–8% of neutrophils express OLFM4. Thus, in only 6–8% of neutrophils would have OLFM4-dependent, increased cathepsin C activity and the protective phenotype would be attributable to this small number of neutrophils. These data suggest OLFM4 has other functions in immune responses that are yet to be described. The finding that mice are similar to humans in that OLFM4 expression is limited to a subset of neutrophils, adds to the likelihood that the murine model may be informative to the function and regulation of OLFM4 expression in mammalian neutrophils.
Previous studies evaluating the role of OLFM4 in murine neutrophils have been under the assumption that all neutrophils express OLFM4 protein.21,22,34,35 Furthermore, these studies were done on the C57Bl6 background, which we show here to only express OLFM4 in 6–8% of neutrophils. This perhaps suggests that some of the observed phenotype differences, such as differences in superoxide production and increased killing of bacteria, may be due to secreted OLFM4 rather than the intrinsically expressed OLFM4, because differences reported would be attributable to only 6–8% of neutrophils. It could also be due to OLFM4 originating from a source other than neutrophils. These questions will wait for the development of a conditional OLFM4 murine line.
A major limitation of our studies is the inability to separate live OLFM4+ and OLFM4− neutrophils from humans or mice. This is because OLFM4 is expressed intracellularly and requires fixation and permeabilization of the cells to identify them. This precludes important experiments like transcriptome comparisons, bacterial killing assays, oxidative burst comparisons and adoptive transfer. The development of an OLFM4 reporter mouse would greatly facilitate these experiments in the future.
Our preliminary data from human patients with septic shock suggests that the percentage of neutrophils that express OLFM4 can change over time.18 Others have reported that during health, the percentage of OLFM4+ neutrophils does not change over time. Here we found that adult mice up-regulate Olfm4 transcript with LPS exposure, but do not increase the percentage of neutrophils that express OLFM4 with LPS stimulation or during sepsis induced by cecal slurry injection. We did appreciate a trend toward decreased percentage of OLFM4+ cells during sepsis and there was a notable decrease in OLFM4 MFI in peritoneal neutrophils. We cannot be sure from these preliminary studies, but we suspect that the decrease in MFI and trend toward decreased percentage of OLFM4+ neutrophils is due to degranulation of neutrophils and loss of intracellular OLFM4. These findings in inbred mouse lines are somewhat different than human studies that show increase in percentage of OLFM4+ neutrophils in some patients. However, it also suggests that this is a consistent subpopulation of neutrophils and that OLFM4 is not simply an activation marker or marker of neutrophil maturation, but represents a separate subpopulation of neutrophils, defined early in neutrophil development.
Supplemental Material
Supplemental Material for Olfactomedin 4 marks a subset of neutrophils in mice by Matthew N Alder, Jaya Mallela, Amy M Opoka, Patrick Lahni, David A Hildeman and Hector R Wong in Innate Immunity
Acknowledgements
We thank Dr Andrew Herr for assistance in producing recombinant OLFM4 protein, CCHMC Research Flow Cytometry Core, CCHMC Confocal Imaging core and the CCHMC Transgenic Animal and Genome Editing Care for assistance in OLFM4 null mouse generation. We thank H Leighton Grimes, Marie-Dominique Filippi and Andre Olsson for assistance and advice.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: NIH grants K08 GM124298, K12 HD028827, T32 GM008478, and R01 GM108025.
References
- 1.Ch Ho E, Buckley KM, Schrankel CS, et al. Perturbation of gut bacteria induces a coordinated cellular immune response in the purple sea urchin larva. Immunol Cell Biol 2016; 94: 861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nauseef WM, Borregaard N. Neutrophils at work. Nat Immunol 2014; 15: 602–611. [DOI] [PubMed] [Google Scholar]
- 3.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol 2013; 13: 159–175. [DOI] [PubMed] [Google Scholar]
- 4.Mayadas TN, Cullere X, Lowell CA. The multifaceted functions of neutrophils. Annu Rev Pathol 2014; 9: 181–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruger P, Saffarzadeh M, Weber AN, et al. Neutrophils: Between host defence, immune modulation, and tissue injury. PLoS Pathog 2015; 11: e1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat Immunol 2011; 12: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyrau M, Bodkin JV, Nourshargh S. Neutrophil heterogeneity in health and disease: A revitalized avenue in inflammation and immunity. Open Biology 2012; 2(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Huang Q, Yang Z, et al. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res 2004; 64: 2474–2481. [DOI] [PubMed] [Google Scholar]
- 9.Rosenbauer F, Wagner K, Zhang P, et al. pDP4, a novel glycoprotein secreted by mature granulocytes, is regulated by transcription factor PU.1. Blood 2004; 103: 4294–4301. [DOI] [PubMed] [Google Scholar]
- 10.Grover PK, Hardingham JE, Cummins AG. Stem cell marker olfactomedin 4: critical appraisal of its characteristics and role in tumorigenesis. Cancer Metastasis Rev 2010; 29: 761–775. [DOI] [PubMed] [Google Scholar]
- 11.Oh HK, Tan AL, Das K, et al. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res 2011; 17: 2657–2667. [DOI] [PubMed] [Google Scholar]
- 12.Yu L, Wang L, Chen S. Olfactomedin 4, a novel marker for the differentiation and progression of gastrointestinal cancers. Neoplasma 2011; 58: 9–13. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Li H, Hong SH, et al. Olfactomedin 4 deletion induces colon adenocarcinoma in Apc(Min/+) mice. Oncogene 2016; 35: 5237–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuijers J, van der Flier LG, van Es J, et al. Robust cre-mediated recombination in small intestinal stem cells utilizing the OLFM4 locus. Stem Cell Reports 2014; 3: 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemmensen SN, Bohr CT, Rorvig S, et al. Olfactomedin 4 defines a subset of human neutrophils. J Leukoc Biol 2012; 91: 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welin A, Amirbeagi F, Christenson K, et al. The human neutrophil subsets defined by the presence or absence of olfm4 both transmigrate into tissue in vivo and give rise to distinct NETs in vitro. Plos One 2013; 8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong HR, Cvijanovich N, Allen GL, et al. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med 2009; 37: 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alder MN, Opoka AM, Lahni P, et al. Olfactomedin-4 is a candidate marker for a pathogenic neutrophil subset in septic shock. Crit Care Med 2017; 45(4): e1929–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang JC, Liu WL, Tang DC, et al. Identification and characterization of a novel member of olfactomedin-related protein family, hGC-1, expressed during myeloid lineage development. Gene 2002; 283: 83–93. [DOI] [PubMed] [Google Scholar]
- 20.Liu WL, Yan M, Liu YQ, et al. Olfactomedin 4 down-regulates innate immunity against Helicobacter pylori infection. Proc Natl Acad Sci USA 2010; 107: 11056–11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, Yan M, Sugui JA, et al. Olfm4 deletion enhances defense against Staphylococcus aureus in chronic granulomatous disease. J Clin Invest 2013; 123: 3751–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu WL, Yan M, Liu YQ, et al. Olfactomedin 4 inhibits cathepsin c-mediated protease activities, thereby modulating neutrophil killing of Staphylococcus aureus and Escherichia coli in mice. J Immunol 2012; 189: 2460–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan CL, Hu YC. A transgenic core facility's experience in genome editing revolution. Adv Exp Med Biol 2017; 1016: 75–90. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swamydas M, Lionakis MS. Isolation, purification and labeling of mouse bone marrow neutrophils for functional studies and adoptive transfer experiments. J Vis Exp 2013; 10(77): e50586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toscano MG, Ganea D, Gamero AM. Cecal ligation puncture procedure. J Vis Exp 2011; 7(51). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr ME, Steele AM, Saito M, et al. A new cecal slurry preparation protocol with improved long-term reproducibility for animal models of sepsis. PLoS One 2014; 9: e115705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brand HK, Ahout IML, de Ridder D, et al. Olfactomedin 4 serves as a marker for disease severity in pediatric respiratory syncytial virus (RSV) infection. Plos One 2015; 10(7): e131927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kangelaris KN, Prakash A, Liu KD, et al. Increased expression of neutrophil-related genes in patients with early sepsis-induced ARDS. Am J Physiol Lung Cell Mol Physiol 2015; 308: L1102–L1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satake S, Hirai H, Hayashi Y, et al. C/EBPbeta is involved in the amplification of early granulocyte precursors during candidemia-induced ‘emergency’ granulopoiesis. J Immunol 2012; 189: 4546–4555. [DOI] [PubMed] [Google Scholar]
- 31.Hu N, Mora-Jensen H, Theilgaard-Monch K, et al. Differential expression of granulopoiesis related genes in neutrophil subsets distinguished by membrane expression of CD177. PLoS One 2014; 9: e99671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z, Liang R, Ohnesorg T, et al. Heterogeneity of human neutrophil CD177 expression results from CD177P1 pseudogene conversion. PLoS Genet 2016; 12: e1006067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang D, Chen G, Manwani D, et al. Neutrophil ageing is regulated by the microbiome. Nature 2015; 525: 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin KL, Aerbajinai W, Zhu J, et al. The regulation of OLFM4 expression in myeloid precursor cells relies on NF-kappaB transcription factor. Br J Haematol 2008; 143: 421–432. [DOI] [PubMed] [Google Scholar]
- 35.Liu W, Liu Y, Li H, et al. Olfactomedin 4 contributes to hydrogen peroxide-induced NADPH oxidase activation and apoptosis in mouse neutrophils. Am J Physiol Cell Physiol 315(4): C494–C501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for Olfactomedin 4 marks a subset of neutrophils in mice by Matthew N Alder, Jaya Mallela, Amy M Opoka, Patrick Lahni, David A Hildeman and Hector R Wong in Innate Immunity