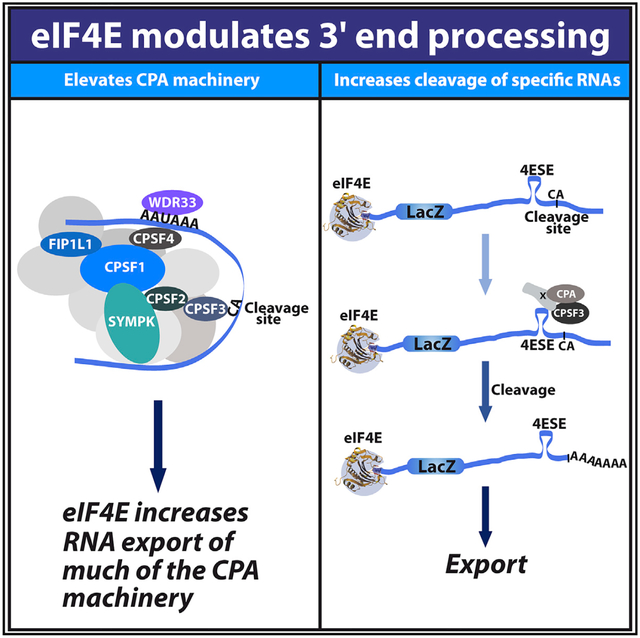
SUMMARY
The eukaryotic translation initiation factor eIF4E is nuclear and cytoplasmic where it plays roles in export and translation of specific transcripts, respectively. When we were studying its mRNA export activity, we unexpectedly discovered that eIF4E drives the protein expression of elements of the 3′-end core cleavage complex involved in cleavage and polyadenylation (CPA), including CPSF3, the enzyme responsible for cleavage, as well as its co-factors CPSF1, CPSF2, CPSF4, Symplekin, WDR33, and FIP1L1. Using multiple strategies, we demonstrate that eIF4E stimulates 3′-end cleavage of selected RNAs. eIF4E physically interacts with CPSF3, CPSF1, and uncleaved target RNA, suggesting it acts directly and indirectly on the pathway. Through these effects, eIF4E can generate better substrates for its mRNA export and translation activities. Thus, we identified an unanticipated function for eIF4E in 3′-end processing of specific target RNAs, and this function could potentially affect the expression of a broad range of oncoproteins.
Graphical Abstract
In Brief
Davis et al. demonstrate that the eukaryotic translation initiation factor eIF4E, which is usually associated with nuclear export and translation of specific transcripts, also acts in 3′-end processing of selected RNAs. Through these effects, eIF4E can generate better substrates for its export and translation activities and, thus, modulate the proteome.
INTRODUCTION
The eukaryotic translation initiation factor eIF4E has important roles in regulating the expression of specific proteins based on its activities in mRNA metabolism. These proteins are the downstream mediators of the physiological activities of eIF4E in proliferation, survival, invasion, and metastasis (Carroll and Borden, 2013). At the biochemical level, eIF4E modulates protein expression through three discrete functions: mRNA translation in the cytoplasm, mRNA stability and/or sequestration in processing bodies (P-bodies, and similar structures), and mRNA export in the nucleus (Borden, 2016). In all three cases, eIF4E targets subsets of N7 methyl guanosine (m7G)-capped RNAs, which also contain specific cis-acting elements typically found in their UTRs (Borden, 2016). These cis-acting elements provide the specificity that underlies eIF4E’s effects on selective substrate RNAs. As predicted by the RNA regulon model (Keene and Tenenbaum, 2002), groups of transcripts containing the same collection of cis-acting elements can be co-regulated by eIF4E, allowing the coordinated processing of groups of transcripts encoding proteins involved in related biochemistries (Culjkovic-Kraljacic et al., 2016; Zahreddine et al., 2017). In this way, eIF4E can potently affect cell physiology. Indeed, targeting these eIF4E activities in the clinic correlated with objective responses, including remissions (Assouline et al., 2009, 2015).
In some cell types, including some cancers, eIF4E is found mainly in the nucleus where its mRNA export activity is highly elevated (Assouline et al., 2009; Carroll and Borden, 2013; Culjkovic-Kraljacic et al., 2016). Further, eIF4E interacts with ~3,500 mRNAs in the nucleus, many encoding proteins that act in the same oncogenic networks (Culjkovic-Kraljacic et al., 2016). Importantly, eIF4E does not bind all RNAs in the nucleus, e.g., ACTIN, Lamin, or glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which are neither export nor translation targets of eIF4E (Rousseau et al., 1996). Further, not all translation targets of eIF4E are nuclear export targets, e.g., vascular endothelial growth factor (VEGF) is not an export target of eIF4E in the nucleus but is a translation target of eIF4E (Culjkovic et al., 2005, 2006; Culjkovic-Kraljacic et al., 2012). RNA-export substrates are capped and contain a ~50-nt element in the 3′ UTR, known as the eIF4E sensitivity element (4ESE) (Culjkovic et al., 2005, 2006). Studies with reporter RNAs, such as LacZ-4ESE, demonstrated that the addition of the 4ESE alone was sufficient to enable transcripts to bind eIF4E in the nucleus and be exported in an eIF4E-dependent manner (Culjkovic et al., 2005, 2006; Culjkovic-Kraljacic et al., 2012). Mutant forms of the 4ESE did not have that activity. By contrast, bulk mRNAs or LacZ alone were not bound to eIF4E in the nucleus and were not transported in an eIF4E-dependent manner but, rather, were exported using the bulk export pathway via NXF1/TAP (Culjkovic et al., 2005, 2006). The 4ESE acts in export by recruiting co-factors involved in eIF4E-dependent mRNA export, including the leucine-rich pentatricopeptide repeat cassette protein (LRPPRC), which directly binds the 4ESE RNA element eIF4E and the nuclear pore receptor CRM1, thereby forming a core export ribonuclear particle (RNP) (Topisirovic et al., 2009; Volpon et al., 2017). eIF4E also reprograms the nuclear pore complex to stimulate export (Culjkovic-Kraljacic et al., 2012), indicating that eIF4E can employ multiple strategies to produce an environment conducive to the export of its target transcripts.
Nuclear mRNA export typically requires transcripts to have a 3′ polyadenylated (polyA) tail, as well as a 5′ m7G cap (Culjkovic-Kraljacic and Borden, 2013). Thus, factors that facilitate mRNA processing indirectly stimulate nuclear export of matured mRNAs. Here, we explored the possibility that eIF4E is involved in maturation of the 3′ end of its export target mRNAs, as yet another strategy to promote mRNA export of its targets. Maturation of the 3′ end of most transcripts involves cleavage and polyadenylation (CPA), a two-step process starting with cleavage of a CA dinucleotide located near to a polyadenylation signal (PAS) by the endonuclease CSPF3, followed by the addition of the polyA tail by polyA polymerases (Proudfoot et al., 2002, 2011). These processes are closely linked and are, indeed, widely considered to occur concurrently (Mayr, 2016). The CPA machinery consists of several essential co-factors, including a core cleavage complex that consists of six key subunits, including cleavage and polyadenylation specific factor 4 (CPSF4), which directly binds the PAS site along with pre-mRNA 3′ end-processing protein (WDR33) (Chan et al., 2014; Schönemann et al., 2014). Further CPSF1 is predicted to recruit additional core subunits, including CPSF2 (Hunter et al., 1995; Murthy and Manley, 1995), pre-mRNA 3′ end-processing factor FIP1 (FIP1L1) (Kaufmann et al., 2004), predicted scaffold protein symplekin (SYMPL) (Kolev and Steitz, 2005), and CPSF3 (Mandel et al., 2006). After that, two key polyadenylation factors, polyA polymerase (PAP) and nuclear PolyA binding protein 1 (PABPN1) are recruited for polyA tail synthesis (Kühn et al., 2009; Chang et al., 2014; Jenal et al., 2012; de Klerk et al., 2012) (Figure 1A, adapted from Neve et al., 2017). Here, we report that eIF4E promotes the expression of several factors in the core cleavage complex, interacts with the CPSF3 cleavage enzyme, and promotes 3′-end cleavage of specific RNAs.
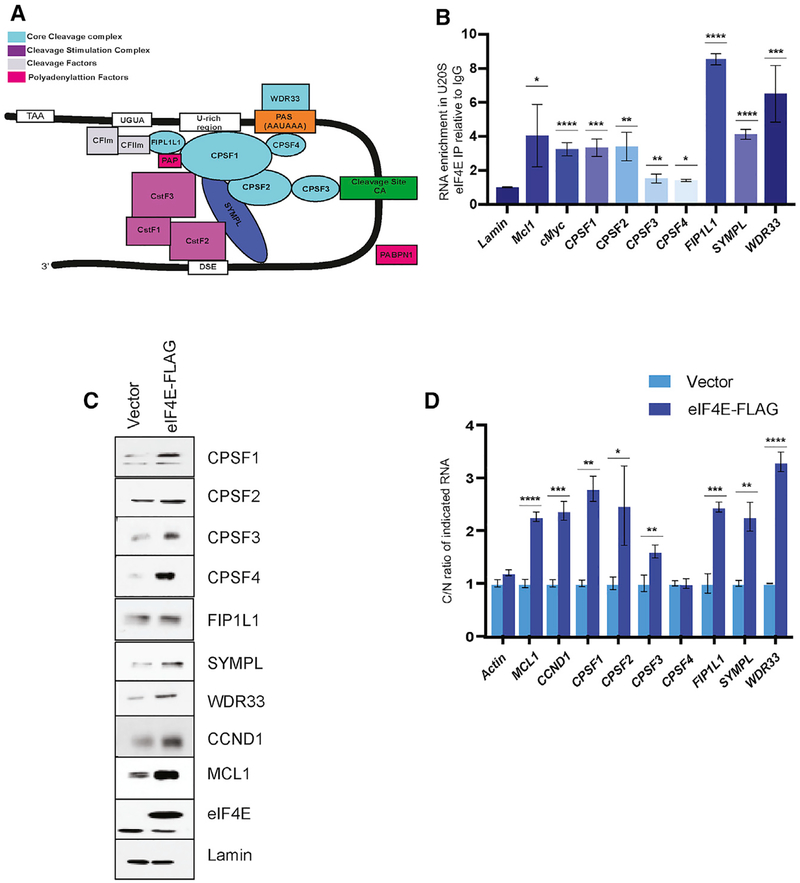
Figure 1. eIF4E Promotes Production of the CPA Machinery.
(A) Model of the cleavage and polyadenylation (CPA) machinery. Only some factors are shown for simplicity. Elements important for this process in the RNA are also shown. Cleavage site is depicted as a green box. See text for details.
(B) Endogenous eIF4E RNA immunoprecipitation in U2OS cells relative to IgG for indicated RNAs using qRT-PCR. Lamin RNA is a negative control and MCL and c-MYC are positive controls. RNAs were normalized to actin RNA.
(C) Analysis of the effects of eIF4E overexpression relative to vector control on the indicated CPA components by western blot analysis. Lamin serves as a loading control; MCL1 and CCND1 are positive controls. Representative results from three biological replicates are shown.
(D) Analysis of RNA export of indicated RNAs by monitoring the cytoplasmic to nuclear (C/N) ratio of RNAs using qRT-PCR. Values are relative to the vector control for each transcript.
RIP experiments were performed in four biological replicates each in technical triplicate for (B) and export experiments performed in three biological replicates each in triplicate in (D). RNAs were normalized to GAPDH. Means ± SD are shown with p values (Student’s t test) as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
RESULTS
eIF4E Drives Production of Core Cleavage Complex
Our previous nuclear eIF4E RNA immunoprecipitation (RIP) RNA sequencing (RNA-seq) studies in B cell non-Hodgkin lymphoma cell line, ODI-LY1, indicated that eIF4E associated with transcripts encoding several core cleavage factor subunits, including CPSF1, CPSF3L, SYMPL, and FIP1L1 (Culjkovic-Kraljacic et al., 2016). This positions these transcripts as potential RNA export targets of eIF4E, and thus, we reasoned that eIF4E could elevate these factors and thereby modulate PAS cleavage for at least some RNAs. Therefore, we investigated whether eIF4E promoted production of these factors and could affect cleavage in a more pliable model cell line, U2OS, which is derived from human osteosarcoma. We generated stable U2OS cell lines overexpressing eIF4E using eIF4E-FLAG or vector-FLAG (vector) constructs as in Culjkovic et al. (2005). We fractionated cells into nuclear and cytoplasmic components and monitored fractionation quality via U6 small nuclear RNA (snRNA) and tRNAlys, respectively, as markers of the nucleus and cytoplasm using semiquantitative PCR (Zahreddine et al., 2017). Here, by means of nuclear, endogenous eIF4E RIP, we show that eIF4E associated with CPSF1, CPSF2, CPSF3, CPSF4, and SYMPL transcripts with a ~2–4-fold enrichment, relative to immunoglobulin G (IgG) controls and notably comparable to positive controls MCL1 and cMYC RNAs (Figure 1B). Additionally, we observed ~7-fold enrichment of PAS recognition co-factor WDR33 and a ~9-fold enrichment of FIP1L1. Lamin was used as a negative control and not enriched over IgG controls (Figure 1B). We observed an upregulation of protein expression in CPSF1, CPSF3, CPSF4, SYMPL, and WDR33, relative to the controls MCL1 and CCND1, with modest, but significant, increase in CPSF2 and FIP1L1 and no effect on the negative control LAMIN (Figure 1C). These data indicate that eIF4E increases levels of at least some components of the CPA machinery.
Next, we set out to define the underlying mechanism that promoted production of the core cleavage-complex factors, in response to eIF4E upregulation. First, eIF4E overexpression affects nuclear export via CRM1 of 4ESE-containing target transcripts, e.g., MCL1, CCND1; thus, we performed export assays (Culjkovic et al., 2005). eIF4E-FLAG and vector cells were fractionated into nuclear and cytoplasmic fractions with U6snRNA and tRNAlys as described above (Figure S1A). RNA levels were monitored via qRT-PCR and normalized to GAPDH, which is not sensitive to eIF4E-driven nuclear export (Culjkovic et al., 2005, 2006; Culjkovic-Kraljacic et al., 2012, 2016; Rousseau et al., 1996). We observed significant increases in nuclear export ranging from ~2 to 3-fold of CPSF1, CPSF2, CPSF3, FIPL1, SYMPL, and WDR33 transcripts comparable to the positive controls CCND1 and MCL1 in eIF4E-overexpressing cells versus vector controls (Figure 1D). There was no change to export activity of CPSF4 relative to the negative control ACTIN (Figure 1D). The total mRNA levels in each case remained unaffected, demonstrating that those effects were post-transcriptional (Figure S1B).
Given the role of eIF4E in translation, we also examined whether eIF4E overexpression enhanced the translation efficiency of those transcripts by performing polysome-loading assays (Figure S1C). Although eIF4E translation is known to target MCL1, displaying a significant shift in polysomal loading, we observed no, or very minor, polysomal shifts for the examined transcripts (CPSF1, CPSF2, CPSF3, CPSF4, FIP1L1, WDR33, and SYMPL) as a result of eIF4E overexpression, comparable to ACTIN, an established negative control (Rousseau et al., 1996) (Figure S1D). Although the dearth of effects from eIF4E on CPSF4 at the export or translation level are apparent, there is a clear increase at the protein level suggesting it is an indirect target of eIF4E. Thus, we concluded that the effects of eIF4E on protein levels were mediated mainly through the ability of eIF4E to promote the export of the corresponding RNAs encoding the 3′-end processing machinery.
In all, our data demonstrate that eIF4E drives the production of key elements of the CPA machinery. This raised two non-exclusive possibilities: eIF4E could modulate cleavage indirectly through its effects on the cleavage machinery, or it could have a yet-to-be-defined, direct role in the processing of its target export transcripts.
eIF4E Interacts with the Key Cleavage Enzyme CPSF3 and Its Co-factor CPSF1
To explore the possibility that eIF4E could have some type of direct role in PAS cleavage, we examined whether eIF4E interacted with the cleavage machinery in the nucleus, focusing on the cleavage enzyme CPSF3 and its co-factor CPSF1. We examined these interactions by immunoprecipitations (IPs) of endogenous eIF4E in nuclear lysates from U2OS cells (Figure 2A). We found that eIF4E physically associated with both CPSF3 and CPSF1 but did not immunoprecipitate with the negative control LAMIN (Figures 2A and S2A). Conversely, we used CPSF1 and CPSF3 antibodies and observed immunoprecipitation with those factors and endogenous eIF4E or eIF4E-FLAG in nuclear lysates from U2OS cells (Figure S2B). Fractionation quality was monitored by western blot using Lamin and RNA polymerase II (PolII) for nuclear fractions and tubulin and GAPDH for cytoplasmic fractions (Figure 2A). Thus, in addition to the ability of eIF4E to promote the production of elements involved in CPA, eIF4E also physically associated with the enzymatically active components of that complex in the nucleus.
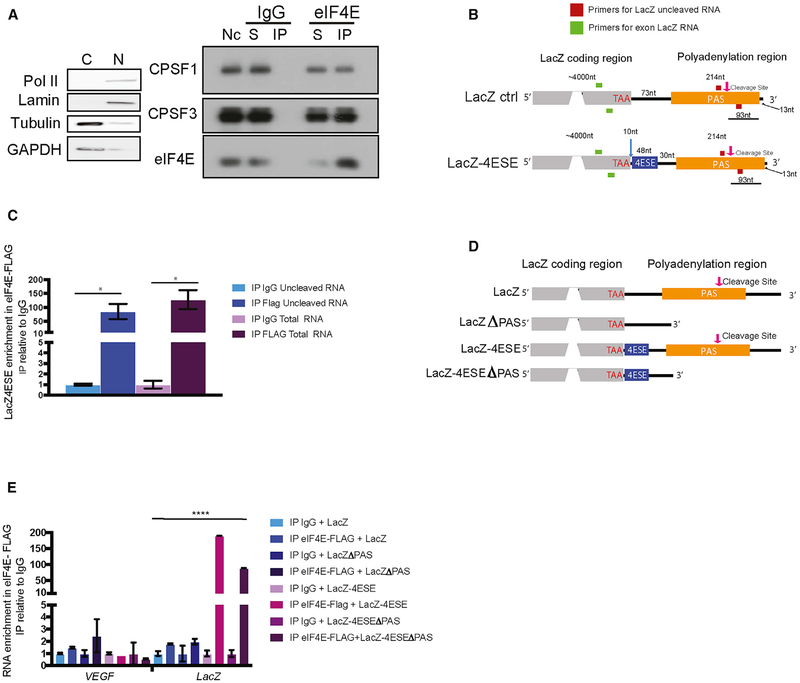
Figure 2. eIF4E Physically Associates with Some of the Cleavage Machinery and RNA Substrates for the Pathway.
(A) Endogenous eIF4E immunoprecipitated (IP) with CPSF1 and CPSF3 in nuclear lysates from U2OS cells. Fractionation controls for nuclear IP were Lamin and RNA polymerase II for the nucleus (N) and GAPDH and tubulin for the cytoplasm (C). Nc, nuclear lysate; S, supernatant.
(B and D) Diagram of constructs, which is not to scale, with the position of primers to detect cleaved (reverse primer immediately downstream the cleavage site (C/A) + 13 thymine nucleotides, shown in pink (qRT-PCR; Figure S3C) and uncleaved LacZ transcripts (primers in red, forward used for both cleaved and uncleaved) (B). (D) depicts constructs used for studies into the deletion of the PAS region.
(C) eIF4E-FLAG RIP using primers that bracket the cleavage site as used for the cleavage assay (uncleaved primers [red], exon “total” primers [green]). Results are relative to IgG controls. These findings indicate that, in the nucleus, eIF4E associated with uncleaved LacZ-4ESE RNA. (E) eIF4E-FLAG RIP from nuclear lysates shows PAS enriched association with LacZ-4ESE by 2-fold relative to LacZ-4ESEΔPAS. As expected, eIF4E did not interact with VEGF RNA in nuclear lysates.
Experiments were three biological replicates each performed in technical duplicates for (C) and (E) and three biological replicates for (A), each performed in technical triplicates. For qRT-PCR experiments, values are means ± SD with p values as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
eIF4E Binds Uncleaved 4ESE RNA
If eIF4E is physically involved in some step of the cleavage process, it would be expected to physically interact with uncleaved RNAs as well as maintaining its conventional interaction with processed RNAs. We focused on LacZ-4ESE RNAs because that is the best-defined RNA export model for eIF4E and directly binds eIF4E in the nucleus, whereas the LacZ-control RNA does not physically associate with nuclear eIF4E (Culjkovic et al., 2005, 2006; Topisirovic et al., 2009; Volpon et al., 2017). Vector and eIF4E-FLAG U2OS cell lines were stably transfected with constructs containing the ~50-nt 4ESE element fused to the 3′ end of LacZ (LacZ-4ESE) or LacZ constructs, without the addition of the 4ESE (LacZ; Figure 2B) (Culjkovic et al., 2005, 2006). Previously generated constructs were used, but briefly, the 48-nt 4ESE from Pim1 was inserted into the LacZ constructs using enzymatic digestion (EcoRI/XbaI), which excised a 33-bp fragment, leaving a net difference in size of 15 bp between LacZ and LacZ-4ESE RNAs (Figure 2B; Culjkovic et al., 2006). Using these constructs, we generated the following cell lines: eIF4E-FLAG + LacZ-4ESE, eIF4E-FLAG + LacZ, vector + LacZ-4ESE, and vector + LacZ. We note that there were no introns in the LacZ constructs.
Using exonic primers to capture the complete population of nuclear LacZ-4ESE RNAs, we found that nuclear eIF4E-FLAG RIPs with LacZ-4ESE more than ~120-fold, relative to IgG controls (Figure 2C), and further, that eIF4E does not interact with LacZ, similar to previous studies (Culjkovic et al., 2005, 2006; Topisirovic et al., 2009) (Figure 2E) We then monitored whether LacZ-4ESE RNA bound eIF4E while it contained an intact PAS cleavage site (uncleaved RNA) by carrying out eIF4E-FLAG RIPs from nuclear lysates and using primers that bracketed the PAS cleavage site to detect RNA before cleavage (Figure 2C). Our RIP data demonstrated that eIF4E also immunoprecipitated uncleaved LacZ-4ESE RNA, ~75-fold more than IgG controls. Thus, eIF4E binds LacZ-4ESE RNA both before and after PAS site cleavage. We note that transcripts must be capped to interact with eIF4E in the nucleus, as in the cytoplasm (Culjkovic et al., 2005).
Given that LacZ-4ESE RNA bound to eIF4E in both its cleaved and uncleaved forms, we investigated whether deletion of the PAS, would modulate the ability of eIF4E to associate with its target RNAs. We monitored LacZ RNA as a negative control. We generated LacZ and LacZ-4ESE constructs with the PAS deleted (ΔPAS) (Figure 2D). In eIF4E RIP studies with exonic primers to detect target RNAs by qRT-PCR, we observed that eIF4E-FLAG associated with LacZ-4ESE RNAs with ~150-fold enrichment and LacZ-4ESEΔPAS RNA with a ~50-fold enrichment, relative to IgG controls (Figure 2E). Thus, although the PAS was not required for the association of eIF4E with 4ESE-containing RNAs, it enhanced the interaction by ~2 fold (Figure 2E). As expected, eIF4E did not associate with LacZ or LacZΔPAS mRNAs relative to IgG controls (Figure 2E). As expected, eIF4E did not bind the negative-control VEGF RNA, given that it was not a nuclear target of eIF4E (Culjkovic-Kraljacic et al., 2012). There was no change to total RNA target levels (Figure S2C). Thus, eIF4E binds LacZ-4ESE RNA with or without PAS, but there is an increase in its association over LacZ-4ESEΔPAS, an increase that could arise because of differences in processing (fewer polyadenylated RNAs would be produced from these transcripts) and/or because, in the nucleus, the PAS element recruits factors that stabilize the eIF4E-RNA interaction. In all, these results show that nuclear eIF4E is positioned to bind the enzyme in the core cleavage complex, the PAS enhances the eIF4E-RNA interaction, and, in binding the uncleaved RNA, eIF4E binds an RNA substrate for the CPA machinery.
eIF4E Modifies Cleavage of 4ESE RNA
Given that eIF4E increased the expression of, and interacted with, many CPA elements as well as with RNA substrates of this machinery (Figures 1 and 2), we investigated whether eIF4E overexpression altered cleavage efficiency (Figure 3). As a direct measure of cleavage, we used an in vitro assay based on reports for histone intronless RNAs (Friend et al., 2007). Biotinylated RNA fragments corresponding to the 3′ end of LacZ and LacZ-4ESE transcripts were generated by in vitro transcription. Specifically, the 4ESE and control (Ctrl) RNA fragments were derived from the LacZ-4ESE and LacZ constructs, respectively, by insertion of a T7 site as shown (Figure 3A). RNAs were incubated with nuclear lysates derived from eIF4E-FLAG or vector control U2OS cells for 30 min. Both a representative assay and quantification from three biological replicates are shown (Figures 3B and 3C). Incubation of 4ESE RNA with lysates from eIF4E-overexpressing cells relative to vector cells corresponded to an ~4-fold increase in cleavage (Figures 3B, lanes 3 versus 4, and 3C, column 1 versus 2). By contrast, there was no substantial difference in the amount of cleavage (~1.1 fold) for Ctrl RNAs incubated with lysates from vector or eIF4E-overexpressing cells (Figures 3B, lanes 1 versus 2, and 3C, middle column). This is despite the fact that ~2.5-fold more Ctrl RNA was loaded relative to 4ESE RNA (Figure 3B, lanes 1 and 2 versus 3 and 4). Finally, we noted that, in eIF4E-overexpressing cells, LacZ-4ESE RNA was better cleaved by ~3.5-fold than LacZ was (Figures 3B, lanes 2 versus 4, and 3C, column 3). Note that, for quantification, the amount of cleaved RNA was normalized to the corresponding uncleaved RNA to account for loading differences that might contribute to the amount of cleaved RNA detected. This can also be analyzed in terms of relative percentage of each species; we observed that ~30% of LacZ-4ESE RNAs were cleaved in eIF4E-FLAG-overexpressing cells compared with ~10% in vector cells. In all, the in vitro cleavage assay supported the hypothesis that eIF4E overexpression increased cleavage of LacZ-4ESE RNAs.
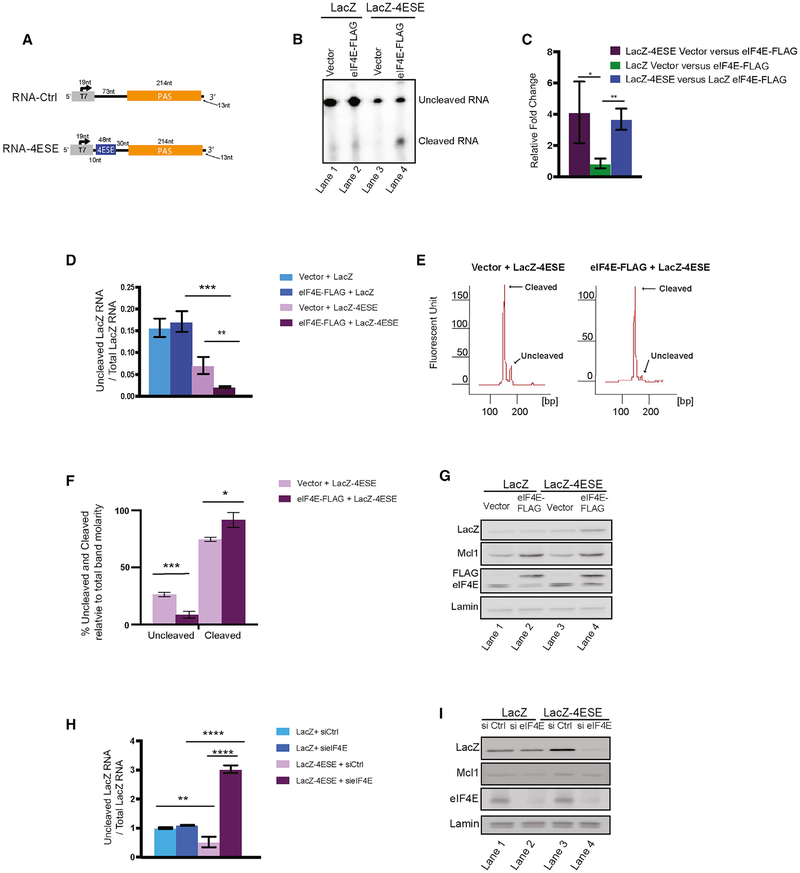
Figure 3. eIF4E Stimulates Cleavage of LacZ-4ESE RNA.
(A) Schematic of LacZ-4ESE and LacZ constructs used to generate respective RNA fragments (RNA-4ESE and RNA-Ctrl) for the in vitro cleavage assays in (B). Color coding is the following: gray, LacZ coding region and T7 site; blue, 4ESE; orange, PAS site. For further description of primer location, see Figure 2B.
(B) In vitro cleavage assay of RNA fragments incubated with nuclear lysates from eIF4E-FLAG or vector controls cells and visualized by northern blot.
(C) Quantification of three biological replicates of the cleavage assays indicated eIF4E increased cleavage of 4ESE-containing, more so than control fragments. Band intensity was quantified using ImageJ.
(D) Uncleaved/total LacZ or LacZ-4ESE transcripts measured in nuclear lysates from vector or eIF4E-FLAG U2OS cells as monitored by qRT-PCR and ratios normalized to total LacZ or LacZ-4ESE from whole-cell lysates. Increased levels of uncleaved transcripts indicated reduced PAS cleavage.
(E) Results from competitive semi-qPCR assays monitored by Bioanalyzer in the same cells as in (D). The relative amount of uncleaved and cleaved RNA can be detected in nuclear lysates in vector controls and eIF4E-FLAG cells. Representative of three biological replicates
(F). Average percentage of uncleaved and cleaved transcripts from Bioanalyzer data in (E). Calculation: (band uncleaved molarity/total of uncleaved and cleaved band molarity) 3×100.
(G) Corresponding western blots from total cell lysates showing LacZ and LacZ-4ESE expression with MCL1 protein serving as a positive control, given it is an established eIF4E target. Lamin is a loading control.
(H) RNAi knockdown of endogenous eIF4E (sieIF4E) increased levels of uncleaved LacZ-4ESE RNA relative to siRNA controls (siCtrl) but not LacZ. Experiments were performed in vector-control U2OS cells. Cleavage was measured in nuclear lysates obtained after appropriate siRNA.
(I) Corresponding western blots of total cell lysates for sieIF4E and controls. Lamin is a loading control.
Results are shown from three (C and D) or four (E and F) biological replicates, each performed in triplicate. Values are means ± SD with p values as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 from Student’s t test. Western blots are representative of three biological replicates.
To assess these effects in intact cells, we first monitored PAS cleavage in nuclear fractions as a function of eIF4E overexpression for LacZ and LacZ-4ESE RNAs using a qRT-PCR strategy monitoring the amount of uncleaved RNA relative to the total RNA. We started with uncleaved RNAs because this species was only present in the nuclear fractions, i.e., none was detected in the cytoplasm, indicating that this RNA species was not an eIF4E export target. By contrast, cleaved RNAs are rapidly polyadenylated and subsequently exported (Huang and Carmichael, 1996), thereby post-cleavage transcripts present a “moving target” in which 3′-end maturation is functionally coupled to nuclear export. Indeed, we observed polyadenylated LacZ and LacZ-4ESE RNAs in both nuclear and cytoplasmic compartments. Thus, measurement of uncleaved transcript levels is the most accurate way to assess cleavage alone without confounding additional factors, such as export. Specifically, we monitored the amount of LacZ or LacZ-4ESE transcripts in the nucleus that contained the intact PAS (i.e., uncleaved RNAs) using primers bracketing the cleavage site (Figure 2B, in red). To detect cleavage status, RNAs were isolated from nuclei in which PAS cleavage occurs (Liu et al., 2014). To confirm fraction quality, we monitored U6 snRNA and tRNAlys as nuclear and cytoplasmic markers, respectively, by semi-qRT-PCR (Figure S3A). To follow levels of all RNA species present, we monitored LacZ or LacZ-4ESE transcripts from whole-cell lysates using exon-specific primers (referred to as total LacZ or LacZ-4ESE RNA) (Figure 2B, in green). Before these studies, we verified the PAS site location for both LacZ constructs using 3′ QuantSeq, which identifies the precise CA cleavage site used (see STAR Methods). These studies demonstrated that there was a single cleavage at the predicted CA site within the bovine growth hormone, canonical PAS site derived from the vector (Goodwin and Rottman, 1992). Importantly, eIF4E did not alter the stability of total mRNAs in the nuclear or cytoplasmic compartments, as assessed by actinomycin D treatment (Culjkovic-Kraljacic et al., 2012). Finally, eIF4E overexpression did not alter LacZ or LacZ-4ESE RNA levels isolated from whole-cell lysates, as assessed using exonic primers (Figure S3B).
For studies quantifying levels of uncleaved RNAs, we observed an ~4-fold reduction in uncleaved LacZ-4ESE RNA levels in eIF4E-overexpressing versus vector LacZ-4ESE cells (Figure 3D); in close agreement with the ~4-fold change observed for these RNAs in the in vitro cleavage assay (Figures 3B and 3C). In addition, we observed an ~8-fold reduction in uncleaved LacZ-4ESE RNA levels relative toLacZ in eIF4E overexpressing cells. We also noted a ~2-fold reduction of uncleaved LacZ-4ESE RNA levels relative to LacZ in vector cells that could arise due to cleavage by endogenous eIF4E (Figure 3D). We note that the levels of uncleaved LacZ RNA were not sensitive to eIF4E levels, consistent with our in vitro cleavage assays (Figures 3D and 3B, respectively). Further, we observed that uncleaved LacZ and LacZ-4ESE RNA were only detected in the nuclear fraction. This is consistent with the literature, i.e., transcripts without 3′-end processing are poor substrates for RNA export (Liu et al., 2014). Indeed, eIF4E overexpression leads to increased mRNA export of LacZ-4ESE RNAs relative to vector controls but not LacZ RNAs (data not shown), as we reported previously (Culjkovic-Kraljacic et al., 2012). This is consistent with eIF4E directly or indirectly producing a population of RNAs that are better export substrates, i.e., by reducing the amount of nuclear, uncleaved RNAs without altering the total RNA levels (as measured by exonic primers, from whole-cell lysates). Thus, eIF4E overexpression led to reduced amounts of uncleaved LacZ-4ESE RNAs supporting the increased cleavage observed in vitro.
Next, we performed the converse assay using primers specific to cleaved and polyadenylated RNAs to measured levels of cleaved LacZ and LacZ-4ESE transcripts as a function of eIF4E overexpression. Here, forward primers were the same as in the uncleaved studies, but the reverse primer bracketed the polyA tail and remaining PAS site (Figure 2B). At first glance, the most obvious experiment was to compare LacZ and LacZ-4ESE cleaved and uncleaved transcripts from the same nuclear fractions as a function of eIF4E overexpression. However, that represents a major technical challenge, as mentioned above, because cleaved RNAs are known to be rapidly polyadenylated and then are excellent export substrates, and their measurement represents a “moving target.” Under those conditions, we observe an ~2-fold increase in cleavage of LacZ-4ESE RNAs isolated from the nuclear fractions of eIF4E-overexpressing cells over vector LacZ RNA control cells (Figure S3C). Although cleavage increased, it was not as substantial as the uncleaved or in vitro assays, likely because of that technical challenge.
To obtain the relative percent of uncleaved and cleaved transcripts, we used competitive, semi-qPCR and Bioanalyzer analysis. We focused on LacZ-4ESE in eIF4E and vector cells, given the interaction with eIF4E and the specific effects we observed, as described above. Using the equivalent forward primer (Figure 2B, in red) and a 1:1 ratio of uncleaved and cleaved reverse primers (Figure 2B, in pink), we determined the levels of both RNA species isolated from the nuclear fraction from our above set of cell lines. Primer efficiency was verified to be nearly identical for the different primers using qRT-PCR standard curve analysis. The uncleaved and cleaved primers generated the expected products at molecular weights of ~170 and ~150 bp. We measured the molarity of the cleaved and uncleaved bands and calculated the relative percent via Bioanalyzer methods. We note that in vector + LacZ-4ESE cells, there was ~25% uncleaved LacZ-4ESE RNA, which was reduced to ~10% in eIF4E+LacZ-4ESE cells (Figures 3E and 3F, left and right). This is similar to the magnitude of effects observed for the above three assays. Although the overall trends are the same, we also note that the percentages of uncleaved RNA are higher in the in vitro cleavage assay, but there are important caveats to consider. First, in vitro cleavage assays have the advantage that RNAs will not be “moving targets,” given the entire experiment is monitored within nuclear lysates. However, these experiments are in vitro and, thus, do not fully reflect the normal organization within intact cells. In the competitive PCR experiments, an important caveat is the potential for differential primer efficiency in the competitive conditions, which could skew the results and, additionally, given these experiments were measuring RNA in intact cells, the “moving target” caveat still exists. These limitations notwith standing, all of our results are consistent with enhanced cleavage of LacZ-4ESE RNAs upon eIF4E overexpression.
We analyzed protein levels to determine whether changes in cleavage had a biological effect. We denote the protein product derived from the LacZ-4ESE or LacZ transcripts as LacZ-4ESE and LacZ proteins, respectively. Consistent with elevated PAS cleavage, we observed a substantial increase in LacZ-4ESE protein levels in eIF4E-overexpressing cells relative to vector conrols (Figure 3G, lanes 3 versus 4). By contrast, LacZ protein levels were not changed in eIF4E-overexpressing versus vector cells (Figure 3G, lanes 1 versus 2). Thus, reduced levels of uncleaved and increased levels of cleaved LacZ-4ESE RNAs in eIF4E-overexpressing cells correlated with increased export of LacZ-4ESE RNAs and corresponding increase of their protein levels. By contrast, there was little change to LacZ RNA export or protein levels as a function of eIF4E overexpression (Culjkovic-Kraljacic et al., 2012), consistent with the in vitro and cell-based cleavage assays.
Effects of Reduced eIF4E Levels on Cleavage
Next, we monitored the effects of eIF4E RNAi knockdown on cleavage using LacZ and LacZ-4ESE cell lines that expressed endogenous eIF4E. We observed an ~6-fold increase in uncleaved RNA levels upon eIF4E knockdown for LacZ-4ESE relative to the small interfering (siRNA) controls indicating reduced PAS cleavage (Figure 3H). These effects were paralleled at the mRNA export level, where eIF4E RNAi led to an ~3-fold reduction in mRNA export, with no changes to total RNA levels (Figures S3D–S3F). Unlike LacZ-4ESE, LacZ RNA was unaffected by eIF4E knockdown with no change in cleavage, total mRNA levels, or mRNA export (Figures 3H and S3D–S3F). By western blot, we observed that eIF4E knockdown reduced LacZ-4ESE protein levels relative to RNAi controls (Figure 3I, lanes 3 versus 4). Importantly, LacZ protein levels did not change consistent with the requirement of the 4ESE for eIF4E-dependent cleavage (Figure 3I, lanes 1 versus 2). Further, reduced levels of endogenous eIF4E correlated with decreased MCL1 protein levels, an established eIF4E mRNA export target, which served as a positive control (Figure 3I, lanes 1 versus 2 and lanes 3 versus 4). eIF4E knockdown was confirmed by western blot, and there were no changes to Lamin protein levels, which served as a loading and negative control (Figure 3I). Thus, decreased eIF4E reduced PAS cleavage, as well as LacZ-4ESE and MCL1 protein levels, but did not affect LacZ or lamin.
Finally, we examined whether the polyA tail length was altered in eIF4E-overexpressing or vector-control conditions. We did not observe any changes in length for LacZ-4ESE in vector versus eIF4E-overexpressing cells (data not shown). In all, results strongly suggest that eIF4E overexpression leads to increased cleavage of specific RNAs using our four cleavage assays. Given the “moving target” caveat of measuring nuclear levels of cleaved RNA in intact cells, we opted to measure the amount of uncleaved RNA for the remainder of this study because the uncleaved RNA population is unaffected by eIF4E-dependent export.
eIF4E Affects Cleavage of Its Endogenous mRNA Export Targets CCND1 and MCL1
We investigated whether eIF4E could modulate the cleavage of endogenous RNAs targets CCND1 and MCL1, which are known to associate with eIF4E in the nucleus and to be targets of eIF4E-dependent RNA export. Initially, we identified cleavage-site locations from the literature and Ensembl (https://ensembl.org), and then, by 3′ QuantSeq, to confirm the CA sites within the PAS for primer design (Chen et al., 2008; Thomas et al., 2010; Jenal et al., 2012; Wang et al., 2018). We observed an ~2-fold reduction in uncleaved RNA upon eIF4E overexpression for both MCL1 and CCND1 transcripts (Figure S3G). We attempted to use northern blot analysis to monitor cleavage, but even theoretically, cleavage is expected to only lead to small alterations in band mobility because of the position of the cleavage sites relative to total transcript size. Indeed, analysis of those northern blots was further confounded by the wide variety of splice variants present and the small expected changes in mobility, making these data uninterpretable.
As an alternative method that was independent of the PAS site used, we monitored the levels of cleaved and then polyadenylated MCL1 and CCND1 RNA, as a function of total RNA for each case. In this assay, we used an oligo-deoxythymidine (dT) resin and performed pulldowns, monitoring RNA by qRT-PCR. We observed an ~1.5- to 2-fold increase in polyadenylated MCL1 and CCND1 RNAs in eIF4E-overexpressing cells, relative to vector control from whole-cell lysates, which were used to avoid the concern over cleaved and polyadenylated RNA being exported from nuclear fractions (Figure S3H). The increase in polyA RNAs for CCND1 was statistically significant, whereas the change for MCL1 transcripts trended toward significance. At the same time, there was no change in total RNA, as measured using exonic primers analyzing RNAs isolated from whole-cell lysates (Figure S3I). In this way, the ratio of cleaved and then polyadenylated RNA was increased by eIF4E overexpression for its targets. By contrast, VEGF and Lamin transcripts did not have increased processing as a function of eIF4E overexpression (Figure S3H). In all, our studies with MCL1 and CCND1 strongly support increased cleavage, as observed by reduced levels of uncleaved transcripts with no alteration to total transcripts (as assessed by exonic primers from whole-cell lysates), and an increase in protein expression in response to elevated eIF4E (Figure S3J). Thus, our results are consistent with a model whereby eIF4E enhances the cleavage of endogenous 4ESE-containing transcripts.
CPSF3 Is Used in eIF4E-Driven 3′ Processing
Given eIF4E immunoprecipitated with CPSF3 protein and elevated CPSF3 protein levels (Figures 1 and 2), we investigated whether eIF4E-dependent PAS cleavage required CPSF3. We compared the effects on LacZ and LacZ-4ESE RNA in eIF4E-FLAG cells to decouple increased production of the CPA machinery by eIF4E overexpression from specific effects on LacZ-4ESE RNA versus LacZ RNAs. If these transcripts were direct targets of CPSF3, we would expect CPSF3 to physically associate with those RNAs in the nucleus. Indeed, CPSF3 RIPs both LacZ and LacZ-4ESE RNAs with similar enrichment relative to IgG controls in nuclear fractions, as observed using exonic primers and qRT-PCR (Figure 4A). Further, CPSF3 RIPs ACTIN mRNA, consistent with its general role in 3′-end processing (Figure 4A). As above, the primers used to detect endogenous RNAs crossed exon boundaries, thereby detecting spliced RNAs.
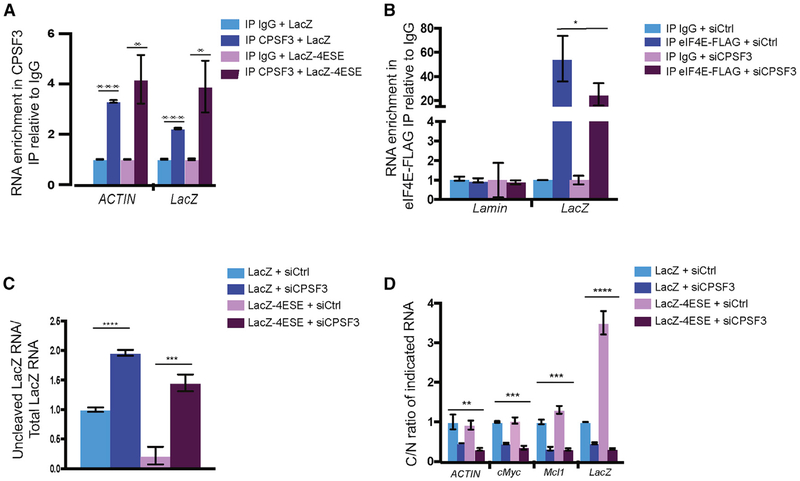
Figure 4. eIF4E Requires CPSF3 Activity for Its Effects on Cleavage.
(A) Endogenous CPSF3 RIPs ACTIN, LacZ, and LacZ-4ESE RNAs relative to IgG controls in nuclear lysates from eIF4E-FLAG U2OS cells.
(B) eIF4E-FLAG RIP as a function of RNA knockdown to CPSF3 (siCPSF3) relative to RNAi controls (siCtrl) showed reduced binding of eIF4E with LacZ-4ESE RNAs compared with siCtrls. RIPs were relative to IgG controls, and Lamin is a negative control.
(C) siCPSF3 increased levels of uncleaved RNA (i.e., reduced cleavage) for both LacZ-4ESE and LacZ RNAs relative to siCtrl by qRT-PCR in nuclear lysates from eIF4E-FLAG U2OS cells.
(D) siCPSF3 impaired nuclear export of all RNAs examined. The cytoplasmic-to-nuclear RNA ratio (C/N ratio) is shown.
(A), (C), and (D) were repeated in three biological replicates and (B) in two biological replicates, all with technical triplicates. Values are means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Student’s t test was used for calculation of p values for for (A)–(C), whereas ANOVA was used for (D).
Knockdown controls and total RNA levels are in Figure S4.
Next, we explored the effects of CPSF3 on the ability of eIF4E to associate with its target RNAs in the nucleus. CPSF3 knockdown reduced the ability of eIF4E-FLAG to RIP LacZ-4ESE RNAs by ~2-fold (Figure 4B). Importantly, the eIF4E RIPs were still enriched ~20-fold over IgG control for LacZ-4ESE RNAs, indicating that CPSF3 is not required for, but is enhanced by, these interactions, using exonic primers for the LacZ transcripts (Figure 4B). The negative-control Lamin RNA did not bind eIF4E-FLAG, demonstrating specificity. In addition, total LacZ-4ESE RNA levels were not altered by CPSF3 knockdown, but its mRNA export was inhibited (Figure S4A). In summary, CPSF3 enhanced the association of eIF4E for RNAs but was not required for it. The knockdown of CPSF3 could either decrease the ability of eIF4E to bind RNAs because of a reduction in 3′-end processing and/or because of the physical interaction between these two proteins.
We then monitored cleavage activity as a function of CPSF3 RNAi knockdown. The levels of uncleaved RNA in the nucleus were increased, indicating reduced cleavage by ~10-fold for LacZ-4ESE and ~2-fold for LacZ RNAs relative to RNAi controls, whereas total RNA levels (as determined using exonic primers from whole-cell lysates) were not affected (Figures 4C and S4A). Indeed, siRNA mediated knockdown of CPSF3 (siCPSF3) repressed export of all RNAs examined, including ACTIN, consistent with the preference of RNA export pathways for 3′-end processing (Figure 4D). Importantly, CPSF3 knockdown impaired export of LacZ-4ESE RNA by ~7-fold relative to RNAi controls and ~2-fold for LacZ RNA, suggesting that 4ESE RNAs were more sensitive to decreased CPSF3 levels (Figure 4D). siCPSF3 was confirmed by qRT-PCR and western blot (Figures S4A and 4B). Consistent with the general role of CPSF3 in PAS cleavage (Proudfoot, 2011), the effects of its knockdown are not restricted to 4ESE-containing RNAs. Thus, CPSF3 acted in both bulk and eIF4E-dependent PAS cleavage.
eIF4E Requires a PAS Site to Modify Cleavage and Nuclear Export
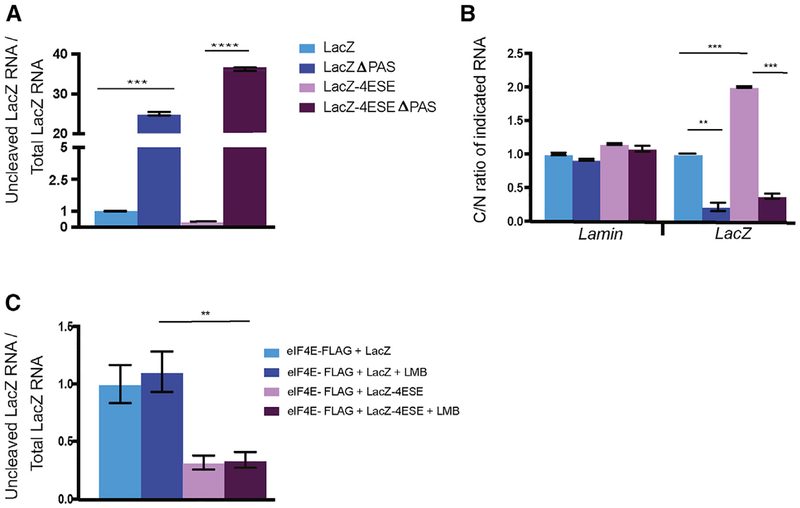
We investigated the relevance of the PAS site for eIF4E-dependent cleavage. This was important to assess both because of the importance of the PAS for bulk RNA cleavage and because, although the PAS was not required, it increased the amount of LacZ-4ESE RNA associating with eIF4E in RIPs (Figure 2E). Considering these points, we predicted that PAS deletion would impair cleavage and subsequent export of target transcripts. All experiments were done in eIF4E-FLAG cells so that levels of CPA machinery were the same across the different LacZ cell lines. To monitor the effects of PAS deletion on uncleaved RNA levels, we designed another set of primers that were downstream of the PAS site and thus, would be present in the PAS-deleted constructs (Figure S5A) The same primers were used for both LacZ and LacZ-4ESE constructs (Figures 2B and S5A). We monitored both the levels of uncleaved RNA from nuclear lysates using the above primers and total RNA using exonic primers from whole-cell lysates to capture the total LacZ RNA populations in the respective cells. We observed that PAS deletion increased levels of uncleaved LacZ-4ESEΔPAS RNA more than ~70-fold relative to LacZ-4ESE RNA and that correlated with impaired LacZ-4ESEΔPAS mRNA export by ~10-fold (Figure 5A). The total levels of LacZ and LacZ-4ESE RNA in whole-cell lysates were not altered by PAS deletion (Figure S5B). Deletion of the PAS from LacZ (LacZΔPAS RNA) also impaired its cleavage and mRNA export, as expected given the relevance of the 3′-end formation for bulk mRNA export (Figure 5B). In parallel, we mutated a key recognition site within the PAS of LacZ-4ESE RNAs, AAUAAA to AAUAAG, and examined the effects using the same primers for detection of uncleaved RNA as for the wild-type LacZ constructs. This mutation decreased cleavage by ~2-fold in nuclear lysates and reduced mRNA export by ~3-fold relative to transcripts with wild-type PAS but did not alter total levels of the RNAs from whole-cell lysates (Figures S5C–S5E). Thus, the PAS had a critical role in the cleavage and mRNA export activities of eIF4E, consistent with recruiting the cleavage machinery as well as stabilizing the eIF4E-4ESE RNA interaction.
Figure 5. The PAS RNA Element Is Required for eIF4E-Dependent Cleavage.
(A) Uncleaved/total RNA levels for LacZ and LacZ-4ESE and their functional PAS deletion (ΔPAS). ΔPAS increased uncleaved levels indicating reduced cleavage using qRT-PCR and primers specific to these ΔPAS constructs to detect uncleaved RNA (seeTable S1).
(B) The PAS was required for nuclear export of both LacZ and LacZ-4ESE constructs. Cytoplasmic-to-nuclear (C/N) ratios of indicated RNAs by qRT-PCR are shown.
(C) Cleavage assay in indicated cell lines demonstrated that Leptomycin B (LMB) did not impair cleavage of eIF4E-dependent cleavage as evidenced by no alteration in levels of uncleaved/total LacZ or LacZ-4ESE RNA levels.
Results are shown from three biological replicates each performed in technical triplicate. Values are means ± SD with p values as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Student’s t test was used for (A), whereas ANOVA was used for (B).
eIF4E-Driven Cleavage and Export Are Distinct Processes
To test whether eIF4E-dependent cleavage and export processes were distinct, we investigated whether those processes could be decoupled. We monitored the effects of Leptomycin B (LMB), a CRM1 inhibitor, on cleavage and export in eIF4E-FLAG cells. LMB did not alter cleavage of LacZ or LacZ-4ESE transcripts (Figure 5C). However, it reduced LacZ-4ESE RNA export by ~3-fold, similar to the effects of eIF4E knockdown but did not affect LacZ RNA, which uses NXF1 for export (Figure S5F) (Culjkovic et al., 2005, 2006). Taken together, eIF4E-mediated cleavage is functionally distinct from mRNA export. However, those eIF4E activities are linked because cleavage increased production of substrates that could be funneled into the export pathway.
DISCUSSION
eIF4E is usually considered to interact with processed RNAs and to act only in mRNA export, translation, and perhaps, stability in cytoplasmic foci (Borden, 2016). Surprisingly, we found that eIF4E can promote 3′-end cleavage of selected RNAs using multiple strategies. The discovery that eIF4E acts as a control point for the production of the CPA machinery and physically interacts (directly or indirectly) with CPA factors has important mechanistic implications. For those transcripts that are sensitive to eIF4E in terms of cleavage, this represents a specialized 3′-end processing pathway. Requirements for this pathway include the CPSF3 cleavage enzyme and the CA cleavage site within the canonical PAS site, the 4ESE (at least for our model RNAs), and the eIF4E. Whether other unanticipated co-factors are also needed for eIF4E-dependent cleavage is an interesting future avenue of study. Further, indirect effects of altering levels of the cleavage machinery could be expected to have broad ranging effects on its own, which would likely have effects beyond 4ESE-containing RNAs. Indeed, these direct and indirect effects may work together to support the impact of eIF4E on cleavage. In support of specificity, eIF4E-mediated increase of cleavage factors was not sufficient to enhance cleavage of all transcripts, e.g., LacZ, and did not increase their export, e.g., LacZ, ACTIN, VEGF, and Lamin RNAs. Clearly, RNAs that are export targets of eIF4E, e.g., CCND1, MCL1, and LacZ-4ESE, appear to be one group that will likely be a target of this form of regulation. Although it is tempting to speculate that the effects of eIF4E could be broad ranging, future studies will be needed to assess the full impact of these activities.
The requirement of the 4ESE in our LacZ-model RNAs for increased cleavage and export strongly suggests that cleavage and nuclear RNA export functions of eIF4E could be functionally connected with cleavage substrates funneling into the eIF4E-dependent RNA export pathway. However, it is formally possible that these processes could be decoupled. For example, if the 4ESE is downstream of the PAS element, the 4ESE would be removed upon cleavage, redirecting the RNA to an export pathway that did not require the 4ESE. Thus, it may be that not all eIF4E-dependent cleavage target RNAs are eIF4E export targets. This would provide further layers of control for the expression of subsets of eIF4E target RNAs. Further, inhibition of eIF4E-dependent mRNA export with Leptomycin B did not inhibit the ability of eIF4E to promote cleavage. This suggests that, although these processes could be related by having increased cleavage produce substrates for export, their biochemistry is distinct.
In summary, we discovered that eIF4E modulates 3′ cleavage of specific transcripts in the nucleus through its effects on the protein levels of the CPA machinery and through its physical interaction with at least some CPA components. Given that ~3,500 transcripts bind eIF4E in the nucleus (Culjkovic-Kraljacic et al., 2016), this cleavage activity is likely implicated in the processing of a broad array of mRNAs. Although other aspects of 3′-end processing can be studied genome wide, i.e., PAS site selection and polyA tail length (Chen et al., 2017; Lianoglou et al., 2013), there is not yet a means to study cleavage efficiency genome wide, i.e., if PAS sites are not altered, but cleavage efficiency is increased uniformly for all sites. This situation appears to be reasonably common as observed by the fact that ~5,300 RNAs have only one PAS site out of the ~10,000 RNAs detected in our CA identification studies in U2OS cells (data not shown). Thus, many transcripts are subject to regulation primarily at the level of cleavage efficiency rather than at the PAS site selection (given they have only one PAS site). PolyA tail length and PAS site selection may also be modulated by eIF4E, at least for a subset of its nuclear targets, which remains to be tested further. As methods evolve, we will develop a fuller understanding of the number of transcripts affected by eIF4E-dependent enhanced 3′-end cleavage. Because eIF4E associates with networks of functionally related RNAs, these studies uncovered a mechanism for the coordination of the 3′-end processing of groups of transcripts. Indeed, increasing 3′-end formation via more efficient cleavage makes these mRNAs better substrates for export and translation and thus, ultimately, increases their protein levels. Thus, this activity provides a funneling framework by which eIF4E stimulates processing of targets to make them better substrates for its export and translation activities. It is likely that the oncogenic potential of eIF4E will ultimately be driven by the combinatorial effect of all its mRNA-processing activities on the cellular proteome.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and request for reagents and resources should be directed to and will be fulfilled by the Lead Contact, Katherine Borden (katherine.borden@umontreal.ca).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The human osteosarcoma cell line U2OS (ATCC), derived from a 15-year-old female, was cultured in 5% CO2 at 37° C in Dulbecco’s modified Eagle’s medium (DMEM) (ThermoFisher Scientific, cat# 11995–065) supplemented with 10% fetal bovine serum (ThermoFisher Scientific, cat# 12483–020) and 1% penicillin-streptomycin (Invitrogen cat #15140–122), referred throughout as DMEM complete. The identity of U2Os cell lines has been authenticated using STR profiling (Montreal EpiTerapia Inc.). Cell lines were routinely checked to verify that there was no mycoplasma contamination using MycoAlert Mycoplasma Detection kit (Lonza,NY, U.S.A, cat# LT07–418). Transfections for stable cell lines were performed using Trans IT-LT1 Transfection Reagent (Mirus, cat# MIR 2300) as specified by the manufacturer, and selected in G418-containing medium, 1 mg/mL, (Wisent-Bioproduct cat # 400–130) for stable eIF4E overexpressing cell lines. For transfection of LacZ constructs, U2Os cells were transfected with the given wild-type or mutant LacZ and LacZ-4ESE constructs and were selected for with 1 mg/mL G418 and 1μg/mL puromycin (Wisent-Bio product cat #450–162). Multiple independent clones (> 3) were used to obtain biological replicates. For eIF4E and CPSF3 knockdowns, U2OS cells were transfected with Lipofectamine 2000 (Invitrogen cat # 11668–019) and 20nM siRNA duplex according to the manufacturer’s instructions (Table S1). Cells were analyzed 96 hr after transfection.
METHODS DETAILS
Constructs
pcDNA-2FLAG-eIF4E (eIF4E-FLAG), vector, LacZ and LacZ-4ESE constructs were described (Culjkovic et al., 2005, 2006; Volpon et al., 2017). Mutations in or deletions of the PAS for LacZ and LacZ-4ESE were generated by PCR-directed mutagenesis (See Table S1) and verified by sequencing (IRIC-Genomic Platform).
Cellular Fractionation and Export Assay
Fractionation protocol was described (Culjkovic-Kraljacic et al., 2016; Rousseau et al., 1996; Topisirovic et al., 2002) Approximately 3 ×107 cells were collected and washed in ice cold PBS (1,200 rpm/ 5 min), then gradually re-suspended in 1 mL of lysis buffer B (10 mM Tris pH 8.4, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP40, 1 mM DTT and 100 U/ml RNase Inhibitors (Invitrogen cat# 10777019)). The whole cell lysate was centrifuged at 1000 g for 3 min at 4°C and supernatant (cytoplasmic fraction) collected. The pellet (nuclear fraction) was re-suspended in 1 mL of lysis buffer B, transferred to round bottom polypropylene tube and 1/10 volume of detergent stock (3.3% (w/v) Sodium Deoxycholate, 6.6% (v/v) Tween 40 in DEPC H20) was added with slow vortexing and incubated on ice for 5 min. The suspension was transferred to a microtube and centrifuged at 1,000 g for 3 min at 4°C. Supernatant (post-nuclear fraction) was collected and the pellet (nuclear fraction) was washed with 1 mL of lysis buffer B and centrifuged at 1,000 g for 3 min at 4°C. The nuclear pellets were re-suspended in 100–200 μL of lysis buffer B and sonicated. Post-nuclear and cytoplasmic fractions were combined for analysis as described in (Lejbkowicz et al., 1992; Volpon et al., 2016). The RNA was extracted using TRIzol reagent (Invitrogen cat # 15596018) according to the manufacturer’s guidelines.
Polysome Assays
Polysomal profiling was complete as described (Tcherkezian et al., 2014; Zahreddine et al., 2017)). In brief, cells were treated with cyclohexamide (100 μg/ml, Sigma Aldrich cat# C7698) 10 minutes prior to harvesting. Lysates were prepared with 1mL of polysome lysis buffer (15 mM Tris pH 7.4, 250 mM NaCl, 15 mM MgCl2, 1% Triton X-100, 100 g/ml cyclohexamide, 1 mM DTT, 400 U/ml RNase inhibitors (Invitrogen cat# 10777019) and protease inhibitors (Sigma Aldrich cat # 111697498001). Equal amounts (5–10 mg) of protein lysates were layered on a 20%–50% linear sucrose gradient (20% and 50% sucrose solutions in 15 mM Tris pH 7.4, 15 mM MgCl2, 150 mM NaCl, 1 mM DTT, 100 μg/Ml cyclohexamide and 20 U/ml RNase inhibitors), mixed on Gradient Station IP Biocomp and centrifuged in a Beckman SW41Ti rotor at 92,000 g for 3 hr at 4°C. Polysome fractions were collected by continuously monitoring and recording the A254 on a Gradient Station IP (Biocomp) attached to a UV-MII (GE Healthcare, CA) spectrophotometer. RNAs were isolated from polysomal fractions using TRIzol reagent. RNA transcripts from each fraction (1–10) were examined via RTqPCR. Calculations were performed as outlined in Chassé et al. (2017) and standard t test analyses were conducted to conclude statistical significance.
Immunoprecipitation (IP) and RNA-IP (RIP)
IP and RIP was performed as described (Culjkovic et al., 2005). The antibodies and respective concentrations used are detailed in the Key Resources Table. For RIP analysis, after antibody and lysate incubation, complexes were eluted in Tris (hydroxymethyl) aminomethane-EDTA containing 1% sodium dodecyl sulfate and 12% β-mercaptoethanol, at 95°C for 5 minutes. RNA was isolated using 1V Phenol/Chloroform extraction (Ambion, cat # AM9720) and cDNA synthesis was completed using SuperScript VILO cDNA (Invitrogen cat n# 11754250).
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Phusion® High-Fidelity DNA Polymerase | NEB | Cat # M0530L |
| Antibodies | ||
| Mouse monoclonal anti-eIF4E | BD Biosciences | Cat # 610270; RRID:AB_397664 |
| Rabbit monoclonal anti- eIF4E | Millipore | Cat # 04–347; RRID:AB_673078 |
| Mouse Monoclonal anti- eIF4E | Santa Cruz | Cat # sc-9976; RRID:AB_627502 |
| Mouse monoclonal anti- FLAG M2 | Sigma Aldrich | Cat # F3165; RRID:AB_259529 |
| Mouse monoclonal anti-α-tubulin | Sigma Aldrich | Cat # T5168; RRID:AB_477579 |
| Mouse monoclonal anti-β-actin | Sigma Aldrich | Cat # A5441; RRID:AB_476744 |
| Rabbit polyclonal anti- CPSF1 antibody | Bethyl | Cat # A301–580A; RRID:AB_1078859 |
| Rabbit polyclonal anti- CPSF2 antibody | Bethyl | Cat # A301–581A; RRID:AB_1078861 |
| Rabbit polyclonal anti- CPSF3 antibody | Bethyl | Cat # A301–090A; RRID:AB_873009 |
| Rabbit polyclonal anti- CPSF4 antibody | Bethyl | Cat # A301–584A; RRID:AB_1078872 |
| Rabbit monoclonal anti-Cyclin D1 | Abcam | Cat # Ab134175; RRID:AB_2750906 |
| Rabbit polyclonal anti- FIP1 (FIP1L1) | Bethyl | Cat # A301–461A; RRID:AB_999564 |
| Rabbit polyclonal anti- GAPDH (FL-335) | Santa Cruz | Cat # sc-25778; RRID:AB_10167668 |
| Rabbit polyclonal anti-Lamin A (C-terminal) | Sigma Aldrich | Cat # L1293; RRID:AB_532254 |
| Rabbit polyclonal anti- Mcl1-I (S-19) | Santa Cruz | Cat # sc-819; RRID:AB_2144105 |
| Mouse monoclonal anti- c-Myc (9E10) | Santa Cruz | Cat # sc-40; RRID:AB_627268 |
| Rabbit polyclonal anti-Pol II N-20 | Santa Cruz | Cat # sc-899; RRID:AB_632359 |
| Rabbit polyclonal anti-Symplekin | Bethyl | Cat # A301–463A; RRID:AB_999670 |
| Rabbit polyclonal anti-WDR33 antibody | Bethyl | Cat # A301–152A; RRID:AB_2215378 |
| Mouse Monoclonal IgG | Millipore | Cat # 12–371; RRID:AB_145840 |
| Rabbit Monoclonal IgG | Millipore | Cat #12–370; RRID:AB_145841 |
| Goat Monoclonal IgG | Santa Cruz | Cat # sc-2028; RRID:AB_737167 |
| Experimental Models: Cell Lines | ||
| U2OS (Female) | ATCC | Cat # HTB-96™; RRID:CVCL_0042 |
| Oligonucleotides | ||
| See Table S1 | ||
| Software and Algorithms | ||
| Bioanalyzer Software, 2100 Expert Software | Agilent | Cat # G2946CA |
| PRISM 7.0 | GraphPad | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Leptomycin B (LMB) | LC Laboratories | Cat # L-6100 |
| Cyclohexamide | Sigma Aldrich | Cat # C7698 |
Western Blot Analysis
Western Blot analysis was performed as described previously with a modified lysis buffer (40 mM HEPES, pH 7.5, 120 mM NaCl, 1 mM EDTA, 10 mM β-glycerophosphate, 50 mM NaF, 0.5 μM NaVO3, and 1% [vol/vol] Triton X-100 supplemented with complete protease inhibitors (all were purchased from Sigma-Aldrich). Further, blots were blocked in 5% milk (in TBS–Tween 20), and primary antibodies were diluted in milk blocking solution. The following HRP conjugated secondary antibodies were all sourced from Jackson Immuno-Research, Monoclonal Mouse Anti-Rabbit (cat # 211-032-171;RRID:AB_2339149), Goat-Anti-Mouse (cat # 115-035-174; RRID:AB_2338512) and Bovine Anti-Goat (cat # 805-035-180; RRID:AB_23408074).
Reverse transcription and Quantitative PCR
DNase treated RNA samples were reverse transcribed using MMLV reverse transcriptase (Invitrogen cat# 28025013). RT-qPCR analyses were performed using SensiFAST Sybr Lo-Rox Mix (BIOLINE, cat # CSA-01195) in AB StepOne thermal cycler using the relative standard curve method (Applied Biosystems User Bulletin #2). All conditions were described previously Culjkovic et al. (2005). All primers were purchased from IDT Technologies unless otherwise noted (Table S1).
In vitro cleavage assay
Biotinylated RNA fragments were generated using in vitro transcription MegaScript T7 (ThermoFisher Scientific cat # AM1334) and biotinylated-14CTP (ThermoFisher Scientific cat # 19519016) as per manufacturer’s instructions. Fragment sizes and purity were confirmed by Northern blot. RNAs were incubated with nuclear lysates produced as outlined in Friend et al. (2007). Incubations shown are for 30 minutes. The following buffer was used: 1.5 mM MgCl2, 0.25 mM ATP, 1 mM cordycepin, 20 mM phosphcreatine, 2 units of RNase out. The reaction was stopped with G50 (20 mM TRIS, pH 7.5, 2 mM EDTA 200 mM NH4OAc, 0.25% SDS and RNase out). This was followed by proteinase K treatment for 30 min at 55C (ThermoFisher Scientific cat # AM2548). RNA was extracted using phenol / chloroform (ThermoFisher Scientific cat # 15596026) and Northern blots were carried out as described in Culjkovic et al. (2005); Friend et al. (2007), and Topisirovic et al. (2002).
RT-qPCR Cleavage Assay
eIF4E-FLAG+ LacZ or LacZ-4ESE cells were plated in a 15 cm tissue culture dish (GIBCO) to 70% confluent (36–48 hours) and were trypsinized with 0.5% trypsin-EDTA (ThermoFisher Scientific cat # 15400–054). Cells were washed twice in ice cold PBS and fractionated as detailed above. RNA was extracted using TRIzol (ThermoFisher Scientific cat # 15596026), and cDNA was synthesized using MMLV (ThermoFisher Scientific cat. # 28025013) (RNA (300ng-1μg) and DNase treated (Ambion, cat # AM2238). NA (300ng-1μg). RT-qPCR was performed (as described above) on nuclear lysates and whole cell lysates using described primers for LacZ and LacZ-4ESE to bracket the cleavage site to measure uncleaved and cleaved pre-mRNAs, LacZ uncleaved Fwd 5′CCCGTGCCTTCCTTGAC3′, LacZ uncleaved Rvs 5′ATGACACCTACTCAGACAATG3′, LacZ cleavage Rvs 5′TTTTTTTTTTTTGCGATGCAA3′. Cleavage was calculated as the nuclear uncleaved or cleaved expression of LacZ (or LacZ-4ESE) / total cell lysate LacZ (or LacZ-4ESE) expression, respectively. LacZ transcript (LacZ and LacZ-4ESE) cleavage and PAS site location was verified using Quant Seq 3′ mRNA-Seq Library Prep Kit REV for Illumina (LEXOGEN cat # SKU: 016.96.), as per manufacturer’s instructions. Sequencing was performed via Miseq, 1 Flowcell v3 (25M de fragments), 150 cycles, Paired-End (maximum 2 × 85 nt) in three biological replicates.
Endogenous Targets
To measure cleavage of endogenous CCND1 and MCL1, in vector and eIF4E-FLAG cells, primers flanked the CCND1 3′UTR PAS from Chen et al. (2008), CCND1_ uncleaved FWD:5′TCCCTCCTCTCCGGAGCATTT 3′, CCND1_uncleaved, RVS: 5′ATCATCTGTAGCACAACCCTCCTC 3′. CCND1 internal primers were previously described (Culjkovic et al., 2006) (Primers purchased from IDT). MCL1 primers included, MCL1_uncleaved FWD: 5′TCACATCAGGTGGATGGAGA 3′, MCL1_uncleaved RVS: 5′GCTTTATTGACATACTAGGCTTAGACC 3′ (Primers purchased from IDT). Cleavage was calculated as the nuclear uncleaved expression of MCL1 or CCND1 / total cell lysate MCL1 and CCND1 expression (Table 3), respectively. CCND1 and MCL1 transcript cleavage and PAS site location was verified using Quant Seq 3′ mRNA-Seq Library Prep Kit REV for Illumina (LEXOGEN cat # SKU: 016.96.), as per manufacture instructions. Sequencing was run by IRIC Genomics Platform via Nexseq500, 1 Flowcell High Output (400M fragments), 75 cycles, single-end read (maximum 1×85 nt) in 5 biological replicates.
Competitive PCR
cDNA was synthesized via MMLV reverse transcriptase (ThermoFisher Scientific cat. # 28025013) using Random Hexamers (ThermoFisher Scientific cat # N8080127). Competitive PCR was performed via Phusionâ High-Fidelity DNA Polymerase (NEB cat # M0530L) with a 1:1:1 concentration of the following primers: LacZ FWD O/H primers: 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCG TGCCTTCCTTGACCCTG 3′, LacZ RVS uncleaved O/H primers: 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTAGAATGACACCTACTCAGACAA 3′ and LacZ RVS cleaved O/H primers: 5′ GTCTCGTGGGCTCGGAGATGTGTA TAAGAGACAGTTTTTTTTTTTGCGATGCAATTTC 3′ All oligos were purchased from IDT. PCR conditions as follows: 98.0°C for 00:30, 98.0°C for 00:10, 60.0°C for 00:30, 72.0°C for 01:00, 25 cycles and 72.0°C for 00:10. PCR samples were run on Agilent’s Bioanalyzer, DNA1000, and results were analyzed using Bioanalyzer Software, 2100 Expert Software. Percent uncleaved calculated: (uncleaved band molarity) / (sum of cleaved + uncleaved molarity) X 100. See Table S1 for primers.
Oligo dT pulldown
Vector and eIF4E-FLAG cells were harvested at 70% confluent and total cell lysate RNA was extracted using TRIzol (ThermoFisher Scientific cat # 15596026). Samples were processed using Dynabeads Oligo (dT)25 (ThermoFisher Scientific cat # 61005) and final RNA extracted via TRIzol (ThermoFisher Scientific cat # 15596026). cDNA synthesis was done using MMLV (ThermoFisher Scientific cat. # 28025013). PCR at 55°C annealing and 30 cycles was performed for quality control using ACTIN and VEGF, and final analysis was done using RT-qPCR.
PolyA tail length
Polyadenylation length determination of ACTIN, LacZ and MCL1 in vector and eIF4E-FLAG cells was done using Affymetrix USB Poly(A) Tail-length Assay kit (Affymetrix, cat # 76435), as per manufactures instructions. PCR amplification was conducted using the following 10μM primers: ACTIN FWD: 5′ TCTCTCTAAGGAGAATGGC 3′, ACTIN polyA reverse: 5′ AAGTGCACACCTTAAAAATG 3′, LacZ FWD: 5′ CGTCAGTATCGGCGGAGTTC 3′, LacZ polyA reverse: 5′ TATCCCCACGCGCCCTGTA 3′, MCL1 FWD: 5′ AAAGGGAGGATTTGCTTAG 3′ and MCL1 polA reverse: 5′ CTAACTTTCTGTTTTTCCTGAG 3′. All primers purchased from IDT, see Table S1.
Leptomycin treatment
Leptomycin B (LMB) studies were conducted in U2OS cells expressing eIF4E-FLAG + LacZ or eIF4E+ LacZ-4ESE cell lines, treated with 50nM LMB (LC Laboratories, cat # L-6100) at ~70% confluent and incubated for 4 hours at 37°C. RNAs were isolated andanalyzed using RT-qPCR as described below.
QUANTIFICATION AND STATISTICAL ANALYSIS
Values from PCR-based experiments, bioanalyzer data or Northern blots from the given number of biological replicates were combined in PRISM 7.0 (GraphPad Software) to calculate means, standard deviations and P values. When required, ImageJ was used to calculate band intensities. Unless noted, analyses used to determine statistical significance were Student’s t test or ANOVA and reported as P values.
Supplementary Material
Highlights.
eIF4E drives the expression of the 3′-end processing machinery
eIF4E binds the cleavage machinery and promotes cleavage of specific RNAs
eIF4E-dependent PAS cleavage and mRNA export are distinct processes
eIF4E functions beyond export and translation to include RNA processing
ACKNOWLEDGMENTS
Many thanks for the technical assistance of Drs. Biljana Culjkovic-Kraljacic, Luce Skrabanek, Laurent Volpon, and Heloise Chasse. Research reported in this publication was supported by the National Cancer Institute of the NIH under grants R01CA080728 and RO1CA098571. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was also supported by Canadian Institutes of Health Research PJT159785. K.L.B.B. holds a Canada Research Chair and M.R.D. holds a Cole Foundation Postdoctoral Fellowship and an Institute for Research in Immunology and Cancer Postdoctoral Fellowship Award. Figure 1A derivative of Figure 2 in Neve et al. (2017), used under CC BY NC 4 (https://creativecommons.org/licenses/by-nc/4.0/) and is licensed under CC BY NC 4 by M.R.D., M.D., and K.L.B.B.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.04.008.
DECLARATION OF INTERESTS
The authors have no competing interests.
REFERENCES
- Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, Caplan S, Leber B, Roy DC, Miller WH Jr., and Borden KL (2009). Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood 114, 257–260. [DOI] [PubMed] [Google Scholar]
- Assouline S, Culjkovic-Kraljacic B, Bergeron J, Caplan S, Cocolakis E, Lambert C, Lau CJ, Zahreddine HA, Miller WH Jr., and Borden KL (2015). A phase I trial of ribavirin and low-dose cytarabine for the treatment of relapsed and refractory acute myeloid leukemia with elevated eIF4E. Haematologica 100, e7–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL (2016). The eukaryotic translation initiation factor eIF4E wears a “cap” for many occasions. Translation (Austin) 4, e1220899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M, and Borden KL (2013). The oncogene eIF4E: using biochemical insights to target cancer. J. Interferon Cytokine Res 33, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR 3rd, Ule J, Manley JL, and Shi Y (2014). CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev. 28, 2370–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, and Kim VN (2014). TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell 53, 1044–1052. [DOI] [PubMed] [Google Scholar]
- Chassé H, Boulben S, Costache V, Cormier P, and Morales J (2017). Analysis of translation using polysome profiling. Nucleic Acids Res. 45, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RW, Bemis LT, Amato CM, Myint H, Tran H, Birks DK, Eckhardt SG, and Robinson WA (2008). Truncation in CCND1 mRNA alters miR-16–1 regulation in mantle cell lymphoma. Blood 112, 822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jia Q, Song Y, Fu H, Wei G, and Ni T (2017). Alternative polyadenylation: methods, findings, and impacts. Genomics Proteomics Bioinformatics 15, 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, and Borden KL (2005). eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3′ UTR. J. Cell Biol 169, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, and Borden KL (2006). eIF4E is a central node of an RNA regulon that governs cellular proliferation. J. Cell Biol 175, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjkovic-Kraljacic B, and Borden KL (2013). Aiding and abetting cancer: mRNA export and the nuclear pore. Trends Cell Biol. 23, 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjkovic-Kraljacic B, Baguet A, Volpon L, Amri A, and Borden KL (2012). The oncogene eIF4E reprograms the nuclear pore complex to promote mRNA export and oncogenic transformation. Cell Rep. 2, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjkovic-Kraljacic B, Fernando TM, Marullo R, Calvo-Vidal N, Verma A, Yang S, Tabbò F, Gaudiano M, Zahreddine H, Goldstein RL, et al. (2016). Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood 127, 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klerk E, Venema A, Anvar SY, Goeman JJ, Hu O, Trollet C, Dickson G, den Dunnen JT, van der Maarel SM, Raz V, and ‘t Hoen PA (2012). Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 40, 9089–9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Lovejoy AF, and Steitz JA (2007). U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3′ end formation.Mol. Cell 28, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, and Rottman FM (1992). The 3′-flanking sequence of the bovine growth hormone gene contains novel elements required for efficient and accurate polyadenylation. J. Biol. Chem 267, 16330–16334. [PubMed] [Google Scholar]
- Huang Y, and Carmichael GG (1996). Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol 16, 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter KM, Charon R, and Coulehan JL (1995). The study of literature in medical education. Acad. Med 70, 787–794. [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kühn U, Menzies FM, Vrielink Oude J.A., Bos AJ, Drost J, Rooijers K, et al. (2012). The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538–553. [DOI] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, and Keller W (2004). Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23, 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, and Tenenbaum SA (2002). Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9, 1161–1167. [DOI] [PubMed] [Google Scholar]
- Kolev NG, and Steitz JA (2005). Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev.19, 2583–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U, Gündel M, Knoth A, Kerwitz Y, Rüdel S, and Wahle E (2009). Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem 284, 22803–22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejbkowicz F, Goyer C, Darveau A, Neron S, Lemieux R, and Sonenberg N (1992). A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. USA 89, 9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, and Mayr C (2013). Ubiquitously transcribed genes use alternative polyadenylation to achieve tissuespecific expression. Genes Dev. 27, 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Luo M, and Wen JK (2014). mRNA stability in the nucleus.J. Zhejiang Univ. Sci. B 15, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, and Tong L (2006). Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444, 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C (2016). Evolution and biological roles of alternative 3′ UTRs. Trends Cell Biol. 26, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KG, and Manley JL (1995). The 160-kD subunit of human cleavage polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 9, 2672–2683. [DOI] [PubMed] [Google Scholar]
- Neve J, Patel R, Wang Z, Louey A, and Furger AM (2017). Cleavage and polyadenylation: ending the message expands gene regulation. RNA Biol. 14, 865–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ (2011). Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, and Dye MJ (2002). Integrating mRNA processing with transcription. Cell 108, 501–512. [DOI] [PubMed] [Google Scholar]
- Rousseau D, Kaspar R, Rosenwald I, Gehrke L, and Sonenberg N (1996). Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 93, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönemann L, Kuhn U, Martin G, Schäfer P, Gruber AR, Keller W, Zavolan M, and Wahle E (2014). Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 28, 2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, and Roux PP (2014). Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5’TOP mRNA translation. Genes Dev. 28, 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LW, Lam C, and Edwards SW (2010). Mcl-1; the molecular regulation of protein function. FEBS Lett. 584, 2981–2989. [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Capili AD, and Borden KL (2002). Gamma interferon and cadmium treatments modulate eukaryotic initiation factor 4E-dependent mRNA transport of cyclin D1 in a PML-dependent manner. Mol. Cell. Biol 22, 6183–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Siddiqui N, Lapointe VL, Trost M, Thibault P, Bangeranye C, Piñol-Roma S, and Borden KL (2009). Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 28, 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpon L, Culjkovic-Kraljacic B, Osborne MJ, Ramteke A, Sun Q, Niesman A, Chook YM, and Borden KL (2016). Importin 8 mediates m7G capsensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc. Natl. Acad. Sci. USA 113, 5263–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpon L, Culjkovic-Kraljacic B, Sohn HS, Blanchet-Cohen A, Osborne MJ, and Borden KLB (2017). A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA 23, 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, He G, Hou M, Chen L, Chen S, Xu A, and Fu Y (2018). Cell cycle regulation by alternative polyadenylation of CCND1. Sci. Rep 8, 6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahreddine HA, Culjkovic-Kraljacic B, Emond A, Pettersson F, Midura R, Lauer M, Rincon Del S., Cali V, Assouline S, Miller WH, et al. (2017). The eukaryotic translation initiation factor eIF4E harnesses hyaluronan production to drive its malignant activity. eLife 6, e29830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.