Summary
Forkhead box A1 (FOXA1) is a pioneer transcription factor that is essential for the normal development of several endoderm-derived organs, including the prostate gland1,2. FOXA1 is frequently mutated in the hormone-receptor driven prostate, breast, bladder, and salivary gland tumors3–8. However, how FOXA1 alterations affect cancer development is unclear, with FOXA1 previously ascribed both tumor suppressive9–11 and oncogenic12–14 roles. Here we assemble an aggregate cohort of 1546 prostate cancers (PCa) and show that FOXA1 alterations fall into three distinct structural classes that diverge in clinical incidence and genetic co-alteration profiles, with a collective prevalence of 35%. Class1 activating mutations originate in early PCa without ETS/SPOP alterations, selectively recur within the Wing2-region of the DNA-binding Forkhead domain (FKHD), enable enhanced chromatin mobility and binding frequency, and strongly transactivate a luminal androgen receptor (AR) program of prostate oncogenesis. By contrast, class2 activating mutations are acquired in metastatic PCa, truncate the C-terminal domain of FOXA1, enable dominant chromatin binding by increasing DNA affinity, and through TLE3 inactivation promote WNT-pathway driven metastasis. Finally, class3 genomic rearrangements are enriched in metastatic PCa, comprise of duplications and translocations within the FOXA1 locus, and structurally reposition a conserved regulatory element, herein denoted FOXA1 Mastermind (FOXMIND), to drive overexpression of FOXA1 or other oncogenes. Our study reaffirms the central role of FOXA1 in mediating AR-driven oncogenesis, and provides mechanistic insights into how different classes of FOXA1 alterations uniquely promote PCa initiation and/or metastatic progression. Furthermore, these results have direct implications in understanding the pathobiology of other hormone-receptor driven cancers and rationalize therapeutic co-targeting of FOXA1 activity.
Keywords: FOXA1 mutations, FOXA1 locus rearrangements, FOXA1 alteration classes, androgen receptor (AR), AR cofactor, prostate cancer, hormone-receptor oncogenesis
FOXA1 independently binds to and de-compacts condensed chromatin to reveal binding sites of partnering nuclear hormone-receptors15,16. In prostate luminal epithelial cells, FOXA1 delimits tissue-specific enhancers17, and reprograms AR-activity in prostate cancer (PCa)14. Accordingly, FOXA1 and AR are co-expressed in PCa cells, wherein FOXA1 activity is indispensable for cell survival and proliferation14 (Extended Data Fig. 1a–i). Thus, it is intriguing that FOXA1 is the third most-highly mutated gene4,5 and, as shown here for the first time, among the most-highly rearranged genomic loci in AR-dependent PCa. Counterintuitively, recent studies have suggested these alterations to be inactivating18,19 and have described FOXA1 as a tumor suppressor in AR-driven metastatic PCa9–11. However, FOXA1 alterations have not been fully characterized or experimentally investigated in cancer.
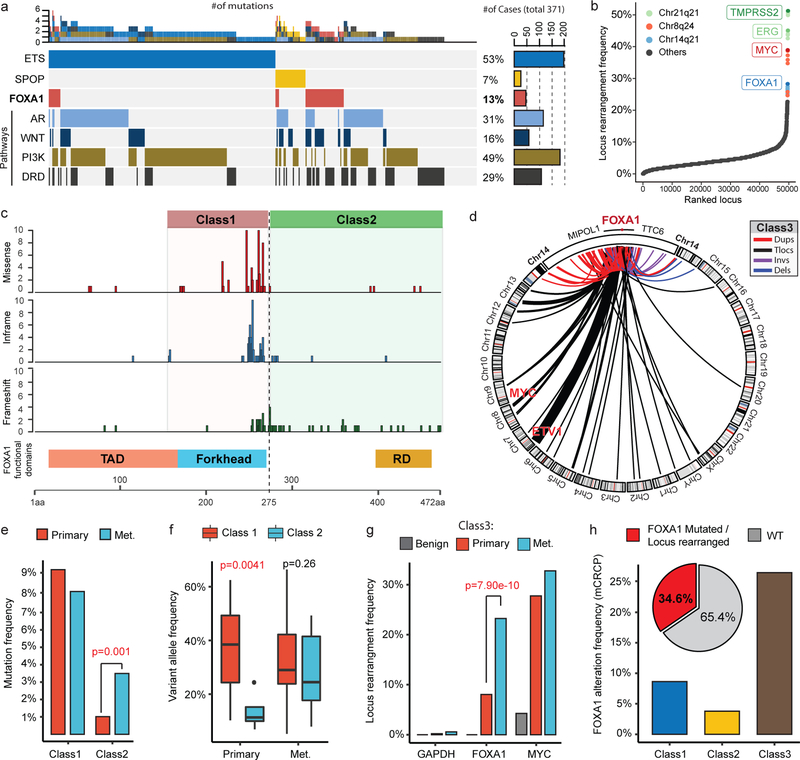
First, we curated an aggregate PCa cohort comprising of 888 localized and 658 metastatic samples4,5,8,20, of which 498 and 357 had matched RNA-sequencing (RNA-seq) data, respectively. Here, FOXA1 mutations recurred at a frequency of 8–9% in the primary disease that increased to 12–13% in metastatic castration-resistant PCa (mCRPC; Fig. 1a and Extended Data Fig. 1j). RNA-seq calls of structural variants (SVs) revealed a high prevalence (Fig. 1b and Supplementary Table 1) and density (Extended Data Fig. 1k) of rearrangements within the FOXA1 locus. The presence of SVs was confirmed by whole-exome and whole-genome sequencing (Extended Data Fig. 1l,m and Supplementary Table 2,3). Overall, we estimated the recurrence of FOXA1 locus rearrangements at 20%−30% in mCRPC (Extended Data Fig. 1n). All FOXA1 mutations were heterozygous and FOXA1 itself was copy-amplified in over 50% of cases with no biallelic deletions (Extended Data Fig. 2a,b). We also found a stage-wise increase in FOXA1 expression in PCa (Supplementary Discussion and Extended Data Fig. 2c).
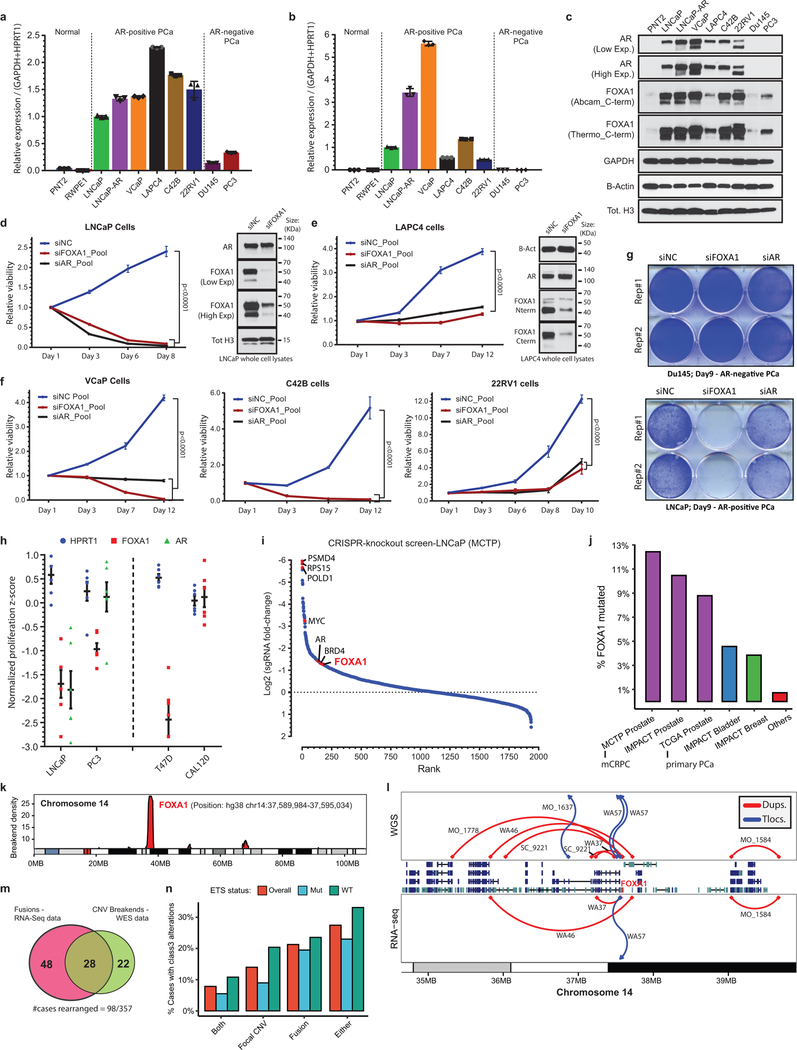
Figure 1. Distinct structural classes of FOXA1 aberrations.
a) FOXA1 mutations and key alterations in mCRPC. Mutations in ETS, AR, WNT, PI3K, DNA repair (DRD) were aggregated at the pathway/group level. b) Locus-level recurrence of RNA-seq structural variations (SVs). c) Structural classification of FOXA1 mutations. TAD, transactivation domain; Forkhead, Forkhead DNA-binding domain; RD, regulatory domain d) Structural classification of FOXA1 locus rearrangements. Dups, Tandem duplications; Tlocs, translocations; Invs, inversions; Dels, deletions. e) Frequency of FOXA1 mutational classes by PCa stage (n=888 primary, 658 metastatic) (two-sided Fisher’s exact test; tFET). f) Variant allele frequency by stage and class (two-sided t-test). Boxplot center: median, box: Q1/Q3, whiskers: Q1/Q3±1.5xIQR. g) Locus-level recurrence of SVs based on RNA-seq by PCa stage (tFET). h) Integrated (RNA-seq and WES) recurrence of FOXA1 alterations classes in mCRPC (SU2C-MCTP, n=370).
Next, on mapping mutations onto the protein domains of FOXA1, we found two structural patterns: 1) missense and in-frame indel mutations were clustered at the C-terminal end of the FKHD, while 2) truncating frameshift mutations were restricted to the C-terminal half of the protein (Fig. 1c). FOXA1 SVs predominantly comprised of tandem-duplications and translocations, which clustered in close proximity to the FOXA1 gene without disrupting its coding sequence (Fig. 1d). Thus, we categorized FOXA1 aberrations into three structural classes: class1 comprising of all the mutations within the FKHD, class2 comprising of mutations in the C-terminal end following the FKHD, and class3 comprising of SVs within the FOXA1 locus (Fig. 1c,d and Extended Data Fig. 2d). Similar classes of FOXA1 alterations were also found in breast cancer. (Extended Data Fig. 2e,f).
Remarkably, we found that the majority of FOXA1 mutations in primary PCa belonged to class1, which showed no enrichment in the metastatic disease (Fig. 1e). Conversely, class2 mutations were significantly enriched in metastatic PCa; and in the rare primary cases with class2 mutations, the mutant allele was detected at sub-clonal frequencies (Fig. 1e,f and Extended Data Fig. 2g,h). We found no cases with both class1 and class2 mutations. Class3 SVs were also significantly enriched in mCRPC (odds ratio (OR)=3.46; Fig. 1g). Overall, we found the cumulative frequency of FOXA1 alterations to be over 34% in mCRPC (Fig. 1h). Assessment of concurrent alterations revealed class1 mutations to be mutually exclusive with other primary events, namely ETS fusions (OR=0.078), while class2-mutant mCRPC to be enriched for RB1 deletions (OR=4.17) (Extended Data Fig. 2i,j). Both mutational classes were further enriched for alterations in DNA repair, mismatch repair, and WNT signaling pathways (Extended Data Fig. 2i,k), and had higher FOXA1 mRNA expression relative to the WT cases (Extended Data Fig. 2l). Together, these data suggest that class1 mutations emerge in localized PCa, while class2 and class3 aberrations are acquired or enriched, respectively, in the course of disease progression.
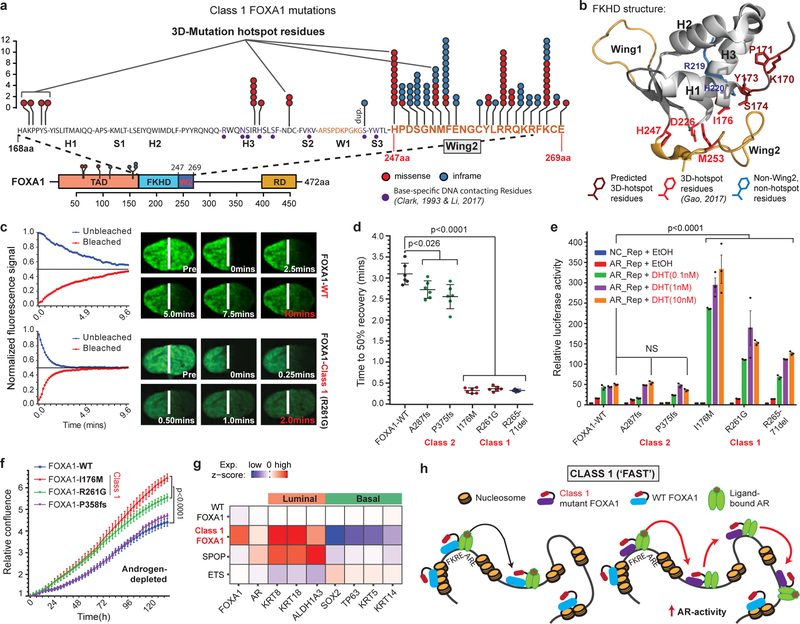
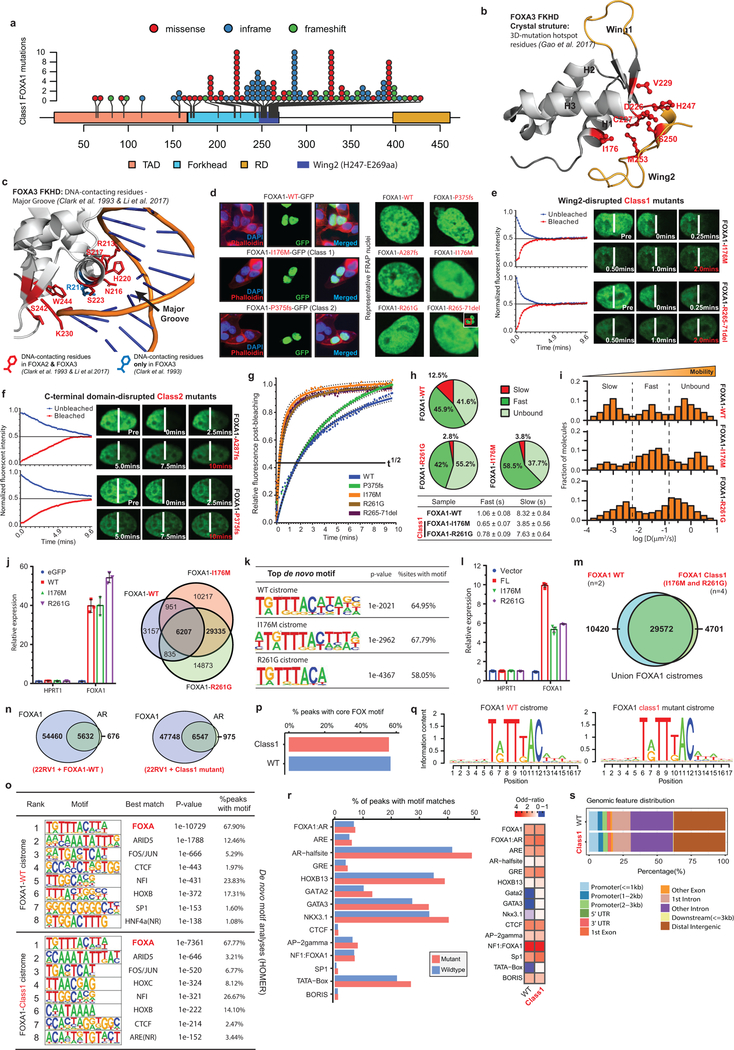
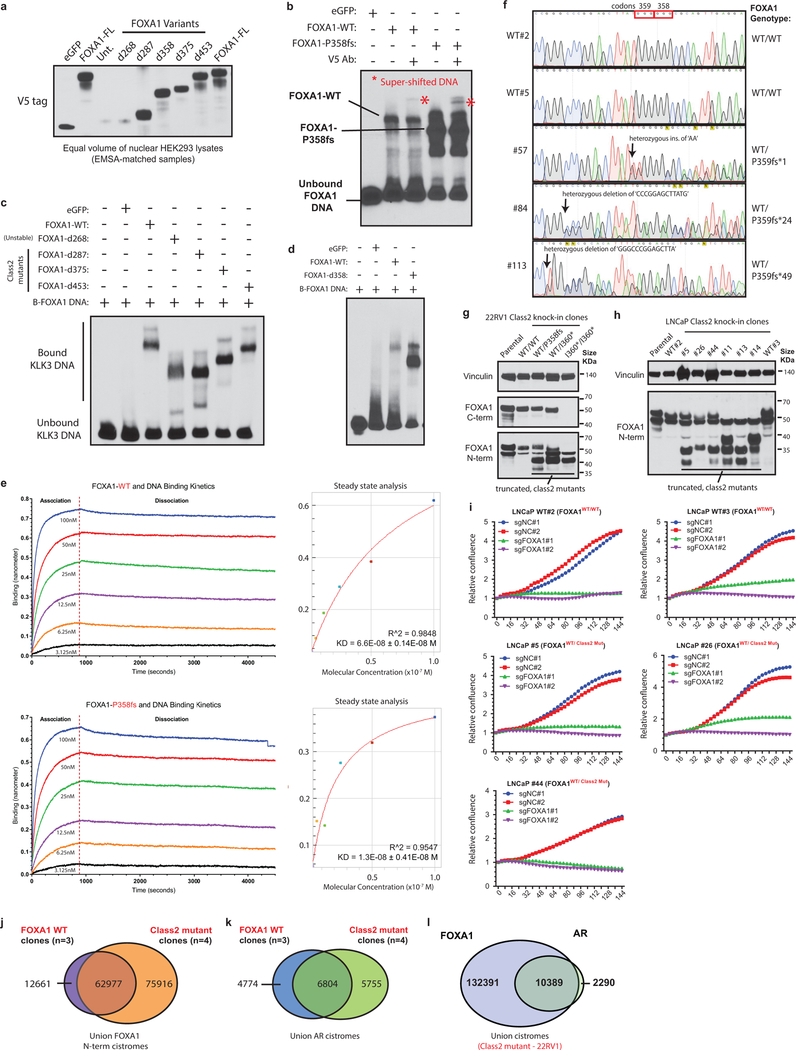
Class1 mutations comprise of missense and in-frame indels that cluster at the C-terminal edge of the winged-helix DNA-binding FKHD. Intriguingly, the majority of the class1 mutations were located either within the Wing2 region (residues 247–269) or a 3D-hotspot spatially protruding towards Wing2 (Fig. 2a,b and Extended Fig. 3a,b)21. Notably, these mutations did not alter FKHD residues that make base-specific interactions with the DNA22,23 (Fig. 2a and Extended Data Fig. 3c). In FOXA proteins, Wing2 residues make base-independent (i.e. non-specific) contacts with the DNA-backbone23,24, which reportedly impede its nuclear movement24. Thus, we hypothesized that the Wing2-altered class1 mutants would display faster nuclear mobility.
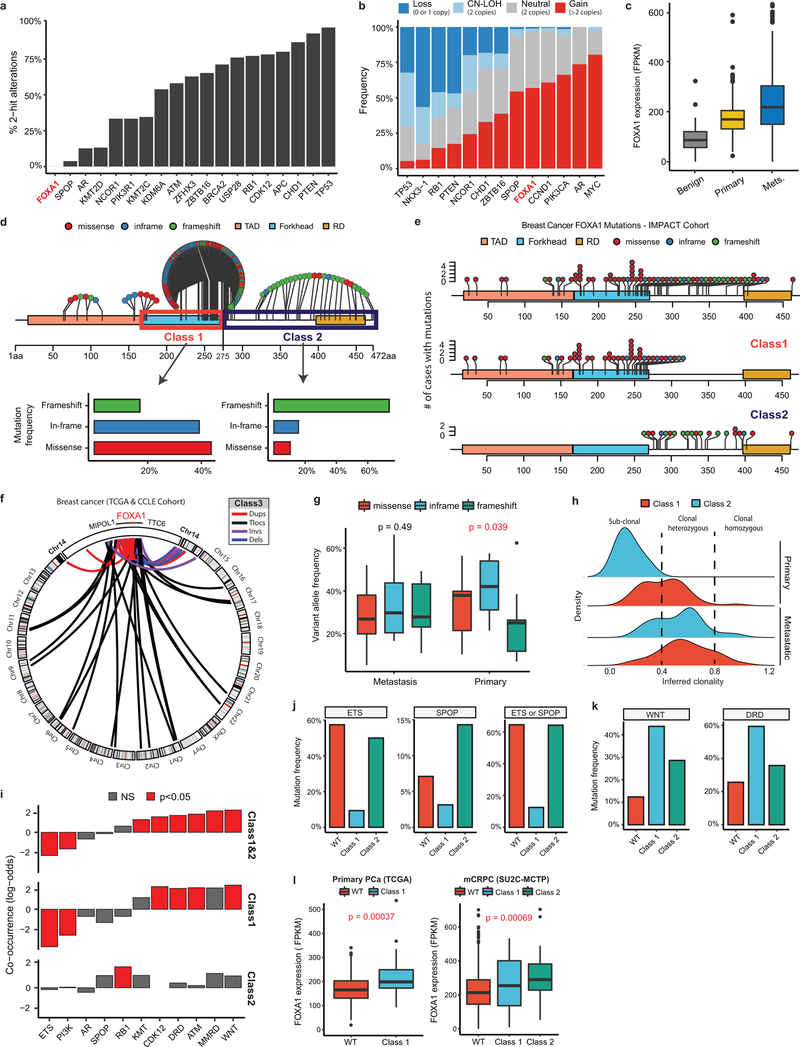
Figure 2. Functional characterization of Class1 mutations of FOXA1.
a) Distribution of class1 mutations on the protein map of FOXA1 functional domains and FKHD secondary structures. b) Crystal structure of the FKHD with visualization of non-Wing2 (i.e. outside of 247–269aa) mutations. 3D-hotspot mutations are in red. c) FRAP kinetic plots (left) and representative time-lapse images from pre-bleaching to the equilibrated state (n=6 biological replicates). Images are uniformly brightened for signal visualization. d) FRAP durations till 50% recovery (n=6 nuclei/variant). e) AR reporter activity with overexpression of FOXA1 variants and DHT stimulation (n=3 biological replicates). f) Growth (IncuCyte) of 22RV1 cells overexpressing FOXA1 variants in androgen-depleted medium (n=5 biological replicates). In d-f, means ± s.e.m are shown, and p-values are from two-way ANOVA and Tukey’s test. g) Relative expression of luminal and basal markers in class1 (n=38) tumors compared with WT (n=457), SPOP (n=48), and ETS (n=243) primary PCa tumors. h) Class1 model: Wing2-disrupted FOXA1 shows increased chromatin mobility and chromatin sampling frequency, resulting in stronger transcriptional activation of oncogenic AR-signaling. FKRE, forkhead responsive element; ARE, androgen responsive element.
We cloned representative class1 mutants: I176M (3D-hotspot mutation), R261G (missense) and R265–71del (in-frame deletion), all of which retained their nuclear localization (Extended Data Fig. 3d). Remarkably, in fluorescence-recovery after photobleaching (FRAP) assays, we found class1 mutants to have 5–6 times faster nuclear mobility, irrespective of the mutation type (Fig. 2c,d and Extended Data Fig. 3e,g). In contrast, Wing2-intact class2 mutants were still sluggish in their nuclear movement (Fig. 2d and Extended Data Fig. 3f,g). Using single particle tracking (SPT), we verified class1 mutants to have higher overall rate of nuclear diffusion, with 3–4 fold fewer slow particles and shorter chromatin dwell times (Extended Data Fig. 3h,i). Next, in chromatin immunoprecipitation with parallel DNA-sequencing (ChIP-seq) assays, we found ectopically expressed class1 mutants in HEK293 cells to bind DNA at the consensus FOXA1 motif (Extended Data Fig. 3j,k). In PCa cells, the class1 cistrome entirely overlapped with WT binding sites, with similar enrichment for FOXA1 and AR cofactor motifs, AR-binding sites, and genomic distribution (Extended Data Fig. 3l–s). Furthermore, in growth rescue experiments using UTR-specific siRNAs targeting the endogenous FOXA1 transcript, we found exogenous class1 mutants to be able to fully compensate for the WT protein (Extended Data Fig. 4a).
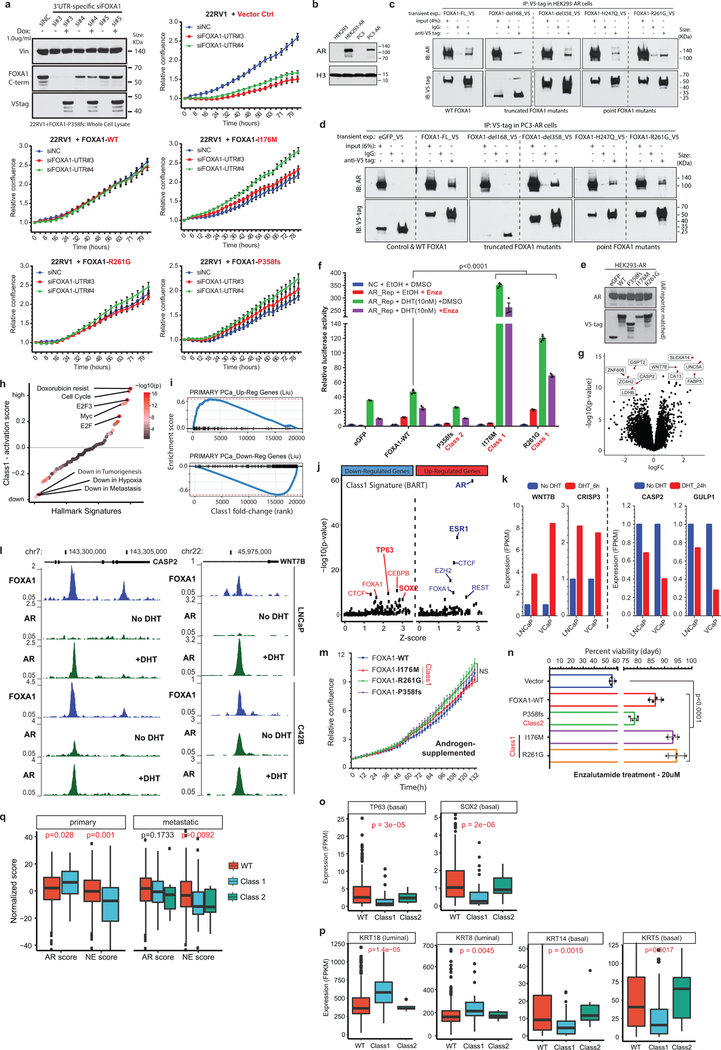
Next, we asked how class1 mutations affect AR-signaling. Like WT FOXA1, both class1 and class2 mutants interacted with the AR-signaling complex (Extended Data Fig. 4b–d). Strikingly, in reporter assays, class1 mutants induced 3–6 fold higher activation of AR-signaling (Fig. 2e), which was evident even under castrate-levels of androgen and enzalutamide treatment (Extended Data Fig. 4e.f). In parallel assays, class2 mutants showed no differences relative to WT FOXA1 (Fig. 2e). Transcriptomic analyses of class1 patient tumors revealed activation of hyper-proliferative and pro-tumorigenesis pathways, and further enrichment of primary PCa genes (Extended Data Fig. 4g–i). Notably, AR was predicted25 as the driver TF for class1 up-regulated genes, which we experimentally confirmed for several targets (Extended Data Fig. 4j–l). Concordantly, overexpression of class1 mutants in 22RV1 cells increased growth in androgen-depleted medium (Fig. 2f), but not in androgen-supplemented medium, as well as rescued proliferation upon enzalutamide treatment (Extended Data Fig. 4m,n). Interestingly, for class1 down-regulated genes, basal TFs TP63 and SOX2 were predicted as transcriptional drivers (Extended Data Fig. 4j). Consistently, in class1 patient specimens, both TFs were significantly downregulated with concomitant downregulation of basal and upregulation of luminal markers (Fig. 2g and Extended Data Fig. 4o,p). Additionally, class1 tumors had a higher AR and a lower neuroendocrine transcriptional signature (Extended Data Fig. 4q). Together, these data suggest that Wing2 mutations increase the nuclear speed and genome-scanning efficiency of FOXA1 without affecting its DNA sequence specificity (Supplementary Discussion), and drive a luminal AR program of prostate oncogenesis (Fig. 2h).
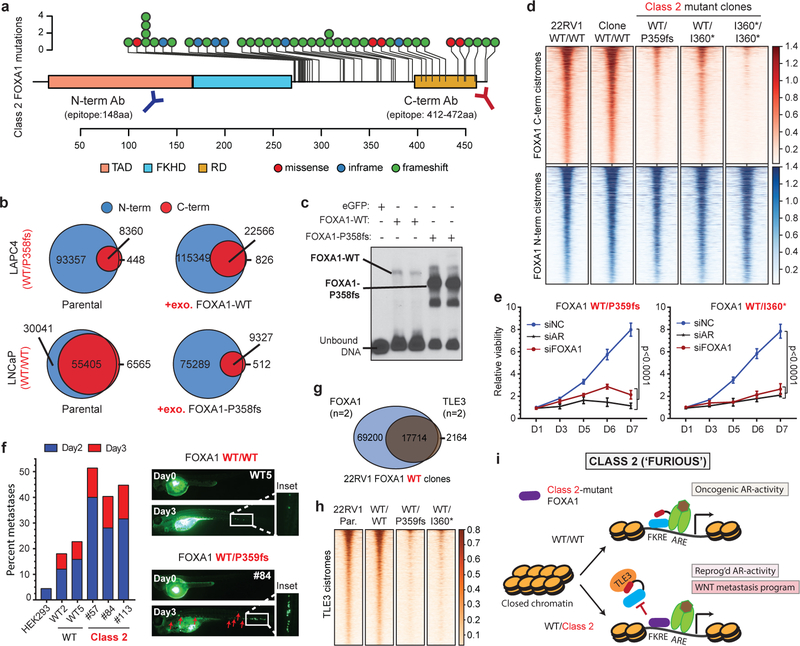
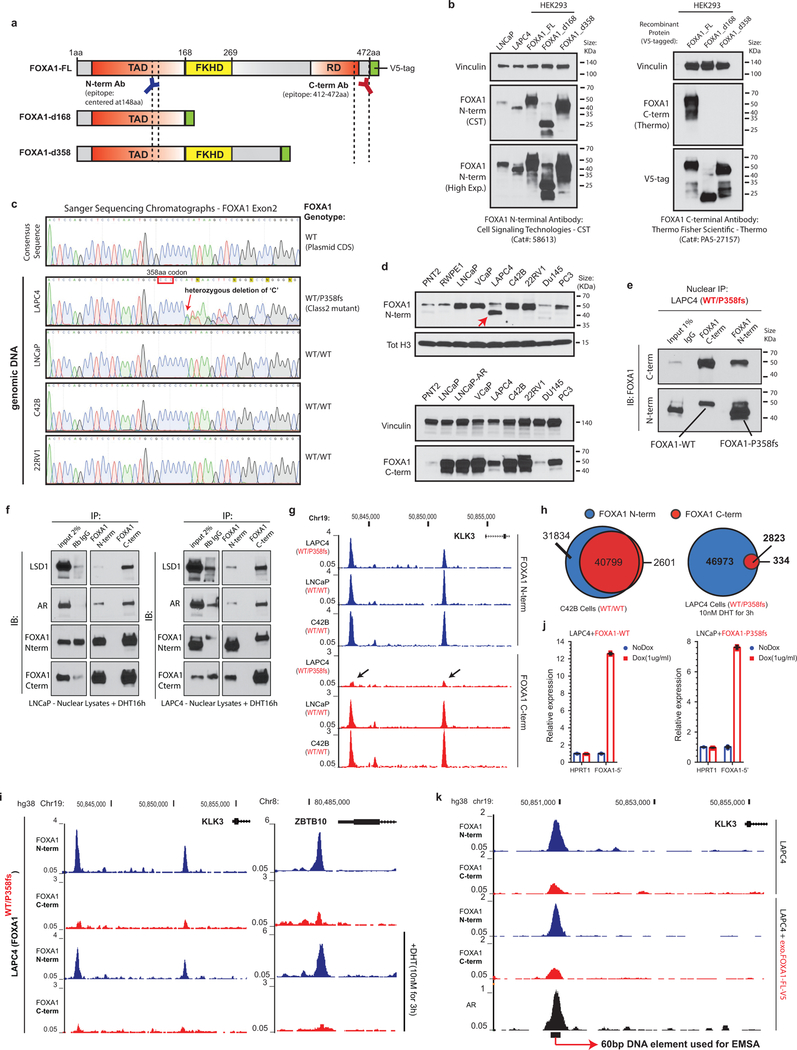
Class2 mutations comprise of frameshifting alterations that truncate the C-terminal regulatory domain of FOXA1 (Fig. 3a). Thus, we used N-terminal and C-terminal antibodies to characterize the class2 cistrome, with the latter exclusively binding to WT FOXA1 (Extended Data Fig. 5a,b). Notably, mCRPC-derived LAPC4 cells endogenously harbor a FOXA1 class2 mutation (i.e. P358fs), and both WT and mutant variants interacted with the AR complex (Extended Data Fig. 5c–f). Strikingly however, in ChIP-seq assays only the N-terminal antibody detected FOXA1 binding to the DNA. In contrast, N-terminal and C-terminal FOXA1 cistromes significantly overlapped in WT PCa cells (Fig. 3b and Extended Data Fig. 5g–i). Even with 13-fold overexpression of WT FOXA1 in LAPC4, the endogenous class2 mutant retained its binding dominance (Fig. 3b and Extended Data Fig. 5j,k). Conversely, overexpression of the P358fs mutant in LNCaP cells markedly diminished the endogenous WT cistrome (Fig. 3b). In in-vitro assays, class2 mutants showed markedly stronger binding to the KLK3 enhancer element (Fig. 3c and Extended Data Fig. 6a–d), and biolayer interferometry confirmed the P358fs mutant to have ~5-fold higher DNA-binding affinity (Extended Data Fig. 6e). Next, in CRISPR-engineered class2-mutant 22RV1 clones (Extended Data Fig. 6f,g), FOXA1 ChIP-seqs reaffirmed the cistromic-dominance of distinct class2 mutants (Fig. 3d). More importantly, knockdown of either mutant FOXA1 or AR in 22RV1 or LNCaP class2 CRISPR-clones significantly attenuated proliferation (Fig. 3e and Extended Data Fig. 6h,i). Consistently, in rescue experiments, the P358fs mutant fully compensated for the loss of WT FOXA1 (Extended Data Fig. 4a).
Figure 3. Functional characterization of Class2 mutations of FOXA1.
a) Class2 mutations and antibody epitopes on the protein map of FOXA1. b) N-term and C-term FOXA1 cistromes in PCa cells that are (right) untreated or (left) have exogenous overexpression of FOXA1 variants. c) Electromobility shift of FOXA1 variants bound to the KLK3-enhancer (n=3 biological replicates). For gel source data, see Supplementary Figure 1. d) FOXA1 ChIP-seq read-density heatmaps in independent class2-mutant 22RV1 CRISPR clones. e) Growth of class2-mutant 22RV1 clones treated with non-targeting (siNC), AR or FOXA1 targeting siRNAs (n=5 biological replicates; two-way ANOVA and Tukey’s test). Mean ± s.e.m. are shown. f) Left, Metastasis frequency in zebrafish embryos injected with HEK293 (negative control), WT, or class2-mutant 22RV1 clones (n≥30 embryos/group); Right, representative embryo images showing the disseminated PCa cells. g) Overlap of WT FOXA1 and TLE3 binding sites in 22RV1 CRISPR clones (n=2 biological replicates each). h) TLE3 ChIP-seq read-density heatmaps in two distinct FOXA1 WT and class2-mutant 22RV1 clones. i) Class2 model: Truncated FOXA1 shows dominant chromatin binding and displaces WT FOXA1 and TLE3 from the chromatin, resulting in increased WNT-signaling. FKRE, forkhead responsive element; ARE, androgen responsive element.
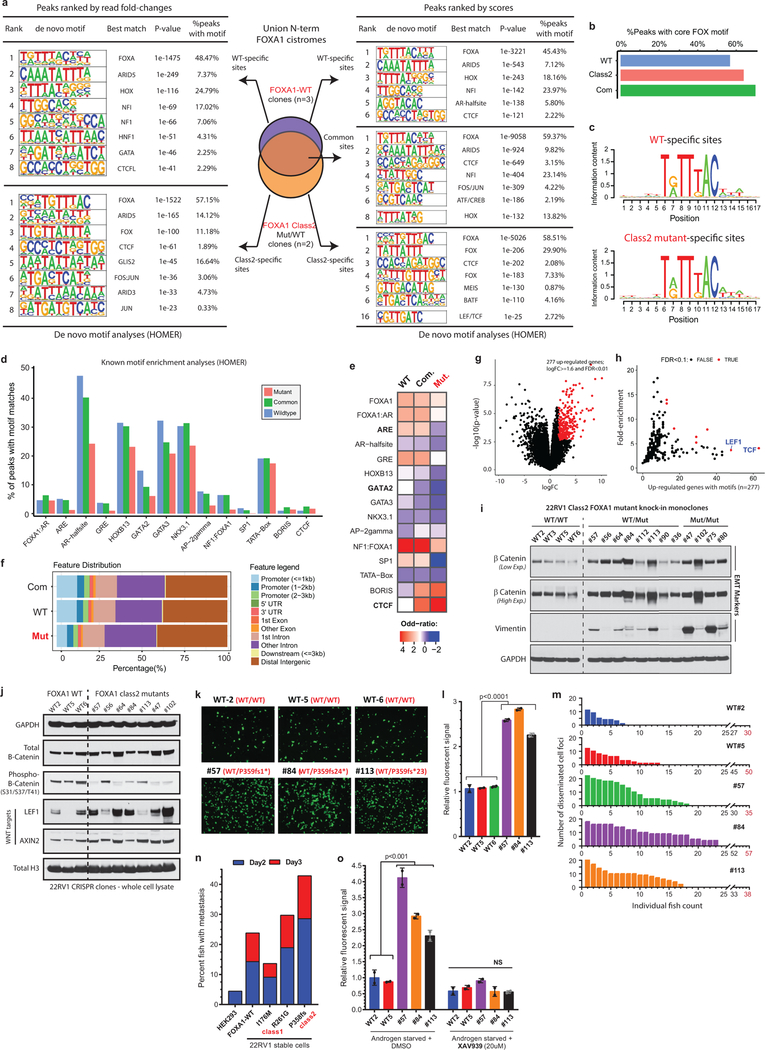
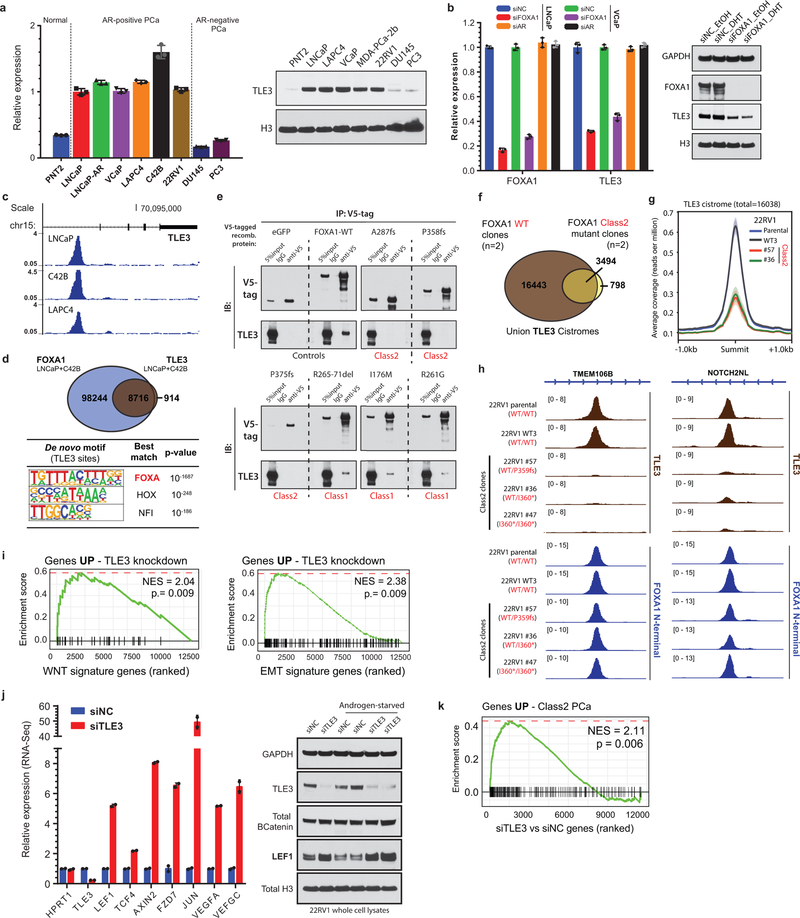
Intriguingly, the class2 cistrome was considerably larger with the acquired sites being enriched for the CTCF motif and distal regulatory regions (Supplementary Discussion and Extended Data Fig. 6j–l, 7a–e). In transcriptomic and motif analyses of the class2 clones, LEF and TCF were predicted as the top regulatory TFs for the up-regulated genes (Extended Data Fig. 7g,h). The LEF/TCF complex is the primary nuclear effector of WNT-signaling and remains inactive until bound by β-Catenin26. Consistently, we found marked accumulation of transcriptionally-active, S31/S37/T41 non-phosphorylated β-Catenin in distinct mutant clones, as well as a concomitant increase in expression of WNT targets LEF1 and AXIN2 (Extended Data Fig. 7i,j). In Boyden chamber assays, class2 clones showed 2–3 fold higher invasiveness (Extended Data Fig. 7k,l), and strikingly, in zebrafish embryos showed a higher rate as well as extent of metastatic dissemination (Fig. 3f and Extended Data Fig. 7m). In these assays, class1 mutant cells showed no differences relative to the WT cells (Extended Data Fig. 7n). Further, treatment with a WNT inhibitor (XAV939) completely abrogated the class2 invasive phenotype (Extended Data Fig. 7o). Investigating the mechanism, we found FOXA1 to transcriptionally activate and, through its C-terminal domain, recruit TLE3 (a bonafide WNT corepressor27) to the chromatin (Extended Data Fig. 8a–e). Distinct class2 mutants had lost this interaction, which remarkably led to TLE3 chromatin-untethering and downstream activation of WNT-signaling (Fig. 3g,h, Extended Data Fig. 8e–k, and Supplementary Discussion). Together, these data suggest that class2 mutations confer cistromic-dominance and abolish TLE3-mediated repression of the WNT program of metastasis (Fig. 3i).
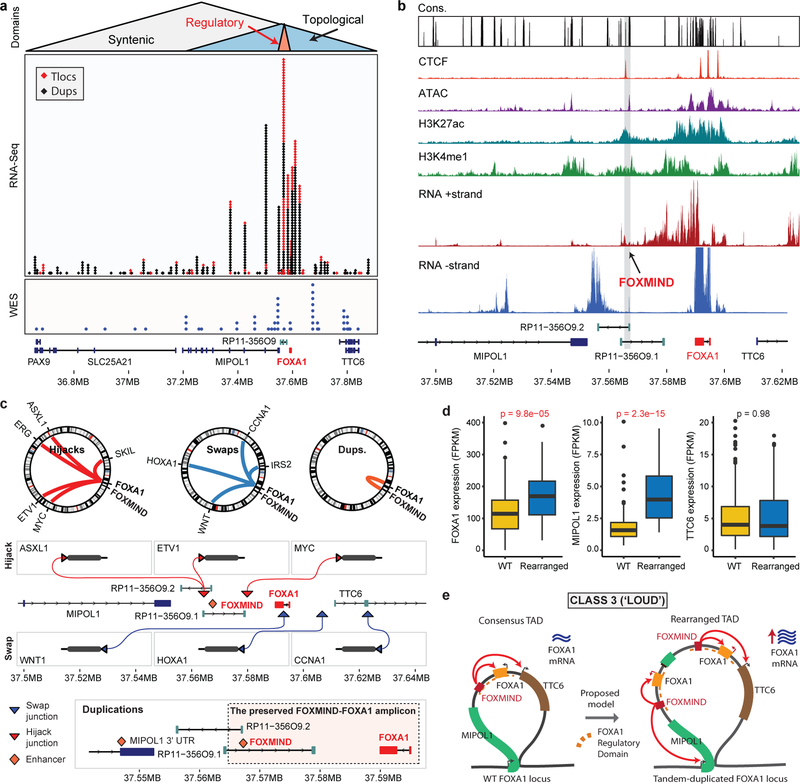
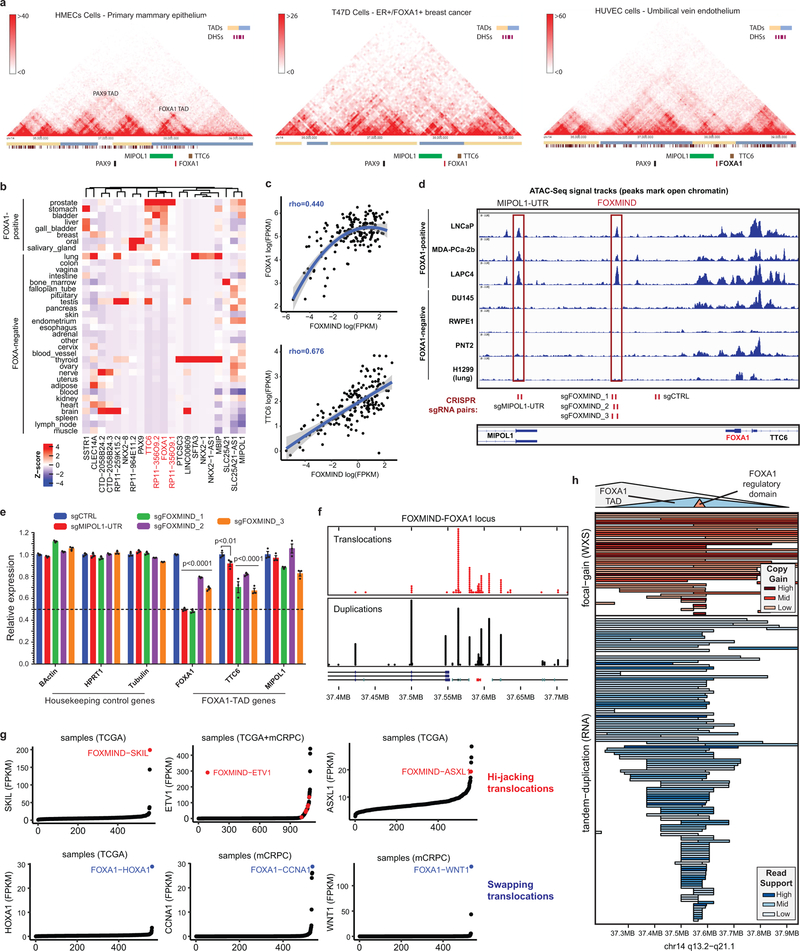
Class3 rearrangements occur within the PAX9/FOXA1 locus that is linearly conserved across the deuterostome superphylum28 (Fig. 4a). Intriguingly, almost all breakends were clustered within the FOXA1 topologically associating domain (TAD) (Extended Data Fig. 9a), suggesting class3 SVs to alter its transcriptional regulation. We found the FOXA1 TAD genes to have highest expression in the normal prostate, and the non-coding RP11–356O9.1 transcript to have a prostate-specific expression (Extended Data Fig. 9b). Furthermore, in patient tumors, expression of RP11–356O9.1 was strongly correlated with FOXA1 and TTC6 expression (Extended Data Fig. 9c). Thus, to identify prostate-specific enhancers of the FOXA1 TAD, we performed the assay for transposase-accessible chromatin using sequencing (ATAC-seq) and interrogated chromatin features in AR+ and AR- prostate cells. Notably, a CTCF-bound intronic site in RP11–356O9.1, hereafter denoted as FOXA1 Mastermind (FOXMIND), and a site within the 3’UTR of MIPOL1 were accessible and marked with active enhancer modifications in only AR+/FOXA1+ PCa cells (Fig 4b and Extended Data Fig. 9d). This strongly suggested these conserved sites to be enhancer elements. Consistently, CRISPR knock-out of these loci in VCaP cells led to a significant decrease in the expression of FOXA1 and TTC6, but not MIPOL1, which has its promoter outside of the FOXA1 TAD (Extended Data Fig. 9d,e).
Figure 4. Genomic characterization of Class3 rearrangements of the FOXA1 locus.
a) Breakends in relation to the FOXA1 syntenic, topological, and regulatory domains. b) Representative functional genomic tracks at the FOXA1 locus. Base level conservation (Cons), DNA accessibility (ATAC), enhancer-associated histone modifications (H3K27me1 and H3K27Ac), CTCF chromatin binding, and stranded RNA-seq read densities are visualized. FOXMIND enhancer is highlighted. c) Structural patterns of translocations and duplications. Hijacks occur between FOXMIND and FOXA1; swaps occur upstream of FOXA1. Duplications amplify the highlighted FOXMIND-FOXA1 regulatory domain. d) Transcriptional changes in FOXA1 TAD gene in locus WT (n=320) and rearranged (n=50) cases (two-sided t-test). Boxplot center: median, box: Q1/Q3, whiskers: Q1/Q3±1.5xIQR. e) Class3 model: Tandem duplications within the FOXA1 TAD amplify FOXMIND to drive FOXA1 overexpression.
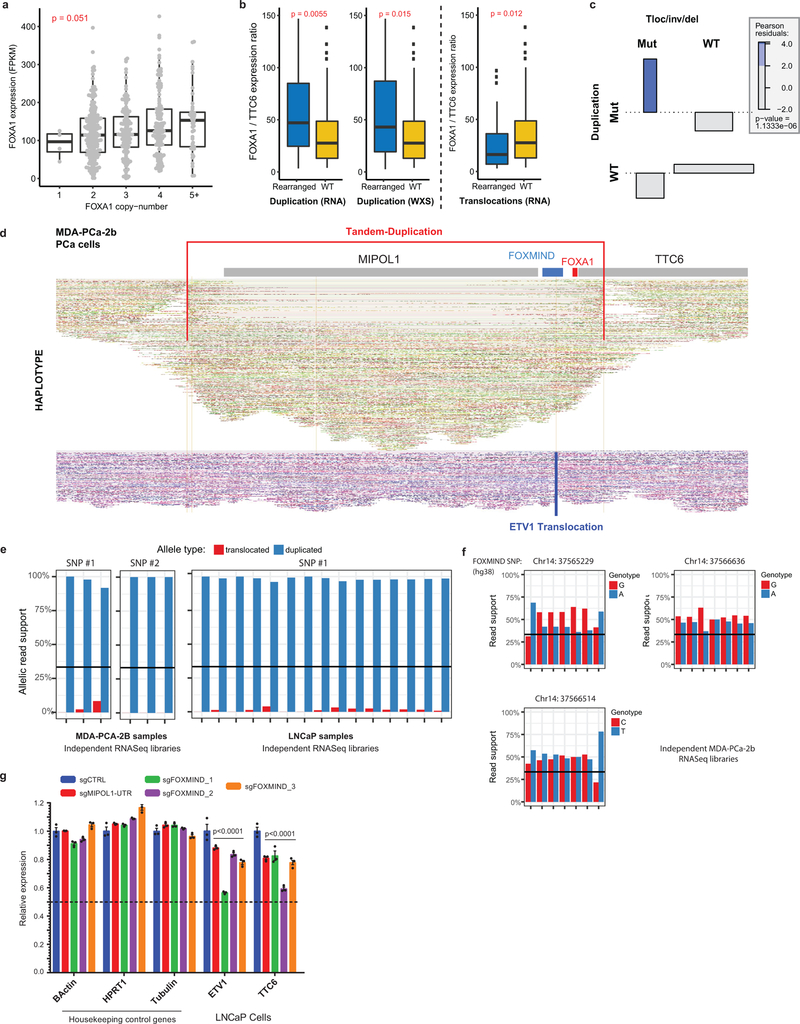
Strikingly, we found that translocations were largely within a 50 kb region between FOXA1 and 3’ UTR of MIPOL1, while breakend junctions from duplications mostly flanked the FOXMIND-FOXA1 region (Fig. 4a and Extended Data Fig. 9f). For translocations, we delineated two patterns: 1) hijacking of the FOXMIND enhancer and 2) inserting upstream of the FOXA1 promoter (Fig. 4c). The first pattern subsumes previously reported in-frame fusions transcripts involving RP11–356O9.1 and ETV129 / SKIL30, as well as a novel ASXL1 fusion (Supplementary Table 4). The second pattern inserts an oncogene, such as CCNA1, upstream of FOXA1 (Fig. 4c). Notably, both mechanisms resulted in outlier expression of the translocated gene (Extended Data Fig. 9g). For duplications, which constitute 70% of all rearranged cases, we found FOXMIND and FOXA1 to be typically co-amplified (89%) and never separated (bottom, Fig. 4c and Extended Data Fig 9h), thus preserving the FOXMIND-FOXA1 regulatory domain.
Next, while assessing the transcriptional impact of duplications, we found FOXA1 mRNA levels to be poorly correlated with copy-number (Extended Data Fig. 10a), but highly sensitive to focal SVs. Tandem duplications, ascertained at the RNA and DNA levels, significantly increased FOXA1 and MIPOL1 expression, but not TTC6 expression (Fig. 4d). Surprisingly, translocations resulted in a modest decrease in FOXA1 levels (Extended Data Fig. 10b), despite a significant co-occurrence with tandem-duplications (OR=3.89, Extended Data Fig. 10c). To explore this further, we carried out haplotype-resolved, linked-read sequencing of MDA-PCA-2b cells, which harbor a FOXMIND-ETV1 translocation. Here, ETV1 translocation was accompanied by a focal tandem-duplication in the non-translocated FOXA1 allele (Extended Data Fig. 10d). Intriguingly, the translocated FOXA1 allele was inactivated, resulting in monoallelic transcription (Extended Data Fig. 10e); but without a net-loss in FOXA1 expression (266 FPKM, 95th percentile in mCRPC). Contrarily, RP11–356O9.1 retained biallelic expression (Extended Data Fig. 10f). In LNCaP cells, which also harbor ETV1-translocation into the FOXA1 locus, deletion of FOXMIND caused a significant reduction in ETV1 expression (Extended Data Fig. 10g). Thus, translocations result in the loss of FOXA1 expression from the allele in cis, which is rescued by tandem-duplications of the allele in trans. Altogether, we propose a coalescent model wherein class3 SVs duplicate or re-position FOXMIND to drive overexpression of FOXA1 or other oncogenes (Fig. 4e).
In summary, we identify three previously undescribed structural classes of FOXA1 alterations that differ in genetic associations and oncogenic mechanisms. We establish FOXA1 as a principal oncogene in AR-dependent PCa, altered in over 34.6% of mCRPC. Given the distinct pathogenic features, we propose to refer to these classes as the ‘FAST’ (class1), ‘FURIOUS’ (class2), and ‘LOUD’ (class3) aberrations of FOXA1 (Fig 2h, 3i, 4e, Supplementary Table 5 and Supplementary Discussion). Structurally equivalent FOXA1 alterations are also found in other hormone-receptor driven cancers, thereby positioning FOXA1 as a promising therapeutic target in these malignancies.
Methods
Cell Culture
Most cell lines were originally purchased from the American Type Culture Collection (ATCC) and were cultured as per the standard ATCC protocols. LNCaP-AR and LAPC4 cells were gifts from Dr. Charles Sawyers lab (Memorial Sloan-Kettering Cancer Center, New York, NY). Until otherwise stated, for all the experiments LNCaP, PNT2, LNCaP-AR, C42B, 22RV1, DU145, PC3 cells were grown in the RPMI 1640 medium (Gibco) and VCaP cells in the DMEM with Glutamax (Gibco) medium supplemented with 10% Full Bovine Serum (FBS; Invitrogen). LAPC4 cells were grown in IMEM (Gibco) medium supplemented with 15%FBS and 1nM of R1881. Immortalized normal prostate cells: RWPE1 were grown in keratinocyte media with regular supplements (Lonza); PNT2 were grown in RPMI medium with 10%FBS. HEK293 cells were grown in DMEM (Gibco) medium with 10% FBS. All cells were grown in a humidified 5%CO2 incubator at 37℃. All cell lines were biweekly tested to be free of mycoplasma contamination and genotyped every month at the University of Michigan Sequencing Core using Profiler Plus (Applied Biosystems) and compared with corresponding short tandem repeat (STR) profiles in the ATCC database to authenticate their identity in culture between passages and experiments.
Antibodies
For immunoblotting, the following antibodies were used: FOXA1_N-terminal (Cell Signaling Technologies: 58613S; Sigma-Aldrich: SAB2100835); FOXA1_C-terminal (ThermoFisher Scientific: PA5–27157; Abcam: ab23738); AR (Millipore: 06–680); LSD1 (Cell Signaling Technologies: 2139S); Vinculin (Sigma Aldrich: V9131); H3 (Cell Signaling Technologies: 3638S); GAPDH (Cell Signaling Technologies: 3683); B-Actin (Sigma Aldrich: A5316); B-Catenin (Cell Signaling Technologies: 8480S); Vimentin (Cell Signaling Technologies: 5741S); Phospho(S33/S37/T41)-B-Catenin (Cell Signaling Technologies: 8814S); LEF1 (Cell Signaling Technologies: 2230S); AXIN2 (Abcam: ab32197), and TLE3 (Proteintech: 11372–1-AP).
For co-immunoprecipitation and ChIP-seq experiments, the following antibodies were used: FOXA1_N-terminal (Cell Signaling Technologies: 58613S); FOXA1_C-terminal (ThermoFisher Scientific: PA5–27157); AR (Millipore: 06–680); V5-tag (R960–25); TLE3 (Proteintech: 11372–1-AP).
Immunoblotting and nuclear co-immunoprecipitation
Cell lysates were prepared using the RIPA lysis buffer (ThermoFisher Scientific; Cat#: 89900) and denatured in the complete NuPage 1X LDS/reducing agent buffer (Invitrogen) with 10 minutes heating at 70C. 10–25ug of total protein was loaded per well, separated on 4–12% SDS polyacrylamide gels (Novex) and transferred onto 0.45-micron nitrocellulose membrane (Thermo Fisher Scientific; Cat#: 88018) using a semi-dry transfer system (Trans-blot Turbo System; BioRad) at 25V for 1h. The membrane was incubated for 1 hour in blocking buffer (Tris-buffered saline, 0.1% Tween (TBS-T), 5% nonfat dry milk) and incubated overnight at 4°C with primary antibodies. If samples were run on multiple gels for an experiment, then multiple loading control proteins (i.e. GAPDH, BActin, Total H3, and Vinculin) were probed on each membrane separately. Host species-matched secondary antibodies conjugated to horseradish peroxidase (HRP; BioRad) were used at ½0,000 dilution to detect primary antibodies and blots were developed using enhanced chemiluminescence (ECL Prime, Thermo Fisher Scientific) following the manufacturer’s protocol.
For nuclear co-immunoprecipitation assays, 8–10 million cells ectopically overexpressing different V5-tagged FOXA1 variants and WT AR (or TLE3) were fractionated to isolate intact nuclei using the NE-PER kit reagents (Thermo Fisher Scientific; Cat#: 78835) and lysed in the complete IP lysis buffer (Thermo Fisher Scientific; Cat#: 87788). Nuclear lysates were incubated for 2 hours at 4C with 30ul of magnetic Protein-G Dynabeads (Thermo Fisher Scientific; Cat#: 10004D) for pre-clearing. A fraction of the pre-cleared lysate was saved as input and the remainder was incubated overnight (12–16 hours) with 10ug of target protein antibody at 4C with gentle mixing. Next day, 50ul of Dynabeads Protein-G beads were added to the lysate-antibody mixture and incubated for 2h at 4C. Beads were washed 3 times with IP buffer (150nM NaCl; Thermo Fisher Scientific) and directly boiled in 1X NuPage LDS/reducing agent buffer (ThermoFisher Scientific; Cat#: NP0007 and NP0009) to elute and denature the precipitated proteins. These samples were then immunoblotted as described above with the exception of using protein A-HRP secondary (GE HealthCare, Cat#: NA9120–1ML) antibody for detection.
RNA extraction and quantitative polymerase chain reaction (qPCR)
Total RNA was extracted using the the miRNeasy Mini Kit (Qaigen), with the inclusion of on-column genomic DNA digestion step using the RNase-free DNase Kit (Qaigen), following the standard protocols. RNA was quantified using the NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific) and 1ug of total RNA was used for complementary DNA (cDNA) synthesis using the SuperScript III Reverse Transcriptase enzyme (ThermoFisher Scientific) following manufacturer’s instructions. 20ng of cDNA was inputted per polymerase chain reaction (PCR) using the FAST SYBR Green Universal Master Mix (ThermoFisher Scientific) and every sample was quantified in triplicates. Gene expression was calculated relative to GAPDH and HPRT1 (loading control) using the delta-delta Ct method and normalized to the control group for graphing. quantitative PCR (qPCR) primers were designed using the Primer3Plus tool (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and synthesized by Integrated DNA Technologies.
Primer used in this study are listed below:
GAPDH: F, TGCACCACCAACTGCTTAGC and R, GGCATGGACTGTGGTCATGAG;
HPRT1: F, AGGCGAACCTCTCGGCTTTC and R, CTAATCACGACGCCAGGGCT;
B-Actin: F, AGGATGCAGAAGGAGATCACTG and R, AGTACTTGCGCTCAGGAGGAG;
AR: F, CAGTGGATGGGCTGAAAAAT and R, GGAGCTTGGTGAGCTGGTAG;
FOXA1–3’: F, GAAGACTCCAGCCTCCTCAACTG and R, TGCCTTGAAGTCCAGCTTATGC;
FOXA1–5’: F, CTACTACGCAGACACGCAGG and R, CCGCTCGTAGTCATGGTGTT;
TLE3: F, AAGGACAGCTTGAGCCGATA and R, TTTGGTCTTGGAGGAAGGTG;
TTC6: F, CGAACAGAGCCAGGAGGTAG and R, GTTCTCCCTGGGCTCCTAAC;
MIPOL1: F, GCAAACGGTTAGAGCAGGAG and R, GGGTCTGGATTTCCTCTTCC;
ETV1: F, TACCCCATGGACCACAGATT and R, CACTGGGTCGTGGTACTCCT;
B-Tubulin: F, CTGGACCGCATCTCTGTGTACT and R,GCCAAAAGGACCTGAGCGAACA.
siRNA-mediated gene knockdown
Cells were seeded in a 6-well plate at the density of 100,000–250,000 cells per well. After 12 hours, cells were transfected with 25nM of gene-targeting ON-TARGETplus SMARTpool siRNAs or non-targeting pool siRNAs as negative control (Dharmacon) using the RNAiMAX reagent (Life Technologies; Cat#: 13778075) on two consecutive days, following manufacturer’s instructions. Both total RNA or protein was extracted on day 3 (total 72h) to confirm efficient (>80%) knockdown of the target genes. For crystal violet staining, at day 9 growth medium was aspirated and cells were first fixed with 4% formaldehyde solution, followed by a 30 minute incubation in 0.5% crystal violet solution in 20% methanol and scanned. Catalogue numbers and guide sequences (5’ to 3’) of siRNA SMARTpools (Dharmacon) used are:
Non-targeting control (Cat#: D-001810–10-05; UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA, UGGUUUACAUGUUUUCCUA);
AR (Cat#: L-003400–00-0005; GAGCGUGGACUUUCCGGAA, UCAAGGAACUCGAUCGUAU, CGAGAGAGCUGCAUCAGUU, CAGAAAUGAUUGCACUAUU);
FOXA1 (Cat#: L-010319–00-0005; GCACUGCAAUACUCGCCUU, CCUCGGAGCAGCAGCAUAA, GAACAGCUACUACGCAGAC, CCUAAACACUUCCUAGCUC);
TLE3 (Cat#: L-019929–00-0005; GCCAUUAUGUGAUGUACUA, GCAUGGACCCGAUAGGUAU, GAACCACCAUGAACUCGAU, UCAGGUCGAUGCCGGGUAA).
The FOXA1 SMARTpool comprises of siRNAs targeting 5’ as well as 3’ ends of the FOXA1 transcript. Thus, both WT and class2 mutant transcripts are degraded using the SMARTpool siRNAs. This was experimentally confirmed in LAPC4 cells that endogenously harbor a FOXA1 class2 mutation (Extended Data Fig. 1d, e).
CRISPR-Cas9-mediated gene or enhancer knockout
Cells were seeded in a 6-well plate at the density of 200,000–300,000 cells per well and infected with viral particles with lentiCRISPR-V2 plasmids coding either non-targeting (sgNC) or sgRNAs targeting the Exon1 or the Forkhead domain of FOXA1 (both ensuing in FOXA1 inactivation). This was followed by 3 days of puromycin selection after which proliferation assays were carried out as described below. The lentiCRISPR-V2 vector was a gift from Dr. Feng Zhang’s lab (Addgene plasmid # 52961).
sgRNA sequences used are as follows:
sgNC#1 5’-GTAGCGAACGTGTCCGGCGT-3’;
sgNC#2; 5’-GACCGGAACGATCTCGCGTA-3’
sgFOXA1_Exon1: 5’-GTAGTAGCTGTTCCAGTCGC-3’;
sgFOXA1_Forkhead: 5’-GCCGTTCTCGAACATGTTGC-3’.
Alternatively, for functional interrogation of the FOXA1 TAD enhancer elements, VCaP or LNCaP cells were transfected with pairs of sgRNAs targeting the MIPOL1-UTR or FOXMIND or a control locus within the FOXA1 topologically associating domain (TAD). Transfected cells were then selected with puromycin (1.0ug/ml) for 48h, followed by incubation for an additional 72h. Total RNA was extracted and qPCR was performed as described above.
Pair-wise sgRNA sequences are as follows (5’ to 3’):
sgCtrl: CACCGATTAGCCTCAACTATACCA & CACCGTGCAATATCTGAATCACACG;
sgMIPOL1-UTR: CACCGTGAAAAAAAACGACAGTCTG & CACCGAACTCAAGTCAGCAGCAAAG;
sgFOXMIND_1: CACCGCTTTAATAAAGCTATTTGC & CACCGATAGAGTGACTAATGCCCTG;
sgFOXMIND_2: CACCGTAACAGTTGACCTACTAAC & CACCGATTTAGATAAGGGGATAGAA;
sgFOXMIND_3: CACCGCTTTAATAAAGCTATTTGC & CACCGATTTAGATAAGGGGATAGAA.
CRISPR knock-out screen
For the genome-wide CRISPR knock-out screen, a two vector system was employed. First, LNCaP cells were engineered to stably overexpress the enzymatically active Cas9 protein. These cells were then treated with the human GeCKO knockout sgRNA library (GecKO V2) that was a gift from the Zhang Lab (Addgene, Cat#: 1000000049). This was followed by puromycin selection for 48h after which fraction of these cells were processed to isolated genomic DNA as the input sample. The remaining cells were then cultured for 30 days, and genomic DNA was extracted at this time point. sgRNA sequences were amplified using common adaptor primers and sequenced on the Illumina HiSeq 2500 (125-nucleotide read length). Sequencing data was analyzed as described31 and depletion or enrichment of individual sgRNAs at 30 days was calculated relative to the input sample. Note: Only a subset of genes including essential controls, epigenetic regulators and transcription factors from the GeCKO-V2 screen were plotted in Extended Data Fig. 1i.
Proliferation assays
For siRNA growth assays, cells were directly plated in a 96-well plate at the density of 2,500–8,000 per well and transfected with gene-specific or non-targeting siRNAs as described above on Day 0 and Day 1. Every treatment was carried out in six independent replicate wells. CellTiter-Glo reagent (Promega) was used to assess cell viability at multiple timepoints post-tranfections following manufacturer’s protocol. Data was normalized to siNC-Day 1 readings and plotted as relative cell viability to generate growth curves.
Alternatively, for CRISPR-sgRNA growth assays, cells were treated as described above for target gene inactivation and seeded into a 24-well plate at 20,000 cells/well density with 2 replicates per group. After 12 hours, plates were placed into the IncuCyte live cell imaging machine (IncuCyte) set at the phase contrast option to record cell confluence every 3 hours for upto 7–9 days. Similarly, for class1 growth assays (Fig. 2f), stable doxycycline-inducible 22RV1 cells were grown in 10% charcoal-stripped serum (CSS)-supplemented medium for 48 hours. Androgen starved cells were then seeded into a 96-well plate at 5000 cells/well density in 10%CSS medium with or without addition of doxycycline (1ug/ml) to induce control or mutant protein expression (6 replicates/group). Once adherent, treated cells were placed in the IncuCyte live cell imaging machine set at phase contrast to record cell confluence every 3 hours for upto 7–9 days. In all IncuCyte assays, confluence measurements from all time points were normalized to the matched measurement at 0 hours and plotted as relative confluence to generate growth curves.
Cloning of representative FOXA1 mutants
WT FOXA1 coding sequence was purchased from Origene (Cat#: SC108256) and cloned into the pLenti6/V5 lentiviral vector (ThermoFisher Scientific; Cat#: K4955–10) using the standard TOPO cloning protocol. Class1 missense mutations (I176M; H247Q and R261G) were engineered from the WT FOXA1 vector using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Tech) as per manufacturer’s instructions. All point mutations were confirmed using Sanger sequencing through the University of Michigan Sequencing Core Facility. Engineered mutant plasmids were further transfected in HEK293 cells to confirm expression of the mutant protein. For truncated class2 variants, the WT coding sequence upto the amino acid before the intended mutation was cloned. All FOXA1 variants had the V5-tag fused on the C-terminus. Also, select mutants were cloned into a doxycycline-inducible vector (Addgene: pCW57.1; Cat# 41393) to generate stable lines. For FRAP and SPT assays, the pCW57.1 vector was edited to incorporate an in-frame GFP or Halo coding sequences at the C-terminal end, respectively.
Fluorescent recovery after photobleaching (FRAP) assay and data quantification
PNT2 cells were seeded in a 6 well plate at 200,000 cells/well density and transfected with 2ug of doxycycline-inducible vectors coding different FOXA1 variants fused to GFP on the C-terminal end. After 24 hours, cells were plated in the glass-bottom microwell dishes (MatTek: #P35G-1.5–14-C) in phenol-free growth medium supplemented with doxycycline (1ug/ml). Cells were then incubated for 48 hours to allow for robust expression of the exogenous GFP-tagged protein and strong adherence to the glass surface. Microwell dishes were placed in humidity control chamber set at 37C (Tokai-Hit) and mounted on the SP5 Inverted 2-Photon FLIM Confocal microscope (Leica). FRAP Wizard from the Leica Microsystems software suite was used to conduct and analyze FRAP experiments. Fluorescent signals were automatically computed in regions-of-interest using in-built tools in the FRAP Wizard. Roughly half of the nucleus was photobleached using the Argon-laser at 488nm and 100% intensity for 20–30 iterative frames at 1.2 second intervals. Laser intensity was reduced to 1% for imaging post bleaching. Immediately after photobleaching, 2 consecutive images were collected at 1.2 second intervals followed by images taken at 10 seconds intervals for 60 frames (i.e. 10 minutes).
For data analyses, recovery of signal in the bleached half and loss of signal in the unbleached half were measured as average fluorescence intensities in at least 80% of the respective areas, excluding the immediate regions flanking the separating border. All intensity curves were generated from background-subtracted images. The fluorescence signal measured in a region-of-interest (ROI) was normalized to the signal prior to bleaching using the following formula32:
where, ‘Io’ is the average intensity in the ROI before bleaching, ‘It’ is the average intensity in the ROI at any time-point post-bleaching, and ‘Ibg’ is the background fluorescence signal in a region outside of the cell nucleus. Raw recovery kinetic data from above were fitted with best hyperbolic curves using the GraphPad Prism software and time to 50% recovery were calculated from the resulting best fit equations. Please note for representative time-lapse nuclei images shown in the FRAP figures, the fluorescence signal was uniformly brightened for the easy of visualization.
Single particle tracking (SPT) experiment and data quantification
PNT2 cells were transiently transfected with doxycycline-inducible vectors encoding C-terminal Halo-tagged WT or class1 mutant variants of FOXA1. Transfected cells were seeded in glass bottom DeltaT culture dishes (Bioptechs, Cat# 04200417C) and incubated for 24 h with 0.01ug/ml of doxycycline. Cells were then treated with phenol-red free medium containing 2% FBS and 5 nM cell permeable JF549 Halo ligand dye (Grimm et al, Nat. Methods, 2015) for 30 min at 37 oC. Cells were subsequently washed 2 times, 10 min per wash at 37 oC, with phenol-red free medium containing 2% FBS. Prior to imaging cells were washed once with the 1X HBSS buffer and were imaged in the buffer.
SPT was performed on an Olympus IX81 microscope via HILO illumination, as described33 at a spatial accuracy of 30 nm and temporal resolution of 33 ms. Image analysis was performed as described34. Briefly, tracking was done in Imaris (bitplane) and particles that were at least visible for four continuous frames were used for further analysis. Diffusion constants were calculated as described35, assuming a Brownian diffusion model under steady-state conditions. Dwell time histograms were fit to a double-exponential function to extract fast and slow dwell times of “bound” particles that displayed a frame-to-frame displacement of < 300 nm. All particles that were visible for less than 4 consecutive frames or those that moved > 300 nm between frames were counted as “unbound” particles. At least five cells were imaged for each transcription factor variant and >500 particles were tracked to extract diffusion constants and dwell time.
Dual luciferase AR reporter assay
HEK293 cells stably overexpressing the WT AR protein (i.e. HEK293-AR) were used for the AR reporter assays. HEK293-AR cells were seeded in a 12-well plate at 300,000 cell/well density and transfected with 2ug of the pLenti6/V5 vector coding different FOXA1 variants or GFP (control). After 8 hours, medium was replaced with 10%-CSS-supplemented phenol-free medium (androgen depleted) and cells were transfected with the AR-reporter Firefly luciferase or negative control constructs from the Cignal AR-Reporter(luc) kit (QAIGEN; Cat# CCS-1019L) as per manufacturer’s instructions. Both constructs were premixed with constitutive Rinella luciferase vector as control. After 12 hours, cells were treated with different dosages of dihydrotestosterone (DHT) or enzalutamide (at 10uM dosage); and additional 24 hours later dual luciferase activity was recorded for every sample using the Dual-Glo Luciferase assay (Promega; E2980) and luminescence plate reader (Promega-GLOMAX-Multi Detection System). Each treatment condition had 4 independent replicates. Firefly luciferase signals were normalized with the matched Rinella luciferase signals to control for variable cell number and/or transfection efficiencies, and normalized signals were plotted relative to the negative control reporter constructs.
Electrophoretic mobility shift assay (EMSA)
HEK293 cells were plated in 10cm dishes at 1M/plate density and transfected with 10ug of the pLenti6/V5 vector coding GFP (control) or different FOXA1 variants. After 48 hours, cells were trypsinized and nuclear lysates were prepared using the NE-PER kit reagents (ThermoFisher Scientific). Immunoblots were run to confirm comparable expression of recombinant FOXA1 variants in 2ul (i.e. equal volume) of final nuclear lysates. Next, FOXA1 and AR ChIP-seq data was used to identify the KLK3 enhancer element. 60bp of the KLK3 enhancer, centered at the FOXA1 consensus motif 5’-GTAAACAA-3’, was synthesized as single stranded oligos (IDT) and biotin-labeled using the Biotin 3’-End DNA labeling kit (ThermoFisher Scientific) and then annealed to generate a labeled double-stranded DNA duplex.
Binding reactions were carried out in 20ul volumes containing 2ul of the nuclear lysates, 50ng/uL poly(dI.dC), 1.25% glycerol, 0.025% Nonidet P-40 and 5mM MgCl2. 10fmol of biotin-labeled KLK3 enhancer probe was added at the very end with gentle mixing. Reactions were incubated for 1h at room temperature, size-separated on a 6% DNA retardation gels (100V for 1h; Invitrogen) in 0.5X TBE buffer, and transferred on the Biodyne Nylon membrane (0.45um; ThermoFisher Scientific) using a semi-dry system (BioRad). Transferred DNA was crosslinked to the membrane using the UV-light at 120mJ/cm2 for 1 minute. Biotin-labeled free and protein-bound DNA was detected using HRP-conjugated streptavidin (ThermoFisher Scientific) and developed using chemiluminescence according to the manufacturer’s protocol.
Protein synthesis and purification
First, WT and P358fs mutant FOXA1 proteins were purified using the E. coli bacterial expression system and Nickle-affinity chromatography. Briefly, WT of P358fs coding sequences were cloned into the pFC7A (HQ) Flexi vector (Promega, Cat#: C8531) with a C-terminal HQ-tag, following manufacturer’s protocol. These expression constructs were used to transform the Single Step (KRX) Competent E. coli cells (Promega, Cat#: L3002), which have been modified for synthesis of mammalian proteins. A starter broth of 2 ml was inoculated with a single colony of transformed bacterial cells and incubated at 37C with constant shaking at 250 rpm until the OD600 of 0.4–0.5 was reached. The starter brother was then used to inoculate 1000 ml of LB broth containing Ampicillin, and protein synthesis was induced using 0.1% v/v of rhamanose. Induced culture was incubated at 20C for 16h with constant shaking at 250 rpm. Bacterial cells were then pelleted by centrifugation at 4,000 rpm for 30 mins and mechanically lysed through sonication in 50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 1% glycerol) in the presence of protease inhibitors (Roche). HisLink Purification Resin (Promega, Cat#: V8821) was used to purify untagged recombinant proteins from the crude bacterial lysates as per manufacturer’s protocol (this includes removal of the His tag as well). Purified protein fractions were then tested for purity by Coomaisse staining relative to the crude input lysates, and purified protein concentrations were estimated using protein standards of known concentrations (ThermoFisher Scientific, Cat#: 23208). Also, identity of purified proteins were confirmed via immunoblotting using an N-terminal FOXA1 antibody (Cell Signaling Technology: Cat# 58613S).
Biolayer interferometry (BLI) assay
BLI assays were carried out using the Octet-RED96 system (PALL ForteBio) and in-built analyses softwares. Briefly, biotin-labelled, 60bp KLK3 enhancer element centered at the FOXA1 consensus motif was immobilized on the Super Streptavidin Biosensors (PALL ForteBio, Part#: 18–5057) with the loading step carried out for 1000 seconds with shaking at 500 rpm. This was followed by baseline measurements for 120 seconds and association for 900 seconds using varying concentrations of purified FOXA1 proteins (3.125–100 nM; two replicate biosensors per concentration). A control DNA element with no FOXA1 motif was used in the negative control reaction with 100 nM of the protein. The association step was followed by the dissociation step for 3000 seconds. Signal from all the biosensors was adjusted for the background signal from the control sensors and normalized data of DNA binding kinetics was analyzed using the Octet-RED96 (PALL ForteBio) analysis softwares, as described previously36.
Generation of CRISPR clones and stable lines
22RV1 or LNCaP cells were seeded in a 6-wells plate at 200,000 cells/well density and transiently transfected with 2.5ug of lentiCRISPR-V2 (Addgene: #52961) vector using the Lipofectamine 3000 reagent (Cat#: L3000008), encoding the Cas9 protein and sgRNA that cuts either at amino acid 271 (5’-GTCAAGTGCGAGAAGCAGCCG-3’) or 359 (5’-GCCGGGCCCGGAGCTTATGGG-3’) in Exon2 of FOXA1. Cells were treated with non-targeting control sgRNA (5’-GACCGGAACGATCTCGCGTA-3’) vector to generate isogenic WT clones. Transfected cells were selected with puromycin (Gibco) for 3–4 days and FACS-sorted as single cells into 96-well plates. Cells were maintained in 96-wells for 4–6 weeks with replacement of the growth medium every 7days to allow for the expansion of clonal lines. Clones that successfully seeded, were further expanded and genotyped for FOXA1 using Sanger sequencing and immunoblotting with the N-terminal FOXA1 antibody. Sequence and expression validated 22RV1 and LNCaP clones with distinct class2 mutations were used for growth, invasion and metastasis assays as described.
To generate stable cells, doxycyclline-inducible vectors coding different FOXA1 variants or GFP (control) were packaged into viral particle through the University of Michigan Vector Core. PCa cells were seeded in a 6-well plate at 100,000–250,000cells/well density and infected with 0.5ml of 10X viral titres packaged at the UofM Vector Core. This was followed by 3–4 days of puromycin (Gibco) selection to generate stable lines.
Rescue growth and functional compensation experiments
Stable 22RV1 cells with doxycycline-inducible expression of empty vector (control), FOXA1 WT, or distinct FOXA1 mutants were seeded in a 6-well plate in the completed growth medium supplemented with 1.0ug/ml of doxycycline. Notably, the exogenous genes only contain the coding sequence of FOXA1 without its intron and UTRs. After 24h, cells were transfected with 30nM of either distinct 3’UTR-specific FOXA1-targeting siRNAs or a non-targeting control siRNA using the RNAiMAX (Life Technologies; Cat#: 13778075) reagent. FOXA1 UTR-specific siRNAs were purchased from ThermoFisher Scientific [Cat#: siNC – 4390844 (sequence is proprietary); si#3 - s6687 (sense sequence: 5’-GCAAUACUCUUAACCAUAA-3’); si#4 – 5278 (sense sequence: 5’-AACACATAAAATTAGTTTC-3’) and si#5 – 107428 (sense sequence: 5’-AAGTTATAGGGAGCTGGAT-3’)]. On the following day, cells were counted and seeded in a 96-well plate at a density of 5000 cells/well with six replicates for each treatment condition. Cell growth was then assessed using the incucyte assay, as described above.
Testing the GFP-tagged WT FOXA1 variant:
22RV1 cells were seeded in 10 cm dishes and transfected with 8ug of mammalian expression plasmids encoding either FOXA1-WT or FOXA1-WT-GFP (the exact construct was used in the FRAP assay) using the Lipofectamine 3000 (Life Technologies; Cat#: L3000008) reagent, as per the manufacturer’s protocol. Transgene expression was induced using 1.0ug/ml of doxycycline and cells were cultured for 96h with doxycycline-replenishment every 48h. Total RNA was extracted and RNA-Seq was performed as described. A portion of these cells were used for the rescue growth experiments using UTR-specific FOXA1 siRNAs as described earlier.
Matrigel invasion assay
22RV1 CRISPR clones were grown in 10% CSS-supplemented medium for 48 hour for androgen starvation. Special matrigel-coated invasion chamber were used that were additionally coated with a light-tight polyethylene terephthalate membrane to allow for fluorescent quantification of the invaded cells (Biocoat: 24-well format, #354166). 50,000 starved cells were resuspended in serum-free medium and were added to each invasion chamber. 20% FBS-supplemented medium was added to the bottom wells to serve as a chemoattractant. After 12 hours, medium from the bottom well was aspirated and replaced with 2ug/ml Calcein-green AM dye (ThermoFisher Scientific; C3100MP) in 1X Hank’s Balanced Salt solution (Gibco) and incubated for 30 minutes at 37C. Invasion chambers were then placed in a fluorescent plate reader (Tecan-Infinite M1000 PRO) and fluorescent signal from the invaded cells at the bottom was averaged across 16 distinct regions/chamber to determine the extent of invasion.
Chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing
ChIP experiments were carried out using the HighCell# ChIP-Protein G kit (Diagenode) as per manufacturer’s protocol. Chromatin from 5M cells were used per ChIP reaction with 6.5ug of the target protein antibody. Briefly, cells were trypsinized and washed twice with 1XPBS, followed by crosslinking for 8 min in 1% formaldehyde solution. Crosslinking was terminated by the addition of 1/10 volume 1.25 M glycine for 5 min at room temperature followed by cell lysis and sonication (Bioruptor, Diagenode), resulting in an average chromatin fragment size of 200 bp. Fragmented chromatin was then used for immunoprecipitation using various antibodies with overnight incubation at 4C. ChIP DNA was de-crosslinked and purified using the iPure Kit V2 (Diagenode) using the standard protocol. Purified DNA was then prepared for sequencing as per manufacturer’s instructions (Illumina). ChIP samples (1–10 ng) were converted to blunt-ended fragments using T4 DNA polymerase, E. coli DNA polymerase I large fragment (Klenow polymerase) and T4 polynucleotide kinase (New England BioLabs (NEB)). A single A base was added to fragment ends by Klenow fragment (3′ to 5′ exo minus; NEB) followed by ligation of Illumina adaptors (Quick ligase, NEB). The adaptor-ligated DNA fragments were enriched by PCR using the Illumina Barcode primers and Phusion DNA polymerase (NEB). PCR products were size selected using 3% NuSieve agarose gels (Lonza) followed by gel extraction using QIAEX II reagents (Qiagen). Libraries were quantified and quality checked using the Bioanalyzer 2100 (Agilent) and sequenced on the Illumina HiSeq 2500 Sequencer (125-nucleotide read length).
Zebrafish embryo metastasis experiment
Wild type ABTL zebrafish were maintained in aquaria according to standard protocols. Embryos were generated by natural pairwise mating and raised at 28.5°C on a 14h light/10h dark cycle in a 100 mm petri dish containing aquarium water with methylene blue to prevent fungal growth. All experiments were performed with 2 to 7 days old embryos post-fertilization and were done in approved University of Michigan fish facilities using protocols approved from the University of Michigan Institutional Animal Care and Use Committee (UM-IACUC). Cell injections were carried out as described in this study37. Briefly, GFP-expressing normal (control) or cancer cells were resuspended in PBS at the concentration of 1×107 cells/ml. 48 hours post-fertilization, wild-type embryos were dechorionated and anaesthetized with 0.04 mg/ml tricaine. Approximately 10 nl (approx. 100 cancer cells) were microinjected into the perivitelline space using a borosilliac micropipette tip with filament. Embryos were returned to aquarium water and washed twice to remove tricaine, then moved to a 96 well plate with one embryo per well and kept at 35°C for the duration of the experiment. All embryos were imaged at 24 hour intervals to follow metastatic dissemination of injection cells. Water was changed daily to fresh aquarium water. More than 30 fish were injected for each condition (WT#2, n=30; WT#5, n=50; #57, n=35; #84, n=57; #113, n=38) and metastasis was visually assessed daily up to 5 days after injection (i.e. a total of 7 days post-fertilization) by counting the total number of distinct cellular foci in the body of the embryos. All of the metastasis studies were terminated at 7 days post-fertilization in accordance with the approved embryo protocols. Embryos were either imaged directly in the 96 well plates or placed onto a concave glass slide to capture representative images using a fluorescent microscope (Olympus-IX71). For quantification, evidently distinct cell foci in the embryo body were counted 72 hours after the injections.
For all these experiments, relevant ethical regulations were carefully followed. No statistical methods were used to predetermine sample size for any of the cohort analyses or experiments. The experiments were not randomized and investigators were not blinded to allocation during experiments and outcome assessment unless otherwise stated.
Assay for Transposase Accessible Chromatin using sequencing (ATAC-seq) and data analysis
ATAC-seq was performed as previously described38. Briefly, 25,000 normal prostate or prostate cancer cells were washed in cold PBS and resuspended in cytoplasmic lysis buffer (i.e. CER-I from of the NE-PER kit, Invitrogen, Cat. # 78833). This single cell suspension was incubated on ice for 10 mins with gentle mixing by pipetting at every 2 mins. The lysate was centrifuged at 1300 g for 5 mins at 4℃. Nuclei were resuspended in 2X TD buffer, then incubated with Tn5 enzyme for 30 mins at 37℃ (Nextera DNA Library Preparation Kit, Cat. # FC-121–1031). Samples were immediately purified by Qiagen minElute column and PCR amplified with the NEBNext High-Fidelity 2X PCR Master Mix (NEB, Cat. # M0541L). qPCR was utilized to determine the optimal PCR cycles to prevent over-amplification. The amplified library was further purified by Qiagen minElute column and SPRI beads (Beckman Coulter, Cat. # A63881). ATAC-seq libraries were sequenced on the Illumina HiSeq 2500 (125-nucleotide read length).
Paired-end fastq files were uniquely aligned to hg38 human genome assembly using Novoalign (Novocraft, Inc) with the following parameters: -r None -k -q 13 -k -t 60 -o sam –a CTGTCTCTTATACACATCT, and converted to bam files using SAMtools (version 1.3.1). Reads mapped to mitochondrial or duplicated reads were removed by SAMtools and PICARD MarkDuplicates (version 2.9.0), respectively. Filtered Bam files from replicates were merged for downstream analysis. MACS2 (2.1.1.20160309) was used to call ATAC-seq peaks. The coverage tracks were generated using the program bam2wig (http://search.cpan.org/dist/Bio-ToolBox/) with the following parameters: --pe --rpm --span --bw. Bigwig files were then visualized using the IGV (Broad Institute) open source genome browser.
ChIP-seq data analysis
Paired-end 125bp reads were trimmed and aligned to the GRCh38 human reference using the STAR (version 2.4.0g1) aligner with splicing disabled, the resulting reads were filtered using samtools “samtools view -@ 8 -S −1 -F 384”. The resulting BAM file was sorted and duplicate marked using novosort and converted into a bigwig files for visualization using “bedtools genomecov -bg -split -ibam” and “bedGraphToBigWig”. The coverage signal was normalized to total sequencing depth / 1e6 reads. Peak calling was performed using MACS2 with the following settings “macs2 callpeak --call-summits --verbose 3 -g hs -f BAM -n OUT --qvalue 0.05”. ChIP peak profile plots and read-density heatmaps were generated using deepTool239 and cistrome overlap analyses were carried out using the ChIPpeakAnno40 package in R. It is important to note that given the cistromic dominance of class2 mutants, in heterozygous class2 mutant clones, part of the FOXA1 protein antibody binds to the WT protein that does not interact with, or immunoprecipitate, the DNA. This confounds all analyses involving peak read density comparisons between the WT and class2 mutant FOXA1 ChIP-seqs, and thus this strategy was largely avoided in our study. Due to the same reason, the read densities from only the heterozygous clones were multiplied by a factor of 1.5 for heatmap generation in Fig. 3d.
De novo and known motif enrichment analysis
All de novo and known motif enrichment analyses were performed using the HOMER (v4.10) suite of algorithms41. Peaks were called by the findPeaks function (-style factor -o auto) at 0.1% false discovery rate; de novo motif discovery and enrichment analysis of known motifs were performed with findMotifsGenome.pl (-size 200 -mask). For motif analysis of peaks segmented into common, WT- and MT-specific sections, top 5000 peaks ranked by score were used as input, and a common set background sequences were generated by di-nucleotide shuffling the input sequences using fasta-shuffle-letters function from MEME42. Alternatively, we ranked peaks by the relative signal fold-change between MT and WT, and selected top and bottom 5000 peaks (while keeping the requirement that MT-specific peaks are not called in the WT-cistrome and vice-versa) for motif discovery. For class2 mutants, only heterozygous 22RV1 clones were used that more accurately recapitulate the clinical presentation of FOXA1 mutations. Also, for both mutational classes, cistromes from biological replicates were merged to define a union cistrome that was compared to the union WT cistrome generated from matched FOXA1 WT cells. For the supervised motif analyses, we identified all instances of the FOXA canonical motif (5’-T[G/A]TT[T/G]AC-3’) within cistromes (ChIP-seq peaks) of class1 and WT FOXA1 proteins using motifmatchR, and calculated nucleotide frequencies in the flanking positions.
Utilized cohorts, data sets and resources
This study levarages previously published public or restricted patient genetic data. Genetic calls for primary PCa and breast cancer (BCa) were obtained from the Genomic Data Commons (GDC)43 for the PCa-PRAD5 and BCa-BRCA6,44 cohorts, respectively. Raw RNA-seq data (paired-end reads from unstranded polyA libraries) for those samples was downloaded from the GDC and processed with our standard Clinical RNA-seq Pipeline CRISPR/CODAC (see below). For the TCGA PRAD and BRCA cohorts we downloaded mutational calls from multiple sources (GDC, cBio Portal, UCSC Xena) and additionally used the BAM-slicing tool to download sequence alignments from whole-exome sequencing libraries to the FOXA1 locus. We then used our internal pipeline (see below) to call SNVs and indels within FOXA1. We also used the downloaded aligned data for manual review of FOXA1 mutation calls. Mutation calls for advanced primary and metastatic cases were obtained from the MSK-IMPACT cohort (downloaded from the cBio portal45). The main MCTP mCRPC cohort includes 360 cases reported previously (the location of all raw BAM files is provided in (Wu et al., 2018 in press), the 10 additional mCRPC cases included herein but not in Wu et al. are being included Database of Genotypes and Phenotypes (dbGaP): phs000673.v3.p1, and belong to a continuous sequencing program with the same IRB-approved protocol (MI-Oncoseq program, University of Michigan Clinical Sequencing Exploratory Research). The genetic sequencing data (WXS) for rapid autopsy cases is available from dbGaP: hs000554.v1.p1and phs000567.v1.p1. De-identified somatic mutation calls, RNA-seq fusion calls, processed/segmented copy-number data, and RNA-seq expression matrices across the full MCTP mCRPC 370 case cohort is available on request from the authors.
Preparation of WES and RNA-seq libraries
Integrative clinical sequencing, comprising exome sequencing and polya and/or capture RNA-seq, was performed using standard protocols in our Clinical Laboratory Improvement Amendments (CLIA) compliant sequencing lab. In brief, tumor genomic DNA and total RNA were purified from the same sample using the AllPrep DNA/RNA/miRNA kit (QIAGEN). Matched normal genomic DNA from blood, buccal swab, or saliva was isolated using the DNeasy Blood & Tissue Kit (QIAGEN). RNA sequencing was performed by exome-capture transcriptome platform46. Exome libraries of matched pairs of tumor/normal DNAs were prepared as described before47, using the Agilent SureSelect Human All Exon v4 platform (Agilent). All the samples were sequenced on the Illumina HiSeq 2000 or HiSeq 2500 (Illumina Inc) in paired-end mode. The primary base call files were converted into FASTQ sequence files using the bcl2fastq converter tool bcl2fastq-1.8.4 in the CASAVA 1.8 pipeline.
Analysis of whole-exome sequencing data
The FASTQ sequence files from whole exome libraries were processed through an in-house pipeline constructed for analysis of paired tumor/normal data. The sequencing reads were aligned to the GRCh37 reference genome using Novoalign (version 3.02.08) (Novocraft) and converted into BAM files using SAMtools (version 0.1.19). Sorting, indexing, and duplicate marking of BAM files used Novosort (version 1.03.02). Mutation analysis was performed using freebayes (version 1.0.1) and pindel (version 0.2.5b9). Variants were annotated to RefSeq (via the UCSC genome browser, retrieved on 8/22/2016), as well as COSMIC v79, dbSNP v146, ExAC v0.3, and 1000 Genomes phase 3 databases using snpEff and snpSift (version 4.1g). SNVs and indels were called as somatic if they were present with at least 6 variant reads and 5% allelic fraction in the tumor sample, and present at no more than 2% allelic fraction in the normal sample with at least 20X coverage; additionally, the ratio of variant allelic fractions between tumor and normal samples was required to be at least six in order to avoid sequencing and alignment artifacts at low allelic fractions. Minimum thresholds were increased for indels observed to be recurrent across a pool of hundreds of platform- and protocol-matched normal samples. Specifically, for each such indel, a logistic regression model was used to model variant and total read counts across the normal pool using PCR duplication rate as a covariate, and the results of this model were used to estimate a predicted number of variant reads (and therefore allelic fraction) for this indel in the sample of interest, treating the total observed coverage at this genomic position as fixed. The variant read count and allelic fraction thresholds were increased by these respective predicted values. This filter eliminates most recurrent indel artifacts without affecting our ability to detect variants in homopolymer regions from tumors exhibiting microsatellite instability. Germline variants were called using ten variant reads and 20% allelic fraction as minimum thresholds, and were classified as rare if they had less than 1% observed population frequency in both the 1000 Genomes and ExAC databases. Exome data was analyzed for copy number aberrations and loss of heterozygosity by jointly segmenting B-allele frequencies and log2-transformed tumor/normal coverage ratios across targeted regions using the DNAcopy (version 1.48.0) implementation of the Circular Binary Segmentation algorithm. The Expectation-Maximization Algorithm was used to jointly estimate tumor purity and classify regions by copy number status. Additive adjustments were made to the log2-transformed coverage ratios to allow for the possibility of non-diploid tumor genomes; the adjustment resulting in the best fit to the data using minimum mean-squared error was chosen automatically and manually overridden if necessary.
Detection of copy-number breakends from Whole Exome Sequencing
The output of our clinical WES pipeline includes segmented copy-number data, inferred absolute copy-numbers and predicted parent-specific genotypes (e.g. AAB), detection of loss-of-heterozygosity (LOH), and detection of copy-neutral LOH (uniparental disomy). Together these data enable the detection of joint discontinuities in the copy-number profile (log-ratio and B-allele frequencies) at exon level resolution. A subset of genomic rearrangements results in changes in copy-number or allelic shifts, and hence the presence of such discontinuities in paired tumor-normal WES data is strongly indicative of a somatic breakpoint. For example, 1 copy-gains will result in a segment with an increased log-ratio, and a corresponding zygosity deviation (see above). This segment will be discontinuous with adjacent segments, which will result in the call of a WES breakend (discontinuity) on either side of the copy-gain. The size of the breakend depends on the density of covered exons and in general the resolution is better in genic vs. intergenic regions. We assessed the presence of such breakpoints within the gene-dense and exon-dense FOXA1 locus, all copy-number breakends met statistical thresholds of the CBS algorithm (see above) at either the log-ratio or B-allele level.
Genetic characterization of mCRPC tumors samples at the Pathway Level
The co-occurrence or mutual exclusivity of FOXA1 aberrations with other previously described genetic events in PCa has been carried out at the pathway level, but grouping putative functionally equivalent (and largely genetically mutually exclusive) events. All known types of ETS fusion (ERG, ETV1, FLI1, ETV4, ETV5) were considered as ETS-positive tumors, PI3K alterations included PTEN homozygous los, PIK3CA activating mutations and PIK3R1 inactivating mutations, AR pathway alterations included AR, NCOR1, NCOR2, and ZBTB16 mutations/deletions, but excluded AR amplifications / copy-gains. The KMT category included mutations in all recurrently mutated lysine methyltransferases. The WNT category included inactivating aberrations in APC and activating mutations in CTNNB1. DRD included cases with mutations in: BRCA1, BRCA2, PALB2, ATM, all common mismatch repair genes, and CDK12.
Assessment of two-hit (biallelic) alterations
To assess the frequency of genetic inactivations of both alleles we integrated mutational, copy-number, and RNA-seq (fusion) data. A gene was considered having both alleles inactivated for any combination (pair) of the following events: copy-loss, mutation, truncating fusion, copy-number breakpoint, in addition to homozygous deletion of both copies and two independent mutations. Ambiguous cases were manually reviewed to increase the accuracy and ascertain whether both events e.g. copy-number breakpoint and gene fusion are likely independent events.
Unified mutation calling and variant classification of FOXA1
Mutation calls for FOXA1 obtained / downloaded from the GDC, TCGA flagship manuscripts5,6, and our internal pipelines were lifted over to GRCh38 (using the Bioconductor package rtracklayer) and annotated with respect to the canonical RefSeq FOXA1 isoform. For TCGA samples/cases multiple call-sets were available and we manually reviewed all discrepancies in FOXA1 mutation calls resulting in a union call set with improved sensitivity and specificity. Mutational impact (consequence) was simplified into 3 categories: missense, inframe indel, and frameshift the latter category included stop-gain, stop-loss, and splice-site mutations. The resulting mutations were dichotomized into Class1 and Class2 based on their position relative to residue 275aa. Variant allele frequencies (VAF) were only available for TCGA and the in-house mCRPC cohorts.
Analysis of whole-genome sequencing data
The bcbio-nextgen pipeline version 1.0.3 was used for the initial steps of tumor whole-genome data analysis. Paired-end reads were aligned to the GRCh38 reference using BWA (bcbio default settings), and structural variant calling was done using LUMPY 48 (bcbio default settings), with the following post-filtering criteria: ‘‘(SR>=1 & PE>=1 & SU>=7) & (abs(SVLEN)>5e4) & DP<1000 & FILTER==‘‘PASS’’.’’ The following settings were chosen to minimize the number of expected germline variants: (FDR < 0.05 for germline status for both deletions and duplications), additionally common structural germline variants were filtered.
Analysis of 10X genomics long-read sequencing data
High-molecular weight (HMW) DNA from MDA-PCA-2B and LNCaP cell lines was isolated and processed into linked-read NGS libraries per manufacturer’s instructions (10X WGS v2 kit). The resulting paired-end sequencing data were sequenced on an Illumina Hi-Seq 2500 instrument and analyzed (demultiplexing, alignment, phasing, structural variant calls) using the longranger 2.2.1 pipeline with all default settings. The resulting libraries met all 10X-recommended QC parameters including molecule size, average phasing length, and sequencing coverage (~50X). Here, we focused on structural variant calls within the FOXA1 TAD and confirmed the presence of the previously reported FOXMIND-ETV1 fusions i.e. translocation for MDA-PCA-2B, and balanced insertional translocation for LNCaP. Both cell lines were confirmed to harbor three copies of FOXA1 i.e. one translocated allele and two duplicated alleles.
RNA-seq data pre-processing and primary analysis
RNA-seq data processing, including quality control, read trimming, alignment, and expression quantification by read counting, was carried out as described previously 47, using our standard clinical RNA-seq pipeline ‘‘CRISP’’ (available at https://github.com/mcieslik-mctp/bootstrap-rnascape). The pipeline was run with default settings for paired-end RNA-seq data of at least 75bp. The only changes were made for unstranded transcriptome libraries sequenced at the Broad Institute and the TCGA/CCLE/CCLE cohorts, for which quantification using ‘‘featureCounts’’ (Liao et al., 2014) was used in unstranded mode ‘‘-s0.’’. The resulting counts were transformed into FPKMs using upper-quartile normalizations as implemented in EdgeR 49. For mCRPC samples FOXA1 expression estimates were adjusted by tumor-content estimated from WES (see above) given the highly prostate specific FOXA1 expression profile. For the quantification of FOXMIND expression levels, a custom approach was necessary given the poor-annotation and unspliced nature of this transcript. First, we delineated regions of sense and antisense transcription from the FOXMIND ultra-conserved regulatory elements, chr14:37564150–37591250:+ and chr14:37547900–37567150:-, respectively. Next in order to make the expression estimates reliable in unstranded libraries we identified region of significant overlap between the sense/antisense FOXMIND transcripts and FOXA1 and MIPOL1. These overlaps have been excluded from quantification, resulting in the following trimmed target regions: chr14:37564150–37589500, and chr14:37553500–37567150. Within those regions the average base-level coverage coverage normalized to sequencing depth was computed as an expression estimate.
Differential expression analyses
All differential expression analyses were done using limma R-package50, with the default settings for the ‘‘voom’’51, ‘‘lmFit,’’ ‘‘eBayes,’’ and ‘‘topTable’’ functions. The contrasts were designed as follows, to identify transcriptional signatures of Class1 mutants: 1) given the mutual exclusivity of the genotypes in primary and metastatic tumors the overall MCTP mCRPC 371 cohort was partitioned into 4 groups: ETS/SPOP mutant tumors, Class 1 mutant tumors, Class 2 mutant tumors, tumors WT for ETS/SPOP/FOXA1. To avoid confounding effects, the Class2 and ETS/SPOP groups were excluded from Class1 transcriptional analyses. Next, the Class1 samples were contrasted with the WT samples with additional independent regressors for assay type (Capture vs polyA, as described previously), and mutational status (see above) for the following genes/pathways: PI3K, WNT, DRD, RB1, TP53. In other words, we constructed a design matrix with coefficients for Class 1 mutational status, in addition to coefficients for confounding variables and recurrent genetic heterogeneity. This allowed us to estimate the log fold-changes and adjusted p-values associated with FOXA1 mutations and other genotypes i.e. PI3K status. An analogous procedure was carried out for the primary class1 samples (TCGA) and for class2 mutations in mCRPC (MCTP), but given the lack of mutual-exclusivity between Class2 mutations and ETS/SPOP only Class1 mutations were excluded.
Pathway and signature enrichment analyses
The Molecular Signature Database (MSigDB)52 has been used as a source of gene sets comprising cancer hallmarks, molecular pathways, oncogenic signatures, and transcription factor targets. The enrichment of signatures was assessed using the parametric Random-Set method53, and visualized using the GSEA enrichment statistic54 and barcode plots. All p-values have been adjusted for multiple-hypothesis testing using FDR correction. To identify putative transcription factors regulating differentially expressed genes, we used the transcription-factor prediction tool BART 25. BART was run with all default settings, and provided TF databases. We used voom/limma-based gene-level fold-changes as input to the algorithm.
Detections of structural variants from RNA-seq
The detections of chimeric RNAs (gene fusions, structural variants, circular RNAs, read-through events) was carried out using our in-house toolkit for the comprehensive detection of chimeric RNAs ‘‘CODAC’’ (available at https://github.com/mctp/codac), and introduced previously 47. Briefly, three separate alignment passes (STAR 2.4.0g1) against the GRCh38 (hg38) reference with known splice-junctions provided by the (Gencode 27) are made for the purposes of expression quantification and fusion discovery. The first pass is a standard paired-end alignment followed by gene expression quantification. The second and third pass are for the purpose of gene fusion discovery and enable STAR’s chimeric alignment mode (chimSegmentMin: 10, chimJunctionOverhangMin: 1, alignIntronMax: 150000, chimScoreMin: 1). Fusion detection was carried out using CODAC with default parameters to balance sensitivity and specificity (annotation preset:balanced). CODAC uses MOTR v2 a custom reference transcriptome based on a subset of Gencode 27 (available with CODAC). Prediction of topology (inversion, duplication, deletion, translocation), and distance (adjacent – breakpoints in two directly adjacent loci, cytoband – breakpoints within the same cytoband based on UCSC genome browser, arm – breakpoints within the same chromosome arm). The high specificity of our pipeline has been assessed through Sanger sequencing 47. To create fusion circos plots, we have color coded the CODAC variants based on the inferred topology of the breakpoints. Unbiased discovery of recurrently rearranged loci has been carried out by breaking the genome into 1.5Mb windows with a step of 0.5Mb. For each window the percentage of patients with at least one RNA breakend has been calculated. The resulting genomic windows were ranked and clustered by proximity for visualization. CODAC has the ability to make fusion calls independent of known transcriptome references/annotations and hence is capable of detecting fusions involving intergenic or poorly annotated regions.
Classification of FOXA1 locus genomic rearrangements
Structural variants within the FOXA1 locus have been partitioned into two broad topological patterns: 1) translocations (including inversions and deletions involving distal loci on the same chromosome), and 2) focal duplications. The translocations have been further subdivided into Hijacking and Swapping events based on their position relative to FOXMIND (GRCh38: chr14:37564150–37591250) and FOXA1. Hijacking translocations position a translocation partner within the FOXMIND-FOXA1 regulatory domain (defined as GRCh38: chr14:37547501–37592000, based on manual review of HI-C, CTCF, H3K4me1, H3K27ac, and evolutionary/syntenic data). Swapping translocations preserve the FOXMIND-FOXA1 regulatory domain but insert the translocation partner upstream of the FOXA1 promoter, frequently “swapping-out” the TTC6 gene. Notably, one isoform of TTC6 gene can be transcribed from the bi-directional FOXA1 promoter. Focal duplications within the FOXA1 locus have been derived from the CODAC structural-variant output file. Briefly, for each case independently, all RNA-seq fusion junctions annotated by CODAC as tandem-duplications and overlapping the FOXA1 topological domain (GRCh38: chr14:37210001–37907919) have been collated and used to infer the minimal duplicated region (MDR). Since RNA-seq chimeric junctions are generally coinciding with splice junctions (limited resolution) and generally cannot be phased (ambiguous haplotype), the inference of MDRs makes the necessary and parsimonious assumption that overlapping tandem-duplications are due to a single somatic genetic event and not multiple independent events.
Data Availability
All raw data for the graphs, immunoblot and gel electrophoresis figures are included in matched Source Data files or Supplementary Information. All materials are available from authors upon reasonable request. All the raw next-generation sequencing data generated in this study has been deposited into the Gene Expression Omnibus (GEO) repository at NCBI (accession code: GSE123625). All custom data analysis software and bioinformatics algorithms used in this study are publically available on Github:
Extended Data
Extended Data Figure 1. Functional essentiality and recurrent alterations of FOXA1 in AR-positive prostate cancer.
AR (a) and FOXA1 (b) mRNA (qPCR) and (c) protein expression in a panel of PCa cells (n=3 technical replicates). Mean ± s.e.m are shown and dots are individual data points. d-f) Growth curves of AR-positive PCa cells treated with non-targeting control (siNC), AR, or FOXA1 targeting siRNAs (25nM at Day 0 and 1; n=6 biological replicates). Immunoblots confirm knockdown of FOXA1 protein in LNCaP and LAPC4 72h after siRNA-treatment. For all gel source data, see Supplementary Figure 1. g) Crystal violet stain of AR-negative, DU145 PCa and LNCaP (control) cells treated with siNC, AR or FOXA1 targeting siRNAs. Results represent 3 independent experiments (n=2 biological replicates). h) Averaged proliferation Z-scores for 6 independent FOXA1-targeting sgRNAs extracted from publically available CRISPR Project Achilles data (BROAD Institute) in prostate and breast cancer cells. HPRT1 and AR data serve as negative and positive controls, respectively. Mean ± s.e.m are shown; dots are proliferative z-score for independent sgRNAs. i) Ranked depletion or enrichment of sgRNA read counts from GeCKO-V2 CRISPR knockout screen in LNCaP cells (at day 30) relative to the input sample. Only a subset of genes, including essential controls, chromatin modifiers and transcription factors, are visualized. j) Recurrence of FOXA1 mutations across TCGA, MSK-IMPACT, and SU2C cohorts. k) Density of breakends (RNA-seq chimeric junctions) within overlapping 1.5Mb windows along chr14 in mCRPC tumors. l) Whole-genome sequencing of 7 mCRPC index cases with distinct patterns FOXA1 translocations (Tlocs) and duplications (Dups), nominated by RNA-seq (WA46, WA37, WA57, MO_1584) or WES (MO_1778, SC_9221, MO_1637). m) Concordance of RNA-seq (chimeric junctions) and WES based FOXA1 locus rearrangements calls (mCRPC cohort). n) Frequency of FOXA1 locus rearrangements in mCRPC based on RNA-seq and WES.
Extended Data Figure 2. Genomic characteristics of the three classes of FOXA1 alterations in prostate and breast cancer.
a) Bi-allelic inactivation and b) copy-number variations of FOXA1 across mCRPC (n=371). c) FOXA1 expression in benign (n=51), primary (n=501), and metastatic (n=535) prostate RNA-seq libraries. d) Distribution and functional categorization of FOXA1 mutations (all cases in the aggregate cohort) on the protein map of FOXA1. TAD, trans-activating domain; RD, regulatory domain. e) Aggregate and class-specific distribution of FOXA1 mutations in advanced breast cancer (MSK-Impact cohort). f) Structural classification of FOXA1 locus rearrangements in breast cancer (TCGA and CCLE cell lines). g) Variant allele frequency of FOXA1 mutations by tumor stage, and h) clonality estimates of class1 and class2 mutations in tumor content corrected primary PCa (n=500) and mCRPC (n=370) specimens. i) Mutual exclusivity or co-occurrence of FOXA1 mutations (two-sided Fisher’s exact test). Mutations in AR, WNT, and PI3K were aggregated at the pathway level. ETS, ETS gene fusions; DRD, DNA repair defects and included alterations in BRCA½, ATM and CDK12; MMRD, mismatch repair deficiency (total n=371). j) Mutual exclusivity of ETS and/or SPOP (n=26) aberrations with FOXA1 (n=46) alterations distinguished by class in mCRPC (n=371). k) Co-occurrence of WNT (n=58) and DRD (n=107) pathway alterations with FOXA1 alteration classes in mCRPC (n=371). l) Stage and class-specific increase in FOXA1 expression levels in primary (n=500) and metastatic PCa (n=357). Right: two-sided t-test. Left: two-way ANOVA. For all boxplots: center shows median, box extends from Q1 to Q3, and whiskers span Q1/Q3±1.5xIQR.
Extended Data Figure 3. Biophysical and cistromic characteristics of the Class1 FOXA1 mutants.
a) Distribution of class1 mutations on the protein map of FOXA1. b) 3D-structure of FKHD (FOXA3) with visualization of all mutated residues collectively identified as the 3D-mutational hotspot in FOXA1 across cancers. c) DNA-bound 3D structure of FKHD with visualization of all residues shown through crystallography to make direct base-specific contacts with the DNA in FOXA2 and FOXA3 proteins. FKHD; Forkhead domain. d) Representative fluorescent images of nuclei expressing different FOXA1 variants fused to GFP at the C-termini. e, f) FRAP recovery kinetic plots (left) and representative time-lapse images (right) from pre-bleaching (‘Pre’) to 100% recovery (red timestamps) for (e) wing2-altered class1 and (f) truncated class2 mutants (i.e. A287fs and P375fs), respectively (n=6 nuclei/variants; quantified in Fig 2d). White lines indicate the border between bleached and unbleached areas. g) Representative FRAP fluorescence recovery kinetics in the bleached area for indicated FOXA1 variants. t½ line indicates the time to 50% recovery. Colored dots show raw data; superimposed solid curves show a hyperbolic fit with 95% confidence intervals. h) SPT quantification of chromatin bound (slow and fast) and unbound (freely diffusing) particles of WT and class1 FOXA1 variants, and average chromatin dwell times (mean ± s.d.) for the bound fractions (n≥500 particles/variant). i) Diffusion constant histograms of single particles of WT or distinct class1 FOXA1 mutants. Particles were categorized into chromatin bound (slow and fast) or unbound fractions using cutoffs marked by dashed lines (n≥500 particles/variant imaged in 3–5 distinct nuclei). j) Left, mRNA expression (qPCR) of labeled FOXA1 variants in stable, isogenic HEK293 cells (n=3 technical replicates). Right, overlaps between FOXA1 WT and class1 mutant cistromes from these cells (n=2 biological replicates). k) Top de novo motifs identified from the three FOXA1 cistromes from HEK293 cells (HOMER: hypergeometric test). l) mRNA expression (qPCR) of labeled FOXA1 variants in stable, isogenic 22RV1 cells (n=3 technical replicates). For j and l, centers show mean values and lines mark s.e.m. m) Overlap between WT (n=2 biological replicates) and class1 (n=4 biological replicates) cistromes from stable 22RV1 overexpression models. n) Overlap between the FOXA1 WT and AR union cistromes generated from 22RV1 cells overexpressing WT (n=2 biological replicates) or class1 mutant (I176M or R216G; n=2 biological replicates each) FOXA1 variants. o) De novo motif results for the WT or class1 mutant FOXA1 binding sites from PCa cells (HOMER: hypergeometric test). p) Percent of WT or class1 binding sites with perfect match to the core FOXA1 motif (5’-T[G/A]TT[T/G]AC-3’) and q) the consensus FOXA1 motifs identified from these sites. r) Left, Percent of WT or class1 binding sites harboring known motifs of the labelled FOXA1 or AR cofactors; Right, Enrichment of the cofactor motifs in the two cistromes relative to the background (n=top 5000 peaks by score/variant, see Methods). s) Genomic distribution of WT and class1 binding sites in PCa cells.
Extended Data Figure 4. Functional impact of FOXA1 mutations on oncogenic AR-signaling.
a) Immunoblot showing expression of endogenous and V5-tagged exogenous FOXA1 proteins in dox-inducible 22RV1 cells transfected with distinct UTR-specific FOXA1 siRNAs (#3–5) or a non-targeting control siRNA (siNC). These results represent 2 independent experiments. Incucyte growth curves of 22RV1 cells overexpressing empty vector (control), WT, or mutant FOXA1 variants upon treatment with UTR-specific FOXA1-targeting siRNAs (n = 5 biological replicates). Mean ± s.e.m are shown. b) Immunoblots confirming stable overexpression of the WT AR protein in HEK293 and PC3 cells. c, d) Co-immunoprecipitation assay of indicated recombinant FOXA1 variants using a V5-tag antibody in (c) HEK293-AR and (d) PC3-AR cells. eGFP is a negative control; FL is the full-length WT FOXA1. d168 and d358 are truncated FOXA1 variants with only the first 168aa (i.e. before the Forkhead domain) or 358aa of FOXA1 protein. H247Q and R261G are missense class1 mutant variants. e) Immunoblots confirming comparable expression of AR and recombinant FOXA1 variants in AR reporter assay-matched HEK293 lysates. Immunoblots show representative results from 2–3 independent experiments and class1 and class2 mutants serve as biological replicates. For all gel source data (a,b-e), see Supplementary Figure 1. f) AR dual-luciferase reporter assays with transient overexpression of indicated FOXA1 variant in HEK293-AR cells with or without DHT stimulation and Enzalutamide treatment (n=3 biological replicates/group). Mean ± s.e.m are shown (two-way ANOVA and Tukey’s test). g) Genes differentially expressed in class1 patient samples (n=38) compared to FOXA1 WT tumors (see Methods). Most significant genes are shown in red and labeled (Limma two-sided test). h) Differential expression of cancer hallmark signature genes in class1 mutant PCa tumors (GSEA statistical test). i) Localized, primary PCa gene signature showing concordance between class1 tumor and primary PCa genes. j) BART prediction of specific TFs mediating observed transcriptional changes. The significant and strong (Z-score) mediators of transcriptional responses in class1 tumors are labeled (BART: Wilcoxon rank-sum test). k) mRNA expression (RNA-Seq) of class1 signature genes in LNCaP and VCaP cells either starved for androgen (no DHT) or stimulated with DHT (10nM). RNA-seq from two distinct PCa cell lines are shown. l) Representative FOXA1 and AR ChIP-Seq normalized signal tracks at the WNT7B or CASP2 gene loci in LNCaP and VCaP cells. ChIP-seq were carried out in two distinct PCa cell lines with similar results. m) Growth curves (IncuCyte) of 22RV1 cells overexpressing distinct FOXA1 variants in complete, androgen-supplemented growth medium (n = 2 biological replicates). Mean ± s.e.m are shown. n) Percent viable 22RV1 stable cells, overexpressing either empty vector, WT, or mutant FOXA1 variants upon treatment with enzalutamide (20 uM for 6 days; n = 4 biological replicates). Mean ± s.e.m are shown. P-values in m and n were calculated using two-way ANOVA and Tukey’s test. o,p) mRNA expression (RNA-Seq) of labeled basal and luminal TFs or canonical markers in FOXA1 WT, class1, or class2 mutant tumors in primary PCa (total n = 500; two-way ANOVA). q) Extent of AR and NE pathway activation in FOXA1 WT, class1, or class2 mutant cases from both primary (n = 500) and metastatic (n = 370) PCa. Both AR and NE scores were calculated using established gene signatures (see Methods. Left, two-sided t-test; right, two-way ANOVA). For all boxplots: center shows median, box marks Q1/Q3, whiskers span Q1/Q3±1.5xIQR.
Extended Data Figure 5. DNA-binding dominance of the Class2 FOXA1 mutants.
a) FOXA1 protein maps showing the recombinant proteins used to validate the N-terminal (N-term) and C-terminal (C-term) FOXA1 antibodies. TAD, trans-activating domain; FKDH, Forkhead domain; RD, regulatory domain. b) Immunoblots depicting detection of all variants by the N-term antibody (left), and of only the full-length WT FOXA1 protein by the C-term antibody (right). These results were reproducible in 2 independent experiments. Antibody details are included in the Methods. c) Sanger sequencing chromatograms showing the heterozygous class2 mutation in LAPC4 cells after the P358 codon in Exon2 (n=2 technical replicates). All other tested PCa cell lines were WT for FOXA1. d) Immunoblots confirming the expression of the truncated FOXA1 variant in LAPC4 at the expected ~40kDa size (top, red arrow). The short band is detectable only with the N-term (top) FOXA1 antibody and not the C-term (bottom) antibody. These results were reproducible in 2 independent experiments. e) Co-immunoprecipitation and immunoblotting of FOXA1 using a N-term and C-term antibodies from LAPC4 nuclei with species-matched IgG used as control. f) Nuclear co-immunoprecipitation of FOXA1 from LAPC4 or LNCaP cells stimulated with DHT (10nM for 16h) using N-term and C-term antibodies. Species-matched IgG are controls. Immunoprecipitations and immunoblots in d-f were reproducible in 2 and 3 independent experiments, respectively. For gel source data (b,d,e,f), see Supplementary Figure 1. g) FOXA1 N-term and C-term ChIP-seq normalized signal tracks from FOXA1 WT or class2 mutant PCa cells at canonical AR targets KLK3 and ZBTB10. h) Left, Overlap between global N-term and C-term FOXA1 cistromes in untreated C42B cells. Right, Overlap between global N-term and C-term FOXA1 cistromes in LAPC4 cells treated with DHT (10nM for 3h). i) FOXA1 ChIP-seq normalized signal tracks from N-term and C-term antibodies in LAPC4 cells with or without DHT-stimulation (10nM for 3h) at KLK3 and ZBTB10 locus. ChIP-seqs in g and i were carried out in two distinct FOXA1 WT PCa cells. For LAPC4 ChIP-seqs, results were repreproducible in two independent experiments. j) Left, mRNA (qPCR) expression of FOXA1 in LAPC4 cells with exogenous overexpression of WT FOXA1; Right, in LNCaP cells with exogenous overexpression of the P358fs mutant (n=3 technical replicates). Mean ± s.e.m are shown and dots are individual data values. k) FOXA1 ChIP-seq normalized signal tracks from N-term and C-term antibodies in parental LAPC4 cells and LAPC4 cells overexpressing WT FOXA1 at the KLK3 locus. This experiment was independently repeated twice with similar results. The 60bp AR and FOXA1 bound KLK3 enhancer element used for EMSA is shown.
Extended Data Figure 6. DNA-binding affinity and functional essentiality of the Class2 FOXA1 mutants.
a) Immunoblot showing comparable expression of recombinant FOXA1 variants in equal volume of nuclear HEK293 lysates used to perform EMSAs. b) Higher exposure of EMSA with recombinant WT or P358fs mutant and KLK-enhancer element showing the super-shifted band with addition of the V5 antibody (red asterisks; matched to Main Fig. 3f). c, d) EMSA with recombinant WT or different class2 mutants (truncated at 268, 287, 358, 375, and 453aa) and KLK3 enhance element. Class2 mutants display higher affinity vs WT FOXA1. Each class2 mutant serves as a biological replicate and these results were reproducible in two independent experiments. e) DNA association and dissociation kinetics at varying concentrations of purified WT or P358fs class2 FOXA1 mutants from the biolayer-interferometry assay performed using OctetRED system. Overall binding curves and equilibrium dissociation constants (mean± s.d.) are shown. These results were reproducible in 2 independent experiments. f) Sanger sequencing chromatograms from a set of 22RV1 CRISPR clones confirming the introduction of distinct indels in the endogenous FOXA1 allele, resulting in a premature stop codon (n=2 technical replicates). Protein mutations are identified on the right. g) Immunoblots showing the expression of endogenous WT or class2 mutant FOXA1 variants in parental and distinct CRISPR-engineered 22RV1 clones. h) Immunoblots showing expression of FOXA1 (N-term antibody) in parental and CRISPR-engineered LNCaP clones expressing distinct class2 mutants with truncations closer to the Forkhead domain. For gel source data (a,b,c,d,g,h), see Supplementary Figure 1. i) Growth curves of WT or mutant clones upon treatment with the non-targeting or FOXA1-targeting sgRNAs and CRISPR-Cas9 protein (see Methods). For i, distinct class2 clones and distinct sgRNAs serve as biological replicates. j, k) Overlap between union (j) FOXA1 and (k) AR cistromes from WT (n=3 biological replicates) and class2-mutant (n=4 biological replicates) 22RV1 clones. l) Overlap between union FOXA1 and AR cistromes from class2 mutant 22RV1 cells.
Extended Data Figure 7. Cistromic and WNT-driven phenotypic characteristics of the Class2 FOXA1 mutants.
a) De novo motif analyses of the WT-specific, common, and class2-specific FOXA1 binding site subsets defined from either (left) sequencing read fold-changes or (right) peak-calling scores of ChIP-seq data in 7a. WT and class2 cistromes were generated from n=3 and n=2 independent biological replicates, respectively. Only the top 5K or 10K peaks from each subset were used as inputs for motif discovery (see Methods; HOMER: hypergeometric test). b) Percent of WT or class2 binding sites with perfect match to the core FOXA1 motif (5’-T[G/A]TT[T/G]AC-3’) and c) the consensus FOXA1 motifs identified from these sites. d) Percent of binding sites in the three FOXA1 binding site subsets harboring known motifs of the labelled FOXA1 or AR cofactors, and e) enrichment of the cofactor motifs in the three binding site subsets relative to the background. f) Genomic distribution of WT-specific, common and class2-specific binding sites in PCa cells. g) Differential expression of genes in FOXA1 class2 mutant CRISPR clones relative to FOXA1 WT clones (n=2 biological replicates (Limma two-sided test). h) Distinct TF motifs within the promoter (2kb upstream) of differentially expressed genes. TFs with the highest enrichment (fold-change, percent of up-regulated genes with the motif, and significance) are highlighted and labeled (two-tailed Fisher’s exact test). i) Immunoblots showing the expression of B-Catenin and Vimentin in a panel of WT and heterozygous or homozygous class2 mutant 22RV1 CRISPR clones. j) Immunoblots showing the phosphorylation status of B-Catenin and expression of direct WNT target genes in select class2 mutant 22RV1 clones. Immunoblots in i) and j) are representative of two independent experiments; every individual clone serves as a biological replicate. For gel source data, see Supplementary Figure 1. k) Representative images of Boyden chambers showing invaded cells stained with Calcein AM dye. l) Quantified fluorescence signal from invaded cells (n=2 biological replicates/group; two-way ANOVA and Tukey’s test). Mean ± s.e.m are shown and dots are individual data points. n) Percent metastasis at day2 and day3 in zebrafish embryos injected with either the normal HEK293 cells (negative controls) or 22RV1 PCa cells virally overexpressing WT, class1, or class2 mutant FOXA1 variants (n>20 for each group). m) Absolutely counts of disseminated cell foci in individual zebrafish embryos as a measure of metastatic burden. o) Fluorescent signal from the invaded WT or class2-mutant 22RV1 cells after androgen starvation (5% charcoal-stripped serum medium for 72h) or treatment with the WNT inhibitor, XAV939 (20μM for 24h; n=2 biological replicates/group; two-way ANOVA and Tukey’s test). Mean ± s.e.m and individual data points are shown.
Extended Data Figure 8. Functional association of FOXA1 and TLE3 in prostate cancer.
a) mRNA (qPCR) and protein (immunoblot) expression of TLE3 in a panel of PCa cells. Mean ± s.e.m and individual data points are shown. b) Left, mRNA expression of FOXA1 and TLE3 in LNCaP and VCaP cells treated with siRNAs targeting either FOXA1 or AR (n=3 technical replicates). Two FOXA1 WT PCa cells serve as biological replicates. Mean ± s.e.m and individual data points are shown. Right, protein expression of FOXA1 and TLE3 in matched LNCaP lysates. c) FOXA1 N-terminal ChIP-seq normalized signal tracks from LNCaP, C42B and LAPC4 PCa cells at the TLE3 locus. Each cell line serves as a biological replicate. d) Overlap of the union WT FOXA1 and TLE3 binding sites from LNCaP, C42B and 22RV1 PCa cells (n=1 for each), and top de novo motifs discovered (HOMER: hypergeometric test) in the TLE3 cistrome. e) Co-immunoprecipitation assays of labelled recombinant FOXA1 WT, class1, or class2 variants using a V5-tag antibody in HEK293 cells overexpressing the TLE3 protein. V5-tagged GFP protein was used as a negative control. These results were reproducible in two independent experiments and distinct class1 and class2 mutant serve as biological replicates. f) Overlap of union TLE3 cistromes from isogenic WT (n=2 biological replicates) or heterozygous class2-mutant (n=2 biological replicates) 22RV1 CRISPR clones. g) ChIP peak profile plots from TLE3 ChIP-seq in isogenic FOXA1 WT or class2-mutant 22RV1 clones (n=2 biological replicates each). h) Representative TLE3 and FOXA1 ChIP-seq read signal tracks from independent 22RV1 CRISPR clones with or without endogenous FOXA1 class2 mutation (n=2 biological replicates each). i) Gene set enrichment analyses showing significant enrichment of (left) WNT and (right) EMT pathway genes in 22RV1 cells treated with TLE3-targeting siRNAs (n=2 biological replicates for each treatment; GSEA enrichment test). j) Left, mRNA (RNA-seq) expression of direct WNT target genes in 22RV1 upon siRNA-mediated knockdown of TLE3 (n=2 biological replicates). Right, Immunoblot showing LEF1 up-regulation upon TLE3 knockdown in 22RV1 PCa cells with and without androgen starvation (representative of two independent experiments). For gel source data (a-d,j), see Supplementary Figure 1. k) Gene enrichment plots showing significant enrichment of class2 up-regulated genes upon TLE3 knockdown in 22RV1 cells (n=2 biological replicates for each treatment; GSEA enrichment test).
Extended Data Figure 9. Topological, physical, and transcriptional characteristics of the FOXA1 locus in normal tissues and prostate cancer.
a) HI-C data (from: http://promoter.bx.psu.edu/hi-c/view.php) depicting conserved topological domains within the PAX9/FOXA1 syntenic block in normal and FOXA1-positive cancer cell lines. b) Highly tissue-specific patterns of gene expression within the PAX9/FOXA1 syntenic block. Tissues were dichotomized into FOXA1+ and FOXA1- based on FOXA1 expression levels; genes were subject to unsupervised clustering. Z-score normalization was performed for each gene across all tissues. c) Correlation of FOXMIND (Methods) and FOXA1 / TTC6 expression levels across metastatic tissues (n=370; Spearman rank-correlation coefficient). The 95% confidence interval is shown. d) Representative ATAC-seq (n=1) read signal tracks from normal basal epithelial prostate (RWPE1, PNT2) or PCa cells. Cells are grouped based on expression of FOXA1 and differentially pioneered loci are marked with the red boxes. CRISPR sgRNA pairs used for genomic deletion of the labelled elements are shown at the bottom. Distinct FOXA1+ and FOXA1- cell lines serve as biological replicates for ATAC-seq. e) mRNA (qPCR) expression of control, FOXA1 TAD genes, and MIPOL1 in VCaP cells treated with CRISPR-sgRNA pairs targeting a control site (sgCTRL), the FOXMIND, or the MIPOL1-UTR regulatory element (see Extended Data Fig. 2c for sgRNA binding sites). Distinct sgRNA pairs cutting at FOXMIND serve as biological replicates. Mean ± s.e.m are shown (n=3 technical replicates; two-way ANOVA and Tukey’s test). f) Distribution of tandem duplication and translocation breakends (chimeric junctions or copy-number segment boundaries) focused at the FOXMIND-FOXA1 regulatory domain. g) Outlier expression of genes involved in translocations with the FOXA1 locus. Translocations positioning a gene between FOXMIND and FOXA1 (Hi-jacking) are shown on top (red). Translocations positioning a gene upstream of the FOXA1 promoter (Swapping) are shown on the bottom (blue). h) Inferred duplications within the FOXA1 locus based on RNA-seq (tandem breakends) and WES (copy-gains) zoomed-in at the FOXA1 TAD.
Extended Data Figure 10. Transcriptional and genomic characteristics of Class3 FOXA1 rearrangements in prostate cancer.
a) Dosage sensitivity of the FOXA1 gene. Expression of FOXA1 (RNA-seq) across mCRPC tumors (n=370) as a function of gene ploidy (as determined by absolute copy number at the FOXA1 locus (two-way ANOVA). b) Relative expression of FOXA1 (within the minimally amplified region) to TTC6 (outside the amplified region) in rearranged (n=50) (duplication or translocation) vs WT (n=320) FOXA1 loci (two-sided t-test). All boxplot center shows median, box marks Q1/Q3, whiskers span Q1/Q3±1.5xIQR. c) Association plot visualizing the relative enrichment of cases with both translocation and duplications within the FOXA1 locus (n=370). Over-abundance of cases with both events is quantified using Pearson-residuals. Significance of this association is based on the Chi-square test without continuity correction. Tloc, translocation; inv, inversion; del, deletion. d) FOXA1 locus visualization of linked-read (10X platform) whole genome-sequencing of the MDA-PCA-2B cell line. Alignments on the haplotype-resolved genome are shown in green and purple. Translocation and tandem-duplication calls are indicated in blue and red, respectively. e) Monoallelic expression of FOXA1 cell-lines with FOXMIND-ETV1 translocations in MDA-PCA-2b (n=6 biological replicates) and LNCaP (n=15 biological replicates). Phasing of FOXA1 SNPs to structural variants is based on linked-read sequencing (Methods). f) Biallelic expression of RNA from the FOXMIND locus assessed using three distinct SNPs in MDA-PCa-2b cells that harbor ETV1 translocation into the FOXA1 locus (n=7 biological replicates). g) mRNA (qPCR) expression of ETV1 and TTC6 upon sgRNA-mediated disruption of the FOXMIND or the MIPOL1-UTR enhancer in LNCaP cells, which also harbor ETV1 translocation into the FOXA1 locus (see Extended Data Fig. 9d for sgRNA binding sites). Distinct sgRNA pairs cutting at FOXMIND serve as biological replicates. Mean ± s.e.m are shown (n=3 technical replicates; two-way ANOVA and Tukey’s test).
Supplementary Material
Acknowledgements
We thank D. Macha, L. Wang, S. Zelenka-Wang, I. Apel, M. Tan, Y. Qiao, A. Delekta, K. Juckette and J. Tien for technical assistance. We thank S. Gao for assistance with the manuscript. This work was supported by the Prostate Cancer Foundation (PCF), Early Detection Research Network (UO1 CA214170), NCI Prostate SPORE (P50 CA186786), and Stand Up 2 Cancer-PCF Dream Team (SU2C-AACR-DT0712) grants to A.M.C. A.M.C. is an NCI Outstanding Investigator, Howard Hughes Medical Institute Investigator, A. Alfred Taubman Scholar, and American Cancer Society Professor. A.P. is supported by Predoctoral Department of Defense (DoD) - Early Investigator Research Award (W81XWH-17–1-0130). M.C. is supported by DoD - Idea Development Award (W81XWH-17–1-0224) and PCF Young Investigator Award.
Footnotes
Supplementary Information: Includes a detailed discussion of the key genomic, functional, and phenotypic data pertaining to the three classes of FOXA1 alteration as well as raw uncropped scans of the immunoblot and gel electrophoresis figures.
Data deposition
ChIP and RNA sequencing data from this study can be obtained from the GEO repository (GSE123625).
Materials & Correspondence
Correspondence and requests for materials should be addressed to Arul M. Chinnaiyan.
Competing interests
The authors declare no competing financial interests.
References (print only):
- 1.Gao N et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development 132, 3431–3443 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Friedman JR & Kaestner KH The Foxa family of transcription factors in development and metabolism. Cell. Mol. Life Sci 63, 2317–2328 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson D et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciriello G et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 163, 506–519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalin MG et al. Comprehensive Molecular Characterization of Salivary Duct Carcinoma Reveals Actionable Targets and Similarity to Apocrine Breast Cancer. Clin. Cancer Res 22, 4623–4633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zehir A et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin H-J, Zhao JC, Ogden I, Bergan RC & Yu J Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res 73, 3725–3736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin H-J, Zhao JC, Wu L, Kim J & Yu J Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat. Commun 5, 3972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song B et al. Targeting FOXA1-mediated repression of TGF-β signaling suppresses castration-resistant prostate cancer progression. J. Clin. Invest (2018). doi: 10.1172/JCI122367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson JLL et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J 30, 3019–3027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson JLL et al. Elevated levels of FOXA1 facilitate androgen receptor chromatin binding resulting in a CRPC-like phenotype. Oncogene 33, 5666–5674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomerantz MM et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet 47, 1346–1351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cirillo LA et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Iwafuchi-Doi M et al. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol. Cell 62, 79–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupien M et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbieri CE et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet 44, 685–689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang YA & Yu J Current perspectives on FOXA1 regulation of androgen receptor signaling and prostatecancer. Genes Dis 2, 144–151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grasso CS et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J et al. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med 9, 4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark KL, Halay ED, Lai E & Burley SK Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Li J et al. Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site. Biochemistry 56, 3745–3753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekiya T, Muthurajan UM, Luger K, Tulin AV & Zaret KS Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev 23, 804–809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z et al. BART: a transcription factor prediction tool with query gene sets or epigenomic profiles. Bioinformatics (2018). doi: 10.1093/bioinformatics/bty194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behrens J et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382, 638–642 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Daniels DL & Weis WI Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol 12, 364–371 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Zhong J, Su B, Zhou Y & Wang Y-Q Comparison of Pax1/9 Locus Reveals 500-Myr-Old Syntenic Block and Evolutionary Conserved Noncoding Regions. Mol. Biol. Evol 24, 784–791 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Tomlins SA et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448, 595–599 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Annala M et al. Recurrent SKIL-activating rearrangements in ETS-negative prostate cancer. Oncotarget 6, 6235–6250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phair RD et al. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol 24, 6393–6402 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitchiaya S et al. Dynamic recruitment of single RNAs to processing bodies depends on RNA functionality. bioRxiv 375295 (2018). doi: 10.1101/375295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swinstead EE et al. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell 165, 593–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitchiaya S, Androsavich JR & Walter NG Intracellular single molecule microscopy reveals two kinetically distinct pathways for microRNA assembly. EMBO Rep 13, 709–715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah NB & Duncan TM Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. J. Vis. Exp e51383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng Y et al. Evaluating human cancer cell metastasis in zebrafish. BMC Cancer 13, 453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramírez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu LJ et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey TL et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson S et al. Developing Cancer Informatics Applications and Tools Using the NCI Genomic Data Commons API. Cancer Res 77, e15–e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Network, T. C. G. A. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cieslik M et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res 25, 1372–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson DR et al. Integrative clinical genomics of metastatic cancer. Nature 548, 297–303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Layer RM, Chiang C, Quinlan AR & Hall IM LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15, R84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smyth GK limma: Linear Models for Microarray Data. in Bioinformatics and Computational Biology Solutions Using R and Bioconductor 397–420 (Springer, New York, NY, 2005). [Google Scholar]
- 51.Law CW, Chen Y, Shi W & Smyth GK Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liberzon A et al. The Molecular Signatures Database Hallmark Gene Set Collection. cels 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newton MA, Quintana FA, Boon JAD, Sengupta S & Ahlquist P Random-Set Methods Identify Distinct Aspects of the Enrichment Signal in Gene-Set Analysis. Ann. Appl. Stat 1, 85–106 (2007). [Google Scholar]
- 54.Subramanian A, Kuehn H, Gould J, Tamayo P & Mesirov JP GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23, 3251–3253 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data for the graphs, immunoblot and gel electrophoresis figures are included in matched Source Data files or Supplementary Information. All materials are available from authors upon reasonable request. All the raw next-generation sequencing data generated in this study has been deposited into the Gene Expression Omnibus (GEO) repository at NCBI (accession code: GSE123625). All custom data analysis software and bioinformatics algorithms used in this study are publically available on Github: