Abstract
The function of mammalian periodontal tissues depends on the presence of a nonmineralized periodontal ligament (PDL) juxtaposed in between mineralized tooth anchorage tissues alveolar bone (AB) and root cementum. In the present study we have hypothesized that the Wnt antagonist secreted frizzled related protein 1 (SFRP1) is an essential regulator of periodontal tissue mineral homeostasis. Our immunoreactions and western blot data demonstrated that SFRP1 was substantially expressed higher in PDL fibroblasts than in surrounding AB progenitors and cementoblasts. SFRP1 was also detected at higher levels in PDL fibroblasts than in dental follicle (DF) cells, but the difference was less pronounced. Preferential H3K4me3 active histone mark enrichment on the SFRP1 promoter and a lack of H3K27me3 repression were most dramatic in PDL progenitors, to a lesser degree in DF cells, and not detected in AB progenitors and cementoblasts. Selective inhibition of SFRP1 using a small molecule inhibitor WAY-316606 demonstrated that SFRP1 block increased PDL cell mineralization and mineralization gene expression such as β-catenin, alkaline phosphatase, osteocalcin, collagen I, and RUNX2. The effect of SFRP1 inhibition on PDL cell mineral homeostasis was confirmed by RNA silencing. These studies also demonstrated that SFRP1 knockdown promotes PDL differentiation through histone H3K4me3-mediated activation of RUNX2 and SP7. Finally, when SFRP1 inhibition and silencing studies were performed using AB progenitors instead of PDL progenitors, there was little effect on mineralized state control and gene expression, with the exception of osteocalcin, which was dramatically upregulated upon SFRP1 silencing. Together, the results of our study document the highly specific role of the Wnt inhibitor SFRP1 in maintaining the nonmineralized state of PDL progenitors.
Keywords: periodontal ligament, homeostasis, Wnt signaling, SFRP1, epigenetic regulation
Introduction
Highly precise regulation of periodontal homeostasis between a nonmineralized periodontal ligament (PDL) and mineralized surrounding tissues alveolar bone (AB) and cementum (CEM) is essential for the hammock-like attachment of mammalian teeth in their sockets and the physiological function of teeth as resilient partitioning organs for nutrients [1]. The nonmineralized PDL, while exclusive to mammals, crocodilians, and a few teleosts, is rather the exception than the rule among vertebrates: teeth in most fishes, amphibians, and reptiles are attached by ankylosis, a bony connection that directly anchors teeth to the jawbone [2]. The evolved position of mammals among vertebrates suggests that the nonmineralized anchorage of teeth must have arisen at some time during the course of vertebrate evolution. In earlier studies we have demonstrated that key steps toward a nonmineralized PDL occurred in evolutionary transition lineages, including caimans and mosasaurs [3,4]. In mammals, the mineralized state commitment of periodontal lineages occurs during late tooth development before and during tooth root development [5]. Expanding on our earlier studies we now focus on developing periodontal tissues and periodontal progenitor populations to ask how the nonmineralized state of the PDL is regulated and what molecules and signals might be involved.
All three mesenchymal periodontal tissues, CEM, PDL, and AB, are odontogenic neural crest derived tissues that develop from a common precursor, the dental follicle (DF) [6–8]. When the DF gives rise to the periodontal connective tissues, AB progenitors give rise to mineralized lineages to become AB, and cementoblasts give rise to mineralized lineages in close proximity to the root surface to become root CEM. Only the third DF-derived lineage, the PDL, develops into a nonmineralizing tissue layer sandwiched between AB and CEM [2,9]. To determine the molecular signatures that are responsible for the cell fate determination of individual periodontal tissues, we have conducted system biological studies and microarray analyses to identify signature marker genes and factors responsible for periodontal lineage commitment [10]. This analysis resulted in a close association of Jagged 1 and DKK1 with cementoblasts, BSP and IGFBP5 with AB cells, and secreted frizzled related protein 1 (SFRP1), periostin (POSTN), and scleraxis (SCX) with PDL progenitors [10]. Two of the three proteins identified as marker genes for periodontal progenitors are known inhibitors of the Wnt (wingless) signaling pathway SFRP1 and DKK1 [10]. We therefore hypothesized that inhibition of the Wnt pathway is a major prerequisite for the maintenance of the nonmineralized state of the PDL.
Wnts are evolutionarily conserved, cysteine-rich glycoproteins that regulate a multitude of cellular processes through canonical (β-catenin dependent) and other pathways [11,12]. The Wnt signaling pathway plays a major role in periodontal tissue homeostasis through its effect on multiple signaling intermediaries and extracellular matrix Wnt inhibitors [13]. Extracellular matrix Wnt inhibitors affect Wnt function and Wnt-related pattern formation by binding to Wnts and inhibiting their interaction with cell surface receptors [14]. Extracellular matrix Wnt inhibitors involved in mineral homeostasis include Dickkopf (DKK), Wnt inhibitory factor (WIF), sclerostin (SOST), and soluble frizzled-related proteins (SFRPs) [12,14]. When Wnts bind to members of the Frizzled family of transmembrane receptors, GSK-3 activity is downregulated, resulting in increased β-catenin in the cytosol and nucleus, as well as changes in the transcriptional activity of target promoters [15,16]. Earlier microarray-based analyses from our laboratory have identified the Wnt antagonist SFRP1 as the most selective molecule specifically associated with the PDL [10]. In addition, SFRP1 plays a role in the modulation of trabecular bone density modulation and osteoblast function [17,18], while blocking osteoblast-induced osteoclastogenesis [19]. SFRP1 has also been credited with reducing the severity of periodontitis due to its antiapoptotic properties [20].
In the present study we seek to determine the role of the Wnt antagonist SFRP1 as a key regulator of the mineralized state of periodontal cells and tissues. Immunoreactions, western blots, and chromatin immunoprecipitations (ChIPs) on the SFRP1 promoter 1,300 and 400 bp upstream of the transcription start site (TSS) have been applied to ask whether the PDL is associated with higher levels of SFRP1 than surrounding periodontal tissues DF, AB, and CEM. Selective inhibition of SFRP1 by means of a small molecule inhibitor was used as a strategy to assess the effect of SFRP1 block on PDL cell mineralization and mineralization gene expression. Small molecule inhibition studies were confirmed by RNA silencing using short hairpin RNA (shRNA). RNA silencing was also applied to identify the mechanism by which SFRP1 regulates PDL cell differentiation. Finally, the effect of SFRP1 on PDL cell differentiation and gene expression was compared with its effect on AB progenitors to determine whether SFRP1 function is cell type specific. Together, our studies have resulted in a comprehensive analysis of the function of the Wnt inhibitor SFRP1 as it relates to periodontal mineral homeostasis.
Methods
Isolation of dental progenitors and osteogenic induction
Healthy human teeth were extracted from 12- to 15-year-old patients for orthodontic reasons as per the guidelines of the Institutional Review Board of Texas A&M University College of Dentistry. A written informed consent was obtained from patients before surgery. Dissection of dental tissues from tooth organs and the isolation of DF, PDL, AB, and CEM mesenchymal progenitors were done as described previously [10]. Progenitor cells were cultured in DMEM containing 10% fetal bovine serum and 1 × antibiotics. For osteogenic induction, progenitors were seeded at a density of 26,000/cm2 and cultured for 24 to 48 h before differentiation with induction medium (complete media supplemented with 50 μg/mL ascorbic acid 2-phosphate, 10 mM β-glycerophosphate, and 10 nM dexamethasone). The induction medium was changed on alternate days. The SFRP1 inhibitor WAY-316606 (ApexBio, Houston, TX) was prepared in DMSO and diluted to a final concentration of 1 and 2 μM in culture media. Early passage progenitor cells (P3–P6) were used for all experiments.
Animals
All experimental procedures on rodents were approved by the Institutional Animal Care and Use Committee of Texas A&M University College of Dentistry. Adult Sprague-Dawley rats (weight, 125–175 g) were obtained from Charles River Laboratories (Wilmington, MA), and wild-type C57BL/6 mice were from The Jackson Laboratory (Farmington, CT). Animals were housed in a temperature (20°C ± 2°C) and humidity controlled (50%) environment on a regular light–dark schedule and given sterile food and water ad libitum.
Inhibitor treatment in rat maxillae
WAY-316606 was diluted (1 μM) in sterile saline and carefully injected (final volume 10 μL) with a fine needle in the palatal periodontal space of the maxillary molar tooth in anesthetized rats. Control rats were injected with sterile saline. Injections were carried out on alternate days for 7 days. Control and inhibitor treated maxillae were harvested 48 h after the last injection and fixed in 10% formalin for micro-computed tomography (micro-CT) analysis. Inhibitor treatment was repeated four times with three animals in each experimental group.
Tissue preparation and immunohistochemistry
Freshly harvested mouse mandibles from wild-type mice were fixed in 10% neutral buffered formalin at 4°C for 24 h. The samples were then demineralized in 10% EDTA, dehydrated in a graded series of alcohols, and embedded in paraffin wax. Sagittal sections (5 μm) were deparaffinized and rehydrated, and antigen retrieval was performed by 0.25% trypsin treatment in phosphate-buffered saline (PBS) for 30 min at 37°C. Immunoreactions were performed using Histostain-Plus IHC Kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer's instructions. Briefly, the slides were blocked for 10 min at room temperature and incubated with primary antibodies (Rabbit anti SFRP1, 1:100, ab4193; Abcam, Cambridge, MA and Mouse anti β-catenin, 1:100, 610153; BD Transduction, San Jose, CA) overnight at 4°C. Subsequently, the sections were washed and incubated with secondary antibody, and target protein localization was visualized with the help of AEC (Red) substrate. Incubation times were according to the manufacturer's instructions. Cell nuclei were counterstained with Hematoxylin and sections mounted using Hydromount (National diagnostics, Atlanta, GA). Stained sections were imaged under a bright-field microscope (Leica, Germany) and processed using Adobe Photoshop.
Micro-CT analysis
Maxillae from control and WAY-316606 treated rats were scanned using μCT35 (Scanco Medical, Bassersdorf, Switzerland) at 55 kVp, 800 ms integration time with a 6 μm voxel size. Three-dimensional (3D) images were generated from CT scans and sliced through the center of the root to visualize PDL and AB architecture. For AB histomorphometry, a region of interest was first defined in micro-CT two-dimensional sagittal sections in an area between the roots of the first molar. Values corresponding to bone volume fraction [bone volume/tissue volume (BV/TV) %], trabecular number (Tb.N.; 1/mm), mean trabecular thickness (T.Th.; mm), and mean trabecular separation (Tb.Sp.; mm) were measured volumetrically through 50 serial sections (300 μm) for each sample using Scanco Software.
Western blot analysis
Cells were lysed in radioimmunoprecipitation (RIPA) buffer in the presence of protease inhibitors. Protein concentration was quantified using the BCA Protein Assay Kit (Thermo Fisher Scientific), and 50 μg was separated on 4%–20% gradient SDS-polyacrylamide gels. The proteins were electrophoretically transferred to a PVDF membrane, blocked with 5% nonfat milk, and incubated overnight at 4°C with primary antibodies against β-catenin (BD Transduction), RUNX2 (ab76956; abcam), SP7 (ab22552; abcam), BGLAP (250483; Abbiotech, Escondido, CA), IBSP (ab33022; abcam), or β-actin (ab8226; abcam) followed by incubation with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Dallas, TX). After washing, protein bands were visualized using an enhanced chemiluminescence (ECL) detection system (Thermo Fisher Scientific).
Chromatin immunoprecipitation
PDL and AB progenitor cells grown in 100 mm dishes were cross-linked with 1.1% formaldehyde for 10 min at room temperature and quenched with 125 mM Glycine. After washing twice with cold 1 × PBS, cells were harvested and stored at −80°C. ChIP assays were performed as previously described [21]. Briefly, nuclei from 1 × 106 cells were suspended in lysis buffer and chromatin sonicated in a cup horn sonicator (Q Sonica) to a size of 300 bp–1 Kb. The sheared chromatin was centrifuged at 12,000 rpm for 10 min and soluble chromatin incubated overnight with 50 μL of DynaI beads (ThermoFisher Scientific) prebound to 5 μg of antibody against histone H3 tri methyl K4 (ab8580; abcam), histone H3 tri methyl K27 (ab6002; abcam), and β-catenin (BD Transduction). An input fraction corresponding to 5% of soluble chromatin was kept for chromatin normalization. Chromatin incubated beads were washed five times with 1 × RIPA buffer and once with 1 × TE, pH 8.0. Protein-DNA complexes were eluted from beads by incubating with elution buffer followed by cross-link reversal overnight at 65°C. DNA was purified using ChIP DNA Clean and Concentrator columns (Zymo Research, Irvine, CA) and diluted to a concentration of 2 ng/μL. Relative enrichment was measured by real-time quantitative polymerase chain reaction (PCR) as detailed below.
Alkaline phosphatase assay, alizarin red staining, and imaging
To measure levels of alkaline phosphatase (ALP) activity, control and treated cells were fixed in cold methanol and stained with NBT/BCIP solution for 10 min. Mineral deposits were detected by incubating cells with 1% alizarin red S (ARS) (Sigma Aldrich, St. Louis, MO) solution after cold methanol fixation. Freshly stained cells were imaged on a high resolution photo color scanner (Epson) and processed using Adobe Photoshop.
RNA extraction and quantitative real-time PCR analysis
Total RNA was extracted from control and treated cells using the RNeasy Plus Mini Kit (Qiagen, Germantown, MD) according to manufacturer's instructions. Two micrograms of RNA was used for cDNA generation using SuperScript III (Thermo Fisher Scientific). PCR primers for transcription factor genes and mineralization related genes (Table 1) were designed based on the GenBank database (NIH). Transcript levels were quantified by SYBR Green (Applied Biosystems, Foster City, CA) based Real-Time Quantitative PCR analysis on the ABI 7000 sequence detection system (Applied Biosystems). Expression levels were normalized to levels of GAPDH. Relative expression levels were calculated using the 2−ΔΔCt method, and values were represented as the mean expression level ± standard deviation (SD).
Table 1.
Oligonucleotides for ChIP Analysis and Real-Time Expression Analysis
| Oligos for ChIP | ||
|---|---|---|
| hCTNNB1 | Forward | TGGAACCAGATAAAAATGGAATC |
| Reverse | AGCCCGCAATTCAACAAGT | |
| hSFRP1-400 | Forward | GCTTAACCCTCCCTCTCAGG |
| Reverse | GACGGTATCAGCACAACAGC | |
| hSFRP1-1300 | Forward | GCAGTCAGCGGAGATAGC |
| Reverse | GGGGAGCCTGGATCATACTT | |
| hRUNX2-3000 | Forward | GTGGGACTCGAGAGACCTGT |
| Reverse | AGACCGGACCAACTGGAAG | |
| hRUNX2-2000 | Forward | GCGTTTGCACTGAGCAATAA |
| Reverse | CCTCGAAGCATCAAGGAAGA | |
| hRUNX2-1900 | Forward | CGCAAACACGTTTTCAAGC |
| Reverse | GGAAATCCGCTTGAGGCTAT | |
| hSP7-3500 | Forward | TCCCCATCTTTGTTCCTCTG |
| Reverse | GAAGACCAGATGTGTGGCTTTA | |
| hSP7-3100 | Forward | TCTATCAGCCACCTGGTTCC |
| Reverse | CAGCAGGTAGGCACCAATTT | |
| hRUNX2 bCATbind | Forward | CGTAGTAGTACACAACGCCG |
| Reverse | GTTTCGTGTCTGTCTTCCCC | |
| Oligos for real-time RT-PCR analysis | ||
| hBMP2 | Forward | TCAAGCCAAACACAAACAGC |
| Reverse | AGCCACAATCCAGTCATTCC | |
| hBMP4 | Forward | CTTTACCGGCTTCAGTCTGG |
| Reverse | ATGTTCTTCGTGGTGGAAGC | |
| hCTNNB1 | Forward | TCTGAGGACAAGCCACAAGATTACA |
| Reverse | TGGGCACCAATATCAAGTCCAA | |
| hMSX2 | Forward | CGGAAAATTCAGAAGATGGAG |
| Reverse | GAGGAGCTGGGATGTGGTAA | |
| hRUNX2 | Forward | GTGCCTAGGCGCATTTCA |
| Reverse | GCTCTTCTTACTGAGAGTGGAAGG | |
| hSP7 | Forward | TACCCCATCTCCCTTGACTG |
| Reverse | GCAACAGGGGATTAACCTGA | |
| hIBSP | Forward | AACCTACAACCCCACCACAA |
| Reverse | CGTACTCCCCCTCGTATTCA | |
| hBGLAP | Forward | GACTGTGACGAGTTGGCTGA |
| Reverse | AGCAGAGCGACACCCTAGAC | |
| hCOL I | Forward | GGAGCTCCAAGGACAAGAAA |
| Reverse | ATGAAGGCAAGTTGGGTAGC | |
| hALP | Forward | CAACCCTGGGGAGGAGAC |
| Reverse | GCATTGGTGTTGTACGTCTTG | |
| hGAPDH | Forward | ACAGTCAGCCGCATCTTCTT |
| Reverse | ACGACCAAATCCGTTGACTC | |
ChIP, chromatin immunoprecipitation; RT-PCR, real-time polymerase chain reaction.
To quantify enrichment following ChIP analysis, 4 ng of immunoprecipitated DNA was used for each real-time quantitative PCR using specific primers that bind to promoter regions upstream to the TSS (Table 1). DNA from total input was used for the normalization of ChIP enrichment. Enrichment obtained from beads alone served as a negative control. Enrichment data for each primer pair are presented as mean enrichment ± SD after subtracting the values obtained from corresponding negative controls. Reproducibility of the results was confirmed by performing the analyses in triplicates for three independent ChIP experiments.
shRNA mediated knockdown of SFRP1
SureSilencing shRNA plasmids (Qiagen) specific to SFRP1 were used for knockdown studies in PDL and AB progenitors. These plasmids express shRNA under the control of the U1 promoter and also feature a SV40-driven Neomycin resistance gene product for stable selection in cells. shRNAs targeting four different regions of SFRP1 gene were transfected separately into cells using the NanoJuice Transfection Kit (MilliporeSigma, Burlington, MA) as per the manufacturer's instructions. After 48 h, transfected cells were expanded to 100 mm dishes and selected with 500 μg/mL G418 antibiotic (Thermo Fisher Scientific) for 10 days. Stably selected cell colonies were first checked for SFRP1 knockdown efficiency and maintained in culture media containing 200 μg/mL G418. Cells expressing shRNA targeting a nonspecific region were used as a negative control.
Statistical analysis
All quantitative data are derived from at least three independent experiments and presented as mean ± SD. Statistical significance between two groups was evaluated with Student's t-test. The difference between groups was considered statistically significant as follows: *P < 0.05, **P < 0.005, ***P < 0.001.
Results
High expression of the Wnt antagonist SFRP1 in PDL fibroblasts correlated with active H3K4me3 histone marks at its promoter
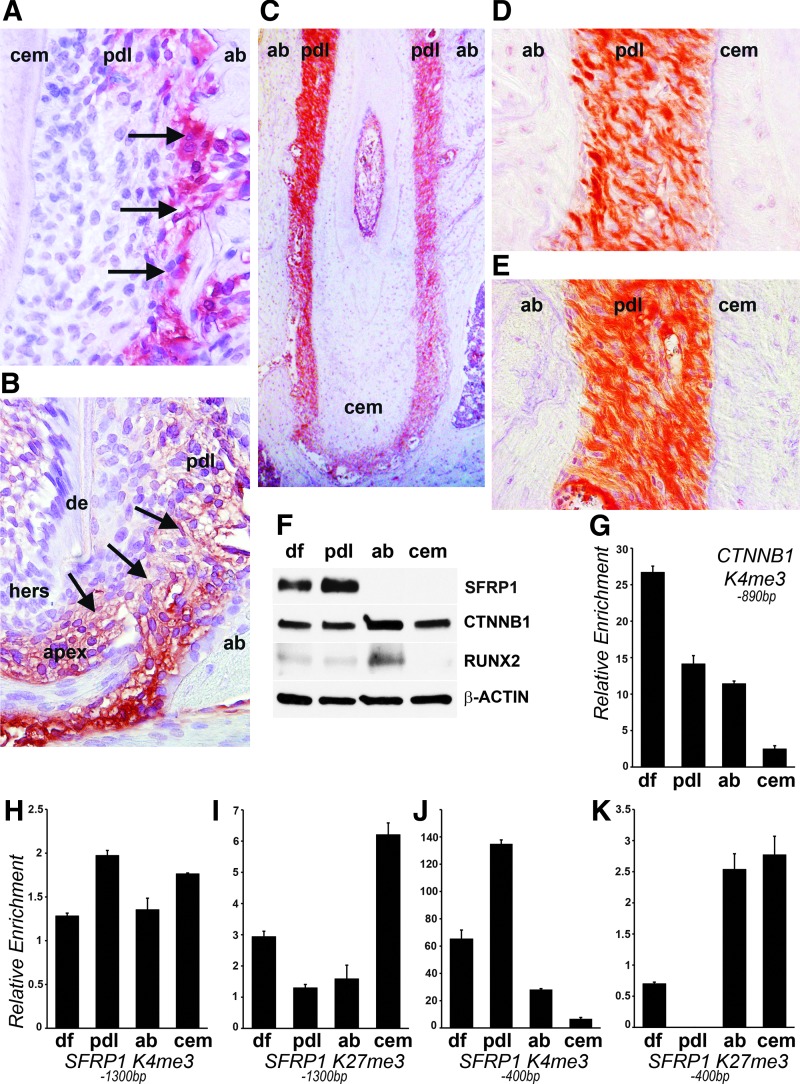
In previous studies we have reported the exceptionally high expression levels of the Wnt antagonist, SFRP1, in PDL cells [10]. To determine and compare SFRP1 localization and expression in mouse periodontal tissues, immunohistochemical analysis was performed using specific antibodies in early postnatal and adult mouse molars. Our analysis demonstrated high levels of SFRP1 expression in the PDL, while the AB and CEM did not exhibit any significant staining (Fig. 1A–E). A closer examination of SFRP1 expression in the PDL of 12- and 15-day postnatal mice revealed that SFRP1 was not uniformly expressed throughout the PDL, but instead preferentially associated with the PDL-AB interface (Fig. 1A, B; arrowheads). Interestingly, high levels of SFRP1 were also detected at the root apex (Fig. 1B; arrowhead). In the adult mouse periodontium (3 weeks and older), SFRP1 was fairly uniformly expressed (Fig. 1C, D) with a slight increase at the PDL-AB interface (Fig. 1D). Interestingly, the Wnt effector protein, β-catenin, was also expressed throughout the PDL, without exhibiting an increase in expression levels at the AB margins (Fig. 1E). We next compared SFRP1 and β-catenin expression in the human periodontal progenitor cell populations, DF, PDL, AB, and CEM. In support of our immunohistological staining experiments, SFRP1 was highly expressed in human PDL progenitors and to a lesser extent in DF progenitors (Fig. 1F). AB and CEM progenitors did not exhibit detectable levels of the protein (Fig. 1F). In contrast, β-catenin was expressed in all four human progenitor populations, with the highest levels detected in AB progenitors (Fig. 1F). Likewise, the mineralization marker, RUNX2, was also highly expressed in AB progenitors (Fig. 1F).
FIG. 1.
Expression and epigenetic profiling of SFRP1 and β-catenin. Immunohistochemical staining for SFRP1 (A–D) and β-catenin (E) in mouse mandibular first molars. SFRP1 expression in 12-day-old molars (A) and 15-day-old molars (B). Note the intense staining for SFRP1 (arrows) along the PDL-bone interface (A) and at the tooth apex (B). (C) Panoramic view of SFRP1 expression in the adult mouse periodontium. Expression of SFRP1 (D) and β-catenin (E) in the mouse PDL. (F) Western blots illustrating expression of SFRP1, β-catenin, and RUNX2 in human periodontal progenitors. β-actin was used as a loading control. Relative enrichment of the H3K4me3 histone modification on β-catenin (G) and SFRP1 (H, J) promoters in human dental progenitors. Relative enrichment of the H3K27me3 histone modification on the SFRP1 promoter (I, K) in human dental progenitors. ChIP regions analyzed are indicated as base pairs upstream to the TSS for respective promoters. Two regions on the SFRP1 promoter were analyzed (−1,300 and −400 bp). ab, alveolar bone; CEM, cementum; ChIP, chromatin immunoprecipitation; de, dentin; df, dental follicle; hers, Hertwig's epithelial root sheath; pdl, periodontal ligament; TSS, transcription start site.
To determine whether epigenetic regulatory mechanisms at promoters of β-catenin and SFRP1 contribute toward their differential expression within periodontal tissues, we performed chromatin immunoprecipitation (ChIP) studies with antibodies against an active (H3K4me3) and a repressive (H3K27me3) histone mark. We have previously provided a detailed introduction into the role of histone methylation marks on nucleosome configuration and gene expression [21]. For these assays we used human periodontal progenitors because of the ease in establishing a large colony of human PDL progenitor cells and since they expressed high levels of SFRP1 similar to the mouse PDL. Based on β-catenin expression comparison data in all four human periodontal progenitor cells, we performed ChIP analysis to detect enrichment levels of the active H3K4me3 histone mark on the β-catenin promoter. Our analysis indicated that H3K4me3 levels at the β-catenin promoter were the highest in DF cells and lowest in CEM cells (Fig. 1G). Even though β-catenin protein levels in AB cells were elevated (Fig. 1F), our analysis did not detect higher enrichment for H3K4me3 on the β-catenin promoter in AB cells compared to DF and PDL progenitors (Fig. 1G). Next we compared the enrichment of both H3K4me3 and H3K27me3 histone marks at two genetic loci (400 and 1,300 bp upstream to the TSS) within the SFRP1 promoter among all four dental progenitors. Matching the high levels of SFRP1 expression in PDL cells, ChIP analysis demonstrated highest levels of the active H3K4me3 histone mark on both promoter regions of SFRP1 in PDL progenitors (Fig. 1H, J). To the contrary, enrichment levels of the repressive histone mark, H3K27me3, were the lowest at both SFRP1 promoter regions in PDL cells (Fig. 1I, K), suggesting that SFRP1 expression in periodontal tissues is epigenetically regulated.
SFRP1 loss of function enhanced the mineralization response in human PDL fibroblasts
The high levels of SFRP1 expression in the PDL compared to AB and CEM point to an important role for this Wnt antagonist in PDL progenitors. The unique position of the PDL in between the two periodontal mineralized tissues, AB and CEM, prompted us to hypothesize that SFRP1 regulates the mineralization state of the PDL. To test our hypothesis we used a small molecule inhibitor, WAY-316606, to specifically inhibit SFRP1 function in PDL and AB cells. WAY-316606 has been demonstrated to bind SFRP1 and inhibit its function, thereby stimulating canonical Wnt signaling [22].
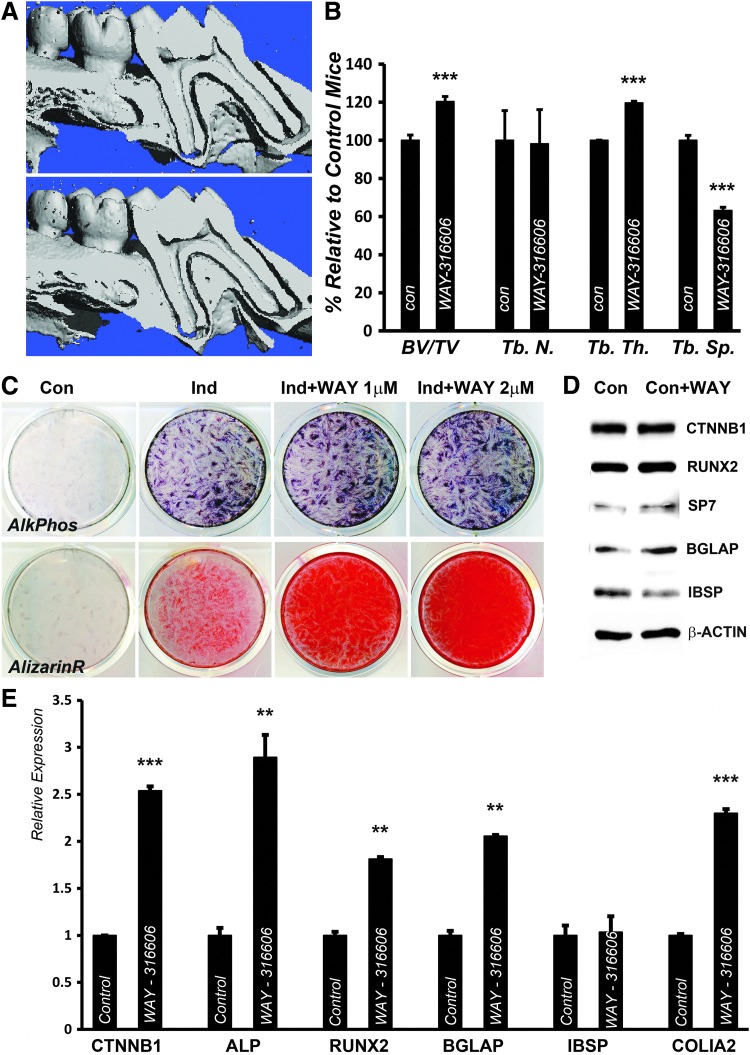
As a means to test the effect of inhibitor mediated SFRP1 inhibition in vivo, we injected WAY-316606 into a surgically created periodontal pocket near the lingual PDL space of first molars in rats with the help of a fine needle. Micro-CT analysis of WAY-316606 treated rat mandibles revealed a substantial gain of interradicular bone mass near the inhibitor application site (Fig. 2A) compared to rat mandibles treated with a vehicle control (Fig. 2A). Our analysis did not detect any apparent differences in the PDL space between the inhibitor treated and control groups (Fig. 2A). Furthermore, bone morphometry analysis of the site proximal to the inhibitor application site corroborated our observations in 3D reconstruction models as indicated by the higher BV in the WAY-316606 treated group compared to controls (Fig. 2B). Our analysis also identified higher trabecular thickness accompanied by a decrease in trabecular separation, while there was no significant difference in trabecular number (Fig. 2B).
FIG. 2.
Small molecule based inhibition of SFRP1 function enhances the mineralization potential of human PDL fibroblasts. (A, B) SFRP1 inhibition improves AB parameters. Representative sagittal section through a 3D micro-CT reconstruction model of a WAY-316606 treated (A, bottom) and control treated (A, top) rat mandible. (B) Comparison of AB morphometric parameters based on a 3D micro-CT comparison between control and WAY-316606 treated molars. micro-CT analysis and bone morphometry are representative of four to five samples analyzed for each experimental group. (C) PDL progenitors were either subjected to osteogenic induction through mineralization media or treated with the SFRP1 inhibitor (WAY-316606) at indicated concentrations. ALP activity was assessed after 7 days of culture (C, top panel), while calcium deposits in treated cells were detected after 14 days of culture using ARS (C, bottom panel). (D) Western blot analysis for expression of mineralization related genes in PDL fibroblasts grown under osteogenic conditions with or without 1 μM WAY-316606 for 7 days. β-actin was used as a loading control. (E) Relative mRNA expression of osteogenic differentiation markers in PDL fibroblasts induced with mineralization media with or without 1 μM WAY-316606 for 7 days. Expression was normalized against the levels of GAPDH and statistical significance determined using Student's t-test. (**P < 0.01, ***P < 0.001). 3D, three-dimensional; AB, alveolar bone; ALP, alkaline phosphatase; ARS, alizarin red S; BV/TV, bone volume/tissue volume; con, control; ind, induced; micro-CT, micro-computed tomography; SFRP1, secreted frizzled related protein 1; Tb.N, trabecular number, Tb.Th, mean trabecular thickness, Tb.Sp, mean trabecular separation; WAY, WAY-316606.
Based on our WAY-316606 in vivo studies and to alleviate the effects of external modifying factors such as occlusal load, we decided to verify the effect of our inhibitor on our human PDL progenitors. Osteoinductive conditions were used as a means to induce in vitro differentiation of human PDL progenitors (Fig. 2C). Under these conditions, PDL cells displayed elevated levels of mineralization markers, including the early osteoblast differentiation marker ALP after 1 week of culture and also displayed mineral deposits when stained with ARS after 2 weeks of induction (Fig. 2C). Inhibition of SFRP1 by addition of WAY-316606 to differentiating PDL cells increased ALP activity and greatly enhanced mineral deposits in a dose dependent manner compared to cells cultured with induction media alone (Fig. 2C). To understand the molecular mechanisms accompanying the elevated mineralization response in PDL cells subsequent to SFRP1 inhibition, expression profiles of key mineralization markers were compared between induced and WAY-316606 treated cells at the mRNA and protein level. After 1 week of treatment, protein levels of key mineralization regulators, RUNX2 and SP7, and of the matrix associated protein osteocalcin (BGLAP) were marginally upregulated in WAY-316606 treated cells (Fig. 2D) compared to control induced cells. Consistent with increased Wnt signaling upon SFRP1 inhibition, β-catenin mRNA levels were upregulated in WAY-316606 treated cells (Fig. 2E), although β-catenin protein levels were not significantly elevated after similar treatment (Fig. 2D). Corroborating our protein expression data and the enhanced differentiation observed in WAY-316606 treated PDL cells, the mRNA levels of ALP, RUNX2, and BGLAP were significantly elevated upon SFRP1 inhibition (Fig. 2E). Supporting our report of heightened mineralized matrix secretion in WAY-316606 treated PDL cells, there was a significant upregulation of mRNA levels for the extracellular matrix protein collagen I in SFRP1 inhibited PDL cells (Fig. 2E). Interestingly, mRNA and protein levels of IBSP were not upregulated in response to WAY-316606 mediated SFRP1 inhibition (Fig. 2D, E).
SFRP1 knockdown spontaneously increased PDL differentiation by elevating H3K4me3 histone marks on RUNX2 and SP7 promoters resulting in increased mineralization gene expression
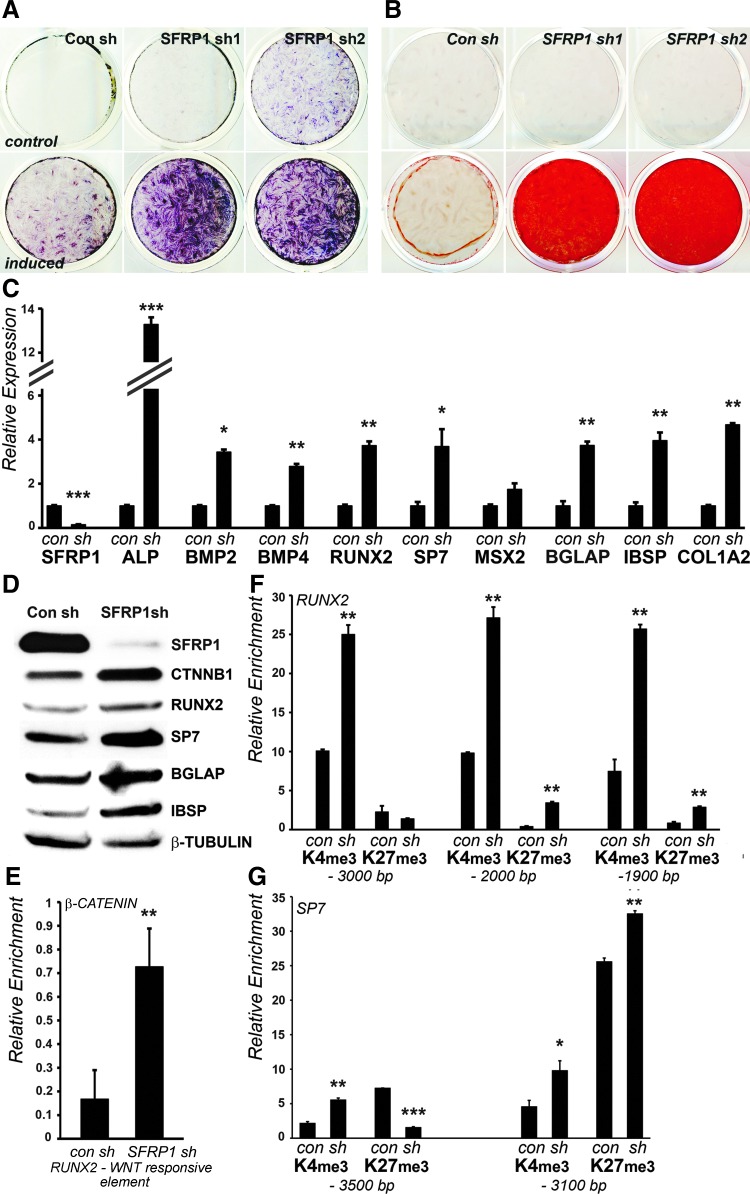
To further verify whether SFRP1 is essential for the maintenance of a nonmineralized state in PDL cells, knockdown studies were performed using a shRNA based strategy. Of the four SFRP1 specific shRNAs transfected in PDL cells, SFRP1 sh1 and SFRP1 sh2 markedly decreased SFRP1 protein levels compared to control shRNA expressing cells (data not shown). Consistent with our observations with SFRP1 function inhibition studies using WAY-316606, PDL cells stably expressing SFRP1 sh1 and sh2 demonstrated a significantly upregulated ALP activity (Fig. 3A) and demonstrated a drastic increase in mineral deposits as revealed by alizarin Red staining (Fig. 3B) after 7 and 14 days of osteogenic induction, respectively, compared to controls. Interestingly, PDL cells expressing SFRP1 sh2 displayed elevated ALP activity even in the absence of mineralization induction after 7 days of culture (Fig. 3A), suggesting that the loss of SFRP1 primes these cells toward osteogenic lineage differentiation. To verify the effect of SFRP1 on osteogenic lineage commitment, expression levels of mineralization marker genes were tested in PDL cells stably expressing SFRP1 sh1 or a control shRNA at both the mRNA and protein levels. Our data demonstrated that the mRNA level of the early mineralization marker, ALP, was upregulated 13-fold in SFRP1 sh1 expressing cells (Fig. 3C). In addition, transcript levels of the bone morphogenetic proteins, BMP2 and BMP4, key upstream regulators of mineralization, RUNX2 and SP7, and the extracellular matrix related genes, OCN, IBSP, and COL1A2, were significantly upregulated (greater than twofold) upon SFRP1 knockdown (Fig. 3C). Although mRNA levels of the MSX2 homeobox gene were elevated upon SFRP1 knockdown (Fig. 3C), the difference was not statistically significant. Our mRNA expression data were further validated by western blot analysis of lysates from SFRP1 sh1 and control shRNA expressing PDL cells. As expected, PDL cells stably expressing SFRP1 shRNA exhibited a dramatic decrease in SFRP1 protein levels (Fig. 3D). Furthermore, the protein levels of the mineralization related transcription factors, RUNX2 and SP7, and of the mineralized matrix associated proteins, BGLAP and IBSP, were significantly increased upon SFRP1 knockdown (Fig. 3D). Consistent with its role as a Wnt antagonist, SFRP1 knockdown stabilized the protein levels of β-catenin (Fig. 3D), indicating an increase in Wnt signaling.
FIG. 3.
SFRP1 knockdown enhances osteo-differentiation in PDL fibroblasts by increasing H3K4me3 marks on RUNX2 and SP7 promoters. PDL cells stably expressing a con sh or shRNA against SFRP1 (SFRP1 sh1 and SFRP1 sh2) were grown with or without osteogenic induction. ALP activity was assayed after 7 days (A) and mineral deposits stained using ARS after 14 days (B). Note the elevated ALP activity in SFRP1 sh2 expressing PDL cells grown under nonmineralizing conditions. (C) mRNA expression of mineralization markers in SFRP1 sh1 expressing cells compared against con sh expressing cells. (D) Western blot analysis for osteogenic markers in lysates from con sh and SFRP1 sh1 expressing cells. (E) β-catenin enrichment on its response element located on the RUNX2 promoter in con sh and SFRP1 sh1 expressing cells. Relative enrichment of H3K4me3 and H3K27me3 histone modifications on the RUNX2 promoter (F) and on the SP7 promoter (G) in con sh and SFRP1 sh1 expressing cells. ChIP regions analyzed are marked as base pairs upstream to the TSS for respective promoters. Semiquantitative real-time RT-PCR for mRNA expression and ChIP analysis data are representative of three independent experiments with similar results. (Statistical significance is depicted as *P < 0.05, **P < 0.01, ***P < 0.001). con sh, control shRNA; RT-PCR, real-time polymerase chain reaction; shRNA, short hairpin RNA.
Runx2 is a known target of β-catenin/TCF1 in mesenchymal osteoprogenitor cells [23,24]. Our previous experiments demonstrated that SFRP1 knockdown in PDL cells stabilized β-catenin levels and upregulated RUNX2 expression (Fig. 3C, D). To test whether RUNX2 upregulation upon SFRP1 knockdown is a β-catenin dependent process, ChIP studies were performed to measure β-catenin enrichment levels on the Wnt-responsive element located within the proximal RUNX2 promoter. These experiments revealed a significantly higher level of β-catenin binding to the RUNX2 promoter in PDL cells expressing SFRP1 sh1 compared to controls (Fig. 3E).
The C-terminal β-catenin activation domain has been demonstrated to promote H3K4 trimethylation possibly through its association with TRRAP/TIP60 and mixed-lineage-leukemia (MLL1/MLL2) SET1-type chromatin modifying complexes [25]. To determine if the increased binding of β-catenin elevates the enrichment of the H3K4me3 histone modification on the RUNX2 promoter, ChIP studies were performed using antibodies specific to active and repressive histone methylation modifications: H3K4me3 and H3K27me3. Three different genetic loci were analyzed along the RUNX2 promoter for histone enrichment as indicated in Fig. 3F. The results from these studies demonstrated a significant increase in H3K4me3 histone modification levels on all three SFRP1 promoter regions tested in SFRP1 sh1 expressing cells compared to controls (Fig. 3F). A minor elevation in H3K27me3 enrichment was also observed at two of the RUNX2 promoter regions tested (−1,900 and −2,000 bp relative to TSS), but the increase in H3K4me3 marks was far more pronounced in SFRP1 knockdown cells (Fig. 3F). In addition, we tested whether the SP7 promoter was epigenetically reprogrammed in response to SFRP1 knockdown. Of the two regions tested within the SP7 promoter, both revealed a higher H3K4me3 level in SFRP1 sh1 expressing cells compared to controls (Fig. 3G). While enrichment was decreased for the suppressive H3K27me3 mark at the −3,500 bp region, the second region at −3,100 bp revealed an increase for the H3K27me3 mark (Fig. 3G). Taken together, the increased expression of RUNX2 and SP7 matched the increased H3K4me3 enrichment at the corresponding promoters in SFRP1 depleted PDL cells.
SFRP1 exerted only minor effects on mineralized state homeostasis in AB progenitors compared to PDL progenitors
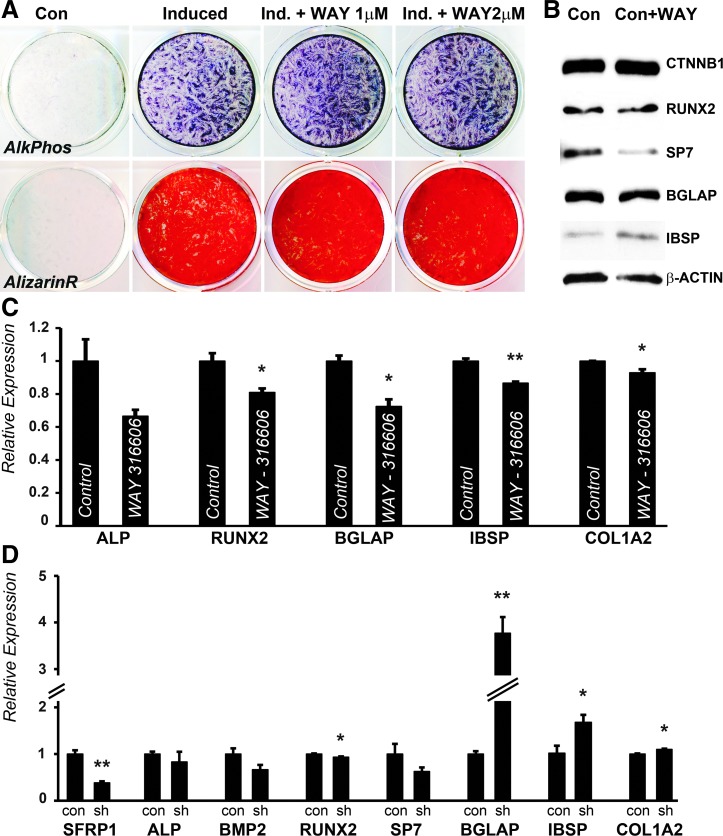
To further verify whether SFRP1-mediated mineral homeostasis regulation is unique to PDL cells, SFRP1 inhibition and knockdown studies were performed in AB progenitors. Both PDL and AB progenitors originate from the DF, which is an ectomesenchymal population of cells derived from the odontogenic neural crest [6,7,26]. Moreover, the fibroblastic cells located within the PDL are a source of osteoblasts required for the constant remodeling of AB in response to tooth eruption and forces of mastication [27]. Osteogenic differentiation and mineralized tissue lineage induction were performed in our AB progenitors as described in our PDL cell studies using osteogenic medium. Under these conditions, there was a substantial increase in ALP activity after 7 days and a massive increase in mineral deposits as revealed by ARS staining after 14 days of induction (Fig. 4A). However, in contrast to our studies in PDL cells, the addition of the SFRP1 inhibitor WAY-316606 did not result in a dramatic increase in ALP activity or the amount of mineral deposits at similar concentrations (Fig. 4A). Western blot analysis for protein levels of mineralization markers also did not reveal any increase in the expression levels of RUNX2, SP7, and BGLAP after 7 days of treatment with WAY-316606 under osteoinductive conditions (Fig. 4B). However, protein levels of the matrix protein IBSP were upregulated upon SFRP1 inhibition (Fig. 4B). There was also a small amount of increase in the levels of β-catenin upon WAY-316606 treatment (Fig. 4B).
FIG. 4.
Periodontal AB fibroblasts upregulate terminal mineralization markers upon SFRP1 inhibition independent of RUNX2 and SP7. In this study, AB fibroblasts were subjected to mineralizing conditions with either induction media or in the presence of the SFRP1 inhibitor WAY-316606 at various concentrations. ALP activity was assessed after 7 days (A, top panel), while mineral deposits were detected using ARS after 14 days of culture (A, bottom panel). (B) Western blot analysis for expression of mineralization related genes in AB progenitors grown under osteogenic conditions with or without 1 μM WAY-316606 for 7 days. β-actin was used as a loading control. (C) Relative mRNA expression of osteogenic differentiation markers in AB fibroblasts grown in mineralization media with or without 1 μM WAY-316606 for 7 days. (D) mRNA expression of mineralization-related marker genes in AB cells stably expressed in con shRNA or SFRP1 sh1 shRNA. Expression was normalized against GAPDH, and statistical significance was determined using Student's t-test. (*P < 0.05, **P < 0.01). ARS, alizarin Red S; ALP, alkaline phosphatase; con, control; ind, induced.
As an alternative approach for the comparison of the mineralization and differentiation response between PDL and AB cells, we conducted a series of shRNA mediated knockdown studies in AB cells. For this comparison, AB cell populations stably expressing shRNA against SFRP1 or a control shRNA were established. For this purpose we used SFRP1 sh1 shRNA, which effectively knocked down SFRP1 protein levels in PDL cells. Similar to our observations with WAY-316606 treatment, there was no increase in mRNA transcript levels of RUNX2 and SP7, and in fact, both genes were downregulated in SFRP1 sh1 expressing AB cells (Fig. 4D). As expected, SFRP1 levels were effectively downregulated, while there was no statistically significant difference in the levels of ALP and BMP2 (Fig. 4D). Unexpectedly, the mRNA levels of all three matrix related genes analyzed, BGLAP, IBSP, and COL1A2, were upregulated in a statistically significant manner upon SFRP1 knockdown in AB cells (Fig. 4D), explaining the subtle increase of ALP activity and alizarin Red staining observed with the WAY-316606 treatment. Surprisingly, there was a fourfold increase in the transcript levels of BGLAP in SFRP1 sh1 expressing AB cells compared to control shRNA expressing cells (Fig. 4D).
Discussion
In the present study we have documented the key role of the Wnt antagonist SFRP1 as a regulator of periodontal tissue mineral homeostasis. Immunoreactions, western blot data, and ChIP studies confirmed high expression levels of SFRP1 in PDL cells compared to cells from surrounding tissues AB and CEM. Selective inhibition of SFRP1 by a small molecule inhibitor or by RNA silencing demonstrated that SFRP1 block promoted PDL cell mineralization and mineralization gene expression such as β-catenin, ALP, osteocalcin, collagen I, and RUNX2. Explaining the effect of SFRP1 inhibition on PDL homeostasis, RNA silencing studies documented that knockdown of SFRP1 protein levels affects PDL differentiation through histone H3K4me3 mediated activation of RUNX2 and SP7. Finally, SFRP1 inhibition and silencing in AB progenitors instead of PDL progenitors suggested that SFRP1 function is unique to PDL progenitors as there was little effect on AB progenitor mineralized state and gene expression. Together, the results from our studies have established SFRP1 as a key regulator of periodontal mineralized state homeostasis.
Our data have demonstrated preferential SFRP1 localization and gene expression in the PDL versus other periodontal tissues, including AB, CEM, and DF. These data match our earlier report identifying SFRP1 as the most prominent marker gene for PDL cells compared to DF progenitors, AB osteoblasts, or cementoblasts in a microarray study based on different human periodontal cell populations [10]. SFRP1 immunoreactions were narrowly confined to the PDL region, while AB and CEM were devoid of any SFRP1 labeling. In contrast, there was a less intense SFRP1 band on western blots in the DF compared to the SFRP1 band in PDL extracts. There was also ∼50% less H3K4m3 promoter enrichment on the proximal SFRP1 promoter in DF cells compared to the H3K4me3 enrichment on the SFRP1 promoter in PDL cells. The strong specificity of SFRP1 for the PDL and a less intense relationship between SFRP1 and the DF are likely due to the DF's role as a precursor tissue for the PDL [7–10,26]. Our data indicated that there was a strong contrast between elevated PDL/DF H3K4me3 enrichment versus AB/CEM H3K27me3 enrichment on the −400 bp proximal promoter region, while H3K4me3 and H3K27me3 enrichment levels at the more distal −1,300 bp promoter region were less clearly defined and mostly characterized by a high H3K27me3 enrichment level for cementoblasts. These data suggest that the mineralization cell fate decision may occur close to the proximal SFRP1 promoter, while additional epigenetic repression of SFRP1 gene expression in cementoblasts may take place on further distal promoter regions.
Our data indicate that SFRP1 loss of function as obtained by administration of the WAY-316606 inhibitor increased AB volume, enhanced trabecular thickness, and decreased trabecular separation in a rodent model and enhanced the mineralization response in human PDL fibroblasts as evidenced by ALP/alizarin red staining and enhanced β-catenin, ALP, osteocalcin, Collagen I, and RUNX2 gene expression. The effectiveness of the WAY-316606 small molecule inhibitor to inhibit SFRP1 function in mouse calvarial tissues and to bind to SFRP1 has been documented in previous studies [22]. In this study, the WAY-316606 small molecule inhibitor demonstrated its efficacy as a promoter of mineralization in periodontal tissues at the gene expression level, the cellular level, and at the tissue level. Together, these studies establish SFRP1 block of function as a powerful mechanism to promote mineralization in periodontal tissues.
Further elaborating on the role of SFRP1 in periodontal tissues, our study demonstrated that SFRP1 knockdown promotes PDL differentiation through histone H3K4me3 mediated activation of RUNX2 and SP7. In general, the results of our SFRP1 RNA silencing studies matched those of our SFRP1 inhibition studies using the WAY-316606 small molecule inhibitor. However, in addition we demonstrated that SFRP1 RNA silencing resulted in a three- to fourfold increase in the enrichment of the active H3K4me3 mark on the RUNX2 promoter, explaining the effect of loss of SFRP1 on osteogenic lineage commitment and mineralization. Moreover, SFRP1 silencing significantly increased more than twofold the enrichment of H3K4me3 on the SP7 −100 to −3,500 bp distal promoter region. The role of Runx2 as a multifunctional regulator of osteoblast differentiation and bone formation has been well established [28–30]. In addition, the zinc finger transcription factor Osterix/Sp7 plays critical roles in osteoblast differentiation, bone growth, and homeostasis [31,32]. By affecting both Runx2 and Sp7 gene expression, SFRP1 has the potential to exert powerful control over osteoblast lineage differentiation and bone formation. Thus, our studies demonstrate that loss of SFRP1 primes PDL cells toward osteogenic lineage differentiation through genetic and epigenetic mechanisms, including increased RUNX2 and SP7 gene expression.
In contrast to the well-defined effects of SFRP1 inhibition on periodontal progenitor cell gene expression and mineralization, SFRP1 exerted only negligible effects on mineralized state homeostasis and mineralization gene expression in AB progenitors. Key mineralization genes such as RUNX2, SP7, ALP, BSP, and collagen I exhibited only subtle changes after SFRP1 inhibition, both in response to the WAY-316606 inhibitor treatment and to SFRP1 RNA silencing. The absence of an obvious mineralization target gene network by which SFRP1 might control AB mineralization might be the result of the fairly low expression levels of SFRP1 in bone and confirmatory of the roles of other regulators in control of bone mineralization. The only exception from the general unresponsiveness of mineralization genes toward SFRP1 inhibition in bone cells was the fourfold increase of osteocalcin gene expression following SFRP1 shRNA treatment, but not in response to the WAY-316606 inhibitor. This finding suggests that silencing of SFRP1 mRNA in bone cells dramatically and directly affected osteocalcin gene expression, while no such effects were detected by binding to the SFRP1 gene product.
Acknowledgments
Generous funding by NIDCR grants DE027930 and DE026198 to T.G.H.D. and DE019463 to X.L. is gratefully acknowledged.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Reed DA. and Diekwisch TGH. (2015). Morphogenesis and wound healing in the periodontium. In: Stem Cell Biology and Tissue Engineering in the Dental Sciences. Vishwakarma A, Shi S, Sharpe P, and Ramalingam M, eds. Elsevier, Philadelphia, PA, pp 445–458 [Google Scholar]
- 2. Diekwisch TG. (2016). Our periodontal tissue: a masterpiece of evolution. J Clin Periodontol 43:320–322 [DOI] [PubMed] [Google Scholar]
- 3. McIntosh JE, Anderton X, Flores-De-Jacoby L, Carlson DS, Shuler CF. and Diekwisch TG. (2002). Caiman periodontium as an intermediate between basal vertebrate ankylosis-type attachment and mammalian “true” periodontium. Microsc Res Tech 59:449–459 [DOI] [PubMed] [Google Scholar]
- 4. Luan X, Walker C, Dangaria S, Ito Y, Druzinsky R, Jarosius K, Lesot H. and Rieppel O. (2009). The mosasaur tooth attachment apparatus as paradigm for the evolution of the gnathostome periodontium. Evol Dev 11:247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luan X, Ito Y. and Diekwisch TG. (2006). Evolution and development of Hertwig's epithelial root sheath. Dev Dyn 235:1167–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diekwisch TG. (2001). The developmental biology of cementum. Int J Dev Biol 45:695–706 [PubMed] [Google Scholar]
- 7. Diekwisch TG. (2002). Pathways and fate of migratory cells during late tooth organogenesis. Connect Tissue Res 43:246–256 [PubMed] [Google Scholar]
- 8. Luan X, Dangaria S, Ito Y, Walker CG, Jin T, Schmidt MK, Galang MT. and Druzinsky R. (2009). Neural crest lineage segregation: a blueprint for periodontal regeneration. J Dent Res 88:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dangaria SJ, Ito Y, Luan X. and Diekwisch TG. (2011). Successful periodontal ligament regeneration by periodontal progenitor preseeding on natural tooth root surfaces. Stem Cells Dev 20:1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dangaria SJ, Ito Y, Luan X. and Diekwisch TG. (2010). Differentiation of neural-crest-derived intermediate pluripotent progenitors into committed periodontal populations involves unique molecular signature changes, cohort shifts, and epigenetic modifications. Stem Cells Dev 20:39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Niehrs C. (2012). The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol 13:767–779 [DOI] [PubMed] [Google Scholar]
- 12. Baron R. and Kneissel M. (2013). WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med 19:179–192 [DOI] [PubMed] [Google Scholar]
- 13. Lim WH, Liu B, Cheng D, Williams BO, Mah SJ. and Helms JA. (2014). Wnt signaling regulates homeostasis of the periodontal ligament. J Periodontal Res 49:751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komiya Y. and Habas R. (2008). Wnt signal transduction pathways. Organogenesis 4:68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moon RT, Kohn AD, De Ferrari GV. and Kaykas A. (2004). WNT and β-catenin signalling: diseases and therapies. Nat Rev Genet 5:691–701 [DOI] [PubMed] [Google Scholar]
- 16. Zhong Z, Ethen NJ. and Williams BO. (2014). WNT signaling in bone development and homeostasis. Wiley Interdiscip Rev Dev Biol 3:489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bodine PV, Zhao W, Kharode YP, Bex FJ, Lambert AJ, Goad MB, Gaur T, Stein GS, Lian JB. and Komm BS. (2004). The Wnt antagonist secreted frizzled-related protein-1 is a negative regulator of trabecular bone formation in adult mice. Mol Endocrinol 18:1222–1237 [DOI] [PubMed] [Google Scholar]
- 18. Yao W, Cheng Z, Shahnazari M, Dai W, Johnson ML. and Lane NE. (2010). Overexpression of secreted frizzled-related protein 1 inhibits bone formation and attenuates parathyroid hormone bone anabolic effects. J Bone Miner Res 25:190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Häusler KD, Horwood NJ, Chuman Y, Fisher JL, Ellis J, Martin TJ, Rubin JS. and Gillespie MT. (2004). Secreted frizzled-related protein-1 inhibits RANKL-dependent osteoclast formation. J Bone Miner Res 19:1873–1881 [DOI] [PubMed] [Google Scholar]
- 20. Li CH. and Amar S. (2007). Inhibition of SFRP1 reduces severity of periodontitis. J Dent Res 86:873–877 [DOI] [PubMed] [Google Scholar]
- 21. Gopinathan G, Kolokythas A, Luan X. and Diekwisch TG. (2013). Epigenetic marks define the lineage and differentiation potential of two distinct neural crest-derived intermediate odontogenic progenitor populations. Stem Cells Dev 22:1763–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bodine PV, Stauffer B, Ponce-de-Leon H, Bhat RA, Mangine A, Seestaller-Wehr LM, Moran RA, Billiard J, Fukayama S, Komm BS. and Pitts K. (2009). A small molecule inhibitor of the Wnt antagonist secreted frizzled-related protein-1 stimulates bone formation. Bone 44:1063–1068 [DOI] [PubMed] [Google Scholar]
- 23. Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, Van Wijnen AJ, Stein JL, Stein GS. and Lian JB. (2005). Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem 280:33132–33140 [DOI] [PubMed] [Google Scholar]
- 24. Han N, Zheng Y, Li R, Li X, Zhou M, Niu Y. and Zhang Q. (2014). β-catenin enhances odontoblastic differentiation of dental pulp cells through activation of Runx2. PLoS One 9:e88890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sierra J, Yoshida T, Joazeiro CA. and Jones KA. (2006). The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev 20:586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luan X, Ito Y, Dangaria S. and Diekwisch TG. (2006). Dental follicle progenitor cell heterogeneity in the developing mouse periodontium. Stem Cells Dev 15:595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walker CG, Dangaria S, Ito Y, Luan X. and Diekwisch TG. (2010). Osteopontin is required for unloading-induced osteoclast recruitment and modulation of RANKL expression during tooth drift-associated bone remodeling, but not for super-eruption. Bone 47:1020–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karsenty G. (2001). Minireview: transcriptional control of osteoblast differentiation. Endocrinology 142:2731–2733 [DOI] [PubMed] [Google Scholar]
- 29. Franceschi RT, Xiao G, Jiang D, Gopalakrishnan R, Yang S. and Reith E. (2003). Multiple signaling pathways converge on the Cbfa1/Runx2 transcription factor to regulate osteoblast differentiation. Connect Tissue Res 44:109–116 [PMC free article] [PubMed] [Google Scholar]
- 30. Lian JB. and Stein GS. (2003). Runx2/Cbfa1: a multifunctional regulator of bone formation. Curr Pharm Des 9:2677–2685 [DOI] [PubMed] [Google Scholar]
- 31. Zhou X, Zhang Z, Feng JQ, Dusevich VM, Sinha K, Zhang H, Darnay BG. and de Crombrugghe B. (2010). Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A 107:12919–12924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Renn J. and Winkler C. (2014). Osterix/Sp7 regulates biomineralization of otoliths and bone in medaka (Oryzias latipes). Matrix Biol 34:193–204 [DOI] [PubMed] [Google Scholar]