Abstract
Functionalized copolymers were synthesized and are proposed as hosts of RNA. The copolymers are based on carboxymethyl cellulose and poly-(ethylene glycol)-OH. These copolymers were functionalized with two amino acids, either lysine or histidine, through amide bond formation. The functionalized copolymer was then used to adsorb ribosomal RNA. The RNA loading was based on the nature of the amino acid functionalization of the copolymer. The array of RNA-copolymers was observed to be soft sphere-like, where the density of spheres was a function of the molecular weight of the carboxymethyl cellulose and the nature of the amino acid. Such RNA-copolymer systems are very sensitive to changes in pH.
Introduction
Nanoparticles are already being developed as effective carriers of drugs and gene delivery to target regions of the body that were previously hard to access using traditional drug formulation methods. Through manipulation of their elemental composition, charge, size, and chemical functionalization, it may be possible to target particles to specific organs [1].
Gene therapy uses nucleic acids as a powerful tool to cure genetic deficiencies or diseases that currently have no cure. This includes a number of brain diseases (Alzheimer’s and Parkinson’s disease), viral infections and cancer [2, 3]. Therapeutic RNA (SiRNA, ribozymes, and mRNA) and DNA (plasmid DNA; oligonucleotides) delivery have been limited by a number of factors. Naked, single-stranded RNA, degraded by a nuclease, activates the immune system and is negatively charged to passively cross the cell membrane; it must be able to enter the cell and escape from endosomes [4, 5].
Complex DNA nanostructures can be functionalized with different biomolecules or nanomaterials, such as different nanowires, nanoparticles, organic molecules, peptides or proteins, to combine the properties of both DNA and nanomaterials for achieving the aimed functionality [6]. Nucleic acids require encapsulation, protection and stability in nanosized carriers by using viral or nonviral vectors that enable efficient intracellular delivery [6]. The use of nonviral vectors is gaining attention due to their low immunogenicity compared with that of viral vectors [7]. Various nonviral vectors, such as cationic polymers, including polylysine and polyamidoamine, are used to electrostatically balance the negatively charged RNA or DNA; however, excess cationic components cause adverse reactions, such as platelet aggregation and inflammatory reactions [8, 9]. Dendrimers, gold nanoparticles, quantum dots and methacrylate/methacrylamide polymers have also been proposed [10–12]. However, it is necessary to improve these materials and develop new materials to avoid gene delivery problems. Additionally, it is important to consider the protection of DNA, ease of fabrication, ability to target specific cell types, inexpensive synthesis, facile purification, stability, internalization, endolysosomal escape, efficient unpackaging, nontoxicity, and nonimmunogenicity [13].
Given their high degree of chemical flexibility, polymers are commonly used materials for nanoparticle-based delivery [14, 15]. Polysaccharides are used for pharmaceutical and biomedical applications due to their biocompatibility and nonimmunogenic properties. Currently, there is growing interest in applying these polymers for the development of nanomedicines [16]. In this sense, carboxymethyl cellulose sodium salt (CMC) is a derivate of cellulose used in food, cosmetics and pharmaceutical products due to its high biocompatibility, biodegradability and low immunogenicity [17]. Another advantage is the ease of chemical modification due to the availability of various functional groups on the glycosidic units (hydroxyls, carboxylic acids) [18].
Furthermore, a polymer coating with polyethylene glycol (PEG) provides protection from uptake by human monocytes. Surface modification of nanoparticles with PEG prolongs the circulation time of the nanoparticles, temporarily avoiding the mononuclear phagocyte system [19]. This polymer is amphiphilic and soluble in water as well as in many organic solvents. This polymer is nontoxic and is approved by the U.S. Food and Drug Administration for use in different pharmaceutical formulations, cosmetics and foods [20].
The nucleic acid nanocarriers are usually captured by the cells and internalized via an endocytic uptake mechanism [21]; they should preferably escape from the endosomes in the cell cytoplasm to avoid nucleic acid degradation and release the encapsulated or complexed nucleic acid. Several strategies have been explored to improve the escape from endosomes such as the incorporation of fusogenic agents. The incorporation of histidine and imidazole into cationic polymers, cationic lipids or peptides has led to nucleic acid delivery [22]. Lipids/peptides in liposomes and polyplexes, with the introduction of an ionizable group, are efficient systems to generate a proton sponge effect inside endosomes [23, 24]. pH-responsive compounds, such as amino acids, have been incorporated into nanocarriers (conjugated) to achieve efficient intracellular delivery of complexed nucleic acid through electrostatic interactions. In this study, copolymers of carboxymethyl cellulose-polyethylene glycol were prepared and functionalized with histidine and lysine for use as carriers of ribosomal RNA.
Experimental procedures
Carboxymethyl cellulose [CEKOL 700 (7CMC), MW-270,000; CEKOL 30 (3CMC), MW-80,000; degree of substitution (DS) = 0.82] was obtained from CPkelco. Hydrochloric acid, acetonitrile, diethyl ether, methanol, poly(ethylene glycol) methyl ether (mPEG-OH, MW = 2000), 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide HCl (EDC HCl), N-hydroxysuccinimide (NHS), 4-dimethylaminopyridine (DMAP), phosphate-buffered saline (PBS), sodium hydroxide, l-lysine, and l-histidine monohydrochloride monohydrate were purchased from Sigma-Aldrich. Quant-iT RiboGreen RNA Reagent was purchased from Fisher Scientific, Inc.
Materials
Copolymers of carboxymethyl cellulose-polyethylene glycol
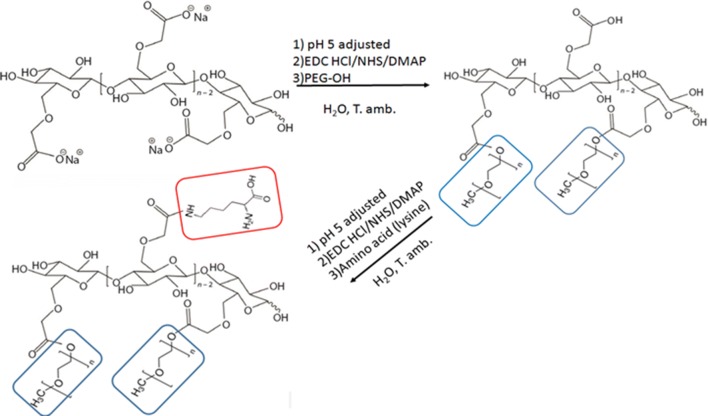
Copolymers of carboxymethyl cellulose-polyethylene glycol were synthesized as follows: CMC (1.2 mmol acid) was dissolved in water (30 ml) and pH adjusted to 5 (HCl solution), to which EDC HCl (1.4 mmol), NHS (1.4 mmol), and DMAP (0.2 mmol) were added. The solution was stirred for 1 h at room temperature under dark conditions. After an hour, PEG-OH (0.36 mmol) reagent was added to the CMC-PEG solution. After overnight reaction, the product was precipitated with methanol and washed with a mixture of acetonitrile and diethyl ether (90:10). It was subsequently dissolved in water and precipitated in methanol, filtered and dried at room temperature. EDC HCl, DMAP, and NHS were dissolved in water (2 ml), and mPEG-OH was dissolved in water (4 ml). Two copolymers were synthesized and labeled 7CMC-PEG or 3CMC-PEG according to the precursor of CMC utilized in synthesis (Scheme 1).
Scheme 1.
Schematic of functionalized copolymer preparation
Functionalization of copolymers with amino acids
Water at pH 5 was used to dissolve 7CMC-PEG or 3CMC-PEG. Then, EDC HCl (1.4 mmol), NHS (1.4 mmol), and DMAP (0.2 mmol) were added. Lastly, after 1 h, 0.36 mmol of amino acid, histidine or lysine was added (Scheme 1).
Preparation of nanoparticles of copolymers
The formulation, composed of copolymers (carboxymethyl cellulose-PEG-amino acid) and Tween 80 solution, was prepared by dissolving 3 mg of copolymer in 2.5 ml of sterile solution. The solution at 6% was prepared by diluting Tween 80 in sterile deionized water. The samples were sonicated for 2 h and centrifuged at 12,000 rpm for 30 min. The nanoparticles were suspended in sterile deionized water, and the size of the nanoparticles was measured by dynamic light scattering.
Characterization
The structures of the materials were characterized by infrared (FTIR) and 13C nuclear magnetic resonance (CP MAS NMR) spectroscopies. FTIR spectra (ATR mode) of samples were acquired using a Bruker Alpha FTIR spectrometer. The spectra were recorded at a resolution of 4 cm−1 in the spectral window 4000–400 cm−1. 13C CP MAS NMR spectra were obtained at a Larmor frequency of 75.4 MHz using a Bruker Avance 300 spectrometer equipped with a 4 mm cross-polarization (CP) MAS probe. The samples were spun at a rate of 5 kHz. Spectra were recorded using a contact time of 5 ms and π/2 pulses of 5 µs. The chemical shifts were referenced to TMS.
The thermal properties of the materials were characterized by thermogravimetric analysis (TGA). TGA profiles were obtained using a TGA 500 system. Samples were heated at a heating rate of 10 °C/min from 30 to 900 °C under a nitrogen atmosphere with a flow rate of 1.0 ml/min.
The average hydrodynamic diameter, zeta potential and polydispersity index (PDI) of the nanoparticles were measured by dynamic light scattering (DLS) analysis using a Zeta sizer Nano ZS90 (Malvern Instruments). All measurements were carried out at 25 °C, at a detection angle of 90° (for size and PDI) and 120° for zeta potential.
Ribosomal RNA onto copolymers
The nanoparticles were suspended in ribosomal RNA solution (2 ml with a 100 ng/ml concentration). The solution was incubated at 8 °C for 10 min. Then, the mixture was centrifuged at 12,000 rpm for 30 min to pellet the nanoparticles. The supernatant was removed and analyzed by the Quant-iT RiboGreen assay (Invitrogen). The amount of ribosomal RNA in the supernatant (w) was then subtracted from the total amount of ribosomal RNA added (w, 100 ng). The experiments were conducted in triplicate. The percentage efficiency (E) of ribosomal RNA entrapment to the nanoparticles was calculated using the following formula:
Measurement data were expressed as the mean ± standard deviation; the comparison between two groups was analyzed using a two-sample t-test. Statistical analysis was performed using GraphPad Prism ver. 5.0 (GraphPad software, USA). P values ≤ 0.05 were considered statistically significant.
The nanoparticles loaded with ribosomal RNA were suspended in sterile phosphate-buffered saline (PBS) at a pH of 5, 6 and 7.4. The pH of the PBS was modified using 0.1 M NaOH or 0.1 M HCl. The mixture was then sonicated for 1 h. The nanoparticles were stored at 8 °C for periods as long as 1 week. The particle size and zeta potential of polymer complexes were measured at various time points (1 h, 24 h and 1 week).
Array of components of hybrid copolymer-RNA
Small-angle X-ray scattering profiles were obtained using a Kratky camera coupled to a copper anode tube. The distance between the sample and the linear proportional counter was 25 cm; a Ni filter selected the Cu KR radiation. The sample was introduced into a capillary tube. The intensity I(q) was measured for 9 min to obtain high-quality statistics. The SAXS data were processed with the ITP program [25–27], where the scattering angle (q) is defined as q = 2πsinθ/λ, where θ and λ are the X-ray scattering angle and the wavelength, respectively. The shape and size distribution function of the scattering objects was estimated from the Kratky plot, q2I(q) against q. Lastly, the fractal dimension of the scattering objects was estimated from the slope of the curve log I(q) vs log(q) [28, 29].
Results and discussion
Functionalization of copolymers
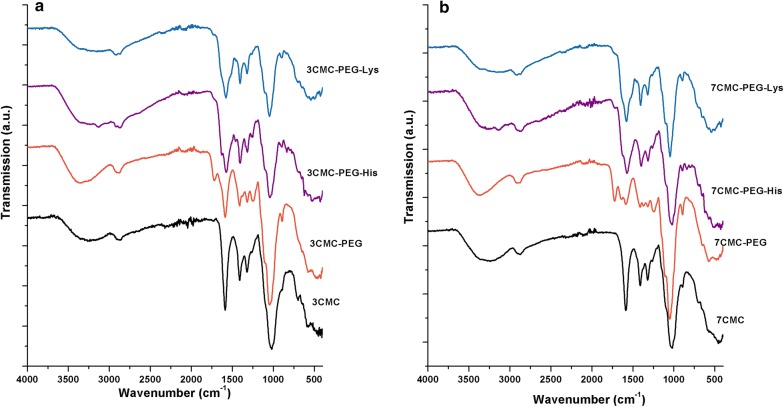
The FTIR spectra of two polymers functionalized with amino acids are shown in Fig. 1. The spectra of carboxymethyl celluloses are also included as a reference. The spectra of both celluloses, 3CMC and 7CMC, present a broad absorption band at 3422 cm−1, which is due to the stretching vibrational mode of the hydroxyl group. The band at 2924 cm−1 is assigned to the νC-H stretching vibration. The absorption band at 1613 cm−1 is assigned to ν C=O present in carboxyl group. The band at 1062 cm−1 is due to C–O–C stretching. The spectra of copolymers 3CMC-PEG and 7CMC-PEG fit relatively well with that of celluloses; however, it must be emphasized that the bands at 1740 cm−1 for 3CMC-PEG and at 1742 cm−1 for 7CMC-PEG are ascribed to the C=O stretching mode of the ester groups [30, 31]. These bands confirm that polyethylene glycol (PEGOH) had been linked to sodium carboxymethyl cellulose. The spectra of copolymers functionalized with histidine and lysine, 3CMC-PEG-His and 3CMC-PEG-Lys, did not show significant differences from that of 3CMC. A similar behavior was observed with the spectra of functionalized copolymers 7CMC. To obtain complementary information about the structure of these materials, solid-state 13C NMR measurements were performed.
Fig. 1.
FTIR spectra of copolymers, a spectra of 3CMC and 3CMC functionalized with PEG and amino acids (lysine and histidine). b Spectra of 7CMC functionalized with PEG and amino acids
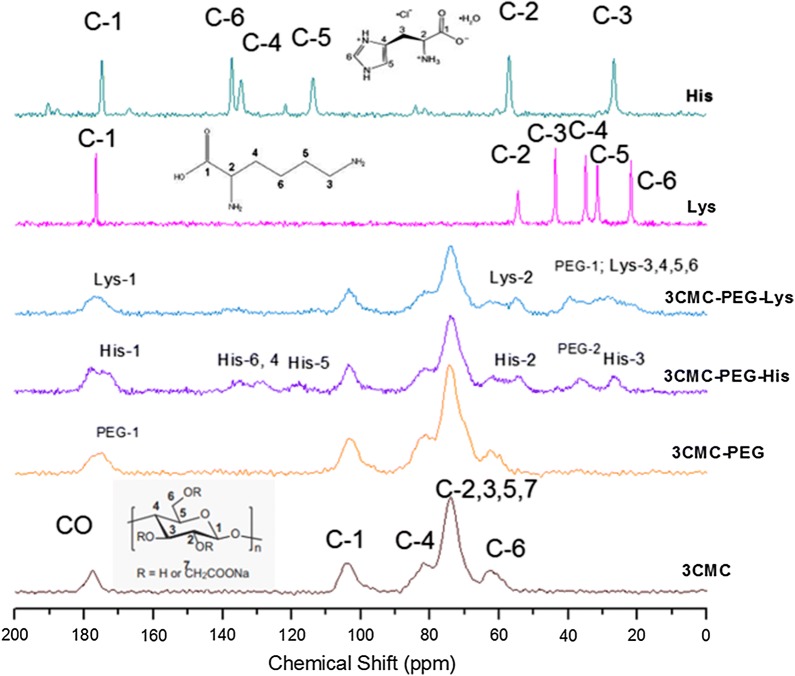
Figure 2 displays the 13C CP MAS NMR spectra of celluloses and amino acids as references and their corresponding copolymers. In the spectra of 3CMC and 7CMC, the broad NMR peaks at 178, 104, 83, 75, and 63 ppm were assigned to the carbonyl carbons, C1 and C4, and the overlap of C2, C3, C5, C7, and C6, respectively [32]. The peaks due to histidine were resolved at 174.94, 137.36, 134.42, 67.06 and 26.86 ppm. The assignment of these peaks is performed for the corresponding spectrum according to the number of carbons in Fig. 2.
Fig. 2.
13C CP MAS NMR spectra of carboxymethylcellulose (low molecular weight, 3CMC) and carboxymethylcellulose functionalized with PEG and amino acids (lysine, histidine)
The spectra of 3CMC-PEG and 7CMC-PEG exhibit a new signal close to 177 ppm, in addition to the resonances of celluloses. This peak suggests that the –COOH groups of carboxymethyl cellulose reacted with the hydroxyl groups of PEGOH to form the ester crosslink. In the spectrum of 3CMC-PEG-His, Fig. 2, the peaks between 110 and 140 ppm, assigned to aromatic carbons, are broader, and a downfield shift was observed for carbons C-2 (carbon bonded to amine in histidine). Furthermore, an additional peak at 174 ppm was observed. These observations can be explained as the aromatic ring of histidine interacting with the 3CMC-PEG surface and CMC-PEG reacting with the amino groups (C-2) of histidine to form the amide link. Regarding the spectra of copolymers containing lysine, the broadness of aliphatic carbons C3, C4, C5, and C6 indicates that lysine interacts with the copolymer through the amine at the end of chain, reducing the motion of four carbons.
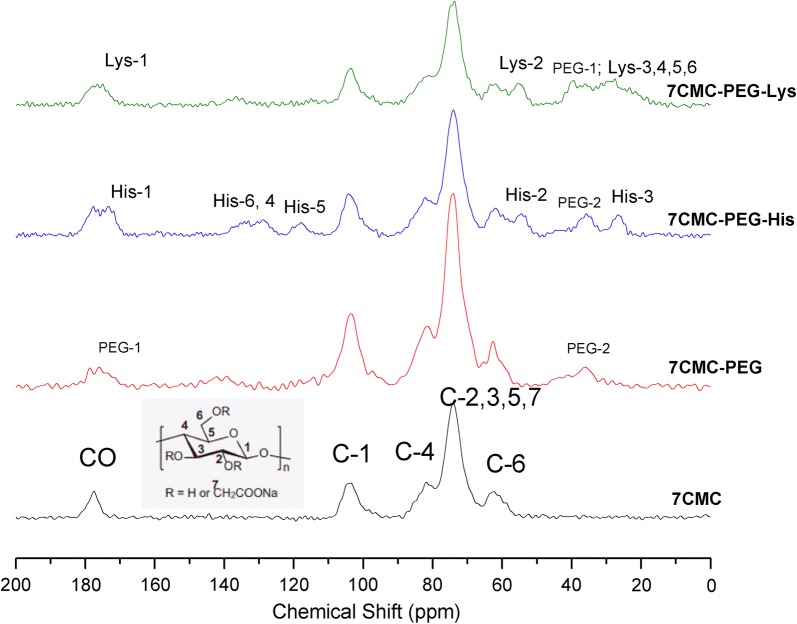
The 13C CP MAS NMR spectra in Fig. 3 correspond to samples synthesized with 7CMC, and a similar behavior to the series of samples with 3CMC was observed.
Fig. 3.
13C CP MAS NMR spectra of carboxymethylcellulose (high molecular weight, 7CMC) and carboxymethylcellulose functionalized with PEG and amino acids (lysine, histidine)
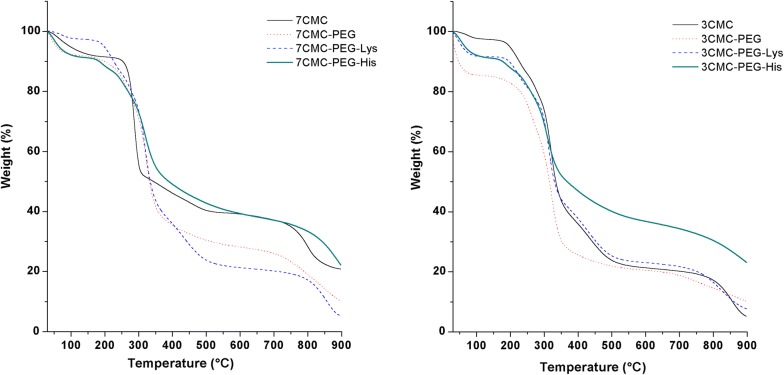
The TG curves of copolymers prepared from 7CMC are shown in Fig. 4. The degradation of polymers occurs in two steps. The first TG step (2–15% mass loss), due to the loss of adsorbed water, occurs in the range from 30 to 150 °C. The second TG step occurs in the 250–460 °C temperature range, with a mass loss (30–40%). This step is attributed to the degradation of the side chain and the loss of CO2 [33, 34]. The copolymers began to degrade at approximately 150 °C, and the final decomposition temperature was found at approximately 450 °C. The higher decomposition temperature indicates that copolymers are more thermally stable than 7CMC and 3CMC. Table 1 compares the degradation temperatures and percentage mass for individual polymers and their blends. The TGA profiles of copolymers with PEG or amino acids show more than two degradation steps. The two mass losses between 150 and 260 can be attributed to the loss of CO2, –OCH3 groups of PEG and –NH2–R groups.
Fig. 4.
TGA curves of copolymers and polymers 7CMC (left), 3CMC (right)
Table 1.
TG data for carboxymethycellulose and copolymers
| Sample | Decomposition step | Temperature (°C) | Weight loss % | |
|---|---|---|---|---|
| Start | End | |||
| 7CMC | 1st | 30 | 175 | 8 |
| 2nd | 175 | 350 | 42 | |
| 7CMC-PEG | 1st | 30 | 150 | 8 |
| 2nd | 150 | 280 | 14 | |
| 3rd | 280 | 420 | 43 | |
| 7CMC-PEG-His | 1st | 30 | 150 | 8 |
| 2nd | 150 | 215 | 5 | |
| 3rd | 215 | 270 | 10 | |
| 4th | 270 | 450 | 30 | |
| 7CMC-PEG-Lys | 1st | 30 | 150 | 2 |
| 2nd | 150 | 250 | 13 | |
| 3rd | 250 | 280 | 9 | |
| 4th | 280 | 400 | 40 | |
| 3CMC | 1st | 30 | 150 | 2 |
| 2nd | 150 | 255 | 14 | |
| 3rd | 250 | 380 | 40 | |
| 3CMC-PEG | 1st | 30 | 150 | 15 |
| 2nd | 150 | 285 | 20 | |
| 3rd | 285 | 450 | 40 | |
| 3CMC-PEG-His | 1st | 30 | 150 | 8 |
| 2nd | 150 | 250 | 8 | |
| 3rd | 250 | 382 | 39 | |
| 3CMC-PEG-Lys | 1st | 30 | 150 | 8 |
| 2nd | 150 | 210 | 6 | |
| 3rd | 210 | 270 | 8 | |
| 4th | 270 | 460 | 34 | |
Particle size distribution
The data reported in Table 2 include the particle size, the polydispersity index (PDI) and the zeta-potential values. The particle size ranged between 9 and 14 nm. Particles of copolymers prepared with 7CMC were smaller than those prepared from 3CMC, suggesting that molecular weight is an important parameter influencing the crosslinking between cellulose and polyethylene glycol. Loading amino acids induces an increase in particle size; however, no direct correlation was observed between amino acids and increasing particle size. It should be mentioned that particles of copolymers containing 7CMC were the materials with the largest PDI values. In fact, a width of 30% PDI is often considered the frontier between mono- and polydisperse. This corresponds to PDI equal to 0.09, which leads to the recommendation that PDI < 0.1 is monodisperse and PDI > 0.1 is multimodal. Thus, all the prepared copolymers are multimodal, except 3CMC-PEG-Hys, and these results are in line with previous results obtained for other polymeric systems [35–37]. Lastly, a remarkable difference was observed between polymers containing 3CMC and 7CMC. When amino acids are loaded onto copolymers with 7CMC, the Z-potential becomes more positive with respect to that of amino acid-free copolymers. In contrast, the opposite trend was observed for the series containing 3CMC. This result can be explained as the degree of substitution between 7CMC and 3CMC polymeric chains being very different because the substitution groups drive the surface interactions, which in turn determine the electrostatic charge.
Table 2.
Particle size (diameter), polydispersity index (PDI) and Zeta potential of samples under study as determined by DLS data
| Sample | Without ribosomal RNA | Loaded with ribosomal RNA | ||||
|---|---|---|---|---|---|---|
| Particle size ± SD (nm) | PDI | Zeta potential (mV) | Particle size (nm) | PDI | Zeta potential (mV) | |
| 7CMC-PEG | 9.83 ± 0.12 | 0.151 | − 25.3 | 12.98 ± 0.07 | 0.22 | − 25.9 |
| 7CMC-PEGLys | 12.24 ± 0.07 | 0.34 | − 25.06 | 9.14 ± 0.11 | 0.16 | − 17.6 |
| 7CMC-PEGHis | 11.39 ± 0.075 | 0.25 | − 24.66 | 11.27 ± 0.09 | 0.22 | − 20.9 |
| 3CMC-PEG | 11.2 ± 0.035 | 0.25 | − 3.22 | 14.18 ± 0.05 | 0.27 | − 27.6 |
| 3CMC-PEGLys | 13.01 ± 0.039 | 0.099 | − 5.95 | 11.92 ± 0.15 | 0.22 | − 19.9 |
| 3CMC-PEGHis | 13.9 ± 0.049 | 0.175 | − 9.84 | 13.96 ± 0.02 | 0.23 | − 23.4 |
RNA as guest in CMC-PEG-amino acid hosts
The histogram of Fig. 5 plots the amount of RNA adsorbed on different copolymers. The loaded RNA amount is expressed as a percentage of the RNA retained by copolymers from the total amount put in contact with particles. The highest fraction of RNA was loaded in both copolymers functionalized with lysine. The copolymer retaining the lowest fraction of RNA was 3CMC-PEG-His. The statistical parameters confirmed the efficiency of copolymers containing lysine. Compared to that in 7CMC-PEG, the incorporation efficiency in 7CMC-Lys was significantly increased (t = 21.94. P = 0.0001), as was the incorporation efficiency of 3 CMC-PEG LYS compared to that of 3 CMC PEG (t = 7.236, P = 0.0019).
Fig. 5.
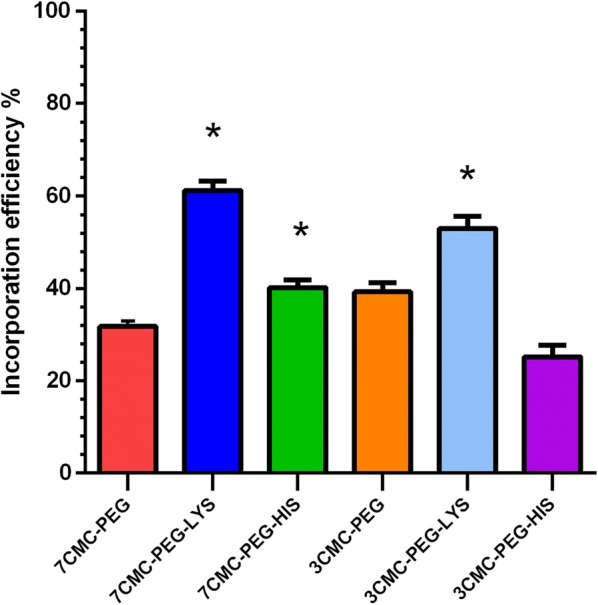
Ribosomal RNA (Maximal loading) on different copolymers. Experiments were performed by triplicate and standard deviation included in graphs
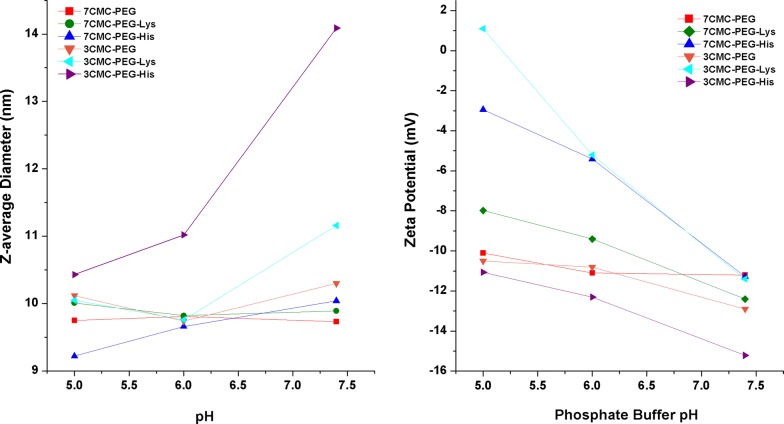
RNA loading, of course, changes the particle size, as shown in Table 2. In both amino acid-free copolymers, the particle size increases approximately 3 nm after RNA incorporation. The PDI value increases slightly upon RNA loading on both 7CMC-PEG and 3CMC-PEG copolymers. This result suggests that as a consequence of RNA loading, particles have a broader molecular weight distribution. This effect was observed earlier in polymeric systems where the RNA loading did not reach a saturation level [38]. The size does not increase with RNA adsorption onto all amino acid-copolymer systems. The copolymers functionalized with histidine had almost the same particle size before and after the RNA loading. The size of particles containing lysine decreases by approximately 2 nm, which is an unexpected result explained as follows: the acidic hydrogen permits a strong interaction between the functionalized surface of the copolymer and RNA. This compactness (or condensation) behavior was also previously observed in other polymeric systems [39, 40], and it will be confirmed below by SAXS results. Furthermore, the presence of RNA on particles significantly changes the Z-potential values. With RNA, the Z-potential becomes more positive for samples prepared from 7CMC and more negative for samples prepared from 3CMC. Before RNA loading, the Z-potential value was very different between samples containing 3CMC and that prepared from 7CMC. However, this difference was clearly lessened after RNA loading. Interestingly, the most positive materials containing RNA are those using lysine in functionalization, and the most negative are those free of amino acids. RNA is anchored to the surface of copolymers, but the surface properties of copolymers are crucial to the resulting hybrid RNA-copolymer materials. Because of these results, it is expected that materials differ significantly in response to changes in pH. The incorporation of pH-sensitive amino acids onto copolymers enables controlled endolysosomal escape. In this context, Fig. 6a plots the particle size of RNA-copolymers as a function of pH. The size of the nanoparticles decreases when the pH becomes acidic. At pH 7.4, the carboxyl groups are present in the form –COOH, and free amino groups (the other amino group forms the ester crosslink with CMC) are present in the protonated form, NH3+. Therefore, the electrostatic interaction between RNA and polymers becomes significantly favorable between the amino group and the negatively charged RNA, Scheme 2; consequently, the size of the particle decreases. No critical changes in the size of 7CMC-PEG and 3CMC-PEG were observed when the pH changed due to the absence of amino acids that give ionizable properties to the polymer.
Fig. 6.
Size of particle and Zeta potential of copolymers as a function of pH variations. Copolymers were stored in phosphate-buffered solutions at pH 5.0, 6.0 and pH 7.4
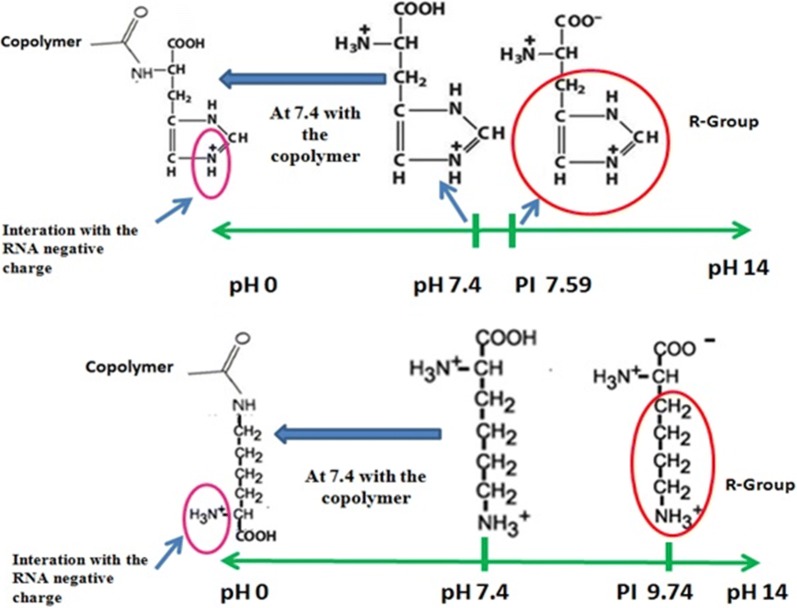
Scheme 2.
Isoelectric point of aminoacids anchored to copolymers. The carboxyl groups have a negative charge (− 1). But one-half of the amino groups and one-half of the R-Groups (–NH3+) have a positive charge, so the sum of their charge is + 1. At pH lower than IP, amine and carboxyl groups are protonated and at values higher than IP, ionizable groups donate protons
To deduce the stability of particles, they were suspended in water at different pH values, and the particle size was measured over time. The results are summarized in Table 3. It is remarkable that at the same pH value, only slight changes were observed for the copolymers functionalized with amino acids, i.e., they were stable for periods as long as 1 week. At same time, the pH value is a parameter that also slightly influences the particle size, but no simple correlation was observed. The samples without amino acids, 7CMC-PEG and 3CMC-PEG, form agglomerates for periods as long as 24 h. It should be concluded that the incorporation of amino acids in the polymer chains generates stability in the aqueous medium.
Table 3.
Particle size (diameter (nm)) of RNA-copolymers under different conditions of pH
| Sample | Deionized water pH 7 | pH 5 | pH 6 | pH 7.4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 h | 24 h | 1 week | 1 h | 24 h | 1 week | 1 h | 24 h | 1 week | 1 h | 24 h | 1 week | |
| 7CMC-PEG | ||||||||||||
| Size | 12.98 | 29.09 | 50.72 | 9.75 | 9.49 | 9.74 | 9.81 | 11.72 | 120.86 | 9.73 | 11.51 | 10.32 |
| PDI | 0.22 | 0.2 | 0.38 | 0.05 | 0.1 | 0.16 | 0.09 | 0.28 | 0.34 | 0.06 | 0.24 | 0.16 |
| 7CMC-PEGLys | ||||||||||||
| Size | 9.34 | 8.97 | 9.53 | 10.01 | 10.04 | 10.02 | 9.82 | 9.81 | 9.89 | 9.89 | 45.95 | 10.04 |
| PDI | 0.16 | 0.14 | 0.15 | 0.13 | 0.19 | 0.13 | 0.1 | 0.16 | 0.09 | 0.13 | 0.15 | 0.18 |
| 7CMC-PEGHis | ||||||||||||
| Size | 11.27 | 11.94 | 13.71 | 9.22 | 9.78 | 9.88 | 9.66 | 9.75 | 9.98 | 10.04 | 9.96 | 10.09 |
| PDI | 0.22 | 0.15 | 0.32 | 0.12 | 0.09 | 0.1 | 0.07 | 0.11 | 0.13 | 0.08 | 0.16 | 0.14 |
| 3CMC-PEG | ||||||||||||
| Size | 14.18 | 26.37 | 176.3 | 10.12 | 10.17 | 10.28 | 9.74 | 10.39 | 9.96 | 10.3 | 10.01 | 11.65 |
| PDI | 0.27 | 0.16 | 0.28 | 0.12 | 0.16 | 0.16 | 0.08 | 0.2 | 0.13 | 0.31 | 0.22 | 0.31 |
| 3CMC-PEGLys | ||||||||||||
| Size | 11.92 | 12.12 | 12.31 | 10.05 | 11.52 | 9.67 | 9.76 | 9.89 | 10.54 | 11.16 | 119.1 | 59.75 |
| PDI | 0.22 | 0.24 | 0.22 | 0.08 | 0.22 | 0.06 | 0.07 | 0.09 | 0.15 | 0.12 | 0.23 | 0.18 |
| 3CMC-PEGHis | ||||||||||||
| Size | 13.96 | 15.39 | 14.62 | 10.43 | 10.58 | 10.41 | 11.02 | 13.62 | 11.0 | 14.09 | 10.04 | 10.51 |
| PDI | 0.23 | 0.4 | 0.45 | 0.17 | 0.16 | 0.18 | 0.24 | 0.35 | 0.26 | 0.21 | 0.21 | 0.25 |
Interestingly, the particle size of the sample with lysine (7CMC-PEG-Lys) remained stable from pH 7.4 to acidic pH. This can be attributed to the value of the isoelectric point (pH where the molecule has a net charge of zero), in the case of lysine, which is 9.74. At pH 7.4, it is probable that the amino groups and the carboxyl group are completely protonated. The histidine has an isoelectric point of 7.59. At physiological pH, it is probable that not all the –COOH and NH2 groups of the copolymer are protonated, Scheme 2.
In Fig. 6, the zeta potential measured in different copolymers stored at different pH values is plotted. The polymers presented a negative charge under a physiological environment (pH 7.4), but the surface charge became slightly more positive at lower pH. The change in zeta potential can be attributed to protonation of the amine group under low-pH conditions. The surface charges of polymers play a role in cell uptake and blood stability. Generally, positively charged polymers promote cellular uptake due to the greater affinity for negatively charged cell membranes, but they tend to undergo rapid clearance from blood due to the strong interaction with the serum. Furthermore, negatively charged polymers can resist protein absorption, leading to a longer blood circulation time [41].
In summary, the results suggest that changes in the pH allow the polymer complexes to escape endosomal entrapment at acidic pH values (5 and 6). Thus, as a consequence of the protonation of carboxyl and amine groups, the polymers could reversibly convert surface charges in response to pH, facilitating endosome escape and cellular uptake. The pH dependence of the charge could be modulated by varying the molar ratio of carboxyl or amino groups.
Array of the RNA-copolymer hybrid system
Figure 7 shows SAXS profiles, more precisely the Kratky modified plots [I(q) × q5/3 vs q], of the RNA-copolymers. The modified Kratky plots have a main peak as well as a number of secondary peaks, and these profiles indicate that scattered objects have a globular shape. No significant differences concerning the shape of scattered particles were observed among the copolymers under study. The copolymer particle sizes were estimated from the SAXS data curves by fitting to a poly-disperse system of spheres. Table 4 shows that the size of RNA-copolymers ranges between 12.42 and 13.88 nm. The highest particle size was achieved when copolymers were prepared from 3CMC, which is in line with results obtained from DLS data. In general, RNA-copolymers functionalized with histidine are larger than those functionalized with lysine, confirming that protonation of amino acids is an important parameter for determining the interaction of copolymers with RNA. In this context, the fractal dimension value is a useful parameter because the loading of RNA onto copolymers without amino acids leads to spherical dense particles. However, the fractal dimension decreases when RNA is incorporated into the copolymers functionalized with amino acids, i.e., the particle size increases but the density decreases. Among the two functionalized amino acids, the polymers containing histidine lead to the most and least dense RNA copolymers; in other words, the spheres are not “hard” but respond to interactions of amino acids with copolymers and with RNA.
Fig. 7.
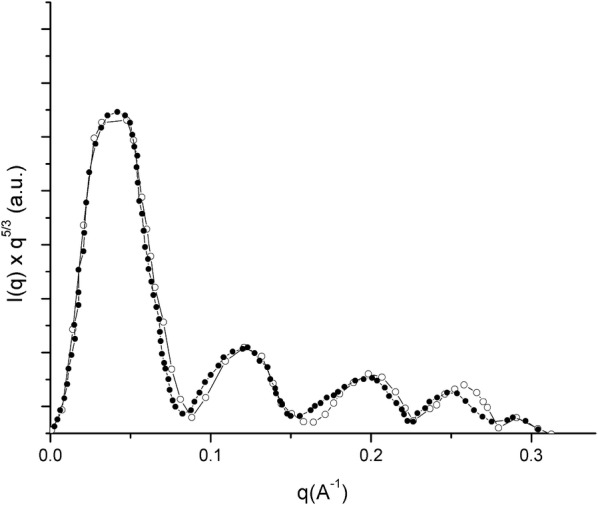
Modified Kratky profile for RNA loaded in 7CMC-PEGLys (empty circles) and, 7CMC-PEGHis (filled circles)
Table 4.
Particle size and fractal dimension of RNA-copolymer particles as determined by SAXS data
| Sample | Loaded with ribosomal RNA | |
|---|---|---|
| Particle size (nm) | Fractal dimension | |
| 7CMC-PEG | 12.42 | 2.81 |
| 7CMC-PEGLys | 12.61 | 2.76 |
| 7CMC-PEGHis | 12.97 | 2.62 |
| 3CMC-PEG | 13.01 | 2.89 |
| 3CMC-PEGLys | 13.14 | 2.73 |
| 3CMC-PEGHis | 13.88 | 2.66 |
Conclusion
PEGylated link polymers of carboxymethyl cellulose were effectively synthesized by EDC-NHS reaction and coupling amino acids onto the side groups of CMC-PEG copolymers. Copolymers were formed as stable soft spheres. Functionalization with lysine and histidine is crucial to obtaining copolymers prone to adsorbing ribosomal RNA; the type of amino acid determines the electric charge on the external surface of the spherical copolymer where RNA is incorporated. RNA-copolymers are partially stable as the amino acid-RNA interactions vary with pH, which could facilitate the release of RNA; this is a promising result because this RNA-copolymer system could then be used for RNA delivery.
Authors’ contributions
MH and GL conducted the preparation of copolymers and physicochemical characterization; MH and AG achieved the functionalization of copolymers; MH and JM performed the RNA loading onto copolymers. VL and CF performed SAXS characterization. MH and EL: Designed the experiments and manuscript writing. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to G. Cedillo and A. Tejeda for their technical assistance.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All the data are mentioned. Not applicable.
Funding
This work was financially supported by CONACYT (Grant 220436) and PAPIIT-IN106517.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Parlea L, Puri A, Kasprzak W, Bindewald E, Zakrevsky P, Satterwhite E, Joseph K, Afonin KA, Shapiro BA. Cellular delivery of RNA nanoparticles. ACS Comb Sci. 2016;18:527–547. doi: 10.1021/acscombsci.6b00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchida S, Kinoh H, Ishii T, Matsui A, Tockary T, Takeda K, Uchida H, Osada K, Itaka K, Kataoka K. Systemic delivery of messenger RNA for the treatment of pancreatic cancer using polyplex nanomicelles with a cholesterol moiety. Biomaterials. 2016;82:221–228. doi: 10.1016/j.biomaterials.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 3.Zheng X, Pang X, Yang P, Wan X, Wei Y, Guo Q, Zhang Q, Jiang X. A Hybrid siRNA delivery complex for enhanced brain penetration and precise amyloid plaque targeting in Alzheimer’s disease mice. Acta Biomater. 2017;49:388–401. doi: 10.1016/j.actbio.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Lu Z, Wientjes G, Au J. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Zhao Y, Zhao X, Lee R, Teng L, Zhou C. Enhancing the therapeutic delivery of oligonucleotides by chemical modification and nanoparticles encapsulation. Molecules. 2017;22:1724. doi: 10.3390/molecules22101724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Arrabito G. Hybrid, multiplexed, functional DNA nanotechnology for bioanalysis. Analyst. 2015;140:5821–5848. doi: 10.1039/C5AN00861A. [DOI] [PubMed] [Google Scholar]
- 7.Hardee C, Arevalo L, Hornstein B, Zechiedrich L. Advances in non-viral DNA vectors for gene therapy. Genes. 2017;8:65. doi: 10.3390/genes8020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai W. Cyclodextrins in non-viral gene delivery. Biomaterials. 2014;35:401–411. doi: 10.1016/j.biomaterials.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kodama Y, Kuramoto H, Mieda Y, Muro T, Nakagawa H, Kurosaki T, Sakaguchi M, Nakamura T, Kitahara T, Sasaki H. Application of biodegradable dendrigraft poly-l-lysine to a small interfering RNA delivery system. J Drug Target. 2017;25:49–57. doi: 10.1080/1061186X.2016.1184670. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez B, Howard K. Polycation based nanoparticle delivery of RNAi therapeutics: adverse effects and solutions. Adv Drug Deliv Rev. 2012;64:1717–1729. doi: 10.1016/j.addr.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Swati B, Torchilin V. Dendrimers for SiRNA delivery. Pharmaceuticals. 2013;6:161–183. doi: 10.3390/ph6020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding Y, Jiang Z, Saha K, Chang S, Sung T, Ryan L, Rotello V. Gold nanoparticles for nucleic acid delivery. Mol Ther. 2014;22:1075–1083. doi: 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Getz T, Mendintz I, Delehanty J, Susumu K, Dawson P, Dawson G. Quantum Dot-mediated delivery of siRNA to inhibit sphingomyelinase aCtivities in brain-derived cells. J Neuro Chem. 2016;139:872–885. doi: 10.1111/jnc.13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pack D, Hoffman A, Pun S, Staylon P. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 15.Kamaly N, Yameen B, Wu Y, Farokhzad OC. Degradable controlled-release polymers and polymeric nanoparticles: mechanisms of controlling drug release. Chem Rev. 2016;116:2602–2663. doi: 10.1021/acs.chemrev.5b00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remdonck K, Martens T, Braeckmans K, Demeester J, Smedt S. Polisaccharide-based nucleic acid nanoformulations. Adv Drug Deliv Rev. 2013;65:1123–1147. doi: 10.1016/j.addr.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Kono H. Characterization and properties of carboxymethyl cellulose hydrogels crosslinked by polyethylene glycol. Carbohydr Polym. 2014;106:84–93. doi: 10.1016/j.carbpol.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Fekete T, Borsa J, Takács E, Wojnárovits L. Synthesis of carboxymethylcellulose/acrylic acid hydrogels with superabsorbent properties by radiation-initiated crosslinking. Radiat Phys Chem. 2016;124:135–139. doi: 10.1016/j.radphyschem.2015.09.018. [DOI] [Google Scholar]
- 19.Nelson C, Kintzing J, Hanna A, Shannon J, Gupta M, Duvall C. Balancing cationic and hydrophobic content of PEGylated siRNA polyplexes enhances endosome escape, stability, blood circulation time, and bioactivity in vivo. ACS Nano. 2013;7:8870–8880. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuertges F, Abuchowski A. The clinical efficacy of poly(ethylene glycol)-modified proteins. J Control Release. 1990;11:139–148. doi: 10.1016/0168-3659(90)90127-F. [DOI] [Google Scholar]
- 21.Ma D. Enhancing endosomal scape for nanoparticles. Nanoscale. 2014;6:6415–6425. doi: 10.1039/c4nr00018h. [DOI] [PubMed] [Google Scholar]
- 22.Hakamatani T, Asayama S, Kawakami H. Synthesis of alkylated poly(1-vinylimidazole) for a new pH-sensitive DNA carrier. Nucleic Acids Symp Ser (Oxf). 2018;52:677–678. doi: 10.1093/nass/nrn342. [DOI] [PubMed] [Google Scholar]
- 23.Malamas S, Gujrati M, Kummitha CM, Xu R, Lu ZR. Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery. J Control Release. 2013;171:296–307. doi: 10.1016/j.jconrel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benns JM, Choi J, Mahato R, Park J, Kim S. pH sensitive cationic polymers gene delivery vehicle: N-Ac-poly (l-histidine)-graft-poly(l-lysine)comb shaped polymer. Bioconjug Chem. 2000;5:637–645. doi: 10.1021/bc0000177. [DOI] [PubMed] [Google Scholar]
- 25.Glatter O. Data evaluation in small angle scattering: calculation of the radial electron density. Acta Phys Aust. 1977;47:83–102. [Google Scholar]
- 26.Glatter O. A new method for the evaluation of small-angle scattering data. J Appl Crystallogr. 1977;10:415–421. doi: 10.1107/S0021889877013879. [DOI] [Google Scholar]
- 27.Glatter O. Evaluation of small-angle scattering data from lamellar and cylindrical particles by the indirect transformation method. J Appl Crystallogr. 1980;13:7–577. doi: 10.1107/S0021889880011429. [DOI] [Google Scholar]
- 28.Guinier A, Fournet G. Small angle scattering of X-rays. New York: Wiley; 1955. [Google Scholar]
- 29.Glatter O, Kratky O. Small angle X-ray scattering. New York: Academic Press; 1982. [Google Scholar]
- 30.Biswall D, Singh R. Characterisation of carboxymethyl cellulose and polyacrylamide graft copolymer. Carbohydr Polym. 2004;57:379–387. doi: 10.1016/j.carbpol.2004.04.020. [DOI] [Google Scholar]
- 31.Barbucci R, Magnani A, Consumis M. Swelling behavior of carboxymethylcellulose hydrogels in relation to cross-linking, pH, and charge density. Macromolecules. 2000;33:7475–7480. doi: 10.1021/ma0007029. [DOI] [Google Scholar]
- 32.Kono H, Onishi K, Nakamura T. Characterization and bisphenol adsorption capacity of β-cyclodextrin–carboxymethylcellulose-based hydrogels. Carbohydr Polym. 2013;15:784–792. doi: 10.1016/j.carbpol.2013.06.065. [DOI] [PubMed] [Google Scholar]
- 33.Fang Y, Gang L, Yan-Gang H, Feng-Xia R, Gui-Xiang W. Synthesis, characterization, and applied properties of carboxymethyl cellulose and polyacrylamide graft copolymer. Carbohydr Polym. 2009;78:95–99. doi: 10.1016/j.carbpol.2009.04.004. [DOI] [Google Scholar]
- 34.Jasaswini T, Dinesh K, Kunj B. Graft copolymerization of N-vinylformamide onto sodium carboxymethylcellulose and study of its swelling, metal ion sorption and flocculation behaviour. Carbohydr Polym. 2009;75:604–611. doi: 10.1016/j.carbpol.2008.09.019. [DOI] [Google Scholar]
- 35.Zhou Z, Xia X, Bong D. Synthetic polymer hybridization with DNA and RNA directs nanoparticle loading, silencing delivery, and aptamer function. JACS. 2015;137:8920–8923. doi: 10.1021/jacs.5b05481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng C, Convertine A, Stayton P, Bryers J. Multifunctional triblock copolymers for intracellular messenger RNA delivery. Biomaterials. 2012;33:6868–6876. doi: 10.1016/j.biomaterials.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piotr S, Kowalski U, Palmiero Y, Arnab R, Langer R, Anderson D. Ionizable amino-polyesters synthesized via ring opening polymerization of tertiary amino-alcohols for tissue selective mRNA delivery. Adv Mater. 2018;30:1801151. doi: 10.1002/adma.201801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Contr Rel. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Katayose S, Kataoka K. Water-soluble polyion complex associates of DNA and poly(ethylene glycol)–poly(l-lysine) blockcopolymer. Bioconjug Chem. 1997;8:702–707. doi: 10.1021/bc9701306. [DOI] [PubMed] [Google Scholar]
- 40.Itaka K, Yamauchi K, Harada A, Nakamura K, Kawaguchi H, Kataoka K. Polyion complex micelles from plasmid DNA and poly(ethylene glycol)–poly(l-lysine) block copolymer as serum-tolerable polyplex system: physicochemical properties of micelles relevant to gene transfection efficiency. Biomaterials. 2003;24:4495–4506. doi: 10.1016/S0142-9612(03)00347-8. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y, Miyata K, Oba M, Ishii T, Fukushima S, Han M, Koyama M, Nishiyama N, Kataoka K. Charge-convension ternary polyplex with endosome disruption moiety: a technique for efficient and safe gene delivery. Angew Chem. 2008;47:5163–5166. doi: 10.1002/anie.200800963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data are mentioned. Not applicable.