Summary
Co‐expression of protease inhibitors like the tomato cystatin Sl CYS8 is useful to increase recombinant protein production in plants, but key proteases involved in protein proteolysis are still unknown. Here, we performed activity‐based protein profiling to identify proteases that are inhibited by Sl CYS8 in agroinfiltrated Nicotiana benthamiana. We discovered that Sl CYS8 selectively suppresses papain‐like cysteine protease (PLCP) activity in both apoplastic fluids and total leaf extracts, while not affecting vacuolar‐processing enzyme and serine hydrolase activity. A robust concentration‐dependent inhibition of PLCPs occurred in vitro when purified Sl CYS8 was added to leaf extracts, indicating direct cystatin–PLCP interactions. Activity‐based proteomics revealed that nine different Cathepsin‐L/‐F‐like PLCPs are strongly inhibited by Sl CYS8 in leaves. By contrast, the activity of five other Cathepsin‐B/‐H‐like PLCPs, as well as 87 Ser hydrolases, was unaffected by Sl CYS8. Sl CYS8 expression prevented protein degradation by inhibiting intermediate and mature isoforms of granulin‐containing proteases from the Resistant‐to‐Desiccation‐21 (RD21) PLCP subfamily. Our data underline the key role of endogenous PLCPs on recombinant protein degradation and reveal candidate proteases for depletion strategies.
Keywords: activity‐based protein profiling, protease inhibitor, cystatin, Sl CYS8, Nicotiana benthamiana, proteomics, papain‐like cysteine proteases
Introduction
Plant cells are increasingly used as alternative expression hosts for the production of recombinant proteins (Daniell et al., 2015; Lomonossoff and D'Aoust, 2016; Ma et al., 2015). However, protein proteolytic processing leads to either partial or complete degradation of recombinant proteins (Mandal et al., 2016; Pillay et al., 2014). Hundreds of genes code for proteolytic enzymes of diverse classes involved in many physiological processes in plants (Grosse‐Holz et al., 2018a; van der Hoorn, 2008). Controlling plant protease activity is a worthwhile approach to improve accumulation of recombinant proteins (Benchabane et al., 2008; Mandal et al., 2016). Unintended proteolysis has been mitigated in plant‐based expression systems by (i) targeting proteins to alternative cellular compartments (Benchabane et al., 2009); (ii) the addition of translational fusion partners to increase protein stability (Sainsbury et al., 2013); or (iii) the removal of protein domains and sequences targeted by endogenous plant proteases (Hehle et al., 2016; Zischewski et al., 2015). In addition, protease activity‐depleted environments have been created using gene silencing with RNAi strategies (Duwadi et al., 2015; Mandal et al., 2014) and inhibition of proteases using ectopic inhibitors in stable or transient plant expression systems (Goulet et al., 2010; Komarnytsky et al., 2006; Robert et al., 2013). Yet proteolytic degradation remains a major problem in the exploitation of plants as biofactories.
Although plants have diverse protease families, cysteine (Cys) proteases are a major constituent of the recombinant protein degradation machinery (Jutras et al., 2016; Niemer et al., 2014). Papain‐like cysteine proteases (PLCPs, C1A family) can be inhibited by cystatins (I12 family). Cystatins harbour a conserved motif Gln‐Xaa‐Val‐Xaa‐Gly (QxVxG) in the central region of the polypeptide chain and are competitive protease inhibitors that have strong affinity for the active site of target proteases (Benchabane et al., 2010; Martínez et al., 2012). Transgenic tobacco plants constitutively expressing a rice cystatin (OC‐I) have reduced protease activity, correlated with a higher accumulation of recombinant proteins (Pillay et al., 2012). Transient expression of tomato cystatin SlCYS8 (Girard et al., 2007) in the cell secretory pathway of agroinfiltrated Nicotiana benthamiana leaves also prevents degradation of recombinant proteins, notably leading to higher yields of fully assembled human antibodies (Grosse‐Holz et al., 2018b; Jutras et al., 2016; Robert et al., 2013). An inactive version of SlCYS8, which bears a proline (P) instead of a glutamine (Q) in the inhibitory loop QxVxG motif (Q47P SlCYS8), has no stabilizing effect on recombinant proteins (Grosse‐Holz et al., 2018b; Jutras et al., 2016; Sainsbury et al., 2013), indicating that SlCYS8 stabilizes recombinant proteins through inhibiting proteases. However, target proteases of SlCYS8 and other inhibitors have not been identified to date. A better understanding of which plant proteases are inhibited by SlCYS8 is needed to identify proteases that degrade recombinant proteins in molecular pharming.
Our goal was to identify the proteases that are suppressed by SlCYS8 in planta. In tomato, the SlCYS8 encoding gene is induced by jasmonic acid. SlCYS8 is the eighth domain of a multicystatin that accumulates in the cytosol of leaf cells upon herbivory challenge, presumably to inhibit digestive cysteine proteases in the midgut of herbivorous arthropods (Girard et al., 2007). In this study, we characterized the impact of SlCYS8 expression on protease activity profiles in agroinfiltrated leaves of N. benthamiana. We performed activity‐based protein profiling (ABPP) to determine the inhibitory spectrum of SlCYS8 and proteomics to identify target proteases. ABPP is increasingly used in plant science to monitor the activity of hundreds of proteins using tagged chemical probes that react covalently and irreversibly with the active site of proteins (Morimoto and van der Hoorn, 2016). Here, we used ABPP to show that SlCYS8 selectively inhibits nine papain‐like cysteine proteases in N. benthamiana. These proteases are likely involved in recombinant protein degradation.
Results and discussion
SlCYS8 inhibits papain‐like cysteine proteases in the apoplast
Most recombinant proteins are targeted to the secretory pathway for post‐translational modifications and are secreted in the apoplast. Recently, peptides corresponding to 196 proteases have been identified in the extracellular space of agroinfiltrated N. benthamiana leaves (Grosse‐Holz et al., 2018a). We thus focused on extracellular proteases as protein degradation in the apoplastic space limits the accumulation of recombinant proteins (Hehle et al., 2011). Apoplastic fluids were isolated from plants agroinfiltrated with a SlCYS8 transgene‐harbouring vector, or with an ‘empty’ vector. As negative control, we expressed the inactive version of the inhibitor, Q47P SlCYS8 (Sainsbury et al., 2013).
Activity‐based protein profiling (ABPP) was first conducted to assess whether the expression of a secreted version of SlCYS8 could alter papain‐like cysteine protease (PLCP) activity in the extracellular space of N. benthamiana leaves (Sainsbury et al., 2013). PLCP activity was characterized by incubating apoplastic fluids with MV201, a fluorescent derivative of the chemical PLCP inhibitor E‐64 which has been frequently used in plants (Richau et al., 2012). MV201 carries a bodipy fluorophore and targets PLCPs by carrying P2 = Leu. Labelled proteases are detected and quantified using protein in‐gel fluorescent scanning. SDS‐PAGE gel analysis showed a significant reduction of PLCP activity in leaves expressing SlCYS8 when compared to the empty vector control (Figure 1). The main fluorescent signals detected at ~38 and ~33 kDa are significantly decreased in the apoplast from leaves expressing SlCYS8 (arrows, Figure 1a), indicating that PLCP activity was depleted. Strong signals at ~33 kDa (open arrow, Figure 1a) are likely caused by mature isoforms of various PLCP subfamilies, including RD21‐like proteases and Cathepsin B, as those proteases are active in the extracellular space of N. benthamiana leaves (Grosse‐Holz et al., 2018a). No significant effect on fluorescence intensity was detected in apoplastic fluids from plants expressing the inactive Q47P SlCYS8 mutant (Figure 1a), confirming that the effect of SlCYS8 is related to protease inhibition. Pre‐incubation with the PLCP inhibitor E‐64 suppressed MV201 labelling (Figure 1a), thus confirming that the detected band signals originate from PLCPs.
Figure 1.
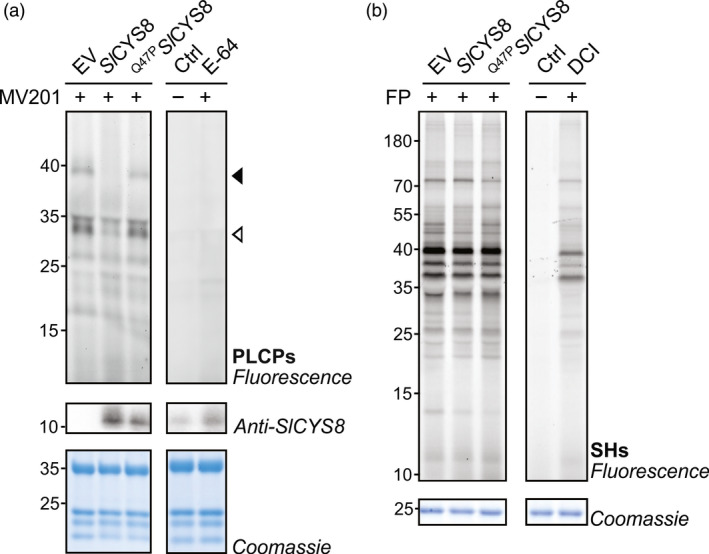
Sl CYS8 inhibits papain‐like cysteine proteases (PLCPs) in the apoplast of N. benthamiana leaves. Apoplastic fluids from plants agroinfiltrated with an empty vector control (EV) or transiently expressing Sl CYS8 or its inactive mutant Q47 P S l CYS8 were isolated and incubated with MV201 or fluorophosphonate (FP)‐TAMRA probes to target (a) papain‐like cysteine proteases (PLCPs) and (b) Ser hydrolases (SHs), respectively. Labelled proteins were detected by in‐gel fluorescence scanning. Proteins were electrotransferred for immunodetection of Sl CYS8 in the apoplast. Arrows show signals with different fluorescence intensities in plants expressing Sl CYS8. As controls, apoplastic fluids were mixed and pre‐incubated with specific chemical inhibitors (E‐64 for PLCPs and DCI for SHs) before incubation with or without (Ctrl) the probes. Coomassie blue‐stained gels are shown as loading controls.
To determine the specificity of SlCYS8 and to study potential indirect regulation of proteases by SlCYS8, ABPP was performed on apoplastic fluids with an activity‐based fluorescent probe targeting Ser hydrolases. Ser hydrolases, which include subtilases (S8 family) and Ser carboxypeptidases (S10 family), are present in the extracellular space of agroinfiltrated N. benthamiana leaves and are involved in proteolytic processing of recombinant proteins (Grosse‐Holz et al., 2018a; Komarnytsky et al., 2006). Ser hydrolases were labelled with a fluorophosphonate (FP)‐TAMRA probe, which labels Ser residues in the active site of Ser hydrolases (Kaschani et al., 2009; Liu et al., 1999). Gel fluorescence scanning of labelled apoplastic fluids showed that activity profiles of Ser hydrolases were unchanged upon the expression of SlCYS8 or the inactive Q47P SlCYS8 mutant, confirming a specificity of SlCYS8 to PLCPs and indicating that these Ser proteases are not regulated by SlCYS8‐sensitive PLCPs (Figure 1b). These data show the alteration of extracellular PLCPs upon SlCYS8 expression and identification of PLCPs as potential proteases involved in apoplastic protein degradation.
SlCYS8 targets papain‐like cysteine proteases in total leaf extracts
Although many proteases enter the endomembrane system, not all of them accumulate in the apoplast. Some accumulate in the secretory pathway or in the vacuole and are potentially involved in proteolytic processing of recombinant proteins. To assess how N. benthamiana intracellular proteases respond to SlCYS8 expression, total soluble proteins were extracted from plants transiently expressing SlCYS8 or the inactive Q47P SlCYS8 mutant. Leaf extracts were incubated with fluorescent MV201 probe to monitor PLCP activity. In contrast to apoplastic fluids, granulin‐containing proteases dominate PLCP activity in total extracts, causing strong signals at ~38 kDa (closed arrow, Figure 2a). Fluorescence intensity of these signals drastically decreased in the presence of SlCYS8 (closed arrow, Figure 2a). Similar to the apoplast, the signal at ~33 kDa also decreased in leaves expressing SlCYS8 (open arrow, Figure 2a). SlCYS8 expression prevents the degradation of the large subunit of ribulose‐1,5‐bisphosphate carboxylase/oxygenase (RbcL) and other proteins in total extracts during labelling, causing fluorescent signals that cannot be competed by E‐64, indicating that these fluorescent signals are caused by non‐specific labelling (Figure 2a). The inactive Q47P SlCYS8 mutant did not affect labelling or RbcL degradation. Taken together, these results confirm a strong inhibition of endogenous PLCP activity, both inside and outside the cells, upon SlCYS8 expression in N. benthamiana leaves.
Figure 2.
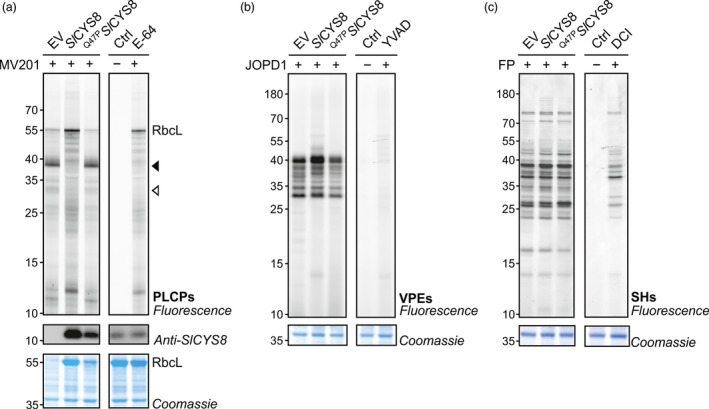
PLCPs are suppressed upon Sl CYS8 expression in total leaf extracts. Activity profiles of PLCPs (a), vacuolar‐processing enzymes (b) and Ser hydrolases (c) displayed by labelling with MV201, JOPD1 and FP‐TAMRA, respectively. Plants were agroinfiltrated with an empty vector control (EV) or Sl CYS8 or its inactive mutant Q47 P S l CYS8. Total leaf extracts isolated at 6 dpi were labelled with respective probes and fluorescent‐labelled proteins were detected by in‐gel fluorescence scanning. Arrows show signals with different fluorescence intensities in plants expressing Sl CYS8. The large subunit of ribulose‐1,5‐bisphosphate carboxylase/oxygenase (RbcL) is stabilized during labelling in Sl CYS8‐expressing leaves. Proteins were electrotransferred for immunodetection of Sl CYS8. As controls, leaf extracts were mixed and pre‐incubated with specific chemical inhibitors (E‐64 for PLCPs, YVAD for VPEs and DCI for SHs) before incubation with or without (Ctrl) the probes. Coomassie blue‐stained gels are shown as loading controls.
Legumain‐like Cys proteases (C13 family), or vacuolar‐processing enzymes (VPEs), are a group of Asn/Asp‐specific proteinases and are primarily located in the vacuole (Wang et al., 2018). VPEs are essential for the maturation of various vacuolar proteins and can be responsible for the degradation of recombinant proteins in plants (Outchkourov et al., 2003; Pillay et al., 2016). Although PLCPs and VPEs are two distinct types of Cys proteases, their activity can both be inhibited by cystatins (Martinez et al., 2007). We performed ABPP on leaf extracts to investigate if VPEs are inhibited by SlCYS8. The fluorescent JOPD1 probe was used to monitor VPE activity upon SlCYS8 expression as it carries an Asp residue at the P1 position and a Pro residue at the P2 position that prevents the labelling of PLCPs (Lu et al., 2015). Labelling by JOPD1 was unchanged upon SlCYS8 expression, except for minor increases at ~40 kDa in SlCYS8‐expressing leaves that might be the result of a stabilizing effect initiated by the depletion of most PLCPs (Figure 2b). As many Ser hydrolases also accumulate intracellularly, ABPP was performed on total leaf extracts using (FP)‐TAMRA probe. Activity profiles of Ser hydrolases were unaffected by SlCYS8 expression or the inactive Q47P SlCYS8 mutant (Figure 2c). Taken together, these results suggest that SlCYS8 inhibits exclusively PLCPs.
In vitro assays confirm direct inhibition of PLCPs by SlCYS8
To test if the impact of SlCYS8 on PLCP activity is direct, total leaf extracts from agroinfiltrated plants were pre‐incubated with 0–150 nm purified SlCYS8 produced in Escherichia coli, prior to labelling with MV201. Addition of purified SlCYS8 suppressed the two major PLCP signals (Figure 3a), similar to what was observed in vivo (Figure 2a). Fluorescence intensities were quantified and plotted against SlCYS8 concentration, showing that activity depletion is dependent on the SlCYS8 concentration (Figure 3b). Inhibition assays were also performed on apoplastic fluids isolated from agroinfiltrated plants pre‐incubated with 0–1500 nm purified SlCYS8 (Figure 3c). The data showed a similar concentration‐dependent inhibition of PLCPs in apoplastic extracts, reaching 90% and 65% inhibition of the major ~38 and ~33 kDa signals, respectively (Figure 3d). These results confirm a direct inhibitory effect of SlCYS8 on endogenous N. benthamiana PLCPs.
Figure 3.
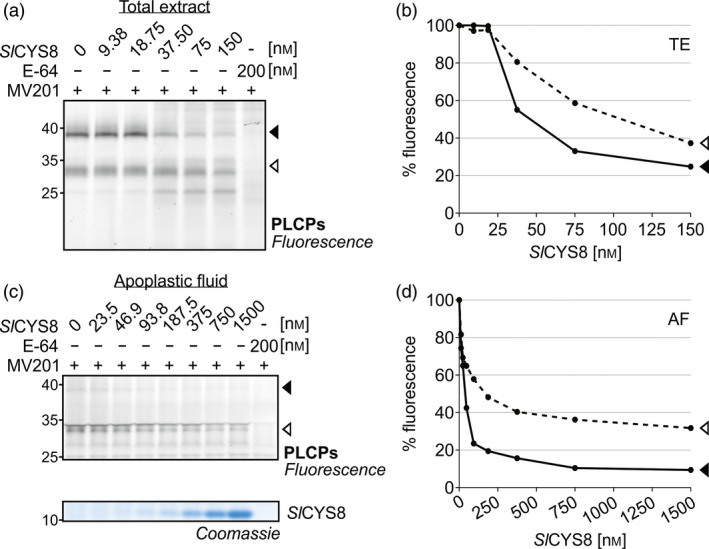
Purified Sl CYS8 suppresses labelling of PLCPs in total leaf extracts and apoplastic fluids. (a) Total extracts (TE) and (c) apoplastic fluids (AF) from plants agroinfiltrated with an empty vector control were isolated and pre‐incubated with various concentrations of Sl CYS8 purified from E. coli, prior to PLCP labelling with MV201. Quantification of fluorescent band intensity (b and d) shows that PLCP activity is dependent on Sl CYS8 concentration. As a control, plant extracts were mixed and pre‐incubated with E‐64 inhibitor before MV201 labelling. A Coomassie blue‐stained gel shows Sl CYS8 in the samples.
Activity‐based proteomics reveals N. benthamiana PLCPs inhibited by SlCYS8
To identify the proteases that are inhibited by SlCYS8, we performed activity‐based proteomics (ABPP‐MS) on leaf extracts. Total extracts from agroinfiltrated leaves expressing SlCYS8 (in vivo) or empty vector control were pre‐incubated with or without 280 nm of purified SlCYS8 and labelled with a mix of two biotinylated probes to label both PLCPs and Ser hydrolases (Greenbaum et al., 2000; Liu et al., 1999). Labelled proteins were captured on avidin beads, digested with trypsin and quantified by mass spectrometry (MS). 14 PLCPs and 87 Ser hydrolases were enriched at a confidence level of 95% (P < 0.05) compared to the no‐probe control, indicating that they were active. No Ser hydrolase was convincingly affected by the presence of SlCYS8 in vivo or in vitro (Figure 4a and Table S1), consistent with the fluorescent profiling data (Figures 1 and 2).
Figure 4.
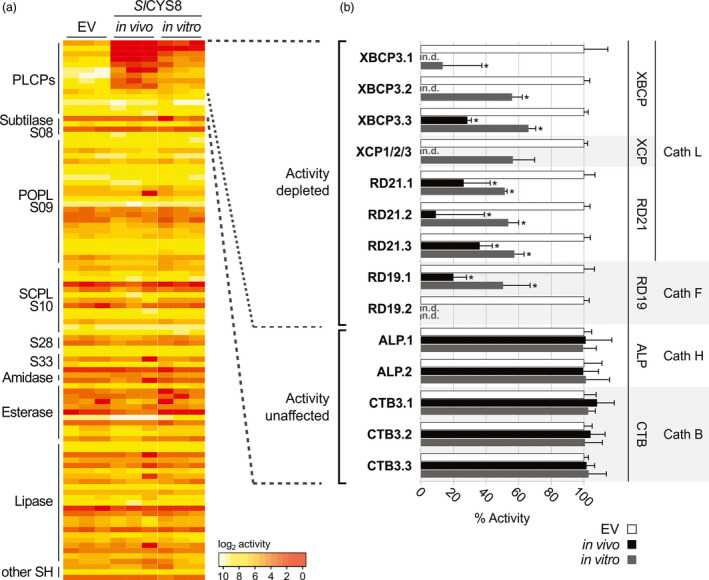
Activity‐based proteomics reveals nine endogenous PLCPs that are inhibited by Sl CYS8. Total leaf extracts were isolated from plants expressing Sl CYS8 (in vivo) or expressing an empty vector (EV), pre‐incubated with or without 280 nm of purified Sl CYS8 (in vitro). Proteins were labelled with DCG04 and FP‐biotin to label PLCPs and Ser hydrolases, respectively. Labelled proteins were purified and identified by MS. (a) Heatmap showing active enzymes that were significantly enriched (FDR < 0.05) compared to a no‐probe control, fold inhibition serves as a proxy for activity. Most PLCP activities were depleted upon Sl CYS8 expression, while no Ser hydrolase was inhibited. (b) Identified PLCPs were grouped by subfamilies and the activity was normalized to the EV control (100%). Bars are the means of three biological replicates ± SE. Star indicates significant differences from the EV control (Student's t test, P < 0.05). PLCP subfamilies: Xylem and bark Cys proteases (XBCPs), Xylem‐specific Cys proteases (XCPs), Resistant‐to‐Desiccation‐21 proteases (RD21s), Resistant‐to‐Desiccation‐19 proteases (RD19s), Aleurain‐Like proteases (ALPs) and Cathepsin‐B‐like proteases (CTBs). n.d.; not detected. MS dataset is presented in Table S1.
Nine PLCPs were significantly less labelled in leaves expressing SlCYS8 when compared to the empty vector control, suggesting that their activity was depleted by SlCYS8 both in vivo and in vitro. These nine inhibited PLCPs belong to four subfamilies: Xylem and bark Cys proteases (XBCPs), Xylem‐specific Cys proteases (XCPs), Resistant‐to‐Desiccation‐19 proteases (RD19s) and Resistant‐to‐Desiccation‐21 proteases (RD21s). Two XBCP‐like and one RD21‐like PLCPs were more inhibited upon SlCYS8 expression in vivo compared to extracts treated with SlCYS8 in vitro (Figure 4b), potentially due to incomplete inhibition of the proteases using 280 nm SlCYS8. These nine inhibited PLCPs are strong candidates to be implicated in proteolytic processing of recombinant proteins in agroinfiltrated leaves.
Our data are consistent with the literature. Some of the PLCPs identified in total extracts may cause the signals in apoplastic fluids, as earlier work revealed that the extracellular space contains XBCP, XCP, RD21 and RD19 proteases (Grosse‐Holz et al., 2018a). A recent study showed that purified RD21‐like NbCYSP6 and XCP‐like NbCYSP7 proteases rapidly degrade antibodies in vitro (Paireder et al., 2017). Silencing the gene encoding a RD21‐like protease (NtCYSP6) in tobacco also significantly enhanced accumulation of recombinant proteins (Duwadi et al., 2015).
In contrast, labelling of Aleurain‐like proteases (ALPs) and Cathepsin‐B‐like proteases (CTBs) was unaffected either by SlCYS8 expressed in plants (in vivo) or purified SlCYS8 (in vitro; Figure 4b). This is consistent with ALPs being cathepsin‐H‐like aminopeptidase rather than endopeptidase, as one side of the substrate‐binding groove is occupied by a minichain (Gunčar et al., 1998; Holwerda and Rogers, 1992). The shorter substrate‐binding groove of ALPs may not accommodate the tripartite wedge of SlCYS8. The ALP subfamily might have a minor contribution in proteolytic processing of recombinant proteins, as previous in vitro assays showed negligible effect of NbALP on antibody stability (Niemer et al., 2016). CTBs are peptidyl‐peptidases with an occluding loop that blocks part of the substrate‐binding groove (Musil et al., 1991; Tsuji et al., 2008). The occluding loop may prevent its inhibition by SlCYS8. Thus, even though NbCathB protease, a sequence 93% identical to the identified NbCTB3.3 protease, can degrade antibodies in vitro (Niemer et al., 2016), these CTBs probably do not contribute to recombinant protein degradation. Taken together, these data confirm overall the impact of SlCYS8 expression on endogenous proteases and indicate that XBCP, XCP, RD21 and RD19‐like proteases may constitute the core of the recombinant protein degradation machinery that is blocked by SlCYS8.
SlCYS8 inhibits different isoforms of granulin‐containing proteases
To confirm that SlCYS8 inhibits both the intermediate and the mature forms of granulin‐containing proteases, we tested C14, the tomato orthologue of the N. benthamiana RD21.1 (Kaschani et al., 2010; Shabab et al., 2008). Activity‐based profiling of leaves transiently expressing C14 displayed a strong ~38 kDa signal of the intermediate form (iC14) carrying the granulin domain and a weaker signal at ~33 kDa corresponding to the mature protein (mC14) with no granulin domain (Figure 5a). Co‐expressing SlCYS8 significantly suppressed the activity of both active C14 isoforms and endogenous NbRD21‐like proteases, while not affecting C14 accumulation in leaves (Figure 5a and b). A Coomassie gel showed that protein degradation occurs faster upon C14 expression and co‐expression of C14 with SlCYS8 prevented this degradation (Figure 5c). Staining protein gels is a simple assay to monitor proteolytic activity of RD21‐like proteases during extraction (Gu et al., 2012). These results underline the impact of SlCYS8 on protease isoforms and confirm the broad inhibition of RD21‐like proteases by SlCYS8.
Figure 5.
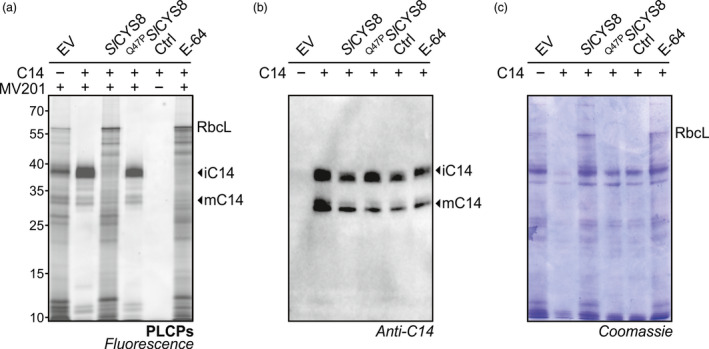
Sl CYS8 inhibits different isoforms of granulin‐containing proteases. (a) Leaf extracts from agroinfiltrated plants overexpressing C14, the tomato RD21‐like protease, with or without Sl CYS8 were labelled with MV201 to monitor PLCP activity. In‐gel fluorescence confirmed inhibition of C14 upon Sl CYS8 co‐expression, while no effect was observed when co‐expressed with the inactive Q47 P S l CYS8 mutant. As a control, plant extracts were mixed and pre‐incubated with E‐64 inhibitor before incubation with or without (Ctrl) the probe. (b) Immunoblot shows equal amount of the intermediate protease (iC14) and the mature (mC14) form of C14 protease. (c) Coomassie gel shows that protein degradation occurred upon C14 expression and is prevented by co‐expressing Sl CYS8. The large subunit of ribulose‐1,5‐bisphosphate carboxylase/oxygenase (RbcL) is stabilized in Sl CYS8‐expressing leaves.
Conclusion
Proteolytic degradation of recombinant proteins by endogenous plant proteases is a major limiting factor in molecular pharming. Different strategies have been used to suppress unwanted proteolysis in planta (Mandal et al., 2016), but the responsible proteases have not yet been identified. The tomato cystatin SlCYS8 is a stabilizing co‐expression partner or a fusion partner for proteins targeted to the plant cell secretory pathway (Grosse‐Holz et al., 2018b; Jutras et al., 2016; Sainsbury et al., 2013). In this study, we used SlCYS8 to identify target proteases that may contribute to low yields of foreign recombinant proteins in N. benthamiana. Our data highlight that SlCYS8 specifically inhibits PLCPs, inside and outside the cells, with no impact on the activity of vacuolar‐processing enzymes and Ser hydrolases. Proteomics revealed nine inhibited PLCPs and protein gels confirm that SlCYS8 inhibits different isoforms of an RD21‐like protease, preventing protein degradation. We thus discovered unique proteases in agroinfiltrated leaves that are promising targets to knockout. Depletion of these PLCPs by genome editing could reduce protein proteolysis in the secretory pathway of N. benthamiana.
Experimental procedures
Plasmid constructs
Gene expression constructs for secreted SlCYS8 (GenBank Accession No. AF198390) and its inactive variant Q47P SlCYS8 were described previously (Sainsbury et al., 2013). An ‘empty’ vector (bearing the coding sequence of gene silencing suppressor P19) was used as a negative control. The vectors were transformed into Agrobacterium tumefaciens strain LBA4404 and used for agroinfiltration. Gene expression construct for secreted tomato C14 was described previously (Kaschani et al., 2010). All gene constructs were proof checked by automatic DNA sequencing.
Transient expression in leaves
Nicotiana benthamiana plants were kept in a growth room at 21 °C under a 16/8 h light/dark regime. Agrobacteria containing the binary expression plasmids were grown for 21 h at 28 °C with agitation in LB containing appropriate antibiotics. Bacteria were collected by centrifugation at 2000 g for 5 min at room temperature (RT), resuspended in infiltration buffer (10 mm 2‐(N‐morpholino) ethanesulfone (MES), 10 mm MgCl2, pH 5.7, 100 μm acetosyringone) to OD600 = 0.5 and left for 2 h at 28 °C with agitation to recover. All bacterial suspensions were mixed in a 1 : 1 ratio with a suspension of A. tumefaciens harbouring an expression cassette for the silencing suppressor protein P19 of artichoke mottled crinkle virus (Lombardi et al., 2009). The first and second fully expanded leaves of pre‐flowering N. benthamiana (4–5 weeks old) were infiltrated with the bacteria suspensions using a needleless syringe (D'Aoust et al., 2009). Leaf tissue was harvested 6 days post‐infiltration (dpi) to allow for maximum protein accumulation in the presence of the P19 silencing suppressor. Three independent replicates including leaves of three plants were used for each treatment to minimize variation of protein expression levels and to allow for statistical analysis of the data.
Protein extraction
For total leaf extracts, infiltrated leaf tissue was harvested as leaf discs, flash‐frozen in liquid nitrogen and pulverized using a pestle and a mortar. Proteins were extracted in three volumes (v/fresh weight) of cold 50 mm sodium acetate (NaAc), pH 5, containing 5 mm DTT, and centrifuged at 16 000 g for 20 min at 4 °C. For apoplastic fluid extraction, six N. benthamiana leaves per sample were detached and vacuum‐infiltrated with ice‐cold water, dried on the surface and placed in a needle/plunger‐less syringe inserted into a 50 mL Falcon tube. Apoplastic fluids were collected by centrifugation at 2000 g , 4 °C for 25 min and used immediately for protein analysis.
Activity‐based protein profiling
Forty‐eight μL of leaf extract or apoplastic fluid adjusted to 50 mm NaAc, pH 5, 5 mm DTT, was pre‐incubated with or without 0.2 mm of inhibitor (E‐64; DCI, 3,4‐Dichloroisocoumarin; YVAD, Ac‐Tyr‐Val‐Ala‐Asp‐Chloromethylketone) for 30 min, and then incubated for 3.5 h at room temperature with fluorescent probes targeting papain‐like cysteine proteases (MV201; Richau et al., 2012) or vacuolar‐processing enzymes (JOPD1; Lu et al., 2015) or incubated for 1 h with an FP‐TAMRA probe targeting Ser hydrolase activity. FP‐TAMRA was obtained from Thermo (88318); MV201 and JOPD1 were described before (Lu et al., 2015; Richau et al., 2012). ABPP reactions were ended by adding 1 mL cold acetone. Samples were centrifuged for 3 min at 13 000 rpm and the supernatant was discarded. Proteins were resuspended in gel loading buffer (100 mm Tris‐HCl, pH 6.8, 200 mm DTT, 4% w/v SDS, 0.02% w/v bromophenol blue, 25% v/v glycerol), heated for 5 min at 95 °C, and resolved by 12% w/v SDS‐PAGE in reducing conditions. SDS‐PAGE gels were scanned on a Typhoon scanner (Amersham/GE Healthcare, Little Chalfont, UK) using Cy3 settings to detect in‐gel fluorescence. Image data were analysed using the Open source software ImageJ (http://rsb.info.nih.gov/ij/) to quantify band fluorescence intensity. Background values were subtracted from each image based on the average value of images acquired from no‐probe controls. Bands from three biological replicates were used in each case.
Immunoblotting
Proteins resolved by 12% w/v SDS‐PAGE gel were electrotransferred onto a PVDF membrane using the TransBlot Turbo system (Bio‐rad, Hercules, CA). SlCYS8 was detected using custom‐made primary IgG raised in rabbit against anti‐SlCYS8 (Robert et al., 2013; Agrisera, Sweden, 1/5000) and goat anti‐rabbit horseradish peroxidase (HRP)‐conjugated secondary antibodies (Agrisera, Sweden, 1/10 000). C14 was detected with anti‐C14 polyclonal IgG (1/1000) raised in rabbit (Kaschani et al., 2010) and goat anti‐rabbit HRP secondary antibodies (Agrisera, Sweden, 1/5000). Non‐specific binding sites on PVDF membranes were blocked for 1 h with 5% w/v skimmed milk powder in PBS buffer containing 0.025% v/v Tween‐20, which also served as antibody dilution buffer. Chemiluminescent signals were detected using the Clarity ECL Western blotting detection kit (Bio‐rad), and protein–antibody complex signals were captured on a Gel Doc imager (Bio‐rad).
Bacterial expression of recombinant SlCYS8
Glutathione S‐transferase (GST)‐tagged SlCYS8 expression in E. coli was carried out as reported (Goulet et al., 2008). Briefly, 5 mL pre‐cultures of BL21 E. coli cells were incubated overnight in LB medium at 37 °C, with carbenicillin added in the culture medium. The pre‐cultures were transferred into 250 mL LB cultures and incubated at 37 °C until an OD600 of 0.6. Protein expression was induced by the addition of 0.5 mm of isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) and the cultures were incubated for another 6 h at 30 °C. Bacteria were collected by centrifugation at 3000 g for 10 min, submitted to four freeze‐thaw cycles, resuspended in 3 mL of lysis buffer (50 mm Tris pH 8.0, 5% m/v sucrose, 50 mm EDTA, 5% v/v Triton X‐100, 1 mm PMSF), incubated on ice for 5 min and centrifuged at 13 000 g for 10 min 4 °C. The supernatant was collected and incubated with 250 μL of Glutathione Sepharose‐4B beads (GE Healthcare Life Sciences, Chicago, IL) for 60 min with low agitation. Samples were centrifuged 5 min at 500 g to spin down the beads and the supernatant was discarded. Beads were washed three times using 5 mL of 50 mm Tris pH 8.0 and centrifuged 5 min at 500 g. The last wash was with 5 mL of Factor Xa cleavage buffer (20 mm Tris‐HCl, 100 mm NaCl, 2 mm CaCl2, pH 8.0). After centrifugation (5 min at 500 g ), the supernatant was discarded and 300 μL of Factor Xa cleavage buffer was added to the beads. The GST fusion partner was removed by adding 5 μL of Factor Xa enzyme (NEB, Ipswich, MA). The samples were incubated with Factor Xa for 16 h at room temperature with gentle agitation, and centrifuged at 6000 g for 5 min. Proteins in the supernatant were assayed using the standard Bradford protocol and maintained at −80 °C until further use.
In vitro inhibition assays
Total leaf extracts (TE) or apoplastic fluids (AF) from leaves agroinfiltrated with the empty vector control were extracted in sodium acetate buffer and purified SlCYS8 protein variants were added at different concentrations (0–150 nm for TE and 0–1500 nm for AF) for a 30‐min incubation at room temperature prior to PLCP labelling as previously described. The same amount of factor Xa cleavage buffer was added in the no SlCYS8 control. Inhibition rates were determined by the quantification of band fluorescence intensity compared to the no SlCYS8 control. Incremental amounts of purified SlCYS8 added in the samples were visualized on Coomassie blue‐stained polyacrylamide gels following 12% SDS‐PAGE.
Activity‐based proteomics
Leaves from different plants were infiltrated with agrobacterial suspensions (OD600 = 0.5) harbouring the P19 vector or 1:1 (v/v) SlCYS8:P19 vectors. Proteins were extracted at 6 dpi as described above and diluted to 1.5 mg/mL. In the ‘SlCYS8 in vitro’ samples, 3 ng of purified SlCYS8 from E. coli were added per 1 μL volume. All samples were incubated at RT on a rotator for 45 min. An amount of 1 mL of each sample was then incubated with 5 μm FP‐biotin (Sigma 88317) and 5 μm DCG04 (Greenbaum et al., 2000) for 5 h on a rotator at RT in darkness. A no‐probe control contained a mix of equal volumes of each sample and DMSO. Samples were transferred to 15 mL falcon tubes and 4 mL ice‐cold methanol, 1 mL ice‐cold chloroform and 3 mL ice‐cold water were added subsequently, vortexing the sample after each step. Samples were centrifuged for 30 min at 3000 g (4 °C). The upper aqueous layer was carefully removed, 4 mL of methanol was added and the samples were centrifuged for 30 min at 3000 g (4 °C). The supernatant was discarded and the pellet was resuspended in 2 mL PBS containing 1.2% w/v sodium dodecyl sulphate (SDS) and then diluted using 5 mL PBS. Proteins were denatured by heating at 90 °C for 8 min and cooled on ice, before dilution with 3 mL of PBS and stored at −20 °C until the next day. After thawing, 130 μL avidin beads (Sigma A9207, pre‐washed three times in 1X PBS) was added to each sample. Samples were incubated on a rotator for 1 h at room temperature and centrifuged for 7 min at 400 g. The supernatant was discarded and the beads were washed four times in 1% w/v SDS and twice in water after centrifuging for 7 min at 400 g each time. Purified proteins on the beads were reduced in 256 μL 50 mm Tris‐HCl pH 8.0, 8 m urea, 10 mm DTT for 15 min at 65 °C with agitation in darkness, then cooled to 35 °C and alkylated by adding 12.5 μL of 400 mm iodoacetamide in 50 mm Tris‐HCl pH 8.0 and incubating for 30 min with agitation in darkness. An amount of 4 μL of Trypsin‐LysC (Promega V5071) was added to each sample and the LysC digest was performed for 3 h at 37 °C under gentle agitation. Samples were then diluted in 50 mm Tris‐HCl pH 8.0 to reach a final urea concentration of less than 1 m and trypsin digestion was performed for 16 h at 37 °C with agitation. The samples were centrifuged for 3 min at 1000 g , and 0.1% v/v trifluoracetic acid added to the supernatant in new tubes. The peptides were purified using Sep‐Pak C18 cartridges (Waters, 610 Centennial Park, Herts, UK) according to the manufacturer's instructions.
Mass spectrometry
Mass spectrometry analysis is described in Appendix S1.
Bioinformatics tools for leaf proteome analysis
Peptide spectra were annotated using Andromeda (Cox et al., 2011). Included modifications were carbamidomethylation (static) and oxidation, N‐terminal acetylation and carbamylation of Lysines and N‐termini (dynamic). Protein quantification was performed using MaxQuant version 1.5.5.30 (Tyanova et al., 2016a), including all modifications. Filtering and imputation of missing values using default settings were performed in Perseus (Tyanova et al., 2016b) and further data analysis carried out in R using the data.table and ggplot packages (Ihaka and Gentleman, 1996; Wickham, 2016). Sequence analyses and plasmid design were performed in Geneious (Kearse et al., 2012).
Conflicts of interest
The authors declare no conflicts of interest.
Supporting information
Table S1 Proteases identified by ABPP‐MS.
Appendix S1 Mass spectrometry analysis.
Acknowledgements
We thank Urszula Pyzio, Caroline O’ Brien and Sarah Rodgers for excellent technical support. This work was financially supported by ERC consolidator grant 616449 ‘GreenProteases’ and H2020 project ‘Newcotiana’ (R.H. and P.V.J., grant No. 760331), ERC starting grant (M.K., grant No. 258413) and the Deutsche Forschungsgemeinschaft (M.K., grant no. INST 20876/127‐1 FUGG). This work was also supported by Discovery and CRD grants from the Natural Science and Engineering Research Council (NSERC) of Canada to D.M. P.V.J. was the recipient of an AgroPhytoSciences NSERC–CREATE scholarship and of a BMP graduate scholarship jointly funded by Medicago inc., NSERC and Québec Government's research funding body FRQNT.
References
- Benchabane, M. , Goulet, C. , Rivard, D. , Faye, L. , Gomord, V. and Michaud, D. (2008) Preventing unintended proteolysis in plant protein biofactories. Plant Biotechnol. J. 6, 633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchabane, M. , Saint‐Jore‐Dupas, C. , Bardor, M. , Faye, L. , Michaud, D. and Gomord, V. (2009) Targeting and post‐translational processing of human alpha1‐antichymotrypsin in BY‐2 tobacco cultured cells. Plant Biotechnol. J. 7, 146–160. [DOI] [PubMed] [Google Scholar]
- Benchabane, M. , Schlüter, U. , Vorster, J. , Goulet, M.‐C. and Michaud, D. (2010) Plant cystatins. Biochimie, 92, 1657–1666. [DOI] [PubMed] [Google Scholar]
- Cox, J. , Neuhauser, N. , Michalski, A. , Scheltema, R.A. , Olsen, J.V. and Mann, M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Daniell, H. , Streatfield, S.J. and Rybicki, E.P. (2015) Advances in molecular farming: key technologies, scaled up production and lead targets. Plant Biotechnol. J. 13, 1011–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust, M.‐A. , Lavoie, P.‐O. , Belles‐Isles, J. , Bechtold, N. , Martel, M. and Vézina, L.‐P. (2009) Transient expression of antibodies in plants using syringe agroinfiltration. Methods Mol. Biol. 483, 41–50. [DOI] [PubMed] [Google Scholar]
- Duwadi, K. , Chen, L. , Menassa, R. and Dhaubhadel, S. (2015) Identification, characterization and down‐regulation of cysteine protease genes in tobacco for use in recombinant protein production. PLoS ONE, 10, e0130556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, C. , Rivard, D. , Kiggundu, A. , Kunert, K. , Gleddie, S.C. , Cloutier, C. and Michaud, D. (2007) A multicomponent, elicitor‐inducible cystatin complex in tomato, Solanum lycopersicum . New Phytol. 173, 841–851. [DOI] [PubMed] [Google Scholar]
- Goulet, M.‐C. , Dallaire, C. , Vaillancourt, L.‐P. , Khalf, M. , Badri, A.M. , Preradov, A. , Duceppe, M.‐O. et al. (2008) Tailoring the specificity of a plant cystatin toward herbivorous insect digestive cysteine proteases by single mutations at positively selected amino acid sites. Plant Physiol. 146, 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet, C. , Benchabane, M. , Anguenot, R. , Brunelle, F. , Khalf, M. and Michaud, D. (2010) A companion protease inhibitor for the protection of cytosol‐targeted recombinant proteins in plants. Plant Biotechnol. J. 8, 142–154. [DOI] [PubMed] [Google Scholar]
- Greenbaum, D. , Medzihradszky, K.F. , Burlingame, A. and Bogyo, M. (2000) Epoxide electrophiles as activity‐dependent cysteine protease profiling and discovery tools. Chem. Biol. 7, 569–581. [DOI] [PubMed] [Google Scholar]
- Grosse‐Holz, F. , Kelly, S. , Blaskowski, S. , Kaschani, F. , Kaiser, M. and van der Hoorn, R.A.L. (2018a) The transcriptome, extracellular proteome and active secretome of agroinfiltrated Nicotiana benthamiana uncover a large, diverse protease repertoire. Plant Biotechnol. J. 16, 1068–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse‐Holz, F. , Madeira, L. , Zahid, M.A. , Songer, M. , Kourelis, J. , Fesenko, M. , Ninck, S. et al. (2018b) Three unrelated protease inhibitors enhance accumulation of pharmaceutical recombinant proteins in Nicotiana benthamiana . Plant Biotechnol. J. 16, 1797–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, C. , Shabab, M. , Strasser, R. , Wolters, P.J. , Shindo, T. , Niemer, M. , Kaschani, F. et al. (2012) Post‐translational regulation and trafficking of the granulin‐containing protease RD21 of Arabidopsis thaliana . PLoS ONE, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunčar, G. , Podobnik, M. , Pungerčar, J. , Štrukelj, B. , Turk, V. and Turk, D. (1998) Crystal structure of porcine cathepsin H determined at 2.1 Å resolution: location of the mini‐chain C‐terminal carboxyl group defines cathepsin H aminopeptidase function. Structure, 6, 51–61. [DOI] [PubMed] [Google Scholar]
- Hehle, V.K. , Paul, M.J. , Drake, P.M. , Ma, J.K. and van Dolleweerd, C.J. (2011) Antibody degradation in tobacco plants: a predominantly apoplastic process. BMC Biotechnol. 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehle, V.K. , Paul, M.J. , Roberts, V.A. , van Dolleweerd, C.J. and Ma, J.K.‐C.C. (2016) Site‐targeted mutagenesis for stabilization of recombinant monoclonal antibody expressed in tobacco (Nicotiana tabacum) plants. FASEB J. 30, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holwerda, B.C. and Rogers, J.C. (1992) Purification and characterization of aleurain : a plant thiol protease functionally homologous to Mammalian cathepsin H. Plant Physiol. 99, 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59, 191–223. [DOI] [PubMed] [Google Scholar]
- Ihaka, R. and Gentleman, R. (1996) R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314. [Google Scholar]
- Jutras, P.V. , Marusic, C. , Lonoce, C. , Deflers, C. , Goulet, M.C. , Benvenuto, E. , Michaud, D. et al. (2016) An accessory protease inhibitor to increase the yield and quality of a tumour‐targeting mAb in Nicotiana benthamiana leaves. PLoS ONE, 11, e0167086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani, F. , Gu, C. , Niessen, S. , Hoover, H. , Cravatt, B.F. and van der Hoorn, R.A.L. (2009) Diversity of serine hydrolase activities of unchallenged and Botrytis‐infected Arabidopsis thaliana . Mol. Cell Proteom. 8, 1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani, F. , Shabab, M. , Bozkurt, T. , Shindo, T. , Schornack, S. , Gu, C. , Ilyas, M. et al. (2010) An effector‐targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154, 1794–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , Buxton, S. et al. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnytsky, S. , Borisjuk, N. , Yakoby, N. , Garvey, A. and Raskin, I. (2006) Cosecretion of protease inhibitor stabilizes antibodies produced by plant roots. Plant Physiol. 141, 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Patricelli, M.P. and Cravatt, B.F. (1999) Activity‐based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci. 96, 14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi, R. , Circelli, P. , Villani, M.E. , Buriani, G. , Nardi, L. , Coppola, V. , Bianco, L. et al. (2009) High‐level HIV‐1 Nef transient expression in Nicotiana benthamiana using the P19 gene silencing suppressor protein of Artichoke Mottled Crinckle Virus. BMC Biotechnol. 9, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff, G.P. and D'Aoust, M.‐A. (2016) Plant‐produced biopharmaceuticals: a case of technical developments driving clinical deployment. Science, 353, 1237–1240. [DOI] [PubMed] [Google Scholar]
- Lu, H. , Chandrasekar, B. , Oeljeklaus, J. , Misas‐Villamil, J.C. , Wang, Z. , Shindo, T. , Bogyo, M. et al. (2015) Subfamily‐specific fluorescent probes for cysteine proteases display dynamic protease activities during seed germination. Plant Physiol. 168, 1462–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.K.‐C. , Drossard, J. , Lewis, D. , Altmann, F. , Boyle, J. , Christou, P. , Cole, T. et al. (2015) Regulatory approval and a first‐in‐human phase I clinical trial of a monoclonal antibody produced in transgenic tobacco plants. Plant Biotechnol. J. 13, 1106–1120. [DOI] [PubMed] [Google Scholar]
- Mandal, M.K. , Fischer, R. , Schillberg, S. and Schiermeyer, A. (2014) Inhibition of protease activity by antisense RNA improves recombinant protein production in Nicotiana tabacum cv. Bright Yellow 2 (BY‐2) suspension cells. Biotechnol. J. 9, 1065–1073. [DOI] [PubMed] [Google Scholar]
- Mandal, M.K. , Ahvari, H. , Schillberg, S. and Schiermeyer, A. (2016) Tackling unwanted proteolysis in plant production hosts used for molecular farming. Front. Plant Sci. 7, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, M. , Diaz‐Mendoza, M. , Carrillo, L. and Diaz, I. (2007) Carboxy terminal extended phytocystatins are bifunctional inhibitors of papain and legumain cysteine proteinases. FEBS Lett. 581, 2914–2918. [DOI] [PubMed] [Google Scholar]
- Martínez, M. , Cambra, I. , González‐Melendi, P. , Santamaría, M.E. and Díaz, I. (2012) C1A cysteine‐proteases and their inhibitors in plants. Physiol. Plant. 145, 85–94. [DOI] [PubMed] [Google Scholar]
- Morimoto, K. and van der Hoorn, R.A.L. (2016) The increasing impact of activity‐based protein profiling in plant science. Plant Cell Physiol. 57, 446–461. [DOI] [PubMed] [Google Scholar]
- Musil, D. , Zucic, D. , Turk, D. , Engh, R.A. , Mayr, I. , Huber, R. , Popovic, T. et al. (1991) The refined 2.15 A X‐ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO J. 10, 2321–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemer, M. , Mehofer, U. , Torres Acosta, J.A. , Verdianz, M. , Henkel, T. , Loos, A. , Strasser, R. et al. (2014) The human anti‐HIV antibodies 2F5, 2G12, and PG9 differ in their susceptibility to proteolytic degradation: down‐regulation of endogenous serine and cysteine proteinase activities could improve antibody production in plant‐based expression platforms. Biotechnol. J. 9, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemer, M. , Mehofer, U. , Verdianz, M. , Porodko, A. , Schähs, P. , Kracher, D. , Lenarcic, B. et al. (2016) Nicotiana benthamiana cathepsin B displays distinct enzymatic features which differ from its human relative and aleurain‐like protease. Biochimie, 122, 119–125. [DOI] [PubMed] [Google Scholar]
- Outchkourov, N. , Rogelj, B. , Strukelj, B. and Jongsma, M.A. (2003) Expression of sea anemone equistatin in potato. Effects of plant proteases on heterologous protein production. Plant Physiol. 133, 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paireder, M. , Tholen, S. , Porodko, A. , Biniossek, M.L. , Mayer, B. , Novinec, M. , Schilling, O. et al. (2017) The papain‐like cysteine proteinases NbCysP6 and NbCysP7 are highly processive enzymes with substrate specificities complementary to Nicotiana benthamiana cathepsin B. Biochim. Biophys. Acta ‐ Proteins Proteom. 1865, 444–452. [DOI] [PubMed] [Google Scholar]
- Pillay, P. , Kibido, T. , Du Plessis, M. , Van Der Vyver, C. , Beyene, G. , Vorster, B.J. , Kunert, K.J. et al. (2012) Use of transgenic oryzacystatin‐I‐expressing plants enhances recombinant protein production. Appl. Biochem. Biotechnol. 168, 1608–1620. [DOI] [PubMed] [Google Scholar]
- Pillay, P. , Schlüter, U. , van Wyk, S. , Kunert, K.J. and Vorster, B.J. (2014) Proteolysis of recombinant proteins in bioengineered plant cells. Bioengineered, 5, 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay, P. , Kunert, K.J. , van Wyk, S. , Makgopa, M.E. , Cullis, C.A. and Vorster, B.J. (2016) Agroinfiltration contributes to VP1 recombinant protein degradation. Bioengineered, 7, 459–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richau, K.H. , Kaschani, F. , Verdoes, M. , Pansuriya, T.C. , Niessen, S. , Stuber, K. , Colby, T. et al. (2012) Subclassification and biochemical analysis of plant papain‐like cysteine proteases displays subfamily‐specific characteristics. Plant Physiol. 158, 1583–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, S. , Khalf, M. , Goulet, M.‐C. , D'Aoust, M.‐A. , Sainsbury, F. and Michaud, D. (2013) Protection of recombinant mammalian antibodies from development‐dependent proteolysis in leaves of Nicotiana benthamiana . PLoS ONE, 8, e70203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury, F. , Varennes‐Jutras, P. , Goulet, M.‐C. , D'Aoust, M.‐A. and Michaud, D. (2013) Tomato cystatin SlCYS8 as a stabilizing fusion partner for human serpin expression in plants. Plant Biotechnol. J. 11, 1058–1068. [DOI] [PubMed] [Google Scholar]
- Shabab, M. , Shindo, T. , Gu, C. , Kaschani, F. , Pansuriya, T. , Chintha, R. , Harzen, A. et al. (2008) Fungal effector protein AVR2 targets diversifying defense‐related Cys proteases of tomato. Plant Cell, 20, 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, A. , Kikuchi, Y. , Ogawa, K. , Saika, H. , Yuasa, K. and Nagahama, M. (2008) Purification and characterization of cathepsin B‐like cysteine protease from cotyledons of daikon radish, Raphanus sativus . FEBS J. 275, 5429–5443. [DOI] [PubMed] [Google Scholar]
- Tyanova, S. , Temu, T. and Cox, J. (2016a) The MaxQuant computational platform for mass spectrometry‐based shotgun proteomics. Nat. Protoc. 11, 2301–2319. [DOI] [PubMed] [Google Scholar]
- Tyanova, S. , Temu, T. , Sinitcyn, P. , Carlson, A. , Hein, M.Y. , Geiger, T. , Mann, M. et al. (2016b) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods, 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Zhou, X.M. , Xiong, H.X. , Mao, W.Y. , Zhao, P. and Sun, M.X. (2018) Papain‐like and legumain‐like proteases in rice: genome‐wide identification, comprehensive gene feature characterization and expression analysis. BMC Plant Biol. 18, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H . (2016) Ggplot2: elegant graphics for data analysis, 2nd edn. New York, NY: Springer.
- Zischewski, J. , Sack, M. and Fischer, R. (2015) Overcoming low yields of plant‐made antibodies by a protein engineering approach. Biotechnol. J. 11, 107–116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Proteases identified by ABPP‐MS.
Appendix S1 Mass spectrometry analysis.


