Summary
Rice (Oryza sativa L.) cultivars harbour morphological and physiological traits different from those of wild rice (O. rufipogon Griff.), but the molecular mechanisms underlying domestication remain controversial. Here, we show that awn and long grain traits in the near‐isogenic NIL‐GLA are separately controlled by variations within the GLA (Grain Length and Awn Development) gene, a new allele of GAD1/RAE2, which encodes one member of the EFPL (epidermal patterning factor‐like protein) family. Haplotype analyses and transgenic studies revealed that InDel1 (variation for grain length, VGL) in the promoter region of GLA (GLAVGL) increases grain length by promoting transcription of GLA. Absence of InDel3 (variation for awn formation, VA) in the coding region (CDS) of GLA (GLA va) results in short awn or no awn phenotypes. Analyses of minimum spanning trees and introgression regions demonstrated that An‐1, an important gene for awn formation, was preferentially domesticated and its mutation to an‐1 was followed by GLA and An‐2. Gene flow then occurred between the evolved japonica and indica populations. Quality analysis showed that GLA causes poor grain quality. During genetic improvement, awnlessness was selected in ssp. indica, whereas short–grained and awnless phenotypes with good quality were selected in japonica. Our findings facilitate an understanding of rice domestication and provide a favourable allele for rice breeding.
Keywords: allelic frequency, awn development, domestication, grain length, natural variation
Introduction
Rice (Oryza sativa L.), one of the earliest domesticated, primary food crops, feeds more than half of the world population (Khush, 1997). It originated from wild rice (O. rufipogon Griff.) through domestication (Fuller et al., 2010; Huang et al., 2012; Zong et al., 2007). Wild rice has unfavourable agronomic traits, such as prostrate growth, seed shattering, a spreading panicle, long awns and low number of grains per panicle, all of which changed dramatically under domestication. Compared with its wild ancestors cultivated rice typically exhibits reduced seed shattering and dormancy, increased grain number per panicle, erect growth, closed panicles and no or short awns (Kovach et al., 2007; Sweeney and McCouch, 2007). All of these features that differ between wild rice and cultivars are known as domestication traits.
To date, some domesticated genes are functionally characterized, such as seed shattering genes Sh4 (Li et al., 2006) and qSH1 (Konishi et al., 2006; Li et al., 2006), prog1 controlling straight growth (Jin et al., 2008; Tan et al., 2008), OsLG1 regulating closed panicles (Ishii et al., 2013; Zhu et al., 2013), grain size genes GW5/GSE5 (Duan et al., 2017; Liu et al., 2017), GS3 (Mao et al., 2010), OsLG3 (Yu et al., 2017) and OsLG3b (Yu et al., 2018), seed dormancy gene Sdr4 (Sugimoto et al., 2010), seed hull colour genes Bh4 (Zhu et al., 2011) and Rc (Sweeney et al., 2006), NOG1 increasing grain production (Huo et al., 2017) and awn development genes An‐1 (Luo et al., 2013), An‐2/LABA1 (Gu et al., 2015; Hua et al., 2015) and GAD1/RAE2 (Bessho‐Uehara et al., 2016; Jin et al., 2016).
Awn, one of the most important domestication traits in cereal crops, is common in graminaceous crops such as sorghum (Sorghum bicolor L.), wheat (Triticum aestivum L.), oat (Avena sativa L.) and barley (Hordeum vulgare L.). Long awns, extensions at the tip of the lemmas, help seed dispersal and prevent birds and mammals from preying on the grains (Elbaum et al., 2007; Hu et al., 2011; Kulic et al., 2009). In some cereal crops, such as wheat and barley, awns also have photosynthetic capability that contributes to grain yield (Abebe et al., 2010). However, rice awns have no chloroplasts and are inconvenient for harvesting and post‐harvest processing (Takahashi et al., 1986). Hence, domesticated rice usually possesses no or short awns (Toriba et al., 2010).
The awn is a complex character controlled by multiple genes. Several awn‐related genes in rice have been cloned. An‐1, encoding a bHLH transcription factor, positively controls the formation of awn primordia, cell division and grain length in wild rice. The an‐1 allele conferring shorter awns, shorter grains and higher number of grains per panicle was selected during domestication (Luo et al., 2013). An‐2/LABA1 encodes a cytokinin synthesis enzyme and increases the cytokinin concentration in awn primordia, thus promoting awn elongation. It also negatively regulates grain number per panicle and tiller number per plant (Gu et al., 2015; Hua et al., 2015). RAE2/GAD1 encodes a secreted signal protein member of the epidermal patterning factor‐like family. The precursor peptide of RAE2/GAD1 must be ruptured under the action of lyase to form a mature polypeptide. RAE2/GAD1 regulates awn development, as well as grain number per panicle and grain length (Bessho‐Uehara et al., 2016; Jin et al., 2016). The above reports demonstrated that genes controlling awn phenotype have pleiotropic effects on yield‐related traits. Long awn and grain length are often closely associated, suggesting that alleles conferring longer grain might be neglected because of the selective sweep of awn traits. Currently, the relationship between these traits is not clear and the genetic basis and molecular mechanisms conferring the selective forces involved in awn formation and grain length during domestication need to be further examined.
Here, we isolated a multifunctional gene GLA (Grain Length and Awn Development), which is an allele of GAD1/RAE2. Using GLA‐gene association analysis, haplotype analysis and transgenic studies, we analysed the functional variations of GLA for grain length and awn formation. Evolutionary studies explained the relationship among the An‐1, An‐2 and GLA genes during the domestication process. In addition, quality evaluation and allele utilization were used to analyse the selection histories of japonica and indica rice in the process of genetic improvement. These studies not only improve our understanding of crop domestication, but also provide a valuable genetic resource for future molecular breeding of rice.
Results
Phenotypic characterization of NIL‐GLA and NIL‐gla
The backcross populations segregated at a single locus for the awn trait (Table S1). Long awns and long grains were completely associated in the BC3F4 population (Figure S1). The NIL‐GLA line with long awns and grains and NIL‐gla line with no awn and short grains were selected from the BC3F4 population (Figure S2). SEM of spikelet development at the Sp8I growth stage (Itoh et al., 2005) showed that the awn primordia of NIL‐GLA spikelets were more developed than in NIL‐gla spikelets (Figure 1a,b). NIL‐gla also developed much shorter grains and more grains per panicle than NIL‐GLA, but there was no significant difference in grain width (Figure 1c,d,h–j).
Figure 1.
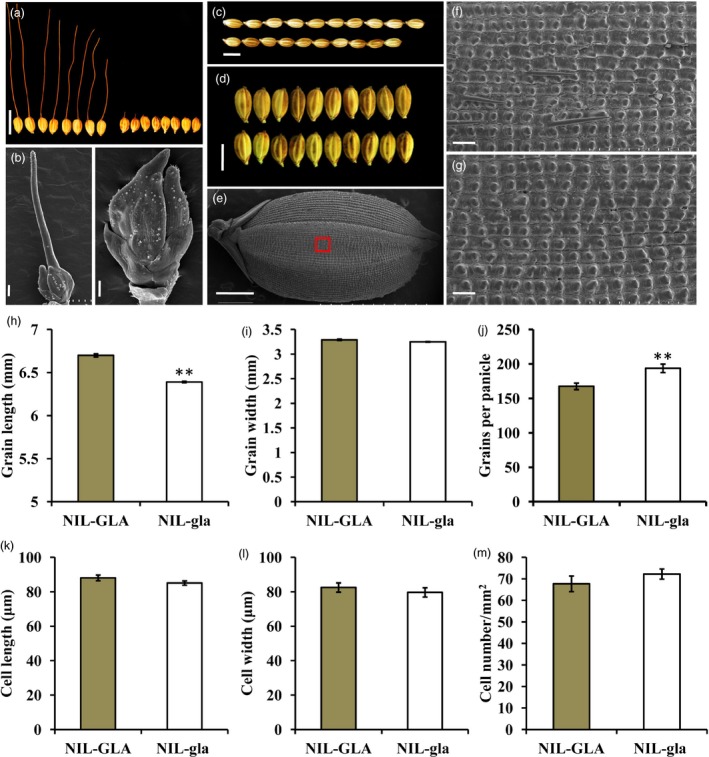
Phenotypes of NIL‐GLA and NIL‐gla NIL plants. (a) Awn phenotypes of NIL‐GLA (left) and NIL‐gla (right). Bar, 10 mm. (b) SEM images of NIL‐GLA (left) and NIL‐gla (right) spikelets at the Sp8 l growth stage. Bar, 200 μm. (c,d) and (h,i) Comparison of grain length (c,h) and width (d,i) between NIL‐GLA (upper) and NIL‐gla (lower) NILs. Bar, 5 mm. (e) SEM image of a whole grain. Bar, 1 mm. The red boxed region is enlarged in SEM images of the outer epidermal cells of the lemma shown in NIL‐GLA (f) and NIL‐gla (g) grains. Bar, 100 μm. (j) Comparison of grain numbers per panicle between NIL‐GLA and NIL‐gla. (k–m) Comparisons of average cell length (k), average cell width (l) and average cell number per mm2 (m) on the outer surface of the lemmas of NIL‐GLA and NIL‐gla grains. Data are represented as means ± SE (n = 15), **P < 0.01 based on Student's t‐tests.
As awn development and grain elongation are related to cell division (Hua et al., 2015; Jin et al., 2016; Luo et al., 2013) the outer lemma surfaces of grains were examined using SEM (Figure 1e). There was no significant difference in average cell number per unit area, cell length or cell width between NIL‐GLA and NIL‐gla, suggesting that cell number rather than cell size was mainly responsible for the longer grains in NIL‐GLA (Figure 1f,g,k–m). These results demonstrated that the gene controlling awn phenotype in NIL‐GLA had pleiotropic effects on grain length and grain number per panicle. It was named GLA (Grain Length and Awn Development).
GLA is a new allele at the RAE2/GAD1 locus
To isolate GLA 477 SSR markers were subjected to BSA (Zhang et al., 1994). Using 203 BC3F3 individuals for preliminary analysis, GLA was mapped between SSR markers RM56 and RM81 (Data S1) on the long arm of chromosome 8 (Figure 2a). Next, one SSR marker (RM37) and five STS markers (In84, In86, In26, S3, S4; Data S1) were used for fine mapping. Among 12 204 BC3F4 individuals, 2981 were awnless and GLA was delimited to a 26.32 kb genomic region between markers RM37 and S3 (Figure 2b).
Figure 2.
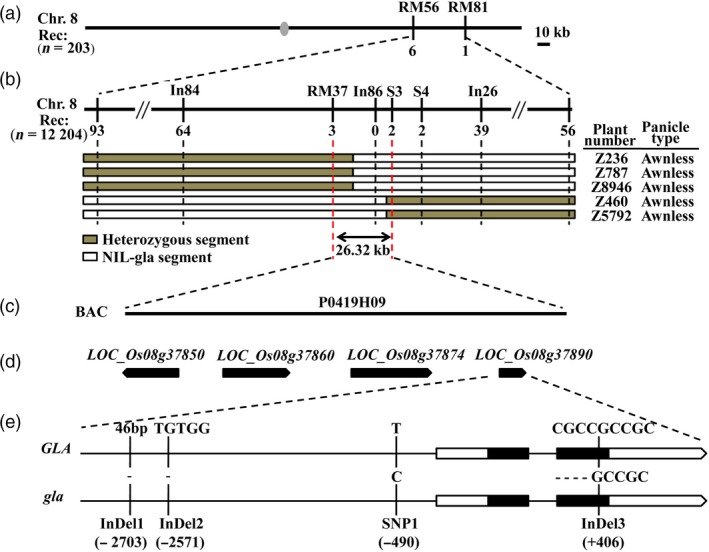
Fine mapping of GLA. (a) GLA was located between markers RM56 and RM81 on rice chromosome 8. Numbers of recombinants between GLA and molecular markers are shown below the bars. Grey ellipse represents the centromere. (b) GLA was fine mapped to a 26.32 kb region between markers RM37 and S3. Numbers of recombinants are given below the markers. Chromosome composition and panicle type of five recombinant plants are shown. White and grey bars represent chromosomal segment of NIL‐gla and heterozygotes respectively. (c) Nipponbare genomic BAC (P0419H09) containing this region. (d) Gene prediction by the Rice Annotation Project Database (http://rice.plantbiology.msu.edu/index.shtml). (e) Sequence variations between GLA in NIL‐GLA and gla in NIL‐gla. Black lines represent 5′ upstream regions and introns. White rectangles represent 5′ and 3′ UTR regions. Black rectangles represent coding regions. One SNP (SNP1) and three InDels (InDel1, InDel2 and InDel3) along with their positions were identified.
Four genes (LOC_Os08 g37850, LOC_Os08 g37860, LOC_Os08 g37874 and LOC_Os08 g37890) on BAC P0419H09 from this region were predicted by the Rice Annotation Project Database (http://rice.plantbiology.msu.edu/index.shtml) (Figure 2c,d, Table S2). Sequence analyses showed that differences between NIL‐GLA and NIL‐gla were present only in LOC_Os08 g37890; there were four variations, including three InDels and one SNP, namely, a 46 bp deletion (−2703, AGGTGTGGCATGGCAAACAAAGTGTGGCCAACAAATTGTTGGCCAC, InDel1), a 5 bp deletion (−2571, TGTGG, InDel2), a nucleotide substitution (−490, from T to C, named SNP1) in the promoter and a 4 bp nucleotide deletion (+406, CGCC, InDel3) in the second exon (Figures 2e and S3). Amino acid sequence analysis indicated that the InDel3 deletion caused a premature stop codon and truncated protein (Figure S4). LOC_Os08 g37890 is the same locus as Hap.A/B/C (Yano et al., 2016), GAD1 (Jin et al., 2016) and RAE2 (Bessho‐Uehara et al., 2016; Jin et al., 2016; Yano et al., 2016) that belong to the epidermal patterning factor‐like protein (EPFL) family and regulate awn development. Thus, LOC_Os08 g37890 was postulated to be the most likely candidate gene for GLA. Amino acid sequence alignments showed that GLA was a new allele of RAE2/GAD1 (Figure S5).
In the previous studies, GLA encoded a pre‐propeptide with an N‐terminal signal peptide and a C‐terminal mature peptide that is specifically cleaved by SLP1 in rice (Bessho‐Uehara et al., 2016; Jin et al., 2016). However, the sub‐cellular localization of GLA remains unknown. Interestingly, we found that fluorescent signals of both GLA‐GFP and gla‐GFP fusion proteins could be detected in the cell membrane, cytoplasm and nuclei of rice protoplasts (Figure S6). It may be related to SLP1 cleavage in rice protoplasts. In addition, although gla encoded a truncated protein, there was no effect on sub‐cellular localization (Figure S6).
Grain length and awn development are regulated by different functional variations of GLA
GLA was shown to affect both grain length and awn traits and the functional loci for each trait were still not clear. GLA‐based association analyses were performed on the basis of 33 SNPs and 13 InDels in 358 cultivated accessions (Data S2). The results indicated that grain length and awn development were significantly associated with different variations. InDel1 and InDel3 made significant contributions to grain length, whereas SNP1 and InDel3 were significantly associated with awn formation (Figure 3a).
Figure 3.
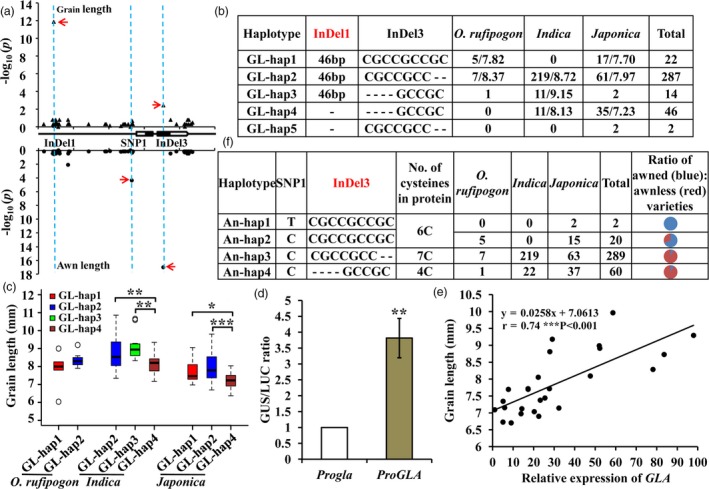
Variations in GLA were significantly associated with grain length and awn development. (a) Gene‐based association analysis of GLA for grain length (upper) and awns (lower) using a naïve model (t‐test; P = 2.17 × 10−2). Red arrows represent significant loci. (b,f) Haplotype analysis of GLA for grain length (b) and awns (f). No., number. Average grain lengths of rice varieties harbouring corresponding haplotypes are indicated following the slash. (c) Comparison of grain lengths in different haplotypes. (d) Transient expression assays of promoter activity in tobacco leaves. ProGLA represents the presence of InDel1 in the promoter. Progla represents the deletion of InDel1 in the promoter. Each experiment consisted of three biological repeats with four technical replications. (e) Correlation analysis between GLA mRNA levels and grain length in 26 rice varieties assessed at Sanya (Hainan province).
According to the significant variation associated with grain length, the sequences of 371 accessions including 358 cultivated and 13 wild rice varieties (Data S2) were divided into five haplotypes named GL‐hap1 to GL‐hap5. The wild rice accessions were mainly GL‐hap1 and GL‐hap2 types, indica accessions were GL‐hap2, GL‐hap3 and GL‐hap4 and japonica accessions were GL‐hap1, GL‐hap2 and GL‐hap4 (Figure 3b, Data S2). There was no significant difference in grain length between GL‐hap1 and GL‐hap2 in O. rufipogon and japonica or between GL‐hap2 and GL‐hap3 in indica. However, compared to GL‐hap1, GL‐hap2 or GL‐hap3, grain length of GL‐hap4 accessions was significantly reduced in both japonica and indica (Figure 3c, Data S2). These results suggested that grain length was influenced by the InDel1 (variation for grain length, VGL) in GLA (GLA VGL).
Transient assays were performed in tobacco leaves to determine whether the promoter activity was influenced by GLA VGL, which is located in the promoter. Relative promoter activities were significantly increased in ProGLA transformants containing the functional InDel1 (Figure 3d) suggesting that InDel1 increased the promoter activity of GLA. Moreover, transcriptional levels of GLA were positively correlated with grain length in 26 rice varieties randomly selected from the 358 cultivated varieties (Figure 3e, Data S3). These results indicated that GLA VGL regulated grain length by affecting the expression of GLA.
To determine the functional variation regulating awn formation on the basis of significant SNP/InDels, we classified the sequences of rice accessions into haplotypes An‐hap1, An‐hap2, An‐hap3 and An‐hap4. Comparing the numbers of cysteines (C) in GLA (Bessho‐Uehara et al., 2016) it became evident that An‐hap1 and An‐hap2 were 6C type, whereas An‐hap3 and An‐hap4 were 7C and 4C types respectively. The rare SNP1 variant in An‐hap1 occurred only in japonica. The InDel3 locus had three haplotypes. Compared with varieties of An‐hap2 (five awned varieties in O. rufipogon and nine awned and six awnless varieties in japonica), most varieties with An‐hap3 (54 of 63 varieties in japonica, 214 of 219 varieties in indica) and An‐hap4 (33 of 37 varieties in japonica, 22 varieties in indica) were awnless (Figure 3f, Data S2). This provided further evidence that InDel3 (variation for awn, VA) in GLA (GLA VA) confirmed the functional variation in awn development, a result that is consistent with the previous results for GAD1 (Bessho‐Uehara et al., 2016; Jin et al., 2016).
To further confirm the functions of VGL and VA, ProGLA VGL::GLA VA, ProGLA VGL::GLA va and ProGLA vgl::GLA VA complementary constructs were introduced into Nip with GLA vgl/va; the transgenic lines were named as NipI, NipII and NipIII respectively (Figure 4a). Compared to Nip, lines NipI‐1, NipI‐2, NipII‐1 and NipII‐2 showed longer grains, whereas the grain lengths of lines NipIII‐1 and NipIII‐2 were unchanged (Figure 4b,d). There was no significant change in grain width between transgenic lines (Figure S7c,f). Moreover, the phenotypes for awn length and awn proportion in NipI‐1, NipI‐2, NipIII‐1 and NipIII‐2 lines were restored (Figures 4c,e and S7a). This further proved that InDel1 and InDel3 in GLA were the functional sites for variation in grain length and the awn trait respectively. To further identify the function of GLA, a CRISPR‐Cas9 vector (CR) was constructed and transformed into NIL‐GLA, producing a CR9 transgenic line that had no awn and no effect on grain length and width (Figures 4b,c,h–j and S7b,d,e,g). Compared to Nip, lines NipI‐1, NipI‐2, NipIII‐1 and NipIII‐2 had lower grain number per panicle (Figure 4g). Compared to NIL‐GLA, the grain number per panicle of line CR9 was significantly increased (Figure 4k), thus suggesting that GLA va increased grain number per panicle. All of these results collectively verified that the functional variation of GLA in regulating grain length was different from that affecting awn development and that natural alleles of GLA in the VGL and VA loci might be responsible.
Figure 4.
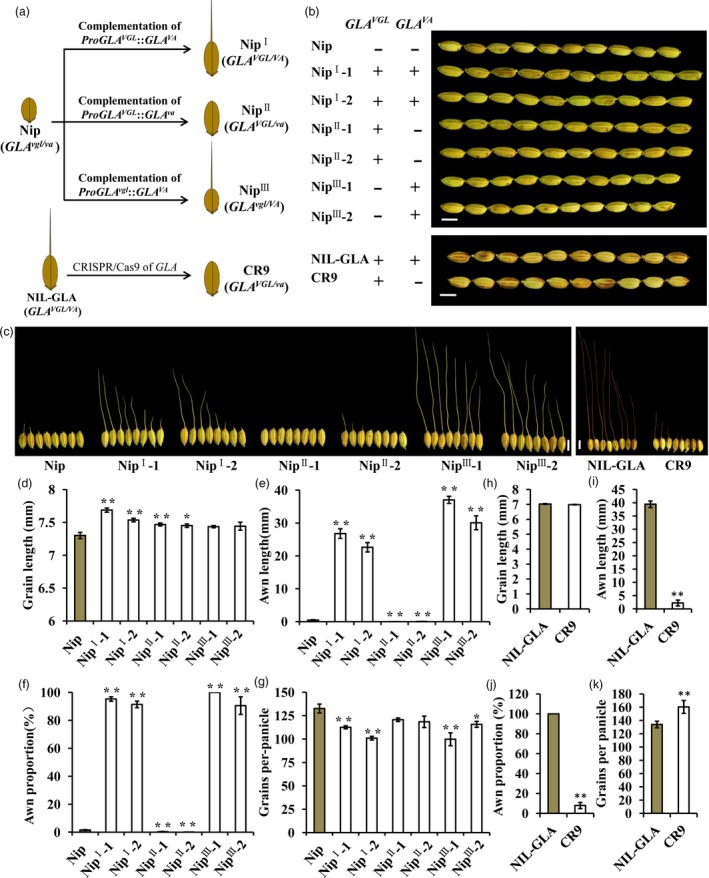
Confirmation of GLA in controlling grain length and awn development. (a) Schematic of transgenosis. VGL, variation for grain length. VA, variation for awn development. (b,c) Grain morphology of Nip, NIL‐GLA, six complementation transgenic lines in Nipponbare (Nip) background, and a CRISPR/Cas9 edited line in NIL‐GLA background. NipI‐1 and NipI‐2 are independent transgenic lines containing the Pro GLAVGL ::GLAVA vector. NipII‐1 and NipII‐2 are independent transgenic lines carrying the Pro GLAVGL::GLA va construct. NipIII‐1 and NipIII‐2 are independent transgenic lines carrying the ProGLA vgl::GLAVA construct. CR9 is a CRISPR/Cas9 edited line. ‘+’ and ‘−’ represent functional and non‐functional genotypes. Bar, 5 mm. (d–k) Comparison of grain lengths (d,h), awn lengths (e,i), awn proportions (f,j) and grain numbers per panicle among transgenic lines. Data are represented as means ± SE (n = 15), *P < 0.05, **P < 0.01 based on Student's t‐tests.
Allele GLA va was selected before GLA vgl
We categorized 371 rice accessions (358 cultivated and 13 wild rice) into four groups according to non‐functional (−) or functional (+) alleles of GLA VGL and GLA VA. There was no accession in group III. Most accessions in group I (five O. rufipogon, 11 of 17 in japonica) with GLA VA had awned phenotypes and most accessions in groups II (53 of 63 in japonica, 225 of 230 in indica) and IV (11 in indica, 34 of 38 in japonica) with GLA va were awnless (Figure 5a, Data S2). Grain lengths of wild rice accessions with GLA VGL showed no significant difference between groups I and II. However, compared to grain lengths of group IV in japonica and indica, respectively, varieties with GLA VGL in group I in japonica, group II in japonica and group II in indica had longer grains (Figure 5b, Data S2). These results indicated that the grain length and awn phenotype of various rice varieties were consistent across different haplotype combinations. Moreover, most cultivated rice accessions belonged to group II with GLA VGL/va.
Figure 5.
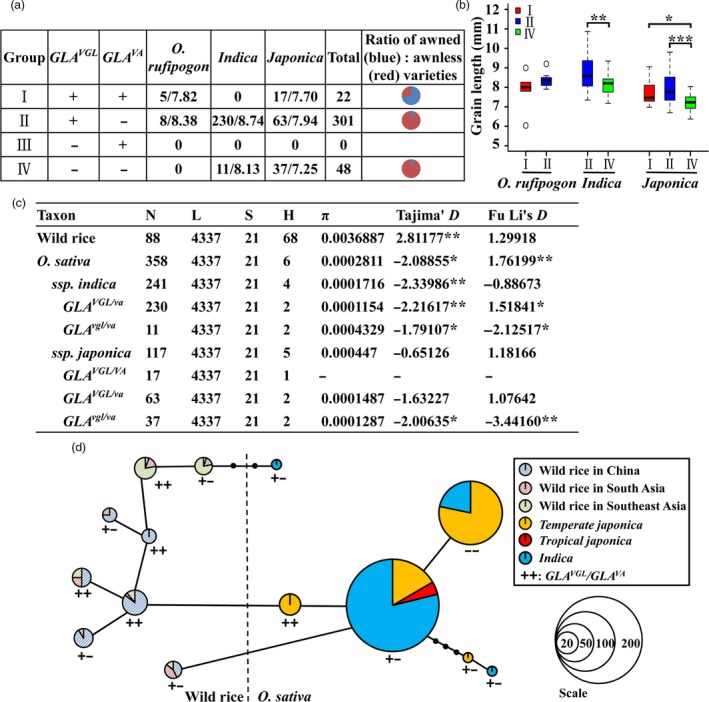
Evolutionary relationships of GLAVGL / VA for grain length and awn traits. (a,b) Haplotype analyses of GLAVGL and GLAVA in 371 rice accessions for grain length and awns. ‘+’ and ‘‐’ represent functional and non‐functional genotypes. (c) Nucleotide variation and neutrality test of GLA. N, L, S, H and π represent total number of sequences, average length (bp) of the sequences per taxon, number of polymorphic sites, number of haplotypes and average number of pairwise nucleotide differences per site calculated based on the total number of polymorphic sites respectively. *P < 0.05, **P < 0.01. (d) Minimum spanning tree for the GLA region based on 358 cultivated and 88 diverse wild rice sequences.
GLA is considered a domestication gene (Bessho‐Uehara et al., 2016; Jin et al., 2016) that controls grain length and awn development. The question then was what genotypes of GLA VGL/VA were selected artificially. A nucleic acid diversity analysis was conducted and a neutrality test was carried out based on the variant sites (~4.4 kb). The nucleotide diversity of gla in cultivated rice was significantly lower than that of GLA in wild rice. Tajima's D test (P < 0.01) revealed that GLA in cultivated and wild rice deviated significantly from neutrality (Figure 5c) indicating that GLA in wild rice was subjected to balanced natural selection and that gla in cultivated rice had undergone directional artificial selection. Thus selection in wild and cultivated rice led to different phenotypes. Natural selection in wild rice favoured awned phenotypes, whereas artificial selection favoured awnlessness (GLA va) in indica and short grains plus awnlessness (GLA vgl/va) in japonica (Figure 5c).
In order to explore evolutionary relationships between GLA VGL and GLA VA, 358 cultivated and 88 wild rice accessions were used to produce a minimum spanning tree. Wild rice had both GLA VGL/VA (documented as ++) and GLA VGL/va (+−) genotypes. However, temperate japonica rice that differentiated from wild rice in China did not acquire the GLA va allele. Temperate japonica with GLA VGL/VA first produced the natural variant GLA va. The long grain, awnless genotype GLA VGL/va was obtained by artificial selection. Subsequently, the GLA VGL/va allele changed to GLA vgl/va (− −) in cultivated rice. These results indicated that the GLA va allele was selected before GLA vgl. Moreover, indica rice first appeared in a long‐grain, awnless combination and japonica and indica had different selection histories, leading to short‐grain, awnless and long‐grain, awnless genotypes respectively (Figure 5d). Thus, we hypothesize that GLA va domestication occurred before divergence of the indica subspecies.
An‐1 was preferentially domesticated before GLA and An‐2
The currently cloned genes controlling awn traits are An‐1, An‐2 and GLA/GAD1/RAE2. To determine the evolutionary relationships among these genes during domestication a minimum spanning tree was constructed using 145 cultivated and 69 wild rice accessions. The results showed that a change in An‐1 was the first step in domestication. This produced temperate japonica with an‐1/An‐2/GLA VGL /GLA VA (symbolized −+++). Then, temperate japonica with an‐1 was domesticated at the functional position GLA VA leading to GLA va (−++−). By this time, indica rice had evolved. Part of the cultivated population with an‐1/An‐2/GLA VGL /GLA va (−++−) underwent natural variation in GLA VGL leading to the an‐1/An‐2/GLA vgl/GLA va (−+−−) genotype. Other cultivated rice (−++−) lines were domesticated by change in An‐2, and an‐1/An‐2/GLA VGL/GLA va (–+−) appeared (Figure 6a). Thus An‐1 was domesticated first, and was subsequently followed by changes in GLA and An‐2. Variation of GLA VGL occurred after the GLA VA domestication step.
Figure 6.
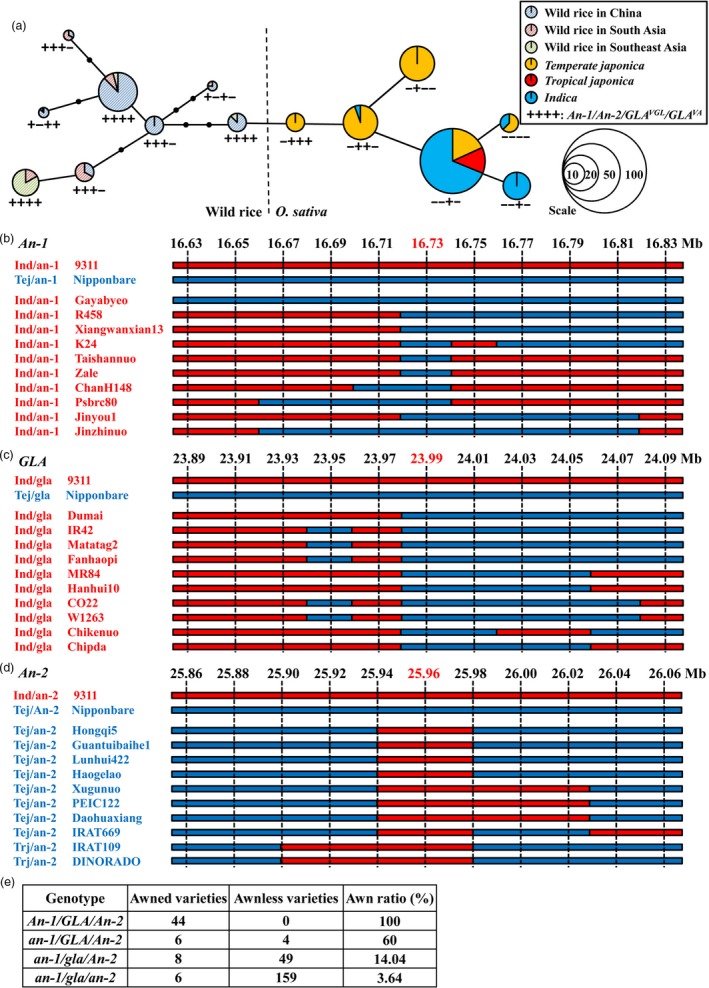
Evolutionary relationships among An‐1,GLA and An‐2. (a) Minimum spanning tree for the An‐1,GLA and An‐2 regions among 145 cultivated and 69 wild rice accessions. (b–d) Analyses of introgressed regions for An‐1 (b), GLA (c) and An‐2 (d). Ten SNP markers in 100 kb intervals upstream and downstream of these genes were used. Ind, indica. Tej, temperate japonica. Trj, tropical japonica. Nipponbare and 9311 were reference sequences. Red and blue bars represent indica and japonica genotypes respectively. Markers at 16.73, 23.99 and 25.96 Mb were the closest to An‐1,GLA and An‐2 respectively. (e) Comparison of rates of contribution to awn traits among An‐1, An‐2 and GLA.
The minimum spanning tree of GLA or three domestication genes controlling awn development clustered indica and japonica together, similar to previous studies on gene flow (Choi et al., 2017; Huang et al., 2012). To determine whether these genes were associated with gene flow of domestication alleles we performed analyses of introgressed regions. The an‐1 and gla domestication alleles were transferred from japonica to the indica population, whereas the an‐2 allele was transferred from indica to japonica (Figure 6b–d). These results demonstrated significant gene flow between the japonica and indica populations.
We then evaluated the contributions of An‐1, An‐2 and GLA to awn development. The genotypes and phenotypes of 232 cultivated and 44 wild rice accessions revealed that after domestication of An‐1, the frequency of accessions with awns decreased from 100% to 60%, and awn proportion fell to 14.04% and 3.64% after GLA and An‐2 domestication respectively (Figure 6e). We concluded that An‐1 made the largest contribution to awn phenotype, followed by GLA and An‐2 respectively.
The selection to GLA vgl is beneficial to rice quality improvement
Grain length is closely related to grain quality (Zhao et al., 2018). We evaluated the grain quality of NILs and found that rice milled from NIL‐GLA had high percentages of chalky grain and chalky area with loose and irregular starch granules, unlike that from NIL‐gla, which displayed compactly arranged and largely sharp‐edged polygonal starch granules (Figure 7a–d). These results indicated that GLA caused poor quality. The grain qualities of transgenic plants were also investigated. Compared to Nip, NipI‐1, NipI‐2, NipII‐1 and NipII‐2, transgenic plants had a higher proportion of chalky grain and small, messy starch granules, whereas Nip, the NipIII‐1 and NipIII‐2 lines had similar phenotypes (Figure 7a,b,e,f). Thus GLA VGL caused poor quality whereas GLA VA had no influence on rice grain quality.
Figure 7.
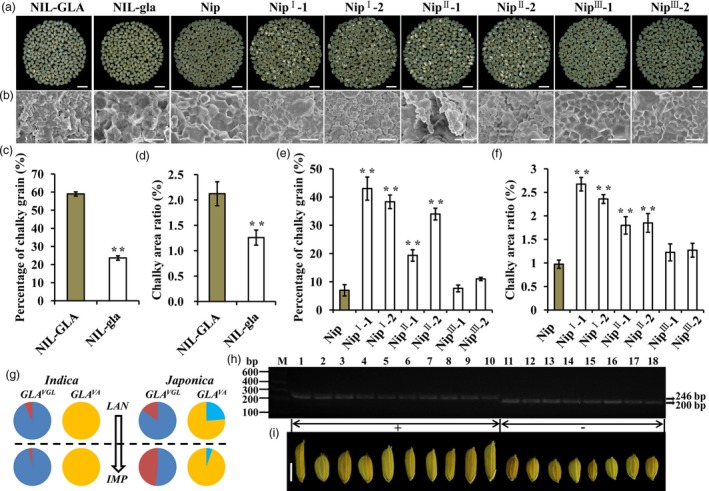
Improved grain quality produced by null GLAVGL. (a) Comparison of milled rice in NILs, Nip and complementation lines. Bar, 1 cm. (b) SEM images of starch granules in transverse sections of grains. Bar, 10 μm. (c–f) Percentages of chalky grain (c,e) and chalky area ratio (d,f). Data are represented as means ± SE. (n = 15), *P < 0.05, **P < 0.01 based on Student's t‐tests. (g) Allele frequency analysis of GLAVGL and GLAVA. Blue and red colors represent functional (GLAVGL) and non‐functional (GLA vgl) alleles of the VGL locus. Skyblue and orange colors represent functional (GLAVA) and non‐functional (GLA va) alleles of the VA locus. Pie charts above and below the horizontal line represent landraces (LAN) and improved varieties (IMP). (h) Genotyping analyses of GLAVGL in 18 diverse accessions of cultivated rice. We used a pair of primers (Table S2) to amplify gene segments containing the variable InDel1 site. The length of the PCR product amplified from accessions with GLAVGL (+) was 246 bp and that from accessions with GLA vgl (−) was 200 bp. (i) Grains of the 18 cultivated rice accessions. Bar, 5 mm. *P < 0.05, **P < 0.01, ***P < 0.001 based on Student's t‐tests.
Given that GLA VGL and GLA VA affected grain length, grain quality and awn development the question was how to make use of GLA in genetic improvement. We surveyed the functional allelic frequencies of GLA VGL/VA in the panel of 358 varieties. Compared with landrace groups (LAN), the non‐functional allele frequencies of GLA vgl and GLA va in improved accessions (IMP) were significantly increased in japonica, unlike indica in which the functional allele GLA VA did not exist and the functional allele frequency of GLA VGL was increased (Figure 7g). These results implied that indica and japonica had different selection histories during genetic improvement. The GLA VGL allele for long grain phenotype was enriched in indica with GLA va, whereas the GLA vgl/va allele for good quality, awnlessness and short grain was utilized in japonica. Additionally, a functional molecular marker (J2) for the VGL locus was developed (Data S1). Genotyping analyses using the J2 marker in 18 cultivars further confirmed that GLA VGL was most likely responsible for long‐grain phenotype (Figure 7h,i, Data S6). The J2 marker should be applied in future molecular marker assisted breeding.
Discussion
GLA has pleiotropic effects on grain length, awn development, grain quality and grain number
Wild rice exhibits long, barbed awns that prevent granivore predation, but is detrimental for harvesting and post‐harvest processing. Hence awnless varieties were produced during domestication of wild rice (Kovach et al., 2007; Sweeney and McCouch, 2007). Several genes associated with awn development have been identified. However, various genes for yield‐related traits, such as grain length, grain number and yield per plant were often closely associated with awn length and hence were neglected because of the selective sweep of awn traits. Thus, the molecular mechanisms of these genes needed to be investigated individually.
In this study, we identified Os08 g37890 as GLA, a new allele of RAE2/GAD1. GLA exhibited pleiotropic effects on grain length, grain quality, grain number and awn development, and had different functional sites. Our study identified a new functional site, GLA VGL, that regulated grain length and grain quality and further verified the GLA VA functional site controlling awn development. In addition, GLA VGL affected grain length at the transcriptional level, consistent with RNAi results in GAD1 studies (Jin et al., 2016). During genetic improvement, japonica rice underwent improved quality, shorter grain and awnlessness, whereas indica became long grained and awnless. A functional molecular marker developed for GLA VGL can be used in breeding programs. On the basis of the long‐grain, awnless phenotype in indica and short‐grain, awnless traits in japonica, grain quality and grain length can be further improved.
Origin and evolution of awn‐associated domestication loci
An‐1 influences the formation of awn primordia by regulating cell division (Luo et al., 2013). An‐2 induces awn elongation by increasing the cytokinin concentration in awn primordial (Gu et al., 2015; Hua et al., 2015). GAD1/RAE2, an EPFL family member, regulates awn length (Bessho‐Uehara et al., 2016; Jin et al., 2016). Evidence from this study indicated that An‐1 was the first of these genes to undergo domestication, followed by GLA and then An‐2. This also suggested that An‐1 is the most important gene controlling awn development. Minimal spanning tree and introgression analyses implied that An‐1 and GLA might have been domesticated preferentially in japonica before the divergence of indica, but subsequently, the An‐2 allele was transferred from indica to japonica. Hence, some questions are worth pondering. Firstly, given that An‐2 makes the least contribution to awn formation why did introgression of An‐2 actually occur. It may be that An‐2 was linked to other genes and that transfer was due to linkage drag. The next issue is whether awn–related domestication genes arose only once and were dispersed by introgressive hybridization or whether they arose more than once. Several domestication‐related loci, such as OsLG3b, Ghd7, LG1 and LABA1, show introgression signals between the subspecies, but others (Sh4, qSH3, qSH1, Prog1, and Rc) lack such signals (Civan and Brown, 2018; Yu et al., 2018).
The origin of cultivated rice is a continuing, controversial issue with variable lines of evidence for single and multiple origins. Multiple origins are currently supported by most academics (Choi et al., 2017; Londo et al., 2006; Sun et al., 2018). Therefore, by combing previous studies and our results, we speculate that proto‐japonica from wild rice in Southern China went through the An‐1 and GLA domestication steps prior to japonica/indica differentiation and produced domesticated proto‐japonica varieties with the an‐1/gla genotype. After emergence of proto‐indica varieties from O. rufipogon in Southeast and South Asia they underwent domestication of An‐2. Later, populations of proto‐japonica with an‐1/gla and proto‐indica with an‐2 merged with each other and hybridization led to the current japonica and indica forms with genotype an‐1/gla/an‐2 (Figure S8). Nevertheless, the detailed evolutionary relationships based on awn traits need to be further analysed. Nonetheless, discovery of the new GLA allele will be of value for understanding the domestication and genetic improvement of rice.
Experimental procedures
Plant materials
Mapping populations were constructed from a cross between japonica cv. SYL (awned and long grains) as donor parent and japonica cv. Nipponbare (Nip, awnless and short grains) as recurrent parent. Recessive awnless and dominant awned individuals in the BC3F4 generation with short and long grains respectively, were chosen as near isogenic lines (NILs), named NIL‐gla and NIL‐GLA. A segregating BC3F3 population containing 203 individuals was used for preliminary mapping of GLA. A BC3F4 population with 12 204 individuals was used for fine mapping. Plant materials were grown under natural paddy conditions at Beijing, or at Sanya in Hainan province.
Fine mapping and candidate gene analysis
Four hundred and seventy‐seven SSR markers distributed across all 12 rice chromosomes were screened in a bulked segregant analysis (BSA) to identify the linked markers associated with the awn trait (Zhang et al., 1994) and were also used for preliminary mapping. Sequence‐tagged site (STS) markers were then developed for fine mapping using DNASTAR software.
Candidate gene annotations were made on the basis of the Rice Annotation Project Database (http://rice.plantbiology.msu.edu/index.shtml). Genomic DNA fragments of four predicted genes were amplified from the DNA of NIL‐gla and NIL‐GLA and sequenced, respectively. Sequences were analysed using the SeqMan program in DNASTAR software. Primers are listed in Data S1.
cDNA and quantitative RT‐PCR
Total RNA was extracted from 5 cm panicles of NIL‐GLA and NIL‐gla using Trizol reagent (Invitrogen, Carlsbad, USA). To eliminate contamination by genomic DNA 50 μg of RNA was digested with Recombinant DNase I (RNase‐free) (Takara, Japan) as described by the manufacturer. Two microgram of DNaseI‐treated RNA was reversely transcribed using M‐MLV Reverse Transcriptase (Takara) with an oligo (dT18) primer. The reaction product was diluted three times as template. Quantitative RT‐PCR (qRT‐PCR) was carried out as previously reported (Sun et al., 2018). OsActin1 was used for internal reference (Data S1).
Vector construction and rice transformation
To make the genomic DNA complementation construct (ProGLA VGL::GLA VA) containing functional InDel1 and InDel3 a 4717 bp genomic DNA fragment of GLA harbouring a 3292 bp region upstream of the initiation codon and a 919 bp region downstream of the start codon was amplified from NIL‐GLA using primers LA and cloned into the PmeI and SacI sites of the binary plant expression vector pMDC163 (Curtis and Grossniklaus, 2003). Similarly, ProGLA VGL::GLA va and ProGLA vgl::GLA VA vectors were constructed using primers LA/la and la/LA respectively. The ProGLA VGL::GLA va vector contained a 3292 bp corresponding region upstream of the initiation codon of GLA from NIL‐GLA line and a 1421 bp region downstream of the initiation codon of GLA from NIL‐gla line. The ProGLA vgl::GLA VA vector consisted of a 3241 bp region upstream of the initiation codon of GLA from NIL‐gla and a 1425 bp region downstream of the start codon of GLA from NIL‐GLA. A CRISPR‐Cas9 construct (CR) targeting the 74th–93th nt (5′‐ GCTGCTACAGCAAGTGCTAC‐3′) of the second exon of GLA was made as previously described (Feng et al., 2013; Mao et al., 2013; Zhang et al., 2014). To generate transgenic plants, the constructs were introduced into Agrobacterium tumefaciens strain EHA105 and subsequently transformed into Nipponbare (Nip) or NIL‐GLA by Agrobacterium‐mediated transformation (Hiei et al., 1994).
Phenotypic evaluation
Three main panicles of each plant were collected for analysis of awn length (>1 mm), awn proportion and grains per panicle. The awn length of the panicle was represented by the average of the apical spikelet on each primary branch. Awn proportion was estimated as the number of awned spikelets per panicle. One hundred fully mature grains were randomly chosen for measurement of grain length and width. The grains were placed on a scanner for scanning. Grain length and width were calculated by analysing the scanned images (Luo et al., 2013). Grain weight per plant was represented by the dry weight of all grains from one plant. At least 15 individuals were used for phenotyping of NIL‐GLA, NIL‐gla and Nip transgenic plants.
Scanning electron microscopy
Young spikelets from NIL‐GLA and NIL‐gla were fixed in 2.5% glutaraldehyde‐phosphate buffer saline fixative solution and dehydrated through an ethanol series (30, 50, 70, 80, 90, 95 and 100%). After dehydration the samples were dried with a carbon dioxide critical‐point dryer. Mature grains were cleaned with 1% Tween 20 and dried in a 45 °C oven. Dried grains and spikelets were gold plated and observed using a Hitachi S‐2460 scanning electron microscope at 15 kV. Cell number, cell length and cell width of lemmas were quantified from scanning electron microscopy images. Quantification was represented by the average of 15 grains.
Transient expression assays of promoter activity
To make constructs ProGLA and Progla, GLA promoters with and without InDel1 were amplified using primers TEGLA and TEgla respectively (Data S1) and cloned into the PmeI and SacI sites of the pMDC162 vector. ProGLA, Progla and Pro35S::LUC plasmid used as an internal control were co‐transferred into tobacco leaves. The ratios of GUS/LUC (luciferase) were used as relative promoter activities (Sun et al., 2018; Zhang et al., 2017). Three biological repeats, each with four technical replicates were analysed for each vector.
Sub‐cellular localization of GLA
GLA and gla cDNAs were amplified and cloned into pSuper1300 vector, containing a Super promoter (Ni et al., 1995; Yang et al., 2010), to obtain ProSuper::GLA‐GFP and ProSuper::gla‐GFP constructs respectively. The resulting vectors and CD3‐1007, a plasma membrane marker (Nelson et al., 2007) were co‐transfected into rice protoplasts. After culturing for 16 h at 28 °C fluorescence signals were observed as described (Zhang et al., 2011).
GLA‐based association analysis and haplotype analysis
A worldwide set of 358 cultivated rice accessions selected from the 3K‐Rice Project (Rice Functional Genomics and Breeding database, RFGB) (Zheng et al., 2015) and 13 Asian wild rice accessions provided by the National Germplasm Nanning Wild Rice Nursery were used for the study (Data S2). Genomic DNA fragments of GLA from different accessions were amplified using the V1, V2, V3 and V4 primers (Data S1 and S2), sequenced and analysed by DNASTAR software. Variable sites, including 33 SNPs (single nucleotide polymorphisms) and 13 InDels (insertion/deletion polymorphisms), were used for GLA‐based association analysis (Liu et al., 2016; Wang et al., 2016; Xiong et al., 2018). Significant sites based on the –log10 (P) values were used for haplotype analyses (Data S2). The InDel1 and InDel3 alleles of GLA in 371 rice accessions (Data S2) were analysed from the sequencing results and then subjected to genotyping. Group analyses were carried out as previously reported (Sun et al., 2018).
Allelic frequencies and neutrality tests
The 358 accessions (143 landraces and 215 improved accessions; Data S2 and S5) were analysed for the An‐1, An‐2 and GLA functional loci controlling grain length or awn development and genotyped as non‐functional or functional alleles (Sun et al., 2018).
Eighty eight wild rice accessions provided by Dr Song Ge and 358 cultivars (241 indica and 117 japonica) varieties were used to evaluate nucleic acid diversity (π) and neutrality (Tajima's D values) of GLA (Data S4). InDels and SNPs in GLA were identified. The data were processed by DnaSP 5.10 software (Librado and Rozas, 2009).
Analyses of minimum spanning trees
Rich sequence diversity for An‐1, An‐2 and GLA was present in both wild and cultivated rice (Data S5) and some haplotypes were represented by only one accession. We chose 9, 3 and 15 variable sites in An‐1 An‐2 and GLA respectively, and integrated haplotypes containing single varieties in developing the minimum spanning tree (Sun et al., 2018). The 358 cultivated and 88 wild rice accessions were used for analysis of the GLA region; and 145 cultivated varieties and 69 wild rice accessions were used to determine evolutionary relationships among the three genes. A minimum spanning tree was generated following a previous procedure (Liu et al., 2015; Yu et al., 2018).
Competing interests
The authors declare that they have no competing interests.
Author contribution
ZL, ZZ, HZ, JL and XS conceived the experiment; BL and CC produced the genetic populations; YZ and BL performed experiments; YZ, XZ, YP and XS conducted the phenotypings; YZ, XZ, XS and HG analysed the data; YZ, XZ, HG, ZZ and ZL wrote and revised the manuscript.
Supporting information
Figure S1 Co‐segregation of awn and long grain traits.
Figure S2 Comparison of awn length and awn proportion between NIL‐GLA and NIL‐gla NILs.
Figure S3 GLA genomic sequences in NIL‐GLA and NIL‐gla.
Figure S4 Sequence alignment of GLA proteins of NIL‐GLA and NIL‐gla.
Figure S5 Comparison of the amino acid sequences in GLA alleles.
Figure S6 Sub‐cellular localization of GLA protein in rice protoplasts.
Figure S7 Phenotypic analysis of transgenic plants.
Figure S8 Proposed evolutionary pathway of awns in O. sativa.
Table S1 Genetic analysis of BC3F3 and BC3F4 populations.
Table S2 Putative genes in the 26.32 kb GLA region.
Data S1 Primers used in this study.
Data S2 Information of cultivated and wild rice.
Data S3 Information of cultivated rice used in the correlation analysis between GLA mRNA levels and grain length.
Data S4 Information of cultivated rice and wild rice used in GLA analyses of nucleic acid diversity, neutral test and a minimum spanning tree.
Data S5 Information of cultivated rice and wild rice used in the phylogenetic tree and allele frequency analyses.
Data S6 Information of cultivated rice used in the genotyping analysis.
Acknowledgements
We thank Professor Robert A McIntosh, University of Sydney, for critical reading of the manuscript. We thank the National Germplasm Nanning Wild Rice Nursery for providing 13 wild rice accessions, Song Ge for providing 88 wild rice accessions, and Dr Jiankang Zhu for providing the sgRNA‐Cas9 vectors of the CRISPR‐Cas9 system. The research was supported by grants from the Ministry of Science and Technology in China (2016YFD0100101‐09, 2015BAD02B01), the National Natural Science Foundation of China (31271689).
References
- Abebe, T. , Melmaiee, K. , Berg, V. and Wise, R.P. (2010) Drought response in the spikes of barley: gene expression in the lemma, palea, awn, and seed. Funct. Integr. Genomics, 10, 191–205. [DOI] [PubMed] [Google Scholar]
- Bessho‐Uehara, K. , Wang, D.R. , Furuta, T. , Minami, A. , Nagai, K. , Gamuyao, R. , Asano, K. et al (2016) Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc. Natl Acad. Sci. USA, 113, 8969–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J.Y. , Platts, A.E. , Fuller, D.Q. , Hsing, Y.I. , Wing, R.A. and Purugganan, M.D. (2017) The rice paradox: multiple origins but single domestication in Asian rice. Mol. Biol. Evol. 34, 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civan, P. and Brown, T.A. (2018) Role of genetic introgression during the evolution of cultivated rice (Oryza sativa L.). BMC Evol. Biol., 18, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, P. , Xu, J. , Zeng, D. , Zhang, B. , Geng, M. , Zhang, G. , Huang, K. et al (2017) Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant, 10, 685–694. [DOI] [PubMed] [Google Scholar]
- Elbaum, R. , Zaltzman, L. , Burgert, I. and Fratzl, P. (2007) The role of wheat awns in the seed dispersal unit. Science, 316, 884–886. [DOI] [PubMed] [Google Scholar]
- Feng, Z. , Zhang, B. , Ding, W. , Liu, X. , Yang, D.L. , Wei, P. , Cao, F. et al (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, D.Q. , Sato, Y.I. , Castillo, C. , Qin, L. , Weisskopf, A.R. , Kingwellbanham, E.J. , Song, J. et al (2010) Consilience of genetics and archaeobotany in the entangled history of rice. Archaeol. Anthropol. Sci. 2, 115–131. [Google Scholar]
- Gu, B. , Zhou, T. , Luo, J. , Liu, H. , Wang, Y. , Shangguan, Y. , Zhu, J. et al (2015) An‐2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Mol. Plant, 8, 1635–1650. [DOI] [PubMed] [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hu, G. , Zhang, D. , Pan, H. , Li, B. , Wu, J. , Zhou, X. , Zhang, Q. et al (2011) Fine mapping of the awn gene on chromosome 4 in rice by association and linkage analyses. Chin. Sci. Bull. 56, 835–839. [Google Scholar]
- Hua, L. , Wang, D. , Tan, L. , Fu, Y. , Liu, F. , Xiao, L. , Zhu, Z. et al (2015) LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell, 27, 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Kurata, N. , Wei, X. , Wang, Z. , Wang, A. , Zhao, Q. , Zhao, Y. et al (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature, 490, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo, X. , Wu, S. , Zhu, Z. , Liu, F. , Fu, Y. and Cai, H. (2017) NOG1 increases grain production in rice. Nat. Commun. 8, 1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, T. , Numaguchi, K. , Miura, K. , Yoshida, K. , Thanh, P.T. , Htun, T.M. , Yamasaki, M. et al (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet., 45, 462–465, 465e1‐2. [DOI] [PubMed] [Google Scholar]
- Itoh, J. , Nonomura, K. , Ikeda, K. , Yamaki, S. , Inukai, Y. , Yamagishi, H. , Kitano, H. et al (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46, 23–47. [DOI] [PubMed] [Google Scholar]
- Jin, J. , Huang, W. , Gao, J. , Yang, J. , Shi, M. , Zhu, M. , Luo, D. et al (2008) Genetic control of rice plant architecture under domestication. Nat. Genet. 40, 1365–1369. [DOI] [PubMed] [Google Scholar]
- Jin, J. , Hua, L. and Zhu, Z. (2016) GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication. Plant Cell, 28, 2453–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush, G.S. (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35, 25–34. [PubMed] [Google Scholar]
- Konishi, S. , Izawa, T. , Lin, S. , Ebana, K. , Fukuta, Y. , Sasaki, T. and Yano, M. (2006) An SNP caused loss of seed shattering during rice domestication. Science, 312, 1392–1396. [DOI] [PubMed] [Google Scholar]
- Kovach, M.J. , Sweeney, M.T. and McCouch, S.R. (2007) New insights into the history of rice domestication. Trends Genet. 23, 578–587. [DOI] [PubMed] [Google Scholar]
- Kulic, I.M. , Mani, M. , Mohrbach, H. , Thaokar, R. and Mahadevan, L. (2009) Botanical ratchets. Proc. Biol. Sci. 276, 2243–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Zhou, A. and Sang, T. (2006) Rice domestication by reducing shattering. Science, 311, 1936–1939. [DOI] [PubMed] [Google Scholar]
- Librado, P. and Rozas, J. (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Liu, H. , Zhou, L. , Zhang, Z. , Zhang, X. , Wang, M. , Li, H. et al (2015) Parallel domestication of the heading date 1 gene in cereals. Mol. Biol. Evol. 32, 2726–2737. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Huang, M. , Fan, B. , Buckler, E.S. and Zhang, Z. (2016) Iterative usage of fixed and random effect models for powerful and efficient genome‐wide association studies. PLoS Genet. 12, e1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Chen, J. , Zheng, X. , Wu, F. , Lin, Q. , Heng, Y. , Tian, P. et al (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants, 3, 17043. [DOI] [PubMed] [Google Scholar]
- Londo, J.P. , Chiang, Y.C. , Hung, K.H. , Chiang, T.Y. and Schaal, B.A. (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa . Proc. Natl Acad. Sci. USA, 103, 9578–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J. , Liu, H. , Zhou, T. , Gu, B. , Huang, X. , Shangguan, Y. , Zhu, J. et al (2013) An‐1 encodes a basic helix‐loop‐helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell, 25, 3360–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, H. , Sun, S. , Yao, J. , Wang, C. , Yu, S. , Xu, C. , Li, X. et al (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl Acad. Sci. USA, 107, 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y. , Zhang, H. , Xu, N. , Zhang, B. , Gou, F. and Zhu, J. (2013) Application of the CRISPR‐Cas system for efficient genome engineering in plants. Mol. Plant, 6, 2008–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B.K. , Cai, X. and Nebenfuhr, A. (2007) A multicolored set of in vivo organelle markers for co‐localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ni, M. , Cui, D. , Einstein, J. , Narasimhulu, S. , Vergara, C.E. and Gelvin, S.B. (1995) Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 7, 661–676. [Google Scholar]
- Sugimoto, K. , Takeuchi, Y. , Ebana, K. , Miyao, A. , Hirochika, H. , Hara, N. , Ishiyama, K. et al (2010) Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl Acad. Sci. USA, 107, 5792–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Zhang, Z. , Chen, C. , Wu, W. , Ren, N. , Jiang, C. , Yu, J. et al (2018) The C‐S‐A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice. J. Exp. Bot. 69, 1485–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, M. and McCouch, S. (2007) The complex history of the domestication of rice. Ann. Bot. 100, 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, M.T. , Thomson, M.J. , Pfeil, B.E. and McCouch, S. (2006) Caught red‐handed: Rc encodes a basic helix‐loop‐helix protein conditioning red pericarp in rice. Plant Cell, 18, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, N. , Alterfa, H.A.H. and Sato, T. (1986) Significant role of awn in rice plants (1): a survey of agricultural value of rice awn. FEMS Microbiol. Lett. 301, 232–240. [Google Scholar]
- Tan, L. , Li, X. , Liu, F. , Sun, X. , Li, C. , Zhu, Z. , Fu, Y. et al (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. [DOI] [PubMed] [Google Scholar]
- Toriba, T. , Suzaki, T. , Yamaguchi, T. , Ohmori, Y. , Tsukaya, H. and Hirano, H.Y. (2010) Distinct regulation of adaxial‐abaxial polarity in anther patterning in rice. Plant Cell, 22, 1452–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Wang, H. , Liu, S. , Ferjani, A. , Li, J. , Yan, J. , Yang, X. et al (2016) Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48, 1233–1241. [DOI] [PubMed] [Google Scholar]
- Xiong, H. , Yu, J. , Miao, J. , Li, J. , Zhang, H. , Wang, X. , Liu, P. et al (2018) Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol. 178, 451–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Shi, Y. , Liu, J. , Guo, L. , Zhang, X. and Yang, S. (2010) A mutant CHS3 protein with TIR‐NB‐LRR‐LIM domains modulates growth, cell death and freezing tolerance in a temperature‐dependent manner in Arabidopsis. Plant J. 63, 283–296. [DOI] [PubMed] [Google Scholar]
- Yano, K. , Yamamoto, E. , Aya, K. , Takeuchi, H. , Lo, P.C. , Hu, L. , Yamasaki, M. et al (2016) Genome‐wide association study using whole‐genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 48, 927–934. [DOI] [PubMed] [Google Scholar]
- Yu, J. , Xiong, H. , Zhu, X. , Zhang, H. , Li, H. , Miao, J. , Wang, W. et al (2017) OsLG3 contributing to rice grain length and yield was mined by Ho‐LAMap. BMC Biol. 15, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Miao, J. , Zhang, Z. , Xiong, H. , Zhu, X. , Sun, X. , Pan, Y. et al (2018) Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 16, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Shen, B. , Dai, X. , Mei, M. , Saghai Maroof, M.A. and Li, Z. (1994) Using bulked extremes and recessive class to map genes for photoperiod‐sensitive genic male sterility in rice. Proc. Natl Acad. Sci. USA, 91, 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Su, J. , Duan, S. , Ao, Y. , Dai, J. , Liu, J. , Wang, P. et al (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast‐related processes. Plant Methods, 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Li, J. , Pan, Y. , Li, J. , Zhou, L. , Shi, H. , Zeng, Y. et al (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 8, 14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D. , Li, Q. , Zhang, C. , Zhang, C. , Yang, Q. , Pan, L. , Ren, X. et al (2018) GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9, 1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, T. , Hong, Y. , Zhang, H. , Wu, Z. , Wang, W. , Tai, S. , Lu, C. et al (2015) Rice functional genomics and breeding database (RFGB): 3K‐rice SNP and InDel sub‐database. Chin. Sci. Bull. 60, 367. [Google Scholar]
- Zhu, B. , Si, L. , Wang, Z. , Zhou, Y. , Zhu, J. , Shangguan, Y. , Lu, D. et al (2011) Genetic control of a transition from black to straw‐white seed hull in rice domestication. Plant Physiol. 155, 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z. , Tan, L. , Fu, Y. , Liu, F. , Cai, H. , Xie, D. , Wu, F. et al (2013) Genetic control of inflorescence architecture during rice domestication. Nat. Commun. 4, 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, Y. , Chen, Z. , Innes, J. , Chen, C. , Wang, Z. and Wang, H. (2007) Fire and flood management of coastal swamp enabled first rice paddy cultivation in east China. Nature, 449, 459–462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Co‐segregation of awn and long grain traits.
Figure S2 Comparison of awn length and awn proportion between NIL‐GLA and NIL‐gla NILs.
Figure S3 GLA genomic sequences in NIL‐GLA and NIL‐gla.
Figure S4 Sequence alignment of GLA proteins of NIL‐GLA and NIL‐gla.
Figure S5 Comparison of the amino acid sequences in GLA alleles.
Figure S6 Sub‐cellular localization of GLA protein in rice protoplasts.
Figure S7 Phenotypic analysis of transgenic plants.
Figure S8 Proposed evolutionary pathway of awns in O. sativa.
Table S1 Genetic analysis of BC3F3 and BC3F4 populations.
Table S2 Putative genes in the 26.32 kb GLA region.
Data S1 Primers used in this study.
Data S2 Information of cultivated and wild rice.
Data S3 Information of cultivated rice used in the correlation analysis between GLA mRNA levels and grain length.
Data S4 Information of cultivated rice and wild rice used in GLA analyses of nucleic acid diversity, neutral test and a minimum spanning tree.
Data S5 Information of cultivated rice and wild rice used in the phylogenetic tree and allele frequency analyses.
Data S6 Information of cultivated rice used in the genotyping analysis.


