Big Data resemble people: interrogate them just so, and they will tell you whatever you want to hear.
Perhaps you, gentle reader, have noticed that there seems to be an awful lot more data in recent years, but perhaps not a lot more knowledge. Welcome to the world of Big Data. Just what is Big Data, and how is it changing the world of cardiovascular medicine? Big Data may be defined as large sets of data that are given to analytic approaches that may reveal underlying patterns, associations, or trends. Big Data has also been characterized by the 4 Vs of volume (a lot of data), variety (data from different sources and in different forms), velocity (data are accumulated rapidly), and veracity (uncertainty as to whether the data are correct). However, these characteristics do not adequately define Big Data; one might say that if you have seen one set of Big Data you have seen one set of Big Data. It is perhaps more useful to think about Big Data as it relates to sources, repositories, and use (Figure 1, Table). Where do such data come from? How are they stored? How can they be analyzed and visualized? What can we learn from Big Data?
Figure 1.
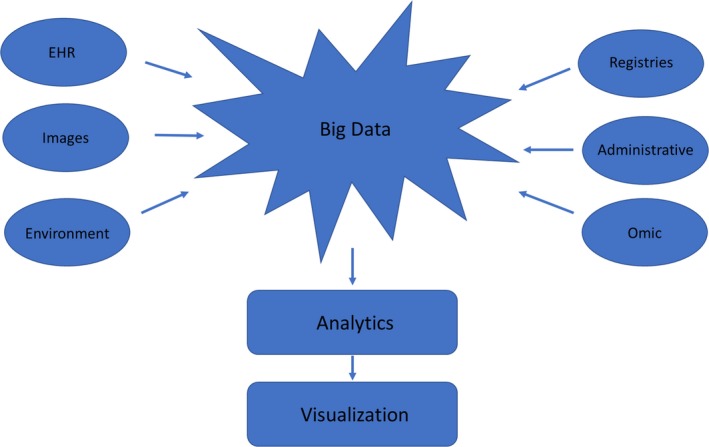
Flow of Big Data from sources to storage, analytics, and visualization. EHR indicates electronic health record.
Table 1.
Examples of Big Data Studies From Various Sources
| Type | Specific Source | Example |
|---|---|---|
| EHR | Academic medical center | EHR data used to develop and validate a prediction mode for adverse outcomes3 |
| Administrative data | Medicare Part A | Comparative effectiveness of percutaneous coronary intervention and coronary bypass surgery21 |
| National registry | Get With The Guidelines‐Resuscitation Registry | Derivation and validation of a mortality prediction tool after pediatric cardiac arrest25 |
| Imaging | MRIs | Cardiac MRI acquitition plane recognition30 |
| Integration of multiple data types | MRI, genetic, biomarker, and clinical registry | Hypertrophic Cardiomyopathy Registry31 |
| Social media/internet | Smart watch and internet application | Identification of cardiac arrhythmias using a smartwatch: the Apple Heart Study33 |
| Embedded clinical trial | National Cardiovascular Data Registry | Comparison of radial and femoral approaches for percutaneous coronary intervention in women: SAFE‐PCI for Women trial45 |
| Machine learning | Hospital heart failure registry | Classification of patients with heart failure57 |
EHR indicates electronic health record; MRI, magnetic resonance imaging; SAFE‐PCI, Study of Access Site for Enhancement of Percutaneous Coronary Intervention.
The Electronic Health Record
Perhaps the most ubiquitous source of Big Data in the realm of human health is the electronic health record (EHR). After several decades of discussion, financial incentives resulted in the relatively rapid conversion from handwritten charts to EHRs.1 The EHR can improve communication and facilitate much in the way of care, such as electronic pharmaceutical prescribing and communication between providers, both locally and regionally, via health information exchanges. However, perhaps the area with greatest potential is for the EHR to be a source of data that may be aggregated as Big Data.2, 3 Much of these data, such as laboratory testing, is readily available in digital, structured form. However, much of the information, such as admission summaries and procedural notes, generally remains in the form of text. For such data to be analyzable, it generally, although perhaps not always, needs to be converted from text to a more structured form, with or without natural language processing, an area of information science concerned with computer recognition and analysis of human language.4 The potential of such data is certainly grand, allowing patient‐level data collected at the point of care to be aggregated into Big Data. Such data can be from a single healthcare provider, such as an outpatient clinic or hospital, but may also be aggregated across healthcare systems. This can be scaled to impressive levels. For instance, the EHR data from the entire Department of Veterans Affairs Health System have been, for several years, on a single EHR platform, permitting querying across the entire system.
Such data can be used for evaluation of quality, business intelligence, and medical research. However, EHR data are not collected with the aim of creating data sets for scientific discovery. Rather, the purpose remains to document and coordinate care and perform administrative functions. EHR data lack the particular rigor of prospectively collected data sets, in which the design is focused on creating data sets for analysis. Thus, there can be serious limitations to EHR data, including missing data and misclassification, as well as use of nonstandard definitions. As the data were not collected for analytical purposes with predefined hypotheses and end points, analysis of EHR data is given to asking open‐ended questions, more of a “fishing expedition,” that may lead to unreliable, nonreproducible results.5, 6
Much can be done to improve Big Data sets coming from EHRs and, thus, provide information that can result in better analytics and more meaningful studies.7 The first is to move toward more structured data and less free text. This would limit the amount of natural language processing that needs to be done, but also can result in less missing data, depending on how the processes for collecting structure data are implemented. The use of structured data collection can also foster the use of data standards, such as those developed by the American Heart Association/American College of Cardiology Task Force on Data Standards.8 Laboratory data are already largely standardized by LOINC, and pharmaceutical data are standardized by RxNorm.9, 10 Using standard definitions for terms allows improved interpretation and more meaningful aggregation of data. Behind the clinical definitions lie the metadata (detailed description of the data) of how the data standards are translated into code that can be programmed into EHRs. Careful implementation of such standards can greatly improve interoperability (ie, data with shared metadata, content, and meaning across systems), facilitating communication; an example would be diabetes mellitus having the same name, definition, and computer representation across systems.11 Data from EHRs can also be retrieved using formal structure to the data, such as Fast Healthcare Interoperability Resources (FHIR), which was developed by the Health Level 7 (HL7) International healthcare standards organization.12, 13 There are also structured approaches for aggregating data from EHRs, such as the Observational Medical Outcomes Partnership (OMOP) Common Data Model, which can provide a framework for integrating data from disparate sources that would then facilitate analysis.14 A Common Data Model is a metadata system providing consistency of data and their meaning across applications and processes that store data in conformance with the model. Although tools such as the FHIR and the OMOP can greatly facilitate analysis of electronic health data, they do not substitute entirely for the use of structured using data standards within EHRs. However, collecting structured data within workflow is difficult to implement, requiring a substantial investment in informatics resources.15 There is also the danger in EHRs of structured data being carelessly copied from note to note without proper evaluation.16
Administrative Data
The counterpart to the EHR is the administrative data set. Such data are collected by healthcare providers largely for billing. The foremost example in the United States is the UB‐04, which is a standardized billing form for institutional providers, containing diagnostic, procedural, and charge data. The counterpart of the UB‐04 for professional billing is the Medicare Professional Claim Form (CMS‐1500). The primary purpose of the UB‐04 and the CMS‐1500 is for billing, by Medicare, Medicaid, and other payers. These data can also be aggregated, meeting anybody's definition of Big Data, for analytic purposes. The most important aggregation of these data sets is the Medicare set of databases. Medicare is also linked to the National Death Index, greatly facilitating analysis. There probably has been more analytic work, both published in the peer‐reviewed literature as well as used for assessment of quality and outcome, from Medicare databases than from any other source. In particular, Medicare data have been used to develop risk models, alone or linked to registry data, for clinical and economic outcomes.17, 18, 19 Medicare data have also been used in comparative effectiveness studies of observational evaluations of therapy.20, 21 However, administrative data have also been criticized for misclassification and missing data (eg, hypertension not coded when it should have been), for not capturing severity of illness, and for generally being less accurate than clinical registries.22
Clinical Registries
Beginning in the 1980s, professional societies began to organize clinical registries. The best known are from the Society of Thoracic Surgeons, the American College of Cardiology (National Cardiovascular Data Registry), and the American Heart Association (Get With The Guidelines).23, 24, 25 These registries collect structured data in various clinical settings (eg, percutaneous coronary intervention or cardiac surgery) using standardized definitions. However, the informatics metadata for interoperability of these registries are generally not available. These registries have grown tremendously, such that the National Cardiovascular Data Registry now has >60 million records, qualifying as Big Data. These databases are used for benchmarking quality at participating institutions and have resulted in many hundreds of publications. As noted above, these databases have also been linked to the Medicare databases, permitting assessment of long‐term outcome and economic evaluations.18, 20, 26 A major limitation of the society registries is that data collection is not generally linked to workflow. The data are often collected at a later time, meaning that data must be abstracted either partially or totally by hand from the EHR. In principal, it would be possible to build a collection of structured data into the workflow, so that data would be collected once, and then parsed where they need to go (Figure 2).15 However, in most healthcare systems, this remains aspirational, and implementation would require considerable investment in informatics to develop and deploy. Thus, data end up being collected several times and generally at least partially hand transposed from one system to another. It is not clear that such an approach will remain viable.
Figure 2.
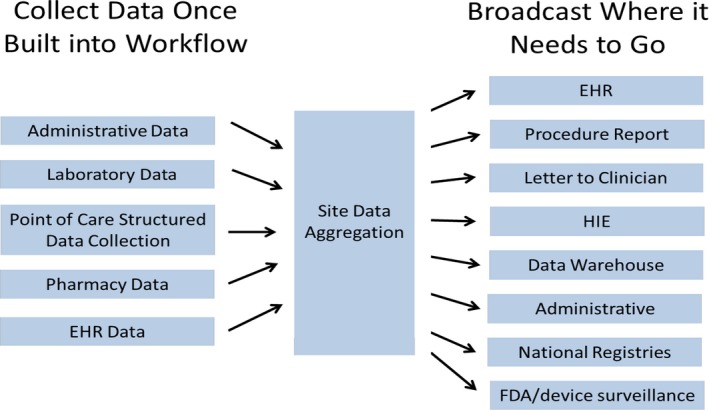
Structured data collection and reporting. EHR indicates electronic health record; FDA, US Food and Drug Administration; HIE, health information exchange.
Imaging Data
We are accustomed to thinking about data as existing in a structure such as flat file spreadsheets or relational databases, where the data are transformed from the original format to structured fields for analysis by classic statistical approaches, whether frequentist or bayesian. However, today, almost all medical images, whether static or moving, are stored in digital rather than analog formats. Images are stored as pixels or voxels describing a small area or volume, with millions of pixels or voxels forming one image. Such data can be analyzed and interpreted using careful image annotation and artificial intelligence approaches, such as neural networks.27, 28, 29 This fits the paradigm of variability of data type in the Big Data framework. Cardiac magnetic resonance imaging yields large data sets, both for image analysis as well as incorporation with other clinical data into registries.30, 31
Genomic and Other ‘Omic Data
Perhaps the prototypical form of Big Data in medical research comes from large‐scale databases of genomic, proteomic, and metabolomic data. This has produced the entire field of bioinformatics, which seeks to develop methods to store, retrieve, and analyze such data sets. This has been made possible by ever more rapid approaches to genetic sequencing. The human genome has >3 billion base pairs, each subject to variation. Furthermore, variation need not involve a single base pair within a single gene. Thus, the potential variation is large. Databases are now available with the full genome on millions of variations. Some 88 million have been identified, with ≈12 million being common. There currently are efforts to sequence 1 million genomes.32
We can expect increasing integration of ‘omic databases with EHR, registry, imaging, and other clinical data, again providing the variety of Big Data (Figure 1).31
Big Healthcare Data From Outside the Healthcare System
Healthcare providers may be forgiven for thinking that they would remain the host of healthcare data. However, it is now entirely possible to assemble large healthcare studies developed partially or totally independent of the healthcare system, with subjects recruited from the internet, often by means of an application and/or social media. This will include data from wearable devices as well as environmental data, such as air pollution. Perhaps the best known example is the Apple Heart Study, with 400 000 participants self‐enrolling, to study the ability of the Apple Watch to detect atrial fibrillation.33 Although this study may not have offered much insight into atrial fibrillation, it was certainly proof of concept that this could be done. We can expect all the major technology companies to be involved in Big Data healthcare projects involving data gathering, storage, and analytics.
It is of historical interest that the first scientific epidemiological study was of environmental exposure, in this case to cholera in London, UK, in the 1850s. Studies of environmental exposure to air and water pollution, toxic exposure (eg, lead or asbestos), and infectious disease (influenza) can result in large data sets, especially as the studies become more detailed.34, 35
What Type of Health and Healthcare Issues Are Addressed by Big Data?
We see successful application of Big Data in our daily lives when internet map direction finders constantly update the information to alert us to time of travel, whether there are accidents, and even if there are speed detectors along the way. We see this when doing an internet search and being rewarded often with a highly informative response within a fraction of a second. The issues on medicine, however, can be a bit more complicated.
The types of issues that can be addressed by Big Data are not necessarily different conceptually from other types of data, in that they can be descriptive, predictive, or prescriptive. The major advantages of Big Data relate to the size of the data sets, their variety, and speed of accumulation. Thus, we could see descriptive data about healthcare use within a hospital or healthcare system, with updating on an essentially instantaneous level. We can imagine an integrated rapid response system, in which a patient experiencing an ST‐segment–elevation myocardial infarction outside the hospital would have the diagnosis made in the field with the ECG compared with previous ones for the patient. Then, the ambulance would be directed to not just the closest hospital, but also to the one with an available catheterization laboratory, ensuring the shortest door‐to‐balloon time, thus maximizing overall efficiency of the system.
Predictive models have been created from Big Data, where the size and reach of the data sets permit assessment of the influence of covariates, not otherwise available, in predicting outcome.19, 26 The size of the data sets also permits many more risk factors to be considered. This offers the potential of identifying patients in whom intervention may be warranted. In principal, comparative effectiveness studies with hundreds of thousands or even millions of patients could be performed at relatively low cost, permitting detailed assessment of subgroups.20 That is, Big Data offers the potential for precision medicine. There are, however, significant barriers and limitations. In particular, using Big Data for comparative effectiveness studies comes with unique, particular difficulties. When comparing nonrandomized diagnostic or therapeutic strategies, there is the potential for treatment selection bias, or bias by indication, resulting in the comparison of groups who are not similar, perhaps varying in critical ways. Various statistical techniques are used to try to overcome this bias, but none can account for unmeasured confounders.36 Furthermore, treatment selection bias will not be overcome or even reduced by the size of the data set. In addition, Big Data may have more misclassification and missing baseline covariate data, which cannot be assumed to be missing at random, making the analytic problems greater. Simply put, it is necessary to be skeptical of observational nonrandomized comparisons of diagnostic or therapeutic strategies, whether the source is from a small number of patients or from Big Data.
Clinicians and medical scientists are used to thinking about data as they pertain to understanding disease and patient care. Data used for health services research are also being mined and analyzed by other entities concerned with health and the business of health care. Big Data in large healthcare systems is now used and increasingly in the future will be used for business intelligence analysis. Decisions will be made on salaries and staffing, capital purchases, and strategic planning. Such decisions can have a profound impact on healthcare delivery and public health. Physician leadership, especially in resource‐intensive areas such as cardiovascular medicine, will need to become skilled in interpreting such studies and understanding their impact on patient care. Big Data is currently used by multiple entities involved in health care, especially insurance companies and the Centers for Medicare and Medicaid Services. Health insurance companies use Big Data to guide rates and analyze markets for strategic investments (eg, the merger of Aetna and CVS). The Centers for Medicare and Medicaid Services uses Big Data to assess quality, watch for fraud and abuse, and guide incentives.
Embedded Randomized Clinical Trials
Big Data would seem to be the ideal platform on which to conduct randomized clinical trials (RCTs).37 Embedded clinical trials, compared with stand‐alone trials, offer the economy of capitalizing on infrastructure and data collection already in place and ongoing, reducing development time and data collection effort. The main, perhaps only, advantage to RCTs is the ability to overcome treatment selection bias. Observational Big Data can have the most bias when considering alternative strategies.38 As noted above, size cannot overcome bias. What Big Data does offer is data that come from wide sources, perhaps offering greater generalizability. The size of Big Data sets will reduce stochastic error, and may offer power to examine subsets, a frequent limitation of even large mega trials. However, the rules still apply.39 Subgroups should be defined in advance, there should be a small number, and there should be a physiologic reason for selecting the subgroup.40 Similarly, the choice of end points and analytic approaches should also be defined in advance. Multiplicity, where investigators consider multiple end points and subgroups, must be accounted for.41, 42 When good design principals are not carefully followed, assessment of alternative therapies or strategies can devolve into an unreliable “fishing expedition.”
An area that has already demonstrated that RCTs can effectively be conducted within larger data gathering environments is the RCT embedded in a clinical registry.39, 43 Examples of include the TASTE (Thrombus Aspiration in Myocardial Infarction) trial,44 embedded in the SWEDEHEART Registry; and the SAFE‐PCI (Study of Access Site for Enhancement of Percutaneous Coronary Intervention) for Women trial,45 embedded in the National Cardiovascular Data Registry. In the TASTE trial, 7244 patients with ST‐segment–elevation myocardial infarction were randomized to thrombus aspiration with PCI versus PCI alone. Thrombus aspiration was not shown to reduce the incidence of death or the composite of death, recurrent myocardial infarction, and stent thrombosis. In the SAFE‐PCI for Women trial, 1718 women undergoing diagnostic cardiac catheterization or PCI were randomized to radial versus femoral arterial access. No advantage to radial access was noted for bleeding or vascular complications. Although these trials themselves are not particularly remarkable, they are excellent examples of proof of concept of RCTs embedded in registries.
Another platform for RCTs is the EHR.46 This offers the possibility of using the EHR both to screen for patients and as a platform for data gathering. Although appealing in principle, it has to date been relatively unusual to be put into practice. EHRs are not designed to capture data in the way RCTs are, and adapting each to the needs of the other so that EHRs can enable embedded RCTs remains something of a challenge. An example of full integration is a point‐of‐care trial comparing insulin administered by a sliding scale versus a weight‐based approach, which was conducted in the Veterans Affairs Healthcare System.47, 48
Data Storage, Integration, and Retrieval
Hospitals can be expected to produce on the order of a petabyte of data, or a million, million bytes, a year. Nobody knows, but systems concerned with human health can be expected to generate multiple petabytes of data a day. How can such voluminous, rapidly accumulating data of multiple types be managed? The systems to store data must also allow ready access and integrate with applications to analyze Big Data. The systems also need to be able to scale to large sizes. Increasingly, health data, much like all other data, are stored in the cloud, with 3 companies leading: Amazon Web Services (https://aws.amazon.com), Azure from Microsoft (https://azure.microsoft.com/en-us), and Google Cloud (https://cloud.google.com), among others (https://www.softwaretestinghelp.com/cloud-computing-service-providers) (Figure 3). Cloud services from these companies are widely and publicly available. Much effort must go into curating data to make them retrievable and understandable. Sophisticated software, such as Hadoop, has been developed over the past several decades, which permits distribution of Big Data across clusters of computers, allowing failure of subsystems without loss of data integrity or availability.49
Figure 3.
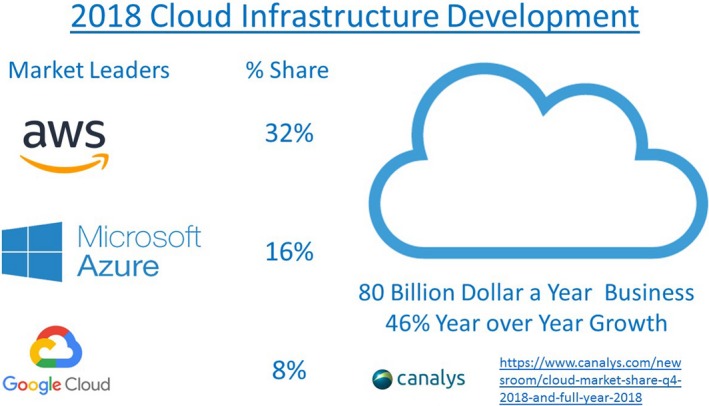
Leading providers of cloud data storage. aws indicates Amazon Web Services.
Analytic Approaches
Large, complicated data sets may not be readily approachable with the types of statistical methods that have been developed over the past 50+ years. This is especially true for predictive or prescriptive studies. Such typical methods include logistic regression and Cox model analysis. These models are generally hand coded by statistical programmers. If hundreds of variables or subgroups are to be considered, this type of statistical programming becomes impractical. To address this issue, several data mining and statistical approaches have been developed that permit analysis of large data sets. An example is random forest, a machine‐learning approach to classification and prediction.50 Random forest can be seen as an extension of decision tree approaches, such as classification and regression trees).51 Classification and regression tree models can be assembled by hand coding, yet offer the ability to more rapidly consider interactions that logistic regression or Cox model. In random forest, multiple decision trees are created from a large data set with replacement, a process that has been called bagging or bootstrap aggregation. Random forest is a method for both classification and regression. This approach can reduce overfitting by averaging multiple trees, reducing the chance finding of a predictive variable that performs well in a test data set, but poorly in validation.
Predictive models may be further developed from covariates developed from data‐mining approaches using advanced regression approaches. The first step might be to take variables generated from data mining and then use classic statistical methods like logistic regression and the Cox model. However, these methods may still not work well when there are potentially many variables and potential interactions. In such settings, machine‐learning approaches, such ridge, lasso, and elastic net allow “penalized” approaches to systematically restrict the number of variables and interactions in models.52 Ridge regression is most useful when there is potential for multicollinearity between variables, bringing variables into a model, but penalizing or limiting their influence in a continuous manner, rather than keeping them or eliminating them completely as in logistic regression. Lasso regression is conceptually similar, penalizing variables, but with many coefficients eliminated; thus, unlike ridge regression, this method allows reduction in the number of retained variables. Lasso allows selection of a small number of variables, whereas ridge works best when there are many predictors. However, in general, the best parameter estimates for a model are unknown; and both lasso and ridge may be too limiting, each in their own way. Elastic net integrates the approaches of lasso and ridge, allowing the degree of penalization of variables to vary between these extremes and allowing both some degree of penalization and some degree of elimination of variables. This can require multiple simulations to find the best model, which may not be easily defined by an algorithm (ie, it ultimately may require expert appraisal of a final model). There are potentially completely different approaches to modeling, such as support vector machine and bayesian machine learning, but the point should be clear. These methods are complex and require considerable experience and expertise to implement and use appropriately.
There is a point of view that analysis of Big Data sets, by systematically searching, may uncover previously unknown predictive variables; or by allowing the incorporation of more variables the precision of the modeling process may be enhanced, in particular by incorporating EHRs, imaging, and environmental and genomic or other ‘omic data (Figure 1). This may, at times, enhance prediction, and in particular genomic data have been shown to enhance cardiovascular risk prediction.53 However, at times, great precision may not be particularly helpful, given the uncertainties in any model, which become greater the further we try to predict into the future. Rather than the model with all statistically significant variables, a more parsimonious model with a small number of variables may be considerably easier to implement and may capture most of the ability to predict outcome.
Visualization
Large complicated data sets that may change rapidly over time offer particular challenges and opportunities for visualization. The static bar graph in black and white will not work in this new type of environment. It is also no longer reasonable and often not possible to visualize data using tools that require analysis first and then graphing the data, often done by hand. Thus, visualization tools also have sophisticated analytic engines permitting integration of analysis and display. Visualization has moved beyond 2‐dimensional printed graphs to 3‐dimensional or multidimensional graphs and may involve rotation on any axis or graphs that move over time. The results can make complex sets of data that are uninterpretable in raw form into easily understandable information.
Although the results can be dramatic, and it is relatively easy to begin using certain tools, creating and optimizing visual displays requires considerable expertise. Thus, there is a whole new field for professionals who specialize in Big Data visualization. In addition, considerable high‐speed computer processing may be required to transform data into the desired display. Some Big Data displays are suitable for many personal computers, whereas others in clinical medicine may require specific workstations with specialized software. Big Data visualization often requires both the ability to manipulate the specialized software as well as thorough understanding of the medical issues.
As with any issue related to data in general and Big Data in particular, the display is only as accurate and meaningful as the underlying data. Thus, before analysis display, careful curation is generally necessary. This may be difficult when the data sets are large and difficult to understand in raw form and the velocity is high. Visualization, once developed for an application, may be easy to run, giving what may appear to be simple, visually appealing results that are actually erroneous. Nonetheless, we should expect that, on a regular basis, data will be offered in visual format and increasingly embedded into health care with displays presented to us from multiple sources and in multiple ways.
Software products for Big Data visualization are widely available from multiple companies, including Cognos Analytics from IBM (https://www.ibm.com/in-en/analytics), Sense and View from Qlik (https://www.qlik.com/us/products), PowerBI from Microsoft (https://powerbi.microsoft.com/en-us), Visual Analyzer from Oracle (https://docs.oracle.com/en/cloud/paas/bi-cloud/bilug/toc.htm), Lumira from SAP (https://www.sap.com/products/lumira.html), Visual Analytics from SAS (https://www.sas.com/en_us/software/visual-analytics.html), the Tableau family of products (https://www.tableau.com/products), and Spotfire from TIBCO (https://www.tibco.com/products/tibco-spotfire).
Remarkable examples of Big Data visualization can often be found in the media. Here is an example from the New York Times 54 https://www.nytimes.com/interactive/2018/03/19/upshot/race-class-white-and-black-men.html?action=click&contentCollection=The%20Upshot®ion=Footer&module=WhatsNext&version=WhatsNext&contentID=WhatsNext&moduleDetail=undefined&pgtype=Multimedia. This article was based on a Big Data study of the entire population of the United States by Chetty et al55 https://opportunityinsights.org/wp-content/uploads/2018/10/atlas_paper.pdf.
Artificial Intelligence and Machine Learning
The concepts of Big Data and artificial intelligence are often conflated but are conceptually distinct. Artificial intelligence is a somewhat difficult concept to adequately define (given our limited ability to define human intelligence), but may be viewed as a set of tasks performed by information systems that normally require human cognitive activity, such as decision making, speech recognition, language translation, and visual perception. Examples abound. Speech recognition personal digital assistants, such as Alexa by Amazon, Google Assistant, and Siri by Apple, are now well known and ubiquitous. Driverless cars will combine speech recognition, visual perception, and decision making.
Of particular relevance to Big Data is how artificial intelligence can use data to drive medical decision making. This type of information system decision making is driven by a specific type of artificial intelligence called machine learning. Machine learning may be defined as software that becomes more accurate in predicting outcomes without being explicitly reprogrammed. The premise is to continually and with increasing accuracy and precision predict outcome as new data become available. Machine learning algorithms can be divided into categories of supervised, unsupervised, and reinforcement.56 Supervised learning requires considerable effort on the part of the investigator, whereas unsupervised learning is used to allow the information system to search for patterns in the data without feedback from the investigators. Reinforcement is a combination, in which the investigators provide guidance to unsupervised learning. Artificial neural networks are probably the most commonly used approach, and can be used for both unsupervised and supervised learning. Alternative approaches to artificial neural networks include support vector machines, random forests, and other decision tree models, naive Bayes classifier, fuzzy logic, and K‐nearest neighbor, each with advantages and disadvantages, as discussed by Krittanawong et al.56 For all analyses, models should be built with a training data set and then validated with a test data set. Whatever the approach, it is critical to avoid both underfitting and overfitting data. Underfitting occurs when the data are suboptimal for the analysis, resulting in predictive variables not being found, whereas overfitting occurs when the model is too complex for the data, and the model developed on a training data set will not prove valid on a test data set.
What may seem to be true artificial intelligence involves deep learning, in which the information system seeks to mimic human cognition, most commonly using multiple layers of artificial neural networks. Rather famously, Google used a deep learning neural network to develop AlphaGo, which defeated human champion Go players. Unsupervised deep learning can be used to develop algorithms to classify groups into clusters or develop rule‐learning algorithms. Shah et al57 published an unsupervised machine‐learning approach using 46 variables to classify patients with heart failure with preserved ejection fraction into 3 phenotypically similar groups. Although the machine learning may have been unsupervised, this was a particularly sophisticated analysis (ie, the real cognitive work was done by the investigators).
Applications of deep learning have been developed to evaluate images, now becoming common in facial recognition. In medicine, there are now several applications of deep learning for image recognition. In a notable example, Gulshan et al58 used neural networks to develop and validate an algorithm for detection of diabetic retinopathy from retinal fundus photographs, an application of artificial intelligence that has now been approved by the US Food and Drug Administration. In this study, a specific type of neural network optimized for image classification, called a deep convolutional neural network, analyzed a training set of 128 175 retinal images that were compared with results from a panel of ophthalmologists. The resultant algorithm was validated in separate data sets, with sensitivity and specificity generally >90%. Image recognition has also been applied to cardiac magnetic resonance imaging scans.30 Deep‐learning programs such as this require considerable time and expertise as well as computer systems with adequate computational power and graphics capabilities.
The next step up from deep learning is “cognitive computing,” in which the information systems are self‐learning and can mimic human cognition, in principal without human assistance. IBM would have its program Watson to be seen as a step in this direction. Cognitive computing would use Big Data and extend deep learning to solve more complicated problems.
Privacy
An obvious concern about Big Data is the effect on privacy. The basic safeguards in the United States remain the Health Insurance Portability and Accountability Act, which guards unauthorized violations of patient privacy for medical records and other health‐related information. Institutional review boards also function to protect patients according to the Common Rule for ethical conduct of research involving human subjects. In the European Union and the European Economic Area, the General Data Protection Regulation is designed to protect all aspects of personal privacy both within the European Union and the European Economic Area as well as export of data to outside the European Union and the European Economic Area. Whether protection of privacy will be sufficient to provide protection in some cases and too restrictive in others will remain to be seen. The general sense in the lay press that personal privacy is compromised by the ready access to data is almost certainly valid in medicine.
Conclusions
Big Data will be increasingly ubiquitous and will incorporate increasingly sophisticated analytic and visualization approaches in ways that can be made meaningful to humans in multiple domains in our lives, including medicine. Indeed, Big Data analytics and artificial intelligence are being used to make decisions already. Although humans must set the algorithms up, once in place they can run without human intervention. This may make life safer and remove much unwanted tedium, but there are clear dangers as well. For the foreseeable future in medicine and medical sciences, it will remain critical for humans to be deeply engaged in the process for rigorous science and humane, ethical medical care. What can be said with reasonable certainty is that Big Data will change medicine over the coming decades in both predictable and unpredictable ways.
Sources of Funding
This work was supported by the National Heart, Lung, and Blood Institute U01HL117006‐01A1 and the Oxford National Institute for Health Research Biomedical Research Centre.
Disclosures
None.
J Am Heart Assoc. 2019;8:e012791 DOI: 10.1161/JAHA.119.012791.
References
- 1. Ratwani RM, Fairbanks RJ, Hettinger AZ, Benda NC. Electronic health record usability: analysis of the user‐centered design processes of eleven electronic health record vendors. J Am Med Inform Assoc. 2015;22:1179–1182. [DOI] [PubMed] [Google Scholar]
- 2. Hemingway H, Asselbergs FW, Danesh J, Dobson R, Maniadakis N, Maggioni A, van Thiel GJM, Cronin M, Brobert G, Vardas P, Anker SD, Grobbee DE, Denaxas S; Innovative Medicines Initiative 2nd programme Big Data for Better Outcomes, Big Data@Heart Consortium of 20 academic industry partners including ESC . Big data from electronic health records for early and late translational cardiovascular research: challenges and potential. Eur Heart J. 2018;39:1481–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Churpek MM, Yuen TC, Park SY, Gibbons R, Edelson DP. Using electronic health record data to develop and validate a prediction model for adverse outcomes in the wards. Crit Care Med. 2014;42:841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koleck TA, Dreisbach C, Bourne PE, Bakken S. Natural language processing of symptoms documented in free‐text narratives of electronic health records: a systematic review. J Am Med Inform Assoc. 2019;26:364–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gill J, Prasad V. Improving observational studies in the era of big data. Lancet. 2018;392:716–717. [DOI] [PubMed] [Google Scholar]
- 6. Bishop D. Rein in the four horsemen of irreproducibility. Nature. 2019;568:435. [DOI] [PubMed] [Google Scholar]
- 7. Hayrinen K, Saranto K, Nykanen P. Definition, structure, content, use and impacts of electronic health records: a review of the research literature. Int J Med Inform. 2008;77:291–304. [DOI] [PubMed] [Google Scholar]
- 8. Hicks KA, Tcheng JE, Bozkurt B, Chaitman BR, Cutlip DE, Farb A, Fonarow GC, Jacobs JP, Jaff MR, Lichtman JH, Limacher MC, Mahaffey KW, Mehran R, Nissen SE, Smith EE, Targum SL; American College of Cardiology and American Heart Association . 2014 ACC/AHA key data elements and definitions for cardiovascular endpoint events in clinical trials: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Data Standards (Writing Committee to Develop Cardiovascular Endpoints Data Standards). Circulation. 2015;132:302–361. [DOI] [PubMed] [Google Scholar]
- 9. McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, Forrey A, Mercer K, DeMoor G, Hook J, Williams W, Case J, Maloney P. LOINC, a universal standard for identifying laboratory observations: a 5‐year update. Clin Chem. 2003;49:624–633. [DOI] [PubMed] [Google Scholar]
- 10. Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hammond WE. The making and adoption of health data standards. Health Aff (Millwood). 2005;24:1205–1213. [DOI] [PubMed] [Google Scholar]
- 12. Mandel JC, Kreda DA, Mandl KD, Kohane IS, Ramoni RB. SMART on FHIR: a standards‐based, interoperable apps platform for electronic health records. J Am Med Inform Assoc. 2016;23:899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saripalle R, Runyan C, Russell M. Using Hl7 FHIR to achieve interoperability in patient health record. J Biomed Inform. 2019. Available at: 10.1016/j.jbi.2019.103188. Accessed June 25, 2019. [DOI] [PubMed] [Google Scholar]
- 14. Klann JG, Joss MAH, Embree K, Murphy SN. Data model harmonization for the All Of Us Research Program: transforming i2b2 data into the OMOP common data model. PLoS One. 2019;14:e0212463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanborn TA, Tcheng JE, Anderson HV, Chambers CE, Cheatham SL, DeCaro MV, Durack JC, Everett AD, Gordon JB, Hammond WE, Hijazi ZM, Kashyap VS, Knudtson M, Landzberg MJ, Martinez‐Rios MA, Riggs LA, Sim KH, Slotwiner DJ, Solomon H, Szeto WY, Weiner BH, Weintraub WS, Windle JR. ACC/AHA/SCAI 2014 health policy statement on structured reporting for the cardiac catheterization laboratory: a report of the American College of Cardiology Clinical Quality Committee. Circulation. 2014;129:2578–2609. [DOI] [PubMed] [Google Scholar]
- 16. Weis JM, Levy PC. Copy, paste, and cloned notes in electronic health records. Chest. 2014;145:632–638. [DOI] [PubMed] [Google Scholar]
- 17. Frizzell JD, Liang L, Schulte PJ, Yancy CW, Heidenreich PA, Hernandez AF, Bhatt DL, Fonarow GC, Laskey WK. Prediction of 30‐day all‐cause readmissions in patients hospitalized for heart failure: comparison of machine learning and other statistical approaches. JAMA Cardiol. 2017;2:204–209. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Z, Kolm P, Grau‐Sepulveda MV, Ponirakis A, O'Brien SM, Klein LW, Shaw RE, McKay C, Shahian DM, Grover FL, Mayer JE, Garratt KN, Hlatky M, Edwards FH, Weintraub WS. Cost‐effectiveness of revascularization strategies: the ASCERT study. J Am Coll Cardiol. 2015;65:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weintraub WS, Grau‐Sepulveda MV, Weiss JM, Delong ER, Peterson ED, O'Brien SM, Kolm P, Klein LW, Shaw RE, McKay C, Ritzenthaler LL, Popma JJ, Messenger JC, Shahian DM, Grover FL, Mayer JE, Garratt KN, Moussa ID, Edwards FH, Dangas GD. Prediction of long‐term mortality after percutaneous coronary intervention in older adults: results from the National Cardiovascular Data Registry. Circulation. 2012;125:1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weintraub WS, Grau‐Sepulveda MV, Weiss JM, O'Brien SM, Peterson ED, Kolm P, Zhang Z, Klein LW, Shaw RE, McKay C, Ritzenthaler LL, Popma JJ, Messenger JC, Shahian DM, Grover FL, Mayer JE, Shewan CM, Garratt KN, Moussa ID, Dangas GD, Edwards FH. Comparative effectiveness of revascularization strategies. N Engl J Med. 2012;366:1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hlatky MA, Boothroyd DB, Baker L, Kazi DS, Solomon MD, Chang TI, Shilane D, Go AS. Comparative effectiveness of multivessel coronary bypass surgery and multivessel percutaneous coronary intervention: a cohort study. Ann Intern Med. 2013;158:727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shahian DM, Silverstein T, Lovett AF, Wolf RE, Normand SL. Comparison of clinical and administrative data sources for hospital coronary artery bypass graft surgery report cards. Circulation. 2007;115:1518–1527. [DOI] [PubMed] [Google Scholar]
- 23. Jacobs JP, Shahian DM, Prager RL, Edwards FH, McDonald D, Han JM, D'Agostino RS, Jacobs ML, Kozower BD, Badhwar V, Thourani VH, Gaissert HA, Fernandez FG, Wright C, Fann JI, Paone G, Sanchez JA, Cleveland JC Jr, Brennan JM, Dokholyan RS, O'Brien SM, Peterson ED, Grover FL, Patterson GA. Introduction to the STS national database series: outcomes analysis, quality improvement, and patient safety. Ann Thorac Surg. 2015;100:1992–2000. [DOI] [PubMed] [Google Scholar]
- 24. Brindis RG, Fitzgerald S, Anderson HV, Shaw RE, Weintraub WS, Williams JF. The American College of Cardiology‐National Cardiovascular Data Registry (ACC‐NCDR): building a national clinical data repository. J Am Coll Cardiol. 2001;37:2240–2245. [DOI] [PubMed] [Google Scholar]
- 25. Holmberg MJ, Moskowitz A, Raymond TT, Berg RA, Nadkarni VM, Topjian AA, Grossestreuer AV, Donnino MW, Andersen LW; American Heart Association's Get With The Guidelines‐Resuscitation Investigators . Derivation and internal validation of a mortality prediction tool for initial survivors of pediatric in‐hospital cardiac arrest. Pediatr Crit Care Med. 2018;19:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shahian DM, O'Brien SM, Sheng S, Grover FL, Mayer JE, Jacobs JP, Weiss JM, Delong ER, Peterson ED, Weintraub WS, Grau‐Sepulveda MV, Klein LW, Shaw RE, Garratt KN, Moussa ID, Shewan CM, Dangas GD, Edwards FH. Predictors of long‐term survival after coronary artery bypass grafting surgery: results from the Society of Thoracic Surgeons Adult Cardiac Surgery Database (the ASCERT study). Circulation. 2012;125:1491–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schlegl T, Waldstein SM, Vogl WD, Schmidt‐Erfurth U, Langs G. Predicting semantic descriptions from medical images with convolutional neural networks. Inf Process Med Imaging. 2015;24:437–448. [DOI] [PubMed] [Google Scholar]
- 28. Seifert S, Kelm M, Moeller M, Mukherjee S, Calallaro A, Huber M, Comaniciu D. Semantic annotation of medical images. Proc SPIE 7628, Medical Imaging 2010: Advanced PACS‐based Imaging Informatics and Therapeutic Applications, 762808. https://pdfs.semanticscholar.org/3c35/ed69ad9602680919d2cca0c41a70f1b307db.pdf. Accessed June 19, 2019.
- 29. Harvey H, Glocker B. A standardized approach for preparing imaging data for machine learning tasks in radiology In: Ranschaert ER, Morozov S, Algra RR, eds. Artificial Intelligence in Medical Imaging. New York: Springer; 2019:61–72. [Google Scholar]
- 30. Margeta J, Criminisi A, Lozoya RC, Lee DC, Ayache N. Fine‐tuned convolutional neural nets for cardiac MRI acquisition plane recognition. Comput Methods Biomech Biomed Eng Imaging Vis. 2015;5:339–349. [Google Scholar]
- 31. Kramer CM, Appelbaum E, Desai MY, Desvigne‐Nickens P, DiMarco JP, Friedrich MG, Geller N, Heckler S, Ho CY, Jerosch‐Herold M, Ivey EA, Keleti J, Kim DY, Kolm P, Kwong RY, Maron MS, Schulz‐Menger J, Piechnik S, Watkins H, Weintraub WS, Wu P, Neubauer S. Hypertrophic Cardiomyopathy Registry: the rationale and design of an international, observational study of hypertrophic cardiomyopathy. Am Heart J. 2015;170:223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, Whitbourne S, Deen J, Shannon C, Humphries D, Guarino P, Aslan M, Anderson D, LaFleur R, Hammond T, Schaa K, Moser J, Huang G, Muralidhar S, Przygodzki R, O'Leary TJ. Million Veteran Program: a mega‐biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. [DOI] [PubMed] [Google Scholar]
- 33. Turakhia MP, Desai M, Hedlin H, Rajmane A, Talati N, Ferris T, Desai S, Nag D, Patel M, Kowey P, Rumsfeld JS, Russo AM, Hills MT, Granger CB, Mahaffey KW, Perez MV. Rationale and design of a large‐scale, app‐based study to identify cardiac arrhythmias using a smartwatch: the Apple Heart Study. Am Heart J. 2019;207:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Apte JS, Messier KP, Gani S, Brauer M, Kirchstetter TW, Lunden MM, Marshall JD, Portier CJ, Vermeulen RCH, Hamburg SP. High‐resolution air pollution mapping with Google street view cars: exploiting big data. Environ Sci Technol. 2017;51:6999–7008. [DOI] [PubMed] [Google Scholar]
- 35. Simonsen L, Gog JR, Olson D, Viboud C. Infectious disease surveillance in the big data era: towards faster and locally relevant systems. J Infect Dis. 2016;214:S380–S385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weintraub WS, Yeh RW. Challenges of assessing therapeutic or diagnostic outcomes with observational data. Am J Med. 2018;131:206–210. [DOI] [PubMed] [Google Scholar]
- 37. Mayo CS, Matuszak MM, Schipper MJ, Jolly S, Hayman JA, Ten Haken RK. Big data in designing clinical trials: opportunities and challenges. Front Oncol. 2017;7:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaplan RM, Chambers DA, Glasgow RE. Big data and large sample size: a cautionary note on the potential for bias. Clin Transl Sci. 2014;7:342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nyberg K, Hedman P. Swedish guidelines for registry‐based randomized clinical trials. Upsala J Med Sci. 2019;124:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yusuf S, Wittes J, Probstfield J, Tyroler HA. Analysis and interpretation of treatment effects in subgroups of patients in randomized clinical trials. JAMA. 1991;266:93–98. [PubMed] [Google Scholar]
- 41. Schulz KF, Grimes DA. Multiplicity in randomised trials I: endpoints and treatments. Lancet. 2005;365:1591–1595. [DOI] [PubMed] [Google Scholar]
- 42. Schulz KF, Grimes DA. Multiplicity in randomised trials II: subgroup and interim analyses. Lancet. 2005;365:1657–1661. [DOI] [PubMed] [Google Scholar]
- 43. James S, Rao SV, Granger CB. Registry‐based randomized clinical trials: a new clinical trial paradigm. Nat Rev Cardiol. 2015;12:312–316. [DOI] [PubMed] [Google Scholar]
- 44. Lagerqvist B, Frobert O, Olivecrona GK, Gudnason T, Maeng M, Alstrom P, Andersson J, Calais F, Carlsson J, Collste O, Gotberg M, Hardhammar P, Ioanes D, Kallryd A, Linder R, Lundin A, Odenstedt J, Omerovic E, Puskar V, Todt T, Zelleroth E, Ostlund O, James SK. Outcomes 1 year after thrombus aspiration for myocardial infarction. N Engl J Med. 2014;371:1111–1120. [DOI] [PubMed] [Google Scholar]
- 45. Rao SV, Hess CN, Barham B, Aberle LH, Anstrom KJ, Patel TB, Jorgensen JP, Mazzaferri EL Jr, Jolly SS, Jacobs A, Newby LK, Gibson CM, Kong DF, Mehran R, Waksman R, Gilchrist IC, McCourt BJ, Messenger JC, Peterson ED, Harrington RA, Krucoff MW. A registry‐based randomized trial comparing radial and femoral approaches in women undergoing percutaneous coronary intervention: the SAFE‐PCI for Women (Study of Access Site for Enhancement of PCI for Women) trial. JACC Cardiovasc Interv. 2014;7:857–867. [DOI] [PubMed] [Google Scholar]
- 46. Angus DC. Fusing randomized trials with big data: the key to self‐learning health care systems? JAMA. 2015;314:767–768. [DOI] [PubMed] [Google Scholar]
- 47. Fiore LD, Brophy M, Ferguson RE, D'Avolio L, Hermos JA, Lew RA, Doros G, Conrad CH, O'Neil JA Jr, Sabin TP, Kaufman J, Swartz SL, Lawler E, Liang MH, Gaziano JM, Lavori PW. A point‐of‐care clinical trial comparing insulin administered using a sliding scale versus a weight‐based regimen. Clin Trials. 2011;8:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fiore LD, Lavori PW. Integrating randomized comparative effectiveness research with patient care. N Engl J Med. 2016;374:2152–2158. [DOI] [PubMed] [Google Scholar]
- 49. Landset S, Khoshgoftaar TM, Richter AN, et al. Journal of Big Data. 2015;2:24 10.1186/s40537-015-0032-1. [DOI] [Google Scholar]
- 50. Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 51. Gareth J, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning. New York: Springer; 2015. [Google Scholar]
- 52. Waldron L, Pintilie M, Tsao MS, Shepherd FA, Huttenhower C, Jurisica I. Optimized application of penalized regression methods to diverse genomic data. Bioinformatics. 2011;27:3399–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Abraham G, Havulinna AS, Bhalala OG, Byars SG, De Livera AM, Yetukuri L, Tikkanen E, Perola M, Schunkert H, Sijbrands EJ, Palotie A, Samani NJ, Salomaa V, Ripatti S, Inouye M. Genomic prediction of coronary heart disease. Eur Heart J. 2016;37:3267–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Badger E, Cain Miller C, Pearce A, Quealy K. Extensive data shows punishing reach of racism for black boys. New York Times. 2018. Available at: https://www.nytimes.com/interactive/2018/03/19/upshot/race-class-white-and-black-men.html?action=click&contentCollection=The%20Upshot®ion=Footer&module=WhatsNext&version=WhatsNext&contentID=WhatsNext&moduleDetail=undefined&pgtype=Multimedia. Accessed April 29, 2019. [Google Scholar]
- 55. Chetty R, Friedman JN, Hendren N, Jones MR, Porter SR. The opportunity atlas: mapping the childhood roots of social mobility. 2018. Available at: https://opportunityinsights.org/wp-content/uploads/2018/10/atlas_paper.pdf. Accessed April 29, 2019.
- 56. Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol. 2017;69:2657–2664. [DOI] [PubMed] [Google Scholar]
- 57. Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang CC, Deo RC. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131:269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. [DOI] [PubMed] [Google Scholar]


