Abstract
Background
We recently found that ARMC5 variants may be associated with primary aldosteronism in blacks. We investigated a cohort from the MH‐GRID (Minority Health Genomics and Translational Research Bio‐Repository Database) and tested the association between ARMC5 variants and blood pressure in blacks.
Methods and Results
Whole exome sequencing data of 1377 blacks were analyzed. Target single‐variant and gene‐based association analyses of hypertension were performed for ARMC5, and replicated in a subset of 3015 individuals of African descent from the UK Biobank cohort. Sixteen rare variants were significantly associated with hypertension (P=0.0402) in the gene‐based (optimized sequenced kernel association test) analysis; the 16 and one other, rs116201073, together, showed a strong association (P=0.0003) with blood pressure in this data set. The presence of the rs116201073 variant was associated with lower blood pressure. We then used human embryonic kidney 293 and adrenocortical H295R cells transfected with an ARMC5 construct containing rs116201073 (c.*920T>C). The latter was common in both the discovery (MH‐GRID) and replication (UK Biobank) data and reached statistical significance (P=0.044 [odds ratio, 0.7] and P=0.007 [odds ratio, 0.76], respectively). The allele carrying rs116201073 increased levels of ARMC5 mRNA, consistent with its protective effect in the epidemiological data.
Conclusions
ARMC5 shows an association with hypertension in blacks when rare variants within the gene are considered. We also identified a protective variant of the ARMC5 gene with an effect on ARMC5 expression confirmed in vitro. These results extend our previous report of ARMC5’s possible involvement in the determination of blood pressure in blacks.
Keywords: adrenocortical adenoma, ARMC5, black, Conn syndrome, genetics, hypertension, primary aldosteronism
Subject Categories: Hypertension; Genetic, Association Studies; Genetics; Epigenetics; Functional Genomics
Clinical Perspective
What Is New?
Germline variants in ARMC5 may in part be responsible for regulation of blood pressure in blacks through their in adrenocortical function.
What Are the Clinical Implications?
Recognition of hyperaldosteronism and/or its genetics among blacks may lead to earlier and more effective treatments that prevent cardiovascular and renal consequences of hypertension in this population.
Introduction
Hypertension is one of the preventable risk factors for cardiovascular disease and death. It is estimated that by the year 2030, over 23 million Americans will die from cardiovascular disease.1 According to the Centers for Disease Control and Prevention, up to 32.5% of Americans older than 20 years have hypertension,2 with varying rates across various ethnicities. Blacks have a disproportionately increased prevalence, earlier age of onset, and greater morbidity related to hypertension. The National Health and Nutrition Examination Survey found that 42.1% of non‐Hispanic black individuals have hypertension.3 The predisposition of hypertension in blacks has been linked to retention of salt and water, either by excess aldosterone secretion, or to excess sensitivity to aldosterone, and genetic variants that may result in overactivity of the epithelial sodium channel.4 Nevertheless, increased risk of hypertension in blacks is likely related to complex interactions between genetic, behavioral, and social‐environmental determinants that are yet to be determined.
Hyperaldosteronism is associated with insulin resistance,5 diabetes mellitus,6, 7 metabolic syndrome,7, 8 and cardiovascular inflammation and fibrosis,9 suggesting that aldosterone plays an important role in the development of cardiovascular disease. In young adult blacks, hyperaldosteronism has been linked to insulin resistance that is independent of age, sex, and blood pressure (BP).10 Primary aldosteronism (PA) is the most common cause of endocrine hypertension and leads to significant morbidity and mortality across all ethnicities. PA is characterized by an autonomous secretion of aldosterone that is independent of renin and sodium status, usually attributable to bilateral adrenocortical hyperplasia. Blacks are more likely to have PA because of bilateral adrenocortical hyperplasia,4, 11, 12, 13, 14, 15, 16, 17 although one report suggests a similar prevalence in whites and blacks.18 Several genetic defects have been identified in PA, although their link to the increased ethnic predisposition to hypertension has not been fully studied or understood.
Our group recently found an association between ARMC5 gene variants, predicted to be damaging, in patients with PA of black descent.19 The ARMC5 gene is a tumor suppressor implicated in cortisol and/or aldosterone‐producing primary macronodular adrenal hyperplasia, a rare form of endogenous hypercortisolemia. We identified 12 germline ARMC5 genetic alterations in 20 unrelated and 2 related individuals (39.3%), in which all affected patients carrying a variant predicted to be damaging were black. This study provided the first evidence of a germline genetic alteration in association with PA specifically for the black population. This genetic association could in part explain the increased predisposition of blacks to low‐renin hypertension. Recognition of genetic causes of low‐renin hypertension and/or PA and its appropriate treatment may lead to a significant reduction of morbidity and mortality from cardiovascular disease in black individiduals.
To date, over 1000 genetic variants contribute to hypertension, explaining in aggregate ≈6% of the trait variance.20, 21, 22, 23, 24, 25, 26, 27, 28 However, none of these studies demonstrated an association with ARMC5. Moreover, investigations of genetic and transcriptome alterations in black patients with hypertension is limited.29, 30, 31 Several genetic variants have been described as associated with hypertension32, 33 and compromised arterial elasticity34, 35, 36 in blacks.4 Several studies failed to discover any relationship.37, 38 Population‐specific genetic variants, variation in allele frequency, and small statistical power were among reasons why some of the genetic loci associated with hypertension lacked the replicability.
Given the paucity of proven genetic drivers of hypertension in this population at risk, we sought to investigate ARMC5's involvement in the regulation of BP among participants in the MH‐GRID (Minority Health Genomics and Translational Research Bio‐Repository Database) study.39
Methods
The data that support the findings of this study are available from the corresponding author upon request.
MH‐GRID Data
The MH‐GRID project is a study of hypertension with data collected across 8 sites in the United States. The data included in this analysis consist of genotype, from whole exome sequencing, and phenotype information of self‐identified black men or women aged 30 to 55 years. Cases are individuals taking ≥2 antihypertensive drugs on a stable regimen (≥6 months) including a diuretic, and controls are individuals with optimal BP (≤120/80 mm Hg) without antihypertension medication and with normal kidney function (estimated glomerular filtration rate >90 mL/min). Patients with kidney disease, diabetes mellitus, heart failure, HIV, and liver disease were excluded. More details of inclusion and exclusion criteria for the MH‐GRID are available in Table S1.
UK Biobank Data
The data included in this analysis for the purpose of replication is from the UK Biobank, a large prospective study of 502 628 participants recruited between 2006 and 2010.40 The subset used in the replication analysis consists of 3015 participants identified as African. Participants provided their medical history, medication information, and lifestyle/behavior factors. BP was measured as the mean of 2 sitting systolic and diastolic BP measurements, taken at baseline using the Omron HEM‐7015IT digital BP monitor (Omron Healthcare). For the purpose of replication, participants with any form of cancer were excluded. Cases were defined as individuals with BP ≥140/90 mm Hg regardless of medication status, and controls were defined as individuals with optimal BP (≤120/80 mm Hg) without BP medication. The genetic data are from the June 2017 release. Details of the design of the arrays and quality control have been described elsewhere.40 The participants were genotyped on the UK Biobank Axiom array, which has 805 426 markers. For this analysis, the genotypes were imputed, via the Michigan Imputation Server,41 using the “freeze5b” release of the Trans‐Omics for Precision Medicine (TOPMed) whole‐genome sequencing data. TOPMed is an initiative sponsored by the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) and one of its goals is to achieve ancestral and ethnic diversity and as such it is currently composed of about 60% of participants with substantial non‐European ancestry. That diversity was the rationale for the choice of TOPMed as the imputation platform for this analysis.
GENE‐FORECAST Data
Plasma renin activity (PRA) data were not available for the MH‐GRID or UK Biobank data sets included in this analysis. Therefore, to investigate the relationship between a common ARMC5 variant and PRA, a smaller data set of 299 samples was used from the GENE‐FORECAST (Genomics, Environmental Factors and the Social Determinants of Cardiovascular Disease in African Americans Study). This is a research platform that establishes a strategic, multiomics systems biology approach amenable to the deep, multidimensional characterization of minority health and disease in blacks (ClinicalTrials.gov identifier: NCT02055209). GENE‐FORECAST is an ongoing study designed to create a cohort established on a community‐based sampling frame of US‐born, black men and women (aged 21–65 years) to be recruited from the metropolitan Washington, DC, area. GENE‐FORECAST samples were genotyped on a customized Illumina Infinium Multi‐Ethnic Global‐8 array platform (Illumina, Inc).
Ethics
The study was approved by the institutional review boards of Morehouse School of Medicine, Kaiser Permanente, Grady Health System Research Oversight Committee, and the NIH (ClinicalTrials.gov identifier: NCT02290392). The institutional review boards of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (until 2010) and National Institute of Diabetes and Digestive and Kidney Diseases (2010–present) at NIH approved the research for other protocols (ClinicalTrials.gov identifier: NCT00005927 and NCT00001595). All research participants signed informed consent.
Genetic Association Analyses
Two analyses were performed using the ARMC5 locus information from genome‐built GRCh37 1: chromosome=16, start position=3146941, and end position=31478484. Single‐variant analysis of common variants (minor allele frequency [MAF] ≥0.05) within ARMC5 was conducted in PLINK 1.9.42, 43 Gene‐based analyses combining common, low frequency (MAF ≥0.01 and <0.05) and rare variants (MAF <0.01) within ARMC5 was performed using the optimized sequence kernel association test (SKAT‐O),44 a method recommended when the genetic architecture of a locus of interest is not known.45 SKAT‐O “collapses” variants into 1 single nucleotide polymorphism set, which is then assigned a single score used to predict trait values. Quality controls for the whole exome data are summarized in Figure S1. We used 1377 patients and 44 variants within ARMC5 (3 common, 4 low frequency, and 37 rare variants) for analysis. All of the association analyses were performed with the genetic variants treated as an additive and the linear model adjusted for potential confounders.
Statistical Analysis
Previous work has shown that known heritable traits (eg, body mass index) can reduce power when included as covariates in regression models.46, 47, 48 Therefore, rather than adjusting indiscriminately for all of the covariates associated with hypertension (Table 1, we used the method of Pirinen et al48 to identify the model that maximizes power (ie, the model that provides the lowest standard error and P value). Our preliminary investigations showed that the optimal model is the one adjusted for age, sex, high‐density lipoprotein, low‐density lipoprotein, smoking, and the relevant principal components (PCs) of the PC analyses (PCA) performed to investigate admixture.
Table 1.
Baseline Characteristics of Patients From the MH‐GRID Study
| Characteristics | Cases (n=623) | Controls (n=754) | P Value |
|---|---|---|---|
| Age, y | 48.25±6.06 | 43.35±7.23 | 1.17×10−40 |
| Sex | |||
| Women, % | 57.11 | 67.18 | 1.10×10−4 |
| Men, % | 42.89 | 32.82 | |
| Current smoker (no/yes) | 451/165 | 449/302 | 2.59×10−7 |
| BMI, kg/m2 | 33.92±7.5 | 28.8±7.46 | 1.25×10−34 |
| SBP | 140±16 | 109±7 | <2.2×10−16 |
| DBP | 89±10 | 70±7 | <2.2×10−16 |
| HDL, mg/dL | 53.28±15.18 | 55.42±16.55 | 1.45×10−2 |
| LDL, mg/dL | 120.01±34.57 | 112.32±34.19 | 5.81×10−5 |
| Triglycerides, mg/dL | 106.95±57.04 | 87.01±52.24 | 8.10×10−11 |
BMI indicates body mass index; DBP, diastolic blood pressure; HDL, high‐density lipoprotein; LDL, low‐density lipoprotein; MH‐GRID, Minority Health Genomics and Translational Research Bio‐Repository Database; SBP, systolic blood pressure.
For MH‐GRID, the PCA results show that PC1 separates the 2 main continental ancestries (West African and European) of blacks (Figure S2). Thus, PC1 was added to the model to adjust for global ancestry.
We chose the “African” subset that had MAFs similar to those observed in MH‐GRID for the replicated loci. This was done to avoid population stratification, which decreases statistical power. For UK Biobank, the PCA results show, since the analysis was restricted to the African subset, PC2 and PC3, which account for admixture in the African populations were added to the model to adjust for global ancestry (Figure S3).
Molecular Analysis
The DNA sequence of ARMC5‐203 isoform (NM_024742) was cloned in pUCminusMCS plasmid (Blue Heron). The rs116201073 (c.*920T>C) DNA change was introduced by targeted mutagenesis following the manufacturer's instructions (200555‐12, Agilent Technologies UK Ltd).
Human embryonic kidney 293 (HEK293) cells were grown in Dulbecco's modified Eagle medium (DMEM) (11995, Gibco) enriched with 10% fetal bovine serum (FBS) (900, Gemini Bio‐Products Inc), GlutaMAX (35050, Gibco), and Anti (15240, Gibco), whereas human adrenocarcinoma (H295R) cells were maintained in DMEM‐F12 (11320, Gibco) containing 10% FBS (900, Gemini Bio‐Products Inc), GlutaMAX (35050, Gibco), Anti (15240, Gibco), and Insulin‐Transferrin‐Selenium (41400, Thermo Fisher Scientific). A total of 300 000 HEK293 or 400 000 H295R cells were seeded in a well of 6‐well plate and were transfected the next day with 1 μg of empty, wild‐type or mutant plasmid using Lipofectamine LTX (15338100, Thermo Fisher Scientific) or Effectene (301425, Qiagen), respectively. Where indicated, cells were treated with 10 ng of cycloheximide (C4859, Sigma‐Aldrich) or DMSO (34869, Sigma‐Aldrich) for the control for 1, 2, or 3 hours before collection. Cells were harvested in Trizol (15596018, Ambion, Inc) 48 hours after transfection for RNA extraction following the manufacturer's protocol. Five hundred thousand nanograms of RNA were then reverse transcribed (11753‐050, Thermo Fisher Scientific) as indicated in the manufacturer's instructions. One microliter of a one‐twentieth dilution of cDNA was amplified by quantitative polymerase chain reaction conducted with SybrGreen (4364344, Thermo Fisher Scientific). The primers used to analyze ARMC5, ARMC5‐203 expression were previously described.49
Results
Demographics
The baseline characteristics of the 1377 individuals who passed quality controls are reported in Table 1. The hypertensive group was older and contained more men and had a larger body mass index. The proportion of smokers was higher in the control group and lipid profiles were better (lower low‐density lipoprotein and triglycerides and higher high‐density lipoprotein) in the control group. The strong association between BP pressure (systolic BP and diastolic BP) and hypertension reported in Table 1 is attributable to the design of MH‐GRID, which focused on the tails of hypertension distribution (controls with optimal BP versus cases taking ≥2 BP medications).
Single‐Variant Analysis
In the discovery analysis (MH‐GRID data), ARMC5 variant rs116201073 reached nominal significance (P=0.044; odds ratio, 0.7), suggesting a protective effect for this variant (Table 2). For the replication analysis (GENE‐FORECAST data), the variant rs116201073 was imputed with high confidence (R 2=0.96) and its MAF (0.077) was similar to what was observed in MH‐GRID. For the results presented in Table 2, the association, in UK Biobank, was adjusted for age, sex, smoking, alcohol, and PC2 and PC3. The other variables we adjusted for in MH‐GRID were not available in UK Biobank.
Table 2.
Single‐Variant Analysis Results and Details of the SNPs
| Variant | MH‐GRID Study | UK Biobank | ||
|---|---|---|---|---|
| rs116201073 | rs11863886 | rs11150624 | rs116201073 | |
| Position on chromosome 16 | 31477442 | 31477460 | 31476458 | 31477442 |
| Alleles (minor/major) | Cytosine/thymine | Adenine/guanine | Thymine/cytosine | Cytosine/thymine |
| MAF | 0.071 | 0.182 | 0.096 | 0.077 |
| OR | 0.7 | 1.21 | 1.08 | 0.76 |
| SD | 0.18 | 0.11 | 0.14 | 0.1 |
| P value | 0.044 | 0.089 | 0.580 | 0.0068 |
| Frequency in cases, % | 5.50 | 18.70 | 10.10 | 7.61 |
| Frequency in controls, % | 8.20 | 16.90 | 9.50 | 8.85 |
| Functional information | Synonymous | Synonymous | Missense | Synonymous |
MAF indicates minor allele frequency; MH‐GRID, Minority Health Genomics and Translational Research Bio‐Repository Database; OR, odds ratio; SNP, single nucleotide polymorphism.
Gene‐Based Analysis
Discovery (MH‐GRID)
The gene‐based analysis was adjusted for the same covariates as in the single‐variant analysis and conducted by combining all 37 rare variants and then applying conditional analysis in SKAT‐O to sift out noise variants50 and identify the variants that truly contribute to the effect (ie, those that decrease the P value of the association). That process identified 16 rare variants that together are associated with hypertension (P=0.0011). SKAT‐O of a set that consists of those 16 variants and the common variant identified in the single‐variant analysis was more strongly associated with hypertension (Table 3). Subsequent SKAT‐O analyses considering low‐frequency variants alone or in combination with the rare variants were not conclusive. The SKAT‐O results for MH‐GRID are summarized in Table S2.
Table 3.
SKAT‐O Gene‐Based Analysis Results
| Variant | P Value | No. of Variants Collapsed |
|---|---|---|
| Rare variants | 0.0011 | 16 |
| Rare variants+rs116201073 | 0.0003 | 17 |
SKAT‐O indicates optimized sequence kernel association test.
All of the 16 rare variants associated with hypertension in the SKAT‐O and outlined in Table 4 have the same effect as the common variant rs116201073 and this explains the stronger association for the set of 17 (16 rare+1 common). Figure 1 reports the number of mutant alleles across the 16 rare variants by cases and control. Across the 1377 individuals, only 17 were heterozygous and 1 was homozygous for the mutant allele and all are controls.
Table 4.
List of the Set of 17 Variants Significantly Associated With Hypertension and MAF in the MH‐GRID Study
| Variant | POS (GRCh37) | MAF | Functional Information (dbSNP) | Under Selection (GERP/SiPhy) |
|---|---|---|---|---|
| 16:31473335 | 31473335 | 0.0004 | ||
| 16:31473823 | 31473823 | 0.0004 | ||
| rs141923065 | 31474091 | 0.0004 | Missense (glutamine ⇒ arginine) | Yes |
| 16:31474095 | 31474095 | 0.0007 | ||
| rs202103062 | 31476361 | 0.0004 | Missense (glycine ⇒ cysteine) | No |
| rs370836071 | 31477234 | 0.0004 | Missense (threonine ⇒ methionine) | |
| 16:31477236 | 31477236 | 0.0004 | ||
| rs116201073 | 31477442 | 0.0711 | Synonymous | Yes |
| 16:31477452 | 31477452 | 0.0004 | ||
| 16:31477486 | 31477486 | 0.0004 | ||
| 16:31477569 | 31477569 | 0.0004 | ||
| 16:31477574 | 31477574 | 0.0004 | ||
| rs181967284 | 31477834 | 0.0004 | Missense (arginine ⇒ glutamine) | Yes |
| rs367810854 | 31477859 | 0.0004 | Synonymous | |
| rs372567714 | 31477945 | 0.0004 | Missense (glutamic acid ⇒ valine) | |
| 16:31478013 | 31478013 | 0.0004 | ||
| rs61734240 | 31478192 | 0.0004 | Synonymous | Yes |
MAF indicates minor allele frequency; MH‐GRID, Minority Health Genomics and Translational Research Bio‐Repository Database; POS, position; GRCh37, Genome Reference Consortium Human Build 37; dbSNP, The Single Nucleotide Polymorphism Database; GERP, Genomic Evolutionary Rate Profiling; SiPhy, SIte‐specific PHYlogenetic analysis.
Figure 1.
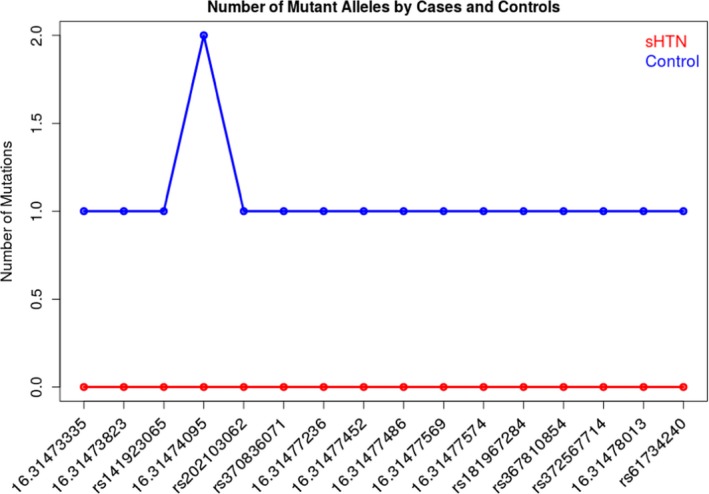
Number of mutant alleles by cases and controls across the Minority Health Genomics and Translational Research Bio‐Repository Database. sHTN indicates severe hypertension.
Table 4 lists the 17 variants. Seven of the 16 rare variants have been previously reported while the remainder, along with 4 of 7 variants known as rare and nonsynonymous, are novel from the MH‐GRID exome data. Overall, 6 variants, including the common single nucleotide polymorphism, exhibit evidence of selective constraint as computed by 2 mammalian conservation algorithms, the Genomic Evolutionary Rate Profiling51 and SiPhy52 as reported in the HaploReg v4.1 database.53
Replication (UK Biobank)
Because of the known challenge of imputing rare variants, only 4 of the 16 rare could be imputed and 2 of those were monomorphic in the UK Biobank data. Therefore, SKAT‐O replication was attempted with 2 rare variants imputed with respective R 2 values of 0.92 (rs367810854. MAF=0.0008 in UK Biobank) and 0.67 (rs141923065, MAF=0.0012 in UK Biobank). The results of the gene‐based analysis with the 3 variants, adjusted for age, sex, smoking, and alcohol, was significant in UK Biobank (P=0.0083). The frequency of the replicated variants in various populations is reported in Table S3.
The common variant rs116201073 seems to be specific to Africans, where it is present only in Africans or African‐admixed populations included in the 1000 Genomes Project (Table 4). In the TOPMed data available from the BRAVO portal (University of Michigan), specific allele frequency are not available for some minority populations and the same info as in the 1000 Genomes Project. In the Genome Aggregation Database (gnomAD), the frequencies reported are 0.075 for African, 0.002 for Latino, 0.002 for “other,” and essentially 0 (<0.00001) for the other populations.
As for the rare variants included in the replication analysis, in the 1000 Genomes Project: rs141923065 is observed only in African‐Caribbean in Barbados, African American from the Southwest (ASW), and Han Chinese. Intriguingly, rs367810854 is observed only in South Asian populations (gnomAD provides similar information with 0.0499 for the category “South Asian” and <0.0001 or other continental populations). Additional details are available in Table S3.
ARMC5 Variant rs116201073 and PRA
First, we evaluated the relationship between hypertension and PRA across the 299 samples (115 with hypertension versus 184 controls). The results showed lower PRA in the hypertensive group (mean=1.40 ng/mL per hour in patients with hypertension versus 2.04 ng/mL per hour in controls), but the difference was not statistically significant (t statistic=1.50, P=0.13 [95% CI, −0.20 to 1.47]). Then we estimated the association between the variant and PRA dichotomized using a cutoff of 0.65 ng/mL per hour54 to have, respectively, 82 and 134 patients in the low and high renin groups across the 216 samples for which genotype data were available in GENE‐FORECAST. The variant was less frequent in the low renin group (6% versus 7% in the rest of the sample); however, the association was not statistically significant (odds ratio, 0.87; P>0.05).
Effect of ARMC5 Variant, rs116201073
The rs116201073 variant is a synonymous variant in most of the ARMC5 isoforms except in the NM_024742 isoform (referred to as ARMC5‐203 in the Ensembl database) in which it is located in the 3′‐untranslated transcribed region (UTR). To determine its potential effect on ARMC5's expression, we transfected wild‐type and mutant ARMC5‐203 plasmids in HEK293 cells and analyzed ARMC5 presence by real‐time quantitative polymerase chain reaction, 24 and 48 hours after transfection. Whereas at 24 hours, no difference was observed between the 2 groups, at 48 hours, there was a significant increase in ARMC5 mRNA accumulation when ARMC5 carried the variant allele (Figure 2).
Figure 2.
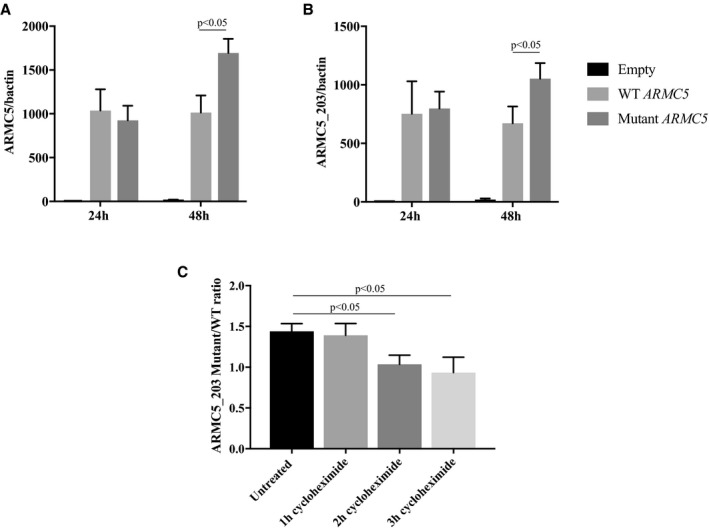
Comparison of wild‐type (WT) and mutant ARMC5 (rs116201073) expression in human embryonic kidney 293 (HEK293) cell line. A and B, Real‐time quantitative polymerase chain reaction (RTqPCR) using primers recognizing all ARMC5 isoforms (A) or only the ARMC5‐203 isoform (B) on HEK293 cells transfected with WT or mutant (variant‐carrying) ARMC5‐203 plasmid for 24 and 48 hours. C, The ratio of the mutant and the WT ARMC5‐203 expression detected by RTqPCR on HEK293 cells untreated or treated 1, 2, or 3 hours with a translation blocker, cycloheximide. The graph represents the means of at least 3 independent experiments±standard error of the mean (SEM).
Similar results were obtained using either primers targeting all ARMC5 isoforms (Figure 2A) or specifically the ARMC5‐203 isoform that was overexpressed (Figure 2B). The treatment of the transfected cells with a translation inhibitor, cycloheximide, for 2 or 3 hours before collection led to a normalization of the ratio of ARMC5‐203 mutant and wild‐type mRNA demonstrating that the elevation of mutant ARMC5 mRNA was the result of a decrease of its translation rate. A similar elevation of ARMC5‐203 mRNA was found at 48 and 72 hours after transfection in an adrenocortical cell line H295R, but this increase was not significant (Figure S4).
Discussion
Analysis of our data identified one common variant (rs116201073) located in the 3′UTR end of the ARMC5 gene that was associated with decreased risk of hypertension (odds ratio, 0.7) in a sample set of 1377 blacks from the MH‐GRID study. That single‐variant association was replicated with a smaller P value in a UK Biobank sample set of 3015 African participants. Gene‐based SKAT‐O analysis, in MH‐GRID, also revealed a set of 16 rare variants associated with hypertension in blacks with the same protective effect as the common variant rs116201073. Together, these 16 rare and 1 common variant (a set of 17 variants) were significantly associated with BP in a subsequent gene‐based test. The gene‐based results were also replicated in the UK Biobank data with the rare variants that could be imputed. These results confirm our previous report of ARMC5's possible involvement in regulating BP in blacks, possibly as a result of its role in determining the presence of bilateral adrenocortical hyperplasia and/or hyperaldosteronism.19
The ARMC5 gene is a putative tumor suppressor that is located on chromosome 16p11.2 and belongs to the family of armadillo‐repeat–containing proteins.55, 56 In humans, ARMC5 consists of 8 exons and has an unknown function,19, 49 although recent evidence suggests that it plays a critical role for fetal development and immune responses through interactions with proteins from different pathways.57, 58 Four ARMC5 isoforms exist, with a different pattern of expression, although all 4 are expressed in the adrenal glands.55 The ARMC5 gene has been recently implicated in endogenous hypercortisolemia due to primary macronodular adrenal hyperplasia,59, 60, 61, 62, 63, 64 which is characterized by multiple nodules (>1 cm) in the adrenal cortex and hypercortisolemia.19 Biallelic inactivation of ARMC5 (germline and somatic) is required for the development of adrenocortical hyperplasia, which is consistent with the 2‐hit hypothesis of tumorigenesis.59 Most disease‐causing variants in ARMC5 are frameshift and/or nonsense, and lead to loss of function of the gene.62 Overexpression of ARMC5 in adrenocortical carcinoma cell line H295R leads to increased cell death59 while silencing of the gene in nonmutated primary macronodular adrenal hyperplasia cell cultures leads to a decrease of apoptosis.65
Genetic variants in ARMC5 have rarely been implicated in PA. The largest study to date examined 56 patients with PA and found 12 different germline variants in ARMC5 (6 predicted to be damaging by in silico analysis) in 20 unrelated and 2 related individuals (39%).19 These variants were exclusively found in black individuals and silencing of ARMC5 in H295R cells decreased CYP11B2 expression.19
A recent study in a different cohort of patients with PA reported 18 ARMC5 variants (5 rare with an allele frequency <1%) and 2 new variants that were not predicted to be damaging.66 Variants in ARMC5 are difficult to identify as pathogenic because ARMC5's function remains unclear; however, some missense variants fail to induce apoptosis after transfection in a human adrenocortical cancer cell line H295R.59 Although the link between ARMC5 and PA is yet to be explained, our data support a potential link between ARMC5 variants and hypertension in people of African descent.
PRA and aldosterone are often used as a screening test for PA. We studied possible associations between PRA and hypertension in patients from the GENE‐FORECAST study. Although these results were not significant, perhaps because of small sample size, the directions of the relationships were consistent with the seemingly protective effect of the variant (rs116201073) reported in the genetic associations’ analyses of MH‐GRID and UK Biobank data sets.
Interestingly, the rs116201073 variant seems to be specific to the African population with the C allele present only in African and African admixed populations included in the 1000 Genomes Project. Given the significantly higher prevalence of the variant in the control we can hypothesize that either the minor C allele has a direct protective effect against hypertension in this population predisposed to low‐renin hypertension or this variant is a genetic marker of a protector factor. This variant predicted to be benign is synonymous in 3 ARMC5 isoforms: ARMC5‐201, ARMC5‐202, and ARMC5‐205. In the ARMC5‐203 isoform, which is ubiquitously expressed49 and even overexpressed in our hypertensive cohort compared with controls, the rs116201073 variant is located in the 3′‐UTR. The 3′‐UTR region is essential for the regulation of mRNA stability, expression, and localization.67 Indeed, our in vitro experiments in HEK293 cells demonstrate that the ARMC5‐203 carrying the variant mRNA is accumulated compared with the wild‐type mRNA and this is the result, at least in part, of a reduction in its translation rate.
ARMC5‐203 function has not yet been studied but it is noteworthy that it is the only protein isoform that has the Armadillo domain but not the BTB (BR‐C, ttk, and bab)/POZ (Pox virus and Zinc finger) domain. This suggests a specific and nonredundant role between this isoform and the 3 other ARMC5 isoforms.49 Altogether, these data suggest that the ARMC5‐203 protein would promote hypertension, as the variant decreasing its protein translation is protective against hypertension.
Predisposition to low‐renin hypertension in blacks has been broadly studied. Part of this predisposition is attributed to the activation of the renin‐angiotensin‐aldosterone system by its promotion of sodium retention.4, 68 It could be explained by genetic variants that predispose to inappropriate secretion of aldosterone, or genetic variants that affect the epithelial sodium channel (eg, Liddle syndrome phenotype), ultimately resulting in water preservation.4, 69 The following genetic variants were found to predispose to PA and/or inappropriate secretion of aldosterone: CYP11B2, KCNJ5, ATP1A1, ATP2B3, CACNA1D, and ARMC5.19, 70, 71, 72 The Liddle syndrome phenotype could be caused by GRK, NEDD4L, CYP4A11, NPPA, UMOD, and perhaps other, yet to be discovered genetic variants.4, 70, 73
Why do people of African origin have more hypertension than foreign‐born Africans in the United States? It is possible, that natural selection for salt and water retention created a survival advantage for people transported across the Atlantic Ocean in hot conditions. This phenomenon is known as the African Diaspora hypothesis.4, 74, 75, 76
Study Limitations
Our study has limitations and should be interpreted with caution. First, the MH‐GRID hypertensive case definition was based on hypertension diagnosis by a clinician and the participant being prescribed ≥2 antihypertensive medications for at least 6 months before enrollment into the study. The available UK Biobank data set did not have information about the number of antihypertensive medications that the study participants were taking, as the cases were defined as those with hypertension diagnosed by a clinician regardless of medication status and/or those with average BP ≥140/90 mm Hg across several measures at the visit;. Second, a “healthy volunteer” effect was demonstrated in the UK Biobank study,77 thus generalization of the present study is likely limited. Third, we do not have information about the duration of antihypertensive therapy, and it is possible that the duration of medication use may be considered to be a proxy for disease severity. Fourth, we have limited data on the biochemical phenotype of the participants: the association between the common genetic variant and low renin could not be reliably established because of the small sample. Fifth, we do not know whether patients had the Liddle syndrome phenotype (low renin and low aldosterone) or PA (low renin and high aldosterone). Finally, we did not consider other variants/loci of interest for an additive effect on hypertension.
Conclusions
We identified one common variant (rs116201073) of the ARMC5 gene that was associated with a decreased risk of hypertension in blacks and a set of 16 rare variants associated with hypertension. These results extend our previous report of germline ARMC5 variants that may be linked to hypertension in blacks. Although not conclusive, the evaluation of the main variant with respect to PRA may suggest a link to low‐renin hypertension. Further genetic and molecular studies are needed to confirm and complement these findings.
Sources of Funding
This research was supported in part by the Intramural Research Program of Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH Grant Z1A HD008920‐08 to Constantine A. Stratakis. Grant 1RC4MD005964‐0 to Gary Gibbons.
Disclosures
None.
Supporting information
Table S1. ARMC5 Variants and Risk of Hypertension in Blacks: Minority Health‐GRID Study
Table S2. Gene‐Based Analysis Results in the MH‐GRID Study
Table S3. MAF of rs116201073, rs141923065, and rs367810854 in the 1000 Genomes Project Populations
Figure S1. Summary of quality controls (QCs) for MH‐GRID (Minority Health Genomics and Translational Research Bio‐Repository Database) exome‐wide sequencing data.
Figure S2. Principal component analyses of MH‐GRID (Minority Health Genomics and Translational Research Bio‐Repository Database) data with 1000 Genomes Project samples.
Figure S3. Graphs of principal component analyses (PCA) of UK Biobank genotype data.
Figure S4. Comparison of wild‐type (WT) and mutant ARMC5 (rs116201073) expression in human embryonic kidney 293 (HEK293) cell line.
Acknowledgments
We thank Diane Cooper, MSLS, NIH Library, for providing assistance in writing this article. We thank patients and researchers from the MH‐GRID Network.
(J Am Heart Assoc. 2019;8:e012508 DOI: 10.1161/JAHA.119.012508.)
References
- 1. Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER III, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB; American Heart Association Statistics Committee and Stroke Statistics Subcommittee . Heart disease and stroke statistics—2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–e322. [DOI] [PubMed] [Google Scholar]
- 2. Statistics NCfH . Health, United States, 2014: selected health conditions and risk factors, by age: United States, selected years 1988–1994 through 2011–2012. 2014.
- 3. Nwankwo T, Yoon SS, Burt V, Gu Q. Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011–2012. NCHS Data Brief. 2013;(133):1–8. [PubMed] [Google Scholar]
- 4. Spence JD, Rayner BL. Hypertension in blacks: individualized therapy based on renin/aldosterone phenotyping. Hypertension. 2018;72:263–269. [DOI] [PubMed] [Google Scholar]
- 5. Garg R, Hurwitz S, Williams GH, Hopkins PN, Adler GK. Aldosterone production and insulin resistance in healthy adults. J Clin Endocrinol Metab. 2010;95:1986–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu VC, Chueh SJ, Chen L, Chang CH, Hu YH, Lin YH, Wu KD, Yang WS; TAIPAI Study Group . Risk of new‐onset diabetes mellitus in primary aldosteronism: a population study over 5 years. J Hypertens. 2017;35:1698–1708. [DOI] [PubMed] [Google Scholar]
- 7. Hanslik G, Wallaschofski H, Dietz A, Riester A, Reincke M, Allolio B, Lang K, Quack I, Rump LC, Willenberg HS, Beuschlein F, Quinkler M, Hannemann A; Participants of the German Conn's Registry . Increased prevalence of diabetes mellitus and the metabolic syndrome in patients with primary aldosteronism of the German Conn's Registry. Eur J Endocrinol. 2015;173:665–675. [DOI] [PubMed] [Google Scholar]
- 8. Bochud M, Nussberger J, Bovet P, Maillard MR, Elston RC, Paccaud F, Shamlaye C, Burnier M. Plasma aldosterone is independently associated with the metabolic syndrome. Hypertension. 2006;48:239–245. [DOI] [PubMed] [Google Scholar]
- 9. de Rita O, Hackam DG, Spence JD. Effects of aldosterone on human atherosclerosis: plasma aldosterone and progression of carotid plaque. Can J Cardiol. 2012;28:706–711. [DOI] [PubMed] [Google Scholar]
- 10. Huan Y, Deloach S, Keith SW, Goodfriend TL, Falkner B. Aldosterone and aldosterone: renin ratio associations with insulin resistance and blood pressure in African Americans. J Am Soc Hypertens. 2012;6:56–65. [DOI] [PubMed] [Google Scholar]
- 11. Spence JD. The current epidemic of primary aldosteronism: causes and consequences. J Hypertens. 2004;22:2038–2039; author reply 2039. [DOI] [PubMed] [Google Scholar]
- 12. Russell RP, Masi AT. The prevalence of adrenal cortical hyperplasia at autopsy and its association with hypertension. Ann Intern Med. 1970;73:195–205. [DOI] [PubMed] [Google Scholar]
- 13. Russell RP, Masi AT, Richter ED. Adrenal cortical adenomas and hypertension. A clinical pathologic analysis of 690 cases with matched controls and a review of the literature. Medicine (Baltimore). 1972;51:211–225. [PubMed] [Google Scholar]
- 14. Russell RP, Masi AT. Significant associations of adrenal cortical abnormalities with “essential” hypertension. Am J Med. 1973;54:44–51. [DOI] [PubMed] [Google Scholar]
- 15. Kidambi S, Kotchen JM, Grim CE, Raff H, Mao J, Singh RJ, Kotchen TA. Association of adrenal steroids with hypertension and the metabolic syndrome in blacks. Hypertension. 2007;49:704–711. [DOI] [PubMed] [Google Scholar]
- 16. Spence JD. Physiologic tailoring of therapy for resistant hypertension: 20 years’ experience with stimulated renin profiling. Am J Hypertens. 1999;12:1077–1083. [PubMed] [Google Scholar]
- 17. Tu W, Eckert GJ, Hannon TS, Liu H, Pratt LM, Wagner MA, Dimeglio LA, Jung J, Pratt JH. Racial differences in sensitivity of blood pressure to aldosterone. Hypertension. 2014;63:1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calhoun DA, Nishizaka MK, Zaman MA, Thakkar RB, Weissmann P. Hyperaldosteronism among black and white subjects with resistant hypertension. Hypertension. 2002;40:892–896. [DOI] [PubMed] [Google Scholar]
- 19. Zilbermint M, Xekouki P, Faucz FR, Berthon A, Gkourogianni A, Schernthaner‐Reiter MH, Batsis M, Sinaii N, Quezado MM, Merino M, Hodes A, Abraham SB, Libe R, Assie G, Espiard S, Drougat L, Ragazzon B, Davis A, Gebreab SY, Neff R, Kebebew E, Bertherat J, Lodish MB, Stratakis CA. Primary aldosteronism and ARMC5 variants. J Clin Endocrinol Metab. 2015;100:E900–E909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Evangelou E, Warren HR, Mosen‐Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, Ng FL, Evangelou M, Witkowska K, Tzanis E, Hellwege JN, Giri A, Velez Edwards DR, Sun YV, Cho K, Gaziano JM, Wilson PWF, Tsao PS, Kovesdy CP, Esko T, Magi R, Milani L, Almgren P, Boutin T, Debette S, Ding J, Giulianini F, Holliday EG, Jackson AU, Li‐Gao R, Lin WY, Luan J, Mangino M, Oldmeadow C, Prins BP, Qian Y, Sargurupremraj M, Shah N, Surendran P, Theriault S, Verweij N, Willems SM, Zhao JH, Amouyel P, Connell J, de Mutsert R, Doney ASF, Farrall M, Menni C, Morris AD, Noordam R, Pare G, Poulter NR, Shields DC, Stanton A, Thom S, Abecasis G, Amin N, Arking DE, Ayers KL, Barbieri CM, Batini C, Bis JC, Blake T, Bochud M, Boehnke M, Boerwinkle E, Boomsma DI, Bottinger EP, Braund PS, Brumat M, Campbell A, Campbell H, Chakravarti A, Chambers JC, Chauhan G, Ciullo M, Cocca M, Collins F, Cordell HJ, Davies G, Borst MH, Geus EJ, Deary IJ, Deelen J, Del Greco MF, Demirkale CY, Dorr M, Ehret GB, Elosua R, Enroth S, Erzurumluoglu AM, Ferreira T, Franberg M, Franco OH, Gandin I, Gasparini P, Giedraitis V, Gieger C, Girotto G, Goel A, Gow AJ, Gudnason V, Guo X, Gyllensten U, Hamsten A, Harris TB, Harris SE, Hartman CA, Havulinna AS, Hicks AA, Hofer E, Hofman A, Hottenga JJ, Huffman JE, Hwang SJ, Ingelsson E, James A, Jansen R, Jarvelin MR, Joehanes R, Johansson A, Johnson AD, Joshi PK, Jousilahti P, Jukema JW, Jula A, Kahonen M, Kathiresan S, Keavney BD, Khaw KT, Knekt P, Knight J, Kolcic I, Kooner JS, Koskinen S, Kristiansson K, Kutalik Z, Laan M, Larson M, Launer LJ, Lehne B, Lehtimaki T, Liewald DCM, Lin L, Lind L, Lindgren CM, Liu Y, Loos RJF, Lopez LM, Lu Y, Lyytikainen LP, Mahajan A, Mamasoula C, Marrugat J, Marten J, Milaneschi Y, Morgan A, Morris AP, Morrison AC, Munson PJ, Nalls MA, Nandakumar P, Nelson CP, Niiranen T, Nolte IM, Nutile T, Oldehinkel AJ, Oostra BA, O'Reilly PF, Org E, Padmanabhan S, Palmas W, Palotie A, Pattie A, Penninx B, Perola M, Peters A, Polasek O, Pramstaller PP, Nguyen QT, Raitakari OT, Ren M, Rettig R, Rice K, Ridker PM, Ried JS, Riese H, Ripatti S, Robino A, Rose LM, Rotter JI, Rudan I, Ruggiero D, Saba Y, Sala CF, Salomaa V, Samani NJ, Sarin AP, Schmidt R, Schmidt H, Shrine N, Siscovick D, Smith AV, Snieder H, Sober S, Sorice R, Starr JM, Stott DJ, Strachan DP, Strawbridge RJ, Sundstrom J, Swertz MA, Taylor KD, Teumer A, Tobin MD, Tomaszewski M, Toniolo D, Traglia M, Trompet S, Tuomilehto J, Tzourio C, Uitterlinden AG, Vaez A, van der Most PJ, van Duijn CM, Vergnaud AC, Verwoert GC, Vitart V, Volker U, Vollenweider P, Vuckovic D, Watkins H, Wild SH, Willemsen G, Wilson JF, Wright AF, Yao J, Zemunik T, Zhang W, Attia JR, Butterworth AS, Chasman DI, Conen D, Cucca F, Danesh J, Hayward C, Howson JMM, Laakso M, Lakatta EG, Langenberg C, Melander O, Mook‐Kanamori DO, Palmer CNA, Risch L, Scott RA, Scott RJ, Sever P, Spector TD, van der Harst P, Wareham NJ, Zeggini E, Levy D, Munroe PB, Newton‐Cheh C, Brown MJ, Metspalu A, Hung AM, O'Donnell CJ, Edwards TL, Million Veteran Program, Psaty BM, Tzoulaki I, Barnes MR, Wain LV, Elliott P, Caulfield MJ. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabrera CP, Ng FL, Warren HR, Barnes MR, Munroe PB, Caulfield MJ. Exploring hypertension genome‐wide association studies findings and impact on pathophysiology, pathways, and pharmacogenetics. Wiley Interdiscip Rev Syst Biol Med. 2015;7:73–90. [DOI] [PubMed] [Google Scholar]
- 22. Ehret GB, Ferreira T, Chasman DI, Jackson AU, Schmidt EM, Johnson T, Thorleifsson G, Luan J, Donnelly LA, Kanoni S, Petersen AK, Pihur V, Strawbridge RJ, Shungin D, Hughes MF, Meirelles O, Kaakinen M, Bouatia‐Naji N, Kristiansson K, Shah S, Kleber ME, Guo X, Lyytikainen LP, Fava C, Eriksson N, Nolte IM, Magnusson PK, Salfati EL, Rallidis LS, Theusch E, Smith AJP, Folkersen L, Witkowska K, Pers TH, Joehanes R, Kim SK, Lataniotis L, Jansen R, Johnson AD, Warren H, Kim YJ, Zhao W, Wu Y, Tayo BO, Bochud M; Consortium CH‐E, Consortium C‐H, Wellcome Trust Case Control C , Absher D, Adair LS, Amin N, Arking DE, Axelsson T, Baldassarre D, Balkau B, Bandinelli S, Barnes MR, Barroso I, Bevan S, Bis JC, Bjornsdottir G, Boehnke M, Boerwinkle E, Bonnycastle LL, Boomsma DI, Bornstein SR, Brown MJ, Burnier M, Cabrera CP, Chambers JC, Chang IS, Cheng CY, Chines PS, Chung RH, Collins FS, Connell JM, Doring A, Dallongeville J, Danesh J, de Faire U, Delgado G, Dominiczak AF, Doney ASF, Drenos F, Edkins S, Eicher JD, Elosua R, Enroth S, Erdmann J, Eriksson P, Esko T, Evangelou E, Evans A, Fall T, Farrall M, Felix JF, Ferrieres J, Ferrucci L, Fornage M, Forrester T, Franceschini N, Duran OHF, Franco‐Cereceda A, Fraser RM, Ganesh SK, Gao H, Gertow K, Gianfagna F, Gigante B, Giulianini F, Goel A, Goodall AH, Goodarzi MO, Gorski M, Grassler J, Groves C, Gudnason V, Gyllensten U, Hallmans G, Hartikainen AL, Hassinen M, Havulinna AS, Hayward C, Hercberg S, Herzig KH, Hicks AA, Hingorani AD, Hirschhorn JN, Hofman A, Holmen J, Holmen OL, Hottenga JJ, Howard P, Hsiung CA, Hunt SC, Ikram MA, Illig T, Iribarren C, Jensen RA, Kahonen M, Kang H, Kathiresan S, Keating BJ, Khaw KT, Kim YK, Kim E, Kivimaki M, Klopp N, Kolovou G, Komulainen P, Kooner JS, Kosova G, Krauss RM, Kuh D, Kutalik Z, Kuusisto J, Kvaloy K, Lakka TA, Lee NR, Lee IT, Lee WJ, Levy D, Li X, Liang KW, Lin H, Lin L, Lindstrom J, Lobbens S, Mannisto S, Muller G, Muller‐Nurasyid M, Mach F, Markus HS, Marouli E, McCarthy MI, McKenzie CA, Meneton P, Menni C, Metspalu A, Mijatovic V, Moilanen L, Montasser ME, Morris AD, Morrison AC, Mulas A, Nagaraja R, Narisu N, Nikus K, O'Donnell CJ, O'Reilly PF, Ong KK, Paccaud F, Palmer CD, Parsa A, Pedersen NL, Penninx BW, Perola M, Peters A, Poulter N, Pramstaller PP, Psaty BM, Quertermous T, Rao DC, Rasheed A, Rayner N, Renstrom F, Rettig R, Rice KM, Roberts R, Rose LM, Rossouw J, Samani NJ, Sanna S, Saramies J, Schunkert H, Sebert S, Sheu WH, Shin YA, Sim X, Smit JH, Smith AV, Sosa MX, Spector TD, Stancakova A, Stanton A, Stirrups KE, Stringham HM, Sundstrom J, Swift AJ, Syvanen AC, Tai ES, Tanaka T, Tarasov KV, Teumer A, Thorsteinsdottir U, Tobin MD, Tremoli E, Uitterlinden AG, Uusitupa M, Vaez A, Vaidya D, van Duijn CM, van Iperen EPA, Vasan RS, Verwoert GC, Virtamo J, Vitart V, Voight BF, Vollenweider P, Wagner A, Wain LV, Wareham NJ, Watkins H, Weder AB, Westra HJ, Wilks R, Wilsgaard T, Wilson JF, Wong TY, Yang TP, Yao J, Yengo L, Zhang W, Zhao JH, Zhu X, Bovet P, Cooper RS, Mohlke KL, Saleheen D, Lee JY, Elliott P, Gierman HJ, Willer CJ, Franke L, Hovingh GK, Taylor KD, Dedoussis G, Sever P, Wong A, Lind L, Assimes TL, Njolstad I, Schwarz PE, Langenberg C, Snieder H, Caulfield MJ, Melander O, Laakso M, Saltevo J, Rauramaa R, Tuomilehto J, Ingelsson E, Lehtimaki T, Hveem K, Palmas W, Marz W, Kumari M, Salomaa V, Chen YI, Rotter JI, Froguel P, Jarvelin MR, Lakatta EG, Kuulasmaa K, Franks PW, Hamsten A, Wichmann HE, Palmer CNA, Stefansson K, Ridker PM, Loos RJF, Chakravarti A, Deloukas P, Morris AP, Newton‐Cheh C, Munroe PB. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48:1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Surendran P, Drenos F, Young R, Warren H, Cook JP, Manning AK, Grarup N, Sim X, Barnes DR, Witkowska K, Staley JR, Tragante V, Tukiainen T, Yaghootkar H, Masca N, Freitag DF, Ferreira T, Giannakopoulou O, Tinker A, Harakalova M, Mihailov E, Liu C, Kraja AT, Fallgaard Nielsen S, Rasheed A, Samuel M, Zhao W, Bonnycastle LL, Jackson AU, Narisu N, Swift AJ, Southam L, Marten J, Huyghe JR, Stancakova A, Fava C, Ohlsson T, Matchan A, Stirrups KE, Bork‐Jensen J, Gjesing AP, Kontto J, Perola M, Shaw‐Hawkins S, Havulinna AS, Zhang H, Donnelly LA, Groves CJ, Rayner NW, Neville MJ, Robertson NR, Yiorkas AM, Herzig KH, Kajantie E, Zhang W, Willems SM, Lannfelt L, Malerba G, Soranzo N, Trabetti E, Verweij N, Evangelou E, Moayyeri A, Vergnaud AC, Nelson CP, Poveda A, Varga TV, Caslake M, de Craen AJ, Trompet S, Luan J, Scott RA, Harris SE, Liewald DC, Marioni R, Menni C, Farmaki AE, Hallmans G, Renstrom F, Huffman JE, Hassinen M, Burgess S, Vasan RS, Felix JF; CHARGE‐HF Consortium , Uria‐Nickelsen M, Malarstig A, Reily DF, Hoek M, Vogt T, Lin H, Lieb W; EchoGen Consortium , Traylor M, Markus HF; METASTROKE Consortium , Highland HM, Justice AE, Marouli E; GIANT Consortium , Lindstrom J, Uusitupa M, Komulainen P, Lakka TA, Rauramaa R, Polasek O, Rudan I, Rolandsson O, Franks PW, Dedoussis G, Spector TD; EPIC‐InterAct Consortium , Jousilahti P, Mannisto S, Deary IJ, Starr JM, Langenberg C, Wareham NJ, Brown MJ, Dominiczak AF, Connell JM, Jukema JW, Sattar N, Ford I, Packard CJ, Esko T, Magi R, Metspalu A, de Boer RA, van der Meer P, van der Harst P; Lifelines Cohort Study , Gambaro G, Ingelsson E, Lind L, de Bakker PI, Numans ME, Brandslund I, Christensen C, Petersen ER, Korpi‐Hyovalti E, Oksa H, Chambers JC, Kooner JS, Blakemore AI, Franks S, Jarvelin MR, Husemoen LL, Linneberg A, Skaaby T, Thuesen B, Karpe F, Tuomilehto J, Doney AS, Morris AD, Palmer CN, Holmen OL, Hveem K, Willer CJ, Tuomi T, Groop L, Karajamaki A, Palotie A, Ripatti S, Salomaa V, Alam DS, Shafi Majumder AA, Di Angelantonio E, Chowdhury R, McCarthy MI, Poulter N, Stanton AV, Sever P, Amouyel P, Arveiler D, Blankenberg S, Ferrieres J, Kee F, Kuulasmaa K, Muller‐Nurasyid M, Veronesi G, Virtamo J, Deloukas P; Wellcome Trust Case Control C , Elliott P; Understanding Society Scientific Group , Zeggini E, Kathiresan S, Melander O, Kuusisto J, Laakso M, Padmanabhan S, Porteous D, Hayward C, Scotland G, Collins FS, Mohlke KL, Hansen T, Pedersen O, Boehnke M, Stringham HM; EPIC‐CVD Consortium, Frossard P, Newton‐Cheh C; CHARGE+ Exome Chip Blood Pressure Consortium, Tobin MD, Nordestgaard BG; T2D‐GENES Consortium; GoT2DGenes Consortium; ExomeBP Consortium; CHD Exome+ Consortium , Caulfield MJ, Mahajan A, Morris AP, Tomaszewski M, Samani NJ, Saleheen D, Asselbergs FW, Lindgren CM, Danesh J, Wain LV, Butterworth AS, Howson JM, Munroe PB. Trans‐ancestry meta‐analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu C, Kraja AT, Smith JA, Brody JA, Franceschini N, Bis JC, Rice K, Morrison AC, Lu Y, Weiss S, Guo X, Palmas W, Martin LW, Chen YD, Surendran P, Drenos F, Cook JP, Auer PL, Chu AY, Giri A, Zhao W, Jakobsdottir J, Lin LA, Stafford JM, Amin N, Mei H, Yao J, Voorman A; Consortium CHDE, Exome BPC, Go TDC, Consortium TDG , Larson MG, Grove ML, Smith AV, Hwang SJ, Chen H, Huan T, Kosova G, Stitziel NO, Kathiresan S, Samani N, Schunkert H, Deloukas P; Myocardial Infarction G, Consortia CAE , Li M, Fuchsberger C, Pattaro C, Gorski M; Consortium CK , Kooperberg C, Papanicolaou GJ, Rossouw JE, Faul JD, Kardia SL, Bouchard C, Raffel LJ, Uitterlinden AG, Franco OH, Vasan RS, O'Donnell CJ, Taylor KD, Liu K, Bottinger EP, Gottesman O, Daw EW, Giulianini F, Ganesh S, Salfati E, Harris TB, Launer LJ, Dorr M, Felix SB, Rettig R, Volzke H, Kim E, Lee WJ, Lee IT, Sheu WH, Tsosie KS, Edwards DR, Liu Y, Correa A, Weir DR, Volker U, Ridker PM, Boerwinkle E, Gudnason V, Reiner AP, van Duijn CM, Borecki IB, Edwards TL, Chakravarti A, Rotter JI, Psaty BM, Loos RJ, Fornage M, Ehret GB, Newton‐Cheh C, Levy D, Chasman DI. Meta‐analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet. 2016;48:1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY, Iribarren C, Chakravarti A, Risch N. Genome‐wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2017;49:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. International Consortium for Blood Pressure Genome‐Wide Association S , Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O'Reilly PF, Amin N, Bragg‐Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw‐Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland‐Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND; Consortium CA, Consortium CK, KidneyGen C, EchoGen C, Consortium C‐H , Aspelund T, Garcia M, Chang YP, O'Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O'Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace‐Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala‐Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska‐Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton‐Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wain LV, Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, O'Reilly PF, Cabrera CP, Warren HR, Rose LM, Verwoert GC, Hottenga JJ, Strawbridge RJ, Esko T, Arking DE, Hwang SJ, Guo X, Kutalik Z, Trompet S, Shrine N, Teumer A, Ried JS, Bis JC, Smith AV, Amin N, Nolte IM, Lyytikainen LP, Mahajan A, Wareham NJ, Hofer E, Joshi PK, Kristiansson K, Traglia M, Havulinna AS, Goel A, Nalls MA, Sober S, Vuckovic D, Luan J, Del Greco MF, Ayers KL, Marrugat J, Ruggiero D, Lopez LM, Niiranen T, Enroth S, Jackson AU, Nelson CP, Huffman JE, Zhang W, Marten J, Gandin I, Harris SE, Zemunik T, Lu Y, Evangelou E, Shah N, de Borst MH, Mangino M, Prins BP, Campbell A, Li‐Gao R, Chauhan G, Oldmeadow C, Abecasis G, Abedi M, Barbieri CM, Barnes MR, Batini C, Beilby J, Blake T, Boehnke M, Bottinger EP, Braund PS, Brown M, Brumat M, Campbell H, Chambers JC, Cocca M, Collins F, Connell J, Cordell HJ, Damman JJ, Davies G, de Geus EJ, de Mutsert R, Deelen J, Demirkale Y, Doney ASF, Dorr M, Farrall M, Ferreira T, Franberg M, Gao H, Giedraitis V, Gieger C, Giulianini F, Gow AJ, Hamsten A, Harris TB, Hofman A, Holliday EG, Hui J, Jarvelin MR, Johansson A, Johnson AD, Jousilahti P, Jula A, Kahonen M, Kathiresan S, Khaw KT, Kolcic I, Koskinen S, Langenberg C, Larson M, Launer LJ, Lehne B, Liewald DCM, Lin L, Lind L, Mach F, Mamasoula C, Menni C, Mifsud B, Milaneschi Y, Morgan A, Morris AD, Morrison AC, Munson PJ, Nandakumar P, Nguyen QT, Nutile T, Oldehinkel AJ, Oostra BA, Org E, Padmanabhan S, Palotie A, Pare G, Pattie A, Penninx B, Poulter N, Pramstaller PP, Raitakari OT, Ren M, Rice K, Ridker PM, Riese H, Ripatti S, Robino A, Rotter JI, Rudan I, Saba Y, Saint Pierre A, Sala CF, Sarin AP, Schmidt R, Scott R, Seelen MA, Shields DC, Siscovick D, Sorice R, Stanton A, Stott DJ, Sundstrom J, Swertz M, Taylor KD, Thom S, Tzoulaki I, Tzourio C, Uitterlinden AG, Volker U, Vollenweider P, Wild S, Willemsen G, Wright AF, Yao J, Theriault S, Conen D, Attia J, Sever P, Debette S, Mook‐Kanamori DO, Zeggini E, Spector TD, van der Harst P, Palmer CNA, Vergnaud AC, Loos RJF, Polasek O, Starr JM, Girotto G, Hayward C, Kooner JS, Lindgren CM, Vitart V, Samani NJ, Tuomilehto J, Gyllensten U, Knekt P, Deary IJ, Ciullo M, Elosua R, Keavney BD, Hicks AA, Scott RA, Gasparini P, Laan M, Liu Y, Watkins H, Hartman CA, Salomaa V, Toniolo D, Perola M, Wilson JF, Schmidt H, Zhao JH, Lehtimaki T, van Duijn CM, Gudnason V, Psaty BM, Peters A, Rettig R, James A, Jukema JW, Strachan DP, Palmas W, Metspalu A, Ingelsson E, Boomsma DI, Franco OH, Bochud M, Newton‐Cheh C, Munroe PB, Elliott P, Chasman DI, Chakravarti A, Knight J, Morris AP, Levy D, Tobin MD, Snieder H, Caulfield MJ, Ehret GB. Novel blood pressure locus and gene discovery using genome‐wide association study and expression data sets from blood and the kidney. Hypertension. 2017;70:e4–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, Ntalla I, Surendran P, Liu C, Cook JP, Kraja AT, Drenos F, Loh M, Verweij N, Marten J, Karaman I, Lepe MP, O'Reilly PF, Knight J, Snieder H, Kato N, He J, Tai ES, Said MA, Porteous D, Alver M, Poulter N, Farrall M, Gansevoort RT, Padmanabhan S, Magi R, Stanton A, Connell J, Bakker SJ, Metspalu A, Shields DC, Thom S, Brown M, Sever P, Esko T, Hayward C, van der Harst P, Saleheen D, Chowdhury R, Chambers JC, Chasman DI, Chakravarti A, Newton‐Cheh C, Lindgren CM, Levy D, Kooner JS, Keavney B, Tomaszewski M, Samani NJ, Howson JM, Tobin MD, Munroe PB, Ehret GB, Wain LV; International Consortium of Blood Pressure (ICBP) 1000G Analyses; BIOS Consortium; Lifelines Cohort Study; Understanding Society Scientific group; CHD Exome+ Consortium; ExomeBP Consortium; T2D‐GENES Consortium; GoT2DGenes Consortium; Cohorts for Heart and Ageing Research in Genome Epidemiology (CHARGE) BP Exome Consortium; International Genomics of Blood Pressure (iGEN‐BP) Consortium; UK Biobank CardioMetabolic Consortium BP working group . Genome‐wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415.28135244 [Google Scholar]
- 29. Duarte JD, Zineh I, Burkley B, Gong Y, Langaee TY, Turner ST, Chapman AB, Boerwinkle E, Gums JG, Cooper‐Dehoff RM, Beitelshees AL, Bailey KR, Fillingim RB, Kone BC, Johnson JA. Effects of genetic variation in H3K79 methylation regulatory genes on clinical blood pressure and blood pressure response to hydrochlorothiazide. J Transl Med. 2012;10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar A, Li Y, Patil S, Jain S. A haplotype of the angiotensinogen gene is associated with hypertension in African Americans. Clin Exp Pharmacol Physiol. 2005;32:495–502. [DOI] [PubMed] [Google Scholar]
- 31. Wen G, Wessel J, Zhou W, Ehret GB, Rao F, Stridsberg M, Mahata SK, Gent PM, Das M, Cooper RS, Chakravarti A, Zhou H, Schork NJ, O'Connor DT, Hamilton BA. An ancestral variant of secretogranin II confers regulation by PHOX2 transcription factors and association with hypertension. Hum Mol Genet. 2007;16:1752–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Darlu P, Sagnier PP, Bois E. Genealogical and genetical African admixture estimations, blood pressure and hypertension in a Caribbean community. Ann Hum Biol. 1990;17:387–397. [DOI] [PubMed] [Google Scholar]
- 33. Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A. Admixture mapping for hypertension loci with genome‐scan markers. Nat Genet. 2005;37:177–181. [DOI] [PubMed] [Google Scholar]
- 34. Ge D, Young TW, Wang X, Kapuku GK, Treiber FA, Snieder H. Heritability of arterial stiffness in black and white American youth and young adults. Am J Hypertens. 2007;20:1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sherva R, Miller MB, Lynch AI, Devereux RB, Rao DC, Oberman A, Hopkins PN, Kitzman DW, Atwood LD, Arnett DK. A whole genome scan for pulse pressure/stroke volume ratio in African Americans: the HyperGEN study. Am J Hypertens. 2007;20:398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen W, Srinivasan SR, Boerwinkle E, Berenson GS. Beta‐adrenergic receptor genes are associated with arterial stiffness in black and white adults: the Bogalusa Heart Study. Am J Hypertens. 2007;20:1251–1257. [DOI] [PubMed] [Google Scholar]
- 37. Reiner AP, Ziv E, Lind DL, Nievergelt CM, Schork NJ, Cummings SR, Phong A, Burchard EG, Harris TB, Psaty BM, Kwok PY. Population structure, admixture, and aging‐related phenotypes in African American adults: the Cardiovascular Health Study. Am J Hum Genet. 2005;76:463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang H, Jorgenson E, Gadde M, Kardia SL, Rao DC, Zhu X, Schork NJ, Hanis CL, Risch N. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Hum Genet. 2006;119:624–633. [DOI] [PubMed] [Google Scholar]
- 39. ClinicalTrials.gov . Minority health genomics and translational research bio‐repository database (MH‐GRID)‐2.0. 2017.
- 40. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C. Next‐generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second‐generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Purcell S, Neale B, Todd‐Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA; Team NGESP‐ELP , Christiani DC, Wurfel MM, Lin X. Optimal unified approach for rare‐variant association testing with application to small‐sample case‐control whole‐exome sequencing studies. Am J Hum Genet. 2012;91:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee S, Abecasis GR, Boehnke M, Lin X. Rare‐variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aschard H, Vilhjalmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome‐wide association studies. Am J Hum Genet. 2015;96:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mefford J, Witte JS. The covariate's dilemma. PLoS Genet. 2012;8:e1003096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pirinen M, Donnelly P, Spencer CC. Including known covariates can reduce power to detect genetic effects in case‐control studies. Nat Genet. 2012;44:848–851. [DOI] [PubMed] [Google Scholar]
- 49. Berthon A, Faucz F, Bertherat J, Stratakis CA. Analysis of ARMC5 expression in human tissues. Mol Cell Endocrinol. 2017;441:140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Petersen A, Alvarez C, DeClaire S, Tintle NL. Assessing methods for assigning SNPs to genes in gene‐based tests of association using common variants. PLoS One. 2013;8:e62161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010;6:e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Alderman MH, Cohen HW, Sealey JE, Laragh JH. Plasma renin activity levels in hypertensive persons: their wide range and lack of suppression in diabetic and in most elderly patients. Am J Hypertens. 2004;17:1–7. [DOI] [PubMed] [Google Scholar]
- 55. Berthon A, Stratakis CA. From beta‐catenin to ARM‐repeat proteins in adrenocortical disorders. Horm Metab Res. 2014;46:889–896. [DOI] [PubMed] [Google Scholar]
- 56. Tewari R, Bailes E, Bunting KA, Coates JC. Armadillo‐repeat protein functions: questions for little creatures. Trends Cell Biol. 2010;20:470–481. [DOI] [PubMed] [Google Scholar]
- 57. Hu Y, Lao L, Mao J, Jin W, Luo H, Charpentier T, Qi S, Peng J, Hu B, Marcinkiewicz MM, Lamarre A, Wu J. Armc5 deletion causes developmental defects and compromises T‐cell immune responses. Nat Commun. 2017;8:13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Berthon A, Faucz FR, Espiard S, Drougat L, Bertherat J, Stratakis CA. Age‐dependent effects of Armc5 haploinsufficiency on adrenocortical function. Hum Mol Genet. 2017;26:3495–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Assie G, Libe R, Espiard S, Rizk‐Rabin M, Guimier A, Luscap W, Barreau O, Lefevre L, Sibony M, Guignat L, Rodriguez S, Perlemoine K, Rene‐Corail F, Letourneur F, Trabulsi B, Poussier A, Chabbert‐Buffet N, Borson‐Chazot F, Groussin L, Bertagna X, Stratakis CA, Ragazzon B, Bertherat J. ARMC5 mutations in macronodular adrenal hyperplasia with Cushing's syndrome. N Engl J Med. 2013;369:2105–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Albiger NM, Regazzo D, Rubin B, Ferrara AM, Rizzati S, Taschin E, Ceccato F, Arnaldi G, Pecori Giraldi F, Stigliano A, Cerquetti L, Grimaldi F, De Menis E, Boscaro M, Iacobone M, Occhi G, Scaroni C. A multicenter experience on the prevalence of ARMC5 mutations in patients with primary bilateral macronodular adrenal hyperplasia: from genetic characterization to clinical phenotype. Endocrine. 2017;55:959–968. [DOI] [PubMed] [Google Scholar]
- 61. Bourdeau I, Oble S, Magne F, Levesque I, Caceres‐Gorriti KY, Nolet S, Awadalla P, Tremblay J, Hamet P, Fragoso MC, Lacroix A. ARMC5 mutations in a large French‐Canadian family with cortisol‐secreting beta‐adrenergic/vasopressin responsive bilateral macronodular adrenal hyperplasia. Eur J Endocrinol. 2016;174:85–96. [DOI] [PubMed] [Google Scholar]
- 62. Espiard S, Drougat L, Libe R, Assie G, Perlemoine K, Guignat L, Barrande G, Brucker‐Davis F, Doullay F, Lopez S, Sonnet E, Torremocha F, Pinsard D, Chabbert‐Buffet N, Raffin‐Sanson ML, Groussin L, Borson‐Chazot F, Coste J, Bertagna X, Stratakis CA, Beuschlein F, Ragazzon B, Bertherat J. ARMC5 mutations in a large cohort of primary macronodular adrenal hyperplasia: clinical and functional consequences. J Clin Endocrinol Metab. 2015;100:E926–E935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Faucz FR, Zilbermint M, Lodish MB, Szarek E, Trivellin G, Sinaii N, Berthon A, Libe R, Assie G, Espiard S, Drougat L, Ragazzon B, Bertherat J, Stratakis CA. Macronodular adrenal hyperplasia due to mutations in an armadillo repeat containing 5 (ARMC5) gene: a clinical and genetic investigation. J Clin Endocrinol Metab. 2014;99:E1113–E1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Correa R, Zilbermint M, Berthon A, Espiard S, Batsis M, Papadakis GZ, Xekouki P, Lodish MB, Bertherat J, Faucz FR, Stratakis CA. The ARMC5 gene shows extensive genetic variance in primary macronodular adrenocortical hyperplasia. Eur J Endocrinol. 2015;173:435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cavalcante IP, Nishi M, Zerbini MCN, Almeida MQ, Brondani VB, Botelho M, Tanno FY, Srougi V, Chambo JL, Mendonca BB, Bertherat J, Lotfi CF, Fragoso M. The role of ARMC5 in human cell cultures from nodules of primary macronodular adrenocortical hyperplasia (PMAH). Mol Cell Endocrinol. 2018;460:36–46. [DOI] [PubMed] [Google Scholar]
- 66. Mulatero P, Schiavi F, Williams TA, Monticone S, Barbon G, Opocher G, Fallo F. ARMC5 mutation analysis in patients with primary aldosteronism and bilateral adrenal lesions. J Hum Hypertens. 2016;30:374–378. [DOI] [PubMed] [Google Scholar]
- 67. Pal JK, Chatterjee S, Rao SJ. Pathological variations in 3′‐untranslated regions of human genes. eLS. 2016;1–11. [Google Scholar]
- 68. Vasan RS, Evans JC, Larson MG, Wilson PW, Meigs JB, Rifai N, Benjamin EJ, Levy D. Serum aldosterone and the incidence of hypertension in nonhypertensive persons. N Engl J Med. 2004;351:33–41. [DOI] [PubMed] [Google Scholar]
- 69. Rayner BL, Spence JD. Hypertension in blacks: insights from Africa. J Hypertens. 2017;35:234–239. [DOI] [PubMed] [Google Scholar]
- 70. Jones ES, Spence JD, McIntyre AD, Nondi J, Gogo K, Akintunde A, Hackam DG, Rayner BL. High frequency of variants of candidate genes in black Africans with low renin‐resistant hypertension. Am J Hypertens. 2017;30:478–483. [DOI] [PubMed] [Google Scholar]
- 71. Nanba K, Omata K, Gomez‐Sanchez CE, Stratakis CA, Demidowich AP, Suzuki M, Thompson LD, Cohen DL, Luther JM, Gellert L, Vaidya A, Barletta JA, Else T, Giordano TJ, Tomlins SA, Rainey WE. Genetic characteristics of aldosterone‐producing adenomas in blacks. Hypertension. 2019;73:885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zilbermint M, Hannah‐Shmouni F, Stratakis CA. Genetics of hypertension in African Americans and others of African descent. Int J Mol Sci. 2019;20:1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Laffer CL, Elijovich F, Eckert GJ, Tu W, Pratt JH, Brown NJ. Genetic variation in CYP4A11 and blood pressure response to mineralocorticoid receptor antagonism or ENaC inhibition: an exploratory pilot study in African Americans. J Am Soc Hypertens. 2014;8:475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wilson TW, Grim CE. Biohistory of slavery and blood pressure differences in blacks today. A hypothesis. Hypertension. 1991;17:I122–I128. [DOI] [PubMed] [Google Scholar]
- 75. Grim CE, Robinson M. Salt, slavery and survival‐ hypertension in the African diaspora. Epidemiology. 2003;14:120–122. discussion 124–126. [DOI] [PubMed] [Google Scholar]
- 76. Spence JD. Hypertension in US‐born vs. foreign‐born African‐Americans. J Hypertens. 2017;35:2369–2371. [DOI] [PubMed] [Google Scholar]
- 77. Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, Collins R, Allen NE. Comparison of sociodemographic and health‐related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. ARMC5 Variants and Risk of Hypertension in Blacks: Minority Health‐GRID Study
Table S2. Gene‐Based Analysis Results in the MH‐GRID Study
Table S3. MAF of rs116201073, rs141923065, and rs367810854 in the 1000 Genomes Project Populations
Figure S1. Summary of quality controls (QCs) for MH‐GRID (Minority Health Genomics and Translational Research Bio‐Repository Database) exome‐wide sequencing data.
Figure S2. Principal component analyses of MH‐GRID (Minority Health Genomics and Translational Research Bio‐Repository Database) data with 1000 Genomes Project samples.
Figure S3. Graphs of principal component analyses (PCA) of UK Biobank genotype data.
Figure S4. Comparison of wild‐type (WT) and mutant ARMC5 (rs116201073) expression in human embryonic kidney 293 (HEK293) cell line.


