Abstract
Phosphoregulation – in which the addition of a negatively charged phosphate group modulates protein activity – is a common feature of proteins that enables dynamic cellular responses. To understand how new phosphoregulation might be acquired, we mutationally scanned the surface of a prototypical yeast kinase (Kss1) to identify potential regulatory sites. The data reveal a set of spatially distributed “hotspots” that coevolve with the active site and preferentially modulate kinase activity. By engineering simple consensus phosphorylation sites at these hotspots we rewired cell signaling in yeast. Following the same approach in a homolog (Hog1), we introduced new phosphoregulation that modifies localization and signaling dynamics. Beyond synthetic biology, the hotspots are used by the diversity of natural allosteric regulatory mechanisms in the kinase family and exploited in human disease.
ONE SENTENCE SUMMARY
Cell signaling is easily rewired by introducing new phosphoregulation at latent allosteric surface sites.
INTRODUCTION
Phosphoregulation provides a dynamic and reversible means for the allosteric regulation of proteins. The introduction of new phosphoregulation (either by engineering or evolution) would seem to require satisfaction of two main properties. First, like any form of allostery, phosphoregulation requires the cooperative action of multiple amino acids to functionally link the phosphorylated site to a spatially distinct active site. Second, the addition of a phosphate group has to somehow engage or activate this underlying cooperative network. Regarding the former, several lines of work indicate that proteins possess a latent capacity for allosteric regulation at a diversity of surfaces. For example, it is possible to engineer synthetic allosteric switches through domain insertion at certain surface sites (1–5), and screens for small molecules that modify protein function sometimes identify cryptic allosteric regulatory sites (6, 7). In addition, experimental analysis of regulation in orthologs of the yeast MAP kinase Fus3 indicates that the capacity for allosteric regulation existed well before the regulatory mechanism evolved (8). Taken together, these findings suggest that proteins have an internal architecture in which multiple sites on the protein surface are functionally “pre-wired” to provide control of protein activity, and that these sites could serve as hot spots for the introduction of new regulation (5).
The question then becomes how placing a phosphate at one of these surfaces might engage the underlying allostery. Previous work from Ferrell and colleagues provides a potential solution: phosphoregulation might evolve simply by mutating an allosterically pre-coupled negatively charged residue (Asp/Glu) to a phosphorylatable residue (Ser/Thr/Tyr) (9). Thus, a constitutive negative charge at a latent allosteric site can be transformed into a regulated negative charge in a potentially stepwise manner (10). Here we experimentally test the proposal that new phospho-regulation can be introduced at negatively charged surface sites, and conduct sequence analyses to understand what properties distinguish sites with regulatory potential.
RESULTS
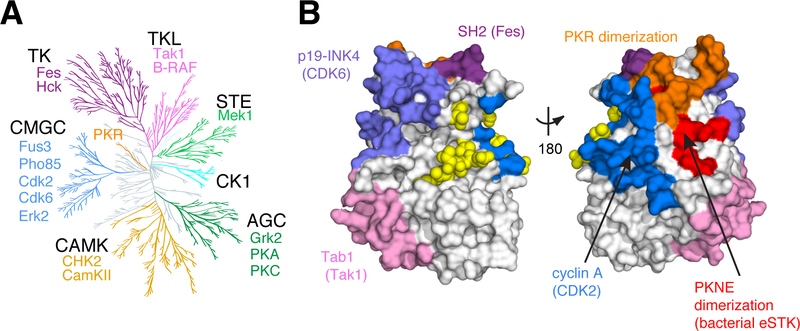
An excellent model to test this proposal is the eukaryotic protein kinases (EPKs), a protein family that has diversified to control a vast array of cellular signaling activities. The EPKs themselves catalyze the transfer of a phosphate group from adenosine triphosphate (ATP) onto a Ser/Thr/Tyr residue of a substrate protein, and are regulated by different mechanisms at distinct surface regions. To illustrate this, we mapped known regulatory sites from a diversity of kinases to a single representative kinase structure (Fig. 1A, B). Sites for regulation are distributed across the kinase surface, and mechanistically include protein-protein interactions, auto-inhibition, dimerization, and post-translational modification (11). This indicates that despite the complex intramolecular cooperativity required, allostery evolves readily at multiple distinct locations in the kinases (12). The diversity of regulation that has evolved across the kinome suggests the possibility that individual kinases might harbor a latent capacity for regulation at many surfaces.
Figure 1. Regulatory Diversity in the Eukaryotic Protein Kinases.
A. Unanchored dendrogram of the human kinome illustrating the diversity of the EPK superfamily and subfamilies. Individual subfamily members with functional mutations shown in Fig. 4c and included in Supplementary Table 7 are listed. TK: tyrosine kinase; TKL: TK-like; STE: STE7/11/20; CK1: Casein Kinase 1; AGC: protein kinase A/G/C; CAMK: Calmodulin kinase; CMGC: cyclin dependent kinase (CDK)/mitogen activated protein kinase (MAPK)/glycogen synthase kinase (GSK)/CDK-like kinase (CLK).
B. Allosteric regulatory sites from diverse kinases mapped to a single representative structure - yeast CDK Pho85 (PDB: 2PK9, shown as space-filled surface). Regulatory surfaces were identified by structural alignment of the kinase of interest to Pho85; all Pho85 positions within 4Å of the interaction surface are colored. Color coding is the same as in (A). Bright yellow spheres indicate known phosphoregulatory sites.
An alanine scan of negatively charged, solvent-accessible residues identifies functionally coupled positions distributed across the surface of Kss1.
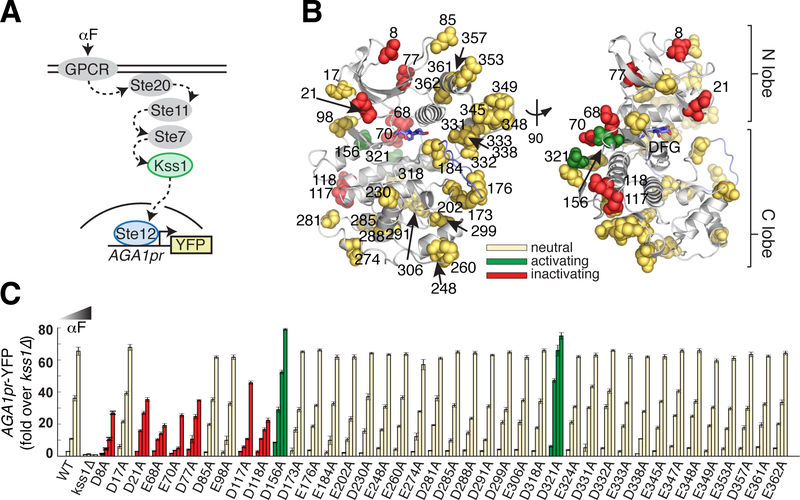
We chose the prototypical yeast CMGC kinase Kss1 as our experimental model (see Supplementary Text for extended Kss1 background). Kss1 is a homolog of human ERK and is involved in signal transduction pathways that regulate yeast filamentous growth and the mating response (13–16). Kss1 activity can be quantitatively monitored in living yeast cells by its ability to specifically activate fluorescent transcriptional reporters of the mating pheromone response in the absence of its paralog, Fus3 (Fig. 2A). We conducted an unbiased alanine scan of all 40 Asp/Glu residues on the surface of Kss1 to determine which positions are functionally coupled to kinase activity. We integrated the resulting 40 Kss1 mutants as the only copy of Kss1 in the yeast genome, tagged at their C-terminus with a 3xFLAG epitope (Supplementary Tables 1 and 2). To test their activity, we assayed for induction of the pheromone-responsive AGA1pr-YFP reporter at four concentrations of the alpha factor mating pheromone (αF) by flow cytometry. Though all mutants maintained wild type-like expression levels, nine mutations altered kinase activity in yeast (Fig. 2B, C, Supplementary Fig. 1A,B). Three of these positions were identified as Kss1 mutants with a functional effect in previous studies (Asp117, Asp156, Asp321, Supplementary Table 3) (17, 18). Though enriched in the N-terminal half of the primary Kss1 sequence, these nine mutations occur at positions distributed broadly over the Kss1 atomic structure – consistent with the notion that multiple, specific surface sites are pre-wired to allosterically influence active site function.
Figure 2. Alanine scan of acidic residues on the solvent accessible surface of yeast MAPK Kss1.
A. Schematic of the Kss1-dependent yeast pheromone pathway. The alpha factor (αF) mating pheromone binds to a G-protein coupled receptor (GPCR), leading to activation of a signaling cascade culminating at the MAPK Kss1. Kss1 then activates the Ste12 transcription factor to induce the mating transcriptional program, which can be monitored by fusing the promoter of the target gene AGA1 to a YFP reporter.
B. Ribbon diagram of a Kss1 homology model (30) with the 40 solvent accessible Asp/Glu residues shown as spheres. The DFG motif and activation loop are indicated in light blue. All 40 positions were mutated individually to alanine to remove negative charge.
C. The 40 resulting yeast strains along with wild type and kss1Δ controls were assayed for activation of the AGA1pr-YFP reporter by flow cytometry following treatment with 0, 0.01, 0.1 and 1 μM αF for 4 hours. Bars represent the average of the median YFP fluorescence from 3 biological replicates normalized to the untreated kss1Δ cells, and error bars are the standard deviation of the biological replicates. Mutations at red and green positions resulted in significantly reduced or increased YFP expression (P < 0.05, as scored by one-way ANOVA) in response to at least two doses of αF, respectively. Yellow positions indicate that the mutation had no effect in this assay. The color coding is identical in (B). Nine acidic positions on the solvent accessible surface are functionally coupled to kinase activity.
Negatively charged and functionally coupled surface sites can be used to engineer new regulation of Kss1 activity.
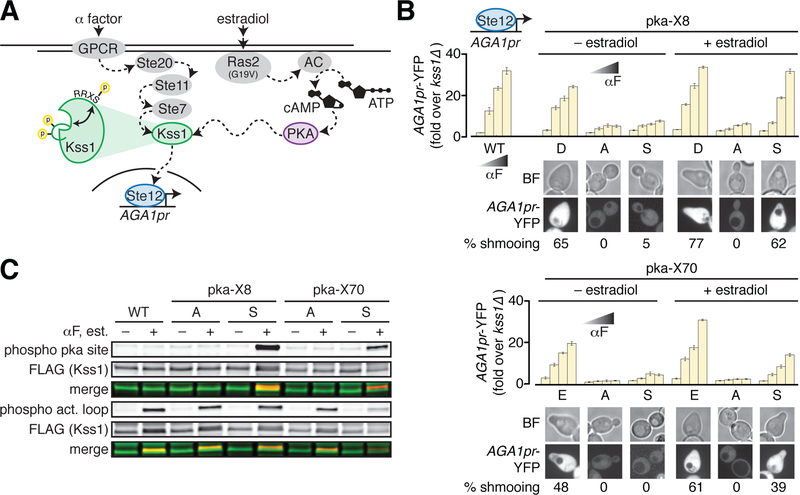
We next tested whether these positions can support new regulation of Kss1 through phosphorylation by another yeast kinase in vivo. In principle, this gain of function can effectively rewire signaling through the mating response pathway. We chose to engineer regulation of Kss1 by protein kinase A (PKA) because the PKA substrate consensus motif, RRxS/T requires minimal local modifications, its activity in yeast cells is orthogonal to the pheromone pathway, and it can be hyper-activated in yeast via ectopic expression of Ras2G19V (Fig. 3A, Supplementary Fig. 2A, B). We selected three of the nine mutationally sensitive positions (Asp8, Glu68 and Glu70) with the highest PKA substrate scores predicted by the computational tool pkaPS (19). To claim PKA-mediated allosteric regulation of Kss1, we must demonstrate: 1) that Kss1 retains functionality following introduction of a local PKA consensus motif (RRxD/E, termed pka-D/E); 2) that Kss1 loses activity when the charge is neutralized (RRxA, termed pka-A); and 3) that Kss1 now displays PKA-dependent activity in yeast with introduction of a phosphorylatable residue (RRxS, termed pka-S) (Supplementary Fig. 2C). In this manner, a functional surface negative charged residue might neutrally acquire a substrate consensus sequence for a kinase and become a phosphoregulatory site with one step of variation.
Figure 3. Engineering allosteric control of Kss1 by PKA phosphorylation.
A. Cartoon of the engineered PKA- and Kss1-dependent yeast pheromone pathway. In this schematic, Kss1 activation requires both activation loop phosphorylation by the upstream MAP2K Ste7, and phosphorylation by PKA at an allosterically coupled surface. To experimentally increase PKA activity, expression of constitutively activated Ras2(G19V) is induced by addition of estradiol, which in turn activates adenylate cyclase (AC) to generate cyclic AMP (cAMP) from ATP to activate PKA.
B. Kss1 mutants with PKA phosphorylation site consensus motifs introduced near position 8 (pka-X8, upper panel) or position 70 (pka-X70, lower panel) were assayed for expression of the AGA1pr-YFP reporter as in Fig. 2c. Again, bars represent the average of the median YFP fluorescence from 3 biological replicates normalized to the untreated kss1Δ cells, and error bars are the standard deviation of the biological replicates. “X” stands for the amino acid at position 8 or 70 as denoted under the bar graphs. The images below the bar graphs show morphology and expression of the AGA1pr-YFP reporter in yeast cells bearing the indicated Kss1 mutants in the presence of 1 μM alpha factor following growth in the presence or absence of 20 nM estradiol. The percent cells shmooing is indicated and was quantified by counting cells in three 512 × 512 pixel frames in each condition. N > 40 cells for each sample.
C. 3xFLAG-tagged wild type Kss1 and pka-X8 and -X70 mutants were immunoprecipitated from untreated cells or cells that had been treated with both 20 nM estradiol and 1 μM alpha factor. IP eluates were analyzed by Western blotting for total Kss1 as well as Kss1 phosphorylated on its activation loop (phospho act. loop) or at the engineered PKA site (phospho pka site). Merged images show that all mutants can be phosphorylated on their activation loop in the presence of alpha factor, but only pka-S8 and pka-S70 can be phosphorylated by PKA in the presence of estradiol.
Introduction of pka-E at position 68 resulted in Kss1 loss-of-function (Supplementary Fig. 2D, E), indicating that in this instance, the mutation of positions 65–66 to arginine to introduce the PKA site was not neutral. However, introducing the PKA consensus motif at positions D8 and E70 showed the complete expected pattern of activity for gain of phosphoregulation. For both sites, introduction of the two arginine residues upstream was near neutral, mutation of the negatively charged residue caused loss of function, and Kss1 pka-S activity depended on enhanced PKA activity via estradiol-induced expression of Ras2G19V (Fig. 3B). Immunoprecipitation of the 3xFLAG-tagged Kss1 mutants followed by Western blot analysis supports this finding. Both the pka-A and pka-S variants displayed activation loop phosphorylation when treated with alpha factor, indicating that they remain substrates of the upstream MAP2K Ste7. However, only Kss1-pka-S8 and Kss1-pka-S70 were recognized by an antibody specific for phosphorylated PKA substrates when purified from cells treated with estradiol (Fig. 3C). Moreover, both Kss1-pka-S8 and Kss1-pka-S70 were able to induce the morphological response to pheromone – the mating projection known as the “shmoo” – in an αF- and PKA activity-dependent fashion (Fig. 3B). Thus, the transcriptional and physiological outputs of Kss1 can be rewired to depend on an orthogonal input by a stepwise process of introducing a phosphorylation site at latent allosteric surface sites.
Functionally coupled Kss1 positions are associated with a network of conserved, co-evolving amino acids in the kinases.
What distinguishes the nine surface negatively charged amino acids that are allosterically coupled to Kss1 activity? Are the functionally coupled positions idiosyncratic to Kss1 or conserved across the kinase family? To address this, we examined the relationship between our mutational data and the pattern of amino acid conservation and co-evolution in protein kinases. The motivation is that conservation should provide an indication of functionally important positions, and co-evolution can provide insights into the pattern of coupling or interaction between amino acids. More specifically, prior work using an approach called the Statistical Coupling Analysis (SCA) showed that coevolving networks of amino acids form sparse yet physically contiguous networks in the protein tertiary structure, termed ‘sectors’. Sectors tend to link protein active sites to distantly positioned allosteric surface sites, and are proposed to represent a cooperative physical mechanism embedded within the protein (20–24). Experiments demonstrated that sector-connected surface sites are “hotspots” for introducing new allosteric regulation by domain insertion, suggesting that the sector endows particular locations on the protein surface with a latent capacity for allosteric regulation (5, 25). Following from this prior work, we: 1) hypothesize that multiple sites on the protein kinase surface are sector connected and allosterically coupled, 2) expect that the precise subset of coupled sites at which regulation is achieved will depend on details of the perturbation (choice of mutation or regulation introduced), and propose that 3) functional mutations will be found with strong statistical preference at sector-connected surface sites. Thus, we wanted to test if the functionally important D/E residues identified in Kss1 had a statistically significant association to co-evolving and/or conserved kinase positions. If so, it would suggest that these regulatory sites are not only relevant for Kss1, but represent conserved allosteric “hotspots” across the CMGC kinases or even the entire kinome.
We applied SCA to two multiple sequence alignments: one encompassing all EPK subfamilies, and a second focused alignment of the CMGC subfamily that includes the MAP kinases (Fig. 4A, B). Using SCA, we computed the conservation of individual positions (as a Kullback-Leibler relative entropy, Di) and an estimate of co-evolution between position pairs (as a conservation-weighted covariance matrix). The resulting pattern of co-evolution was analyzed using standard approaches from linear algebra to identify the sector (see also methods). Regardless of alignment choice, the sector defined for the kinases includes ~30% of amino acid positions and forms a physically contiguous network within the three-dimensional structure (Fig. 4C–E, Supplementary Table 4). The sector defined for the CMGC alignment includes the MAPK insert, a feature that is not conserved across the kinome-wide alignment (Fig. 4C). Because the SCA calculation of co-evolution emphasizes correlations at conserved positions, we see a strong overlap between individually conserved amino acids and co-evolving sector positions (Fig. 4D, see supplemental materials for additional discussion).
Figure 4. Statistical Coupling Analysis (SCA) of the Eukaryotic Protein Kinases.
The analysis was performed for two different multiple sequence alignments of the kinase catalytic domain: one specific to the CMGC kinases (635 sequences), and one containing 7128 kinases sampled across the kinome.
A. Histogram showing the distribution of pairwise sequence identities computed across all pairs of sequences in the CMGC alignment.
B. As in (A) but for the kinome wide alignment. Both alignments show a unimodal distribution with a mean pairwise sequence identity near ~25%.
C. Sector positions derived from the CMGC alignment (blue) or kinome-wide alignment (yellow) are distributed along the primary and secondary structure of the CMGC/MAPK ERK2. Subfamily-specific regions, such as the MAPK-insert, are only part of the sector derived from the CMGC alignment.
D. The relationship between the sector and positional conservation (computed as the Kullback-Leibler relative entropy, Di) for both the CMGC and kinome-wide alignments. Sector positions are highlighted in blue or yellow for the CMGC and kinome-wide alignments respectively. Red stars indicate highly conserved positions (defined as Di > 2.0 in the kinome-wide alignment).
E. The kinome-wide and CMGC-specific sectors (yellow and blue transparent surfaces, respectively) mapped on human ERK2 (gray ribbon) (PDB: 2ERK). Conserved positions are shown as red spheres.
Consistent with expectation, both the sector and conserved amino acids are enriched for positions associated with kinase function. The sector encapsulates several structural motifs well known to be associated with kinase activation including the αC-helix, the DFG motif and the catalytic and regulatory spines (Fig. 4C–E, Supplementary Fig. 3A–C) (26, 27). Further, comparison to a deep mutational scan of human ERK2 (28) shows a clear, statistically significant association between sites associated with loss-of-function and either the sector or conserved positions (Supplementary Fig. 4A–B, Supplementary Table 5). Thus, like for other proteins, analysis of conserved coevolution of amino acids in EPKs provides a sparse, distributed model for the functionally relevant energetic connectivity of amino acids (29).
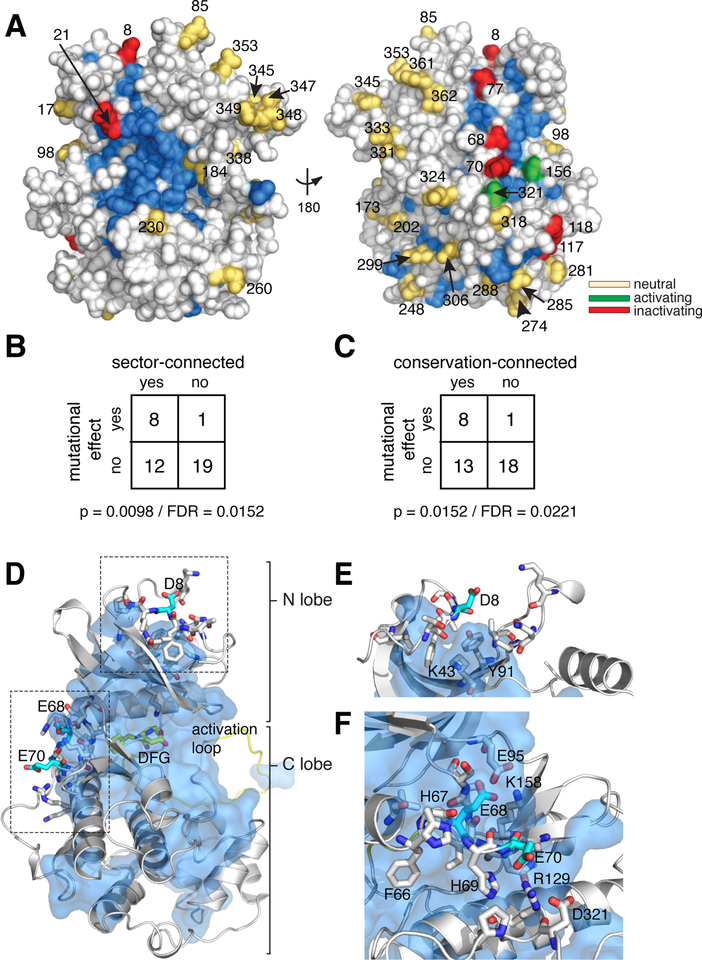
Although there is no crystallographic structure for Kss1, an available homology model permits comparison of our mutational data to the structural distribution of sector and conserved positions (30). Kss1 D/E mutations with a functional effect show a statistically significant association with conserved and co-evolving positions (Fig. 5A–C, Supplementary Fig. 5A,B, Supplementary Table 6). Eight of the nine functionally coupled surface D/E residues in Kss1 are contacting the co-evolving sector, including the two that yield new PKA-dependent phosphoregulation (Asp8 and Glu70). Position 8 is located in the N-terminal loop, and is proximal to two potential hydrogen bonding partners within the sector: Lys43and Tyr91(Fig. 5D,E). Both Lys43 and Tyr91 are moderately conserved (46% of kinases in the CMGC alignment contain K at position 43; 37% contain Y at position 91). Mutation at a position homologous to Kss1 Tyr91 in PKCβ (Y417H) is known to reduce kinase activity and is associated with liver cancer, indicating that this region is functionally important in other kinase family members (31). The second PKA regulatory site – position Glu70 – is contacting the sector, faces outward from the surface of the kinase and appears to interact with Arg129 (Fig. 5D, F). The P124S mutation in MEK2 (homologous to Glu70 in Kss1) is a gain of function and is associated with melanoma, also suggesting a functional role for this region in another kinase (32). Taken together, these data indicate that the gain of new regulatory function in Kss1 occurs at sites that are not idiosyncratic, but that interact with an allosteric network that coevolves across the kinase family.
Figure 5. Association of Kss1 D/E mutations with conserved and co-evolving positions.
A. Space filling diagram of a Kss1 homology model (30). The CMGC sector, defined as positions that co-evolve across the CMGC kinases, is indicated in blue. Acidic surface residues with a neutral, activating, or inactivating effect on kinase function upon mutation to alanine are shown as yellow, green or red spheres respectively.
B. Fisher’s exact table demonstrating statistically significant enrichment of acidic surface residues with a functional effect upon mutation at sector-connected positions. To be sector connected, a position must have at least one atom within 4 Å of the sector. Both a p-value and estimated false discovery rate are indicated.
C. Same as for B, but considering conservation-connected rather than sector-connected positions. The cutoff for conservation was chosen to give a similar number of positions as the sector. A detailed analysis of the effect of conservation and sector cutoffs can be found in the supplement.
D. Homology model of Kss1 illustrating the relationship of positions Asp8, Glu68 and Glu70 (cyan sticks) to the sector (blue surface). The kinase backbone is in grey cartoon; the activation loop and DFG active site motif are colored yellow. Residues proximal to Asp8, Glu68 and Glu70 are shown in grey sticks.
E. Close up view of the region surrounding position 8. Color coding follows from D.
F. Close up view of the region surrounding positions 68–70. Color coding follows from E.
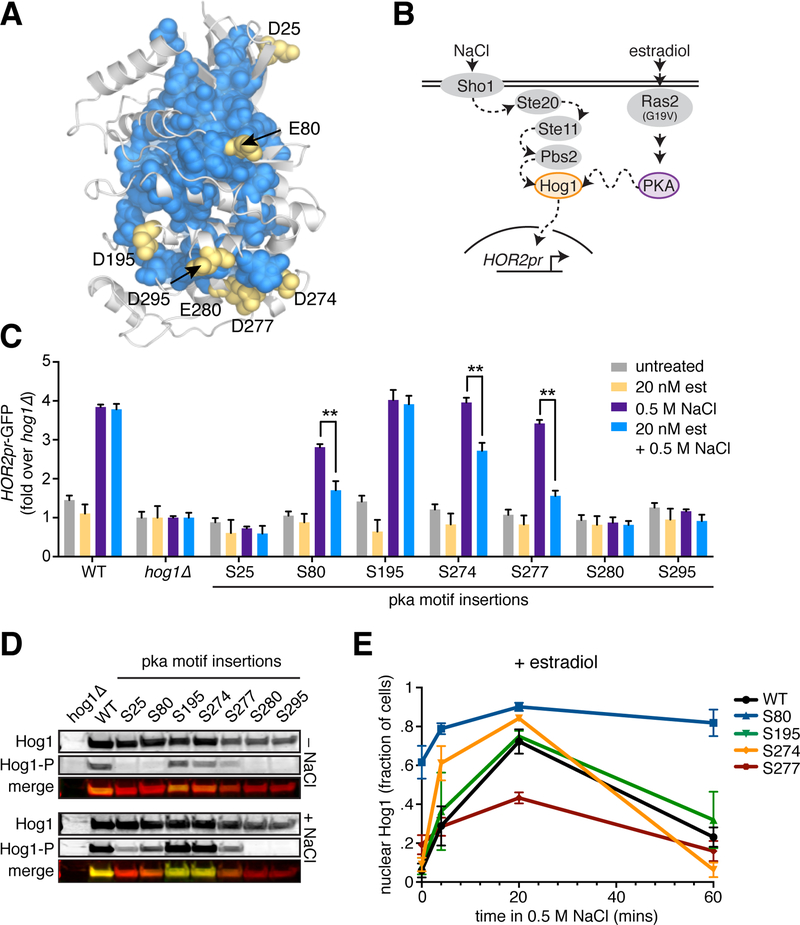
Introduction of phosphorylation sites at sector-connected surfaces on Hog1 can alter signaling dynamics
If the sector reflects a conserved allosteric mechanism within the protein kinases, sector-connected surface sites should be hot spots for the engineering of regulation in other kinase homologs. Further, in addition to simple phosphorylation-mediated restoration of function, we hypothesized that introducing phosphorylation sites at sector-connected surface positions could allow cells to evolve more complex regulatory schemes. For example, phosphorylation could tune the magnitude and dynamics of kinase activation rather than serving as an ON/OFF switch. To test these ideas, we extended our experimental analysis to a kinase that involves an additional layer of regulation: Hog1. Like Kss1, Hog1 is a yeast MAPK (homolog of human p38), but unlike Kss1, it transiently relocalizes from the cytosol to the nucleus upon activation by hyperosmotic stress, presenting the opportunity to modulate its localization and activation dynamics (33, 34).
To attempt to introduce novel functional phospho-regulation, we first identified sector-connected D/E surface residues in Hog1, reasoning that these sites are likely to be enriched for latent regulatory potential and can tolerate negative charge. We directly inserted pka-S motifs (RRxS) at all seven positions that satisfied these requirements and integrated the Hog1-pka mutants as the only copy of Hog1 in the cell (Fig. 6A). To assay for Hog1 activity, we treated cells with 0.5 M NaCl for two hours in the presence or absence of enhanced PKA activity (via estradiol-induced Ras2G19V expression) and measured induction of HOR2pr-GFP, a transcriptional reporter of Hog1 activity, by flow cytometry (Fig. 6B). Six of the seven Hog1-pka mutants showed reduced activity upon addition of NaCl, with three showing PKA-dependence (pka-S80, pka-S274 and pka-S277, Fig. 6C). Importantly, these PKA-dependent mutants displayed altered activity across a range of NaCl concentrations (Fig. S6C). Immunoblot analysis of 3xFLAG-tagged versions demonstrated that all mutants are stably expressed in cells but differentially phosphorylated on their activation loops in response to NaCl as determined by an anti-phospho-p38 antibody (Fig. 6D, Fig. S6A). Notably, activation loop phosphorylation was not perfectly correlated with the transcriptional reporter, suggesting that these normally concerted processes can be decoupled. Moreover, while hog1Δ showed a complete impairment of growth in the presence of NaCl, none of the PKA-dependent mutants showed any differences in proliferation compared to wild type (Fig. S6B). This indicates that Hog1 has retained its essential function in all of these mutants despite their altered patterns of activation loop phosphorylation and transcriptional output. Thus, as in the case of Kss1, the sector connected surface sites are highly functional. However, rather than rescuing the level of the transcriptional reporter, PKA activity reduced Hog1-dependent transcriptional induction.
Figure 6. Modulation of Hog1 activation dynamics by introduction of PKA phosphorylation sites at sector edges.
A. Sector-connected D/E residues are indicated on a homology model of Hog1.
B. Schematic of the engineered PKA/Hog1 pathway. Note that the Sln1/Ypd1 branch of the endogenous HOG pathway is omitted for clarity.
C. The 7 Hog1-pka yeast strains along with wild type and hog1Δ controls were grown in the presence and absence of 20 nM estradiol and assayed for activation of the HOR2pr-GFP reporter by flow cytometry following treatment with 0.5 M NaCl for 2 hours. Bars represent the average of the median GFP fluorescence from 3 biological replicates normalized to the untreated hog1Δ cells, and error bars are the standard deviation of the biological replicates. P-values were calculated by one-way ANOVA, ** denotes P < 0.01.
D. Immunoblots probed with anti-FLAG and anti-phospho-p38 antibodies to show total levels of the Hog1-pka mutants and their levels of activation loop phosphorylation under basal conditions and following treatment with 0.5 M NaCl for 15 minutes.
E. Quantification of Hog1-mKate nuclear localization dynamics in wild type and Hog1-pka mutants. Cells were imaged under basal conditions and following 5, 20 and 60 minutes of treatment with 0.5 M NaCl. N > 50 cells for all mutants at all time points.
To test if these PKA-dependent differences in Hog1 activity could be due to altered signaling dynamics, we fluorescently tagged wild type Hog1, the three PKA-dependent mutants (pka-S80, pka-S274, pka-S277), and the sole mutant that behaved like wild type (pka-S195) to monitor nuclear localization dynamics in single cells by fluorescence microscopy. In the absence of estradiol, all mutants and wild type localized to the nucleus upon acute activation with NaCl (Fig. S6D). In the presence of estradiol, pka-S195 was indistinguishable from wild type in its basal localization and nuclear translocation dynamics upon addition of NaCl. However, each of the three PKA-dependent mutants showed distinct, altered localization patterns (Fig. 6E). Despite not showing constitutive activation of the transcriptional reporter, pka-S80 was enriched in the nucleus in ~60% of cells under basal conditions, then showed persistent nuclear localization upon treatment with NaCl in ~80% of cells for the remainder of the 60-minute time course. By contrast, pka-S277 showed a muted nuclear localization response, maxing out at ~40% of cells with nuclear enrichment and receding with wild type-like kinetics. Perhaps most striking, pka-S274 translocated in and out of the nucleus with faster kinetics than wild type while attaining a maximum value comparable to wild type. Together, these results demonstrate that addition of phosphorylation sites at sector-connected positions can functionally modulate Hog1 signaling dynamics to generate novel regulatory schemes.
Functional mutations sampled across the kinome associate with the sector.
The data for Kss1 and Hog1 support a model that new regulation preferentially emerges in proteins at surface sites that are evolutionarily conserved across protein families. If so, all natural kinases should follow the principle that functionally sensitive and physiologically relevant allosteric sites, regardless of mechanism, should be found with statistical preference at conserved and sector-connected surfaces. The sector-connected surfaces would then provide an explanation for the diversity of regulatory sites observed in extant kinases (Fig. 1). To investigate this, we constructed a curated database of mutations sampled across the kinase superfamily (those listed in Fig 1A, Supplementary Table 7). These mutations were selected because they were experimentally demonstrated to disrupt kinase regulation and/or function, and, in many cases, are also associated with disease. Comparison of these mutations to the sector reveals a clear pattern: the mutations cluster around conserved positions and the sector edges with strong statistical preference (Fig. 7A–C, Supplementary Table 8). These data support the idea that the kinases share a conserved allosteric architecture (Fig. 7D).
Figure 7. Sector connected surface sites are associated with function in multiple kinases.
A. The EPK superfamily-wide sector (blue spheres) mapped to the CMGC yeast kinase Pho85 (PDB: 2PK9, grey cartoon and surface). Red positions are sites collected from the literature known to alter kinase function when mutated in a functional study or human disease context (Supplementary table 7).
B. Fisher’s exact table demonstrating statistically significant enrichment of the functional mutations shown in A at sector-connected positions.
C. Fisher’s exact table demonstrating statistically significant enrichment of the functional mutations shown in A at conservation-connected positions.
D. Model for the evolution of regulatory diversity. Latent allosteric sites distributed across the protein surface (red circles) are connected to the active site via a protein sector (blue arrows). These sites are poised for the acquisition of new regulation via evolutionary, disease, or engineering processes. In any particular family member, only a subset of sites may be used, and the regulatory mechanism need not be conserved across homologs.
DISCUSSION
We show that negatively charged, functionally coupled surface residues in Kss1 can be converted to sites of phosphoregulation by introducing a PKA consensus motif. Further, Kss1 positions with regulatory potential are statistically associated with the sector. This supports the proposal of Ferrell and colleagues that negatively charged surface positions are poised to become sites of phospho-regulation (9), and synthesizes this idea with work indicating that cooperative networks of amino acids within a protein endow certain sites with latent allosteric potential (5, 35). Moreover, in the case of Hog1, we found that more complex regulatory mechanisms can be instantiated by phosphorylation of sites at sector edges. Overall, these results suggest a general strategy for engineering new cell signaling pathways – in vivo phosphoregulation can in principle be introduced into any soluble protein by targeting negatively charged residues at sector-connected surfaces (35).
Beyond Kss1 and Hog1, comparison of the sector to kinome-wide mutational data indicates that sector-connected positions are enriched for mutations that perturb kinase function. The analysis of conservation and co-evolution presented here may thus provide a context for interpreting kinase mutations involved in disease, and suggest possible cryptic sites for the development of allosteric inhibitors (7). The results of our sequence analyses are robust to details of alignment construction and statistical cutoffs for determining sector positions and conservation (Supplementary Fig. 5, Supplementary Table 5,6 and 8). We note that the sector positions and conservation are strongly overlapping and show an equivalent association with the functional data, meaning that it is difficult to distinguish between the functional significance of conserved residues and sector residues (36). However, our goal is not to test the sector as an exclusive model for allosteric networks in proteins, but rather to examine if particular surface sites have a capacity for regulation that might be conserved across the kinase family. The sector provides one way to examine this, and unlike single-site conservation, leads naturally to the interpretation that these positions form a cooperative network embedded within the protein structure.
In terms of regulatory mechanism, one possibility is that mutations at or near highly conserved positions modulate activity through global coupling to kinase stability (e.g. by unfolding the kinase) rather than a more subtle rearrangement of thermodynamic interactions within the network of sector positions. Though our data cannot exclude the first possibility, they show that: 1) the surface D/E mutations do not strongly effect Kss1 expression, 2) the insertion of a PKA consensus motif at 9/10 positions tested on Kss1 and Hog1 does not strongly impact expression, and 3) Kss1-pka-A8 and Kss1-pka-A70 undergo activation loop phosphorylation (though they lack a negative charge at positions 8 and 70 and contain the PKA motif). These observations strongly support the idea that regulation is occurring through allosteric coupling among a defined group of residues, rather than global kinase destabilization at these sites.
Finally, we note that these results are consistent with the model that sector connected surfaces facilitate the evolution of regulatory diversity. In any individual kinase multiple latent allosteric sites are present, and a handful are harnessed by evolution to yield functional regulation. Thus, regulation that appears idiosyncratic across a protein family might be explained by a conserved underlying mechanism (Fig. 7D). Overall, this model provides a path for understanding how complex regulatory systems evolve and suggests that sector edges provide a substrate for generating variation in cellular signaling and communication.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains and plasmids used in this work are described in Supplementary Tables 1 and 2, respectively. All strains are in the W303 genetic background. Gene deletions were performed by one-step PCR as described (37). All Kss1 mutants were integrated into yeast genome as a single copy expressed from the endogenous KSS1 promoter.
Site-directed mutagenesis
Site-directed mutagenesis was performed with QuickChange according to the manufacturer’s directions (Agilent).
Cell growth and treatment with α factor and NaCl
All cells were grown in synthetic complete media with dextrose (SDC). Three single colonies from each Kss1 or Hog1 strain were inoculated in 1 ml SDC in 2 ml 96-well deep well plates and serially diluted 1:5 three times. Plates were incubated overnight at 30°C. In the morning cells from the row that had been diluted 1:25 were typically found to have OD600 ~0.5. These cells were diluted 1:5 in 4 rows of a 96 well U-bottom micro-titer plate in a total volume of 180 μl and incubated for 1 hour at 30°C. For Kss1 strains, in each row, cells were treated with different concentrations of α factor: 0, 0.01, 0.1 and 1 μM (10x stocks of α factor were prepared and 20 μl were added to 180 μl cells). Treated cells were incubated for an additional 4 hours at 30°C before translation was stopped by addition of 50 μg/ml cycloheximide. Cells were incubated for an additional hour at 30°C to allow time for fluorophores to mature. For Hog1 strains, 0, 0.25 0.5 and 0.75 M NaCl was added for 2 hours prior to cycloheximide arrest. For experiments with estradiol, everything is the same except that all media contained 20 nM estradiol for the duration of the overnight growth and throughout the experiment.
Flow cytometry
The AGA1pr-YFP and HOR1pr-GFP reporters were measured by flow cytometry by sampling 10 μl of each sample using a BD LSRFortessa equipped with a 96-well plate high-throughput sampler. Data were left ungated and FlowJo was used to calculate median YFP fluorescence. Bar graphs show the average of the median of the three independent colonies that were assayed, and error bars are the standard deviation.
Confocal microscopy
96 well glass bottom plates were coated with 100 μg/ml concanavalin A in water for 1 hour, washed three times with water and dried at room temperature. For Kss1 experiments, 80 μl of cells that had been treated with pheromone at the indicated concentrations for 3 hours were diluted to OD600 ~0.05 and added to a coated well. Cells were allowed to settle and attach for 15 minutes, and unattached cells were removed and replaced with 80 μl SDC media. Hog-mKate experiments were performed by adding 80 μl of untreated cells to the wells. After the cells were attached and basal images were captured, at t=0, the media was removed and replaced with SDC + 0.5 M NaCl and cells were imaged at 5, 20 and 60 minutes. Nuclear localization was scored manually. Imaging at was performed at the W.M Keck Microscopy Facility at the Whitehead Institute using a Nikon Ti microscope equipped with a 100×, 1.49 NA objective lens, an Andor Revolution spinning disc confocal setup and an Andor EMCCD camera. Images were analyzed in ImageJ.
Immunoprecipitation of 3xFLAG-tagged Kss1 and mutants
2 × 250 ml cultures of each strain were grown to OD600=0.8 at 30°C with shaking, one in SDC and the other in SDC + 20 nM estradiol. The SDC culture was left untreated while the SDC + estradiol culture was treated with 1 μM alpha factor for 30 minutes. Samples were collected by filtration and filters were snap frozen in liquid N2 and stored at −80°C. Cells were lysed frozen on the filters in a coffee grinder with dry ice. After the dry ice was evaporated, lysate was resuspended in 1 ml IP buffer (50 mM Hepes pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% triton x-100, 0.1% DOC, complete protease inhibitors), transferred to a 1.5 ml tube and spun to remove cell debris. Clarified lysate was transferred to a fresh tube and serial IP was performed. First, 25 μl of anti-FLAG magnetic beads (50% slurry, Sigma) were added, and the mixture was incubated for 2 hours at 4°C on a rotator. Beads were separated with a magnet and the supernatant was removed. Beads were washed 5 times with 1 ml IP buffer and bound material eluted 2x with 25 μl of 1 mg/ml 3xFLAG peptide (Sigma) in IP buffer by incubating at room temperature for 10 minutes. Beads were separated with a magnet and the two eluates were pooled in a fresh tube. 10 μl eluate was analyzed by Western blotting.
Western blotting
Total protein was TCA purified from cells as described (38). 10 μl of each sample was loaded into 4–15% gradient SDS-PAGE gels (Bio-Rad). Gels were run at 25 mA for 45 minutes, and blotted to PVDF membrane at 225 mA for 40 minutes. After 1hr blocking in Li-Cor blocking buffer, membranes were incubated with anti-FLAG primary antibody (SIGMA, F3165), anti-phospho-PKA substrate, anti-phospho p38, anti-phospho p44/42 (Cell Signaling, 9101), and/or anti-PGK (22C5D8) overnight at 4°C on a platform rotator (all 1:1000 dilutions in blocking buffer). Membranes were washed three times with TBST and probed by anti-mouse or anti-rabbit IR dye-congugated IgG (Li-Cor, 926–32352, 1:10000 dilution). The fluorescent signal was detected with the Li-Cor/Odyssey system.
Dilution series spot assays (frogging)
Cells were grown to OD600 = 0.3 in SDC, diluted to 0.06 and then serially diluted 1:5. 4 μl of each dilution was spotted on YPD + 20 nM estradiol and YPD + 20 nM estradiol + 0.8 M NaCl plates and incubated for 36 and 72 hours, respectively.
Statistical Coupling Analysis (SCA)
SCA was performed as described in (29) using PySCA 6 (http://reynoldsk.github.io/pySCA/) for two different multiple sequence alignments of the kinase catalytic domain: one specific to the CMGC kinases (635 sequences), and one containing 7128 kinases sampled across the kinome. The CMGC alignment was constructed by searching kinbase (http://kinase.com/kinbase/). Sequences were filtered for a length of 250–350 amino acids, and aligned by Promals3D (39) including the PDBS: 2B9H, 1BI8, 1Q97, 2ERK, 2F49, 2F9G, 2IW8, 2R7I, as reference structures. The kinome-wide alignment was previously constructed by the Shokat lab and was downloaded from http://sequoia.ucsf.edu/ksd/ (40). Following alignment processing and the application of sequence weights (as described in (29)), the alignments contained 464 and 380 total effective sequences for the CMGC and EPK alignments respectively. For both alignments, we followed an identical procedure for defining the sector. Briefly, we compute a conservation-weighted covariance matrix between all pairs of amino acid positions (see Supplementary Text for discussion of the relationship between the sector, conservation, and allosteric hotpots). This matrix provides a statistical description of the “evolutionary coupling” between all pairs of amino acid positions. We then analyze this matrix by conducting principle components analysis (PCA), and rotating the top eigenmodes using independent components analysis (ICA). The top independent components are used to define sectors. For both kinase alignments, we define a single sector that includes all positions contributing to the top 4 independent components (ICs). The group of positions contributing to each IC groups is defined by fitting an empirical statistical distribution to the ICs and choosing positions above a defined cutoff (default, > 95% of the CDF). The full analysis of both families can be downloaded from github (https://github.com/reynoldsk/KinaseAllosterySCA).
Defining sector-connected solvent accessible surface sites
We computed the relative solvent accessible surface area (RSA) over a homology model of Kss1 (30) using Michel Sanner’s MSMS with a probe size of 1.4 Å, excluding all water and heteroatoms (41). A cutoff of 20% RSA was used to define solvent exposed surface positions (5). “Sector-connected” is defined as a position where any atom is within 4.0Å of a sector position.
Supplementary Material
Figure 1. The effect of surface D/E to alanine mutations on Kss1 expression
Figure 2. Experimental approach to introduce a PKA phosphorylation site that controls MAPK Kss1 activity
Figure 3. The kinase sector encompasses the catalytic and regulatory spines
Figure 4. ERK2 mutations within the kinase sector are enriched for loss-of-function
Figure 5. The relationship of negatively charged surface positions to the kinome-wide EPK sector
Figure 6. Transcriptional activity and nuclear localization of Hog1-pka mutants
Table 1. Plasmids
Table 2. Yeast strains
Table 3. Comparison of Kss1 point mutations from the literature with our data and the CMGC sector (cutoff p=0.95)
Table 4. Sector positions mapped to several representative kinase structures
Table 5. Statistical association between the sector, conservation and ERK2 mutational data
Table 6. Statistical association between the sector, conservation and KSS1 D/E surface mutations
Table 7. Functional kinase mutations (sampled across the kinome)
Table 8. Statistical association between the sector, conservation and functional mutations sampled across a diversity of kinases
Acknowledgements:
This collaboration was initiated at the 2013 q-bio conference held at St. Johns College, Santa Fe, NM. We would like to thank R. Ranganathan for discussion and comments on the manuscript, and Dr. H. Zhang (UTSW bioinformatics) for review of our statistical methods. We are grateful to the Whitehead Institute FACS facility and the Keck Microscopy facility for technical assistance.
Funding: This work was supported by an NIH Early Independence Award (DP5 OD017941–01 to D.P.), the Green Center for Systems Biology, and the Gordon and Betty Moore Foundation’s Data-Driven Discovery Initiative (Grant GBMF4557 to K.R.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The SCA analysis for both kinase alignments is available from github (https://github.com/reynoldsk/KinaseAllosterySCA). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES
- 1.Dagliyan O et al. , Rational design of a ligand-controlled protein conformational switch. Proceedings of the National Academy of Sciences of the United States of America 110, 6800 (April 23, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dagliyan O et al. , Engineering extrinsic disorder to control protein activity in living cells. Science 354, 1441 (December 16, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guntas G, Ostermeier M, Creation of an allosteric enzyme by domain insertion. J Mol Biol 336, 263 (February 6, 2004). [DOI] [PubMed] [Google Scholar]
- 4.Nadler DC, Morgan SA, Flamholz A, Kortright KE, Savage DF, Rapid construction of metabolite biosensors using domain-insertion profiling. Nature communications 7, 12266 (July 29, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reynolds KA, McLaughlin RN, Ranganathan R, Hotspots for allosteric regulation on protein surfaces. Cell 147, 1564 (December 23, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA, Discovery of an allosteric site in the caspases. Proceedings of the National Academy of Sciences of the United States of America 101, 12461 (August 24, 2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu P, Clausen MH, Nielsen TE, Allosteric small-molecule kinase inhibitors. Pharmacology & therapeutics 156, 59 (December, 2015). [DOI] [PubMed] [Google Scholar]
- 8.Coyle SM, Flores J, Lim WA, Exploitation of latent allostery enables the evolution of new modes of MAP kinase regulation. Cell 154, 875 (August 15, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearlman SM, Serber Z, Ferrell JE Jr., A mechanism for the evolution of phosphorylation sites. Cell 147, 934 (November 11, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creixell P, Schoof EM, Tan CS, Linding R, Mutational properties of amino acid residues: implications for evolvability of phosphorylatable residues. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 367, 2584 (September 19, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellicena P, Kuriyan J, Protein-protein interactions in the allosteric regulation of protein kinases. Current opinion in structural biology 16, 702 (December, 2006). [DOI] [PubMed] [Google Scholar]
- 12.Kuriyan J, Eisenberg D, The origin of protein interactions and allostery in colocalization. Nature 450, 983 (12/13/print, 2007). [DOI] [PubMed] [Google Scholar]
- 13.Boulton TG et al. , An insulin-stimulated protein kinase similar to yeast kinases involved in cell cycle control. Science 249, 64 (July 06, 1990). [DOI] [PubMed] [Google Scholar]
- 14.Courchesne WE, Kunisawa R, Thorner J, A putative protein kinase overcomes pheromone-induced arrest of cell cycling in S. cerevisiae. Cell 58, 1107 (September 22, 1989). [DOI] [PubMed] [Google Scholar]
- 15.Cook JG, Bardwell L, Thorner J, Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390, 85 (November 6, 1997). [DOI] [PubMed] [Google Scholar]
- 16.Atay O, Skotheim JM, Spatial and temporal signal processing and decision making by MAPK pathways. The Journal of cell biology 216, 317 (February, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusari AB, Molina DM, Sabbagh W Jr., Lau CS, Bardwell L, A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. The Journal of cell biology 164, 267 (January 19, 2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madhani HD, Styles CA, Fink GR, MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91, 673 (November 28, 1997). [DOI] [PubMed] [Google Scholar]
- 19.Neuberger G, Schneider G, Eisenhaber F, pkaPS: prediction of protein kinase A phosphorylation sites with the simplified kinase-substrate binding model. Biology direct 2, 1 (January 12, 2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halabi N, Rivoire O, Leibler S, Ranganathan R, Protein sectors: evolutionary units of three-dimensional structure. Cell 138, 774 (August 21, 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatley ME, Lockless SW, Gibson SK, Gilman AG, Ranganathan R, Allosteric determinants in guanine nucleotide-binding proteins. Proceedings of the National Academy of Sciences of the United States of America 100, 14445 (November 25, 2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R, Structural determinants of allosteric ligand activation in RXR heterodimers. Cell 116, 417 (February 6, 2004). [DOI] [PubMed] [Google Scholar]
- 23.Smock RG et al. , An interdomain sector mediating allostery in Hsp70 molecular chaperones. Molecular systems biology 6, 414 (September 21, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suel GM, Lockless SW, Wall MA, Ranganathan R, Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol 10, 59 (January, 2003). [DOI] [PubMed] [Google Scholar]
- 25.Lee J et al. , Surface sites for engineering allosteric control in proteins. Science 322, 438 (October 17, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornev AP, Haste NM, Taylor SS, Eyck LF, Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proceedings of the National Academy of Sciences of the United States of America 103, 17783 (November 21, 2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornev AP, Taylor SS, Ten Eyck LF, A helix scaffold for the assembly of active protein kinases. Proceedings of the National Academy of Sciences of the United States of America 105, 14377 (September 23, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenan L et al. , Phenotypic Characterization of a Comprehensive Set of MAPK1/ERK2 Missense Mutants. Cell reports 17, 1171 (October 18, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivoire O, Reynolds KA, Ranganathan R, Evolution-Based Functional Decomposition of Proteins. PLoS computational biology 12, e1004817 (June, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T, The SWISS-MODEL Repository and associated resources. Nucleic acids research 37, D387 (January, 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antal CE et al. , Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. Cell 160, 489 (January 29, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikolaev SI et al. , Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nature genetics 44, 133 (December 25, 2011). [DOI] [PubMed] [Google Scholar]
- 33.Ferrigno P, Posas F, Koepp D, Saito H, Silver PA, Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. The EMBO journal 17, 5606 (October 1, 1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westfall PJ, Patterson JC, Chen RE, Thorner J, Stress resistance and signal fidelity independent of nuclear MAPK function. Proceedings of the National Academy of Sciences of the United States of America 105, 12212 (August 26, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pincus D, Resnekov O, Reynolds KA, An evolution-based strategy for engineering allosteric regulation. Physical biology 14, 025002 (April 28, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tesileanu T, Colwell LJ, Leibler S, Protein sectors: statistical coupling analysis versus conservation. PLoS computational biology 11, e1004091 (February, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longtine MS et al. , Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953 (July, 1998). [DOI] [PubMed] [Google Scholar]
- 38.Pincus D, Benjamin K, Burbulis I, Tsong AE, Resnekov O, Reagents for investigating MAPK signalling in model yeast species. Yeast 27, 423 (July, 2010). [DOI] [PubMed] [Google Scholar]
- 39.Pei J, Grishin NV, PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods in molecular biology 1079, 263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buzko O, Shokat KM, A kinase sequence database: sequence alignments and family assignment. Bioinformatics 18, 1274 (September, 2002). [DOI] [PubMed] [Google Scholar]
- 41.Sanner MF, Olson AJ, Spehner JC, Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38, 305 (March, 1996). [DOI] [PubMed] [Google Scholar]
- 42.Ma D, Cook JG, Thorner J, Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Molecular biology of the cell 6, 889 (July, 1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelet S, Nuclear relocation of Kss1 contributes to the specificity of the mating response. Scientific reports 7, 43636 (March 6, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J, Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Molecular and cellular biology 16, 3637 (July, 1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bardwell L et al. , Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes & development 12, 2887 (September 15, 1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bardwell L, Cook JG, Zhu-Shimoni JX, Voora D, Thorner J, Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proceedings of the National Academy of Sciences of the United States of America 95, 15400 (December 22, 1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gartner A, Nasmyth K, Ammerer G, Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes & development 6, 1280 (July, 1992). [DOI] [PubMed] [Google Scholar]
- 48.Beautrait A et al. , Mapping the putative G protein-coupled receptor (GPCR) docking site on GPCR kinase 2: insights from intact cell phosphorylation and recruitment assays. The Journal of biological chemistry 289, 25262 (September 5, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boutet E et al. , UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods in molecular biology 1374, 23 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Chi M et al. , Phosphorylation of calcium/calmodulin-stimulated protein kinase II at T286 enhances invasion and migration of human breast cancer cells. Scientific reports 6, 33132 (September 8, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dey M et al. , Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell 122, 901 (September 23, 2005). [DOI] [PubMed] [Google Scholar]
- 52.Filippakopoulos P et al. , Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell 134, 793 (September 5, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Good M, Tang G, Singleton J, Remenyi A, Lim WA, The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell 136, 1085 (March 20, 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenstein AE, Echols N, Lombana TN, King DS, Alber T, Allosteric activation by dimerization of the PknD receptor Ser/Thr protein kinase from Mycobacterium tuberculosis. The Journal of biological chemistry 282, 11427 (April 13, 2007). [DOI] [PubMed] [Google Scholar]
- 55.Maddika S et al. , Akt-mediated phosphorylation of CDK2 regulates its dual role in cell cycle progression and apoptosis. Journal of cell science 121, 979 (April 1, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP, Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 395, 237 (September 17, 1998). [DOI] [PubMed] [Google Scholar]
- 57.Shaag A et al. , Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Human molecular genetics 14, 555 (February 15, 2005). [DOI] [PubMed] [Google Scholar]
- 58.Wan PT et al. , Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855 (March 19, 2004). [DOI] [PubMed] [Google Scholar]
- 59.Yu Y et al. , Phosphorylation of Thr-178 and Thr-184 in the TAK1 T-loop is required for interleukin (IL)-1-mediated optimal NFkappaB and AP-1 activation as well as IL-6 gene expression. The Journal of biological chemistry 283, 24497 (September 5, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1. The effect of surface D/E to alanine mutations on Kss1 expression
Figure 2. Experimental approach to introduce a PKA phosphorylation site that controls MAPK Kss1 activity
Figure 3. The kinase sector encompasses the catalytic and regulatory spines
Figure 4. ERK2 mutations within the kinase sector are enriched for loss-of-function
Figure 5. The relationship of negatively charged surface positions to the kinome-wide EPK sector
Figure 6. Transcriptional activity and nuclear localization of Hog1-pka mutants
Table 1. Plasmids
Table 2. Yeast strains
Table 3. Comparison of Kss1 point mutations from the literature with our data and the CMGC sector (cutoff p=0.95)
Table 4. Sector positions mapped to several representative kinase structures
Table 5. Statistical association between the sector, conservation and ERK2 mutational data
Table 6. Statistical association between the sector, conservation and KSS1 D/E surface mutations
Table 7. Functional kinase mutations (sampled across the kinome)
Table 8. Statistical association between the sector, conservation and functional mutations sampled across a diversity of kinases