Summary
Lesion mimic mutants that exhibit spontaneous hypersensitive response (HR)‐like necrotic lesions are ideal experimental systems for elucidating molecular mechanisms involved in plant cell death and defence responses. Here we report identification of a rice lesion mimic mutant, spotted leaf 35 (spl35), and cloning of the causal gene by TAIL‐PCR strategy. spl35 exhibited decreased chlorophyll content, higher accumulation of H2O2, up‐regulated expression of defence‐related marker genes, and enhanced resistance to both fungal and bacterial pathogens of rice. The SPL35 gene encodes a novel CUE (coupling of ubiquitin conjugation to ER degradation) domain‐containing protein that is predominantly localized in cytosol, ER and unknown punctate compartment(s). SPL35 is constitutively expressed in all organs, and both overexpression and knockdown of SPL35 cause the lesion mimic phenotype. SPL35 directly interacts with the E2 protein OsUBC5a and the coatomer subunit delta proteins Delta‐COP1 and Delta‐COP2 through the CUE domain, and down‐regulation of these interacting proteins also cause development of HR‐like lesions resembling those in spl35 and activation of defence responses, indicating that SPL35 may be involved in the ubiquitination and vesicular trafficking pathways. Our findings provide insight into a role of SPL35 in regulating cell death and defence response in plants.
Keywords: Oryza sativa, SPL35, CUE domain‐containing protein, cell death, disease resistance
Introduction
Plants have evolved various defence mechanisms to protect themselves from pathogen attack, the most common of which is the hypersensitive response (HR) that acts to inhibit continued invasion or proliferation of pathogens in host plant tissues (Fujiwara et al., 2010; Wang et al., 2017). HR involves bursts of reactive oxygen species (ROS), expression of pathogenesis‐related (PR) genes, accumulation of phytoalexins and cell wall fortification through callose deposition, all of which help to inhibit pathogen proliferation (Shirsekar et al., 2014; Wang et al., 2017). Although HR plays a crucial role in plant disease resistance, its underlying molecular mechanisms are not fully understood.
To gain insights into HR‐mediated cell death in plants, a number of lesion mimic mutants (LMMs), showing typical HR‐like lesions independent of pathogen infection, have been investigated in a range of plant species, including Arabidopsis (Dietrich et al., 1994; Lorrain et al., 2004), maize (Johal et al., 1995; Walbot, 1991) and rice (Wang et al., 2015a, 2017). Many LMMs are known to have activated defence responses and sometimes display significantly enhanced resistance to disease, and thus are considered ideal for studying signalling pathways involved in HR‐mediated cell death (Shirsekar et al., 2014; Wang et al., 2015a,2015b, 2017). Identification and characterization of genes involved in LMMs may facilitate the elucidation of molecular mechanisms underlying plant disease resistance (Shirsekar et al., 2014; Wang et al., 2015b, 2017).
Over the past two decades, a number of lesion mimic genes have been cloned. These genes encode various proteins involved in numerous defence‐related signalling pathways, including those related to ROS generation, disease resistance, calcium influx, and protein ubiquitination or phosphorylation, as well as biosynthesis or metabolism of fatty acids/lipids, porphyrin and phenolic compounds. The identities of these genes indicated that the LMM phenotype could be the result of not only alternations in the HR‐response to pathogens but also due to changes in various physiological pathways essential in response to both biotic and abiotic stresses (Undan et al., 2012; Wang et al., 2015b, 2017).
Protein ubiqutination has emerged as a key mechanism involved in growth and development as well as immunity in animals and plants (Liu et al., 2017; You et al., 2016). Recognition of ubiquitin or specific ubiquitin chains by ubiquitin‐binding domains (UBDs) is vital for deciphering ubiquitin‐mediated signalling pathways (Liu et al., 2012a). Among a handful of types of characterized UBDs, the coupling of ubiquitin conjugation to endoplasmic reticulum (ER) degradation (CUE) domains consisting of 42–43 amino acid sequences have been identified and characterized based on similarity to the region of the yeast Cue1 protein, which is implicated in yeast ER‐associated degradation (ERAD; Shih et al., 2003); however, their roles in ERAD remain poorly understood (Li et al., 2017; Liu et al., 2012a; Shideler et al., 2015). In animals, some CUE domain‐containing (CUEDC) proteins have been implicated in programmed cell death (PCD) and immunity. For examples, the CUEDC protein, ERAD E3 gp78, is a regulator of liver homeostasis and a tumour suppressor in liver, and its CUE domain interacts with ubiquitin and diubiquitin and functions to facilitate substrate binding and processivity in ubiquitination (Liu et al., 2012a; Zhang et al., 2015); another CUEDC protein, CUEDC2, suppresses glioma tumorigenicity by inhibiting the activation of STAT3 and NF‐κB, and its CUE domain is essential for functional interactions with both monoubiquitin and polyubiquitin (Li et al., 2017). In plants, the reported CUECD proteins are the duplicated pair of Arabidopsis RING‐finger E3 ligases, RIN2 and RIN3, which each possess a RING‐finger domain in association with a CUE domain; however, the CUE domain was rarely characterized for its roles in the E3 ligases (Kawasaki et al., 2005). Therefore, the question whether CUEDC proteins are involved in immunity and ubiquitination pathways in plants remains to be answered.
In the present study, we isolated and characterized a rice LMM named spotted leaf 35 (spl35) from a T‐DNA insertion population of cultivar (cv.) Kinmaze. spl35 displayed spotted leaves and enhanced resistance to rice blast and bacterial blight. We cloned SPL35 by TAIL‐PCR (thermal asymmetric interlaced PCR) strategy. SPL35 encodes a novel CUEDC protein and is expressed at all developmental stages and in all tissues examined. SPL35 is mainly localized in cytosol, ER and unknown compartment(s). SPL35 directly interacts with an E2 protein, OsUBC5a, and two coatomer subunit delta proteins, Delta‐COP1 and Delta‐COP2 both in vitro and in vivo. Our results indicate that SPL35 is involved in ubiquitination and vesicular trafficking, and disruption of SPL35 can result in cell death and enhanced defence response in rice.
Results
Phenotypic characteristics of the spl35 mutant
We isolated a LMM, named spotted leaf 35 (spl35), from a T‐DNA insertion population created by transformation of cv. Kinmaze. The spl35 plants began to show small, reddish‐brown lesions or spots from top to bottom of leaves at the three‐leaf stage [about 20 days after seed sowing (DAS)], and such spots gradually developed and scattered on every fully expanded leaf throughout the life cycle (Figure 1a–c). In general, young leaves displayed fewer and smaller lesions, and old leaves had more and larger lesions that frequently merged into each other, in contrast to green healthier leaves in wild‐type (WT, Kinmaze; Figure 1c). As a result, spl35 had significant reduction in plant height, number of effective panicles, number of spikelets per panicle, seed setting rate and 1000‐grain weight by 11.39%, 21.06%, 24.88%, 20.31% and 9.30%, respectively, compared to WT, but no change in heading date and panicle length (Figure 1e,f; Table S2).
Figure 1.

Phenotypes of the spotted leaf 35 (spl35) mutant. (a and b) Leaf blade phenotypes of wild‐type (WT) (a) and spl35 (b) at four‐leaf stage; left to right: fourth, third and second leaf blades respectively. (c) Lesion mimic phenotypes of different leaves in the main culm of spl35 plants at the tillering stage corresponding to (e). From left to right: youngest leaf to oldest leaf blades respectively. (d) Light avoidance assay of lesion mimic flag leaves. Left: leaf blade with no light avoidance; middle and right: leaf blade wrapped with aluminium foil in the middle part for 7 and 14 days respectively. (e) Plants at tillering stage (60 days after seed sowing) in the paddy field. (f) Plants at 15 days after flowering in the paddy field. Scale bars, 5.0 cm.
It is often seen that development of lesions in LMMs is associated with exposure to light (Wang et al., 2017). Thus, we performed light avoidance assays on spl35 as described previously (Wang et al., 2017). The leaf area without exposure to light did not develop lesions of any size, whereas the neighbouring uncovered area did develop numerous lesions (Figure 1d), indicating that formation of lesions in spl35 was light‐dependent.
To examine physiological changes in spl35, we measured chlorophyll (Chl a and Chl b) and carotenoid contents in flag leaves of field‐grown spl35 and WT plants at 10 days after flowering (DAF). Both chlorophyll and carotenoid contents in spl35 were significantly decreased relative to the WT (Figure S1a). We further investigated chloroplast development in spl35 and WT leaves at 10 DAF by TEM. The chloroplast in the spotted leaf area of spl35 was abnormally shaped, with destructed thylakoid stacking, in contrast to elliptical shape of WT chloroplasts with neatly stacked thylakoids and normal grana (Figure S1b,c). Thus, degradation of thylakoids in damaged chloroplasts of spl35 impaired photosynthesis capacity and in turn lead to smaller plant size.
Cell death and H2O2 accumulation in the spl35 mutant
To identify possible biochemical mechanisms underlying the development of HR‐like lesions in spl35, we investigated expression of histochemical markers for cell death and H2O2, in spl35 and WT. Trypan blue staining, a method to detect cell death (Dietrich et al., 1994), showed a number of deep blue spots at lesion sites in spl35, but not in WT (Figure S2a). H2O2 is a membrane‐permeable ROS and one of the major signalling molecules causing cellular damage or triggering PCD (Wang et al., 2017). We examined H2O2 level by 3,3′‐diaminobenzidine (DAB) staining and found consistently higher H2O2 accumulation in spl35 than in WT (Figure S2b). In addition, the H2O2 contents in spl35 seedlings at the two‐ to three‐leaf (before appearance of visible lesions) and at the four‐ to five‐leaf stage (lesions clearly visible) were approximately 1.5‐ and 2‐fold higher than in WT leaves respectively (Figure S2c). These results suggest that the formation of mimic lesions was associated with ROS accumulation and irreversible membrane damage in the spl35 cells.
Activation of defence responses in the spl35 mutant
In some LMMs, activation of defence responses and enhanced disease resistance have been reported (Tang et al., 2011). We first analysed the expression level of ER chaperone genes including OsCNX1, OsBip1 and OsPDIL1‐1 that are known indicators of accumulation of unfolded proteins in ER caused by accumulation of ROS (Cui et al., 2012a,2012b), and found that the RNA expression level of these genes was increased by 1.8‐, 1.5‐ and 2.3‐folds, respectively, in spl35 relative to WT (Figure S3). Then, we investigated resistance of spl35 to blast and bacterial blight diseases with isolates of two Maganaporthe oryzae (M. oryzae) and four Xanthomonas oryzae pv. oryzae (Xoo) pathotypes virulent to WT at the tillering stage, and observed significantly gained disease resistance in spl35 (Figure 2a–d).
Figure 2.
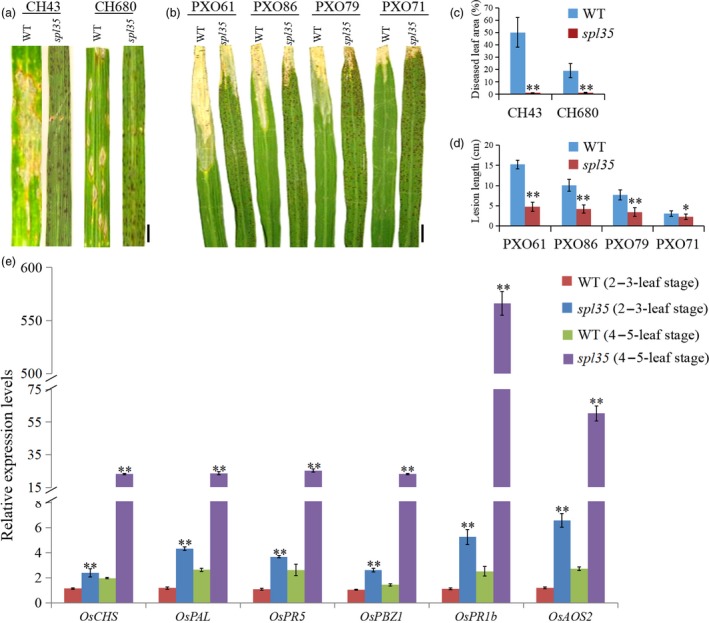
Resistance reactions to Maganaporthe oryzae (M. oryzae) and Xanthomonas oryzae pv. oryzae (Xoo) isolates tested and expression analysis of defence‐related markers in wild‐type (WT) and spl35 plants. (a and b) Disease symptoms of WT and spl35 leaves after challenge with M. oryzae (a) and Xoo (b) isolates. (c) Diseased leaf area of WT and spl35 leaves scanned at 7 days after inoculation with two different M. oryzae isolates using the Chalkiness 1.0 software (Zheng and Wu, 2008). Asterisks indicate significant differences between WT and spl35 leaves (**P < 0.01; Student's t test). Data are means ± SD of at least 10 leaves. (d) Mean lesion length of WT and spl35 leaves measured at 14 days after inoculation with four different Xoo isolates. Asterisks indicate significant differences between WT and spl35 mutant plants (*P < 0.05, **P < 0.01; Student's t test). Data are means ± SD of at least 10 leaves. (e) qRT‐PCR analysis of defence‐related markers in WT and spl35 mutant at two‐ to three‐leaf stage (before appearance of visible lesions in spl35) and four‐ to five‐leaf stage (lesions clearly visible in spl35) respectively. Similar results were obtained in three independent biological replicates. Asterisks indicate significant differences between WT and spl35 seedlings (**P < 0.01; Student's t test). Scale bars, 2.0 cm.
In order to understand the relation of the gained resistance with the development of HR‐like lesions in spl33, we inoculated spl35, WT and LTH (a variety susceptible to a wide range of isolates) seedlings at the two‐ to three‐leaf and four‐ to five‐leaf stages with isolates of six M. oryzae pathotypes virulent to WT. The spl35 seedlings infected at the four‐ to five‐leaf stage exhibited blast resistance as indicated by lack of newly developed lesions, whereas those infected at the two‐ to three‐leaf stage exhibited fewer and smaller blast lesions than WT and LTH, a sign of moderate resistance (Figure S4; Table S3). This observation suggests that the defence response was initiated before the appearance of the visible mimic lesions in spl35.
We then examined the expression of six defence‐related (DR) or pathogenesis‐related (PR) genes, OsCHS, OsPAL, OsPR5, OsPBZ1, OsPR1b and OsAOS2 (Liu et al., 2012b; Tang et al., 2011), in spl35 and WT at two‐ to three‐leaf stages and four‐ to five‐leaf stages by qRT‐PCR. At the two‐ to three‐leaf stage expression level of these six DR/PR genes in spl35 was up‐regulated by 2.1‐, 3.6‐, 3.5‐, 2.4‐, 4.7‐ and 5.5‐folds, respectively, and as development of the seedlings to the four‐ to five‐leaf stage up‐regulation was further increased to 11.7‐, 8.9‐, 9.6‐, 16.2‐, 222.6‐ and 22‐fold higher, respectively, than in WT (Figure 2e). Thus, we speculate that the defence responses are triggered in spl35 and that the uncontrolled continuation of the responses leads to the development of mimic lesions and enhanced disease resistance.
Shi et al. (2013) showed that expression of LOC_Os03g10750 (SPL35) was suggested a role in abiotic stress. In order to evaluate response of SPL35 to other abiotic stresses, we examined expression levels of SPL35 in wild‐type Kinmaze seedlings at different time intervals after treatments with mannitol, H2O2, NaCl, 1‐Amino‐1‐cyclopropanecarboxylic acid (ACC) and abscisic acid (ABA), and showed that SPL35 was significantly induced by these stresses at 4 h post treatment (Figure S5). Based on these aforementioned results, we speculate that SPL35 may be involved in various signalling or regulatory pathways associated with biotic and abiotic stress.
Identification of the spl35 locus
Since spl35 was isolated from a T‐DNA population, we first determined if the lesion mimic phenotype was associated with the presence of the T‐DNA insertion using the hygromycin B marker Hyr‐F/R and number of insertions in this line by Southern blotting. Our analysis indicated that the mutation was recessive, caused by a single T‐DNA insertion (Table S4; Figure 3d). Next, we employed the TAIL‐PCR method and cloned a 750 bp sequence flanking the T‐DNA. BLAST analysis of this sequence (http://blast.ncbi.nlm.nih.gov/Blast.cgi) anchored the insertion to a site 948 bp upstream of the translation initiation site of LOC_Os03g10740 and 458 bp downstream of the translation termination site of LOC_Os03g10750 (Figure 3a,c). Furthermore, using 280 F2 lesion mimic individuals from the cross of spl35 with an indica cv. 93‐11, we were able to delimit the mutation locus to a 760 kb region flanked by markers ID3‐10 and ID3‐11 (location: 4 894 747–5 654 369 bp; Figure 3b). This region spans the T‐DNA site, thus supporting the TAIL‐PCR result.
Figure 3.
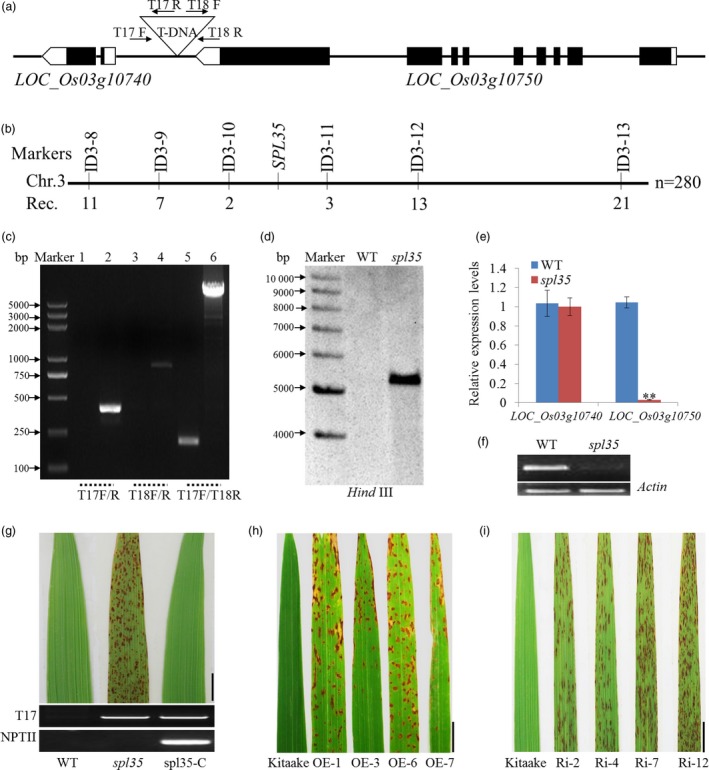
Cloning and molecular analysis of the SPL35 gene. (a) T‐DNA insertion site of the spl35 mutant. The triangle indicates the T‐DNA insertion site in the spl35 mutant. Black boxes indicate exons, lines indicate introns, and white boxes indicate the untranslated regions. (b) Physical map of the SPL35 locus on the short arm of chromosome 3. Numbers below the map indicate the number of recombinants between markers. (c) Flanking sequence was confirmed by PCR using the primers (T17F/R, T18F/R and T17F/T18R) shown in (a). Lanes 1, 3 and 5 represent WT; Lanes 2, 4 and 6 represent spl35; M, DL2000 marker. (d) Copy number determination of T‐DNA insertion by Southern blotting. A fragment of the hygromycin phosphotransferase gene was used as probe. The genomic DNA was digested by Hind III. (e) Expression of two genes within the vicinity of the T‐DNA insertion site by qRT‐PCR analysis. Asterisks indicate significant differences between WT and spl35 plants (**P < 0.01; Student's t test). Data are means ± SD of three independent biological replicates. (f) Expression analysis of LOC_Os03g10750 in WT and spl35 plants by qRT‐PCR. Rice actin gene was used as an internal control. (g) Phenotypes of WT, spl35 and complemented spl35 (spl35‐C) plants at the 15th day after flowering in field conditions. Scale bars, 5.0 cm. (h, i) Lesion mimics on the flag leaves of LOC_Os03g10750‐OE and ‐Ri T1 lines in the cv. Kitaake background. Data are means ± SD of three independent biological replicates. Scale bars, 1.0 cm.
We performed qRT‐PCR and semi‐quantitative RT‐PCR to understand if the T‐DNA insertion caused any change in expression level of LOC_Os03g10750 and LOC_Os03g10740 and found significant down‐regulation of LOC_Os03g10750 (Figure 3e–f). To verify if the down‐regulation of LOC_Os03g10750 was the cause of the mutant phenotype, we introduced the constructs p2300C‐10750 containing a genomic sequence of LOC_Os03g10750, and p2300C‐10740 containing a genomic sequence of LOC_Os03g10740 as a comparison, into spl35, and obtained 22 and 15 independent transgenic plants, respectively. Complementation was found in all the p2300C‐10750, but not p2300C‐10740 plants, suggesting that LOC_Os03g10750 was the candidate gene for SPL35 (Figure 3g; Figure S6).
SPL35 encodes a CUE domain‐containing protein
Sequence comparison between genomic DNA and cDNA showed that SPL35 contains nine exons and eight introns (Figure 3a). The coding sequence of SPL35 consists of 2784 nucleotides encoding a 927 amino acid protein with an estimated molecular weight of 101.95 kD, and a pI of 6.65 (Figure S7a). SPL35 is an unknown protein containing a single CUE domain of 43 amino acids that shares 46.5% similarity to the Cue1p‐CUE domain first identified in yeast (Biederer et al., 1997; Figure S7a,b). The SPL35 protein also contains conserved Phe‐Pro (FP) and Ile‐Leu (IL) motifs that are crucial for efficient ubiquitin binding, suggesting that it may function in ubiquitin binding. The SPL35 CUE domain is also similar to that of the CUEDC proteins Cue2, Tollip and Vps9 which are involved in the trafficking and ubiquitination pathways (Shih et al., 2003; S7b). A Gramene database (http://ensembl.gramene.org/) search showed that there were five other putative CUEDC proteins in rice, LOC_Os02g09180, LOC_Os06g43550, LOC_Os10g16440, LOC_Os06g49200 and LOC_Os07g30100, but they are different from SPL35 in amino acid sequence (Figure S7b). Phylogenetic analysis revealed that SPL35 has the higher similarity to orthologs from monocot plants species such as Brachypodium distachyon (78.9%), Hordeum vulgare (50.2%) Sorghum bicolor (77.4%) and Zea mays (62.8%), but much lower similarity to orthologs in dicot species (29.2%–59.8%) and animals (28.8%–35.3%; Figure S8).
Both overexpression and knockdown of SPL35 cause the lesion mimic phenotype
To examine the expression of SPL35 is tightly regulated for its normal function, we transformed an overexpression vector (p35S:SPL35‐OE) and an RNA interference vector (pUbi:SPL35‐RNAi) into cv. Kitaake, and produced 16 and 21 independent plants, respectively. Overexpression in the p35S:SPL35‐OE plants and down‐regulation in the pUbi:SPL35‐RNAi plants were confirmed by qRT‐PCR (Figure S9). As expected, down‐regulation of SPL35 caused the lesion mimic phenotype, but to our surprise, overexpression of SPL35 also lead to a phenotype similar to spl35 such as decreased tiller number and plant height in addition to the mimic lesions (Figure 3h,i;Figure S10). Those findings suggest that expression of SPL35 must be tightly regulated to ensure its function and that manipulating expression level of SPL35 may be a potential strategy for improving disease resistance in plant breeding.
In addition, we did deletion analysis to SPL35 by overexpressing its N‐terminal (1‐568) containing the CUE domain (p35S:SPL35 1‐568 ‐OE) and its C‐terminal (569‐927) (p35S:SPL35 569‐927 ‐OE) in cv. Kitaake. Sixteen of the 19 p35S:SPL35 1‐568 ‐OE plants, but none of the 15 p35S:SPL35 569‐927 ‐OE plants, showed a phenotype similar to spl35 (Figure S11). This result implies an essential role of the CUE domain for functioning of SPL35.
Expression pattern of SPL35
We investigated the expression pattern of SPL35 at different development stages and in different organs by qRT‐PCR. The transcription level of SPL35 in leaf blades was low at the one‐ to two‐leaf stage, reached a peak at the seven‐ to eight‐leaf stage, and then decreased at the nine‐ to ten‐leaf stage (Figure 4a), indicating developmental regulation of SPL35. SPL35 was expressed in all organs examined, with the strongest expression in leaf blades and panicles (Figure 4b). We also generated transgenic plants carrying the pSPL35:GUS reporter construct. β‐glucuronidase (GUS) staining showed activity of the SPL35 promoter in all tissues examined (Figure 4c–k). These results indicate that SPL35 is ubiquitously expressed in all organs and to certain extent under developmental regulation.
Figure 4.
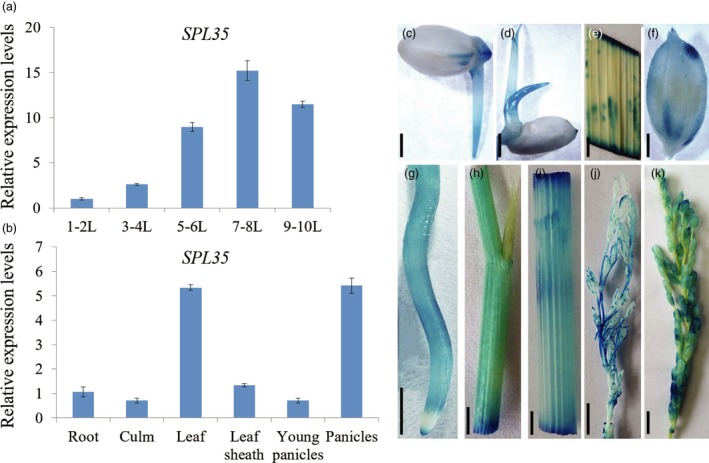
Expression pattern of SPL35 in the wild‐type Kinmaze. (a) Expression levels of SPL35 in leaf blades at different developmental stages. L, leaf blade. Each bar is the mean ± SD of three independent biological replicates. (b) Expression levels of SPL35 in various tissues. Data are means ± SD of three independent biological replicates. (c–k) Gus staining of SPL35pro::GUS transgenic plants. Seed (c), young leaf (d), leaf blade (e), hull (f), root (g), leaf sheath (h), culm (i), 8–10 cm young panicle (j) and mature panicle (k). Scale bars, 1.0 mm.
Subcellular localization of the SPL35 protein
SPL35 has no predicted transmembrane domain except the only CUE domain (http://smart.embl-heidelberg.de/smart). Transient expression of p35S:GFP‐SPL35 in rice protoplasts and immunoblot analysis showed that the GFP‐SPL35 protein was found in both soluble and microsomal fractions, as detected by antibodies specific to the vacuolar membrane marker TIP3‐1 and cytosol marker UGPase (Figure 5a), suggesting that SPL35 could be a protein that locates at multiple places rather than an integral membrane protein.
Figure 5.
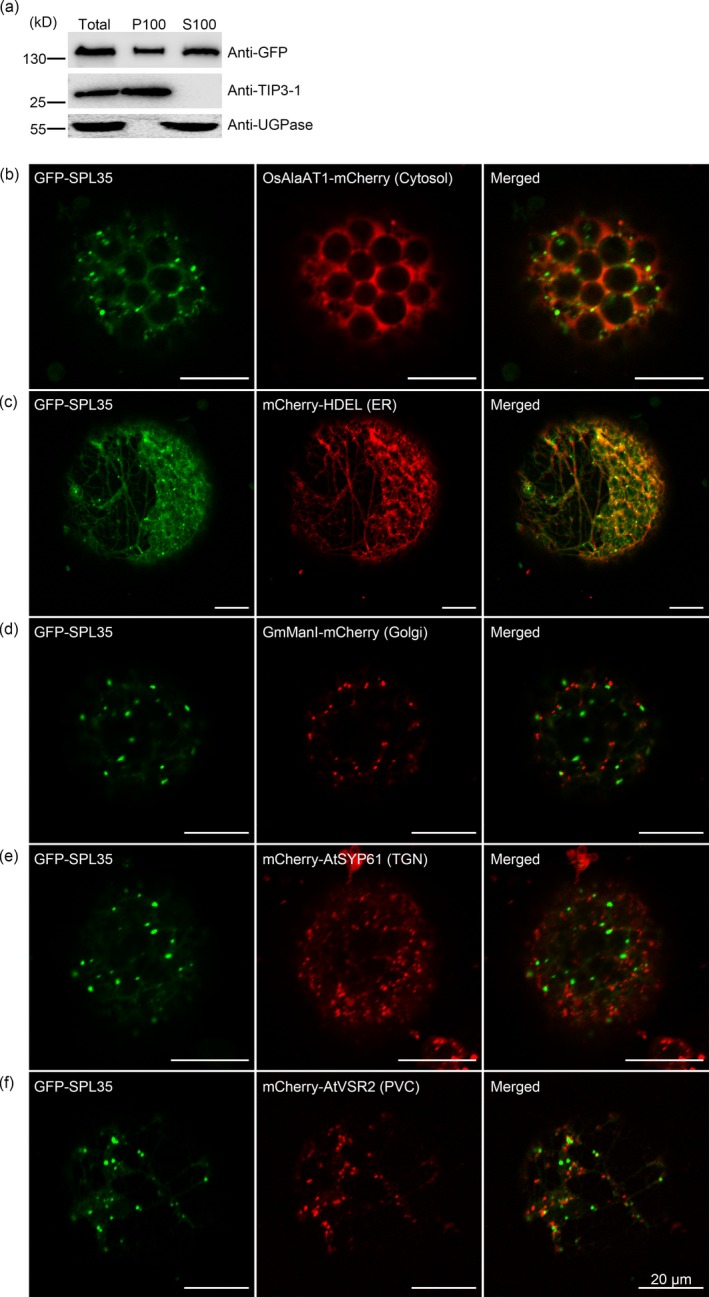
Subcellular localization of SPL35 in Nicotiana benthamiana leaf epidermal cells. (a) SPL35 may be a peripherally membrane‐associated protein. Total extract from rice protoplasts was ultracentrifuged at 100 000 g for 1 h to obtain the pellets (P100) and supernatants (S100) fractions, followed by immunoblot analysis with anti‐GFP and antibodies as indicated. (b–f) Co‐expression of GFP‐SPL35 and OsAlaAT1‐mCherry (Cytosol marker), mCherry‐HDEL (ER marker), GmManI‐mCherry (Golgi marker), mCherry‐AtSYP61 (TGN marker) and mCherry‐AtVSR2 (PVC marker) in Nicotiana benthamiana epidermal cell‐derived protoplasts. Scale bars, 20 μm.
To visualize the intracellular localization of SPL35, we transiently expressed p35S:GFP‐SPL35 in Nicotiana benthamiana (N. benthamiana) epidermal cells. The GFP‐SPL35 fusion protein was predominantly localized in cytosol, ER and punctate compartment(s), as indicated by overlap between GFP signals and red signals from the cytosol marker (OsAlaAT1‐mCherry, Zhong et al., 2019) and the ER marker (mCherry‐HDEL, Nelson et al., 2007; Figure 5b,c). To further understand the nature of the punctate compartment, we coexpressed GFP‐SPL35 with other fluorescent markers, such as for Golgi apparatus (GmManI‐mCherry, Tse et al., 2004), TGN (mCherry‐AtSYP61, Lam et al., 2007) or PVC (mCherry‐VSR2, Miao et al., 2006). As a result, the GFP‐SPL35 punctate structures merged with none of these organelles (Figure 5d–f), indicating that the punctate compartment is not Golgi apparatus, TGN or PVC.
SPL35 interacts with monoubiquitin through the CUE domain
Monoubiquitination is an important cellular regulatory signal that adds single ubiquitin molecules to a target protein, and regulates vesicle budding and histone modification among other functions (Hicke, 2001). Previous studies showed that CUE domains recognize monoubiquitin signals and directly interact with monoubiquitin (Shih et al., 2003). To verify whether the SPL35 protein interacts with ubiquitin, we generated four SPL35 prey vectors containing full‐length or truncated fragments of SPL35, and used the entire coding region of ubiquitin as bait for yeast two‐hybrid assays. Both the full‐length and the CUE domain of SPL35 exhibited strong interaction, but the C‐ or N‐terminal region lacking of the CUE domain showed no interaction with the ubiquitin LOC_Os09g39500 (Figure 6a). This indicates that the CUE domain is required for the interaction of SPL35 with ubiquitin in yeast cells, and thus SPL35 could be involved in monoubiquitination.
Figure 6.
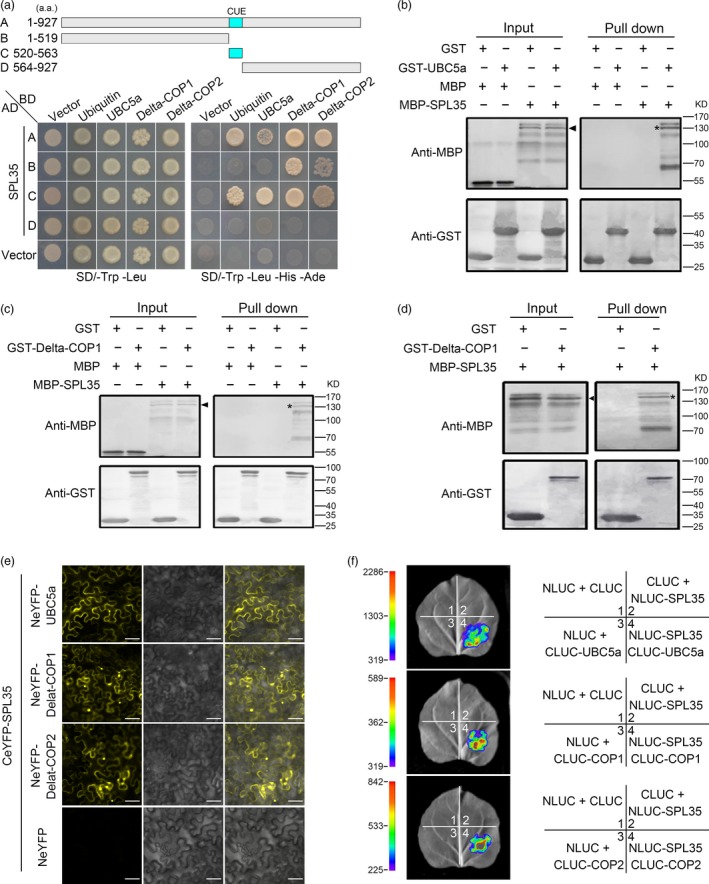
Interaction of SPL35 with OsUBC5a, Delta‐COPI and Delta‐COP2. (a) Yeast two‐hybrid assays for interaction of SPL35 with Ubiquitin, OsUBC5a, Delta‐COP1 or Delta‐COP2. The full‐length and different fragments of SPL35 were inserted to the prey (AD) vector pGADT7, and the ubiquitin, OsUBC5a, Delta‐COP1 and Delta‐COP2 were inserted into vector pGBKT7 as baits. Yeast strains were cultured on the SD/–Trp‐Leu and SD/–Trp‐Leu‐His‐Ade selection medium with 3 mm 3‐amino‐1,2,4‐triazole. (b–d) Pull‐down assays for the interactions of SPL35 with OsUBC5a, Delta‐COP1 and Delta‐COP2 in vitro respectively. GST (26.30 kD), GST‐OsUBC5a (42.87 kD), GST‐ Delta‐COP 1 (84.07 kD), GST‐ Delta‐COP2 (84.07 kD), MBP (42.5 kD) and MBP‐SPL35 (143.25 kD) were expressed in bacteria. Pull‐down was performed using GST binding resin. Proteins were detected with antibodies as indicated. Similar results were obtained in three independent experiments. The asterisk shows the band pulled down, and the arrow shows the immune‐blotting band of MBP‐SPL35 by the anti‐MBP monoclonal antibody. (e) Interactions between SPL35 and OsUBC5a, Delta‐COP1 and Delta‐COP2 shown by BiFC assays in Nicotiana benthamiana leaf epidermal cells. BiFC fluorescence is indicated by the eYFP signal. The signal of eYFP was not detected in the corresponding negative controls (fourth panels). eYFP, enhanced yellow fluorescent protein fluorescence; DIC, differential interference contrast. Scale bars, 10 μm. (f) firefly LUC complementation imaging (LCI) assay detecting the interaction between SPL35 and OsUBC5a, Delta‐COP1 and Delta‐COP2. The coloured scale bar indicates the luminescence intensity. NLUC indicates the N terminus of LUC, while CLUC indicates the C terminus of LUC.
SPL35 directly interacts with OsUBC5a, Delta‐COP1 and Delta‐COP2
We used the full‐length SPL35 in the pGBKT7 vector as bait to screen a rice cDNA library in yeast to look for proteins interacting with SPL35, and identified three genes, LOC_Os01g46926, LOC_Os05g24594 and LOC_Os05g24601. LOC_Os01g46926 encodes a rice ubiquitin conjugation enzyme, OsUBC5a (Takai et al., 2002). LOC_Os05g24594 and LOC_Os05g24601 encode rice clathrin‐associated proteins Delta‐COP1 (Q0DJA0) and Delta‐COP2 (Q0DJ99) respectively (http://www.uniprot.org/uniprot/).
We generated three bait vectors each containing full‐length CDS of OsUBC5a, Delta‐COP1 or Delta‐COP2, to verify their interaction with SPL35. OsUBC5a interacted with both full‐length and the CUE domain‐containing truncation of SPL35; Delta‐COP1 and Delta‐COP2 interacted with the full‐length, N‐terminal region and CUE domain‐containing truncations of SPL35 (Figure 6a). To determine whether they interact directly to each other, we performed glutathione S‐transferase (GST) pull‐down assay in vitro. Purified recombinant proteins GST‐OsUBC5a, GST‐Delta‐COP1 and GST‐Delta‐COP2 were separately immobilized to glutathione‐sepharose beads. The SPL35 protein tagged with maltose‐binding protein (MBP) was purified and incubated with each bead preparation in vitro. MBP and GST alone were used as negative controls. Immunoblot assays showed that a 128.24 kD band was recognized by an antibody against MBP, and MBP‐SPL35 was pulled down (Figure 6b–d), indicating that SPL35 directly and independently interacted with OsUBC5a, Delta‐COP1 or Delta‐COP2 in vitro.
To further verify the interaction between SPL35 and OsUBC5a, Delta‐COP1 or Delta‐COP2, we performed subcellular localization of OsUBC5a, Delta‐COP1 and Delta‐COP2, and BiFC assays. We firstly fused the coding regions of OsUBC5a, Delta‐COP1 or Delta‐COP2 to the C terminus of green fluorescent protein (GFP) to produce the GFP‐OsUBC5a, GFP‐Delta‐COP1 or GFP‐Delta‐COP2 fusion construct under control of the CaMV 35S promoter. We observed fluorescence signals of GFP‐OsUBC5a, GFP‐Delta‐COP1 and GFP‐Delta‐COP2 with similar localization of the SPL35 protein (Figure S12). BiFC assays showed that SPL35 interacted with OsUBC5a, Delta‐COP1 and Delta‐COP2 in N. benthamiana leaf cells (Figure 6e). Furthermore, the firefly LUC complementation imaging (LCI) assay confirmed the in vivo interaction of SPL35 and three interactors in leaf epidermal cells of N. benthamiana (Figure 6f). These results suggest that SPL35 directly interacts with OsUBC5a, Delta‐COP1 and Delta‐COP2 in vivo, and may be involved in the protein ubiquitin and transport pathways.
To understand the functions of OsUBC5a, Delta‐COP1 and Delta‐COP2, we transformed the corresponding RNAi constructs (Table S1) into cv. Kitaake to knock down their expression. We obtained 17, 12 and 15 independent RNAi plants for OsUBC5a, Delta‐COP1 and Delta‐COP2 respectively. All the RNAi plants displayed cell death phenotypes resembling that of spl35 (Figure 7a), suggesting that these SPL35‐interacting proteins function in regulating cell death in rice.
Figure 7.
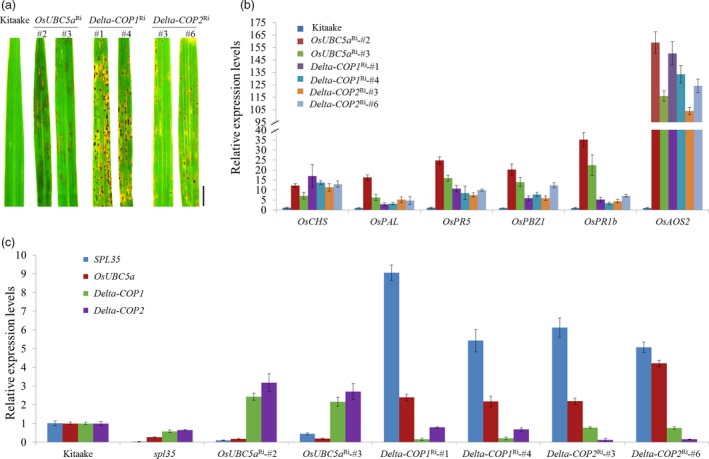
Expression levels of three SPL35‐interacting genes and six defence‐/ pathogenesis‐related genes in wild‐type, spl35 or RNAi transgenic T1 plants showing lesion mimic phenotypes. (a) Lesion mimic phenotypes of the flag leaves of 60‐day‐old plants of the receptor genotype (cv. Kitaake) and RNAi (OsUBC5a Ri, Delta‐COP1 Ri and Delta‐COP2 Ri) T1 transformants in the greenhouse. Scale bars, 1.0 cm. (b) qRT‐PCR analysis of defence‐/ pathogenesis‐related genes in the receptor genotype (cv. Kitaake) and RNAi (OsUBC5a Ri, Delta‐COP1 Ri and Delta‐COP2 Ri) T1 transformants respectively. Similar results were obtained in three independent biological replicates. (c) Expression levels of SPL35, OsUBC5a, Delta‐COP1 and Delta‐COP2 genes in Kitaake, spl35 and RNAi transgenic T1 plants. Data are means ± SD of three independent biological replicates. OsUBC5a Ri‐#2 and OsUBC5aRi‐#3 are OsUBC5a RNAi transgenic plants. Delta‐COP1 Ri‐#1 and Delta‐COP1 Ri‐#4 are Delta‐COP1 RNAi transgenic plants. Delta‐COP2 Ri‐#3 and Delta‐COP2 Ri‐#6 are Delta‐COP2 RNAi transgenic plants.
To probe into the mechanisms that caused the cell death phenotypes in the RNAi plants of OsUBC5a, Delta‐COP1 and Delta‐COP2, we first examined the expression of these genes together with SPL35 in WT during M. oryzae and Xoo infections and in their respective knockdown mutants. The expression patterns of these genes were not obviously affected by the pathogen infections (S13). Then, we examined the expression of six DR/PR genes, OsCHS, OsPAL, OsPR5, OsPBZ1, OsPR1b and OsAOS2, in the OsUBC5a Ri; Delta‐COP1 Ri and Delta‐COP2 Ri plants. The expression levels of these DR/PR genes were all obviously up‐regulated in the RNAi plants, relative to WT (cv. Kitaake; Figure 7b), suggesting an activation of the defence response. The mRNA levels of OsUBC5a, Delta‐COP1 and Delta‐COP2 were dramatically decreased in spl35 relative to WT, and the expression levels of both SPL35 and OsUBC5a were significantly down‐regulated, whereas those of both Delta‐COP1 and Delta‐COP2 were significantly up‐regulated in the OsUBC5a Ri plants; however, the expression levels of both SPL35 and OsUBC5a were significantly up‐regulated in both Delta‐COP1 Ri and Delta‐COP2 Ri plants (Figure 7c). These results reflected the complex mutual influences between SPL35, OsUBC5a, Delta‐COP1 and Delta‐COP2, and suggested that the imbalance in the expression of SPL35 might be a key reason for the cell death phenotypes in the OsUBC5a, Delta‐COP1 and Delta‐COP2 RNAi plants.
Discussion
SPL35 encodes a novel CUE domain‐containing protein in plants
LMMs are ideal materials for investigating HR‐mediated cell death and defence responses in plants. In the present study, we identified a LMM spl35 in a T‐DNA insertion mutant population of rice cv. Kinmaze. This mutant exhibited spontaneous HR‐like lesions on leaves from the three‐leaf stage seedlings through the ripening stage, accompanied by decreased plant height, fewer panicles, lower grain number and weight, and reduced spikelet fertility (Figure 1; Table S2). SPL35 was identified as LOC_Os03g10750, which encodes a CUEDC protein. Functional complementation with wild‐type LOC_Os03g10750 rescued the spl35 phenotype (Figure S6). It is worth noting that both overexpression and down‐regulation of SPL35 resulted in HR‐like cell death and other features typical of spl35 (Figure 3h,i; Figures S9 and S10). This indicates that SPL35 is an important gene, which must be properly regulated to maintain normal plant growth and development.
Five other rice genes are predicted to encode putative CUEDC proteins (Figure S7b). SPL35 has 3.3%–19.8% similarities with those CUEDC proteins; and the CUE domain of SPL35 also differs from those of the other CUEDC proteins (Figure S7b). There are a number of orthologs of SPL35 in other monocot and dicot plant species (Figure S8), but none of them has been functionally characterized. The duplicated pair of Arabidopsis RING‐finger E3 ligases, RIN2 and RIN3, are the only reported CUEDC proteins in plants, but a role of the CUE domains for functioning of RIN2 and RIN3 remains elusive (Kawasaki et al., 2005). The present study demonstrates that the CUE domain of SPL35 is required for functioning of SPL35. We also showed that SPL35 directly interacts with the ubiquitin, OsUBC5a, Delta‐COP1 and Delta‐COP2; its dysfunction can activate cell death and defence responses in rice, and significantly enhances resistance to different diseases. To our knowledge, this is the first report that a CUEDC protein is involved in plant immunity and defence response pathways.
SPL35 regulates cell death and defence responses
HR‐mediated cell death that commonly occurs at invasion sites to restrict further invasion or proliferation of pathogens, is a major hallmark of defence responses in plants (Wang et al., 2017). Identification of LMMs and characterization of the corresponding genes have expanded our understanding of cell death and defence responses in plants. Some CUE domain‐containing proteins were reported to be involved in apoptosis in yeast and mammalian species. For example, CUEDC2 suppresses glioma tumorigenicity by inhibiting the activation of STAT3 and NF‐κB signalling pathway (Li et al., 2017), and gp78 acts as a regulator of normal liver homeostasis and a tumour suppressor in human liver (Zhang et al., 2015). In order to determine whether SPL35 controls cell death in rice, we performed expression analyses of two histochemical markers using two staining methods, DAB staining and trypan blue staining (Wang et al., 2017), and measurement of H2O2 contents in spl35 and WT plants respectively. The results provided evidence that SPL35 controls cell death in plants (Figure S2). Considering that spl35 is characterized by spontaneous cell death in leaves, we conclude that SPL35 protein plays an important role in preventing cell death in rice.
Previous studies suggested that the development of symptoms in LMMs may activate defence response genes, and contribute to enhanced resistance to pathogens (Wang et al., 2015a,2015b, 2017). The present study showed that six defence‐related or pathogenesis‐related genes were rapidly activated (Figure 2e), and resistance to multiple M. oryzae and Xoo isolates was significantly enhanced as HR‐like lesions developed in spl35 plants (Figure 2a–d; Figure S4), indicating that the induced resistance was broad‐spectrum and perhaps nonspecific. Wang et al. (2017) showed that three PR genes, PR1a, PBZ1 and PO‐C1, were significantly up‐regulated in spl33 plants during the development of lesion mimics. A similar result was obtained for spl35 plants, and up‐regulated expression of six DR/PR genes (Figure 2e) occurred regardless of the presence of actual lesion mimics, although the expression levels increased with development after the two‐ to three‐leaf stage. It is worth mentioning that with the up‐regulated expression of these DR/PR genes, resistance to the M. oryzae isolates were coincidentally increased from moderate to high levels in spl35 plants (Figure S4; Table S3). Therefore, we conclude that SPl35 may regulate defence responses correlated with mimic lesion development in spl35.
SPL35 is involved in ubiquitination and vesicular trafficking pathways
Previous studies revealed that the CUE domain has dual roles in mono‐ and poly‐ubiquitination, responsible for protein trafficking and ER‐associated degradation of misfolded proteins in yeasts and mammalian species (Kostova et al., 2009; Man and Zhang, 2011). The present transient expression and two‐hybrid assays showed that SPL35 is predominantly localized to the cytosol and the ER (Figure 5b,c), and can directly interact with ubiquitin (Figure 6a). We identified and validated interaction of SPL35 with the OsUBC5a protein, which belongs to the Ubc4/5 subfamily and functions as an E2 enzyme in rice (Takai et al., 2002), by four types of assays, namely yeast two‐hybrid, GST pull‐down, BiFC and LCI (Figure 6). OsUBC5a was co‐localized with SPL35, and interacted with both full‐length and the CUE domain truncation of SPL35 in vitro and in vivo (Figure 6a,b,e,f). Based on this, we concluded that SPL35 interacts with OsUBC5a through its CUE domain. OsUBC5a catalyses auto‐ubiquitination of EL5, a RING‐type E3 ligase, and regulates cell death in root development (Koiwai et al., 2007; Takai et al., 2002). OsUBC5a also interacted with SPL11, a U‐box type E3 ligase, and regulated plant PCD and defence responses (Bae and Kim, 2013, 2014). Even though further detailed investigation is needed to determine how SPL35 functions in ubiquitination in rice, it is clear that the CUE domain of SPL35 interacts with ubiquitin and OsUBC5a, thus suggesting that SPL35 could participate in the ubiquitination pathway.
Proteins are synthesized by ribosomes in the rough ER, and transported to the Golgi apparatus and TGN, and then to their final destination compartment by carrier vesicles. Three types of coated vesicles have been identified, including coat protein complex I (COPI)‐, COPII‐ and clathrin‐coated vesicles and their adaptor complexes that are important for sorting and directional transport of proteins in the endocytic and secretory pathways (McMahon and Mills, 2004). In yeast, COPI‐coated vesicles seem to mediate anteretrograde transport within the Golgi apparatus, and to retrieve cargos from the Golgi back to the ER (retrograde direction; Béthune et al., 2006). The COPI coat complex consists of at least seven subunits, among which the subunit delta is a stoichiometric component and is essential for eukaryotic cell viability (Faulstich et al., 1996). In plants, COPI‐coated vesicles were identified in situ and isolated. They were localized to microvesicles surrounding or budding from the Golgi apparatus, and involved in retrograde transport from the Golgi apparatus to ER (Pimpl et al., 2000). To date, there have been no reports on functions of the COP complexes in plant cell death. We searched a total of four coatomer subunit delta genes, Delta‐COP1 (LOC_Os05g24594), Delta‐COP2 (LOC_Os05g24601), Delta‐COP3 (LOC_Os01g61710) and Delta‐COP4 (LOC_Os08g28080), in the rice genome (Figure S14; http://www.uniprot.org/uniprot). In this study, we identified and validated SPL35‐interacting proteins Delta‐COP1 and Delta‐COP2 by yeast two‐hybrid, GST pull‐down, BiFC and LCI assays (Figure 6), and found that knockdowns of Delta‐COP1 or Delta‐COP2 resulted in lesion mimic phenotypes (Figure 7a), providing evidence that the COPI protein complex is involved in the regulation of HR‐like cell death in rice. Thus, we speculate that SPL35 directly regulates Delta‐COP1 and Delta‐COP2 by protein–protein interaction and participates in COPI‐mediated protein transport pathways. The facts that overexpression and knockdown of SPL35, and knockdowns of OsUBC5a, Delta‐COP1 and Delta‐COP2 all cause lesion mimic phenotypes (Figure 3h,i; Figure 7a; Figure S10), suggest that imbalance in the expression of SPL35 might inhibit proper complex formation with its regulatory protein, thereby disrupting precise control of downstream events like ubiquitination or vesicular trafficking.
To comprehensively elucidate the regulatory mechanism of SPL35 in prevention of cell death, further analyses of SPL35 and its interacting proteins for their functions, interaction mechanisms and roles in ubiqutination and vesicular trafficking need to be undertaken. Understanding the relationships between SPL35‐mediated protein ubiqutination and/or vesicular trafficking and cell death, in connection with defence response will be part of that effort.
Experimental procedures
Plant materials and growth conditions
The spl35 mutant (line T466) was originally identified among a T‐DNA (vector pCUbi1390) insertion population (T1 plants) of japonica cv. Kinmaze. Among 16 T1 plants derived from T466, 13 were normal and 3 displayed spotted leaves. T3 homozygous plants were obtained by selfing the mutant plants and used in later studies. For a specific light treatment the middle parts of leaf blades of seedlings grown in a growth chamber (12 h of light at 30°C/12 h darkness at 20°C) were wrapped in aluminium foil to block the light. The T2 individuals derived from the T1 mutant plants with normal phenotype but heterozygous genotype of the hygronmycin B marker Hyr‐F/R (Table S1) along with an F2 population from a cross between spl35 and indica cv. 93‐11 were used for genetic analysis. All plants, except specified, were grown in a paddy field at the Changping Experimental Station of Institute of Crop Science during April to October, the normal rice‐growing season.
Transmission electron microscopy
Transverse sections of leaves from plants at 60 DAS grown under the field conditions were used for transmission electron microscopy observation as described previously (Wang et al., 2017).
Pigment and H2O2 measurements
Fresh flag leaves of spl35 and WT at 10 DAF were used to measure chlorophyll and carotenoids contents as described by Arnon (1949). H2O2 contents in leaves were measured in WT and spl35 at two‐ to three‐leaf stage (spl35‐) and four‐ to five‐leaf stage (spl35+) using an H2O2 assay kit (Beyotime Biotech, Shanghai, China) according to the manufacturer's instructions.
Histochemical assay
Fresh leaves of spl35 and WT at 10 DAF were used in histochemical assays. Trypan blue staining for cell death, DAB staining for H2O2 accumulation, and GUS staining were performed as described previously (Wang et al., 2017).
Evaluation of resistance to biotic and abiotic stresses
Two M. oryzae isolates CH43 and CH680, virulent to WT, were used to assess resistance of the spl35 plants to blast disease at the tillering stage by the injection‐inoculation method (Lei et al., 1996). The spl35 along with WT plants were grown at the same time and inoculated leaves of similar development stage with these two isolates. In the extended experiment to investigate when spl35 initiates a resistant response, six M. oryzae isolates, CH43, CH680, CH1971, CH1899, FJ07‐18‐1 and GD02‐15‐1, were separately used to infect spl35 seedlings using a spray‐inoculation method (Li et al., 2008) at the two‐ to three‐leaf and four‐ to five‐leaf stages respectively. Blast disease resistance was scored 7 days post inoculation (Mackill and Bonman, 1992).
Four Xoo isolates PXO61, PXO71, PXO76 and PXO86, virulent to WT, were used to assess resistance of the spl35 plants to blight disease at the booting stage by the scissors‐dipping method (Kauffman et al., 1973) or at the seedling stage by the infiltration‐inoculation method (Wang et al., 2015c). Lesion lengths were measured 2 weeks post inoculation.
To evaluate responses of spl35 to abiotic stresses, 14‐day‐old seedlings growing on three layers of Whatman 3 mm filter paper were used for various treatments. The roots of seedlings were dipped in 300 mm Mannitol, 200 mm NaCl or 200 mm H2O2 solution (Zhang et al., 2007). Seedlings were sprayed with 50 μm ACC or 100 μm ABA [dissolved in 0.01% (v/v) ethanol] (Wan et al., 2011). RNA was extracted from seedling leaves at 0, 1, 2, 4, 6, 8 and 10 h after treatment.
TAIL‐PCR and Southern blotting
TAIL‐PCR (Liu et al., 1995) was used to isolate regions flanking the T‐DNA insertion with Genome Walking Kit (TaKaRa, Tokyo, Japan). PCR‐amplified fragments were cloned into the pEASY‐Blunt vector (TransGen Biotech, China) and then sequenced (Biomed Biotech, Beijing, China). For Southern blotting, about 10 μg genomic DNA from wild‐type and spl35 mutant were digested with HindIII overnight, and digested products were separated in 1% agarose gels and then transferred onto a Hybond‐N+ membrane (Amersham Pharmacia, Biotech, Buckinghamshire, UK). A 481‐bp fragment of the hygromycin phosphotransferase gene amplified from the pCUbi1390 vector by the primer pair Hyr‐F/R was DIG‐labelled and used as a hybridization probe. Southern blotting was performed with DIG high prime DNA labelling and a detection starter kit II according to the manufacturer's instructions (Roche, Mannheim, Germany). Primers used for TAIL‐PCR and Southern blotting are listed in Table S1.
Quantitative real‐time PCR (qRT‐PCR) analysis
RNA was extracted from seedling leaves at the four‐leaf stage, flag leaves, leaf sheaths, culms and young panicles at the booting stage. Reverse transcription and qRT‐PCR were performed as described previously (Ma et al., 2015). The rice Ubiquitin gene (LOC_Os03g13170) was used as an internal control (primer pair Ubi), and the 2−ΔΔCT method was used to calculate relative levels of gene expression (Schmittgen and Livak, 2008). Primer pairs designed using GenScript (https://www.genscript.com/ssl-bin/app/primer) are listed in Table S1.
Vector construction and rice transformation
Two complementation vectors (p2300C‐10750 and p2300C‐10740) were constructed by inserting each of the corresponding genomic DNA fragments from cv. Kinmaze into the SacI and SalI sites of the pCAMBIA2300 vector (http://www.cambia.org/) using In‐Fusion Advantage Cloning Kit (Clontech, Beijing, China). The 8729 bp insertion in p2300C‐10750 consists of a 2369 bp region upstream of ATG of LOC‐Os03g10750 (SPL35), the entire coding region of 5507 bp and an 853 bp region downstream of TGA. The 2414 bp insertion in p2300C‐10740 consists of a 1086 bp region upstream of ATG of LOC‐Os03g10740, the entire gene of 418 and a 910 bp downstream of TGA.
Three overexpression vectors (p35S:SPL35‐OE, p35S:SPL35 1‐568 ‐OE and p35S:SPL35 569‐927 ‐OE) were constructed by inserting each of the corresponding cDNA fragments from cv. Kinmaze into the KpnI site of the vector pCUbi1390 using In‐Fusion Advantage Cloning Kit (Clontech, Beijing, China). p35S:SPL35‐OE contains the full‐length coding DNA sequence (CDS) of SPL35 (2874 bp); and p35S:SPL35 1‐568 ‐OE and p35S:SPL35 569‐927 ‐OE contain a 1704 bp fragment encoding the N‐terminal of SPL35 (amino acids 1‐568) and a 1080 bp fragment encoding the C‐terminal (amino acids 569‐927) respectively.
Five RNAi vectors (pUbi:SPL35‐RNAi, pUbi:LOC_Os03g10740‐RNAi, pUbi:OsUBC5a‐RNAi, pUbi:Delta‐COP1‐RNAi, and pUbi:Delta‐COP2‐RNAi) were constructed by inserting each of corresponding cDNA fragments from cv. Kinmaze into the SacI and SnaBI sites of binary vector pLH‐FAD2‐1390RNAi using In‐Fusion Advantage Cloning Kit (Clontech, Beijing, China). pUbi:SPL35‐RNAi contains a 279 bp cDNA fragment of SPL35 spanning the bases 995 and 1273; pUbi:LOC_Os03g10740‐RNAi contains a 420 bp cDNA fragment of LOC_Os03g10740 spanning the bases ‐6 and 414; pUbi:OsUBC5a‐RNAi contains a 502 bp cDNA fragment of OsUBC5a spanning the bases 1 and 502; pUbi:Delta‐COP1‐RNAi contains a 301 bp cDNA fragment of Delta‐COP1 spanning the bases 63 and 363; and pUbi:Delta‐COP2‐RNAi contains a 300 bp cDNA fragment of Delta‐COP2 spanning the bases −115 and 185.
The reporter construct pSPL35:GUS was created by amplifying a 2382 bp fragment upstream of the ATG start codon of SPL35 from cv. Kinmaze, and cloning it into the pCAMBIA1305 binary vector (EcoRI and NcoI sites) using In‐Fusion Advantage Cloning Kit (Clontech, Beijing, China).
The primers used for building above constructs are listed in Table S1. All the constructs were verified by sequencing and subsequently introduced into spl35, Kinmaze or Kitaake by Agrobacterium tumefaciens‐mediated transformation as described previously (Nishimura et al., 2006).
Subcellular fractionation and immunodectection
Coding DNA sequence of SPL35 without the stop codon was amplified and inserted into the BglII site of the pAN580 vector, generating the fusion construct pAN35S:GFP‐SPL35. pAN35S:GFP‐SPL35 was transformed into callus‐derived rice protoplasts as described previously (Zhou et al., 2013). Subcellular fractionation was performed as described previously (Wang et al., 2016). Total extract, pellets (P100) and supernatants (S100) were separated by SDS‐PAGE and transferred to PVDF membrane (0.22 μm; Millipore, Shanghai, China), followed by incubation with antibodies against GFP (diluted 1:4000; Roche, Mannheim, Germany), TIP3‐1 (diluted 1:2000; Ren et al., 2014) and UGPase (diluted 1:3000; Agrisera, Vännäs, Sweden).
Transient expression analysis in N. benthamiana
The CDS of SPL35, OsUBC5a, Delta‐COP1 or Delta‐COP2 was amplified and inserted into the pCAMBIA1305‐GFP vector (BglII site) to produce p35S:GFP‐SPL35, p35S:GFP‐OsUBC5a, p35S:GFP‐Delta‐COP1 or p35S:GFP‐Delta‐COP2 using In‐Fusion Advantage Cloning Kit (Clontech, Beijing, China) as described previously (Ren et al., 2014). Suspension cultures of the Agrobacterium strain GV3101 carrying above constructs and organelle markers together with the p19 strain carrying the gene‐silencing suppressor, p19 protein, were infiltrated into 5‐week‐old N. benthamiana leaves as described previously (Liu et al., 2010). Nicotiana benthamiana protoplasts were isolated as described previously (Ren et al., 2014). Fluorescence signals were detected using a laser confocal scanning microscopy (ZEISS Microsystems LSM 700, https://www.zeiss.com/microscopy/int/home.html). The primers used for building the above constructs are listed in Table S1.
Yeast two‐hybrid assay
A rice seedling cDNA library was constructed in yeast using the BD Matchmaker library construction and screening kit (Clontech, Beijing, China). The entire ORF of SPL35 was cloned into pGBKT7 for screening the cDNA library. SPL35 CDS fragments were cloned into the pGADT7 vector as preys, and CDSs of LOC_Os09g39500 (ubiquitin gene), OsUBC5a, Delta‐COP1 and Delta‐COP2 were inserted into pGBKT7 as baits, respectively. For two‐hybrid assays, the bait and prey vectors were co‐transformed into yeast AH109‐competent cells. Transformed cells were incubated on SD/‐Leu‐Trp‐plates at 30 °C for 2 days, and then transferred to the restrictive medium [SD/‐His‐Ade‐Leu‐Trp‐plates with 3 mm 3‐amino‐1,2,4‐triazole (3‐AT)] and cultured at 30 °C for another 2–4 days. Three repeats were performed. Primers used are listed in Table S1.
Pull‐down assay
Coding DNA sequence of SPL35 was inserted into the pMAL‐c2x vector to generate the construct MBP‐SPL35. CDSs of OsUBC5a, Delta‐COP1 and Delta‐COP2 were cloned into the pGEX4T‐1 vector to generate GST‐OsUBC5a, GST‐Delta‐COP1 and GST‐Delta‐COP2 respectively. The constructs, together with the empty vector, were transformed into Escherichia coli BL21 competent cells (TransGen Biotech, Beijing, China) to express protein. Recombinant proteins from E. coli lysates were purified using the corresponding resins [amylose resin for MBP purification (New England Biolabs, Beijing, China), and GST‐binding resin for GST purification (Merck, Beijing, China)]. GST or GST fusion proteins coupled to beads were incubated with MBP or MBP‐SPL35. In vitro GST pull‐down analysis was performed as described previously (Wang et al., 2009). For protein detection, anti‐GST antibody (1:1000), anti‐MBP antibody (1:500) and goat anti‐mouse HRP‐conjugated antibody (1:5000; MBL International, Beijing, China) were used. Three independent repeats were performed. Primers used are listed in Table S1.
BiFC and LCI assays
For the BiFC assay, CDS of SPL35 was cloned into the pCCFP‐X (eYFP) vector to construct a CeYFP‐SPL35 fusion. The CDSs of OsUBC5a, Delta‐COP1 and Delta‐COP2 were cloned into the pNYFP‐X (eYFP) vector to construct NeYFP‐OsUBC5a, NeYFP‐Delta‐COP1 and NeYFP‐Delta‐COP2 fusions respectively. BiFC was performed in N. benthamiana leaf epidermis cells as described previously (Liu et al., 2010). eYFP signals were observed 48 h after infiltration with a laser confocal scanning microscope (ZEISS Microsystems LSM 700). Primers used are listed in Table S1.
For the LCI assay, CDS of SPL35 was cloned into the NLUC construct to produce the NLUC‐SPL35 fusion, while the CDSs of OsUBC5a, Delta‐COP1 and Delta‐COP2 were separately cloned into the CLUC construct to generate the CLUC‐OsUBC5a, CLUC‐Delta‐COP1 and CLUC‐Delta‐COP2 fusions. LCI assay was performed in N. benthamiana leaf epidermis cells. LUC activities were analysed 48 h post‐infiltration using Night‐SHADE LB 985 (Berthold, Bad Wildbad, Germany).
Conflicts of interest
The authors declare no conflicts of interest.
Supporting information
Figure S1 Pigment contents and transmission electron microscopy (TEM) analysis of chloroplasts in wild‐type (WT) and spl35 mesophyll cells.
Figure S2 The expression of histochemical markers and measurement of H2O2 in wild‐type (WT) and spl35 mutant.
Figure S3 Expression analysis of three endoplasmic reticulum (ER) chaperone genes in the wild‐type (WT) and spl35 plants by quantitative real‐time PCR (qRT‐PCR).
Figure S4 Disease reactions of the wild‐type (WT) and spl35 mutant to six Maganaporthe oryzae isolates.
Figure S5 Expression of SPL35 in response to different abiotic treatments.
Figure S6 Complementation assays of SPL35 transgenic plants.
Figure S7 SPL35 encodes a CUE domain‐containing protein.
Figure S8 Phylogenetic analysis of SPL35 with other homologues.
Figure S9 Transcription levels of the LOC_Os03g10750 gene in different overexpressed (OE) or RNAi (Ri) transgenic lines (T1) detected by qRT‐PCR.
Figure S10 Morphology of LOC_Os03g10750‐overexpressed (OE) and ‐RNAi (Ri) transgenic lines (T1) in Kitaake background under field conditions. Scale bars, 5.0 cm.
Figure S11 Lesion mimics on the flag leaves of the transgenic lines (T1) overexpressing the N‐terminal 1‐568 amino acids of SPL35 in the cv. Kitaake (wild‐type) background OEN‐2, 3 and 5 represent the leaves of pSpl351‐568‐OE transgenic T1 lines in cv. Kitaake background.
Figure S12 Subcellular localization of GFP‐OsUBC5a, GFP‐Delta‐COP1 and GFP‐Delta‐COP2 fusion proteins in Nicotiana benthamiana leaf epidermal cells. GFP protein itself is distributed throughout the nucleus and cytoplasm (fourth panels). GFP, green fluorescent protein; DIC, differential interference contrast. Scale bars, 10 µm.
Figure S13 qRT‐PCR analysis of transcript levels of SPL35, OsUBC5a, Delta‐COP1 and Delta‐COP2 during Maganaporthe oryzae (M. oryzae) and Xanthomonas oryzae pv. oryzae (Xoo) infections.
Figure S14 The relationship of four coatomer subunit delta proteins in rice by unrooted phylogenetic tree analysis.
Table S1 Sequences of primers used in this study.
Table S2 Comparison of agronomic traits between spl35 mutant and its wild‐type (WT, cv. Kinmaze).
Table S3 Disease reactions of the spl35 mutant to six M. oryzae isolates.
Table S4 Genetic analysis of the spl35 mutant.
Acknowledgements
We thank Dr. Kaijun Zhao and Dr. Chunlian Wang, Institute of Crop Sciences, CAAS, for kindly providing the Xoo strains. This work was supported by grants from National Natural Science Foundation of China (Grant No. 31471758), the Major Science and Technology Project to Create New Crop Cultivars using Gene Transfer Technology (Grant No. 2016ZX08001‐002), and the Agricultural Science and Technology Innovation Program of CAAS “Crop Functional Genomics”.
Dr. Lei is fully responsible for the distribution of all materials associated with this article.
Contributor Information
Chuanyin Wu, Email: wuchuanyin@caas.cn.
Yulong Ren, Email: renyulong@caas.cn.
References
- Arnon, D.I. (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris . Plant Physiol. 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, H. and Kim, W.T. (2013) The N‐terminal tetra‐peptide (IPDE) short extension of the U‐box motif in rice SPL11 E3 is essential for the interaction with E2 and ubiquitin‐ligase activity. Biochem. Biophys. Res. Comm. 433, 266–271. [DOI] [PubMed] [Google Scholar]
- Bae, H. and Kim, W.T. (2014) Classification and interaction modes of 40 rice E2 ubiquitin‐conjugating enzymes with 17 rice ARM‐U‐box E3 ubiquitin ligases. Biochem. Biophys. Res. Comm. 444, 575–580. [DOI] [PubMed] [Google Scholar]
- Béthune, J. , Wieland, F. and Moelleken, J. (2006) COPI‐mediated transport. J. Membr. Biol. 211, 65–79. [DOI] [PubMed] [Google Scholar]
- Biederer, T. , Volkwein, C. and Sommer, T. (1997) Role of Cue1p in ubiquitination and degradation at the ER surface. Science, 278, 1806–1809. [DOI] [PubMed] [Google Scholar]
- Cui, F. , Liu, J. , Li, Q. , Yang, C. and Xie, Q. (2012a) UBC32 mediated oxidative tolerance in Arabidopsis . J. Genet. Genomics, 39, 415–417. [DOI] [PubMed] [Google Scholar]
- Cui, F. , Liu, L. , Zhao, Q. , Zhang, Z. , Li, Q. , Lin, B. , Wu, Y. et al (2012b) Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid‐mediated salt stress tolerance. Plant Cell, 24, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, R.A. , Delaney, T.P. , Uknes, S.J. , Ward, E.R. , Ryals, J.A. and Dangl, J.L. (1994) Arabidopsis mutants simulating disease resistance response. Cell, 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Faulstich, D. , Auerbach, S. , Orci, L. , Ravazzola, M. , Wegchingel, S. , Lottspeich, F. , Stenbeck, G. et al (1996) Architecture of coatomer: molecular characterization of delta‐COP and protein interactions within the complex. J. Cell Biol. 135, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, T. , Maisonneuve, S. , Isshiki, M. , Mizutani, M. , Chen, L. , Wong, H.L. , Kawasaki, T. et al (2010) Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 285, 11308–11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke, L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201. [DOI] [PubMed] [Google Scholar]
- Johal, G.S. , Hulbert, S. and Briggs, S.P. (1995) Disease lesion mimics of maize: a model for cell death in plants. BioEssays, 17, 685–692. [Google Scholar]
- Kauffman, H.E. , Reddy, A.P.K. , Hsieh, S.P.V. and Merca, S.D. (1973) An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae . Plant Dis. Report, 57, 537–541. [Google Scholar]
- Kawasaki, T. , Nam, J. , Boyes, D.C. , Holt, B.F. 3rd , Hubert, D.A. , Wiig, A. and Dangl, J.L. (2005) A duplicated pair of Arabidopsis RING‐finger E3 ligases contribute to the RPM1‐ and RPS2‐mediated hypersensitive response. Plant J. 44, 258–270. [DOI] [PubMed] [Google Scholar]
- Koiwai, H. , Tagiri, A. , Katoh, S. , Katoh, E. , Ichikawa, H. , Minami, E. and Nishizawa, Y. (2007) RING‐H2 type ubiquitin ligase EL5 is involved in root development through the maintenance of cell viability in rice. Plant J. 51, 92–104. [DOI] [PubMed] [Google Scholar]
- Kostova, Z. , Mariano, J. , Scholz, S. , Koenig, C. and Weissman, A.M. (2009) A Ubc7p‐binding domain in Cue1p activates ER‐associated protein degradation. J. Cell Sci. 122, 1374–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, S.K. , Siu, C.L. , Hillmer, S. , Jang, S. , An, G. , Robinson, D.G. and Jiang, L. (2007) Rice SCAMP1 defines clathrin‐coated, trans‐Golgi‐located tubular‐vesicular structures as an early endosome in tobacco BY‐2 cells. Plant Cell, 19, 296–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, C. , Wang, J. , Mao, S. , Jiang, W. , Zhu, L. and Ling, Z. (1996) Genetic analysis of blast resistance in indica variety Zhaiyeqing 8 (ZYQ8). Chin. J. Genet. 23, 287–293. [Google Scholar]
- Li, W. , Lei, C. , Chen, Z. , Jia, Y. , Huang, D. , Wang, J. , Wang, J. et al (2008) Identification of SSR markers for a broad‐spectrum blast resistance gene Pi20(t) for marker‐assisted breeding. Mol. Breeding, 22, 141–149. [Google Scholar]
- Li, F. , Tang, C. , Jin, D. , Guan, L. , Wu, Y. , Liu, X. , Wu, X. et al (2017) CUEDC2 suppresses glioma tumorigenicity by inhibiting the activation of STAT3 and NF‐κB signaling pathway. Int. J. Oncol. 51, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Mitsukawa, N. , Oosumi, T. and Whittier, R.F. (1995) Efficient isolation and mapping of Arabidopsis thaliana T‐DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Zhang, Y. , Tang, S. , Zhao, Q. , Zhang, Z. , Zhang, H. , Dong, L. et al (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana . Plant J. 61, 893–903. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Chen, Y. , Huang, T. , Tarasov, S. , King, A. , Li, J. , Weissman, A.M. et al (2012a) Promiscuous interactions of gp78E3 ligase CUE domain with polyubiquitin chains. Structure, 20, 2138–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Li, F. , Tang, J. , Wang, W. , Zhang, F. , Wang, G. , Chu, J. et al (2012b) Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice. PLoS ONE, 7, e50089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Ning, Y. , Zhang, Y. , Yu, N. , Zhao, C. , Zhan, X. , Wu, W. et al (2017) OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell, 29, 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, S. , Lin, B. , Auriac, M.C. , Kroj, T. , Saindrenan, P. , Nicole, M. , Balague, C. et al (2004) Vascular associated death1, a novel GRAM domain‐containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell, 16, 2217–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. , Lei, C. , Xu, X. , Hao, K. , Wang, J. , Cheng, Z. , Ma, X. et al (2015) Pi64, encoding a novel CC‐NBS‐LRR protein, confers resistance to leaf and neck blast in rice. Mol. Plant Microbe Interact. 28, 558–568. [DOI] [PubMed] [Google Scholar]
- Mackill, D.J. and Bonman, J.M. (1992) Inheritance of blast resistance in near‐isogenic lines of rice. Phytopathology, 82, 746–749. [Google Scholar]
- Man, J. and Zhang, X. (2011) CUEDC2: an emerging key player in inflammation and tumorigenesis. Protein Cell, 2, 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, H.T. and Mills, I.G. (2004) COP and clathrin‐coated vesicle budding: different pathways, common approaches. Curr. Opin. Cell Biol. 16, 379–381. [DOI] [PubMed] [Google Scholar]
- Miao, Y. , Yan, P. , Kim, H. , Hwang, I. and Jiang, L. (2006) Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY‐2 cells. Plant Physiol. 142, 945–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B. , Cai, X. and Nebenfuhr, A. (2007) A multicolored set of in vivo organelle markers for co‐localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nishimura, A. , Aichi, I. and Matsuoka, M. (2006) A protocol for Agrobacterium‐mediated transformation in rice. Nat. Protoc. 1, 2796–2802. [DOI] [PubMed] [Google Scholar]
- Pimpl, P. , Movafeghi, A. , Coughlan, S. , Denecke, J. , Hillmer, S. and Robinson, D.G. (2000) In situ localization and in vitro induction of plant COPI‐coated vesicles. Plant Cell, 12, 2219–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y. , Wang, Y. , Liu, F. , Zhou, K. , Ding, Y. , Zhou, F. , Wang, Y. et al (2014) Glutelin precursor accumulation3 encodes a regulator of post‐Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell, 26, 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shi, W. , Muthurajan, R. , Rahman, H. , Selvam, J. , Peng, S. , Zou, Y. and Jagadish, K.S.V. (2013) Source‐sink dynamics and proteomic reprogramming under elevated night temperature and their impact on rice yield and grain quality. New Phytol. 197, 825–837. [DOI] [PubMed] [Google Scholar]
- Shideler, T. , Nickerson, D.P. , Merz, A.J. and Odorizz, G. (2015) Ubiquitin binding by the CUE domain promotes endosomal localization of the Rab5 GEF Vps9. Mol. Biol. Cell, 26, 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, S.C. , Prag, G. , Francis, S.A. , Sutanto, M.A. , Hurley, J.H. and Hicke, L. (2003) A ubiquitin‐binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 22, 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirsekar, G.S. , Vega‐Sanchez, M.E. , Bordeos, A. , Baraoidan, M. , Swisshelm, A. , Fan, J. , Park, C.H. et al (2014) Identification and characterization of suppressor mutants of spl11‐mediated cell death in rice. Mol. Plant Microbe Interact. 27, 528–536. [DOI] [PubMed] [Google Scholar]
- Takai, R. , Matsuda, N. , Nakano, A. , Hasegawa, K. , Akimoto, C. , Shibuya, N. and Minami, E. (2002) EL5, a rice N‐acetylchitooligosaccharide elicitor‐responsive RING‐H2 finger protein, is a ubiquitin ligase which functions in vitro in co‐operation with an elicitor‐responsive ubiquitin‐conjugating enzyme, OsUBC5b. Plant J. 30, 447–455. [DOI] [PubMed] [Google Scholar]
- Tang, J. , Zhu, X. , Wang, Y. , Liu, L. , Xu, B. , Li, F. , Fang, J. et al (2011) Semi‐dominant mutations in the CC‐NB‐LRR‐type R gene, NLS1, lead to constitutive activation of defense responses in rice. Plant J. 66, 996–1007. [DOI] [PubMed] [Google Scholar]
- Tse, Y. , Mo, B. , Hillmer, S. , Zhao, M. , Lo, S. , Robinson, D. and Jiang, L. (2004) Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY‐2 cells. Plant Cell, 16, 672–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undan, J.R. , Tamiru, M. , Abe, A. , Yoshida, K. , Kosugi, S. , Takagi, H. , Yoshida, K. et al (2012) Mutation in OsLMS, a gene encoding a protein with two double‐stranded RNA binding motifs, causes lesion mimic phenotype and early senescence in rice (Oryza sativa L.). Genes Genet. Syst. 87, 169–179. [DOI] [PubMed] [Google Scholar]
- Walbot, V. (1991) Maize mutants for the 21st century. Plant Cell, 3, 851–856. [Google Scholar]
- Wan, L. , Zhang, J. , Zhang, H. , Zhang, Z. , Quan, R. , Zhou, S. and Huang, R. (2011) Transcriptional activation of OsDERF1 in OsERF3 and OsAP2‐39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE, 6, e25216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Kong, H. and Ma, H. (2009) F‐box proteins regulate ethylene signaling and more. Genes Dev. 23, 391–396. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Ye, B. , Yin, J. , Yuan, C. , Zhou, X. , Li, W. , He, M. et al (2015a) Characterization and fine mapping of a light‐dependent leaf lesion mimic mutant 1 in rice. Plant Physiol. Biochem. 97, 44–51. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Wang, Y. , Hong, X. , Hu, D. , Liu, C. , Yang, J. , Li, Y. et al (2015b) Functional inactivation of UDP‐N‐acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J. Exp. Bot. 66, 973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Zhang, X. , Fan, Y. , Gao, Y. , Zhu, Q. , Zheng, C. , Qin, T. , et al. (2015c) XA23 is an executor R protein and confers broad‐spectrum disease resistance in rice. Mol. Plant 8, 290–302. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Liu, F. , Ren, Y. , Wang, Y. , Liu, X. , Long, W. , Wang, D. et al (2016) GOLGI TRANSPORT 1B regulates protein export from the endoplasmic reticulum in rice endosperm cells. Plant Cell, 28, 91–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Lei, C. , Wang, J. , Ma, J. , Tang, S. , Wang, C. , Zhao, K. et al (2017) SPL33, encoding an eEF1A‐like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 68, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Q. , Zhai, K. , Yang, D. , Yang, W. , Wu, J. , Liu, J. , Pan, W. et al (2016) An E3 ubiquitin ligase‐BAG protein module controls plant immunity and broad‐spectrum disease resistance. Cell Host Microbe, 20, 758–769. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Li, W. , Chen, J. , Yang, Y. , Zhang, Z. , Zhang, H. , Wang, X. et al (2007) Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol. Biol. 63, 63–71. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Kho, D.H. , Wang, Y. , Harazono, Y. , Nakajima, K. , Xie, Y. and Raz, A. (2015) Gp78, an E3 ubiquitin ligase acts as a gatekeeper suppressing nonalcoholic steatohepatitis (NASH) and liver cancer. PLoS ONE, 10, e0118448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. and Wu, W. (2008) A method for measuring relative lesion area on leaves using a rice Chalkiness ratio analysis software. Sci. Agric. Sin. 41, 3405–3409. [Google Scholar]
- Zhong, M. , Liu, X. , Liu, F. , Ren, Y. , Wang, Y. , Zhu, J. , Teng, X. et al (2019) FLOURY ENDOSPERM12 encoding alanine aminotransferase 1 regulates carbon and nitrogen metabolism in rice. J. Plant Biol. 66, 61–73. (accepted). [Google Scholar]
- Zhou, K. , Ren, Y. , Lv, J. , Wang, Y. , Liu, F. , Zhou, F. , Zhao, S. et al (2013) Young Leaf Chlorosis 1, a chloroplast‐localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta, 237, 279–292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Pigment contents and transmission electron microscopy (TEM) analysis of chloroplasts in wild‐type (WT) and spl35 mesophyll cells.
Figure S2 The expression of histochemical markers and measurement of H2O2 in wild‐type (WT) and spl35 mutant.
Figure S3 Expression analysis of three endoplasmic reticulum (ER) chaperone genes in the wild‐type (WT) and spl35 plants by quantitative real‐time PCR (qRT‐PCR).
Figure S4 Disease reactions of the wild‐type (WT) and spl35 mutant to six Maganaporthe oryzae isolates.
Figure S5 Expression of SPL35 in response to different abiotic treatments.
Figure S6 Complementation assays of SPL35 transgenic plants.
Figure S7 SPL35 encodes a CUE domain‐containing protein.
Figure S8 Phylogenetic analysis of SPL35 with other homologues.
Figure S9 Transcription levels of the LOC_Os03g10750 gene in different overexpressed (OE) or RNAi (Ri) transgenic lines (T1) detected by qRT‐PCR.
Figure S10 Morphology of LOC_Os03g10750‐overexpressed (OE) and ‐RNAi (Ri) transgenic lines (T1) in Kitaake background under field conditions. Scale bars, 5.0 cm.
Figure S11 Lesion mimics on the flag leaves of the transgenic lines (T1) overexpressing the N‐terminal 1‐568 amino acids of SPL35 in the cv. Kitaake (wild‐type) background OEN‐2, 3 and 5 represent the leaves of pSpl351‐568‐OE transgenic T1 lines in cv. Kitaake background.
Figure S12 Subcellular localization of GFP‐OsUBC5a, GFP‐Delta‐COP1 and GFP‐Delta‐COP2 fusion proteins in Nicotiana benthamiana leaf epidermal cells. GFP protein itself is distributed throughout the nucleus and cytoplasm (fourth panels). GFP, green fluorescent protein; DIC, differential interference contrast. Scale bars, 10 µm.
Figure S13 qRT‐PCR analysis of transcript levels of SPL35, OsUBC5a, Delta‐COP1 and Delta‐COP2 during Maganaporthe oryzae (M. oryzae) and Xanthomonas oryzae pv. oryzae (Xoo) infections.
Figure S14 The relationship of four coatomer subunit delta proteins in rice by unrooted phylogenetic tree analysis.
Table S1 Sequences of primers used in this study.
Table S2 Comparison of agronomic traits between spl35 mutant and its wild‐type (WT, cv. Kinmaze).
Table S3 Disease reactions of the spl35 mutant to six M. oryzae isolates.
Table S4 Genetic analysis of the spl35 mutant.


