Abstract
Background:
Gene expression changes within the Hippo pathway were found to be associated with large tumor size and metastasis in breast cancer. The combined effect of genetic variants in genes of this pathway may have a causal role in breast cancer development.
Methods:
We examined 7,086 SNPs that were not highly correlated (r2 < 0.8) in 35 Hippo pathway genes using data from the genome-wide association study of breast cancer from the Root Consortium, which includes 3,686 participants of African ancestry from Nigeria, United States of America, and Barbados: 1,657 cases (403 estrogen receptor-positive [ER+], 374 ER−) and 2,029 controls. Gene-level analyses were conducted using improved AdaJoint test for large-scale genetic association studies adjusting for age, study site and the first four eigenvectors from the principal component analysis. SNP-level analyses were conducted with logistic regression.
Results:
The Hippo pathway was significantly associated with risk of ER+ breast cancer (pathway-level P = 0.019), with WWC1 (Padj = 0.04) being the leading gene. The pathway-level significance was lost without WWC1 (P = 0.12). rs147106204 in the WWC1 gene was the most statistically significant SNP after gene-level adjustment for multiple comparisons (OR = 0.53, 95% CI = 0.41–0.70, Padj = 0.025).
Conclusions:
We found evidence of an association between genetic variations in the Hippo pathway and ER+ breast cancer. Moreover, WWC1 was identified as the most important genetic susceptibility locus highlighting the importance of genetic epidemiology studies of breast cancer in understudied populations.
Keywords: Hippo pathway, Breast cancer, SNPs, Women of African ancestry, Adaptive joint multilocus test
1. INTRODUCTION
The Hippo pathway is an evolutionarily conserved regulator of tissue growth and cell fate during development and regeneration.[1] Mutation and deregulation for a subset of Hippo pathway genes have been reported in several malignancies, including breast cancer.[2] Functional studies indicate that repression of this pathway causes uncontrolled cell proliferation and dramatic tissue overgrowth, which are central to carcinogenesis.[1] There are multiple important components of the Hippo pathway, including serine/threonine kinase 4 (STK4) and STK3, large tumor suppressor 1 (LATS1) and LATS2, together with the adaptor proteins Salvador homologue 1 (SAV1), MOB kinase activator 1A (MOB1A) and MOB1B.[1] This pathway has two key downstream effector proteins, Yes-associated protein (YAP, encoded by YAP1) and transcriptional co-activator with PDZ-binding motif (TAZ, encoded by WWTR1),[3] both of which are overexpressed in breast cancer,[2] especially in triple negative breast cancer.[4]
A few studies have examined the risk of breast cancer associated with genetic variations in the Hippo pathway.[5–7] Single nucleotide polymorphisms (SNPs) in YAP1 and STK4 have been reported to be associated with breast cancer risk in Chinese and European populations, respectively.[5,6] A study conducted in African Americans reported a significant association of the whole pathway with estrogen receptor negative (ER−) breast cancer, and it revealed that CDH1 was responsible for the pathway association.[7] E-cadherin, encoded by CDH1, can function as an upstream regulatory element in the Hippo signaling pathway in mammalian cells.[1,8] Genetic polymorphisms in CDH1 have also been reported to be associated with breast cancer susceptibility among Chinese Han women,[9,10] but no association with lobular cancer was found in a large dataset of European population with 6,023 cases and 34,271 controls. A noticeable limitation of previous studies is that only polymorphisms in a single candidate gene or a small subset of genes in this pathway were assessed, while some core elements of this pathway, such as LATS1/2, were omitted.[1,7] Furthermore, in addition to single-SNP association analysis, it has been suggested that examining a group of SNPs within a biological pathway can provide complementary and valuable insights for the genetic architecture of complex diseases.[11,12]
Considering the paucity of data on the genomic basis of disparities in breast cancer outcomes for women of African ancestry and the growing importance of Hippo pathway in breast carcinogenesis,[13] we sought to explore whether this pathway contributes to the aggressive breast cancer phenotypes observed in women of African ancestry.
2. METHODS
2.1. Study participants, SNP genotyping and imputation
The study populations of The Root Consortium have been described previously.[14–20] To summarize, this study included 3,686 participants of African ancestry. Ascertainment of cases and controls occurred in Nigeria (711 cases and 624 controls), Barbados (92 cases and 229 controls), and four sites in the United States (854 cases and 1,176 controls). Patients were histologically and/or clinically diagnosed as having invasive breast cancer by the clinicians at each study site [19,20]. The mean ages of cases and controls were 49.3 and 48.4 years, respectively. The numbers of ER+ patients and ER− patients were 403 and 374, respectively. ER status was not available for the remaining patients.
Information about genome-wide association study (GWAS) genotyping, quality control, imputation and principal component analysis has been introduced in detail previously.[20] Briefly, approximately 2.4 million genetic variants on the Illumina HumanOmni2.5-8v1 array were genotyped. With the 1000 Genomes Project phase 1 integrated variant set as the reference panel, 23,098,723 SNPs were imputed by IMPUTE2[21] and were included in the GWAS analysis. To account for population structure, the first ten principal components were computed with the smartpca program in the EIGENSOFT package.[22]
2.2. Selection of candidate genes and SNPs in the Hippo pathway
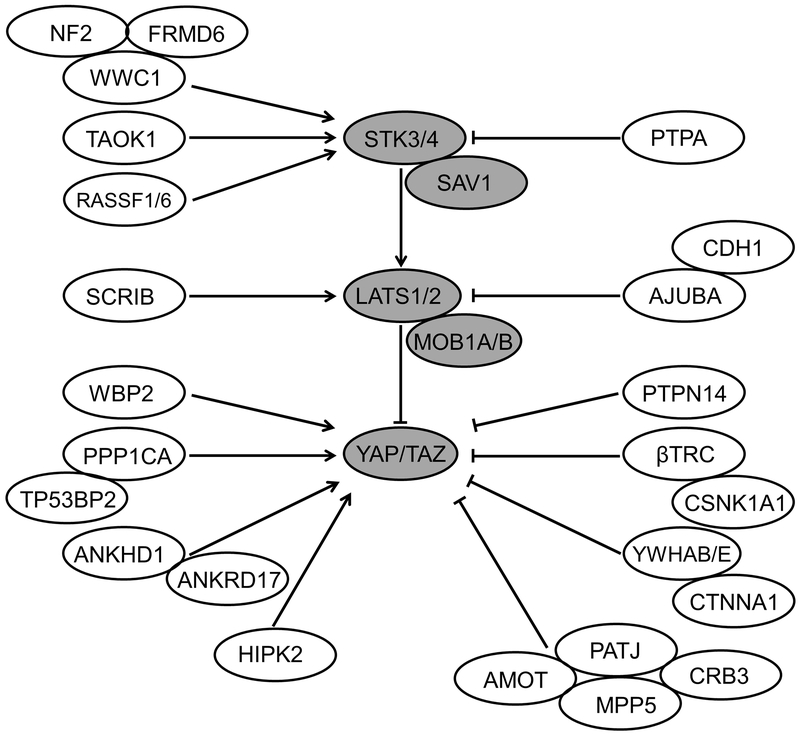
Figure 1 illustrates the 35 genes in the Hippo pathway after a query of the KEGG pathway database (http://www.genome.jp/kegg-bin/show_pathway?hsa04390) and a recent thorough review.[1] For each candidate gene, start and end chromosomal positions ±10kb were determined using the UCSC Genome Browser (https://genome.ucsc.edu). The median of imputation score for SNPs with MAF ≥ 0.01 was above 0.95, suggesting the imputation quality was comparatively good. To be conservative and to better minimize any potential false positives,[23] we excluded SNPs with imputation information scores < 0.7 or minor allele frequency (MAF) < 0.01. SNPs within each gene region were extracted from The Root Consortium GWAS data and a total of 19,889 variants in 35 genes were extracted.
FIGURE 1.
The Hippo pathway. Dark gray ovals indicate the key components of the Hippo pathway.
2.3. AdaJoint pathway analysis
Case-control analyses were conducted for all cases, ER+ cases and ER− cases, compared to cancer-free controls. Analyses were carried out to examine associations of SNPs and breast cancer risk at the pathway, gene and SNP levels.
The pathway- and gene-based analyses were performed using the improved adaptive joint multilocus test (named as AdaJoint) implemented in the R package ARTP2. The AdaJoint multilocus test takes linkage disequilibrium (LD) structure into consideration and adopts a variable selection procedure to identify a subset of genetic markers that jointly show the strongest association signal, and then defines the test statistic based on the selected genetic markers.[11,12,24] Compared with other similar methods including Min-p test, SKAT and ARTP [12], AdaJoint is much more powerful if two functional SNPs are negative correlated and have effects in the same direction; or two functional SNPs are positively correlated and their effects have opposite directions. The SNP with a lower MAF of every pair of SNPs with correlation r2 ≥ 0.8 was excluded from the gene-based tests using the filter in the R package. This prevented the capture of only a few association signals for some genes due to correlations between their top SNPs. After this exclusion, 7,086 SNPs were included in our final set of analysis, and we only examined the effects of those SNPs. Statistical significance for the gene-based analysis was claimed at an alpha level of 0.00143 (= 0.05/35 genes) based on Bonferroni multiple testing correction.
The single-SNP association tests, required as input for the AdaJoint test, were performed for each SNP in genes with nominal P < 0.05. We used logistic regression with case status as outcome under an additive model for genotype, adjusting for age (10-year groups), study site and the first four eigenvectors from PCA using SNPtest.[25] The first four eigenvectors were used to control for population stratification, as only the first four eigenvectors were associated with case status.[20] We corrected for multiple testing with a Bonferroni adjustment for the effective number of independent SNPs included in the final analysis using Gao’s SimpleM approach,[26] and the threshold was set at 0.05/7,086 = 7.06E-06. We also calculated Padj by multiplying the original P value with 7,086 to make the results more straightforward.
2.4. Sensitivity analysis
Four genes with pathway-level P ≤ 0.05 were excluded from AdaJoint analysis one by one to test their individual contribution in the whole Hippo way. We also excluded the 2nd, 3th and 4th genes with the smallest P values together to examine their combined effect to the whole pathway.
2.5. Genotype and gene expression correlation
According to the estimated genetic ancestry,[27] we grouped TCGA breast cancer patients into genomic Black (⩾ 50% African ancestry), genomic White (⩾ 90% European ancestry), and genomic Asian (⩾ 90% Asian ancestry). We imputed The Cancer Genome Atlas (TCGA) breast cancer germline genotypes by IMPUTE2 using the 1000 Genome Project data as the reference panel. Gene expression data (RSEM from RNA-seq V2) was accessed through cBioPortal (Breast Invasive Carcinoma [TCGA, Provisional], http://www.cbioportal.org/study?id=brca_tcga#summary), as of April 10, 2018. Subsequently, for two top genes in the SNP-level association test, we examined the correlation between SNP genotypes and gene expression in TCGA genomic Black breast cancer patients.
2.6. Visualization of association signals and SNP functionality
Single marker associations for top genes were plotted using LocusZoom.[28] Functionality of the significant SNPs and their tagged SNPs were examined in HaploReg v4, in which epigenomic data from the ENCODE (Encyclopedia of DNA Elements), the GTEx (Genotype-Tissue Expression) pilot and RegulomeDB databases were integrated.[29] Searches for expression quantitative trait loci (eQTL) information in breast tissues were also carried out in the GTEx portal and the eQTL Browser (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/). Additional chromosomal and functional annotation for the top SNPs in WWC1 and LATS2 can be found in Supplementary Table S1.
3. RESULTS
Using our unique population of African ancestry that is enriched for women with early age of onset for breast cancer, analysis of germline genetic variations in the Hippo pathway at the gene level did not show a significant association for any gene with overall breast cancer risk or ER− breast cancer, as none of the variants or their combined effect achieved nominal significance (Table 1). However, their combined effect was significantly associated with ER+ breast cancer (pathway-level P = 0.019). Four of the 35 genes in this pathway, WWC1, LATS2, SCRIB, and CDH1, were associated with risk of ER+ breast cancer at nominal significance. After Bonferroni adjustment, only WWC1 gene remained statistically significant (Padj = 0.04), while the LATS2 gene showed a trend towards significance (Padj = 0.07). When WWC1 was removed from the analysis, the pathway-level significance of the assessed germline genetic variations was lost (P = 0.12). It is possible that in addition to WWC1, the major driver, there might be other genes in the Hippo pathway contributing to the association with breast cancer risk. We performed additional analyses excluding CDH1, LATS2, and SCRIB individually or all three together, while keeping WWC1. The P values for the whole Hippo pathway turned out to be 0.075, 0.092, 0.017 and 0.086, respectively. In each analysis the top gene with the most significant P value was consistently WWC1.
TABLE 1.
Pathway- and gene-level tests in association with risk of overall, ER+, and ER− breast cancer
| Gene | Chromosome | Total number of SNPsa | Effective number of SNPsa |
P valuec |
||
|---|---|---|---|---|---|---|
| Overall | ER+ | ER− | ||||
| Pathway | - | 19889 | 7086 | 0.90 | 0.019 | 0.93 |
| Pathway exclude top gene | - | 18699 | 6423 | - | 0.12 | - |
| WWC1 | 5 | 1,190 | 629 | 0.16 | 0.001b | 0.19 |
| LATS2 | 13 | 642 | 275 | 0.43 | 0.002b | 0.41 |
| SCRIB | 8 | 186 | 101 | 0.09 | 0.02b | 0.21 |
| CDH1 | 16 | 614 | 271 | 0.24 | 0.05 | 0.68 |
| TAZ | 23 | 71 | 33 | 0.20 | 0.11 | 0.34 |
| CTNNA1 | 5 | 824 | 138 | 0.17 | 0.16 | 0.24 |
| WBP2 | 17 | 193 | 95 | 0.07 | 0.21 | 0.59 |
| YAP1 | 11 | 697 | 229 | 0.50 | 0.26 | 0.38 |
| MPP5 | 14 | 453 | 103 | 0.38 | 0.27 | 0.93 |
| SAV1 | 14 | 338 | 101 | 0.27 | 0.38 | 0.80 |
| STK3 | 8 | 1,503 | 263 | 0.59 | 0.38 | 0.31 |
| CRB3 | 19 | 130 | 84 | 0.24 | 0.45 | 0.41 |
| TAOK1 | 17 | 626 | 137 | 0.67 | 0.50 | 0.51 |
| PTPA | 9 | 335 | 75 | 0.99 | 0.56 | 0.38 |
| AMOT | 23 | 281 | 128 | 0.24 | 0.59 | 0.22 |
| PATJ | 1 | 2,488 | 1072 | 0.98 | 0.61 | 0.78 |
| NF2 | 22 | 381 | 128 | 0.16 | 0.62 | 0.20 |
| RASSF6 | 4 | 380 | 114 | 0.62 | 0.64 | 0.89 |
| PPP1CA | 11 | 85 | 38 | 0.15 | 0.67 | 0.70 |
| PTPN14 | 1 | 1,435 | 558 | 0.92 | 0.67 | 0.28 |
| YWHAB | 20 | 188 | 50 | 0.70 | 0.71 | 0.82 |
| STK4 | 20 | 623 | 144 | 0.77 | 0.74 | 0.35 |
| HIPK2 | 7 | 1,046 | 472 | 0.22 | 0.78 | 0.23 |
| FRMD6 | 14 | 1,292 | 557 | 0.87 | 0.83 | 0.98 |
| BTRC | 10 | 707 | 159 | 0.93 | 0.84 | 0.66 |
| CSNK1A1 | 5 | 290 | 116 | 0.44 | 0.85 | 0.12 |
| MOB1A | 2 | 254 | 102 | 0.71 | 0.85 | 0.17 |
| YWHAE | 17 | 484 | 225 | 0.66 | 0.85 | 0.68 |
| AJUBA | 14 | 115 | 60 | 0.51 | 0.88 | 0.39 |
| ANKHD1 | 5 | 449 | 125 | 0.41 | 0.94 | 0.81 |
| MOB1B | 4 | 445 | 122 | 0.61 | 0.94 | 0.10 |
| RASSF1 | 3 | 78 | 37 | 0.64 | 0.95 | 0.34 |
| ANKRD17 | 4 | 382 | 144 | 0.71 | 0.96 | 0.76 |
| LATS1 | 6 | 310 | 97 | 0.43 | 0.97 | 0.13 |
| TP53BP2 | 1 | 374 | 104 | 0.87 | 0.97 | 0.44 |
ER−, estrogen receptor-negative; ER+, estrogen receptor-positive; SNP, single nucleotide polymorphism.
For each candidate gene, its start and end chromosomal positions ±10kb were determined using the UCSC Genome Browser. SNPs within each gene region were extracted from The Root Consortium GWAS data and defined as “total SNPs”. “effective number of SNPs” means the number of SNPs after excluding the SNPs with MAF < 0.01 and the SNPs with a lower MAF in every pair of SNPs with correlation r2 ≥ 0.8.
Only P value for WWC1 remained significant after Bonferroni correction. The adjusted P values for WWC1, LATS2, and SCRIB were 0.035, 0.07, and 0.70, respectively. All other adjusted P values were equal to 1.
The numbers of controls and cases were 2,029, and 1,657 respectively. The numbers of ER+ patients and ER− patients were 403 and 374, respectively. ER status was not available for the remaining patients.
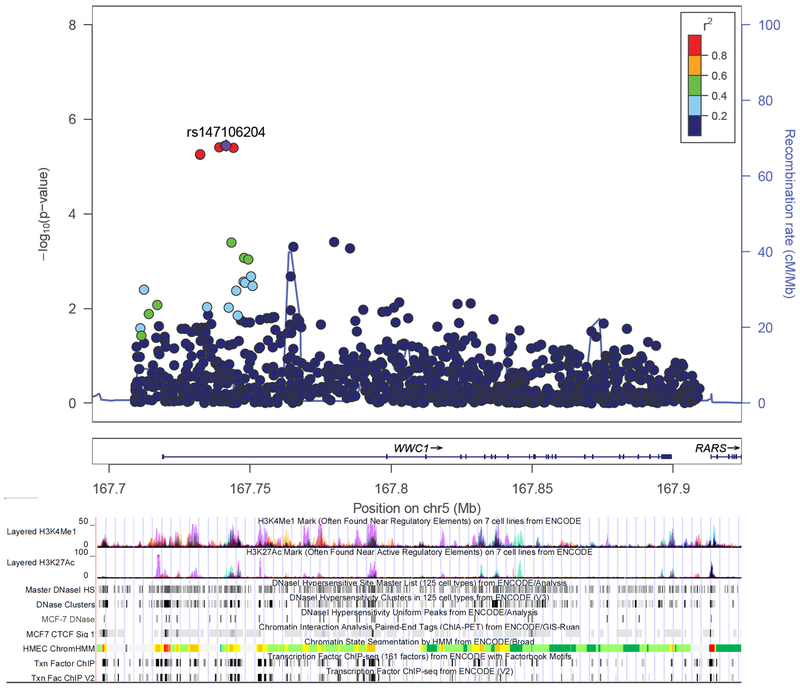
Table 2 shows SNP-level associations with overall, ER+ and ER− breast cancer risk. rs147106204 in the WWC1 gene was the most statistically significant SNP associated with ER+ breast cancer after adjustment for multiple comparisons (odds ratio [OR] = 0.53, 95% confidence interval [CI] = 0.41–0.70, Padj = 0.025). This SNP tags five other SNPs (rs116516633, rs114420099, rs115567889, rs77480437 and rs114592296) with significant Padj values. Additional analysis showed that rs147106204 and its LD-linked SNPs tend to be located in regions with regulatory elements with varying levels of evidence (Figure 2, Supplementary Table S1). The most significant SNP in LATS2 was the genotyped SNP rs73443365 (OR = 0.38, 95% CI = 0.24–0.61, Padj = 0.282). This SNP and five of its tagged SNPs are also located in regions with regulatory elements (Supplementary Figure S1, Supplementary Table S1). In addition, the allele frequencies between ER+ and ER− patients were significantly different for rs7344336 (P = 0.019), while the difference was marginally significant for rs147106204 (P = 0.056).
TABLE 2.
Significant SNPs for ER+ breast cancer
| rs73443365 | rs147106204 | |
|---|---|---|
| Gene | LATS2 | WWC1 |
| Major/minor alleles | A/C | C/A |
| MAFa in controls. | 0.04 | 0.11 |
| MAFa in all cases | 0.02 | 0.08 |
| MAFa in ER+ cases | 0.01 | 0.06 |
| MAFa in ER− cases | 0.03 | 0.09 |
| MAFa in 1000 Genomes Project EUR | 0.002 | 0.001 |
| Imputation information score | Genotyped | 0.991 |
| All cases vs. controlsd | ||
| OR (95% CI)b | 0.74 (0.57-0.96) | 0.82 (0.71-0.96) |
| P | 0.025 | 0.015 |
| Padj | 1.000 | 1.000 |
| ER+ cases vs. controlsd | ||
| OR (95% CI)b | 0.38 (0.24-0.61) | 0.53 (0.41-0.70) |
| P | 4.0E-05 | 3.6E-06 |
| Padj | 0.283 | 0.025 |
| ER− cases vs. controlsd | ||
| OR (95% CI)b | 0.69 (0.45-1.08) | 0.81 (0.62-1.06) |
| P | 0.103 | 0.119 |
| Padj | 1.000 | 1.000 |
CI, confidence interval; ER−, estrogen receptor-negative; ER+, estrogen receptor-positive; EUR, European; MAF, minor allele frequency; OR, odds ratio; SNP, single nucleotide polymorphism.
Minor allele frequency
Additive model with each SNP coded as 0, 1 or 2 copies of the minor allele, adjusting for age (10-year groups), study site and the first four eigenvectors from principal component analysis.
Bold P is significant at the defined cut point level (0.05/7,086 = 7.06E-06). Padj means P value after correction for multiple comparisons.
The numbers of controls and cases were 2,029, and 1,657 respectively. The numbers of ER+ patients and ER− patients were 403 and 374, respectively. ER status was not available for the remaining patients.
FIGURE 2.
LocusZoom plot of log-transformed P-values from single marker analysis for the WWC1 gene in ER+ subgroup test. The labeled single nucleotide polymorphism (SNP) with rs# was the most significant index SNP, and the linkage disequilibrium (LD) between other markers in the gene and the index SNP was color coded, with red color indicating strong LD (r2 > 0.8) and blue color indicating weak LD (r2 < 0.2). The bottom part presents the ENCODE (Encyclopedia of DNA Elements) regulatory tracks through UCSC Genome Browser, including histone modification marks for H3K4Me1 and H3K27Ac of seven cell types, transcription factor binding sites, and DNase hypersensitivity sites of human mammary epithelial cells (HMEC), breast cancer cells (MCF7). Chromosomal coordinates are in NCBI build 37.
Except for rs147106204, no SNP was significantly associated with breast cancer risk after Bonferroni correction (Padj > 0.05), and no eQTL information was available for rs147106204, rs73443365, and their tagged SNPs. Additional information on the SNPs with strongest associations in other non-significant genes is listed in Supplementary Table S2. By investigating the genotype and gene expression data from TCGA genomic Black breast cancer patients, we found that in the Total group (ER− and ER+ combined), the expression level of WWC1 was significantly higher in AA genotype compared with CC and CA of rs147106204, which was the most significant SNP in WWC1 (Supplementary Figure S2). In addition, this trend was consistent in ER− and ER+ groups as well. We did not find a clear genotype-gene expression correlation across different genotypes of LATS2 rs73443365.
4. DISCUSSION
In this relatively large case-control study of women of African ancestry, we found that the combined effect of genetic variations in the Hippo pathway was significantly associated with ER+ breast cancer (P = 0.019). The association was driven primarily by variants in the WWC1 LATS2, and CDH1 genes. Six highly correlated SNPs within WWC1, rs147106204, rs116516633, rs114420099, rs115567889, rs77480437, and rs114592296 were significantly associated with ER+ breast cancer risk at the SNP level.
The protein WW and C2 domain containing 1 (WWC1), also called KIBRA (expression enriched in kidney and brain),[30] interacts with and stimulates the phosphorylation of LATS½ and regulates the phosphorylation of YAP. WWC1 functions as a growth suppressive protein, and a study has reported that WWC1 knockdown enhanced migration and invasion by immortalized breast epithelial cells.[31] We found a significant variant rs147106204 that tags other SNPs that overlap two transcription regulatory marks (enhancers) in breast variant human mammary epithelial cells and breast myoepithelial primary cells (Supplemental Table S1). We also discovered that LATS2 approached a significant association with the development of ER+ breast cancer (Padj = 0.07). At the SNP-level, LATS2 was associated with ER+ breast cancer risk, although the top SNP rs73443365 in this gene became non-significant after Bonferroni correction. LATS2 (also known as KPM), which encodes a serine/threonine protein kinase of the LATS tumor suppressor family and core protein of the Hippo pathway, is down-regulated in many types of cancers including breast cancer.[32] The reduced expression of LATS1/2 has been consistently associated with a biologically aggressive phenotype of breast cancer.[32,33] This indicates that a partial loss of function of these tumor suppressor genes may lead to accelerated cell proliferation, resulting in an increased occurrence of aggressive disease. Given that the mechanisms by which the Hippo pathway becomes dysregulated are still not fully understood,[34] our result from a genetic association study might help to narrow down potential functional elements in the Hippo pathway components, such as WWC1 and LATS2.
It is worthy noting that the associations between the WWC1 and LATS2 genes and breast cancer risk were mainly found in ER+ participants. There might be synergistic effects between WWC1, LATS2 and estrogen receptor.[35,36] The positive synergistic effects between these genes and ER might provide an uncommon opportunity to detect a larger association, whereas it would be challenging to detect a relatively small association attributing only to genes without the interaction with ER. Thus, if the risk factors have positive synergistic effects with ER, it may be more likely to find the associations in ER+ participant. In addition, although the result was not significant in ER− patients, it is possible that this association also exists in ER− patients and the non-significant result in our study could be explained by the difference in effect size and sample size between ER+ and ER− patients. We observed that the WWC1 gene expression level was significantly higher in rs147106204 AA genotype compared with CC and CA genotypes in TCGA Black breast cancer patients. A allele of WWC1 rs147106204 was found to be the protective allele in our study, thus the observation of elevated WWC1 gene expression in AA group is in line with the biological function of WWC1 as a tumor suppressor gene. On the other hand, for LATS2 rs73443365, we could not find an obvious genotype-gene expression correlation across different genotypes due to very limited number of AC genotype and no CC genotype (C allele acts as protective allele) in TCGA Black breast cancer patients (Supplementary Figure S2).
We were not able to compare our association findings in WWC1 and LATS2 with the previous study in African Americans because these two genes were not included in their custom genotyping assay design.[7] In that study, CDH1 was found to be nominally associated with ER− breast cancer risk, and approached the borderline of significance for association with ER+ breast cancer risk, while we obtained a marginal significance with ER+ breast cancer risk. None of these associations survived multiple-testing adjustment. However, in the present study, the pathway-level P value became non-significant after excluding CDH1, which indicated that CDH1 might be one of the driver genes in the Hippo pathway.
We also searched the GAME-ON (Genetic Associations and Mechanisms in Oncology) / DRIVE (Discovery, Biology, and Risk of Inherited Variants in Breast Cancer) (http://gameon.dfci.harvard.edu) for 16 SNPs in WWC1 and LATS2 (Supplementary Table S1) to test whether the associations reported in our study can be replicated in European populations. However, all of these SNPs are absent there. In GAME-ON / DRIVE, both the WWC1 and LATS2 genes contain different SNPs that are associated with breast cancer risks. Overall, the correlations between the significant SNPs in our study and those in the GAME-ON / DRIVE were very low (r2 < 0.1). Actually, it is common to identify different disease-associated index SNPs between European- and African-descent populations, and this phenomenon has been frequently reported in diseases including prostate cancer[37–40] and breast cancer.[7,14,15,41] This inconsistency of association may be due to the different genetic backgrounds, such as LD patterns between African and European ancestries. In addition, although we undertook our GWAS using dense genotyping array with ~2.5 million markers and imputation using the 1000 Genomes Project data, it is worth noting that rare variants (especially very rare ones) were not particularly targeted in array development or reliably imputed, and therefore not included in the analyses. We cannot rule out the possibility that a set of causal rare variants (either coding or regulatory ones), which in general have larger effect size, can be tagged/captured by different index SNPs in different ethnic groups. The combined effect from both common and rare variants, with the same and opposite directions, could confer risk to a disease. Future systematic functional genomic studies are needed to shed light on the genetic architecture of these two genes and breast cancer risk in diverse populations.
Strengths of our study include a well-defined gene list for the Hippo pathway and our inclusion of a unique population of women of African ancestry from Nigeria and Barbados with a relatively young age at diagnosis of breast cancer. In addition, we applied the robust multilocus test (AdaJoint) to detect gene- and pathway-level associations that are often missed in studies only focusing on individual SNPs. However, several limitations need to be noted. First, as one of the largest studies to date on the genetics of breast cancer in women of African ancestry, the present study is still rather small and does not have enough power to detect individual SNP associations of very small magnitude, especially for SNPs with low allele frequency in subtype analyses. Second, our findings require further validation in diverse populations, as the gene-level associations were only marginally significant for WWC1 after correction for multiple tests. Nonetheless, the identified associations provide strong scientific rationale for future experimental studies to characterize the functional effects of these genetic variations.
In conclusion, our findings suggest an association between the Hippo pathway and ER+ breast cancer, with WWC1, LATS2, and CDH1 as possible genes driving this association. Further functional studies on genetic variants in these genes could help understand mechanisms of Hippo pathway in breast carcinogenesis, especially for women of African ancestry who suffer a disproportionate burden of aggressive breast cancers with poor clinical outcomes for reasons that remain unexplained and understudied.
Supplementary Material
SUPPLEMENTARY FIGURE S1 LocusZoom plot of log-transformed P-values from single marker analysis for LATS2 in ER+ subgroup test. The labeled marker with rs# was the most significant SNP (index SNP), and the LD between other markers in the gene and the index SNP was color coded, with red color indicating strong LD (r2 > 0.8) and blue color indicating weak LD (r2 < 0.2). The bottom part presents the ENCODE (Encyclopedia of DNA Elements) regulatory tracks through UCSC Genome Browser, including histone modification marks for H3K4Me1 and H3K27Ac of seven cell types, transcription factor binding sites, and DNase hypersensitivity sites of human mammary epithelial cells (HMEC), breast cancer cells (MCF7). Chromosomal coordinates are in NCBI build 37.
SUPPLEMENTARY FIGURE S2 The correlation between WWC1 and LATS2 gene expression level and genotypes by ER status in TCGA Black breast cancer patients.
The top panel (A) indicated the correlation between WWC1 gene expression level and genotypes grouped by ER status (Negative, Positive, and Total). The bottom panel (B) presents the correlation between LATS2 gene expression level and genotypes grouped by ER status (Negative, Positive, and Total).
SUPPLEMENTARY TABLE S1 Chromosomal and functional annotation for two genotyped risk-associated SNPs rs114592296 (WWC1) and rs73443365 (LATS2) and their tagged SNPs (r2 ≥ 0.8)
SUPPLEMENTARY TABLE S2 Top 10 SNPs (MAF ≥ 0.01) in overall, ER+ and ER− breast cancer
ACKNOWLEDGMENTS
The authors thank all the women who participated in this research. We also thank Walmy Elisabeth Sveen from University of Chicago, for editorial support. SW was supported by a University of Chicago Global Health Fellowship.
Funding information
National Cancer Institute, Grant numbers: CA142996, CA161032; Susan G. Komen for the Cure, Grant number: SAC110026; American Cancer Society, Grant numbers: MRSG-13-063-01-TBG, CRP-10-119-01-CCE; Breast Cancer Research Foundation, Grant numbers: BCRF-17-118, BCRF-17-070.
Abbreviations
- AdaJoint
adaptive joint multilocus test
- CI
confidence interval
- DRIVE
Discovery, Biology, and Risk of Inherited Variants in Breast Cancer
- ENCODE
Encyclopedia of DNA Elements
- eQTL
expression quantitative trait loci
- ER−
estrogen receptor-negative
- ER+
estrogen receptor-positive
- GAME-ON
Genetic Associations and Mechanisms in Oncology
- GTEx
Genotype-Tissue Expression
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- MAF
minor allele frequency
- OR
odds ratio
- PCA
principal component analysis
- SNP
single nucleotide polymorphism
- TCGA
The Cancer Genome Atlas
Footnotes
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature reviews Cancer 2013;13(4):246–257. [DOI] [PubMed] [Google Scholar]
- 2.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nature reviews Cancer 2007;7(3):182–191. [DOI] [PubMed] [Google Scholar]
- 3.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nature cell biology 2011;13(8):877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li YW, Shen H, Frangou C et al. Characterization of TAZ domains important for the induction of breast cancer stem cell properties and tumorigenesis. Cell cycle (Georgetown, Tex) 2015;14(1):146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi H, Bevier M, Johansson R et al. Single nucleotide polymorphisms in the 20q13 amplicon genes in relation to breast cancer risk and clinical outcome. Breast cancer research and treatment 2011;130(3):905–916. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, Wang W, Zhu B et al. A functional variant rs1820453 in YAP1 and breast cancer risk in Chinese population. PloS one 2013;8(11):e79056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Yao S, Hu Q et al. Genetic variations in the Hippo signaling pathway and breast cancer risk in African American women in the AMBER Consortium. Carcinogenesis 2016;37(10):951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proceedings of the National Academy of Sciences of the United States of America 2011;108(29):11930–11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beeghly-Fadiel A, Lu W, Gao YT et al. E-cadherin polymorphisms and breast cancer susceptibility: a report from the Shanghai Breast Cancer Study. Breast cancer research and treatment 2010;121(2):445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia YM, Xie YT, Wang YJ, Han JY, Tian XX, Fang WG. Association of Genetic Polymorphisms in CDH1 and CTNNB1 with Breast Cancer Susceptibility and Patients’ Prognosis among Chinese Han Women. PloS one 2015;10(8):e0135865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu K, Li Q, Bergen AW et al. Pathway analysis by adaptive combination of P-values. Genet Epidemiol 2009;33(8):700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Shi J, Liang F, Wheeler W, Stolzenberg-Solomon R, Yu K. A fast multilocus test with adaptive SNP selection for large-scale genetic-association studies. European journal of human genetics : EJHG 2014;22(5):696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huo D, Ikpatt F, Khramtsov A et al. Population differences in breast cancer: survey in indigenous African women reveals over-representation of triple-negative breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2009;27(27):4515–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huo D, Zheng Y, Ogundiran TO et al. Evaluation of 19 susceptibility loci of breast cancer in women of African ancestry. Carcinogenesis 2012;33(4):835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y, Ogundiran TO, Falusi AG et al. Fine mapping of breast cancer genome-wide association studies loci in women of African ancestry identifies novel susceptibility markers. Carcinogenesis 2013;34(7):1520–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemesure B, Wu SY, Hambleton IR, Leske MC, Hennis AJ, Barbados National Cancer Study G. Risk factors for breast cancer in a black population--the Barbados National Cancer Study. International journal of cancer Journal international du cancer 2009;124(1):174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boersma BJ, Howe TM, Goodman JE et al. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J Natl Cancer Inst 2006;98(13):911–919. [DOI] [PubMed] [Google Scholar]
- 18.Qian F, Ogundiran T, Hou N et al. Alcohol consumption and breast cancer risk among women in three sub-Saharan African countries. PloS one 2014;9(9):e106908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian F, Feng Y, Zheng Y et al. Genetic variants in microRNA and microRNA biogenesis pathway genes and breast cancer risk among women of African ancestry. Human genetics 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huo D, Feng Y, Haddad S et al. Genome-wide association studies in women of African ancestry identified 3q26.21 as a novel susceptibility locus for oestrogen receptor negative breast cancer. Human molecular genetics 2016;25(21):4835–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS genetics 2006;2(12):e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verma SS, de Andrade M, Tromp G et al. Imputation and quality control steps for combining multiple genome-wide datasets. Frontiers in genetics 2014;5:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Wheeler W, Hyland PL et al. A Powerful Procedure for Pathway-Based Meta-analysis Using Summary Statistics Identifies 43 Pathways Associated with Type II Diabetes in European Populations. PLoS genetics 2016;12(6):e1006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nature reviews Genetics 2010;11(7):499–511. [DOI] [PubMed] [Google Scholar]
- 26.Gao X Multiple testing corrections for imputed SNPs. Genet Epidemiol 2011;35(3):154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huo D, Hu H, Rhie SK et al. Comparison of Breast Cancer Molecular Features and Survival by African and European Ancestry in The Cancer Genome Atlas. JAMA oncology 2017;3(12):1654–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruim RJ, Welch RP, Sanna S et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics (Oxford, England) 2010;26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle AP, Hong EL, Hariharan M et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research 2012;22(9):1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kremerskothen J, Plaas C, Büther K et al. Characterization of KIBRA, a novel WW domain-containing protein. Biochemical and biophysical research communications 2003;300(4):862–867. [DOI] [PubMed] [Google Scholar]
- 31.Moleirinho S, Chang N, Sims AH et al. KIBRA exhibits MST-independent functional regulation of the Hippo signaling pathway in mammals. Oncogene 2013;32(14):1821–1830. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi Y, Miyoshi Y, Takahata C et al. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clinical cancer research : an official journal of the American Association for Cancer Research 2005;11(4):1380–1385. [DOI] [PubMed] [Google Scholar]
- 33.Moroishi T, Hayashi T, Pan WW et al. The Hippo Pathway Kinases LATS1/2 Suppress Cancer Immunity. Cell 2016;167(6):1525–1539 e1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plouffe SW, Meng Z, Lin KC et al. Characterization of Hippo Pathway Components by Gene Inactivation. Molecular cell 2016;64(5):993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Britschgi A, Duss S, Kim S et al. The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERalpha. Nature 2017;541(7638):541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rayala SK, den Hollander P, Manavathi B et al. Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells. The Journal of biological chemistry 2006;281(28):19092–19099. [DOI] [PubMed] [Google Scholar]
- 37.Hooker S, Hernandez W, Chen H et al. Replication of prostate cancer risk loci on 8q24, 11q13, 17q12, 19q33, and Xp11 in African Americans. The Prostate 2010;70(3):270–275. [DOI] [PubMed] [Google Scholar]
- 38.Xu Z, Bensen JT, Smith GJ, Mohler JL, Taylor JA. GWAS SNP Replication among African American and European American men in the North Carolina-Louisiana prostate cancer project (PCaP). The Prostate 2011;71(8):881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bensen JT, Xu Z, Smith GJ, Mohler JL, Fontham ET, Taylor JA. Genetic polymorphism and prostate cancer aggressiveness: a case-only study of 1,536 GWAS and candidate SNPs in African-Americans and European-Americans. The Prostate 2013;73(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan DS, Mok TS, Rebbeck TR. Cancer Genomics: Diversity and Disparity Across Ethnicity and Geography. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2016;34(1):91–101. [DOI] [PubMed] [Google Scholar]
- 41.Feng Y, Rhie SK, Huo D et al. Characterizing Genetic Susceptibility to Breast Cancer in Women of African Ancestry. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2017;26(7):1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY FIGURE S1 LocusZoom plot of log-transformed P-values from single marker analysis for LATS2 in ER+ subgroup test. The labeled marker with rs# was the most significant SNP (index SNP), and the LD between other markers in the gene and the index SNP was color coded, with red color indicating strong LD (r2 > 0.8) and blue color indicating weak LD (r2 < 0.2). The bottom part presents the ENCODE (Encyclopedia of DNA Elements) regulatory tracks through UCSC Genome Browser, including histone modification marks for H3K4Me1 and H3K27Ac of seven cell types, transcription factor binding sites, and DNase hypersensitivity sites of human mammary epithelial cells (HMEC), breast cancer cells (MCF7). Chromosomal coordinates are in NCBI build 37.
SUPPLEMENTARY FIGURE S2 The correlation between WWC1 and LATS2 gene expression level and genotypes by ER status in TCGA Black breast cancer patients.
The top panel (A) indicated the correlation between WWC1 gene expression level and genotypes grouped by ER status (Negative, Positive, and Total). The bottom panel (B) presents the correlation between LATS2 gene expression level and genotypes grouped by ER status (Negative, Positive, and Total).
SUPPLEMENTARY TABLE S1 Chromosomal and functional annotation for two genotyped risk-associated SNPs rs114592296 (WWC1) and rs73443365 (LATS2) and their tagged SNPs (r2 ≥ 0.8)
SUPPLEMENTARY TABLE S2 Top 10 SNPs (MAF ≥ 0.01) in overall, ER+ and ER− breast cancer