Abstract
Background
Checkpoint inhibitors have recently been approved for the treatment of patients with hepatocellular carcinoma (HCC). However, biomarkers, which will help identify patients responding to therapy, are missing. We recently tested the combination of anti-CTLA4 treatment (tremelimumab) with loco-regional therapy in patients with HCC and reported a partial response rate of 26%.
Methods
Here, we report updated survival analyses and results from our immune monitoring studies on peripheral blood mononuclear cells (PBMCs) and tumors from these patients.
Results
Tremelimumab therapy increased CD4+-HLA-DR+, CD4+PD-1+, CD8+HLA-DR+, CD8+PD-1+, CD4+ICOS+ and CD8+ICOS+ T cells in the peripheral blood of the treated patients. Patients with higher CD4+PD1+ cell frequency at baseline were more likely to respond to tremelimumab therapy. PD-1 expression was increased on alpha fetal protein (AFP) and survivin-specific CD8 T cells upon tremelimumab treatment. An increase of tumor infiltrating CD3+ T cells were also seen in these patients. Immunosequencing of longitudinal PBMC showed that one cycle of tremelimumab significantly decreased peripheral clonality, while no additional effects were seen after loco-regional therapy.
Conclusion
In summary, we observed a clear activation of T cell responses in HCC patients treated with tremelimumab and identified potential biomarkers which will help identify patients responding to immunotherapy with anti-CTLA4.
Electronic supplementary material
The online version of this article (10.1007/s00262-019-02299-8) contains supplementary material, which is available to authorized users.
Keywords: Hepatocellular carcinoma, Trial, Immunotherapy, Biomarker
Introduction
The first immune checkpoint inhibitor (nivolumab) has recently been approved by the FDA for the treatment of patients with advanced hepatocellular carcinoma, who have progressed on sorafenib therapy. Currently, a number of trials are ongoing to evaluate different immune based approaches in HCC [1]. Nivolumab was approved based on the results from the Checkmate 040 trial. These were patients who had intermediate or advanced HCC with preserved liver function and were candidates for systemic therapy, and had progressed or were intolerant to sorafenib or had refused this drug [2]. After confirming safety and testing different dose levels, an expansion cohort of 214 patients were treated with nivolumab. Similar response rates between 15 and 20% were reported across different etiologies, including both sorafenib-naïve and sorafenib-exposed patients A median overall survival of 28.6 months (95% CI 16.6–NE) in the population naïve to sorafenib, and 15.6 months (95% CI 13.2–18.9) in the population exposed to sorafenib (90% progression) was observed. However, no biomarker was identified to predict or correlate with the outcome and response rates appeared to be independent of PD-L1 expression. Recently, Zhu et al. reported the results from the KEYNOTE 224, in which pembrolizumab was tested in HCC patients who failed prior sorafenib therapy. One complete response and 17 partial responses were observed among 104 treated patients. Again, no biomarker was identified to predict or correlate with outcome and response to immune checkpoint therapy [3].
We have recently published results from a phase II trial, in which we treated 32 HCC patients with anti-CTLA4 (tremelimumab) plus tumor ablation therapy. In this study, we demonstrated a partial response rate of 26.3%. Median time to progression was 7.4 months [4]. Peripheral blood samples were obtained from all patients throughout treatment as well as tumor samples at the time of ablation. We conducted comprehensive immune monitoring with the aim to better understand how checkpoint inhibitors work in patients, why they only work in some patients but not in others, and to identify potential biomarkers, which may predict outcome or at least correlate with outcome and, which should be tested in future studies. Here, we report the results of our correlative studies including flow-cytometry analysis of immune cell subsets from peripheral blood, T-cell receptor sequencing of PBMCs and tumor-infiltrating lymphocytes and immunohistochemistry studies of tumor biopsies derived from patients treated with tremelimumab. Results from this study will help in better understanding immune responses to immune checkpoint blockade in advanced HCC and in enhancing immune monitoring procedures.
Materials and methods
Patients and study design
This was a pilot study to investigate the feasibility of the combination of tremelimumab and loco-regional radiation therapy in refractory advanced HCC. Briefly, a of total 39 adult patients with HCC were enrolled into this study. Tremelimumab was given intravenously every 4 weeks for a total 6 doses, followed by 3-monthly administrations until off-treatment criteria were met. On day 35, patients received subtotal radiofrequency ablation (RFA) or chemo-ablation performed by interventional radiologists. Long term survival data was followed in study participants since the initial data was reported [4]. The eligibility criteria of patients and study design have been previously described in detail. Tumor responses were measured according to RECIST version 1.1 criteria, which helped identify eight patients with a complete (CR) or partial response (PR) and 11 patients who progressed on treatment. However, it has been described that even patients without a radiologic response according to RECIST criteria may benefit from immunotherapy [5] and such analysis would also provide a larger sample number for comparative studies. Therefore, we conducted a second analysis for which patients were classified as responders (R) or non-responders (NR). Responders (n = 19) were defined as those who had an objective response rate as well patients who demonstrated stable disease as their best response and had a progression-free survival greater than 6 months. Non-responders (n = 15) were defined as those who had a best response of progressive disease per RECIST, stable disease less than 6 months or, in the event of non-measurable disease, clear clinical progression of disease within 6 months of study entry.
Flow cytometry
Peripheral blood mononuclear cells were isolated from patients at baseline and after 4, 8 and 12 weeks as previously described [6]. Multi-color flow cytometry was performed to study different immune cell subsets using the following antibodies: CD3 (UCHT1), CD4 (S3.5), CD8 (RPA-T8), PD1 (EH12.1), 4-1BB (4B4-1), TIM3 (F38-2E2), HLA-DR (G46-6), CTLA-4 (BNI3), ICOS (DX29), CD25 (M-A251). Gating strategies were performed as previously described [7]. Antigen-specific T-cell responses were tested in HLA-A2 positive patients using HLA-A2 AFP137–145 and HLA-A2 survivin96–104 tetramers (provided by the NIH Tetramer Core Facility).
Immunohistochemistry
Pre-treatment tumor samples were obtained either from outside of NIH if available and adequate, or before patients received cycle 1 of treatment. Post-treatment tumor samples were obtained at the time of the interventional radiologic procedure (post-treatment, Day 36). In total, there were 19 pre-treatment tumor samples, including 7 from outside of NIH, and 22 post-treatment tumor samples. Among those samples, 11 patients had pre- and post-treatment tumor samples obtained, 8 only had pre-treatment and 9 only had post-treatment tumor samples were available. Formalin-fixed paraffin-embedded tissue was stained with antibodies for T-cell markers CD3 (Ventana 2GV6) and CD8 (Dako 144B), PD-1 (Roche NAT105) and CD68 (Roche KP-1). Immunohistochemically stained sections were imaged at 20X with an Aperio ScanScope XT, and digital imaging analysis was performed with Aperio Positive Pixel Count (V9) on manually annotated regions of tumor as previously described [4]. The algorithm identifies staining intensity of 3, 3-diaminobenzidine chromogen (DAB) in four groups—absent, weak, intermediate and strong. The percentage pixels in the region of interest (ROI) was determined and used as a surrogate for the cell number. Patients were stratified by response. Pre- and post-treatment biopsies were tested. Pre-treatment tumor samples were obtained from 15 patients without available tumor samples before treatment versus 14 patients with available tumor samples from outside of NIH.
T-cell receptor sequencing
DNA from PBMC and tumor samples from the first 26 patients for which adequate material was available was extracted from tumor tissue and PBMC using the DNeasy Blood & Tissue kit from Qiagen. Immunosequencing of the CDR3 regions of human TCR-beta chains was performed using the ImmunoSEQ™ Assay (Adaptive Biotechnologies, Seattle, WA). Extracted genomic DNA was amplified in a bias-controlled multiplex PCR, followed by high-throughput sequencing. Sequences were collapsed and filtered in order to identify and quantitate the absolute abundance of each unique TCR-beta CDR3 region for further analysis as previously described [8–10].
Statistical analysis
Flow cytometry data was analyzed to calculate the medians by cycle or percent changes. The p values for the changes were calculated using Wilcoxon signed rank test corrected for multiple comparisons by the Hochberg method. The method used to test the changes between the different time points was the Wilcoxon signed rank test corrected for the number of tests within each group. The outcomes for trends in the markers were tested across the four and two patient response categories (PR/PD or R/NR categories). Logistic regression was used to analyze the association of various factors with response. Overall and progression-free survival were measured as months since the start of therapy.
Results
Patient characteristics
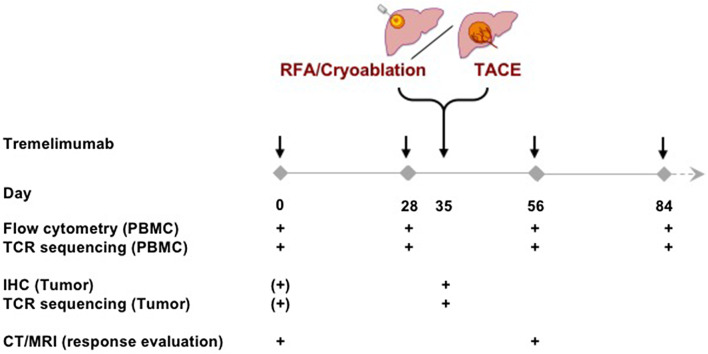
Figure 1 shows the trial design, including the time points when blood and tumor samples were harvested. A total of 39 patients were included in this protocol. Clinical outcome is available for 34 patients. Post-treatment tumor samples were obtained from 30 patients. All patients provided PBMC samples. Table 1 provides a summary of the study patients baseline characteristics including their clinical responses. An updated survival analysis with a median follow-up of 36.6 months is shown in Supplementary Fig. 1 and confirms our previous report with a median OS of 10.9 months (95% CI 8.0–13.7 months). The overall median survival rates in the groups treated with transarterial chemoembolization (TACE), RFA and cryoablation were 13.8 months (95% CI 10.2–17.4 months, N = 17), 9.2 months (95% CI 6.6–11.2 months, N = 10), 15.0 months (95% CI 10.5–19.5 months, N = 9), respectively.
Fig. 1.
Trial design and time points for evaluation of PBMC, tumors and clinical responses. Note that baseline tumor samples may have been obtained at the time of diagnosis. PBMC were harvested on the day of treatment before tremelimumab was given and tumor samples were obtained just prior to loco-regional therapy (IHC immunohistochemistry, RFA radiofrequency ablation, TACE transarterial chemoembolization, PBMC peripheral blood mononuclear cells, TCR T-cell receptor, CT computer tomography, MRI magnet resonance imaging)
Table 1.
Patient characteristics
| Number | 39 |
| Age, median (range) | 63 (36–78) |
| Sex | |
| Male | 34 |
| Female | 5 |
| ECOG | |
| 0 | 10 |
| 1 | 29 |
| Liver cirrhosis | |
| Yes | 26 |
| No | 13 |
| Cause of liver disease | |
| HBV | 7 |
| HCV | 21 |
| Baseline Child–Pugh score | |
| 5 | 16 |
| 6 | 7 |
| 7 | 3 |
| BCLC stage | |
| B | 9 |
| C | 30 |
| Extrahepatic disease | |
| Yes | 17 |
| No | 22 |
| Prior sorafenib | |
| Yes/no | 27/12 |
| Discontinue due to PD/intolerance | 23/4 |
| Other systemic therapies | 10 |
| Other previous interventions | |
| TACE | 13 |
| Surgery | 5 |
| Ablation | 6 |
| Outcome analysis | |
| Non-evaluable | 5 |
| Non-responder | 15 |
| Responder | 19 |
| CR | 1 |
| PR | 7 |
| SD | 15 |
| PD | 11 |
ECOG Eastern Cooperative Oncology Group, HBV Hepatitis B virus, HCV Hepatitis C virus, BCLC Barcelona Clinic Liver Cancer, PD progressive disease, TACE transarterial chemoembolization, CR complete response, PR partial response, SD stable disease
Flow cytometric analysis of cell surface markers in PBMC of patients
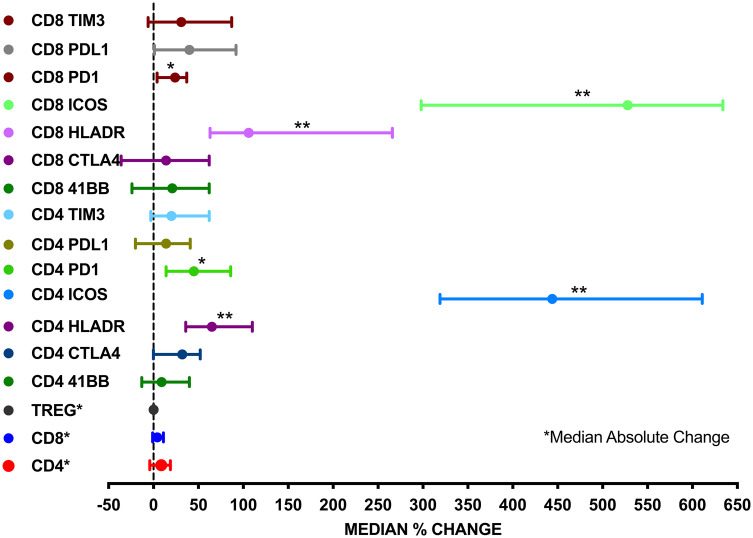
Peripheral blood mononuclear cells were analyzed from all patients at four different time points; before treatment (baseline) and after 4, 8 and 12 weeks. We performed multi-color flow cytometry analysis. Using 11-color flow cytometry, we studied a total of 17 different T-cell populations over time using antibodies to distinguish between T-cell subsets (CD3, CD4 and CD8), costimulatory molecules (PD1, 4-1BB, TIM3, ICOS and CTLA4) and activation markers (CD25 and HLA-DR). A representative staining is shown in Supplementary Fig. 2. We compared the immune cell subsets at baseline and after one dose of tremelimumab. After 4 weeks of therapy, there was a statistically significant increase in the percentage of CD4+HLA-DR+ T cells (median change: 65%, p < 0.0001), CD8+HLA-DR+ T cells (median change: 106%, p < 0.0001); CD4+ICOS+ T cells (median change: 444%, p < 0.0001), CD8+ICOS+ T cells (median change: 528%, p < 0.0001), CD4+PD-1+ T cells (median change: 45%, p = 0.002) and CD8+PD1+ T cells (median change: 24%, p = 0.016) as compared to the baseline (Fig. 2). No significant changes were found for the other immune cells studied including myeloid-derived suppressor cells (MDSC) (data not shown).
Fig. 2.
Forrest plot comparing the absolute (CD4, CD8, Treg) or percent (all others) change of different T-cell subsets in peripheral blood in patients with HCC before and 28 days after tremelimumab treatment (*p < 0.05; **p < 0.0001)
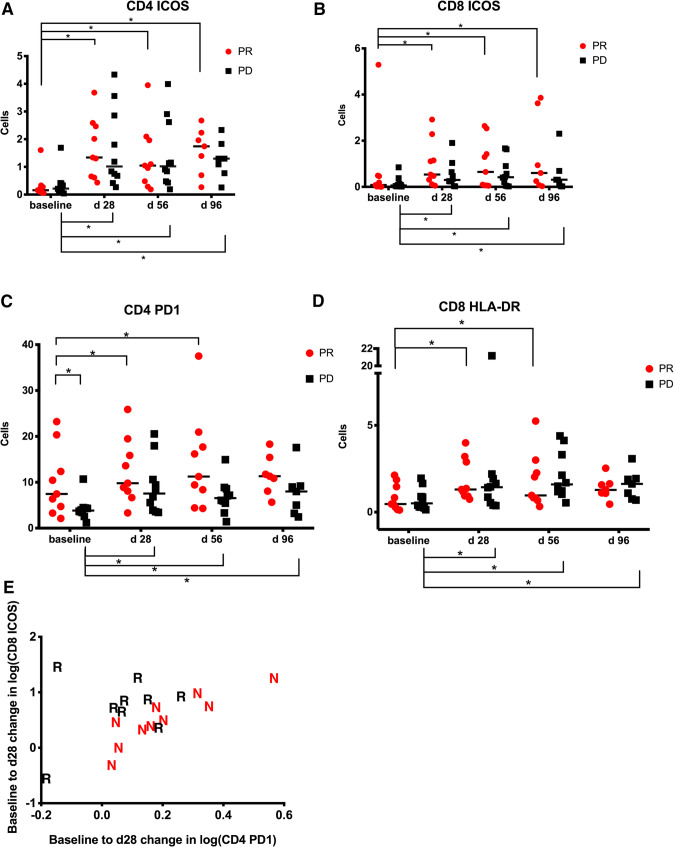
Next, we studied different T-cell subsets and whether any changes correlated with clinical responses. Direct comparison of T cells from baseline and at 4 weeks after the first treatment dose revealed a significant increase in the number of CD4+ICOS+ and CD4+PD-1T cells in patients with a PR (Fig. 3a, c). Next, we studied CD8+ T cells. Again, we noticed an increase in CD8+ICOS+ and CD8+HLA-DR+ T cells in patients demonstrating a PR (Fig. 3b, d). However, similar results were observed in patients without clinical responses.
Fig. 3.
a–d CD4+ICOS+, CD4+PD1+, CD8+ HLA-DR+ and CD8+ICOS+ cells in patients with clinical response or absence of response over time (*p < 0.05) (PR partial response, PD progressive disease). e Combination of CD8 ICOS and CD4 PD-1 suggested that the combination can be relevant to response of the patients to therapy (N non-responders; R responders)
We next correlated changes between the surface markers to determine if there are any combination of factors which were significantly associated with clinical response. The primary outcome to which this method was applied to was the determination of response in responders vs. non-responders. Figure 3e shows that a larger increase in CD8 ICOS combined with a smaller increase in CD4 PD1 makes a patient more likely to achieve a PR. These factors have individual significance levels of approximately p = 0.05, and the combination is significant at the p = 0.03 level.
Finally, we looked for baseline markers which would predict response to treatment. Here, we noticed that the frequency of CD4+PD1+ cells in PBMC was higher in patients responding to therapy at baseline in comparison to patients not responding (Fig. 3c).
Tumor-specific CD8+ T-cell responses
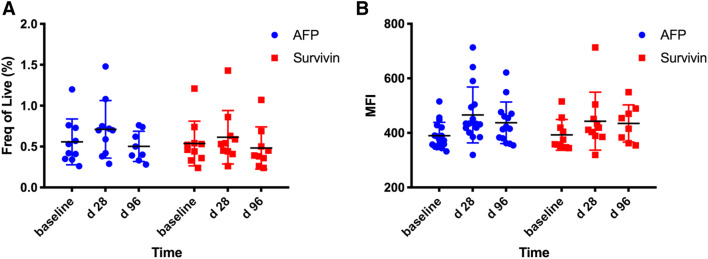
We used HLA-A2 tetramer specific for AFP137–-145 and survivin96–-104 to study tumor-specific immune responses in 15 HLA-A2pos patients. A representative FACS plot is shown in Supplementary Fig. 3. As shown in Fig. 4a, we did not observe a difference in the frequency of AFP137–-145 or survivin96–-104 specific T cells in the peripheral blood from patients treated with tremelimumab plus ablation. In contrast, there was a non-significant increase in PD1 expression on antigen-specific T cells similar to what was observed when peripheral CD8+ T cells were studied without gating on tetramer positive cells, as shown in Fig. 4b.
Fig. 4.
AFP137–145 or survivin96–104 specific T cells in peripheral blood from patients treated with tremelimumab plus ablation
Immune cell infiltration of tumors after tremelimumab treatment
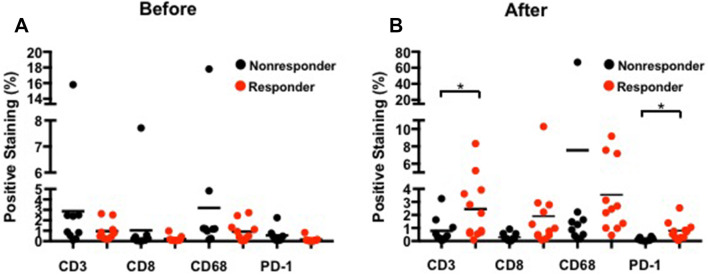
Among 39 cases, 19 pre- and 22 post-treatment tumor biopsies were submitted for immunohistochemistry analysis. Quantitative analysis was performed as shown in Fig. 5. Confirming our previous observation, responders had a significantly increased post-treatment mean positive pixel count of CD3 (number of responders = 12) compared to non-responders (progressive disease, N = 8) 2.45% vs 0.78%, and PD-1 (number of responders = 11) 0.79% vs 0.14% (p = 0.012), respectively. The differences were borderline for post-treatment CD8 counts. Responders (N = 12) and non-responders (N = 10) had a post-treatment mean positive pixel count for CD8 of 1.90% vs. 0.30% (p = 0.075), respectively, which is likely to be attributed to the relatively small number of cases. No difference was found regarding post-treatment KP-1. There were no differences found between responders and non-responders regarding the pre-treatment percentages of total positive pixels of PD-1, CD3, CD8 and KP-1.
Fig. 5.
Quantitative analysis of immunohistochemistry on the biopsied tumor samples from a pre-treatment and b post-treatment (*p < 0.05)
TCR V-beta receptor analysis
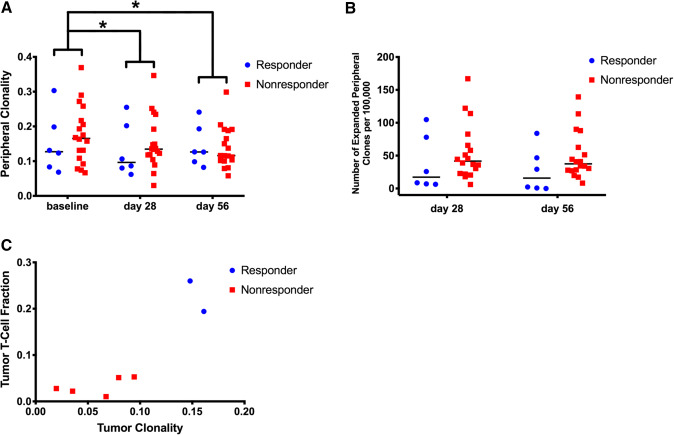
T-cell receptor sequencing is widely used to study the T-cell receptor repertoire and the effect of immune checkpoint blockade on T-cell clonality [11]. Immunosequencing of longitudinal PBMC samples (n = 26) before and on-treatment showed that one cycle of tremelimumab significantly decreased peripheral clonality (Fig. 6a: p = 0.044), indicating a broadening of the T-cell repertoire. We also found that clonality continues to decrease on tremelimumab therapy at the start of cycle 3 after tumor ablation occurred (p = 0.025). Furthermore, subjects who respond to immune checkpoint blockade showed a trend towards lower peripheral clonality pre-treatment, which was similarly observed in PD-L1 blockade of urothelial cancer [12]. Analysis of differentially abundant clones in the peripheral repertoire showed a trend towards non-responders having more expansion than responders while on-treatment, relative to each subject’s pre-treatment baseline (Fig. 6b: p = 0.28 and p = 0.16 at cycles 2 and 3, respectively). Although we are underpowered to determine tumor repertoire features predictive of response (n = 7 with an immunosequenced tumor sample and evaluable response to therapy), Fig. 6c shows marked separation of responders and non-responders based on tumor clonality and T-cell infiltrate: responders tended to have both high tumor clonality and T-cell fraction compared to non-responders, who trended towards low tumor clonality and T-cell fraction.
Fig. 6.
T-cell repertoire characteristics as determined by immunosequencing. a Clonality of PBMC samples before and on-treatment, stratified by responders “R” (CR/PR) and non-responders “NR” (SD less than 6 months/PD). b Expansion of peripheral T-cell clones determined by differential abundance analysis comparing on-treatment to pre-treatment baseline samples. c Tumor T-cell repertoire characteristics showing the relationship between T-cell fraction in the tumor and tumor repertoire clonality (*p < 0.05)
Discussion
With the recent FDA approval of nivolumab for the treatment of patients with HCC after first line sorafenib therapy and a number of ongoing studies evaluating different immune checkpoint inhibitors in HCC, there is a great need to improve our understanding of the immunological responses to immune checkpoint therapy. Biomarkers are needed to identify patients responding to therapy and could play a pivotal role in improving clinical response rates [13].
Comprehensive immune monitoring has not been reported for patients with HCC and most studies do not require collecting samples from patients on treatment. In this report, we present results from the immunological monitoring of a group of 39 patients with advanced HCC that were all treated with anti-CTLA-4 combined with different tumor ablation therapies. The primary objective of this study was to demonstrate safety and feasibility, which we have reported previously in combination with preliminary efficacy data. Here, we report the results from our comprehensive immune monitoring analysis of PBMC using an antibody panel of immune markers as well as T-cell receptor sequencing. In addition, tumor samples from patients, which were obtained at the time of ablation (i.e., after two doses of tremelimumab therapy) were studied. As previously reported, 21% of the patients demonstrated an objective response while 28% of the patients showed progressive disease. This allowed us to study immune response in patients responding to therapy and those who progressed despite treatment. However, due the limited number of patients in this study, we extended our analysis to patients showing a “clinical” response, which we defined as patients with radiological stable disease and lack of clinical disease progression for a period of greater than 6 months. Interestingly, results obtained when our analysis was restricted to patients with partial responses or progressive disease mirrored results obtained when we studied patients with or without clinical responses.
Since HCC is commonly diagnosed by triphasic imaging techniques, one common problem in conducting clinical trials in patients with HCC is the lack of tumor biopsies [14]. We obtained tumor biopsies from patients at the time of ablation and were able to confirm our earlier results that tumor infiltrating CD3+CD8+ T cells were associated with a response to therapy. Additionally, using TCR sequencing analysis, we now provide early data suggesting that responders tend to have both high tumor clonality and T-cell fractions compared to patients not responding to treatment with immune checkpoint inhibitors. A similar pattern of separation between responders and non-responders was observed in immune-sequenced melanoma tumors prior to PD-1 blockade [15]. More data is clearly needed to confirm these findings and we are conducting a follow-up trial in which tumor biopsies are being harvested before and on treatment with immune checkpoint inhibitors.
Obtaining tumor biopsies from patients on treatment may not always be feasible in patients with HCC, therefore, we focused our studies on the analysis of immune cells in peripheral blood. We followed reports, which describe the changes seen in patients with melanoma upon anti-CTLA4 therapy [16].
T-cell receptor sequencing revealed similar responses, which had been described before in other indications. The observed broadening of the T-cell receptor repertoire upon tremelimumab treatment was consistent with previously reported studies of anti-CTLA4 in metastatic prostate cancer and metastatic melanoma [10, 17]. However, no clear differences between responders and non-responders were seen, which may be explained by the limited sample number of this trial. Furthermore, the type of ablation appeared to have no effect on clonality (data not shown), which may be explained by the fact that patients had already received prior tremelimumab therapy and patient cohorts were small. In addition, T-cell receptor sequencing data from tumor infiltrating cells revealed a similar pattern of high tumor clonality and T-cell fractions in tumor biopsies derived from patients responding to immune checkpoint blockade therapy comparable to a pattern, which has been previously described in immunosequenced melanoma tumors prior to PD-1 blockade [15].
One important question to be answered is how to identify HCC patients who benefit from treatment with immune checkpoint inhibitors [18]. In this study, we found preliminary evidence that patients with higher CD4+PD1+ cell counts were more likely to respond to tremelimumab therapy. However, this data still needs to be verified in larger patient cohorts.
In this study, all patients were treated with tremelimumab, a CTLA4-specific antibody. Therefore, it should be noted that our data cannot be immediately translated to patients treated with anti-PD1 or anti-PDL1. As a matter of fact, distinct changes in peripheral blood have been described in melanoma patients treated with PD1–PDL1 targeting antibodies compared to patients treated with anti-CTLA4 [13]. Here, comprehensive genomic analysis of tumor samples may be even more needed. Such information could also be used to confirm a gene signature, which has recently been described to predict patients with highly “immunogenic” tumors, which are more likely to respond to immune based treatment approaches [19].
All patients enrolled in this clinical trial underwent tumor ablation including TACE, radiofrequency ablation and cryoablation, making it impossible to address the question whether tumor ablation may enhance the effect of checkpoint inhibitor therapy. While TCR clonality analysis revealed no differences after tumor ablation (Fig. 6a, b), this does not necessarily rule out that tumor ablation may prime tumor-specific immune response in HCC patients as recently described by others [20] due to the fact that prior tremelimumab therapy may have masked a response. Future studies are needed to specifically address the effect of ablative therapies on anti-tumor immune responses.
In summary, tremelimumab treatment resulted in profound clinical and immunological responses. An activation of tumor-specific T cells and a decrease in T-cell clonality was seen in patients upon tremelimumab treatment. An activation of T cells as well as an influx of CD3+CD8+ T cells in the tumor was associated with better outcome. Future studies are needed to address the question whether similar immunological responses can be observed with nivolumab and to specifically address the immunological effects of local ablative therapy in HCC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- AFP
Alpha fetal protein
- CR
Complete response
- CT
Computer tomography
- FDA
Federal Drug Administration
- HCC
Hepatocellular carcinoma
- MRI
Magnet resonance imaging
- MDSC
Myeloid-derived suppressor cell
- NCI
National Cancer Institute
- NIH
National Institutes of Health
- NR
Non-responders
- OS
Overall survival
- PR
Partial response
- PBMC
Peripheral blood mononuclear cell
- PD
Progressive disease
- RFA
Radiofrequency ablation
- R
Responders
- SD
Stable disease
- TCR
T-cell receptor
- TACE
Transarterial chemoembolization
Author contributions
DA, ME and CX planned and conducted the experiments, analyzed the data, and wrote the manuscript with support from MS, WDF and GB. DP and DEK were responsible for pathology data collection and analysis. DV conduced statistical analysis. JAR, CS and ECY performed TCR sequence analysis. BW was responsible for performing biopsies. AGD was the lead in running the clinical trial. FK and TFG designed and developed the clinical trial and also helped in writing and editing the manuscript for publication.
Funding
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and a Cooperative Research and Development Agreement between NCI and Astra Zeneca and NCI and Adaptive Biotechnologies. Tetramers were provided by the NIH tetramer facility. Tim F. Greten is supported by the Intramural Research Program of the NIH, NCI (ZIA BC 011343).
Compliance with ethical standards
Ethical standards
The study with the ClinicalTrials.gov identifier NCT01853618 was approved by the NCI Institutional Review Board (IRB) on 4/10/2013.
Informed consent
All patients provided written informed consent before enrollment onto the study trial to receive treatment. Patients also provided written informed consent prior to procuring biopsied tissue and blood samples to conduct research analysis understanding the deidentified data collected would be used for publication purposes.
Conflict of interest
Julie A. Rytlewski, Catherine Sanders and Erik C. Yusko all have equity and employment with Adaptive Biotechnologies. All other authors declare that they have no conflict of interest.
Footnotes
David Agdashian, Mei ElGindi and Changqing Xie contributed equally to this paper.
References
- 1.Greten TF, Sangro B. Targets for immunotherapy of liver cancer. J Hepatol. 2018;68:157–166. doi: 10.1016/j.jhep.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu AX, Finn RS, Edeline J, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018 doi: 10.1016/S1470-2045(18)30351-6. [DOI] [PubMed] [Google Scholar]
- 4.Duffy AG, Ulahannan SV, Makorova-Rusher O, et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J Hepatol. 2017;66:545–551. doi: 10.1016/j.jhep.2016.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Khoueiry AB, Melero I, Yau T, et al. Impact of antitumor activity on survival outcomes, and nonconventional benefit, with nivolumab (NIVO) in patients with advanced hepatocellular carcinoma (aHCC): subanalyses of CheckMate-040. J Clin Oncol. 2018;36(suppl 4S–abstract):475. doi: 10.1200/JCO.2018.36.4_suppl.475. [DOI] [Google Scholar]
- 6.Duffy A, Zhao F, Haile L, et al. Comparative analysis of monocytic and granulocytic myeloid-derived suppressor cell subsets in patients with gastrointestinal malignancies. Cancer Immunol Immunother. 2013;62:299–307. doi: 10.1007/s00262-012-1332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12:191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robins H, Desmarais C, Matthis J, et al. Ultra-sensitive detection of rare T cell clones. J Immunol Methods. 2012;375:14–19. doi: 10.1016/j.jim.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robins HS, Campregher PV, Srivastava SK, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cha E, Klinger M, Hou Y, et al. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci Transl Med. 2014;6:238ra70. doi: 10.1126/scitranslmed.3008211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrama D, Ritter C, Becker JC. T cell receptor repertoire usage in cancer as a surrogate marker for immune responses. Semin Immunopathol. 2017;39:255–268. doi: 10.1007/s00281-016-0614-9. [DOI] [PubMed] [Google Scholar]
- 12.Snyder A, Nathanson T, Funt SA, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: an exploratory multi-omic analysis. PLoS Med. 2017;14:e1002309. doi: 10.1371/journal.pmed.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegde PS, Karanikas V, Evers S. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res. 2016;22:1865–1874. doi: 10.1158/1078-0432.CCR-15-1507. [DOI] [PubMed] [Google Scholar]
- 14.Duffy AG, Greten TF. Liver cancer: regorafenib as second-line therapy in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2017;14:141–142. doi: 10.1038/nrgastro.2017.7. [DOI] [PubMed] [Google Scholar]
- 15.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manson G, Norwood J, Marabelle A, et al. Biomarkers associated with checkpoint inhibitors. Ann Oncol. 2016;27:1199–1206. doi: 10.1093/annonc/mdw181. [DOI] [PubMed] [Google Scholar]
- 17.Robert L, Tsoi J, Wang X, et al. CTLA4 blockade broadens the peripheral T-cell receptor repertoire. Clin Cancer Res. 2014;20:2424–2432. doi: 10.1158/1078-0432.CCR-13-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wörns M-A, Galle PR. Immune oncology in hepatocellular carcinoma-hype and hope. Lancet. 2017;389:2448–2449. doi: 10.1016/S0140-6736(17)31044-9. [DOI] [PubMed] [Google Scholar]
- 19.Sia D, Jiao Y, Martinez-Quetglas I, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology. 2017;153:812–826. doi: 10.1053/j.gastro.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greten TF, Duffy AG, Korangy F. Hepatocellular carcinoma from an immunologic perspective. Clin Cancer Res. 2013;19:6678–6685. doi: 10.1158/1078-0432.CCR-13-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.