Abstract
Controlling cellular processes with light can help elucidate their underlying mechanisms. Here we present ZAPALOG, a small-molecule dimerizer that undergoes photolysis when exposed to blue light. Zapalog dimerizes any two proteins tagged with the FKBP and DHFR domains until exposure to light causes its photolysis. Dimerization can be repeatedly restored with uncleaved zapalog. We implement this method to investigate mitochondrial motility and positioning in cultured neurons. Using zapalog, we tether mitochondria to constitutively active kinesin motors, forcing them down the axon towards microtubule (+) ends until their instantaneous release via blue light, which results in full restoration of their endogenous motility. We find that one-third of stationary mitochondria cannot be pulled away from their position and that these firmly anchored mitochondria preferentially localize to VGLUT1-positive presynapses. Furthermore, inhibition of actin polymerization with Latrunculin A reduces this firmly anchored pool. Upon release from exogenous motors, mitochondria are preferentially recaptured at presynapses.
Main
Mitochondria are cells’ primary source of ATP and their motility and localization are fundamental to cellular function. This is particularly true in neurons whose branches can be >1m in length and whose survival and function depend on the presence of healthy mitochondria at key points throughout the cell. Much has been learned from traditional tools, such as knock-outs, mutations, and RNAi, but these methods cause chronic changes that can produce lethality or compensatory changes 1-5. Moreover, they act slowly and therefore are poor probes of rapid or transient events. Our understanding of this dynamic process can benefit from the ability to interact directly with discrete components. What determines which mitochondria move? What mechanisms underlie mitochondrial arrest? If mitochondria could be dragged away from their normal positions, would their motility profile change in response? Tools that can directly (and temporarily) control the movement and position of mitochondria could reveal insights about such fundamental processes.1
Many such cell biological tools have been developed to provide direct control of proteins. Chemical Inducers of Dimerization (CIDs), small cell-permeable molecules such as the widely used ‘rapalogs’, are designed to interact with specific protein tags and quickly induce a physical interaction between two tagged proteins 6. Both chemical and genetically-encoded dimerizers have been valuable for the study of axonal transport of mitochondria and other organelles 7-12.
Here we present a CID of potential value to cell biologists. ‘Zapalog’ is a photocleavable small-molecule hetero-dimerizer that can be used to repeatedly initiate, and instantaneously terminate, a physical interaction between two target proteins. We have used zapalog to probe the molecular underpinnings of mitochondrial motility and positioning in the axon by temporarily forcing their motility through the addition of constitutively active kinesins. Through these manipulations we identified and characterized a population of mitochondria restrained by actin-dependent anchoring.
Results
Design and synthesis of zapalog
Several parameters guided design of zapalog. To allow heterodimerization of engineered proteins, we needed a CID with two orthogonal ligand moieties that exhibit high specificity and affinity for their binding domains and lack alternative endogenous binding partners in mammalian cells at useful concentrations. The CID must be membrane permeable and the linker must be long enough to allow both binding domains to be simultaneously engaged without steric interference. The photocleavable linker must respond to a wavelength that is neither cytotoxic nor interferes with imaging common fluorophores. The compound needs to be synthesized on the milligram scale.
The chosen starting point was the heterodimerizer TMP-SLF, a cell-permeable CID effective at low micromolar concentrations 13. The SLF portion binds the FKBP domain of one chimeric partner; the TMP portion binds the e. coli dihydrofolate reductase (DHFR) domain of another (Figure 1a). To render dimerization reversible with light, the alkyl linker between TMP and SLF was replaced with a dialkoxynitrobenzyl (DANB) moiety (Figure 1b). DANB is closely related to the commonly used dimethoxynitrobenzyl (DMNB) chromophore and is similarly susceptible to photolysis by 405nm light14 (Supplementary Figure S1a). This wavelength is short enough to avoid photolysis during imaging of green or cyan fluorophores but less toxic and more spatially precise than near-UV or UV15-17. Neither TMP nor SLF have endogenous protein targets and they exhibit sub-micromolar affinities for their binding domains, which are relatively compact (DHFR = 158 aa; FKBP = 107aa).
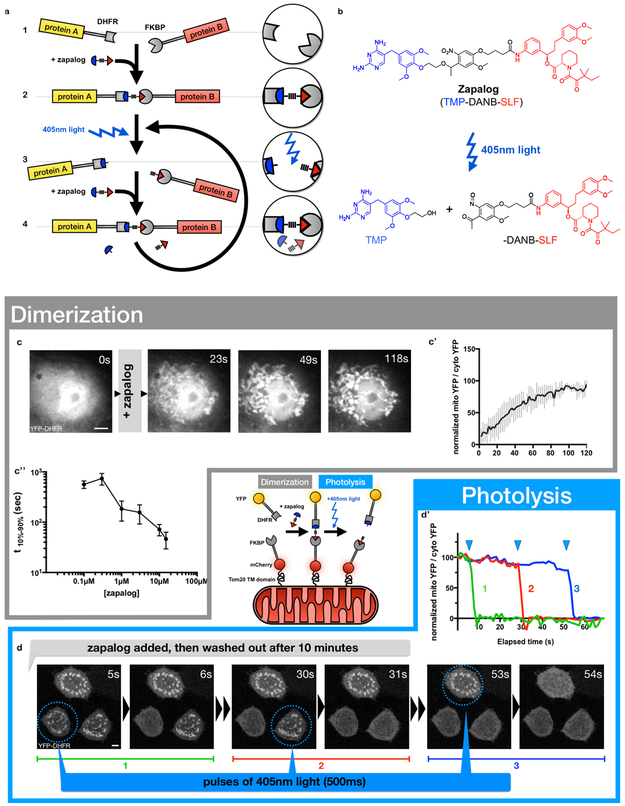
Figure 1. Zapalog, a photocleavable heterodimerizer, can be used to reversibly translocate cytosolic YFP to mitochondria.
a, Schematic illustration of zapalog function: 1. Two proteins of interest are tagged with DHFR and FKBP domains, 2. Addition of zapalog induces dimerization of the tagged proteins, 3. 405nm light photocleaves zapalog, causing rapid dissociation of the dimer, 4. Addition of uncleaved zapalog outcompetes photolyzed zapalog moieties, reestablishing dimerization.
b, Chemical structure of zapalog before and after photolysis of the DANB moiety by 405nm light.
Bottom - Schematic illustration of zapalog-induced translocation of YFP: zapalog attaches cytoplasmic YFP-DHFR-Myc to FKBP domains tethered to mitochondrial outer membranes. Exposure to 405nm light photocleaves zapalog, releasing the YFP-DHFR-Myc back to the cytoplasm.
c, Time-lapsed imaging demonstrates full translocation of YFP-DHFR-Myc onto mitochondria in a COS7 cell, within ~1m after addition of 10μM zapalog to the medium.
c’, Quantification of multiple experiments as in (c) using 10μM zapalog. The ratio of mitochondrial to cytoplasmic YFP-DHFR-Myc was derived from automated image analyses of the datasets, normalized, and average YFP intensity was calculated for each timepoint (n= 5 cells / 3 independent repeats, center value = mean, error bars represent SE, source data in Sup. Table 1)
c”, From assays of YFP translocation to mitochondria, quantified as in (c’), a dose-response curve was generated by plotting time from 10% to 90% of full YFP-DHFR-Myc translocation for each concentration of zapalog administered. (n=98 cells, 6 independent repeats, center value = mean, error bars represent SD, source data in Sup. Table 1)
d, Prior to the series of images shown, YFP-DHFR-Myc translocation to mitochondria was induced by zapalog, as in (c). After wash-out of free zapalog from HeLa cells, the YFP reporter was released from the mitochondria sequentially in each of the 3 cells by exposing the circled region to a brief (500ms) pulse of 405nm light.
d’, Quantification of data shown in (d), demonstrating that photolysis of zapalog is rapid, complete, and spatially localizable.
scale bars = 5µm
Zapalog is photosensitive, cell-permeable, non-toxic and effective as a non-covalent dimerizer.
We commissioned the synthesis of zapalog (detailed in Methods, Supplementary Figure S2, and at Nature Protocol Exchange 18 ) and obtained 10 mg of TLC-purified material. With 405nm light zapalog in solution underwent the expected photocleavage schematized in Figure 1b (Supplementary Figure S1a). Zapalog was not affected by 458nm light, making it suitable for use with CFP imaging (Supplementary Figure S1b).
To assess in living COS7 cells zapalog’s ability to function as a hetero-dimerizer and undergo photocleavage, we modified a fluorophore-translocation assay previously established for rapalogs 19: we tagged the surface of mitochondria with Tom20-mCherry-FKBP and co-expressed cytoplasmic YFP-DHFR-Myc (Figure 1c,d). We used a spinning disk confocal microscope for live imaging of cytoplasmic YFP and mitochondrial mCherry at 2 frames/sec. Addition of zapalog to the media induced dimerization of the constructs and YFP rapidly cleared from the cytoplasm and increased on mitochondria (Figure 1c and Supplementary Movie 1). We quantified the progression of YFP translocation by measuring cytoplasmic and mitochondrial YFP levels in the entirety of each cell (Figure 1c’; see methods) and plotted the time from 10% to 90% YFP translocation as a function of zapalog concentration (Figure 1c”). Zapalog exhibited equivalent on-rate kinetics to previously reported CIDs and an EC50 of ~100nm 13. The time course of binding is likely to be faster than indicated because the measured time-course for YFP recruitment relies mainly on zapalog diffusion in the bath and into the cell, and fluorophore diffusion to the mitochondria. To test the efficiency and speed of reversing the YFP translocation with exposure to 405nm light, we repeated YFP translocation onto mitochondria as above, but removed zapalog from the media after 10min. YFP recruitment to mitochondria persisted after zapalog was washed out, evidence of zapalog retention in the cell. We used a laser scanning confocal to sequentially flash 405nm light separately onto each cell. As shown in Figure 1d, d’ and Supplementary Movie 2, exposure of each cell to a single 500msec flash of blue light proved sufficient to fully dissociate YFP localization from mitochondria in <1sec.
Zapalog can induce multiple rounds of dimerization.
We hypothesized that zapalog could induce multiple rounds of dimerization: if a focused light beam photocleaved a fraction of the total pool of zapalog in the cell, the locally cleaved ligands could potentially be replaced via competitive binding of uncleaved molecules from the surrounding area. The resulting re-dimerization after the photolysis would open the possibility for multiple cycles of cleavage and re-dimerization, potentially on individual mitochondria. Because mitochondria form a highly fused network we first fragmented the network in HeLa cells with a 6hr incubation in CCCP20-22 to prevent lateral diffusion of bound YFP within the network. We then repeated the fluorophore translocation assay on individual mitochondria by using a confocal laser scanning microscope to photocleave zapalog in a 2µm × 2µm region of interest (ROI). Zapalog recruited cytoplasmic YFP-DHFR-Myc back to mitochondria within <1 min. Subsequently, each 500msec pulse of 405nm light caused YFP-DHFR-Myc to dissociate from the mitochondrion within the ROI with subsequent rebinding, resulting in multiple cycles of fluorophore translocation to and release from the mitochondrial surface (Figure 2a and Supplementary Movie 3.) This assay differs in scale from the previous assay: full re-association of YFP with the mitochondrion after photolysis now requires equilibrating only a ~4µm3 volume rather than the entire cell. This results in faster on rates than when zapalog needs to re-enter and reload the entire cytosol. We also repeated this assay on peroxisomes, which do not form a reticulum, by exchanging the Tom20-mCherry-FKBP plasmid with the peroxisome-targeting PEX3-mRFP-FKBP and obtained similar translocation dynamics (Figure 2b and Supplementary Movie 4). These focal assays confirm that zapalog is an effective dimerizer that can be photolyzed fully within <1sec and then replaced quickly and repeatably with unlysed zapalog as needed.
Figure 2. Zapalog can induce multiple rounds of dimerization.
a, (below) HeLa cell expressing mitochondrial Tom20-mCherry-FKBP and YFP-DHFR-Myc incubated with 10μM CCCP for 6hrs to fragment mitochondria, then 2μM zapalog for 5min, which has caused YFP-DHFR-Myc to localize to outer mitochondrial membranes. (above) A blown-up image of the area defined below with a yellow square within which two mitochondrion-containing 2μm × 2μm ROIs (dotted white squares) were defined and imaged at high temporal resolution. In the time series to the right, one of the ROIs, (+) was periodically illuminated with a 405nm laser (~300nW) for 500msec, the other (−) was not. Each flash of blue light causes photolysis of zapalog and immediate reversal of the dimerization in ROI (+) but not (−). Each instance of YFP-DHFR-Myc removal from a mitochondrion is followed by rapid re-dimerization (~30sec) due to influx of unlysed zapalog from outside of the ROI, which outcompetes the lysed zapalog fragments bound to FKBP and DHFR.
a’, schematic illustration of repeated fluorophore translocation mediated by zapalog.
a”, quantification of normalized YFP signal within discrete mitochondrion-containing ROIs (2µm × 2µm) that were illuminated by 405nm laser, and nearby ROIs that were not. (n=7 cells / 3 independent repeats, center value = mean, error bars represent SE, source data in Sup. Table 1)
b, (below) COS7 cell expressing peroxisomal PEX3-mRFP- FKBP and YFP-DHFR-Myc incubated with 2uM zapalog for 5min, which has caused YFP-DHFR-Myc to localize to peroxisomal membranes. (above) Two peroxisome-containing 6μm × 6μm ROIs were defined and imaged at high temporal resolution. In the time series to its right, one of the ROIs, (+) was illuminated with a 405nm laser (10%) for 500msec every 40sec, the other (−) was not. Each flash of blue light causes photolysis of zapalog and immediate reversal of the dimerization in ROI (+) but not (−). Each instance of YFP-DHFR-Myc removal from peroxisomes is followed by rapid re-dimerization (~30sec) due to influx of unlysed zapalog from outside of the ROI, which outcompetes the lysed zapalog fragments bound to FKBP and DHFR.
b’, schematic illustration of repeated fluorophore translocation mediated by zapalog.
b”, quantification of normalized YFP signal within peroxisome-containing ROIs (5µm × 5µm) that were illuminated by 405nm laser, and nearby ROIs that were not. (n=4 cells / 2 independent repeats, center value = mean, error bars represent SE, source data in Sup. Table 1)
Zapalog-induced mitochondrial motility reveals distinctions among stationary axonal mitochondria.
To probe the mechanisms of mitochondrial positioning and movement in axons, we asked whether zapalog could tether a kinesin to mitochondria with sufficient affinity to drag mitochondria towards the (+) ends of microtubules, i.e. away from the soma. We cultured E18 rat hippocampal neurons in spot-groove cultures (see methods), and co-transfected them with Tom20-mCherry-FKBP and Kif1a(1–489)-DHFR-Myc, a truncated, constitutively active version of Kif1a that includes only the motor and neck-linker domains8. Prior to addition of zapalog, ~30% of mitochondria were motile and ~70% stationary, consistent with previous reports 2. Within one minute of 2µM zapalog addition, the majority of mitochondria (60.3% ±12.2) begin to move rapidly anterograde at an average speed of 2.5µm/sec) (Figure 3). Immunocytochemistry with anti-Myc confirmed that zapalog recruits the tagged motors to all mitochondria (Supplementary Figure S3a). The percentage of retrograde moving mitochondria decreased from 13.99% to 3.52%, suggesting that the exogenous kinesin overcomes the dynein motors that had been moving mitochondria retrograde. Despite zapalog-induced recruitment of the kinesin to all mitochondria, 36% of axonal mitochondria remained stationary, seemingly resisting the zapalog-induced pull. The inability of the activated motors to mobilize one-third of the axon’s mitochondria prompts us to expand on the widely used classification of mitochondria as either “motile” or “stationary” - rather this assay subdivides mitochondria into “motile”, “movable” or “immovable” under the conditions of the assay, a distinction that implies different mechanisms or degrees of mitochondrial arrest (Supplementary Figure S3b and Supplementary Movie 5).
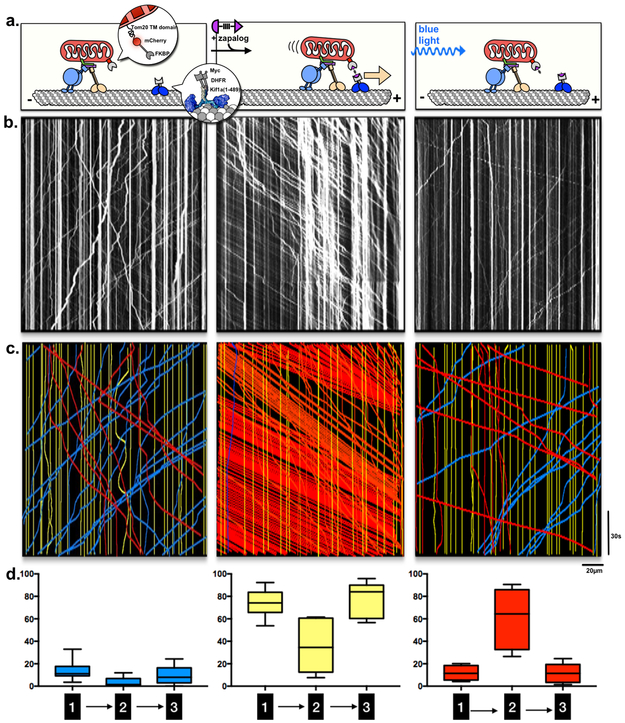
Figure 3. The mitochondrial motor complex retains its activity in the wake of induced translocation.
a, Schematic illustration of zapalog-induced mobilization of axonal mitochondria that shows three phases of the experiment that correspond to the kymographs below:
1. E18 rat hippocampal neurons are transfected with mitochondrial Tom20-mCherry-FKBP and the constitutively active kinesin motor Kif1a(1-489) tagged with DHFR-Myc (enlarged in inserts). Mitochondrial motility is governed by their normal motors and anchors.
2. Addition of zapalog induces attachment of the Kif1a motors to mitochondria and consequently mitochondria are dragged towards the (+) ends of microtubules.
3. Exposure to 405nm light causes the exogenous motors to detach immediately and completely from mitochondria.
b, Representative kymographs. A line-scan of the mCherry signal (x axis, denoting mitochondrial position along an axon) is plotted against time (downwards, y axis, 5 minutes). For each axon imaged, 5min of data were acquired during each of the three phases illustrated above in (a).
c, Motility was analyzed for each kymograph in (b). Vertical yellow lines signify stationary mitochondria; diagonal lines signify mitochondria moving in the retrograde(blue) or anterograde (red) directions.
d, Quantification of motility profiles, plotting “% time spent in motion” for each direction for each of the three phases of the assay. (n=8 / 3 independent repeats; box-and-whisker plots display statistical median (center value), upper and lower quartiles (boxes), and max values (whiskers), source data in Sup. Table 1; Kif1a image adapted from 46)
One necessary consideration in analyzing motility after zapalog is that the “wave” of anterograde-moving mitochondria pile up at growth cones (Supplementary Figure S3c and Supplementary Movie 6) and are gradually depleted elsewhere (Supplementary Figure S3d and Supplementary Movie 7). In some cases, mitochondria piled up at bends or constrictions in mid-axons; such blocked axons appeared unhealthy, fragmented within 10min, and were eliminated from further analysis.
The Mitochondrial Motor Complex Retains its Activity After Release from Zapalog-induced Movement.
We tested the effect of temporarily forced mobilization on endogenous mitochondrial motility. Previous studies used rapalog to attach motors to mitochondria and thereby overpower the endogenous motors10. However, rapalogs are irreversible and therefore cannot be used to determine what happened to the endogenous motor complex when it was superseded. We quantified axonal mitochondrial motility for 5min before zapalog addition, 5min after 2µM zapalog addition, and 5min after zapalog was completely photocleaved (Figure 3 and Supplementary Movie 5). After photocleavage, the parameters of mitochondrial motility to return to their initial values (Figure 3c, d). With Pearson’s chi-squared tests we compared motility before zapalog and after its photolysis; the variability within each parameter (i.e. ‘% retrograde / stationary / anterograde’) originates from axon-to-axon differences (p<0.005) and not as a consequence of zapalog-induced disruption (p>0.1). Thus, the mitochondrial motility profile of an axon is inherently robust, and able to re-establish itself even after a prolonged (5min) zapalog-induced disruption. As mentioned previously, an exception to this robustness is axons with mitochondrial pileups; the pileups did not resolve readily after photocleavage, likely because the induced proximity had caused a hyperfused mass to form that cannot be easily mobilized (Supplementary Figure S3c). Similarly, if prolonged exposure depletes most of the motile mitochondria proximal to the imaged region, anterograde movement into that region will be depressed (Supplementary Figure S3d).
Anchored mitochondria colocalize with VGLUT1.
We were surprised to find that consistently ~20% of mitochondria remain firmly in place even while hundreds of mitochondria are pulled past them by zapalog-enabled exogenous motors. We wished to further characterize this pool of mitochondria that were immoveable in the assay. Previous studies have pointed to several mechanisms promoting arrest of mitochondria specifically at presynaptic loci 23,24. Likely presynaptic loci can be identified in axons as stable puncta of the presynaptic marker VGLUT1 25. We repeated the forced mobilization assay in neurons co-transfected with VGLUT1-Venus (Figure 4a). ~60% of VGLUT1-positive sites colocalized with mitochondria prior to the addition of zapalog, and ~70% of the stationary mitochondria co-localized with stable VGLUT1-Venus puncta. Upon zapalog-induced recruitment of the Kif1a motor, the number of stationary mitochondria occupying VGLUT1-positive loci was unchanged, but the number of stationary mitochondria that were not colocalized with VGLUT1-Venus (Figure 4b) was reduced. Thus, the mitochondria that co-localized with stable VGLUT1 puncta were resistant to the zapalog-enabled pull. This pool of presumably presynaptic mitochondria is therefore not passively stationary, due for example to inactivation of their motors, but are strongly anchored in place, and more strongly than the extra-synaptic stationary mitochondria.
Figure 4. Mitochondria that colocalize with VGLUT1 are unmoved by Kif1a recruitment.
a, Axonal kymographs from live- imaging of neurons co-transfected with Kif1a(1-489)-DHFR-myc, Tom20-mCherry-FKBP and the presynaptic marker VGLUT1-Venus, before addition of 1μM zapalog (1) and after addition of zapalog (2). The mitochondria that are stationary in the presence of zapalog constitute the anchored pool.
b, Quantification of the number of stationary mitochondria before and after addition of zapalog, subdivided into two groups: stationary mitochondria that colocalize with VGLUT1-Venus and stationary mitochondria that do not colocalize with VGLUT1-Venus. (n=28 axons / 4 independent repeats; two-sided paired t-tests; ns = p>0.05, **** =p≤0.0001, source data in Sup. Table 1).
Anchoring of mitochondria involves actin.
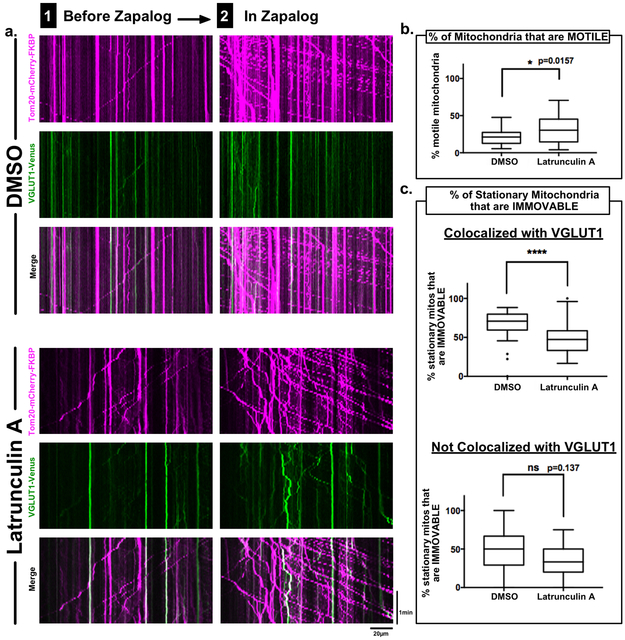
To test a potential anchoring role of the actin cytoskeleton, we pre-treated neurons with Latrunculin A, a blocker of actin polymerization, in a protocol that eliminated all detectible actin filaments without disrupting axonal tubulin or VGLUT1 (Supplementary Figure S4). Prior to addition of zapalog, Latrunculin A had no detectable effect on the number of stationary mitochondria that localized to VGLUT1-Venus-positive sites (Figure 5a). A slight increase in the number of motile mitochondria was found, consistent with other reports that actin can restrain mitochondrial movement (Figure 5b) 26 . However, addition of zapalog led to a significantly smaller proportion of mitochondria in the VGLUT1-associated pool remaining “unmoved” compared to control (Figure 5c). For the smaller pool of stationary mitochondria that did not colocalize with VGLUT1 puncta, the variance of the data was too great to determine if they were also more moveable in Latrunculin A (Figure 5c). Our data strongly suggest that mitochondrial retention at presynaptic sites depends in part on interactions with the actin cytoskeleton.
Figure 5. Actin contributes to anchoring of mitochondria at synapses.
a, Axonal kymographs from live-imaging of neurons co-transfected with Kif1a(1-489)-DHFR-Myc, Tom20-mCherry-FKBP and the presynaptic marker VGLUT1-Venus, incubated for 6hrs with either DMSO or 2.5μM Latrunculin A, and imaged before (1) and after (2) addition of 1μM zapalog.
b, Quantification of the percentage of mitochondria that are motile before addition of zapalog in neurons incubated in DMSO vs neurons incubated with Latrunculin A.
c, Quantification of the percentage of mitochondria that are anchored, i.e. remain stationary even after addition of zapalog, subdivided into two groups: those that colocalize with VGLUT1-Venus and those that do not colocalize with VGLUT1-Venus.
n = 28 axons ea / 4 independent repeats; box-and-whisker plots display statistical median (center value), upper and lower quartiles (boxes), max values (whiskers), and outliers (dots); KS nonparametric tests; ns = p>0.05, * = p≤0.05, **** =p≤0.0001, source data in Sup. Table 1.
Capture of mitochondria at VGLUT1-positive loci.
What happens to mitochondria that were pulled away from their normal positions after they are released from the zapalog-tethered motors? Specifically, in an axon segment in which every moveable mitochondrion has been moved away, do returning mitochondria stop at random places or are they prone to capture at sites that previously held a stationary mitochondrion, such as presynaptic loci? To address this question, we followed mitochondria at high temporal resolution during zapalog-induced mobilization (which was maintained for >10min), and on through 40min after photolysis (Figure 6a). The long zapalog treatment caused most of the moveable mitochondria to be transported to distal axonal regions and therefore most of the movement after release was necessarily retrograde. We marked each event of mitochondrial capture, which we defined as a motile mitochondrion that stops for at least 5min. We normalized the number of capture events to the length of axon that had held stationary mitochondria prior to zapalog treatment (i.e. previously occupied places) vs. the length of axon that hadn’t and observed a clear bias towards previously mitochondria-occupied places (Figure 6b, c) and presynaptic loci (Figure 6d). Because most presynaptic sites had retained an anchored mitochondrion, it was possible that contact with a stationary mitochondrion caused the motile mitochondrion to stop preferentially at those sites. Alternatively, the preference for these sites could be due to an intrinsic factor that made them ‘hotspots’. To distinguish among these possibilities, we normalized the number of capture events to the number of stationary mitochondria occupying nascent presynaptic sites in the axon vs. those that are stationary but not presynaptic. Mitochondria were 7.2-times more likely to stop at the presynaptic locations than elsewhere.
Figure 6. Mitochondria tend to be captured at axonal hotspots that are VGLUT1positive.
- a, Axonal kymographs from live- imaging of neurons co-transfected with Kif1a(1-489)-DHFR-myc, Tom20-mCherry-FKBP and the presynaptic marker VGLUT1-Venus. Axonal mitochondria were imaged: (1) prior to zapalog addition, (2) during zapalog-induced mitochondrial mobilization, and (3) following photolysis as they returned to the imaged region. Mitochondrial capture events were defined as any event wherein a motile mitochondrion becomes stationary for at least 5min.
b, Quantification of the number of capture events that coincided with a space that contained a stationary mitochondrion prior to zapalog addition vs. capture events at all other loci along the segment (normalized to μm of mitochondria-occupied axon and μm of remaining axon).
c, Quantification of the number of capture events that coincided with anchored mitochondria vs. capture events at all other loci along the segment (normalized to μm of anchored mitochondria and μm of remaining axon).
d, Quantification of number of capture events that coincided with presynaptic loci (defined as stationary VGLUT1-Venus in phase 1) vs. capture events at an equal number of randomly generated loci (see methods)
(n=11 axons / 3 independent repeats; box-and-whisker plots display statistical median (center value), upper and lower quartiles (boxes), max values (whiskers), and outliers (dots); two-sided paired t-tests; *** = p≤0.001, **** =p≤0.0001, source data in Sup. Table 1.
Discussion
To date, several systems for controlling the localization and consequently the activity of cellular machinery have been developed6,11,27-30. Rapalogs (i.e. rapamycin-analogs) are prominent; these non-toxic, cell-permeable CIDs force a non-covalent interaction between proteins genetically tagged with the ~11kDa domains FKBP and FRB. Rapalogs, however, lack spatiotemporal control and are irreversible 31. This has prompted the development of additional genetically encoded, photoreactive systems such as TULIP 32, PhyB-PIF 33, and CRY2-CIB1 34. Versatile and useful, these optogenetic methods have some limitations: dimerization can be leaky 29 and reversibility is often slow (TULIP is the fastest at 10s of seconds). Improved CIDs have also appeared. SLF’-TMP is as effective as rapalog but slowly reversible (~10min) when outcompeted by monomeric TMP and therefore repeatable 31. MeNV-HaXS is photocleavable by UV but binds covalently, making its reversal fast but not repeatable 35. We developed zapalog to provide a CID that can bridge some of these gaps and facilitate the manipulation of cargo transport in axons. Derived from TMP-SLF, zapalog can dimerize any two proteins tagged with the FKBP and DHFR domains. Unlike TMP-SLF, zapalog undergoes photolysis by blue light, instantaneously reversing the dimerization. Since binding is not covalent, re-dimerization is then possible via competition with new, non-photocleaved zapalog molecules, making zapalog a CID that is both repeatable and photocleavable.
Zapalog can be incorporated into many experimental paradigms already set up for rapalog, thereby endowing them with precise spatiotemporal control and repeatability. A protein of interest can be forced to repeatedly interact with its target or, conversely, be sequestered away from target sites and then released instantaneously within the confines of a subcellular region of interest. The active domain of a protein of interest can in theory be coupled to its localization domain, recreating the protein’s functional form while rendering it vulnerable to inactivation with light.
We used zapalog to investigate what determines the positioning and movement of axonal mitochondria? Zapalog allowed us to discriminate different classes of stationary mitochondria and to document axonal hotspots for mitochondrial capture and resilient anchoring that are principally at presynaptic specializations.
Zapalog can tether Kif1a motor domains to mitochondria with sufficient affinity to produce highly processive anterograde movement at 2.5 µm/sec. Truncated, constitutively active motors move faster than their full length counterparts8, and the observed velocity is consistent with the known velocity of the Kif1a motor36 and, as expected, faster than the endogenous mitochondrial velocity mediated by the slower Kif5c1,5. Most mitochondria could be dragged from their pre-existing locations by tethering them to the constitutively-active Kif1a motors. Removing these motors restored their endogenous motility profile.
Throughout this study, we divided the mitochondrial motility assay into three phases: 1- prior to zapalog addition, 2- in the presence of active zapalog, 3- following zapalog photolysis. Ideally one might wish to track an individual mitochondrion through all three phases and determine, for example, if a retrogradely moving mitochondrion immediately returned to retrograde movement upon being freed from the tethered Kif1a. However, our ability continuously to track an individual mitochondrion through all three phases was limited. A mitochondrion that cannot be resolved for even one frame can no longer be identified as the same mitochondrion. To reliably follow any single mitochondrion from one time point to the next, we must balance high spatiotemporal resolution with a field of view large enough to ensure that target mitochondria do not exit the field. The average axon segment imaged was 440µm. When tethered to Kif1a and moving 2.5µm/sec, a mitochondrion in the center of the field would exit the field in 88 seconds; this limited our ability to track an individual mitochondrion through all three phases, especially for mitochondria that were motile prior to zapalog addition. Nevertheless, we have successfully recorded many instances of mitochondria that are initially stationary (in phase 1), mobilized by zapalog (in phase 2), and then immediately stationary upon zapalog photolysis (in phase 3). We have recorded only a few instances of mitochondria that are initially moving retrograde (in phase 1), dragged anterograde by zapalog (in phase 2), and then immediately revert to retrograde motility upon zapalog photolysis (in phase 3; see Supplementary Figure S3b). Even these handful of recorded instances carry some significance, as they indicate that when a mitochondrion is moving the retrograde, dragging it in the opposite direction does not necessarily reset its endogenous motor-adapter complex, but merely overpowers it, a manifestation of molecular tug-of-war previously demonstrated in vitro 37. Given the technical limitations in following individual mitochondria, we opted instead to characterize mitochondrial populations during each of the three experimental phases and found that, at the population level, the original balance of anterograde and retrograde motility was restored in the wake of the forced anterograde movement.
Our observations required expanding the standard motile vs. stationary categorization of mitochondria into three categories: motile, immotile but movable, and immovable under our experimental conditions. Mitochondrial motility is mediated by motor adapter complexes that include the motors Kinesin-1 and dynein/dynactin and the adapter proteins Milton/TRAK1/2 and Miro/RhoT1/2 1. Expression of fluorescently tagged Kinesin-1 previously indicated that all axonal mitochondria normally possess this motor 38, regardless of their direction or stationary state. Motility can be terminated by inhibition of a motor protein 38, disassociation of the complex (“motor shedding”) 39, non-processive, anchoring interactions with MTs via syntaphilin 40, and anchoring interactions with myosin and actin microfilaments 26,41. By using zapalog to pull mitochondria, we are able to distinguish between mitochondria that fail to move for passive reasons (i.e. they lack active motors) or have been weakly anchored, and those that are firmly anchored in place and can’t be moved. Mitochondrial may vary in their degrees of resistance to motor pull, rather than binary states of movable and firmly anchored. However, we do find that the ability of a mitochondrion to resist the induced pull correlated with its position: movable mitochondria are scattered along the axon, but firmly anchored mitochondria are mostly found at likely presynaptic sites. This corroborates recent evidence of mitochondrial capture at presynaptic sites 23,42,43 .
While disruption of actin microfilaments with Latrunculin A only slightly increased the motile pool of mitochondria, the number of immoveable mitochondria at VGLUT1-positive presynaptic sites was reduced by 24%. Attachment to actin filaments, either through myosin or another adapter, therefore, plays an important role in presynaptic mitochondrial anchoring. Yet 76% of synaptic mitochondria were still immovable even after actin microfilaments were disrupted; additional mechanisms must therefore underlie mitochondrial anchoring, as expected, such the known role of Syntaphilin in mitochondrial localization to synapses 3.
Upon release of the tethered Kif1a returning mitochondria overwhelmingly tended to stop at sites that had held stationary mitochondria prior to the addition of zapalog, as if there were defined “parking spaces” at which mitochondria will preferentially reside. These parking spaces were largely VGLUT1-positive rather than random locations. The preference of mitochondria for these sites appeared to be intrinsic to the “parking space” rather than attributable to collision with a resident mitochondrion; mitochondria did not show an equivalent propensity to stop at sites that held a mitochondrion but were not VGLUT1 positive. This finding solidifies the notion that the axon is not uniform; rather it contains microenvironments that promote mitochondrial arrest and anchoring. Taken together, an active, ongoing mechanism is implicated for presynaptic mitochondrial anchoring that is in part actin-based. The population of immovable mitochondria are not only stationary, but strongly resistant to the pull of active motors. Long-term restraint of synaptic mitochondria may be an evolutionary adaptation that guarantees adequate ATP production and Ca2+ buffering for the high energy demands of synapses and does so more robustly than would be achieved by transient arrest, such as the response to elevated cytosolic Ca2+ 44,45,38,46.
Cell biological studies often rely on methods that slowly change gene expression or protein activity. Integration of photochemistry into the tool kit of pharmacology and genetics has enabled direct and spatiotemporally precise manipulation of molecules within living cells. By directly and reversibly interfering with axonal transport of mitochondria, zapalog yielded insights into the mechanisms underlying mitochondrial localization. Zapalog, as a repeatable and instantaneously photocleavable dimerizer, should prove to be useful for other inquiries in molecular cell biology.
Methods
Synthesis of compound
Zapalog was prepared by Medicilon Inc. (Shanghai, China) as detailed in Supplementary Figure S1 and in Nature Protocol Exchange48.
Preparation of A - Trimethoprim (500 mg, 1.72 mmol) was suspended in 40% HBr (7 mL) at room temperature. The mixture was then stirred at 100 °C for 30 min. LCMS detected the desired product, the reaction mixture was adjusted to pH 7 with 1N NaOH. The precipitate was collected by filtration and dried to give 180 mg compound A. The reaction was repeated at 5 g scale to give A 1.5 g. 1H NMR (400 MHz, d-DMSO): δ 8.11 (s, 1H), 7.46(s, 1H), 6.49(s, 2H), 6.03(s, 2H), 5.67(s, 2H), 3.70(s, 6H), 3.47(s, 2H).
Preparation of B-2 - To a solution of B-1 (500 mg, 3.0 mmol) in DMF (2 ml) added K2CO3 (632 mg, 4.6 mmol) and ethyl 4-bromobutyrate (585 mg, 3.0 mmol). The reaction mixture was stirred at room temperature overnight, and then heated for 3 h at 50 °C. The solution was extracted with ethyl acetate, washed with H2O and dried over Na2SO4 .The solvent was removed to give 800 mg of B-2 as a white solid in 95% yield. The reaction was repeated at 10 g scale to give B-2 16.2 g.
Preparation of B-3 - A solution of B-2 (500 mg, 1.78 mmol) in 1.5 ml acetic acid was slowly added to a solution of 65% HNO3 (10 ml) and acetic anhydride (2 ml) at 0 °C. The reaction was stirred for 3 h, poured into ice-cold water. The precipitate was immediately collected by filtration, washed extensively with water, dried under vacuum to give 350 mg of B-3 as a pale yellow solid in 61% yield. The reaction was repeated at 16 g scale to give B-3 11 g.
Preparation of B-4 - To a solution of B-3 (350 mg, 1.08 mmol) in 26 ml MeOH at 0 °C was slowly added NaBH4 (105 mg, 2.78 mmol) in portions. The reaction was stirred for 3 h, quenched by addition of 20 ml NH4Cl (aq.). The reaction mixture was extracted with ethyl acetate, washed with brine and dried over Na2SO4. After removal of the solvents in vacuo, the crude product was purified by flash chromatography (petroleum ether: ethyl acetate = 2:1) to give 280 mg of B-4 as a white solid in 79% yield. The reaction was repeated at 11 g scale to give B-4 9.6 g.
Preparation of B-5 - To a solution of B-4 (100 mg, 0.305 mmol) and KI (51 mg, 0.305 mmol) in 2 ml DMF at 0 °C was slowly added NaH (14 mg, 0.336 mmol), the mixture was stirred at r.t. for 10min, allybromide (46 mg, 0.367 mmol) was added. The reaction was stirred at 70 °C overnight, water and EtOAc were added, the organic layer was washed with water and brine, dried by Na2SO4, concentrated and purified by prep-TLC. 1H NMR showed the structure was correct. The reaction was repeated at 1 g scale to give B-5 916 mg.
Preparation of B-6 - To a solution of B-5 (60 mg, 0.163 mmol) and osmium tetraoxide (1 mg,0.004 mmol) in 1 ml THF and 1 ml water was added Sodium periodate (140 mg, 0.653 mmol) at r.t. under Ar. Then the reaction was stirred at 50 °C for 2h, TLC showed the reaction was consumed completely. The mixture was purified by prep-TLC (1H NMR showed the product is not pure). The reaction was repeated at 910 mg scale to give B-6 700 mg.
Preparation of B-7 - To a solution of B-6 (30 mg, 0.081 mmol) in MeOH (3 mL) was added NaBH4 (3 mg, 0.081 mmol) at 0 °C and the reaction was stirred at r.t. for 2h, TLC showed the reaction was consumed completely. The mixture was purified by prep-TLC. 1H NMR showed the structure was correct. The reaction was repeated at 700 mg scale to give B-7 370 mg.
Preparation of B - To a solution of B-7 (100 mg, 0.291 mmol) and CBr4 (116 mg, 0.350 mmol) in DCM was added PPh3 (92 mg, 0.350 mmol) 0 °C under Ar. Then the reaction was stirred at r.t. overnight. The mixture was purified by prep-TLC. 1H NMR showed the structure was correct. The reaction was repeated at 700 mg scale to give B 630 mg. 1H NMR (400 MHz, CDCl3): δ 7.59 (s, 1H), 7.32(s, 1H), 5.23(d, J = 6.0 Hz, 1H), 4.17–4.10(m, 4H), 3.99(s, 3H), 3.70–3.47(m, 4H), 2.54(t, J = 7.2Hz, 2H), 2.19(m, 2H), 1.54(m, 3H), 1.27(t, J = 7.2 Hz, 3H).
Preparation of 5 - To a mixture of A (10 mg, 0.036 mmol) in DMF (0.5 mL) was added K2CO3 (6 mg, 0.040 mmol), and the mixture was stirred at r.t. for 10min. Then B (16.5 mg, 0.040 mmol) was added to this mixture, the mixture was stirred at 40 °C for 3h under Ar, LCMS showed the desired mW ion peak. The mixture was purified by prep-TLC. LCMS and 1H NMR confirmed the structure. The reaction was repeated at 260 mg scale to give compound 5 140 mg. 1H NMR (400 MHz, CDCl3): δ 7.68 (s, 1H), 7.58 (s, 1H), 7.38 (s, 1H), 6.35 (s, 2H), 5.51(br s, 2H, 5.23(m, 1H), 5.00(s, 2H), 4.17–4.10(m, 4H), 3.91(s, 3H), 3.77(s, 6H), 3.59(s, 3H), 3.58(m, 1H), 2.52(t, J = 7.2Hz, 2H), 2.18(m, 2H), 1.51(t, J = 6.0 Hz, 3H), 1.26(t, J = 7.2 Hz, 3H).
Preparation of 6 - To a solution of 5 (20 mg, 0.032 mmol) in 0.5 ml THF and 0.5 ml water was added LiOH.H2O (5 mg, 0.119 mmol) at 0°C under Ar. Then the reaction was stirred at r.t. overnight, LCMS showed that the SM was consumed completely. The mixture was purified by prep-TLC to give 15 mg compound 6. The reaction was repeated at 120 mg scale to give compound 6 70 mg.
Preparation of TMP-DANB-SLF - To a solution of 6 (15 mg, 0.025 mmol) in 0.5 ml DMF was added DIEA (6 mg, 0.045 mmol) and HATU (12 mg, 0.030 mmol) at 0°C under Ar. The mixture was stirred for 10min. Then SLF-NH2 (16 mg, 0.030 mmol) was added to the reaction solution, the mixture was stirred at r.t. overnight. LCMS showed the reaction was difficult to purify. The reaction was repeated at 40 mg scale, and the mixture was purified by prep-TLC to give 30 mg crude product (purity: 82.3%), then the crude was purified by HPLC to give 11.7 mg product (purity: 93.6%). Further purifications by prep-TLC gave the target compound. The reaction was repeated at 30 mg scale, and two batches of product were combined and twice purified via prep-TLC to give 12.0 mg target compound. MS: 1108[M+1]+. HPLC Purity @ 254 nm: 98.2%
Photolysis in Solution
To achieve partial photolysis, a 40µl sample of 1mM Zapalog in phosphate-buffered saline, pH 7.2, contained in a quartz cuvette, were exposed for 5s to ~15 mW of light from a 0.22 NA fiber optic cable (ThorLabs) coupled to a 405nm laser (Vortran Stradus 405–250; Vortran Laser Technology). Both purity and photolytic cleavage of Zapalog were confirmed by reverse-phase high-performance liquid chromatography (Agilent Technologies) on a 4.6 mm 150 mm Zorbax SB-C8 column (5 mm particle size) in H2O, CH3CN, 0.1% TFA at 1 ml/ min. Samples were monitored at 220 nm and 350 nm. A non-photolyzed zapalog sample was prepared and run under the same conditions. (Supplementary Figure S1a)
Testing Sensitivity to 458nm imaging light
To determine if zapalog is photolyzed by 458nM light, a wavelength frequently used for imaging, a 20µL solution of 1mM zapalog dissolved in PBS was placed on a glass coverslip in an upright confocal microscope and illuminated with ~100µW 458 nm laser irradiation through a 10X objective for 1 minute, or simply left in the dark, and then analyzed by HPLC (5% acetonitrile in water with 0.1% TFA gradient to 100% over 10 minutes). (Supplementary Figure S1b)
Molecular cloning
The following DNA constructs were previously published and used in this study: Tom20-mCherry-FKBP 49, PEX-mRFP-FKBP 50, VGLUT1-Venus 51.
Kif1a(1–489)-DHFR-myc (Addgene #117833) was generated by PCR and restriction cloning. The motor and neck linker domains of rat Kif1a were amplified from a plasmid containing full length Kif1a (kindly provided by Kristen Verhey, U. Michigan) using the following primers:
Kif1a(489)-XhoI (30-mer): GTCAAGCTCGAGACCATCTTCCCTCATGGC
BamHI-Kif1a (27-mer): TACAGTGGATCCATGGCTGGGGCCTCT
Afterwards, both the PCR product and pDHFR-myc (Addgene #20214) were cut with BamHI and XhoI and the Kif1a motor fragment was inserted 5’ to the DHFR domain.
YFP-DHFR-Myc (Addgene #118852) was generated by restriction cloning: Both pDHFR-myc (Addgene #20214) and YFP-FRB (Addgene #20148) were cut with XbaI and NdeI and the YFP-encoding fragment was inserted upstream of DHFR.
Culturing COS7 and HeLa cells
COS7 and HeLa cells were cultured in DMEM, high glucose, GlutaMAX™ (ThermoFisher Scientific) supplemented with 1% penicillin/streptomycin (10,000U/mL; ThermoFisher Scientific), and 10% FBS (Atlanta Biological). DNA transfections in COS7 and HeLa cells were performed with GenJet (SignaGen Laboratories).
Culturing rat hippocampal neurons
Rat hippocampal neurons were isolated according to standard procedures and cultured either as Spot-groove cultures52 or standard neuronal cultures as in 53. Rat procedures were approved by the Institutional Animal Care Committee at the Boston Children’s Hospital and complied with all relevant ethical regulations regarding animal research.
Standard culture
Hippocampal neurons were obtained from E18 rat embryos, plated at 120 × 103 on 35mm, 10mm well FluoroDishes (World Precision Instruments) coated with poly-D-lysine and laminin as described above, and maintained in enriched Neurobasal (ThermoFisher Scientific) medium as described above. Neurons were transfected at DIV6-DIV8 using Lipofectamine2000 (Life Technologies) and imaged 1 day later.
Spot-groove culture
To grow straight axons for live imaging of distal axon endings, an optical-quality 24-well plate (Ibidi) was coated with 20 μg/mL poly-L-Lysine (Sigma-Aldrich) and 3.5 μg/mL laminin (ThermoFisher Scientific). After coating, grooves were etched into each well using a pin rake (Tyler Research). Hippocampal neurons were obtained from E18 rat (Charles River) embryos and spotted in the center of the grooves at 15 × 103 cells/well in 7 μL of media. After allowing cells to attach for 1 h in the incubator, media (Neurobasal, 2% B27, 100 U/mL penicillin-streptomycin, 1 mM L-glutamine, 100 ng/μL NGF, 2 ng/μL GDNF) was slowly added. Neurons were transfected at DIV6-DIV8 using Lipofectamine2000 (Life Technologies) and imaged 1 day later.
Live-Cell Imaging and Quantification
All live imaging experiments involving zapalog were conducted in a dark room that was illuminated only with red-filtered lights. Zapalog was kept as a 10mM stock in DMSO and diluted with media before each experiment as needed.
Whole cell fluorophore translocation analysis –
COS7 cells were transfected with Tom20-mCherry-FKBP and YFP-DHFR-Myc, and imaged 24hrs later. To determine time course of zapalog-induced dimerization using different concentrations of zapalog, time-lapsed datasets were acquired on an inverted Andor Revolution spinning disk confocal microscope, equipped with a Piezo Z500CE stage (Prior Scientific) in an incubator maintained at 37˚C / 5%CO2. Images were captured using a 40x / N.A. 1.3 oil UPLFLN objective with excitation at 488nm and 561nm separately, each for 200msec exposures once per second for 300sec. Laser power was set to < 100µW for each channel to minimize damage. Full field photocleavage of zapalog was achieved by activation of a 405nm channel (300µW). In order to calculate fluorophore translocation accurately and without bias at each timepoint, a MATLAB code was written to automatically compute the ratio of fluorescence in mitochondria vs. cytoplasm: As illustrated on Supplementary Figure S5, YFP and mCherry fluorescence were thresholded to generate binary masks of total cell area and of mitochondrial area for every timepoint. A series of masks representing cytoplasmic area in each timepoint was calculated by subtracting the mitochondrial area from total cell area. Finally, mean YFP intensity was measured within the areas of the mitochondrial and cytoplasmic masks, then divided to achieve the final metric of mitochondrial/cytoplasmic YFP ratio. This entire process, including mask creation, was performed separately for each time point, as mitochondrial locations change rapidly in the cell. The code is available for download at https://github.com/ewest11/FTA-Fluorophore-Translocation-Analysis.
Targeted whole cell fluorophore translocation assay –
To determine the time course and efficiency of reversal of zapalog-induced dimerization through photolysis, cells were transfected as above and imaged 24hrs later in a Leica SP8 scanning laser confocal with an HC PL APO 63x/1.40 OIL CS2 objective, set to multichannel imaging of YFP and mCherry with pulsed White Light Laser emission at 514nm and 584nm respectively for a total of 1 frame/sec. 1µM of zapalog was added in dark conditions in order to translocate the YFP from the cytoplasm to the outer membrane of mitochondria, then washed out after 10min by replacing the media with new, pre-warmed media. Using the SP8 FRAP protocol, an ROI of 25µm × 25µm was defined around each cell in the field and a selected cell was then then illuminated separately with a 405nm diode laser (0.5sec @ 30nW), followed by 20 seconds of imaging before repeating the process on the next selected cell.
Repeated, localized fluorophore-translocation assay on non-networked mitochondria –
18hrs after HeLa cells were transfected with Tom20-mCherry-FKBP and YFP-DHFR-Myc, they were incubated for 6hrs in 10µM CCCP. In order to measure repeated fluorophore translocation events in a localized subcellular area, cells were then imaged in a Leica SP8 scanning laser confocal with an HCX PL APO 100x/1.44 oil CORR CS objective, set to multichannel imaging of YFP and mCherry with pulsed White Light Laser emission at 514nm and 584nm respectively for a total of 1 frame/sec. 1µM of zapalog was added in dark conditions in order to translocate the YFP from the cytoplasm to the outer membrane of mitochondria (~5min). Then, using the SP8 FRAP protocol, an ROI of 20×20pixels was defined for repeated rounds of 0.5sec illumination with the 405nm diode laser (30nW), followed by 74 seconds of imaging. Mean YFP intensities in illuminated vs nearby non-illuminated 20×20pxl areas were normalized to range from 0–100, and average YFP intensity was calculated for each timepoint.
Repeated, localized fluorophore translocation assay on Peroxisomes –
COS7 cells were transfected with a peroxisomal outer membrane marker and zapalog receptor (PEX-mRFP-FKBP) and with cytosolic YFP-DHFR-Myc. 24hrs later, cells were imaged in a Leica SP8 scanning laser confocal with a HC PL APO 63x/1.40 OIL CS2 objective, set to multichannel imaging of YFP and mRFP with pulsed White Light Laser emission at 514nm and 584nm respectively for a total of 1 frame/sec. 1µM of zapalog was added in dark conditions in order to translocate the YFP from the cytoplasm to the outer membrane of peroxisomes (~5min). Then, using the SP8 FRAP protocol, an ROI of 50×50pixels was defined for repeated rounds of 0.5sec illumination with the 405nm diode laser (380nW), followed by 30 seconds of imaging. YFP and mRFP mean intensities in illuminated vs nearby non-illuminated 50×50pxl areas were normalized and average YFP intensity was calculated for each timepoint.
Mitochondrial Motility Analysis
Time lapse movies were acquired on an inverted Andor Revolution spinning disk confocal microscope, equipped with a Piezo Z500CE stage (Prior Scientific) in an incubator maintained at 37˚C / 5%CO2. Images were captured using a 40x / N.A. 1.3 oil UPLFLN objective with excitation at 488nm and 561nm separately, each for 200msec exposures once per second for 300sec (except where stated otherwise). Laser power was set to < 30% for each channel to minimize damage. Full field photocleavage of zapalog was achieved by activation of a 405nm channel (300µW). We imaged ~100μm stretches of axon at least 500μm away from the soma; this was done in the middle of the axon such that there were still large populations of mitochondria both upstream and downstream of the field, to prevent a significant skewing of the distribution. Mitochondrial motility was quantified as “% time in motion” using Kymolyzer, a custom ImageJ macro developed in our lab6.
Quantifying presence of stationary mitochondria at presynaptic sites
As illustrated in Supplementary Figure S6a, we analyzed dual-channel kymographs (60 frames in each channel at 5sec intervals for a total imaging of 5min) of axons from neurons expressing both Tom20-mCherry-FKBP (mitochondrial marker) and VGLUT1-Venus (presynaptic marker). We marked all the stationary mitochondria and all the stationary VGLUT1 particles and quantified for each axon the number of stationary mitochondria that coincided with a presynaptic location.
Generation of random locations along the axon
In order to control for possibly coincidental colocalization of mitochondria with the presynaptic marker, random locations were generated for each axon at a comparable density to actual presynaptic sites. The number of random locations generated for each axon equaled the number of presynaptic sites identified for that particular axon, and the algorithm for marking the random sites is illustrated in Supplementary Figure S6b.
Code Availability
The MATLAB code used to analyze the data shown in Figure 1c” is available for download at https://github.com/ewest11/FTA-Fluorophore-Translocation-Analysis.
The ImageJ macro “Kymolyzer” is available upon request.
Statistics and Reproducibility
Fluorophore translocation data (Figs. 1, 2, S2) are expressed as line-connected points (mean) with error bars (SE for assay timecourses 1c’, 2c, S2c; SD for dose response 1c”). Mitochondrial motility and position data are expressed as Tukey box-and-whisker plots (Figs. 4, 6, 7) or scatter plots of pairwise before-and-after datapoints (Fig. 5); median is marked in each. Statistical analyses were performed with Apple Numbers and GraphPad Prism v7.0 for MacOS X. Paired t-tests were used to compare populations made up of pairs of datapoints representing repeated measurements of the same sample (Fig 5). Welch’s t-tests were used to compare populations of unequal variances (Figs. 6,7). Pearson’s chi-squared tests of data shown in Fig.4 were performed with JMP v14 from MacOS X. For all analyses p < 0.05 was considered significant.
Data availability
Source data for Fig. 1-6 and Supplementary Fig. S3 have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We are grateful for plasmids shared with us by Dr. Takanari Inoue (Johns Hopkins University; Tom20-mCherry-FKBP), Dr. Casper Hoogenraad (Utrecht University; PEX-mRFP-FKBP), and Dr. Kristen Verhey (U. Michigan; full length Kif1a). We thank Lala Mkhitaryan for technical support and the members of the T.L. Schwarz lab for fruitful discussions. We are grateful to Sarah Vasquez and Dr. Mustafa Sahin’s laboratory for assistance with primary hippocampal cultures; Drs. Daniel Tom and Lai Ding from Harvard NeuroDiscovery Center’s Enhanced Neuroimaging Core for assistance with live cell imaging (NINDS P30 Core Center Grant #NS072030; Drs. Bernardo Sabatini (HMS) and Gülçin Pekkurnaz (UCSD) for access to equipment. This research was generously supported by the National Institutes of Health grants R01 GM069808 (NIH/NIGMS) and R21 NS87582 (NIH/NINDS) to T.L.S. as well as a BCH Pilot Study Grant. A.G. was supported by F32 GM110984 (NIH/NIGMS) and T32 NS007484 (NIH/NINDS), and M.R.B. by K99/R00 DA034648 (NIH/NIDA).
Footnotes
Competing interests
A US patent for zapalog has been filed by Boston Children’s Hospital and approved (US10053445B2); T.L.S., M.R.B. and A.G. are listed as inventors.
References
- 1.Misgeld T & Schwarz TL Mitostasis in Neurons: Maintaining Mitochondria in an Extended Cellular Architecture. Neuron 96, 651–666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarz TL Mitochondrial trafficking in neurons. Cold Spring Harbor Perspectives in Biology 5, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheng Z-H & Cai Q Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci 13, 77–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxton WM & Hollenbeck PJ The axonal transport of mitochondria. J. Cell. Sci. 125, 2095–2104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misgeld T, Kerschensteiner M, Bareyre FM, Burgess RW & Lichtman JW Imaging axonal transport of mitochondria in vivo. Nat Meth 4, 559–561 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Voss S, Klewer L & Wu Y-W Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells. Current Opinion in Chemical Biology 28, 194–201 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Wilson MH & Holzbaur ELF Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 142, 218–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkins B, Decker H, Bentley M, Luisi J & Banker G A novel split kinesin assay identifies motor proteins that interact with distinct vesicle populations. J. Cell Biol. 198, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.del Castillo U, Winding M, Lu W & Gelfand VI Interplay between kinesin-1 and cortical dynein during axonal outgrowth and microtubule organization in Drosophila neurons. Elife 4, e10140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komatsu T et al. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Meth 7, 206–208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tischer D & Weiner OD Illuminating cell signalling with optogenetic tools. Nature reviews. Molecular cell biology 15, 551–558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Bergeijk P, Adrian M, Hoogenraad CC & Kapitein LC Optogenetic control of organelle transport and positioning. Nature 518, 111–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czlapinski JL et al. Conditional glycosylation in eukaryotic cells using a biocompatible chemical inducer of dimerization. Journal of the American Chemical Society 130, 13186–13187 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lester HA & Nerbonne JM Physiological and Pharmacological Manipulations with Light Flashes. Annu. Rev. Biophys. Bioeng 11, 151175 (1982). [DOI] [PubMed] [Google Scholar]
- 15.Banghart MR, He XJ & Sabatini BL A Caged Enkephalin Optimized for Simultaneously Probing Mu and Delta Opioid Receptors. ACS Chem Neurosci 9, 684–690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trigo FF, Corrie JE & Ogden D Laser photolysis of caged compounds at 405 nm: photochemical advantages, localisation, phototoxicity and methods for calibration. Journal of Neuroscience Methods 180, 9–21 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Lemke EA, Summerer D, Geierstanger BH, Brittain SM & Schultz PG Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat. Chem. Biol. 3, 769–772 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Gutnick A, Banghart MR, , West E,R & Schwarz T,L Synthesis of zapalog (TMP-DNAB-SLF). Nat. Protocol Exchange (2019). [Google Scholar]
- 19.Inoue T, Heo W, Grimley JS, Wandless TJ & Meyer T An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Meth 2, 415–418 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narendra D, Tanaka A, Suen D-F & Youle RJ Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka A et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol 191, 1367–1380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashrafi G, Schlehe JS, LaVoie MJ & Schwarz TL Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J. Cell Biol 206, 655–670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis TL & Polleux F Terminal Axon Branching Is Regulated by the LKB1-NUAK1 Kinase Pathway via Presynaptic Mitochondrial Capture. Cell 153, 1510–1525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang DTW, Honick AS & Reynolds IJ Mitochondrial Trafficking to Synapses in Cultured Primary Cortical Neurons. J. Neurosci 26, 7035–7045 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis TL & Polleux F Terminal axon branching is regulated by the LKB1-NUAK1 kinase pathway via presynaptic mitochondrial capture. Cell 153, 1510–1525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chada SR & Hollenbeck PJ Nerve growth factor signaling regulates motility and docking of axonal mitochondria. Curr Biol 14, 1272–1276 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Ankenbruck N, Courtney T, Naro Y & Deiters A Optochemical Control of Biological Processes in Cells and Animals. Angew. Chem. Int. Ed. Engl 57, 2768–2798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khamo JS, Krishnamurthy VV, Sharum SR, Mondal P & Zhang K Applications of Optobiology in Intact Cells and Multicellular Organisms. J. Mol. Biol 429, 2999–3017 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Pathak GP, Strickland D, Vrana JD & Tucker CL Benchmarking of Optical Dimerizer Systems. ACS synthetic biology (2014). doi: 10.1021/sb500291r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Bergeijk P, Hoogenraad CC & Kapitein LC Right Time, Right Place: Probing the Functions of Organelle Positioning. Trends Cell Biol. (2015). [DOI] [PubMed] [Google Scholar]
- 31.Liu P et al. A bioorthogonal small-molecule-switch system for controlling protein function in live cells. Angew. Chem. Int. Ed. Engl 53, 10049–10055 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Strickland D et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Meth 9, 379–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levskaya A, Weiner OD, Lim WA & Voigt CA Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy MJ et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Meth 7, 973–975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmermann M et al. Cell-permeant and photocleavable chemical inducer of dimerization. Angew. Chem. Int. Ed. Engl. 53, 4717–4720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J-R et al. Characterization of the Movement of the Kinesin Motor KIF1A in Living Cultured Neurons. Journal of Biological Chemistry 278, 26242629 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Derr ND et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science (New York, N.Y.) 338, 662–665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X & Schwarz TL The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell 136, 163–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang J-S et al. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell 132, 137–148 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pathak D, Sepp KJ & Hollenbeck PJ Evidence that myosin activity opposes microtubule-based axonal transport of mitochondria. J. Neurosci 30, 8984–8992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shepherd GM & Harris KM Three-dimensional structure and composition of CA3-->CA1 axons in rat hippocampal slices: implications for presynaptic connectivity and compartmentalization. J. Neurosci 18, 8300–8310 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris KM & Weinberg RJ Ultrastructure of synapses in the mammalian brain. Cold Spring Harbor Perspectives in Biology 4, a005587–a005587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi M, Weaver D & Hajnóczky G Control of mitochondrial motility and distribution by the calcium signal: a homeostatic circuit. J. Cell Biol 167, 661–672 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saotome M et al. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci USA 105, 20728–20733 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Awabdh Al S., et al. Neuronal activity mediated regulation of glutamate transporter GLT‐1 surface diffusion in rat astrocytes in dissociated and slice cultures. Glia 64, 1252–1264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vale RD The molecular motor toolbox for intracellular transport. Cell 112, 467–480 (2003). [DOI] [PubMed] [Google Scholar]
References (Methods)
- 48.Gutnick A, Banghart MR, West ER & Schwarz TL Synthesis of zapalog (TMP-DNAB-SLF). Nature Protocol Exchange [Google Scholar]
- 49.Miyamoto T et al. Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nat. Chem. Biol. 8, 465–470 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kapitein LC et al. Mixed Microtubules Steer Dynein-Driven Cargo Transport into Dendrites. Curr Biol 20, 290–299 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Herzog E et al. In Vivo Imaging of Intersynaptic Vesicle Exchange Using VGLUT1Venus Knock-In Mice. J. Neurosci 31, 15544–15559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gornstein EL & Schwarz TL Neurotoxic mechanisms of paclitaxel are local to the distal axon and independent of transport defects. Exp. Neurol. 288, 153–166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pekkurnaz G, Trinidad JC, Wang X, Kong D & Schwarz TL Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell 158, 54–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Fig. 1-6 and Supplementary Fig. S3 have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.