Abstract
Poly(ADP-ribose) polymerase (PARP) is involved in the pathogenesis of cell dysfunction, inflammation and organ failure during septic shock. The goal of the current study was to investigate the efficacy and safety of the clinically approved PARP inhibitor olaparib in experimental models of oxidative stress in vitro and in sepsis in vivo. In mice subjected to cecal ligation and puncture (CLP) organ injury markers, circulating and splenic immune cell distributions, circulating mediators, DNA integrity and survival was measured. In U937 cells subjected to oxidative stress, cellular bioenergetics, viability and DNA integrity were measured. Olaparib was used to inhibit PARP. The results show that in adult male mice subjected to CLP, olaparib (1–10 mg/kg i.p.) improved multiorgan dysfunction. Olaparib treatment reduced the degree of bacterial CFUs. Olaparib attenuated the increases in the levels of several circulating mediators in the plasma. In the spleen, the number of CD4+ and CD8+ lymphocytes were reduced in response to CLP; this reduction was inhibited by olaparib treatment. Treg but not Th17 lymphocytes increased in response to CLP; these cell populations were reduced in sepsis when the animals received olaparib. The Th17/Treg ratio was lower in CLP-olaparib group than in the CLP control group. Analysis of miRNA expression identified a multitude of changes in spleen and circulating white blood cell miRNA levels after CLP; olaparib treatment selectively modulated these responses. Olaparib extended the survival rate of mice subjected to CLP. In contrast to males, in female mice olaparib did not have significant protective effects in CLP. In aged mice olaparib exerted beneficial effects that were less pronounced than the effects obtained in young adult males. In in vitro experiments in U937 cells subjected to oxidative stress, olaparib (1–100 μM) inhibited PARP activity, protected against the loss of cell viability, preserved NAD+ levels and improved cellular bioenergetics. In none of the in vivo or in vitro experiments did we observe any adverse effects of olaparib on nuclear or mitochondrial DNA integrity. In conclusion, olaparib improves organ function and extends survival in septic shock. Repurposing and eventual clinical introduction of this clinically approved PARP inhibitor may be warranted for the experimental therapy of septic shock.
Keywords: sepsis, shock, multiorgan dysfunction, DNA, mitochondria, cell death, Treg, Th17
1. Introduction
Sepsis is a life-threatening systemic inflammatory disease affecting several million people annually worldwide. It is associated with mortality rates of 20–40%. Its frequency increases every year, in excess of the growth and aging of our population. The outcomes of sepsis are markedly worse in the elderly [1,2]. On the other hand, the female gender (in the pre-menopausal stage) tends to provide some protection against sepsis-associated mortality [3,4]. Despite extensive research and despite a large number of pivotal clinical trials targeting various pathways in immune, inflammatory and coagulation abnormalities in sepsis/septic shock, no specific pharmacological or immunological therapies are approved for the treatment of this condition.
More than two decades ago, the constitutive nuclear and mitochondrial enzyme poly(ADP-ribose) polymerase (PARP) emerged as a potential therapeutic target for various forms of critical illness, including sepsis. Several lines of studies, utilizing PARP inhibitors of various structural classes, as well as PARP-1-deficient mice indicated that PARP inhibition exerts beneficial effects on organ function, vascular function, and survival [5–19]. For instance, PARP-1 knockout mice showed, after cecal ligation and puncture (CLP), lower plasma levels of TNF-α, IL-6 and IL-10 [8]. In an intraperitoneal bacterial sepsis model, pigs treated with PARP inhibitors showed better survival after 24h and 96h, decreased bacteremia, attenuated TNFα plasma levels and protected against the cardiovascular dysfunction when compared to untreated animals [9]. Moreover, treatment with a PARP inhibitor exerted a protective effect against apoptosis in the hearts of rats subjected to CLP [16].
Although PARP activation clearly plays a pathophysiological role in sepsis, clinical translation of this concept was not possible, because there were no clinically approved drugs to inhibit PARP. Recently, several classes of PARP inhibitors have been approved for clinical use, in the field of oncology. This development is based on an independent line of research, one that demonstrated that in cancer cells, PARP inhibition can induce cell death and can potentiate the efficacy of various classes of anticancer agents [20,21]. One of the clinically approved PARP inhibitor drugs is olaparib (Lynparza), a competitive PARP inhibitor that inhibits the binding of NAD+ (the substrate of PARP) into the catalytic site of PARP.
As outlined in our recent position paper, the clinical approval of PARP inhibitors opens the door for potential repurposing of these drugs for various non-oncological indications, with sepsis/septic shock being one of the prime candidates for such repurposing efforts [19]. However, additional preclinical work is needed to lay the groundwork for such repurposing efforts. As described in detail [19], such work needs to focus both on efficacy endpoints, as well as safety endpoints - chiefly, on DNA integrity, considering the fact that PARP has been long considered an enzyme that has an important role in the facilitation of DNA repair. The current report describes the results of such a project. Our experiments incorporated a standard model of sepsis (induced by cecal ligation and puncture) and evaluated multiple efficacy endpoints (primarily focusing on multiorgan injury), as well as safety endpoints (mitochondrial and nuclear DNA integrity). We have also conducted an in vitro sub-study (human monocytic cells subjected to oxidative stress) – once again, both focusing on efficacy endpoints (cell viability, cellular bioenergetics) as well as safety endpoints (DNA integrity). The results of the current study demonstrate the efficacy of olaparib on multiple outcome variables, and do not identify any significant adverse effects of the PARP inhibitor on DNA integrity. Thus, the data presented in the current report lend support for repurposing and clinical introduction of the PARP inhibitor olaparib for the experimental therapy of septic shock.
2. Materials and Methods
2.1. Animals
Male or female C57BL6 mice (8 – 72 weeks old) were obtained from Jackson Laboratories. Animals were kept in a 12 h - 12 h light / dark cycle at 21–23 °C with free access to standard chow diet.
2.2. Cecal ligation and puncture (CLP)
Acute sepsis was induced in mice by cecal ligation and puncture as previously described [22]. Briefly, mice were anesthetized by ketamine/xylazine cocktail (i.p.), the abdomen was shaved, wiped with 70% isopropanol and a midline abdominal incision (1–2 cm) was performed. The cecum was exteriorized, ligated with a sterile silk suture 1 cm from the tip and double punctured with a 20-gauge needle. The cecum was squeezed to assure expression of a small amount of fecal material and returned to the abdominal cavity. The incision was closed with auto-clips and kept clean by povidone-iodine (Betadine). Mice were resuscitated with intraperitoneal injection of 1 ml of lactated Ringer’s solution. Sham-operated mice were treated as described above with the exception of ligation and puncture of the cecum. Buprenorphine (0.1 mg/kg; s.c. 30 minutes before surgery and every 12 hours thereafter) was used for pain management. A sample of whole blood was collected for analysis of organ function using a comprehensive metabolic panel or for circulating mediator measurements using a multiple array system. Major organs were collected and either analyzed for immune cells by flow cytometry or snap frozen and kept at −80 °C until subsequent use (measurement of DNA integrity or MDA or MPO levels) or placed in formalin and processed for histological analysis.
For the survival study, mice were constantly monitored for 48 hours. Mice that survived this period of time were euthanized by cervical dislocation. All animal procedures described in this study have been approved by the respective local Institutional Animal Care and Use Committee of the University of Texas Medical Branch and the University of Sao Paulo.
2.3. Olaparib treatment protocol
All groups of mice received the following intraperitoneal treatment: vehicle (phosphate-buffered saline [PBS] with 4% dimethyl sulfoxide [DMSO] and 5% polyethylene glycol [PEG]); olaparib, 1 mg/kg, 3 mg/kg or 10 mg/kg (dissolved in PBS with 4% DMSO 5% and PEG). In the 24-hour protocol, the animals received two doses of olaparib, the first 30 minutes after the CLP and the second 8 hours after CLP, and the experiment was terminated at 24 hours. In the survival protocol, treatment with the PARP inhibitor was initiated at 30 minutes after the CLP, the second dosing was performed at 8 hours after CLP, and the same dosing was repeated subsequently every 8 hours. Animals were monitored for 48 hours at which time point the experiment was terminated.
2.4. Complete metabolic panel
Samples of whole blood were collected from septic mice, placed in lithium-heparin tubes and immediately processed for the measurement of alanine aminotransferase (ALT), albumin (ALB), alkaline phosphatase (ALP), amylase (AMY) total calcium (Ca2+), creatinine (CRE), glucose (GLU), phosphorus (PHOS), potassium (K+), sodium (Na+), total bilirubin (TBIL), total protein (TP), and urea nitrogen (BUN) using the VetScan Chemistry Analyzer system (Abaxis) [22].
2.5. Detection of circulating mediators
Blood from CLP or sham-operated mice was collected in K2EDTA blood collection tubes and centrifuged at 4 °C for 15 min at 2,000×g within 30 minutes of collection. Plasma was isolated, aliquoted and stored at −80 °C until use. The EMD Millipore’s MILLIPLEX™ MAP Mouse cytokine Magnetic Bead Panel 1 was used as described [22] for the simultaneous quantification of the following analytes: TNFα, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12(p40), IL12(p70), IL-13, IL-15, IL-17, LIF, LIX, eotaxin, G-CSF, GM-CSF, KC, IP-10, MCP-1, RANTES, VEGF, MIP-1α, MIP-1β, MIP-2, M-CSF, MIG and IFNγ. Data were processed using the Luminex xPONENT® acquisition software.
2.6. Determination of tissue lipid peroxidation: malon dialdehyde assay
Tissue malon dialdehyde (MDA) levels, an index of cellular injury/oxidative stress, were measured as described [22] in tissue homogenates from mice subjected to 24 hours of CLP-induced sepsis, using a fluorimetric MDA-specific lipid peroxidation assay kit (Enzo Life Sciences).
2.7. Myeloperoxidase activity assay
Myeloperoxidase activity was measured as described [22] in tissue homogenates from mice subjected to 24 hours of CLP-induced sepsis, using a commercially available MPO fluorometric detection kit (Enzo Life Sciences).
2.8. Measurement of nuclear and mitochondrial DNA integrity
Integrity of the nuclear and mitochondrial DNA, measured by the relative amount of DNA damage, was analyzed by semi-quantitative, long-amplicon PCR assays (LA-PCR) using LongAmp Taq DNA Polymerase (New England BioLabs, Ipswich, MA) in tissue homogenates as described [23]. Total DNA was isolated using DNase Blood and Tissue Kit (QIAGEN, Hilden, Germany). Briefly, damage to nuclear DNA was estimated by quantification of the PCR amplification of the 10kb nuclear-specific DNA fragment using PicoGreen fluorescent dye to detect amplified double-stranded DNA (Quant-iT™ PicoGreen; Life Technologies, Carlsbad, CA). Damage to the mitochondrial DNA was estimated by quantification of the PCR amplification of the 8.9kb mitochondrial-specific DNA fragment using PicoGreen staining. Data were normalized by the secondary PCR amplification of 221bp mitochondrial genome-specific fragment for correction of the multiple copies of the mitochondrial DNA. LA-PCR assay is based on premises that DNA damage inhibits progression of DNA polymerase during PCR reaction and thus amplification of appropriate DNA fragment is negatively correlated with the level of the DNA damage.
2.9. Determination of colony-forming bacterial units (CFUs) after sepsis or bacteremia
Blood was diluted serially in sterile saline. Spleen was homogenized in sterile saline at 100 mg tissue/ml concentration followed by serial dilution. Fifty microliters of each dilution was plated and cultured on LB agar plates at 37°C. After 16 hours of incubation, the number of bacterial colonies was counted and expressed as CFUs per milliliter of blood or as CFUs per milligram or gram tissue.
In an in vitro satellite experiment, we evaluated whether olaparib has any direct effect on bacterial growth. In these experiments DH5α E.coli were cultured in liquid LB growth medium in a shaking incubator set to 37°C and 200 RPM in the presence of various concentrations (1– 100 μM) of olaparib. The culture density was measured at OD600 over 5 hours.
In an in vivo satellite experiment, we evaluated whether olaparib has any direct effect on bacterial clearance in vivo in the absence of septic shock. Male C57BL6 mice (8 weeks old) were obtained from Jackson Laboratories. Animals (either treated with olaparib, 10 mg/kg i.p. at 30 min and 8 hours post-inoculation, or treated with corresponding vehicle control) were injected with 3×108 CFU DH5α E.coli i.p. and tissues harvested 20 hours post-infection. CFUs in blood, spleen and liver were quantified as described above.
2.10. Histological analysis
Tissues were fixed for 1 week in buffered formaldehyde solution (10% in PBS) at room temperature, dehydrated by graded ethanol and embedded in Paraplast (Sherwood Medical, Mahwah, NJ, USA). Tissue sections (thickness: 7 μm) were deparaffinized with xylene stained with hematoxylin/eosin and studied using light microscopy. Slides were evaluated in a blinded fashion.
2.11. Flow cytometry
For phenotypic characterization of cells by flow cytometry from spleen and blood samples, we performed three panels: T cells (panel 1), Treg cells (panel 2) and Th17 cells (panel 3). Cell suspensions were prepared and were re-suspended in flow cytometry buffer (PBS containing 2% BSA). Cells were stained with anti-CD3, CD4, and CD8 (panel 1), anti-CD4, CD25 (panel 2) and anti-CD4 (panel 3) antibodies for 30 minutes in dark at 4°C than were washed twice with flow cytometry buffer. For panel 1, cells were collected and analyzed by flow cytometry. Additionally, macrophages, B cells and neutrophils were similarly analyzed using antibodies against CD11b, CD19, and Ly6G. For panels 2 and 3, cells which were stained with surface markers were fixed and permeabilized with eBioscience’s Permealization Kit, then incubated with anti-Foxp3 (panel 2) and anti-RORγ, IL-17A and IL-17F (panel 3) antibodies for 30 minutes in the dark at 4°C. Cells were then washed and analyzed by flow cytometry (Guava 8HT; Merck-Millipore).
2.12. RT-PCR studies of miRNA expression
Total RNA was extracted from spleen and white blood cells using Trizol reagent (Invitrogen Carlsbad, California, EUA) according to the manufacturer’s specifications. Extracted RNA was eluted in RNase-free water, treated with DNase I, Amplification Grade (ThermoFisher, Waltham, MA, USA) according to the manufacturer’s specifications and quantified by spectrophotometry.
cDNA was synthesized using miScript II RT Kit (Qiagen - Hilden, Germany) using HiSpec buffer chemistry, which exclusively reverse transcribes mature miRNA to cDNA; this mixture was incubated for 60 min at 37 °C and for 5 min at 95 °C to inactivate miScript Reverse transcriptase mix and placed on ice.
A functional quality control was performed on cDNA samples using miScript miRNA QC PCR array. qPCR was performed on a StepOnePlus Real-Time PCR System (ABI, Foster City, California, EUA). Threshold cycles (C) for PPC and RT control (miRTC) were examined to assess PCR and RT efficiencies, respectively. The expression of cel-miR-39 assay was also observed to confirm efficient RNA recovery. C values for all controls were within the manufacturer’s recommended range for a successful quality control (QC).
Large-scale analysis of miRNA expression was performed using miScript miRNA PCR Array Mouse Apoptosis (96 well format) (Qiagen - Hilden, Germany). 12 plaques per organ were made. A set of controls are included on each plate which enables data analysis using ΔΔCT method of relative quantification, assessment of reverse transcription performance, and assessment of PCR performance. The miScript miRNA PCR array enables SYBR Green-based real-time PCR analysis using StepOnePlus Real-Time PCR System (ABI, Foster City, California, EUA) as follows: for 15 min at 95 °C; 40 cycles of 15 s at 94 °C; for 30 s at 55 °C; and for 30 s at 70 °C and its software determined the number of copies of each gene from the microarray plates.
Values of miRNA expression were normalized using the geometric mean calculated from the internal control genes classified according to RefFinder Software and fold changes were calculated by the 2−ΔΔCT method. The analysis of the array was performed by the software available in SaBiosciences’ Data Analysis Center (https://www.qiagen.com/br/shop/genes-and-pathways/data-analysis-center-overview-page/). The most significantly altered miRNAs were confirmed by RT-PCR. The altered miRNAs miR-15a-5p, miR-17–5p, miR-146a-5p and miR-365–3p that were chosen for confirmation from the array; the primers were purchased from Qiagen. The cDNA was synthesized using miScript II RT Kit and the PCR was performed using miScript SYBR Green PCR Kit. Again, the values of miRNA expression were normalized using the geometric mean calculated from the previously internal control genes classified and fold changes were calculated by the 2−ΔΔCT method.
2.13. In vitro studies in U937 cells subjected to oxidative stress
Human monocyte histiocytic lymphoma cells (U937) were obtained from ATCC and maintained in RPMI1640 with 10% fetal bovine serum (Life Technologies). For resazurin and LDH assays, U937 cells were plated in 96-well plates at 2×104 cells/well. For DNA integrity, U937 cells were plated in 12-well plates at 2×105 cells/well. For the NAD+ quantification assay, U937 cells were plated in 12-well plates at 3×105 cells/well.
Cells were differentiated with PMA (100 ng/ml for 48h), then subjected to various concentrations of hydrogen peroxide (H2O2, 300 μM, 600 μM, 1 mM) for 1 hour, in the presence or absence of olaparib (1, 3, 10, 100 μM) pretreatment. After 1 hour, cell viability was measured with the resazurin (7-hydroxy-3H-phenoxazin-3-one 10-oxide) method, or cell necrosis was detected by measuring the release of lactate dehydrogenase (LDH) to the culture medium as described [24]. Mitochondrial and nuclear DNA integrity was measured with the LA-PCR method as described above. Poly-ADP-ribose polymerase-1 (PARP1) enzyme and its product, poly-ADP-ribose (PAR) and actin (a loading control) were detected by Western blotting as described [25]. Total cellular NAD+ was determined using NAD/NADH Quantification Kit (Sigma) according to the manufacturer’s protocol. The amount of NAD+ present in the samples was quantified in a colorimetric assay, measured at 450 nm using a microplate reader. The amount of NAD+ was normalized to protein content of the samples, quantified using the PierceTM BCA Protein Assay.
Cellular bioenergetics was measured by the Extracellular Flux Analysis method as described [26]. Briefly, cells were seeded on cell culture microplates (60,000/well). After 24 h, cells were pretreated with olaparib (1, 3 or 10 μM) or its vehicle for 30 min, followed by exposure to H2O2 (300 μM) for 1 h. For analysis of mitochondrial respiration, cells were washed twice with DMEM medium pH 7.4 supplemented with L-glutamine (2 mM, Gibco), sodium pyruvate (1 mM, Sigma) and glucose (10 mM, Sigma). After 1 h incubation at 37°C in CO2-free incubator, the oxygen consumption rate (OCR) after oligomycin (1 μM) was used to assess ATP production rate and the OCR after carbonyl cyanide-4-trifluoromethoxy phenylhydrazone (FCCP, 0.7 μM) to assess maximal mitochondrial respiratory capacity. Antimycin A (0.5 μM) and rotenone (0.5 μM) were used to inhibit the flux of electrons through complex III and I, to detect residual non-mitochondrial OCR, which is considered to be due to cytosolic oxidase enzymes. For analysis of glycolytic parameters, cells were treated with olaparib and H2O2 (as above), washed twice with phenol red-free DMEM medium pH 7.4 containing L-glutamine (2 mM), sodium pyruvate (1 mM), glucose (10 mM) and HEPES (5 mM, Sigma). After 1 h incubation at 37 °C in CO2-free incubator, proton efflux rate (PER) from basal and compensatory glycolysis was measured. Mitochondria inhibition by rotenone (0.5 μM) and antimycin A (0.5 μM) allows calculation of the mitochondrial-associated acidification. Subsequently, 2-deoxy-D-glucose (50 mM) was used to inhibit glycolysis and stop glycolytic acidification. Glycolytic proton efflux rate (glycoPER) was calculated by subtracting the mitochondrial acidification to the total PER. All the results were normalized by total protein.
2.14. Statistical analysis
Data are shown as mean±SEM. One-way and two-way ANOVA with Bonferroni’s multiple comparison test were used to detect differences between groups. Two main type of comparisons were performed. The first type was to determine whether the insult (CLP in vivo or oxidative stress in vitro) has an effect on a given parameter compared to normal control values (animals not subjected to CLP or cells not subjected to H2O2). Statistically significant differences between these two groups are indicated by *p<0.05 or **p<0.01. The second type was to detect if olaparib affected the response in the groups that were subjected to the insult (CLP in vivo or oxidative stress in vitro), compared to the groups that were subjected to the same insults in the absence of olaparib (i.e. in the presence of olaparib vehicle). Statistically significant differences between these two groups are indicated by #p<0.05 or ##p<0.01. Survival differences were analyzed by the Chi square test. Statistical calculations were performed using Graphpad Prism analysis software.
3. Results
3.1. Olaparib exerts organ protective and anti-inflammatory effects in organs of young adult male mice subjected to CLP, without adversely affecting DNA integrity
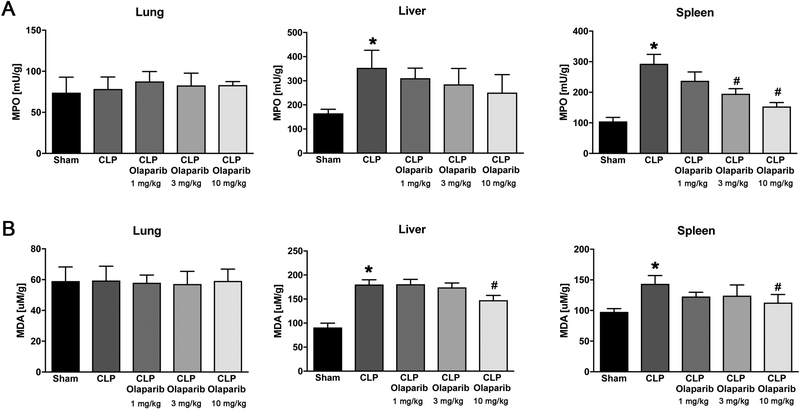
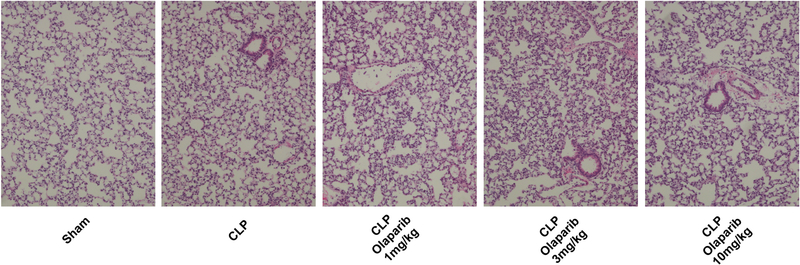
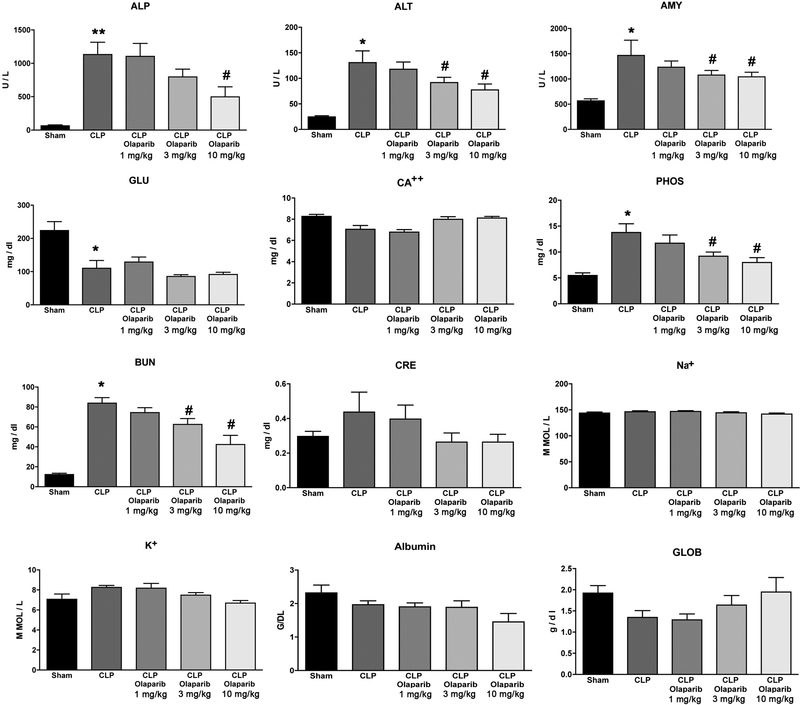
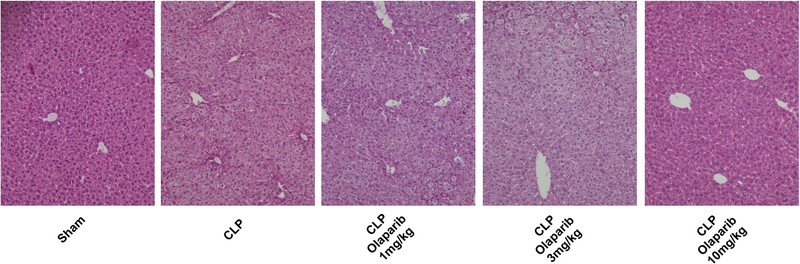
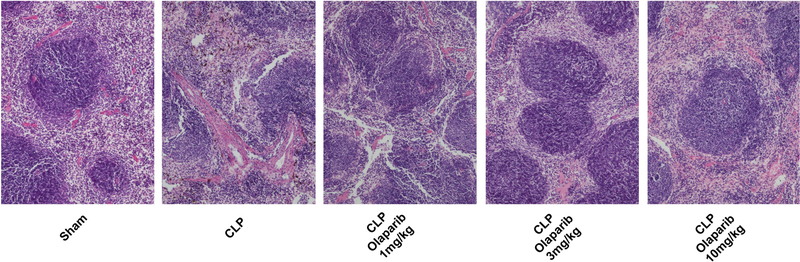
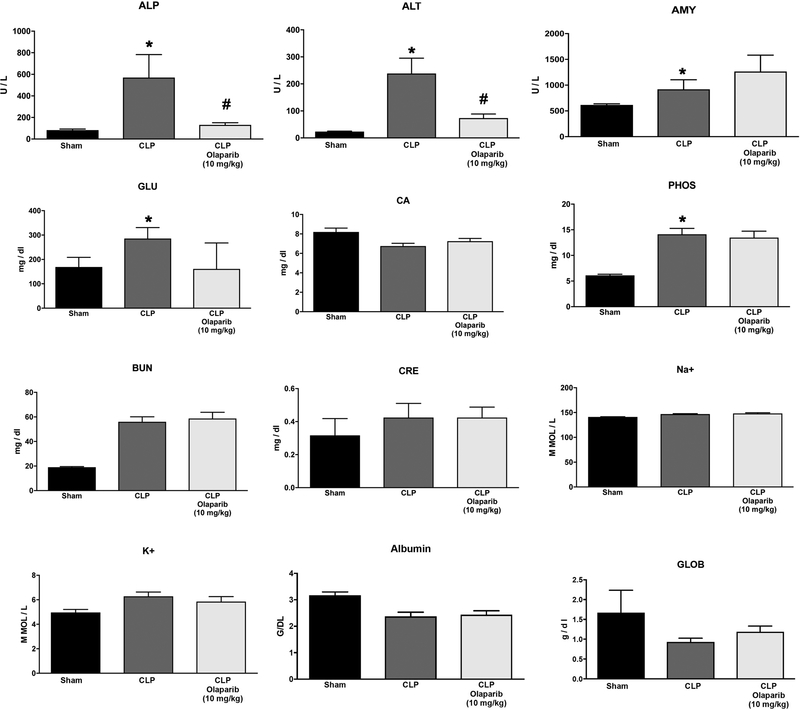
In young adult male mice subjected to CLP, olaparib (1, 3 or 10 mg/kg i.p.) concentration-dependently improved several parameters of multiorgan dysfunction (Figs. 1–3). For instance, the CLP-induced increases in spleen MPO content and liver and spleen MDA levels were attenuated by olaparib (Fig. 1). In addition, the CLP-induced increases in plasma markers of liver and pancreas injury (ALP, ALT, amylase) and renal dysfunction (BUN) were attenuated by olaparib treatment (Fig. 2). The histopathological pictures of the lungs did not show marked alterations in any of the groups, with slight emphysema evident in all CLP groups (Fig. 3). In the liver, foamy degeneration of numerous hepatocytes is evident in the CLP group; olaparib, at the 10 mg/kg dose, normalized the morphology of the hepatocytes (Fig. 4). In the spleen, CLP induced macrophage infiltration, and evidence of hemolysis was evident, with no marked differences between CLP groups with or without olaparib (Fig. 5).
Figure 1. Effect of olaparib on lung, liver and spleen MPO and MDA levels in young male Balb/c mice subjected to CLP.
(A): Lung, liver and spleen MPO levels (expressed as milliunits [mU]/gram tissue) and (B): lung, liver and spleen MDA levels (expressed as μM/gram tissue) are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant increase in MPO in response to CLP, compared to the sham group; #p<0.05 shows significant protective effect of olaparib in CLP mice compared to vehicle-treated CLP mice.
Figure 3. Effect of olaparib on histological alterations injury in the lungs of young male Balb/c mice subjected to CLP.
Representative histological pictures are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Pictures are selected from n=5 animals for each group.
Figure 2. Effect of olaparib on selected parameters of organ injury in young male Balb/c mice subjected to CLP.
Various physiological and organ injury marker levels: alkaline phosphatase (ALP), alanine aminotransferase (ALT), amylase (AMY), plasma glucose (GLU), plasma calcium (CA++), plasma phosphate (PHOS), plasma blood urea nitrogen (BUN), plasma creatinine (CRE), plasma sodium (Na+), plasma potassium (Na+), plasma albumin (ALB) and plasma globulin (GLOB) measured by Vetscan analysis, are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant increase in the respective parameter in response to CLP, compared to the sham group; #p<0.05 shows significant protective effect of olaparib in CLP mice compared to vehicle-treated CLP mice.
Figure 4. Effect of olaparib on histological alterations injury in the liver of young male Balb/c mice subjected to CLP.
Representative histological pictures are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Pictures are selected from n=5 animals for each group.
Figure 5. Effect of olaparib on histological alterations injury in the spleen of young male Balb/c mice subjected to CLP.
Representative histological pictures are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Pictures are selected from n=5 animals for each group.
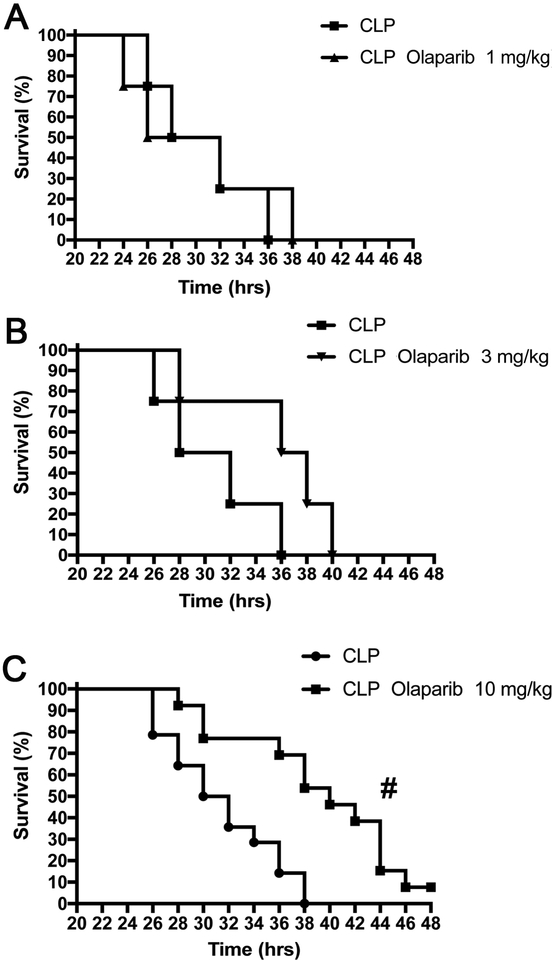
Olaparib, at 10 mg/kg (but not at the two lower doses used), caused a significant prolongation of survival of the animals subjected to CLP (Fig. 6).
Figure 6. Effect of olaparib on the survival rate of young male Balb/c mice subjected to CLP.
Survival rates are shown in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10–12 animals for each group; #p<0.05 shows significant protective effect of olaparib in CLP mice compared to vehicle-treated CLP mice.
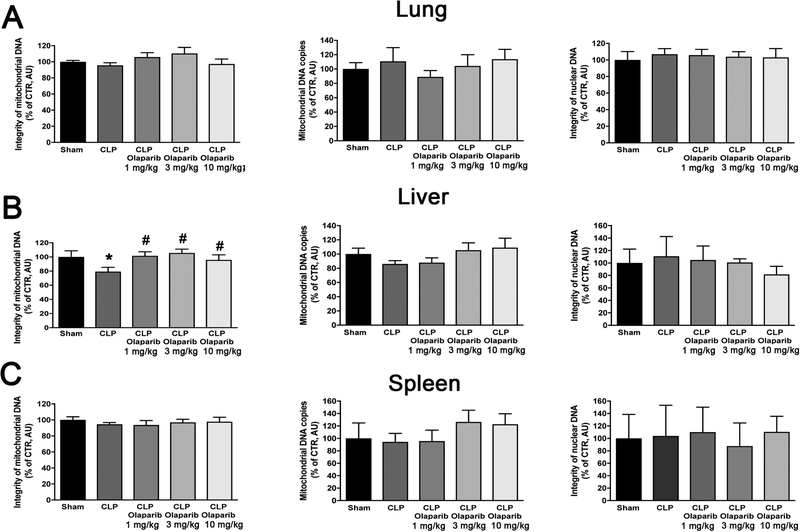
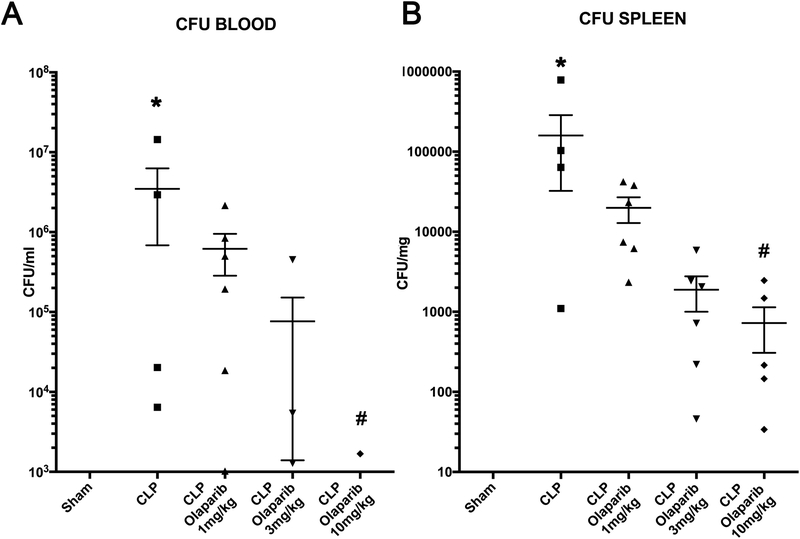
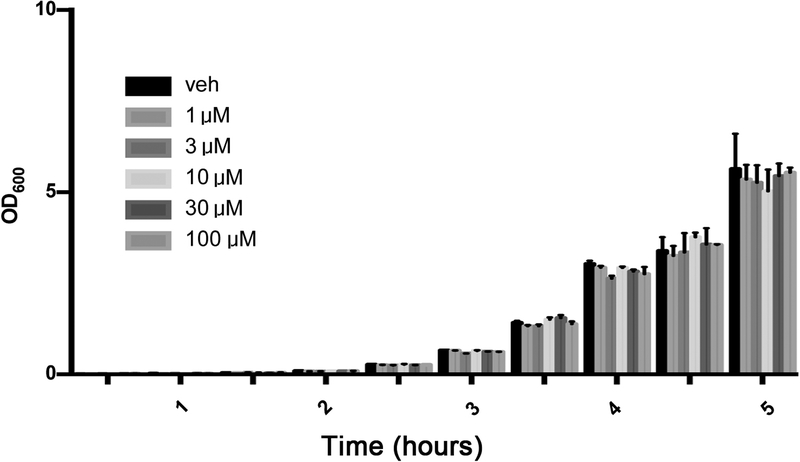
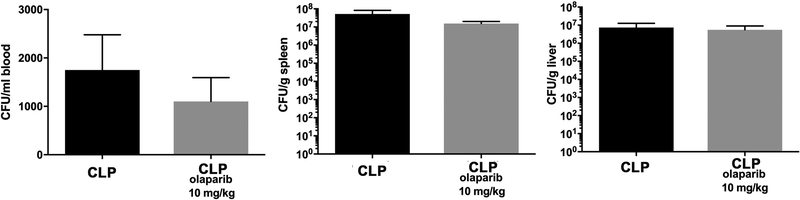
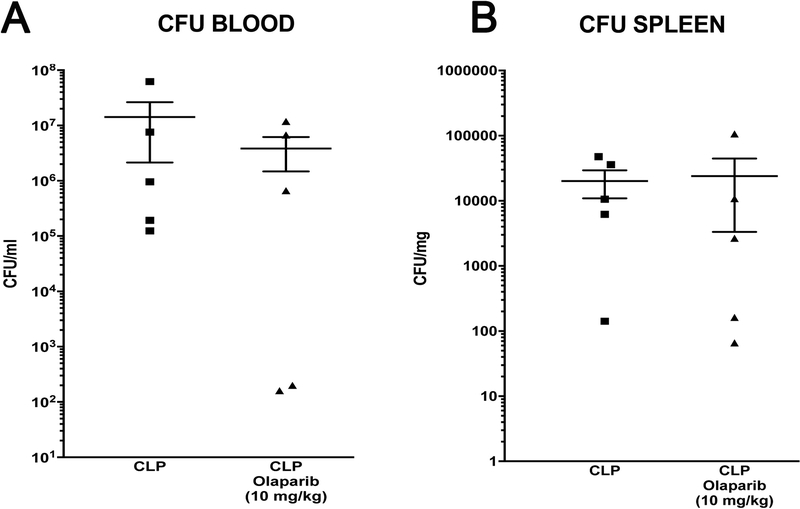
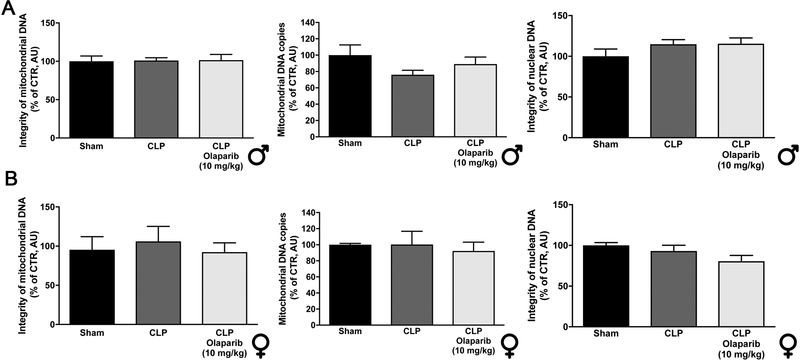
CLP did not induce detectable damage in nuclear DNA in any of the organs (liver, lung, spleen) studied, but in the liver, a significant degree of mitochondrial DNA damage was detected, which was prevented by olaparib treatment (Fig. 7). Olaparib treatment reduced the number of bacteria in the plasma and spleens of mice subjected to CLP (Fig. 8). However, this effect does not appear to be a direct antibacterial action of olaparib, as in vitro incubation of E. coli with various concentrations of olaparib (1–100 μM) did not have any effect on bacterial growth (Fig. 9). Moreover, in a satellite experiment of bacteremia without septic shock, olaparib treatment failed to significantly affect bacterial CFUs in the blood, spleen or liver after an i.p. bolus of E. coli (Fig. 10).
Figure 7. Effect of olaparib on lung, liver and spleen mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity in young male Balb/c mice subjected to CLP.
(A): Lung mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity values (expressed as % of control); (B): liver mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity values (expressed as % of control); (C) spleen mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity values (expressed as % of control) are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant change in a respective parameter in response to CLP, compared to the sham group; #p<0.05 shows significant effect of olaparib on a given parameter in CLP mice compared to vehicle-treated CLP mice.
Figure 8. Effect of olaparib on blood and spleen bacterial colony-forming unit (CFU) numbers in young male Balb/c mice subjected to CLP.
(A): Blood CFUs (expressed as CFU/ml) and (B) spleen CFUs (expressed as CFU/mg) are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant increases in CFUs in response to CLP, compared to the sham group; #p<0.05 shows significant effect of olaparib on CFU values in CLP mice compared to vehicle-treated CLP mice.
Figure 9. Effect of olaparib on E. coli bacterial growth in vitro.
Data show bacterial numbers at various time points in vehicle-treated group, and in the groups of bacteria in the presence or various concentrations (1, 3, 10, 30, 100 μM) olaparib for various time points (1–5 hours). Data are shown as mean ± SEM of 5 determinations per group.
Figure 10. Effect of olaparib on blood, spleen and liver bacterial colony-forming unit (CFU) numbers in young male Balb/c mice subjected to E. coli bacteria in vivo.
Blood (expressed as CFU/ml), spleen and liver CFUs (expressed as CFU/g) are shown in vehicle-treated mice inoculated with E. coli for 24 hours (“CLP”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib. Data are shown as mean ± SEM of 6 animals for each group.
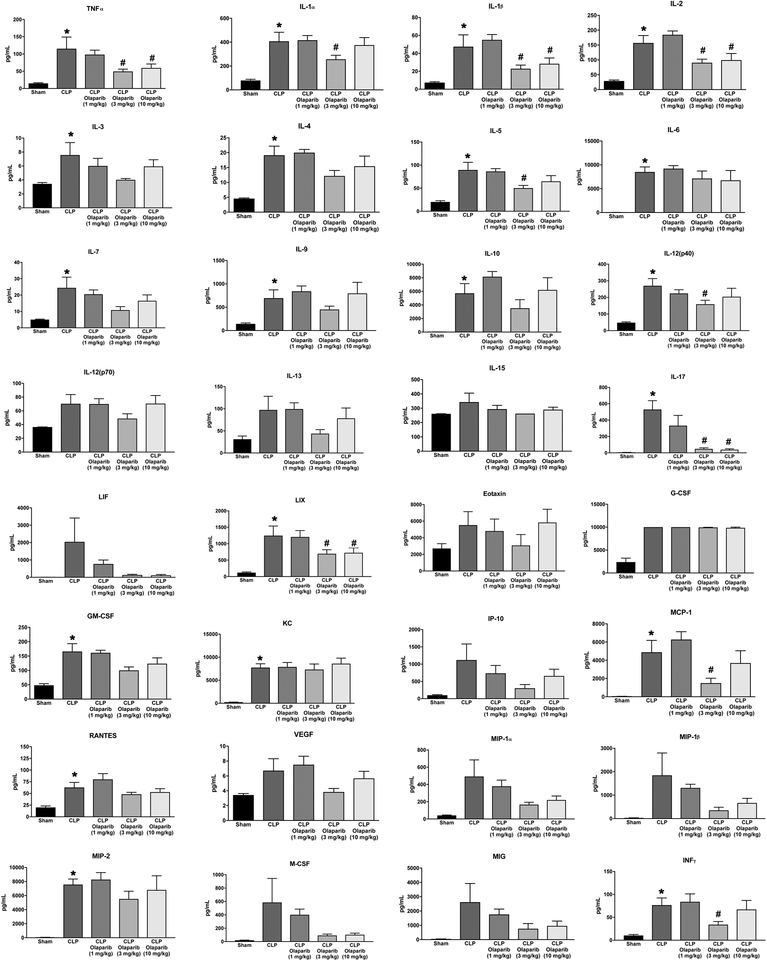
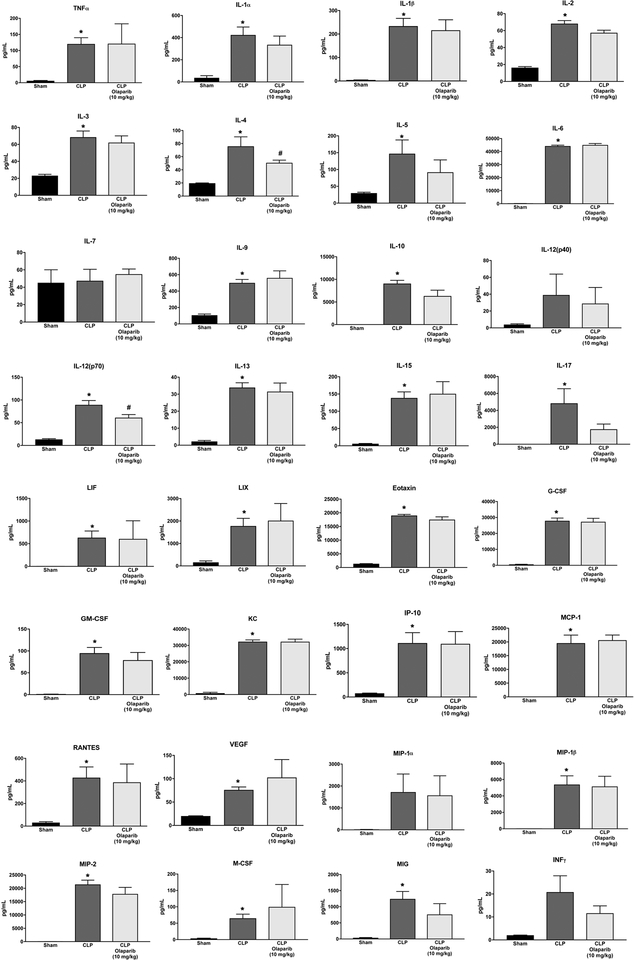
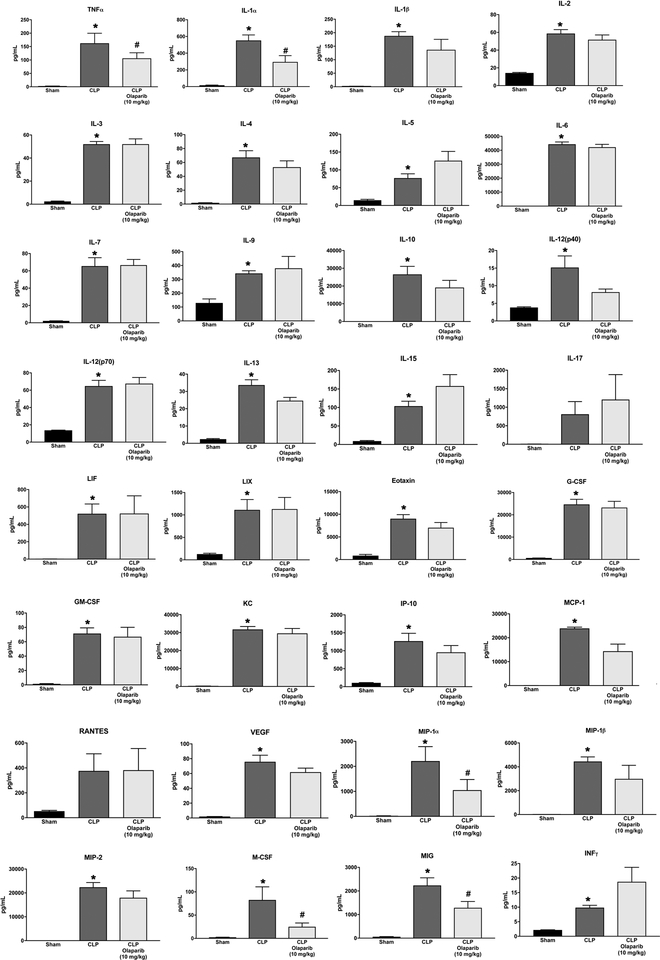
Olaparib treatment in the CLP model attenuated the increases in the levels of several circulating mediators in the plasma (e.g. TNFα, IL-1α, IL-1β, IL-2, IL-4, IL-6, IL-12p40), while others (e.g. IL-10,RANTES, VEGF) were unaffected (Fig. 11).
Figure 11. Effect of olaparib on the levels of various circulating mediators (cytokines, chemokines, growth factors) in young male Balb/c mice subjected to CLP.
Values are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant increase in the respective parameter in response to CLP, compared to the sham group; #p<0.05 shows significant protective effect of olaparib in CLP mice compared to vehicle-treated CLP mice.
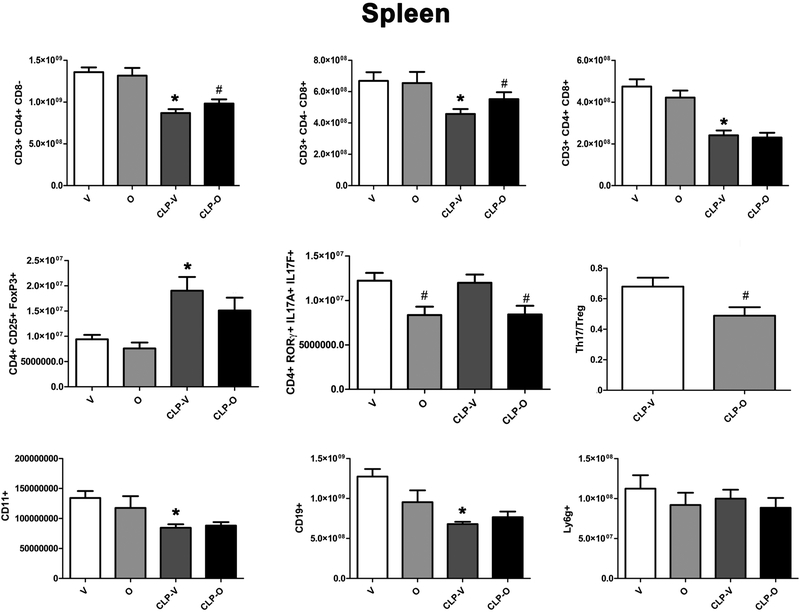
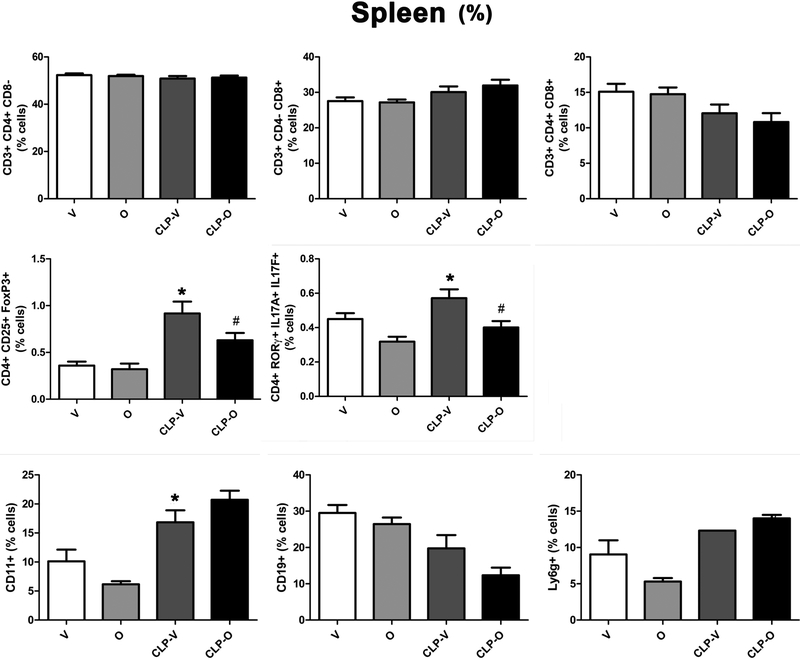
In the spleen, the number of CD4+ and CD8+ lymphocytes were reduced in response to CLP; this reduction was attenuated by olaparib treatment (Fig. 12). Moreover, in the spleen, the number of Treg (CD4+CD25+FoxP3+) but not Th17 (CD4+RORγ+IL17A+IL17F+) lymphocytes increased in response to CLP; the number, as well as percentage of both of these cell populations were reduced in CLP when the animals also received olaparib (Fig. 12, 13). The number of CD11b+ cells in spleen were significantly decreased by CLP, but percentages were significantly increased; olaparib had no significant effect on these responses. The number of CD19+ cells were significantly decreased by CLP and not affected by olaparib, and numbers and percentages of neutrophils were unaffected by CLP and olaparib (Fig. 12, 13).
Figure 12. Effect of olaparib on the numbers of various T-cell populations in the spleen of young male Balb/c mice subjected to CLP.
Values are shown in sham mice (not subjected to CLP, but treated with vehicle for 24 hours) (“V”), in sham mice (not subjected to CLP, but treated with 10 mg/kg olaparib for 24 hours) (“O”) in vehicle-treated mice subjected to CLP for 24 hours (“CLP-V”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib (“CLP-O”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows a significant change in the respective cell population in in comparison to the sham group; #p<0.05 shows significant effect of olaparib under non-CLP conditions (compared to vehicle-treated sham mice) or in CLP mice (compared to vehicle-treated CLP mice).
Figure 13. Effect of olaparib on the percentages of various T-cell populations in the spleen of young male Balb/c mice subjected to CLP.
Values are shown in sham mice (not subjected to CLP, but treated with vehicle for 24 hours) (“V”), in sham mice (not subjected to CLP, but treated with 10 mg/kg olaparib for 24 hours) (“O”) in vehicle-treated mice subjected to CLP for 24 hours (“CLP-V”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib (“CLP-O”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows a significant change in the respective cell population in response to CLP, compared to the sham group; #p<0.05 shows significant effect of olaparib under non-CLP conditions (compared to vehicle-treated sham mice) or in CLP mice (compared to vehicle-treated CLP mice).
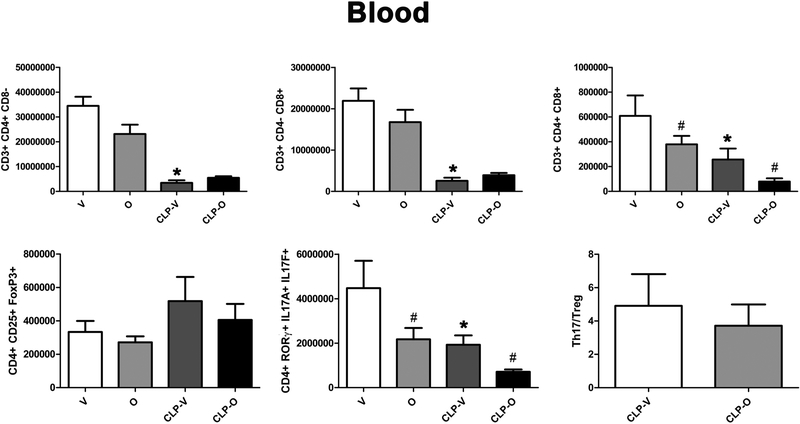
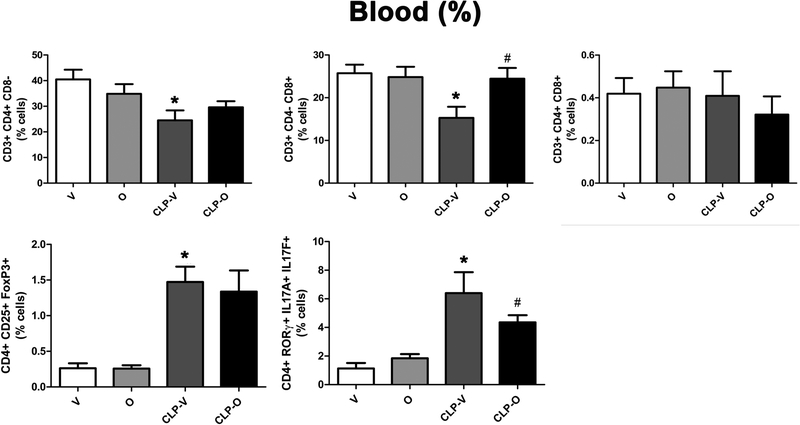
CLP reduced the number of Th17 cells in the blood – but increased as a percentage, since these alterations in Th17 cell numbers occur against the background of an overall lymphopenia - and the number (as well as percentage) of these cells was further reduced by olaparib (Fig. 14, 15). The Th17/Treg ratio is related to SOFA score; elevated ratios have been associated with increases in SOFA scores, an indication of organ damage [27]. In the spleen, this ratio was significantly lower in CLP/olaparib group than in the CLP/vehicle group (Fig. 12) and slightly lower in the blood, although not significantly (Fig. 14).
Figure 14. Effect of olaparib on the numbers of various T-cell populations in the blood of young male Balb/c mice subjected to CLP.
Values are shown in sham mice (not subjected to CLP, but treated with vehicle for 24 hours) (“V”), in sham mice (not subjected to CLP, but treated with 10 mg/kg olaparib for 24 hours) (“O”) in vehicle-treated mice subjected to CLP for 24 hours (“CLP-V”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib (“CLP-O”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows a significant change in the respective cell population in response to CLP, compared to the sham group; #p<0.05 shows significant effect of olaparib under non-CLP conditions (compared to vehicle-treated sham mice) or in CLP mice (compared to vehicle-treated CLP mice).
Figure 15. Effect of olaparib on the percentages of various T-cell populations in the blood of young male Balb/c mice subjected to CLP.
Values are shown in sham mice (not subjected to CLP, but treated with vehicle for 24 hours) (“V”), in sham mice (not subjected to CLP, but treated with 10 mg/kg olaparib for 24 hours) (“O”) in vehicle-treated mice subjected to CLP for 24 hours (“CLP-V”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib (“CLP-O”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows a significant change in the respective cell population in response to CLP, compared to the sham group; #p<0.05 shows significant effect of olaparib under non-CLP conditions (compared to vehicle-treated sham mice) or in CLP mice (compared to vehicle-treated CLP mice).
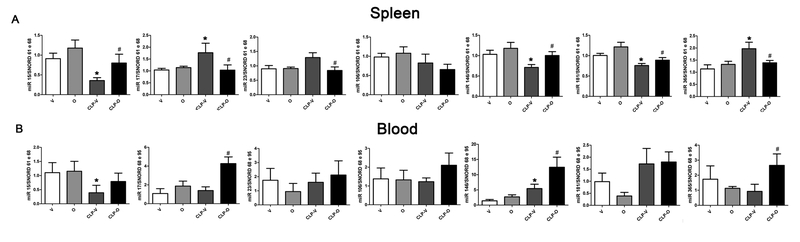
In recent years, miRNAs have been identified as important regulators of a host of immune (as well as parenchymal) cell functions, with each miRNA playing various regulatory roles that are context- and cell-type as well as pathophysiological condition dependent. Large-scale analysis of miRNA expression identified a multitude of changes in splenic miRNA levels after CLP; olaparib treatment attenuated the CLP-induced alterations in miR15, miR17, miR181 and miR365 and levels (Fig. 16). For miR146, in the spleen, olaparib attenuated its CLP-induced downregulation, while in the circulating leukocytes, olaparib enhanced its CLP-induced upregulation (Fig. 16).
Figure 16. Effect of olaparib on spleen and blood T-cell miRNA levels in young male Balb/c mice subjected to CLP.
Values are shown in sham mice (not subjected to CLP, but treated with vehicle for 24 hours) (“V”), in sham mice (not subjected to CLP, but treated with 10 mg/kg olaparib for 24 hours) (“O”) in vehicle-treated mice subjected to CLP for 24 hours (“CLP-V”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib (“CLP-O”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows a significant change in the respective miRNA level in response to CLP, compared to the sham group; #p<0.05 shows significant effect of olaparib under non-CLP conditions (compared to vehicle-treated sham mice) or in CLP mice (compared to vehicle-treated CLP mice).
3.2. The beneficial effects of olaparib are absent in young adult female mice subjected to CLP
In contrast to the findings in male mice, in young adult female mice subjected to CLP, olaparib (10 mg/kg i.p.) did not attenuate blood or splenic CFUs and did not have any significant effect on the various circulating markers of organ injury (Figs. 17,18). This finding confirms prior data showing that PARP inhibitors preferentially exert their beneficial effects in male animals subjected to various forms of critical illness (as overviewed in [19]). The degree of the change in several circulating markers (e.g. amylase, BUN) of injury in female mice tended to be less pronounced in female mice than in male mice after CLP (Fig. 2 vs. Fig. 17), perhaps indicating a lesser degree of baseline CLP-induced multiple organ injury in female mice versus male mice.
Figure 17. Effect of olaparib on selected parameters of organ injury in young female Balb/c mice subjected to CLP.
Various physiological and organ injury marker levels: alkaline phosphatase (ALP), alanine aminotransferase (ALT), amylase (AMY), plasma glucose (GLU), plasma calcium (CA++), plasma phosphate (PHOS), plasma blood urea nitrogen (BUN), plasma creatinine (CRE), plasma sodium (Na+), plasma potassium (Na+), plasma albumin (ALB) and plasma globulin (GLOB) measured by Vetscan analysis, are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant increase in the respective parameter in response to CLP, compared to the sham group.
Figure 18. Effect of olaparib on blood and spleen bacterial colony-forming unit (CFU) numbers in young female Balb/c mice subjected to CLP.
(A): Blood CFUs (expressed as CFU/ml) and (B) spleen CFUs (expressed as CFU/ml) are shown in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib (“CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group.
3.3. Olaparib exerts beneficial effects on some parameters of injury in aged male and female mice subjected to CLP
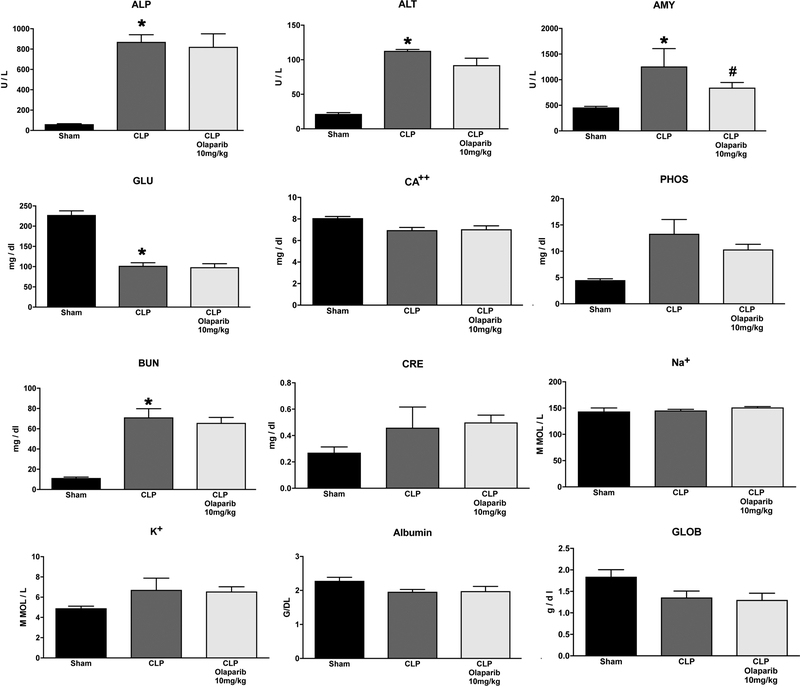
Since septic shock is, to a large extent, a disease of the elderly [1,2], and since in aged organisms some of the pathophysiological mechanisms are not only quantitatively but also qualitatively different from the mechanisms that occur in young adult organisms [2,22,28–30], we have also investigated the effect of olaparib in aged mice (72 weeks old males and in females). Surprisingly, - with the exception of the pancreatic injury marker amylase - olaparib exerted no significant beneficial effects on most organ injury markers in aged male mice (Fig. 19) and only had modulatory effects on a small subset of circulating mediators: IL-4 and IL-12(p70) (Fig. 20).
Figure 19. Effect of olaparib on selected parameters of organ injury in aged male Balb/c mice subjected to CLP.
Various physiological and organ injury marker levels: alkaline phosphatase (ALP), alanine aminotransferase (ALT), amylase (AMY), plasma glucose (GLU), plasma calcium (CA++), plasma phosphate (PHOS), plasma blood urea nitrogen (BUN), plasma creatinine (CRE), plasma sodium (Na+), plasma potassium (Na+), plasma albumin (ALB) and plasma globulin (GLOB) measured by Vetscan analysis, are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant increase in the respective parameter in response to CLP, compared to the sham group; #p<0.05 shows significant effect of olaparib in CLP mice (compared to vehicle-treated CLP mice).
Figure 20. Effect of olaparib on the levels of various circulating mediators (cytokines, chemokines, growth factors) in aged male Balb/c mice subjected to CLP.
Values are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib (“CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant increase in the respective parameter in response to CLP, compared to the sham group; #p<0.05 shows significant protective effect of olaparib in CLP mice compared to vehicle-treated CLP mice.
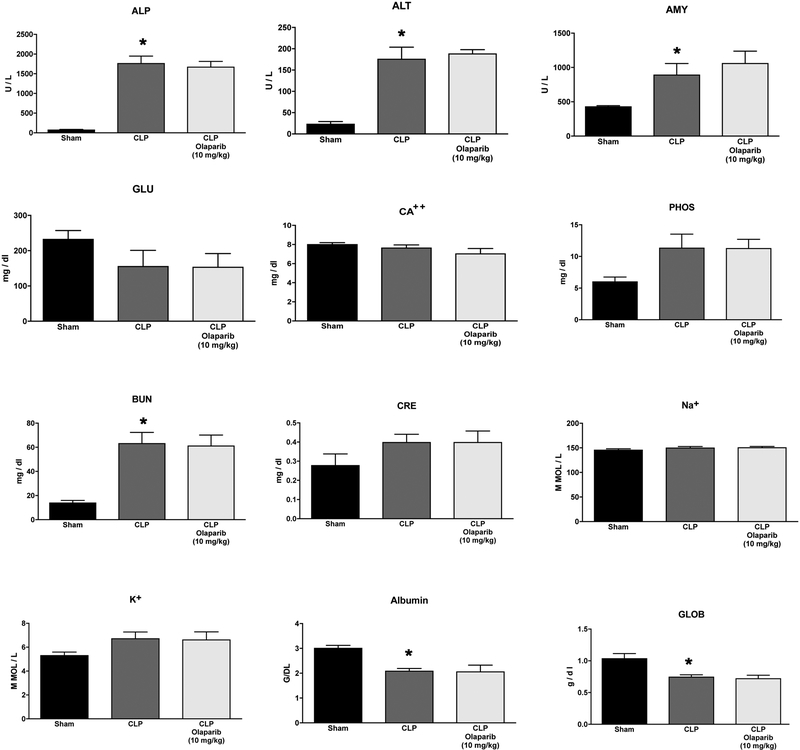
In contrast, in aged female mice, olaparib treatment significantly reduced the CLP-induced liver injury markers ALP and ALT (Fig. 21). However, levels of the kidney injury marker BUN and pancreatic marker amylase were not affected by olaparib after injury. Circulating levels of the CLP-induced mediators TNFα, IL-1α, MIP1α, M-CSF and MIG were significantly reduced by olaparib treatment (Fig. 22).
Figure 21. Effect of olaparib on selected parameters of organ injury in aged female Balb/c mice subjected to CLP.
Various physiological and organ injury marker levels: alkaline phosphatase (ALP), alanine aminotransferase (ALT), amylase (AMY), plasma glucose (GLU), plasma calcium (CA++), plasma phosphate (PHOS), plasma blood urea nitrogen (BUN), plasma creatinine (CRE), plasma sodium (Na+), plasma potassium (Na+), plasma albumin (ALB) and plasma globulin (GLOB) measured by Vetscan analysis, are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of various doses of olaparib (“CLP Olaparib 1 mg/kg”, CLP Olaparib 3 mg/kg” and “CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant increase in the respective parameter in response to CLP, compared to the sham group; #p<0.05 shows significant effect of olaparib in CLP mice (compared to vehicle-treated CLP mice).
Figure 22. Effect of olaparib on the levels of various circulating mediators (cytokines, chemokines, growth factors) in aged female Balb/c mice subjected to CLP.
Values are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib (“CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group; *p<0.05 shows significant increase in the respective parameter in response to CLP, compared to the sham group; #p<0.05 shows significant protective effect of olaparib in CLP mice compared to vehicle-treated CLP mice.
Neither in aged male or female mice subjected to CLP did we observe any adverse effect of olaparib on nuclear or mitochondrial DNA integrity (Fig. 23).
Figure 23. Effect of olaparib on liver mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity in aged male and female Balb/c mice subjected to CLP.
(A): mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity values (expressed as % of control) in aged male mice; (B): mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity values (expressed as % of control) in aged female mice. Data are shown in sham mice (not subjected to CLP) (“Sham”), in vehicle-treated mice subjected to CLP for 24 hours (“CLP”) and in mice subjected to CLP in the presence of 10 mg/kg olaparib (“CLP Olaparib 10 mg/kg”). Data are shown as mean ± SEM of 10 animals for each group.
Overall, the degree of the beneficial effect of the PARP inhibitor tended to be less pronounced in the aged mice than in the young male mice (Figs. 1–2. vs. Figs. 17–22).
3.4. Olaparib exerts cytoprotective and beneficial bioenergetic effects in U937 cells subjected to oxidative stress in vitro, without adversely affecting DNA integrity
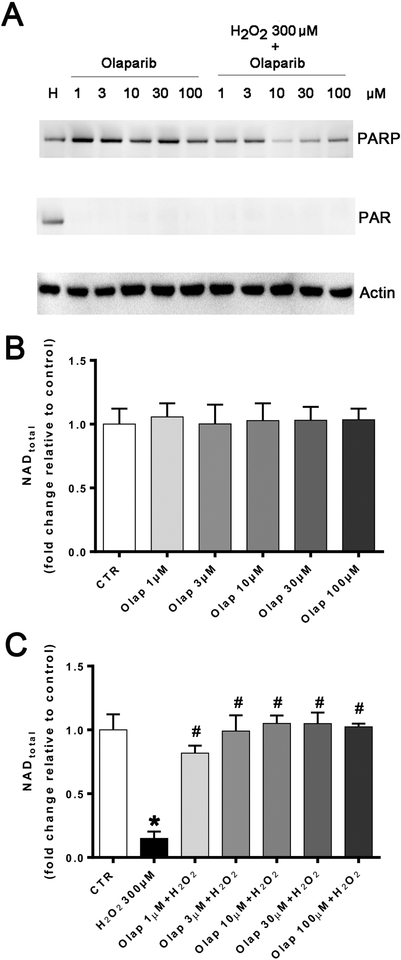
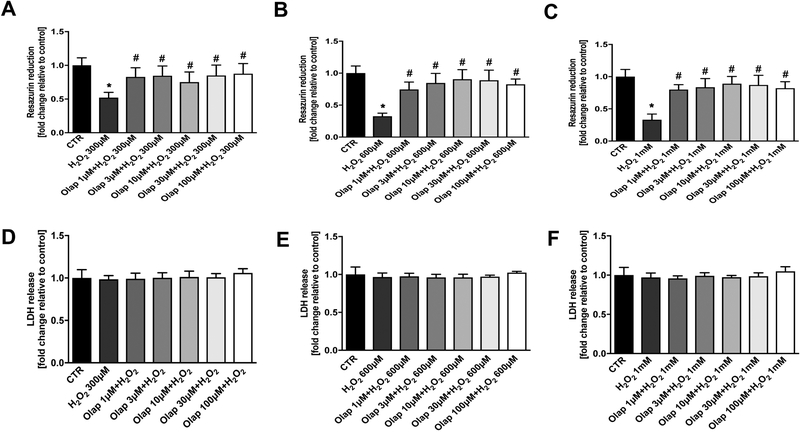
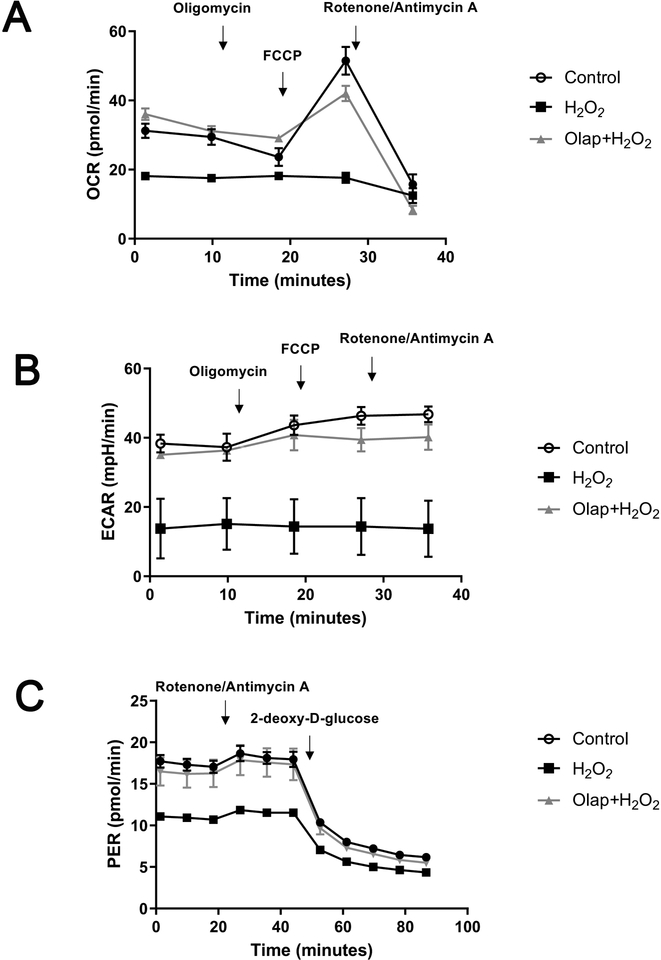
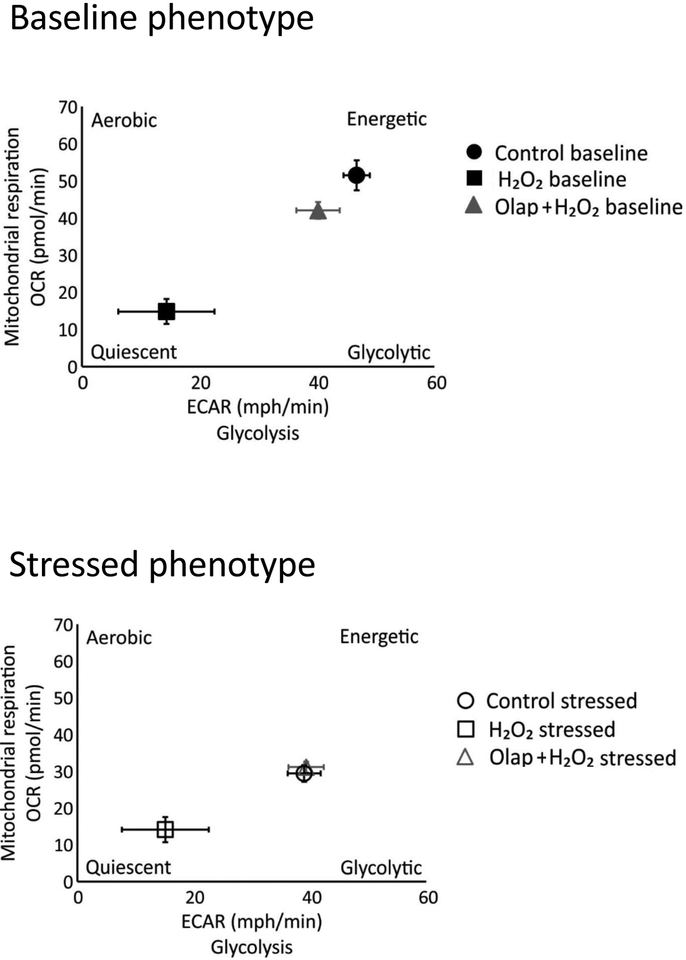
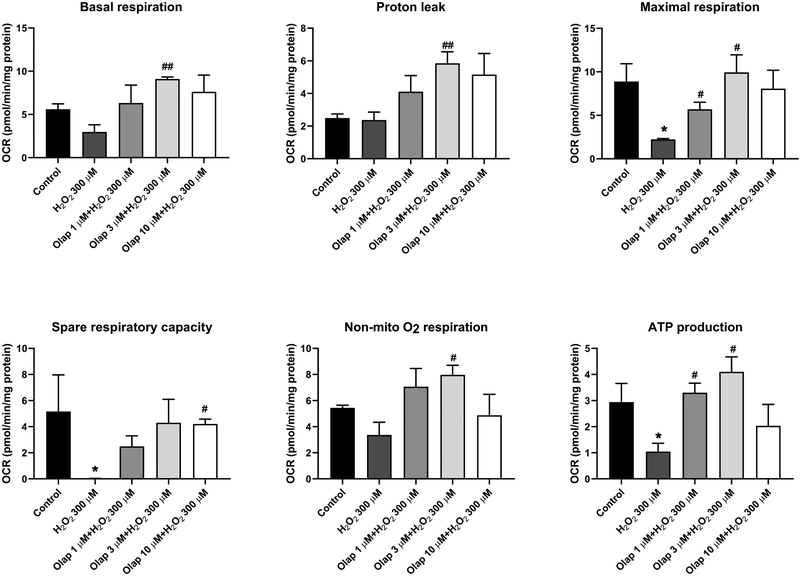
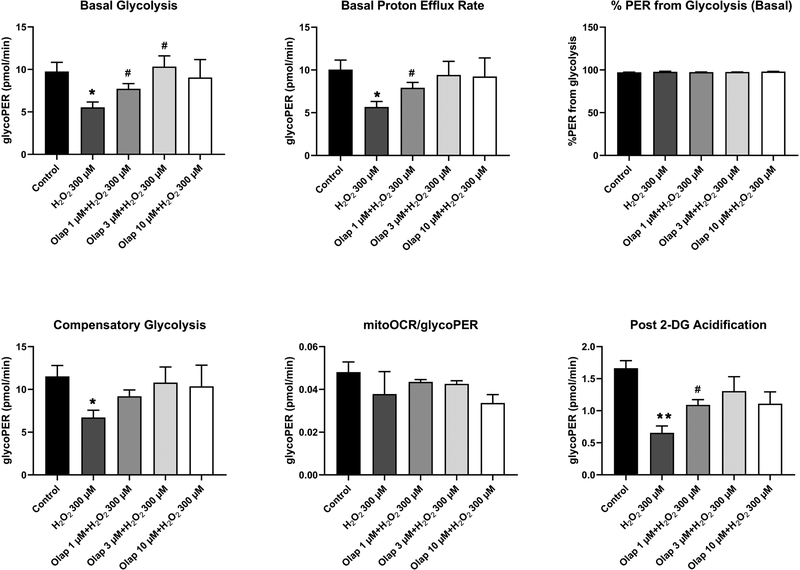
In in vitro experiments in U937 cells subjected to oxidative stress, olaparib (1– 100 μM) completely inhibited PARylation (as measured by the quantification of poly(ADP-ribose) [PAR], the product of the enzyme) already at the lowest concentration (1 μM) used, confirming the well-established [31] potent effect of olaparib to inhibit PARP1 catalytic activity (Fig. 24A). In line with prior findings demonstrating that PARP inhibition prevents the cellular depletion of its substrate, NAD+ [32,33], olaparib also protected against the H2O2-induced loss of cellular NAD+ levels, with the effects already near-maximal at its lowest tested concentration (1 μM) (Fig. 24B,C). Consistent with prior findings demonstrating the protective effect of PARP inhibitors in cells exposed to various oxidants [25,32–35] olaparib protected against the H2O2-induced loss of cell viability, indicated by continued resazurin reduction in the presence of olaparib treatment (Fig. 25). The PARP inhibitor also protected against the H2O2-induced changes in oxidative phosphorylation - as assessed by cellular bioenergetic parameters such as resting and FCCP-stimulated (maximal) mitochondrial respiration - as well as against the H2O2-induced changes in anaerobic respiration (glycolysis) (Figs. 26–29).
Figure 24. Effect of olaparib on PARP activation and cellular NAD+ levels in U937 cells subjected to H2O2-induced oxidative stress in vitro.
(A): Western blotting for PARP1 (top lanes), polyADP-ribose (PAR), the enzymatic product of PARP activation (middle lanes) and actin loading control (bottom lanes) in U937 cells treated with olaparib (1, 3, 10, 30 or 100 μM) in control and in 300 μM H2O2 (“H”) challenged cells at 1 hour. Note the increase in PAR (indicative of PARP activation) in response to oxidative stress, and the full inhibition of this response at all olaparib concentrations tested. The expression of PARP1 was unaffected by olaparib in any of the control conditions tested. However, in the oxidatively stressed cells, in the presence of olaparib, the expression of PARP1 also appeared to be downregulated. Western blots shown are representative of experiments performed on 3 different experimental days. (B): Effect of olaparib on the cellular NAD+ levels in control cells (without oxidative stress challenge). Cells were treated with olaparib (1, 3, 10, 30 or 100 μM) for 1 hour. Data are shown as mean±SEM of n=3 determinations. (C): Effect of olaparib on the cellular NAD+ levels in cells subjected to oxidative (H2O2). Cells were treated with olaparib (1, 3, 10, 30 or 100 μM), followed immediately by 300 μM H2O2 for 1 hour. Data are shown as mean±SEM of n=3 determinations. *p<0.05 shows significant decrease in NAD+ levels in response to H2O2, compared to control cells; #p<0.05 shows significant protective effect of olaparib in H2O2 challenged cells compared to vehicle-treated H2O2 challenged cells.
Figure 25. Effect of olaparib on cellular viability in U937 cells subjected to H2O2-induced oxidative stress in vitro.
(A): Effect of olaparib on cellular resazurin reduction in control cells (without oxidative stress challenge) and in cells subjected to oxidative (H2O2) in the presence of various concentrations of olaparib. Cells were treated with olaparib (1, 3, 10, 30 or 100 μM), followed immediately by 300 μM H2O2 for 1 hour. Data are shown as mean±SEM of n=3 determinations. *p<0.05 shows significant decrease in resazurin conversion in response to H2O2, compared to control cells; #p<0.05 shows significant protective effect of olaparib in H2O2 challenged cells compared to vehicle-treated H2O2 challenged cells. (B): Effect of olaparib on LDH content (a marker of cell necrosis) in the supernatant of control cells (without oxidative stress challenge) and in cells subjected to oxidative (H2O2) in the presence of various concentrations of olaparib. Cells were treated with olaparib (1, 3, 10, 30 or 100 μM), followed immediately by 300 μM H2O2 for 1 hour. Data are shown as mean±SEM of n=3 determinations.
Figure 26. Effect of olaparib on bioenergetic parameters in U937 cells subjected to H2O2-induced oxidative stress in vitro.
(A): Representative oxygen consumption rate (OCR) profiles in control cells, in H2O2 (300 μM) treated cells at 1 hour, and in H2O2 (300 μM) treated cells at 1 hour in the presence of pretreatment with 10 μM olaparib. Arrows indicate the addition of specific mitochondria stressors. Results are indicated as mean ± SEM of n=3 per condition. (B): Representative extracellular acidification rate (ECAR) profiles in control cells, in H2O2 (300 μM) treated cells at 1 hour, and in H2O2 (300 μM) treated cells at 1 hour in the presence of pretreatment with 10 μM olaparib. Arrows indicate the addition of specific mitochondria stressors into the media. Results are indicated as mean ± SEM of n=3 per condition. (C): Representative proton efflux rate (PER) profiles in control cells, in H2O2 (300 μM) treated cells at 1 hour, and in H2O2 (300 μM) treated cells at 1 hour in the presence of pretreatment with 10 μM olaparib. Arrows indicate the addition of specific mitochondria stressors into the media. Results are indicated as mean ± SEM of n=3 per condition.
Figure 29. Effect of olaparib on bioenergetic parameters in U937 cells subjected to H2O2-induced oxidative stress in vitro: energy phenotype graphs.
Representative energy phenotype graph showing control, H2O2 (300 μM) treated cells and H2O2 (300 μM) treated cells in the presence of 10 μM olaparib pretreatment. Open boxes indicate baseline phenotype, filled boxed represent stressed phenotype. Note that H2O2 impairs both basal and stressed energy phenotype, and olaparib provides a complete or near-complete restoration of these alterations. Results are indicated as mean ± SEM of n=3 per condition.
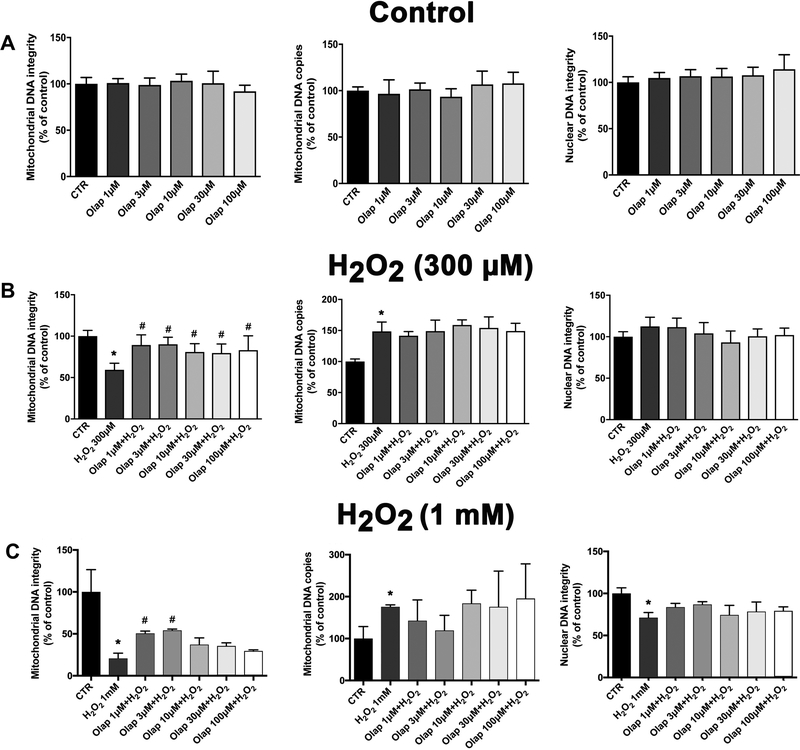
At the entire concentration range tested (1–100 μM), olaparib did not have any adverse effects on nuclear or mitochondrial DNA integrity in control cells (i.e. cells not subjected to oxidative stress) (Fig. 30A) but protected against the development of oxidative stress-induced mitochondrial DNA damage (Fig. 30B,C). This latter finding may be related to effects on the mitochondrial isoform of PARP, which has a qualitatively different role in the regulation of mitochondrial DNA repair than its nuclear counterpart [36]. At the lower levels of oxidative stress -in line with prior observations [36–38] - the H2O2-induced DNA damage was preferential to mitochondrial, as opposed to nuclear DNA (Fig. 30B). At the highest concentration of H2O2 tested (1 mM), nuclear DNA damage was also detected: this damage, however, was not further exacerbated by olaparib (Fig. 30C).
Fig. 30. Effect of olaparib on mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity in U937 cells subjected to H2O2-induced oxidative stress in vitro.
(A): Mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity in control U937 cells (without oxidative stress) subjected to vehicle or various concentrations of (1, 3, 10, 30 or 100 μM) olaparib for 24 hours (data expressed as % of control); (B): Mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity in control U937 cells (without oxidative stress) subjected to vehicle or various concentrations of olaparib (1, 3, 10, 30 or 100 μM), followed by treatment with H2O2 (300 μM) for 24 hours (data expressed as % of control); (C): Mitochondrial DNA integrity, mitochondrial DNA copy number and nuclear DNA integrity in control U937 cells (without oxidative stress) subjected to vehicle or various concentrations of olaparib (1, 3, 10, 30 or 100 μM), followed by treatment with H2O2 (1 mM) for 24 hours (data expressed as % of control). Results are indicated as mean ± SEM of n=3 per condition. *p<0.05 shows a significant change in response to H2O2, compared to control cells; #p<0.05 shows significant protective effect of olaparib in H2O2 challenged cells compared to vehicle-treated H2O2 challenged cells.
Taken together, we have demonstrated cytoprotective and positive bioenergetic effects of olaparib in oxidatively stressed U937 cells, but we did not find any adverse effect of the PARP inhibitor on DNA integrity.
4. Discussion
The overall conclusion of the current study is that in young male mice subjected to CLP the PARP inhibitor olaparib improves organ function, beneficially modulates the inflammatory and immune response, and extends survival in septic shock. Moreover, all of these effects occur without any detectable adverse effects on mitochondrial or nuclear DNA integrity. The in vitro experiments, demonstrating cytoprotective and beneficial cellular bioenergetic effects of olaparib - once again without any detectable adverse effects on mitochondrial or nuclear DNA integrity - are in line with the conclusions of the in vivo experiments.
Prior studies have demonstrated multiple modes of PARP inhibitor’s beneficial effects in various non-oncological models of cell and organ injury and local and systemic inflammation. These effects include direct cytoprotective actions (preserving NAD+ and ATP levels and thereby protecting against cell necrosis) [32,33,39]; suppression of the expression and production of multiple pro-inflammatory mediators and circulating factors [40–42]; prevention of the infiltration of organs with activated mononuclear and polymorphonuclear cells [33,44] and protection against the loss of vascular (especially endothelial) integrity [5–7,33]. Many of the cytoprotective and organ-protective effects of olaparib have also been directly demonstrated in various models, ranging from neuroinjury to endotoxin-induced lung and kidney failure (reviewed in [19]). The current study confirms and extends these observations. However, the current study also demonstrates that not all populations of animals subjected to CLP benefit equally from olaparib. Clearly, the most pronounced effects were noted in the young adult male mice. On the other hand, olaparib exerted no significant benefit in young adult female mice subjected in CLP. This finding is in line with several sets of studies demonstrating that PARP inhibitors (or genetic PARP1 deficiency) tends to favor the male gender in models as diverse as endotoxin-induced organ injury [12] and ischemic stroke [44]. This may be related to a physiological inhibitory effect of estrogen on PARP activation (reviewed in [19]). Because this effect of estrogen is expected to be reduced in aged (i.e. postmenopausal) female organisms, we have expected that in aged female mice, the benefit of olaparib would be reestablished. Indeed, this was the case (at least for some injury markers, such as ALP and ALT, which were suppressed by olaparib in aged female mice. However, other markers, e.g. amylase, or BUN remained unaffected by olaparib in this animal population). Surprisingly, we have observed only a limited degree of beneficial effects of olaparib in aged male mice - with one isolated effect (protection against the increase in the pancreatic injury marker amylase). Overall, the effects of olaparib in aged mice (male or female) tended to be less pronounced than the effects in young adult male mice. This finding is consistent with several lines of studies indicating that in aged organisms, different sets of pathophysiological mechanisms become mobilized than in young adult animals; among many differences, aged organisms appear to lose the ability to mobilize Nrf2, a master switch of antioxidant defense [30]. There are many additional possibilities as well. For instance, it is conceivable that the regulation of PARP1 expression and or activity may be age-dependent, and this should be a subject of further investigations. Indeed, lymphocytes from old subjects show a 50% reduction in the constitutive expression level of both parp 1 and parp 2 genes compared to young subjects [45]. The vast array of mechanisms responsible for multiple organ injury, immune and inflammatory responses and mortality associated with sepsis remain to be further characterized. Nevertheless, the findings reported in the current study clearly indicate that different populations (age and gender) may respond to the same insult with quantitatively different pathophysiological mechanisms, which, in turn, may require different (matching) experimental therapeutic approaches (or a combination of these).
What, then, is the principal mechanism of olaparib in sepsis? As with most in vivo studies, it is difficult to decipher the exact sequence of interrelated effects. We believe that the most likely answer is that olaparib exerts multiple effects at various levels and at multiple interrelated checkpoints of septic shock (Fig. 31). One of these effects may be the preservation of cellular high-energy nucleotides, and inhibition of cell dysfunction (e.g. cellular bioenergetic impairments, or full-scale cell death such as cell necrosis) [19]. In addition - probably through interaction with a number of nuclear signaling mechanisms [33,46,47], olaparib down-regulates the production of multiple circulating mediators [19]. The current results also point to several additional mechanisms that have not been previously documented in the literature (with olaparib or with any of the earlier classes of PARP inhibitors). One of them may be the effect of olaparib on the composition of splenic and circulating immune cells, including Treg and Th17 lymphocytes. Another, likely closely related mechanism may be the effect of olaparib on bacterial clearance: in animals treated with the highest dose of olaparib, there was a marked suppression in circulating and splenic CFUs, indicating that the PARP inhibitor may have, in some way, stimulated bacterial elimination / antibacterial immune responses. This effect of olaparib does not appear to be a direct effect on the bacteria (since olaparib did not affect E. coli growth rates in vitro) and did not affect elimination of an E. coli bolus in an in vivo model of bacteremia without septic shock and without mortality. It is conceivable that the effect of olaparib on T-cell populations may have contributed to a readjustment of the immune responses in response to intestinal tissue damage and polymicrobial sepsis.
Fig. 31. Effects of olaparib in septic shock: a working hypothesis.
Via inhibition of the catalytic activity of PARP, olaparib exerts cellular bioenergetic effects as well as pluripotent modulation of gene transcription. These effects lead to changes in the circulating and tissue-resident immune cell distribution, improve bacterial elimination, protect against multiorgan injury and eventually culminate in improved survival rate. These effects are dependent on gender and age of the animals. See the Discussion section for further context and explanation.
What, then, is the mechanism (and functional consequence) of the changes in T-cell subpopulations reported in the current study? Regulatory T lymphocytes (Tregs) are immunosuppressive cells, and Tregs are elevated in patients with sepsis [48]. Previous studies have demonstrated that PARP-1 negatively regulates the regulatory T lymphocyte (Treg) (CD4+ CD25+FoxP3+) by poly-ADP-ribosylating FoxP3. Its deletion makes the expression of FoxP3 more persistent and stable, conferring a more intense suppressive activity to the Treg lymphocytes. PARP1 deficient mice have been shown to have a higher number of Tregs in peripheral lymphatic organs and in the thymus [49]. In the current study, olaparib, on its own (i.e. in mice not subjected to CLP), did not affect the Treg populations. It is not surprising that long-term, complete deletion of PARP1 has a different effect on cell maturation and selection than a short-term, partial pharmacological inhibition of the same enzyme. However, when the animals were subjected to CLP, olaparib partially counteracted the increase in splenic Treg cell populations (Fig. 12). Tregs are known to reduce amount and activity of immune effector cells, causing a state of immunosuppression that compromises the ability to eliminate bacteria [50]. Therefore, part of our working hypothesis is that the partial prevention of CLP-induced Tregs may prevent immunosuppression and may facilitate the elimination of bacteria during CLP.
Th17 cells are a proinflammatory lineage producing cytokines that increase inflammation, free radical production and organ damage. In the spleen, the number of Th17 cells was unaffected by CLP, but in the blood, the number of Th17 cells decreased, but, in line with prior findings [51], the percentage of these cells increased. From all four groups of mice tested, the lowest number of Th17 populations were observed in the CLP+olaparib group. This may be interpreted as follows: that although – when considering the total number of cells - the animals subjected to CLP are suffering from lymphopenia, the pro-inflammatory lymphocyte profile is more intense. However, importantly, in the olaparib treated CLP mice, Th17 cell populations are reduced, rendering the Th17/Treg ratio lower and thereby reducing the inflammation. It is important to emphasize in this context that (a) the Th17/Treg ratio positively correlates with the SOFA scores and the higher the relation, the greater the adverse outcome in patients with sepsis [27] and (b) in our model, the spleen had a lower Th17/Treg ratio in CLP-olaparib group when compared to CLP-vehicle. Indeed, the reduced Th17/Treg ratio after olaparib treatment was also associated with a reduction in some inflammatory cytokines and also with an improved histopathological picture in the liver. Since Th17 cells are responsible for causing immunopathology during inflammation, the beneficial effects of olaparib on various indices or organ injury may be – directly or indirectly – connected to the effect of olaparib on Th17 cell numbers. However, it is also conceivable that the protective effects of olaparib occur at several different molecular and cellular targets - including the production and/or action of reactive oxidants and free radicals, the development of vascular – e.g. endothelial – dysfunction, the regulation of pro-inflammatory cytokines, chemokines and other soluble mediators, and so on. Taken together, our working hypothesis is that olaparib may help to maintain some degree of immune homeostasis after CLP by, on one hand, reducing inflammation and organ damage (through modulating Th17), while concomitantly preventing some immunosuppression caused by Treg activity (through modulating Tregs), thereby keeping the host able to fight bacteria.
PARP is known to act in conjunction with NF-κB to regulate the activation and transcription of genes responsible for the inflammatory response: PARP knockout animals are known to have reduced production of cytokines and other proinflammatory proteins that are under the control of the NF-κB system [11]. NF-κB - besides activating mRNA production - also activates the epigenetic regulation of the expression of several genes, through the production of microRNAs. Our study suggests that the PARP inhibitor olaparib has an action of epigenetic regulation, since it modifies the production of several miRNA related to the differentiation and activation of Tregs and Th17 cells. Although miRNAs are known to regulate a host of cellular responses in health and disease [52–55], at this time, we don’t know enough about the function of each of the microRNAs studied here to integrate the findings into a coherent mechanistic hypothesis. However, some information related to microRNAs is intriguing in the context of the current findings. For instance, the 15a-5p miRNA is known to serve as a tumor suppressor and is reduced in plasma from sepsis patients who survived when compared to those who did not survive [56,57]. Moreover, overexpression of 17a-5p miRNA reduces the suppressor activity of regulatory T lymphocytes [58–60]. In our study, olaparib treatment tended to normalize miRNA15 and miRNA17 levels in septic animals, and olaparib treatment was also accompanied by a partial normalization of Treg cell counts in CLP animals. Whether the number of Treg cells was regulated through olaparib’s action on miRNA15a and/or miR17 levels remains to be further explored.
The 365–5p miRNA, expressed in patients with sepsis, is an important negative regulator of IL-6 [61,62]. Its expression was elevated in the spleen of CLP-vehicle group, and this was attenuated by olaparib. It is conceivable – but remains to be further tested – that the attenuation of IL-6 levels by olaparib may occur (partially or wholly) through its action on miRNA365.
Although not a DNA repair enzyme per se, PARP1 has been implicated in the maintenance of DNA integrity, chiefly through the recruitment of various DNA repair enzymes and the assembly of nuclear and mitochondrial DNA repair complexes [33,36,63]. In fact, inhibition of DNA repair is the principal mode of action of olaparib’s approved indication in the field of oncology [21]. However, a few things need to be kept in perspective in this regard (these considerations are discussed, in detail, in our recent position paper [19] and are only briefly reiterated here). First of all, the effect of olaparib on DNA repair is especially pronounced when cells harbor specific mutations (e.g. mutations in the BRCA gene), and often in combination with treatment with cytotoxic and/or genotoxic chemotherapeutic agents; in healthy cells, the effects of PARP inhibitors are less pronounced. Second, for the DNA repair effects of PARP inhibitors to be markedly efficient, a high degree of PARP1 inhibition must be achieved: thus, the doses of the PARP inhibitor to be effective as an anticancer agent are substantially higher than the doses of the PARP inhibitor to be effective as a cytoprotective or anti-inflammatory agent in non-oncological indications. This was also clear from the results of the current study, where olaparib at 3–10 mg/kg doses, exerted effects against multiple organ failure, inflammatory and immune dysregulation, and - at 10 mg/kg - also improved survival rates. These doses are at least one order of magnitude lower than the doses of olaparib that exert anticancer effects in vivo in tumor-bearing mice [19].
A distinction should also be made between the role of PARP in the regulation of nuclear and mitochondrial DNA integrity; we have recently demonstrated that PARP1 deficient cells may have some deficits or delays in nuclear DNA repair, but at the same time their mitochondrial DNA repair systems are, paradoxically, more efficient [36]. In line with these findings, we have, in fact, demonstrated that olaparib exerts beneficial effects against the loss of mitochondrial DNA integrity, both in vivo and in vitro. Since the loss of mitochondrial DNA integrity can, subsequently, be associated with the active release of damaged mitochondrial DNA from the cells, and this mitochondrial DNA can, in turn, induce inflammatory responses in adjacent cells [26] the current findings raise the possibility that inhibition of such a response by olaparib may be an additional model of its beneficial action. However, this possibility remains to be tested in future experiments.
The methodology used in the current study to assess mitochondrial and nuclear DNA integrity (the ‘LA-qPCR’ assay) is technically highly reliable and it is widely used in the literature 64,65]. Although we did not observe differences in nuclear DNA and mitochondrial DNA integrity using this assay, this does not necessarily mean that inhibition of PARP does not affect nuclear and mitochondrial DNA repair or DNA integrity in general. One of the limitations of LA-qPCR is that it measures only DNA polymerase protruding DNA lesions such as DNA breaks. However, for example 8-oxoG, which is the most frequently occurring DNA damaged induced by the oxidative stress, will be by-passed during PCR amplification and consequently will not affect the measured DNA integrity. Another limitation of the LA-qPCR is that it will not reflect DNA repair capacity (i.e. the kinetics of the DNA repair process). In addition, since LA-qPCR measures changes in gene-specific integrity it is possible that other DNA regions, particularly in the nuclear DNA, may be affected but not detected by the LA-qPCR. Moreover, the assay does not measure other types of DNA injury (e.g. base modifications, or larger scale aberrations like micronucleus instability or chromosomal alterations) [64,65]. To assess the effect of olaparib on these parameters, further studies needed to be conducted. Nevertheless, it should be pointed out that ours is not the only report that demonstrates that PARP inhibitors can exert beneficial effects in critical illness models without adverse effects. For instance, Hauser and colleagues have demonstrated that a PARP-1 inhibitor of another structural class than olaparib (INO-1001) facilitates hemodynamic stabilization in a porcine aortic cross-clamp model, without adversely affecting DNA integrity (in that particular study, DNA integrity was assessed by the comet assay, i.e. single-cell gel electrophoresis) [14].
Overall, in the context of the experimental therapy of sepsis, it appears that it is possible to find doses or concentrations of olaparib that exert beneficial effects in terms of organ function, immunological parameters, circulating mediators and survival (in vivo) or cell viability and cellular bioenergetics (in vitro), without adversely affecting DNA integrity.
5. Conclusions
Based on the studies in mice subjected to CLP, we conclude that olaparib can improve organ function and extend survival in septic shock. These effects are, at least in part, due to an enhancement of antibacterial clearance, which may be related to the modulation of various splenic, thymic and blood T-lymphocyte subpopulations and/or due to effects on multiple circulating chemokine or cytokine levels. According to our working hypothesis, on one hand, olaparib is able to reduce inflammation and organ damage (perhaps through reduction of Th17 cell numbers in CLP) while, on the other hand, blocking immunosuppression caused by affecting Tregs and thereby preserving host antimicrobial functions. We have also confirmed prior data indicating that the effect of PARP inhibitors is preferentially manifesting in young adult male mice. However, in aged animals - i.e. old male mice and especially in old female mice - olaparib remained partially effective in protecting against multiorgan injury. Based on measurement of mitochondrial and nuclear DNA integrity in vivo (tissues obtained from mice subjected to CLP) and in vitro (human monocytic cell lines subjected to oxidative stress), olaparib, in the dose range (1–10 mg/kg) and concentration range (1–100 μM) tested did not exert adverse effects on nuclear DNA integrity and exerted a protective effect against mitochondrial DNA damage and mitochondrial function. Taken together, the current results suggest that olaparib exerts beneficial effects in sepsis and supports the concept of repurposing and clinical introduction of this PARP inhibitor for the experimental therapy of septic shock.
Figure 27. Effect of olaparib on oxidative phosphorylation-related bioenergetic parameters in U937 cells subjected to H2O2-induced oxidative stress in vitro.
Results are shown in control cells, in H2O2 (300 μM) treated cells at 1 hour, and in H2O2 (300 μM) treated cells at 1 hour in the presence of pretreatment with 1, 3 or 10 μM olaparib. Results are indicated as mean ± SEM of n=3 per condition. *p<0.05 shows a significant change in response to H2O2, compared to control cells; #p<0.05 and ##p<0.01 show significant protective effect of olaparib in H2O2 challenged cells compared to vehicle-treated H2O2 challenged cells.
Figure 28. Effect of olaparib on glycolysis-related bioenergetic parameters in U937 cells subjected to H2O2-induced oxidative stress in vitro.
Results are shown in control cells, in H2O2 (300 μM) treated cells at 1 hour, and in H2O2 (300 μM) treated cells at 1 hour in the presence of pretreatment with 1, 3 or 10 μM olaparib. Results are indicated as mean ± SEM of n=3 per condition. *p<0.05 shows a significant change in response to H2O2, compared to control cells; #p<0.05 shows significant protective effect of olaparib in H2O2 challenged cells compared to vehicle-treated H2O2 challenged cells.
Funding:
This work was supported by grants from the National Institutes of Health (R01GM107876) and the Swiss National Foundation (to C.S.), and by the FAPESP (to R.S. and C.C.)
List of abbreviations:
- ALB
albumin
- ALT
alanine aminotransferase
- AMY
amylase
- BUN
blood urea nitrogen
- Ca2+
total calcium
- CRE
creatinine
- CLP
cecal ligation and puncture
- GLU
glucose
- K+
potassium
- MDA
malon dialdehyde
- MPO
myeloperoxidase
- Na+
sodium
- Nrf2
nuclear factor erythroid 2-related factor 2
- NO
nitric oxide
- PAR
poly(ADP-ribose)
- PARP
poly(ADP-ribose) polymerase
- PHOS
phosphorus
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethics approval and consent to participate: The current report does not contain human studies.
Consent for publication: The current report does not contain human studies or studies that would require consenting.
Availability of data and material: The datasets during and/or analysed during the current study available from the corresponding author on reasonable request.
Competing interests: The authors declare no competing interests.
References
- 1.Martin GS, Mannino DM, Eaton S, Moss M: The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 2001, 348:1546–54. [DOI] [PubMed] [Google Scholar]
- 2.Martin GS, Mannino DM, Moss M: The effect of age on the development and outcome of adult sepsis. Crit Care Med 2006, 34:15–21. [DOI] [PubMed] [Google Scholar]
- 3.Trentzsch H, Nienaber U, Behnke M, Lefering R, Piltz S: Female sex protects from organ failure and sepsis after major trauma haemorrhage & Injury. Int J Care Injured 2014, 45S:20–8. [DOI] [PubMed] [Google Scholar]
- 4.Bösch F, Angele MK, Chaudry IH: Gender differences in trauma, shock and sepsis. Mil Med Res 2018, 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szabó C, Zingarelli B, Salzman AL: Role of poly-ADP ribosyltransferase activation in the vascular contractile and energetic failure elicited by exogenous and endogenous nitric oxide and peroxynitrite. Circ Res 1996, 78:1051–63. [DOI] [PubMed] [Google Scholar]
- 6.Szabo C, Cuzzocrea S, Zingarelli B, O’Connor M, Salzman AL: Endothelial dysfunction in a rat model of endotoxic shock. Importance of the activation of poly (ADP-ribose) synthetase by peroxynitrite. J Clin Invest 1997, 100:723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jagtap P, Soriano FG, Virag L, Liaudet L, Mabley J, Szabo E, Hasko G, Marton A, Lorigados CB, Gallyas F Jr, Sümegi B, Hoyt DG, Baloglu E, VanDuzer J, Salzman AL, Southan GJ, Szabo C: Novel phenanthridinone inhibitors of poly (adenosine 5’-diphosphate-ribose) synthetase: potent cytoprotective and antishock agents. Crit Care Med 2002, 30:1071–82. [DOI] [PubMed] [Google Scholar]
- 8.Soriano FG, Liaudet L, Szabó E, Virág L, Mabley JG, Pacher P, Szabó C: Resistance to acute septic peritonitis in poly(ADP-ribose) polymerase-1-deficient mice. Shock 2002, 17:286–92. [DOI] [PubMed] [Google Scholar]
- 9.Goldfarb RD, Marton A, Szabó E, Virág L, Salzman AL, Glock D, Akhter I, McCarthy R, Parrillo JE, Szabó C: Protective effect of a novel, potent inhibitor of poly(adenosine 5’-diphosphate-ribose) synthetase in a porcine model of severe bacterial sepsis. Crit Care Med 2002, 30:974–80. [DOI] [PubMed] [Google Scholar]
- 10.Murakami K, Enkhbaatar P, Shimoda K, Cox RA, Burke AS, Hawkins HK, Traber LD, Schmalstieg FC, Salzman AL, Mabley JG, Komjati K, Pacher P, Zsengeller Z, Szabo C, Traber DL: Inhibition of poly (ADP-ribose) polymerase attenuates acute lung injury in an ovine model of sepsis. Shock 2004, 21:126–33. [DOI] [PubMed] [Google Scholar]
- 11.Veres B, Radnai B, Gallyas F Jr, Varbiro G, Berente Z, Osz E, Sumegi B: Regulation of kinase cascades and transcription factors by a poly(ADP-ribose) polymerase-1 inhibitor, 4-hydroxyquinazoline, in lipopolysaccharide-induced inflammation in mice. J Pharmacol Exp Ther 2004, 310:247–55. [DOI] [PubMed] [Google Scholar]
- 12.Mabley JG, Horváth EM, Murthy KG, Zsengellér Z, Vaslin A, Benko R, Kollai M, Szabó C: Gender differences in the endotoxin-induced inflammatory and vascular responses: potential role of poly(ADP-ribose) polymerase activation. J Pharmacol Exp Ther 2005, 315:812–20. [DOI] [PubMed] [Google Scholar]
- 13.Soriano FG, Nogueira AC, Caldini EG, Lins MH, Teixeira AC, Cappi SB, Lotufo PA, Bernik MM, Zsengellér Z, Chen M, Szabo C: Potential role of poly(adenosine 5’-diphosphate-ribose) polymerase activation in the pathogenesis of myocardial contractile dysfunction associated with human septic shock. Crit Care Med 2006, 34:1073–9. [DOI] [PubMed] [Google Scholar]
- 14.Hauser B, Gröger M, Ehrmann U, Albicini M, Brückner UB, Schelzig H, Venkatesh B, Li H, Szabó C, Speit G, Radermacher P, Kick J: The PARP-1 inhibitor ino-1001 facilitates hemodynamic stabilization without affecting DNA repair in porcine thoracic aortic cross-clamping-induced ischemia/reperfusion. Shock 2006, 25:633–40. [DOI] [PubMed] [Google Scholar]
- 15.Wang G, Huang X, Li Y, Guo K, Ning P, Zhang Y: PARP-1 inhibitor, DPQ, attenuates LPS-induced acute lung injury through inhibiting NF-κB-mediated inflammatory response. PLoS One 2013, 8:e79757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Yao J, Wang X, Li H, Liu T, Zhao W: Poly (ADP-ribose) synthetase inhibitor has a heart protective effect in a rat model of experimental sepsis. Int J Clin Exp Pathol 2015, 8:9824–35. [PMC free article] [PubMed] [Google Scholar]
- 17.Walko TD 3rd, Di Caro V, Piganelli J, Billiar TR, Clark RS, Aneja RK: Poly(ADP-ribose) polymerase 1-sirtuin 1 functional interplay regulates LPS-mediated high mobility group box 1 secretion. Mol Med 2015, 20:612–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YM, Han RL, Song SG, Yuan XP, Ren XS: Inhibition of PARP overactivation protects acute kidney injury of septic shock. Eur Rev Med Pharmacol Sci 2018, 22:6049–56. [DOI] [PubMed] [Google Scholar]
- 19.Berger NA, Besson VC, Boulares AH, Bürkle A, Chiarugi A, Clark RS, Curtin NJ, Cuzzocrea S, Dawson TM, Dawson VL, Haskó G, Liaudet L, Moroni F, Pacher P, Radermacher P, Salzman AL, Snyder SH, Soriano FG, Strosznajder RP, Sümegi B, Swanson RA, Szabo C: Opportunities for the repurposing of PARP inhibitors for the therapy of non-oncological diseases. Br J Pharmacol 2018, 175:192–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bochum S, Berger S, Martens UM: Olaparib. Recent Results Cancer Res 2018, 211:217–33. [DOI] [PubMed] [Google Scholar]
- 21.Sulai NH, Tan AR: Development of poly(ADP-ribose) polymerase inhibitors in the treatment of BRCA-mutated breast cancer. Clin Adv Hematol Oncol 2018, 16:491–501. [PubMed] [Google Scholar]
- 22.Coletta C, Módis K, Oláh G, Brunyánszki A, Herzig DS, Sherwood ER, Ungvári Z, Szabo C: Endothelial dysfunction is a potential contributor to multiple organ failure and mortality in aged mice subjected to septic shock: preclinical studies in a murine model of cecal ligation and puncture. Crit Care 2014, 18:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szczesny B, Brunyánszki A, Ahmad A, Oláh G, Porter C, Toliver-Kinsky T, Sidossis L, Herndon DN, Szabo C: Time-dependent and organ-specific changes in mitochondrial function, mitochondrial DNA integrity, oxidative stress and mononuclear cell infiltration in a mouse model of burn injury. PLoS One 2015, 10:e0143730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunyanszki A, Erdelyi K, Szczesny B, Olah G, Salomao R, Herndon DN, Szabo C: Upregulation and mitochondrial sequestration of hemoglobin occur in circulating leukocytes during critical illness, conferring a cytoprotective phenotype. Mol Med 2015, 21:666–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korkmaz-Icöz S, Szczesny B, Marcatti M, Li S, Ruppert M, Lasitschka F, Loganathan S, Szabó C, Szabó G: Olaparib protects cardiomyocytes against oxidative stress and improves graft contractility during the early phase after heart transplantation in rats. Br J Pharmacol 2018, 175:246–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szczesny B, Marcatti M, Ahmad A, Montalbano M, Brunyánszki A, Bibli SI, Papapetropoulos A, Szabo C: Mitochondrial DNA damage and subsequent activation of Z-DNA binding protein 1 links oxidative stress to inflammation in epithelial cells. Sci Rep 2018, 8:914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta D, Bhoi S, Mohan T, Galwnkar S, Rao D: Coexistence of Th1/Th2 and Th17/Treg imbalances in patients with post traumatic sepsis. Cytokine 2016, 88:214–21. [DOI] [PubMed] [Google Scholar]
- 28.Tucsek Z, Gautam T, Sonntag WE, Toth P, Saito H, Salomao R, Szabo C, Csiszar A, Ungvari Z: Aging exacerbates microvascular endothelial damage induced by circulating factors present in the serum of septic patients. J Gerontol A Biol Sci Med Sci 2013, 68:652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinheiro da Silva F, Machado MCC: Septic shock and the aging process: A molecular comparison. Front Immunol 2017, 8:1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ungvari Z, Bailey-Downs L, Sosnowska D, Gautam T, Koncz P, Losonczy G, Ballabh P, de Cabo R, Sonntag WE, Csiszar A: Vascular oxidative stress in aging: a homeostatic failure due to dysregulation of NRF2-mediated antioxidant response. Am J Physiol Heart Circ Physiol 2011, 301:H363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorsell AG, Ekblad T, Karlberg T, Löw M, Pinto AF, Trésaugues L, Moche M, Cohen MS, Schüler H: Structural basis for potency and promiscuity in poly(ADP-ribose) polymerase (PARP) and tankyrase inhibitors. J Med Chem 2017, 60:1262–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szabo C, Zingarelli B, O’Connor M, Salzman AL: DNA strand breakage, activation poly (ADP-ribose) synthetase, and cellular energy depletion are involved in cytotoxicity of macrophages and smooth muscle cells exposed to peroxynitrite. Pro Natl Acad Sci USA 1996, 93:1753–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagtap P, Szabo C: Poly(ADP-ribose) polymerase and the therapeutic effects of inhibitors. Nat Rev Drug Discov 2005, 4:421–40. [DOI] [PubMed] [Google Scholar]
- 34.Módis K, Gero D, Erdélyi K, Szoleczky P, DeWitt D, Szabo C: Cellular bioenergetics regulated by PARP1 under resting conditions and during oxidative stress. Biochem Pharmacol 2012, 83:633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukhopadhyay P, Horváth B, Rajesh M, Varga ZV, Gariani K, Ryu D, Cao Z, Holovac Park O, Zhou Z, Xu MJ, Wang W, Godlewski G, Paloczi J, Nemeth BT, Persidsky Liaudet L, Haskó G, Bai P, Boulares AH, Auwerx J, Gao B, Pacher P: ARP inhibition protects against alcoholic and non-alcoholic steatohepatitis. J Hepatol 2017, 66:589–600. [DOI] [PubMed] [Google Scholar]
- 36.Szczesny B, Brunyanszki A, Olah G, Mitra S, Szabo C: Opposing roles of mitochondrial and nuclear PARP1 in the regulation of mitochondrial and nuclear DNA integrity: implications for the regulation of mitochondrial function. Nucleic Acids Res 2014. 42:13161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B: Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role EXOG (5’-EXO/endonuclease) in their repair. J Biol Chem 2011, 286:31975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krainz T, Lamade AM, Du L, Maskrey TS, Calderon MJ, Watkins SC, Epperly Greenberger JS, Bayır H, Wipf P, Clark RSB: Synthesis and evaluation of mitochondria-targeting poly(ADP-ribose) polymerase-1 inhibitor. ACS Chem Biol 2018, 13:2868–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zingarelli B, O’Connor M, Wong H, Salzman AL, Szabo C: Peroxynitrite-mediated strand breakage activates poly-adenosine diphosphate ribosyl synthetase and causes cellular energy depletion in macrophages stimulated with bacterial lipopolysaccharide. J Immunol 1996, 156:350–8. [PubMed] [Google Scholar]
- 40.Szabo C, Virág L, Cuzzocrea S, Scott GS, Hake P, O’Connor MP, Zingarelli B, Salzman A, Kun E: Protection against peroxynitrite-induced fibroblast injury and arthritis development by inhibition of poly(ADP-ribose) synthase. Proc Natl Acad Sci USA 1998. 95:3867–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kunze FA, Hottiger MO: Regulating immunity via ADP-ribosylation: therapeutic implications and beyond. Trends Immunol 2019, in press. [DOI] [PubMed] [Google Scholar]
- 42.Brady PN, Goel A, Johnson MA: Poly(ADP-ribose) polymerases in host-pathogen interactions, inflammation, and immunity. Microbiol Mol Biol Rev 2018, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szabo C, Lim LH, Cuzzocrea S, Getting SJ, Zingarelli B, Flower RJ, Salzman AL, Perretti M: Inhibition of poly (ADP-ribose) synthetase attenuates neutrophil recruitment and exerts antiinflammatory effects. J Exp Med 1997, 186:1041–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan M, Siegel C, Zeng Z, Li J, Liu F, McCullough LD: Sex differences in the response to activation of the poly (ADP-ribose) polymerase pathway after experimental stroke. Exp Neurol 2009, 217:210–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chevanne M, Calia C, Zampieri M, Cecchinelli B, Caldini R, Monti D, Bucci L, Franceschi C, Caiafa P. Oxidative DNA damage repair and parp 1 and parp 2 expression in Epstein-Barr virus-immortalized B lymphocyte cells from young subjects, old subjects, and centenarians. Rejuvenation Res 2007, 10:191–204. [DOI] [PubMed] [Google Scholar]
- 46.Tapodi A, Bognar Z, Szabo C, Gallyas F, Sumegi B, Hocsak E: PARP inhibition induces Akt-mediated cytoprotective effects through the formation of a mitochondria-targeted phospho-ATM-NEMO-Akt-mTOR signalosome. Biochem Pharmacol 2019, in press. [DOI] [PubMed] [Google Scholar]
- 47.Gupte R, Liu Z, Kraus WL: PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 2017, 31:101–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao C, Ma T, Chai Y, Shou S: The role of regulatory T cells in immune dysfunction during sepsis. World J Emerg Med 2015, 6:5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo X, Nie J, Wang S, Chen Z, Chen W, Li D, Li B: Poly(ADP-ribosyl)ation of FOXP3 protein mediated by PARP-1 protein regulates the function of regulatory T cells. J Biol Chem 2015, 290:28675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drewry AM, Samra N, Skrupky LP, Fuller BM, Compton SM, Hotchkiss RS: Persistent lymphopenia after diagnosis of sepsis predicts mortality. Shock 2014, 42:383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunialti MK, Santos MC, Rigato O, Machado FR, Silva E, Salomao R: Increased percentages of T helper cells producing IL-17 and monocytes expressing markers of alternative activation in patients with sepsis. PLoS One. 2012,7:e37393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nachtergaele S, He C: The emerging biology of RNA post-transcriptional modifications. RNA Biol 2017, 14:156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kingsley SMK, Bhat BV: Role of microRNAs in sepsis. Inflamm Res 2017, 66:553–69. [DOI] [PubMed] [Google Scholar]
- 54.Sergi C, Shen F, Lim DW, Liu W, Zhang M, Chiu B, Anand V, Sun Z: Cardiovascular dysfunction in sepsis at the dawn of emerging mediators. Biomed Pharmacother 2017, 95:153–60. [DOI] [PubMed] [Google Scholar]
- 55.Du J, Li M, Huang Q, Liu W, Li WQ, Li YJ, Gong ZC: The critical role of microRNAs in stress response: therapeutic prospect and limitation. Pharmacol Res 2019, in press. [DOI] [PubMed] [Google Scholar]
- 56.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie L: Four serum microRNAs identified as diagnostic biomarkers of sepsis. J Trauma Acute Care Surg 2012, 73:850–4. [DOI] [PubMed] [Google Scholar]
- 57.Long M, Park S, Strickland I, Hayden M, Ghosh S: Nuclear Factor-κB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity 2009, 31:921–31. [DOI] [PubMed] [Google Scholar]
- 58.Yang HY, Barbi J, Wu CY, Zheng Y, Vignali PD, Wu X, Tai JH, Park BV, Bandara S, Novack L, Ni X, Yang X, Chang KY, Wu RC, Zhang J, Yang CW, Pardoll DM, Li H, Pan F: MicroRNA-17 modulates regulatory T cell function by targeting co-regulators of the Foxp3 transcription factor. Immunity 2016, 45:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA: Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med 2008, 205:1983–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B, Wang X, Choi IY, Wang YC, Liu S, Pham AT, Moon H, Smith DJ, Rao DS, Boldin MP, Yang L: miR-146a modulates autoreactive Th17 cell differentiation and regulates organ-specific autoimmunity. J Clin Invest 2017, 127:3702–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu Z, Xiao SB, Xu P, Xie Q, Cao L, Wang D, Luo R, Zhong Y, Chen HC, Fang LR: miR-365, a novel negative regulator of interleukin-6 gene expression, is cooperatively regulated by Sp1 and NF-kappaB. J Biol Chem 2011, 286:21401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gong H, Sheng X, Xue J, Zhu D: MicroRNA-365 regulates the occurrence and immune response of sepsis following multiple trauma via interleukin-6. Exp Ther Med 2018, 16:3745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pascal JM: The comings and goings of PARP-1 in response to DNA damage. DNA Repair (Amst) 2018, 71:177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B: Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods 2000, 22:135–47. [DOI] [PubMed] [Google Scholar]
- 65.van Loon B, Markkanen E, Hubscher U: Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst) 2010, 9:604–16. [DOI] [PubMed] [Google Scholar]