Abstract
The RNA-guided CRISPR-associated (Cas) proteins Cas9 and Cas12a provide adaptive immunity against bacteriophage and function as powerful tools for genome editing in wide-ranging cell types. Here we present a third and fundamentally distinct RNA-guided platform, CRISPR-CasX, which uses a unique structure and mechanism for programmable double-stranded DNA cleavage. Biochemical and in vivo data demonstrate that CasX is active for E. coli and human genome modification. Eight cryo-EM structures of CasX in different states of assembly with its guide RNA and double-stranded DNA substrates reveal an extensive RNA scaffold and an unanticipated domain required for DNA unwinding. These data demonstrate how CasX activity arose through convergent evolution to establish an enzyme family that is functionally separate from both Cas9 and Cas12a.
Archaea and bacteria utilize CRISPR-Cas systems (clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins) for adaptive immunity against invading nucleic acids1,2. CRISPR arrays, consisting of repeated sequences interleaved with sequences acquired from foreign DNA, are templates for CRISPR RNAs (crRNAs) that guide a Cas nuclease to cleave complementary DNA sequences. In addition to their microbial functions, RNA-guided DNA binding and cutting have proven to be transformative tools for genome and epigenome editing across wide-ranging cell types and organisms3–5. Despite extensive effort, just two kinds of CRISPR-Cas nucleases, Cas9 and Cas12a (Cpf1), provide the foundation for this revolutionary technology6,7.
Metagenomic analysis of microbial DNA from groundwater samples revealed a new protein, CasX (a placeholder name pending re-analysis of the class 2 CRISPR-Cas phylogeny) which is also referred as Cas12e5, that prevented bacterial transformation by plasmid DNA when expressed with cognate crRNAs targeting the plasmid8. Sequence analysis of CasX revealed no similarity to other CRISPR-Cas enzymes, except for the presence of a RuvC nuclease domain similar to that found in both Cas9 and Cas12a enzyme families as well as transposases and recombinases8. Phylogenetic analysis suggests that CasX arose from a TnpB-type transposase by an independent insertion event into ancestral CRISPR loci, distinct from Cas12a and the remaining type V effectors (Extended Data Fig. 1a). Consistent with this hypothesis, the CasX RuvC domain shares less than 16% identity to RuvC domains in either Cas9 or Cas12a (Extended Data Fig. 1b). This evolutionary ambiguity of CasX hinted that CasX may have a structure and molecular mechanism distinct from other CRISPR-Cas enzymes. However, without full reconstitution of the CasX enzyme, it was not possible to determine the basis of the previously reported plasmid interference activity.
We demonstrate here that CasX is an RNA-guided DNA endonuclease that generates a staggered double-stranded break in DNA at sequences complementary to a 20-nucleotide segment of its guide RNA. We further find that CasX induces programmable, site-specific genome repression in E. coli and genome editing in human cells. Biochemical data shows that CasX is a hybrid enzyme containing elements of both Cas9 and Cas12a as well as novel RNA folds and protein domains, establishing this enzyme family as the third CRISPR-Cas system effective for genetic manipulation. The small size of CasX (<1000 amino acids), DNA cleavage characteristics and derivation from non-pathogenic microorganisms, offer important advantages over other CRISPR-Cas genome editing enzymes. Eight molecular models of CasX in different states (Supplementary Table 1), determined by cryo-electron microscopy (cryo-EM), reveal an unanticipated quaternary structure in which the RNA scaffold dominates the architecture and organization of the enzyme. Distinct conformational states observed for CasX suggest an ordered non-target and target strand cleavage mechanism that may explain how other CRISPR-Cas enzymes with a single active site, such as Cas12a, achieve double-stranded DNA cleavage5,9,10.
Reconstitution of crRNA-guided CasX cutting of double-stranded DNA
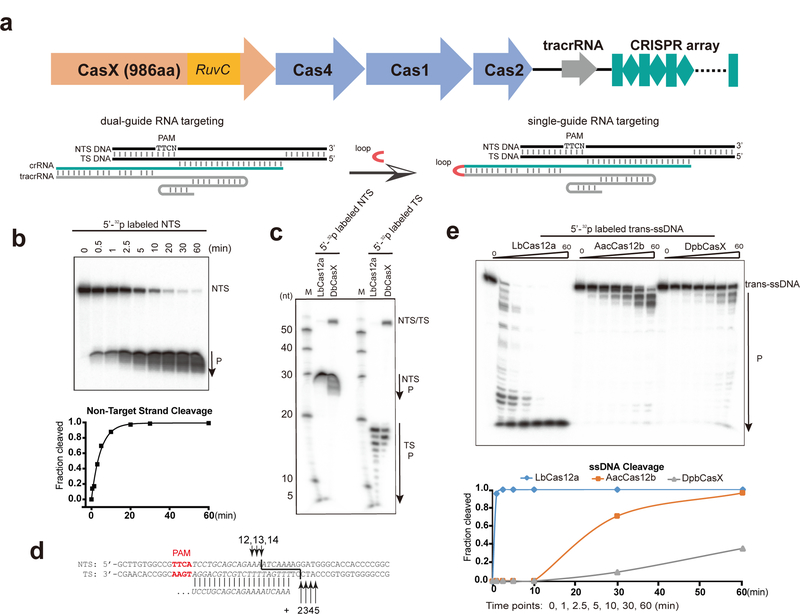
We previously demonstrated that CasX proteins can perform RNA-dependent plasmid interference in bacteria and that the two natural RNAs necessary for this activity (crRNA and trans-activating CRISPR RNA (tracrRNA)) can be combined into a single-guide RNA format8 (Fig. 1a). To determine the molecular function, we undertook biochemical studies of the wild-type CasX from Deltaproteobacteria (DpbCasX). We found that purified DpbCasX with single-guide RNA is capable of cleaving double-stranded DNA bearing a sequence complementary to the 20-nucleotide guide RNA segment and adjacent to a TTCN PAM motif (Fig. 1b). Mapping the cut sites for the target and non-target strands of the DNA showed that DpbCasX generates products with ~10-nucleotide staggered ends due to cleavage 12–14 nucleotides after the PAM on the non-target strand and 22–25 nucleotides after the PAM on the target strand (Fig. 1c, d; Extended Data Fig. 1c). This mode of double-stranded DNA cleavage is consistent with the staggered cuts to DNA observed for Cas12a and Cas12b (C2c1), other CRISPR-Cas enzymes that use a single RuvC active site for DNA cleavage5,10,11.
Figure 1. CasX cuts double stranded DNA with single guide RNA in vitro.
a, A schematic of CRISPR-CasX locus with CasX gene (RuvC domain highlighted) in orange, Cas4, Cas1 and Cas2 in blue, tracrRNA in gray, and CRISPR array in teal. Cartoons are scaled according to the gene size. Schematic of the CasX dual-guide RNA and single guide is shown in the bottom panel-- tracrRNA in gray, crRNA in teal and the target DNA in black. TS and NTS indicate the target strand and non-target strand, respectively. The RNA loop fusing tracrRNA and crRNA is in red. b, DNA cleavage efficiency by DpbCasX. P indicates the cleavage product. The cleavage fraction is calculated based on the NTS band density compared to input NTS band density at reaction time of 0 min. c, Conservation of cleavage specificity of DpbCasx with Cas12a. Lane M shows labeled ladders. d, The cleavage sites for NTS and TS (marked with black arrows). e, Cleavage activity of DpbCasX on trans ssDNA. The cleavage fraction is calculated based on the trans-ssDNA band density compared to input trans-ssDNA band density at reaction time of 0 min. 4 biological repeats for all the assays showed consistent results.
Unlike Cas9, Cas12a becomes a highly active single stranded DNA nuclease after target DNA binding, triggering non-specific single-stranded DNA degradation12,13. To test whether CasX displays similar target-triggered activity, single-stranded phage DNA was incubated with DpbCasX-guide RNA complexes that target a separate, unrelated double-stranded DNA substrate. We found that trans-ssDNA cutting activity was minimal compared to that observed for LbCas12a or for another related enzyme Cas12b (Fig. 1e). These results indicate that the presence of a single active site for double-stranded DNA cleavage does not necessarily correspond to target-dependent trans-cleavage activity, raising the possibility of structural or mechanistic differences between these enzyme families.
CasX triggers genome silencing and editing in bacterial and human cells
To determine whether the RNA-guided DNA cutting activity of CasX can be harnessed for programmed genome targeting, DpbCasX and its single-guide RNA (sgRNA) were expressed in E. coli using a guide sequence complementary to an integrated reporter in the genome of bacterial strain MG165514,15. We found that DpbCasX reduced cell viability in a genome cleavage assay, at close to but slightly less than SpyCas9 activity levels (Fig. 2a). We next tested whether CasX can function as a CRISPRi effector in E. coli. By aligning the RuvC domain among CasX and other Cas proteins, we found three potential active site residues: D672, E769 and D935. Biochemical screening indicated that mutations of these residues to alanines create a deactivated DpbCasX (dDpbCasX) (Extended Data Fig. 1d) that is competent for RNA-guided DNA binding and gene repression rather than cutting (Fig. 2b). Silencing of green fluorescent protein (GFP) expression was observed with this dDpbCasX construct using different guide RNAs targeting multiple sites within the GFP-encoding gene (Extended Data Fig. 1e). Surprisingly, we found that mutation of all three DpbCasX residues was required for maximal gene repression activity (Extended Data Fig. 1f). CasX-based bacterial CRISPRi thus provides an ideal system for rapid, visual and quantitative in vivo characterization of CasX constructs (Extended Data Fig. 1e; Fig. 2c, d).
Figure 2. CasX effectively manipulates genomes in vivo.
a, Genomic cleavage assay in E. coli (n = 3, mean ± s.d.). b, Schematic of E. coli CRISPRi c, E. coli GFP repression as visualized on plates on a dark reader. This assay has been repeated more than 3 times with consistent results. d, Quantitative analysis of E. coli CRISPRi based GFP repression at 12 hrs (n = 4, mean ± s.d.). e, Schematic of CasX human cell assay and readout. f, DpbCasX (Deltaproteobacteria CasX) and PlmCasX (Planctomycetes CasX) GFP disruption in a mammalian cell (HEK293T) assays at two doses of plasmids. g, Sustained GFP disruption of the high dosage mammalian cell GFP disruption assay from f. h, PlmCasX & SpyCas9 GFP disruption at 10 guide sites throughout EGFp (n=3, mean ± s.d.). The average GFP disruption across all EGFP guides for CasX & Cas9 is shown. Cas9-g and cas9-ng indicate targeting and non-targeting RNA guide of Streptococcus pyogenes Cas9 (SpyCas9), respectively. CasX-g and CasX-ng indicate targeting and non-targeting RNA guide of DpbCasX. SpyCas9 or inactive SpyCas9 (dSpyCas9) was used as positive controls.
We next tested whether CasX is capable of inducing cleavage and gene editing of mammalian genomes. Using a previously reported destabilized-GFP disruption assay15 (Fig. 2e), we found that DpbCasX can induce targeted GFP gene disruption in HEK293T cells with limited efficiency using guide RNAs complementary to either the template or coding strand (guide 2 or guide 3 respectively) (Fig. 2f). We also explored the effectiveness of the CasX molecule from Planctomycetes (PlmCasX) that bears ~70% sequence identity to DpbCasX and can utilize the same single guide RNA8. We find that PlmCasX enacts destabilize-GFP gene editing with higher efficiency relative to DpbCasX (up to ~30% in this assay). Since the guide RNA recognizing the coding strand functioned more robustly in each case, we wondered if the additional GFP gene disruption observed for guide 3 could be explained by RNA targeting4,16. However, there was no recovery of GFP expression in these cells over time (Fig. 2g), consistent with genome editing rather than transcript targeting. Furthermore, analysis of DNA derived from the PlmCasX-targeted GFP locus using a T7E1-based assay17 revealed levels of genome editing consistent with the observed GFP disruption (Extended Data Fig. 2a, b). Next, we explored the effect of CasX-sgRNA-encoding plasmid concentration on the extent of genome editing. The highest amounts of transfected PlmCasX plasmid produced GFP locus editing at levels comparable to genome editing levels observed in initial reports for CRISPR-Cas9 and CRISPR-Cas12a (Cpf1) (~34%)6,7,11,18(Extended Data Fig. 2c). Additionally, we developed two additional clonal EGFP HEK 293T reporter lines, 1 and 2, using lentiviral integration. PlmCasX-mediated EGFP disruption in these individual cell lines was comparable to results in the destabilized GFP reporter cell line, with guide 3 producing higher levels of editing than guide 2 (Extended Data Fig. 2d). Sub-cloning PCR-amplified segments of the GFP locus from treated cells revealed a wide variety of indels, many of which map to the cut sites identified in vitro (Extended Data Fig. 2e). Finally, we compared the editing efficiency of PlmCasX to that of SpyCas9, which has been optimized over the past 6 years for high efficiency. Guide RNAs for Cas9 recognize the EGFP locus at same locations to those of CasX guide RNAs but with ~1–3 bp offset to accommodate for PAM specificity differences (Extended Data Fig. 2f). By using an optimized transfection protocol, we observe that 9 out of 10 CasX guide RNAs disrupt GFP and that editing efficacy is, on average, ~30% that of SpyCas9 and in some cases as high as 55% (guide 3) (Fig. 2i; Methods). These results demonstrate that CasX belongs to a third distinct class of CRISPR system capable of targeted genomic regulation and editing, and motivated experiments aimed at determining the structural and mechanistic basis for these activities.
CasX has a unique domain composition
To understand how DpbCasX (hereafter CasX) binds to helical DNA, a ternary complex containing deactivated CasX (D672A-E769A-D935A), sgRNA (122 nt) and a complementary DNA substrate (30 base pairs (bp)) was analyzed by single particle cryo-EM (Extended Data Fig. 1g). Three-dimensional particle classification and refinement revealed two conformational populations of the ternary complex at resolutions of 3.7 Å and 4.2 Å (State I and State II, respectively) (Extended Data Fig. 3). These two conformational states were also observed by cryo-EM analysis of a CasX complex containing a full R-loop (45 bp DNA substrate) and refined at resolutions of 3.2 Å (State I) and 5.2 Å (State II) (Extended Data Fig. 4). With the cryo-EM maps, atomic models of CasX ternary complexes in State I and State II were built ab initio (Extended Data Fig. 5). While structural alignment of the entire modeled polypeptide chain revealed some similarity between CasX and Cas12a (LbCpf1, PDB 5xuu, z-score 15.1, with an rmsd value of 5.3 Å for 671 aligned residues)19, a more detailed analysis of the domains showed that this similarity results exclusively from the RuvC and OBD domains (alignment of RuvC with LbCpf1 PDB 5xut gives a z-score of 13.8 and an rmsd value of 2.5 Å for 173 aligned residues, while alignment of OBD with LbCpf1 PDB 5xh6 gives a z-score of 9.6 and an rmsd value of 3.5 Å for 153 aligned residues)10,20. Although CasX contains other structural elements that appear analogous to those identified in other Cas proteins, including the Helical-I and -II and the REC1 and REC2 domains, these domains have highly distinct folds (Extended Data Fig. 6a, b).
Two unique domains were identified adjacent to the separated DNA strands in the CasX complex that we refer as the non-target strand binding (NTSB) and the target-strand loading (TSL) domains (residues 101–191 (red) and 825–934 (pink) in Fig. 3a and b, respectively). NTSB domain contains a four-stranded beta-sheet and sits next to the non-target strand of the DNA (Fig. 3b). We discuss this domain and its function in depth in the section below. TSL domain is located in a position analogous to that of the so-called “Nuc” domain of other type V CRISPR-Cas enzymes. We propose renaming this type of domain TSL, since “Nuc” was hypothesized incorrectly to be a second nuclease domain responsible for DNA cleavage13,21. Instead, we postulate that the TSL domain is responsible for target strand placement in the RuvC active site10. In the AsCas12a “Nuc” domain, amino acids Arg1226 and Asp1235 aid target strand cleavage and an Arg1226Ala mutation produced an AsCas12a nickase by abolishing Cas12a’s ability to cut the target strand21. In the CasX “Nuc”-analogous domain, the TSL, residues Arg917 and Gln920 interact with DNA (NTS in State I and TS in State II) that is adjacent to the active site (Fig. 3c). Intriguingly, within the CasX full R-loop structure in State I, a TSL loop containing three tyrosines (Tyr867, Tyr868, Tyr870) and three positively charged residues (Arg869, Lys871, Arg872) interacts with the migration point where the RNA-DNA duplex ends and the DNA-DNA duplex reforms (Fig. 3d; Extended Data Fig. 6c). In other enzymes, similar loops or hairpin elements containing a large hydrophobic amino acid (tyrosine or phenylalanine) are thought to be involved in DNA strand separation22–24. Moreover, the TSL domain also contains two CXXC motifs (residues 824–827 and 926–929) that form a Zinc finger/ribbon motif (Extended Data Fig. 6c, d) akin to those found in phage primases, transcription factors and the purported transposase ancestor for Type V CRISPR proteins, TnpB55,25–27. Taken together, it is clear that CasX possesses both domains analogous to other CRISPR-Cas proteins as well as completely novel domains as anticipated by its complete sequence dissimilarity from other CRISPR proteins.
Figure 3. Overall structure of the CasX ternary complex.
a, Domain composition of CasX. CasX contains: NTSB (non-target strand binding, red), Helical-I (yellow), Helical-II (orange), OBD (oligo binding domain, aquamarine), RuvC (green) and TSL (target-strand loading, pink) domains, and a BH (bridge helix, blue). b, Model of CasX ternary complex with 30bp target DNA in State I, shown on side and top views. Different domains are colored as in a. and sgRNA is in teal. For the target DNA, the NTS is in magenta and the TS is in purple. c and d, Models of the CasX ternary complex with 30bp target DNA in State II and State I, shown on top view. Residues Arg917 and Gln920 are shown as red sticks. The TSL-loop is shown as a red ribbon. The positions of the RuvC active site residues are shown as red sticks to illustrate the distance to the active site from the TSL domain elements. The right panels show a zoomed-in views of the TSL domain. e, Schematic of the single guide RNA fold with tracrRNA sequence in gray, crRNA sequence in teal, and the joint loop in red. f, Molecular interactions between CasX and gRNA. Protein residues interacting with gRNA recognition are shown as magenta sticks. Helical-II and OBD are colored in orange and aquamarine, respectively. g, Models of CasX ternary complex in State I and II are aligned and superimposed. CasX is shown as a transparent grey cartoon, and the residues responsible for cleavage activity are shown in red. The nucleic acids are shown as ribbons to emphasize the rotation of the RNA-DNA duplex required for the transition between the two states.
A prominent guide RNA scaffold for CasX
Structural modeling shows that the guide RNA accounts for ~26% of the mass in the CasX-sgRNA binary complex, a value significantly higher than that observed for other type II or V CRISPR-Cas effector complexes (~8% in LbCas12a, ~20% in AacCas12b, and ~16% in SpyCas9; Extended Data Fig. 6e). Dominating the CasX protein complex, the single-guide RNA includes three elements: a triplex stem loop that contacts the OBD, a ‘scaffold’ stem that interacts with the Helical-II domain, and a perpendicular stem loop that projects away from the center of mass of the structure (Fig. 3e, f; Supplementary Video 1). Mutation of the triplex or the scaffold stem diminished CasX activity in vivo, whereas truncated versions of the perpendicular stem loop retained activity (Extended Data Fig. 6f).
In the absence of guide RNA, the CasX protein is poorly resolved by cryo-EM (Extended Data Fig. 7a, d). Consistent with the importance of RNA in CasX protein architecture, CasX cross-linking before and after addition of the sgRNA followed by analytical mass spectrometry (MS) revealed significant RNA-induced CasX domain rearrangements (Extended Data Fig. 7e, g). In line with this analysis, a cryo-EM-derived model of the CasX-sgRNA complex (~7.5 Å resolution map) shows the OBD, RuvC and Helical-II domains assembled along the RNA scaffold, while the NTSB domain associates with the RuvC, Helical-I and Helical-II domains near the nuclease active site (Extended Data Fig.6 b, f). Comparison with the DNA-bound structure shows that upon DNA binding, the NTSB domain moves away from the center of mass of the complex (Extended Data Fig. 7c, h, i).
CasX conformational states suggest a mechanism of sequential DNA cutting
Comparison of the two conformational states (I and II) of the CasX ternary complex revealed a large structural change that alters target DNA strand accessibility to the RuvC domain (Fig. 3c, d; Supplementary Video 2). In State I, the non-target strand DNA sits in the RuvC active site while the target-strand DNA/gRNA duplex engages with the Helical-I and Helical-II domains (Fig. 3b, d). In State II, the target-strand DNA/guide RNA duplex is sharply bent, enabling RuvC access to the target-strand DNA (Fig. 3c). State I is compatible with non-target strand DNA cleavage, while State II is compatible with cleavage of the target strand DNA (Fig. 3g; Extended Data Fig. 7j, k).
Statistical analysis by single particle sorting showed that the majority of particles (~71%) in the 30bp target DNA ternary complex adopted the State I conformation, with the remaining 29% of particles in State II (Extended Data Fig. 3; Fig. 4a). This preference suggests that non-target strand DNA is cleaved by the RuvC domain first, followed by displacement and target strand cleavage. Similar to the 30 bp DNA containing sample, 67% of full R-loop (45 bp) DNA particles adopted State I (Extended Data Fig. 4; Fig. 4b).
Figure 4. Distinct CasX conformational states.
a, Conformational states with 30bp target DNA; b, with a DNA target forming the full R-loop; and c, with the short NTS (20nt) and the 45nt TS. The schematic of the DNA probe used for each data collection is shown on the left, with cleavage sites shown by arrowheads. The top views of the cryo-EM maps for the CasX ternary complex in States I and II are shown on the center panels. The TS density is colored purple, the NTS is colored magenta, and the sgRNA density is colored teal. The RuvC domain is indicated in each map. All the EM maps are low-pass filtered to 4.5 Å. The relative percentage of particles belonging to each state revealed by cryo-EM analysis is shown in the right panel.
In our sequential model of CasX-mediated DNA cleavage, a substrate-bound complex mimicking the intermediate state that occurs after non-target strand cleavage should preferentially adopt State II. To test this idea, we performed cryo-EM analysis on a CasX ternary complex containing sgRNA and a DNA substrate comprising a 45-nt target strand and a post-cleavage-like 20-nt non-target strand (Extended Data Fig. 8). In this intermediate-state sample, the majority of particles (~66.4%) adopted the State II conformation, with the target strand located near the RuvC active site (Fig. 4c). Interestingly, reconstruction of State I showed the 5’ end overhang of the target strand DNA folded back into the RuvC domain. This conformation is incompatible with double-stranded DNA cleavage at position 22 and is unlikely to occur natively (Fig. 1d; Fig. 4c).
The CasX NTSB domain is required for DNA unwinding
The distinct and smaller architecture of CasX relative to other double-stranded DNA targeting CRISPR enzymes implies a unique mechanism of substrate recognition, which requires guide RNA strand invasion into duplex DNA. Observation that the NTSB domain (residues 101–191, red in Fig. 3b) interacts directly with non-target DNA strand both in State I and State II (Fig. 5a) raised the possibility that this unique structure contributes fundamentally to the mechanism of DNA unwinding. To test this hypothesis, we analyzed the behavior and activity of a protein construct lacking the NTSB domain (CasXΔ101–191). Although it showed similar physical behavior to that observed for the wild-type CasX on a size exclusion column (Extended Data Fig. 9a), CasXΔ101–191 was incapable of cleaving a double-stranded DNA substrate (Fig. 5b). However, CasXΔ101–191 retains robust single-stranded DNA cleavage activity, including with mismatched duplex DNA substrates (Fig. 5b; Extended Data Fig. 9b). Of note, CasXΔ101–191 has slightly higher trans- cleavage activity than WT, suggesting that the NTSB domain may also contribute to shielding the RuvC from accessing the trans-DNA substrates (Fig. 5b). Together, these results suggest that the NTSB domain is responsible for initiating or stabilizing DNA duplex unwinding by CasX. This finding also hints at the interesting possibility that the self-contained NTSB domain could be introduced into or acquired by other enzymes to assist with or stabilize double-stranded DNA binding.
Figure 5. Novel domains for target DNA unwinding and loading.
a, Electron density map showing the presence of a domain that directly interacts with the NTS, with models for the CasX ternary complex in State I and II within the cryo-EM map (shown as mesh surface, low-pass filtered to 4.5 Å). CasX is shown in grey with the NTSB domain highlighted in red, the TS in purple and the NTS in magenta. b, Comparison of the cleavage activity of the wild-type CasX and NTSB domain deletion (CasXΔ101–191). The reactions were analyzed at time points from 0 to 120 minutes. Completely base-paired probe and a bubbled probe were used to test the on-target activity, and a random 50nt oligo was used to test the trans- cleavage activity. P indicates the cleavage product. 3 biological repeats for the assays showed consistent results.
Conclusions
Based on functional and structural data, we propose a model of CasX activation and DNA cleavage that includes the following steps: 1) RNA-induced CasX structural stabilization; 2) NTSB-assisted DNA unwinding and R-loop formation, with initial non-target strand binding in the RuvC active site; 3) RNA-DNA hybrid duplex bending with the aid of the proposed TSL domain to position the target DNA strand for cleavage; 4) product release after the cleavage of both DNA strands (Extended Data Fig. 10). Two distinct target DNA-bound states imply that CasX coordinates sequential double-stranded DNA cleavage by its single RuvC nuclease using the zinc-finger containing TSL domain (Extended Data Fig. 10c, d). The TSL domain appears to confer a convergent mechanism of acute target strand DNA bending that is central to all type V single-nuclease CRISPR-Cas enzymes.
These functional insights will enable the continued development of CasX as a third unique platform for RNA-programmed genome editing. The compact size, dominant RNA content, and minimal trans-cleavage activity of CasX differentiate this enzyme family from Cas9 and Cas12a, providing opportunities for therapeutic delivery and safety that may offer important advantages relative to existing genome editing technologies.
Methods
Strains and media
The in vivo CRISPRi28 and cleavage assays described below utilize E. coli MG1655 containing genomically-integrated and constitutively expressed Green fluorescent protein (GFP) and Red fluorescent protein (RFP). Standard cloning techniques were used to create all plasmids. Plasmid construction and retention was ensured with AmpR and CmR as selectable markers15. EZ-rich defined growth media (EZ-RDM, Teknova) was used in all CRISPRi assay fluorescent measurements. 2xYT (LB) with the addition of 1.5% Bacto Agar (BD) was used for all plating assays.
E. coli assays
CRISPRi assays were performed in a similar manner to previous work15. In brief to test CasX’s ability to bind genomic DNA and repress transcription, electrocompetent E. coli were co-transformed with a plasmid encoding the guide RNA and a plasmid encoding the CasX protein as described. They were grown on media containing two antibiotics to ensure selection for both plasmids. Colonies were picked in triplicate from these plates into EZ-RDM liquid media and grown for 12 hours. These saturated cultures were diluted 1:1000 into EZ-RDM media containing 2 nM anhydrotetracycline inducer and 150 μL of this mixture was followed for OD600 and GFP (a.u.) via a 96-well microplate reader (Tecan m1000) every 10 minutes over the course of 12 hours at 37 °C unless otherwise noted.
To perform the bacterial genome targeting assay, 100ng of the CRISPR-Cas protein-encoding plasmid was electroporated into electrocompetent MG1655 E. coli expressing the GFP-targeting sgRNA plasmid using a BTX Harvard apparatus ECM 630 High Throughput Electroporation System in biological triplicate. The guide sequence was moved onto the protein-encoding plasmid and 200ng of this was used in the transformation. The cells were recovered for one hour in 300μL SOC medium at 37°C unless otherwise noted. Two technical replicates of tenfold serial dilutions were spotted onto plates containing antibiotics for plasmid(s) used in the transformation. These grew at either 37°C for 12 hours or 30°C for 16 hours and were used to calculate CFU/mL.
Human cell GFP disruption
The EGFP reporter construct were created in a modified lentivirus backbone with EF1-a promoter driving the EGFP gene of interest and a second PGK promoter driving production of Hygromycin. Transduced 293T cells were selected with hygromycin (used at 250 ug/ml). EGFP clones were isolated by sorting single cells into 96 well plates and characterized by intensity of EGFP. Lentivirus was produced by PEI (Polysciences Inc., 24765) transfection of 293T cells with gene delivery vector co-transfected with packaging vectors pspax2 and pMD2.G essentially as described by (Tiscornia et al., 2006). HEK 293T EGFP experiments were conducted in a similar manner to previous assays. Briefly the EGFP HEK293T reporter cells were seeded into 96 well plates and transfected according to the manufacturer’s protocol with Lipofectamine 2000 (Life Technologies) and the described amount of plasmid DNA encoding the CasX, sgRNA and CasX, P2A-puromycin fusion. The next day cells were selected with 1.5 μg/ml Puromycin for 3 days and analyzed by FACS 8 days after selection to allow for clearance of EGFP protein from the cells. Cells were passaged one time to maintain sub-confluent conditions. EGFP expression was traced using an Attune NxT Flow Cytometer and high-throughput autosampler. For extended assays cells were passaged 1:10 and reanalyzed on the date notes. The PlmCasX vs SpyCas9 EGFP line 1 disruption assays were done as described above but the protocol was improved by using lipofectamine 3000 instead of 2000, according to manufactures protocols, selecting with 1.5ug of Puromycin for 48 hours and analyzing 11 days later.
T7EI assay
T7EI assays were performed as previously described with slight modification15. Briefly, cells were suspended 1:1 in QuickExtract (lucigen) buffer and DNA was extracted using the manufacturer’s protocol. This mixture was used directly in a PCR reaction designed to amplify the GFP locus and ~200ng of PCR product was utilized for denaturing, annealing & digestion with T7E1 (NEB) according to the manufacturer’s protocol. Samples were analyzed on a 2% agarose gel with SYBRsafe (Thermo Fisher).
EGFP fragment sub-cloning and sequencing
The edited GFP locus was amplified via PCR from HEK cells treated with Quick Extract buffer. The PCR fragments were then T4 blunt ligated into a small ColEI plasmid that was digested with PmeI and treated with CIP. Ligation products were transformed into cells and colonies were grown overnight before being picked, miniprepped, and Sanger sequenced at the UC Berkeley DNA Sequencing Facility. Sequencing results were aligned to the target and visualized via Snapgene.
Protein expression, purification and complex reconstitution
The gene encoding CasX was sub-cloned into the 2CT-10 expression vector. CasX-D672A-E769A-D935A and CasXΔ101–191 were obtained by amplifying the CasX plasmid using mutagenetic PCR primers. All the proteins were expressed using Rosetta E. coli cells. Main culture (Terrific broth, containing 100 mg/L ampicillin) was inoculated with 3% of overnight culture grown in Luria broth. The main culture was grown to an OD of 0.5–0.6, cooled down and protein expression was induced by addition of IPTG to a final concentration of 0.5 mM, and expression was allowed to proceed overnight at 16 °C. Cells were harvested, re-suspended in Ni buffer A (500mM sodium chloride, 50 mM HEPES, pH 7.5, 10% glycerol, 0.5 mM TCEP) and frozen at −80 °C For wild type CasX protein preparation, cells were thawed, diluted twice with Ni buffer A, followed by addition of PMSF (final concentration 0.5 mM), and 3 tablets of Roche protease inhibitor cocktail per 100 ml of cell suspension. Cells were lysed by sonication and pelleted at 35000 g for 30 min. Clarified lysate was purified using Ni-NTA agarose beads, using step gradient elution with imidazole-containing buffer (Ni buffer B (highest imidazole concentration): 500mM sodium chloride, 500mM imidazole, 50 mM HEPES, pH 7.5, 10% glycerol, 0.5 mM TCEP). The pure fractions were pooled and TEV protease was added (1 mg protease/20mg purified protein in final concentration). The protein with TEV protease was dialyzed overnight against the following buffer: 500mM sodium chloride, 50 mM HEPES, pH 7.5, 10% glycerol, 0.5 mM TCEP. Then protein was applied to a Maltose Binding Protein (MBP) column and the MBP flow-through was applied to a heparin column. Protein was eluted from the heparin column using a sodium chloride gradient up to 1M sodium chloride. For the wild type protein, there were two peaks containing CasX. The peak that eluted at lower salt concentration was found to contain inactive and aggregated protein and was not pooled; only the second peak contained active protein and only that protein was used for the assays. The active protein from the heparin column was concentrated and applied to a Superdex200 10/300 column in the following buffer: 500mM potassium chloride, 50 mM HEPES, pH 7.5, 10% glycerol, 0.5 mM TCEP. Pure protein was concentrated and flash-frozen. CasXΔ101–191 purification was purified as the same way as wild type CasX. The overall expression yield was similar, but the amount of the well-folded protein (second peak) was lower than in case of wild type protein. For CasX-D672A-E769A-D935A, the purification was similar, except that dialysis buffer was: 300mM sodium chloride, 50 mM HEPES, pH 7.5, 10% glycerol, 0.5 mM TCEP and size-exclusion buffer was 300mM potassium chloride, 50 mM HEPES, pH 7.5, 10% glycerol, 0.5 mM TCEP, and all the protein eluted as a single well-folded protein peak on heparin column.
Single guide RNA was in vitro transcribed using T7 RNA polymerase and purified using 10% UREA-PAGE. In vitro transcription template:
GAAATTAATACGACTCACTATAggCGCGTTTATTCCATTACTTTGGAGCCAGTCCCAGCGACTATGTCGTATGGACGAAGCGCTTATTTATCGGAGAGAAACCGATAAGTAAAACGCATCAAAGTCCTGCAGCAGAAAATCAAA
The CasX-sgRNA complex was assembled by incubating protein with 1.6x-fold stoichiometric excess of sgRNA for 30 min at room temperature. The ternary complexes were assembled by incubating CasX-sgRNA with 1.8x-fold stoichiometric excess of annealed DNA target for 30 min at room temperature. After the complexes were assembled, they were purified by size-exclusion chromatography using Superdex200 10/300 column.
DNA cleavage assays
DNA substrates were 5′-end-labeled with T4 PNK (NEB) in the presence of gamma 32P-ATP. Unless otherwise noted the following conditions were used: proteins were diluted to 4 μM with dilution buffer: 500 mM NaCl, 10% glycerol, 20mM Tris-HCl, pH 7.5, 1 mM magnesium chloride, 0.5 mM TCEP. Single-guide RNA was diluted to 6 μM with reaction buffer: 20 mM HEPES, pH 7.5, 10 mM magnesium chloride, 150mM potassium chloride, 1% glycerol, 0.5 mM TCEP. Resulting stocks of protein and sgRNA were mixed in 1:1 molar ratio and incubated for 10 min at room temperature to produce active complex. Cleavage reactions were conducted in 1x reaction buffer; the radiolabeled probe concentration was 2 nM. Reactions were initiated by addition of CasX-sgRNA to a final concentration of 200 nM. The reactions were conducted at 37 °C, and aliquots were taken at the following time points: 0, 2, 5, 30, 60, 120 minutes. The aliquots were immediately mixed with formamide loading buffer (final concentration 45% formamide and 50 mM EDTA, with trace amount of bromophenol blue) and heated for 10 min at 90°C for quenching. Samples were separated by 10% or 12% UREA-PAGE, gels were dried, and the results were visualized using a phosphoimager (Amersham Typhoon (GE Healthcare)).
In the cleavage assays used to determine the DNA cut sites (Fig. 1b, c), the following concentrations were used: 100 nM Cas protein, 120 nM guide RNA. In the experiment where trans-cleavage activity was compared between different CRISPR-Cas proteins (Fig. 1e) the following concentrations were used: 100 nM Cas proteins, 120 nM guide RNA, 150 nM activator, and M13mp18 ssDNA (New England Biolabs). In the experiments where trans-cleavage activity was compared between CasX and CasXΔ101–191, a random 50 nt oligonucleotide substrate was used. All the nucleotide sequences and plasmids used in this study have been summarized in Supplementary Table 2.
EM sample preparation and data collection
CasX complexes in a buffer containing 20 mM HEPES, pH 7.5, 150 mM KCl, 1 mM DTT, and 0.25% glycerol were used for cryo-EM sample preparation. Immediately after glow-discharging the grid for 14 seconds using a Solaris plasma cleaner, 3.6 μL droplets of the sample (~3μM) were placed onto C-flat grids with 2 μm holes and 2 μm spacing between holes (Protochips Inc.). The grids were rapidly plunged into liquid ethane using a FEI Vitrobot MarkIV maintained at 8 °C and 100% humidity, after being blotted for 4 seconds with a blot force of 8. Data were acquired using an FEI Titan Krios transmission electron microscope operated at 300 keV with a GIF energy filter, at a nominal magnification of ×135,000 (0.9 Å pixel size) for ternary complexes and ×105,000 (1.15 Å pixel size) for binary complex, with defocus ranging from −0.5 to −2 μm. Micrographs were recorded using SerialEM on a Gatan K2 Summit direct electron detector operated in super-resolution mode29. We collected a 4.8s exposure fractionated into 32, 150 ms frames with a dose of 9.58 e- Å−2s-1.
Apo-CasX in a buffer containing 20 mM HEPES, pH 7.5, 500 mM NaCl, 1 mM DTT, and 5% glycerol was used for cryo-EM sample preparation following the same sample protocol used for CasX complexes. Data were acquired using a FEI Titan Krios transmission electron microscope operated at 300 keV with energy filter and Volta Phase plate, at a nominal magnification ×105,000 (1.15 Å pixel size) with defocus of about −0.5μm.
EM data analysis
For CasX binary and ternary complexes, the 28 frames (we skipped the first 2 and last 2 frames) of each image stack in super-resolution model were aligned, decimated, summed and dose-weighted using Motioncor230. CTF values of the summed-micrographs were determined using Gctf31. Initial particle picking to generate template images was performed using EMAN232. About 10,000 particles were selected and then imported into Relion2.0 for reference-free 2D classification33. Particle picking for the complete dataset was carried out using Gautomatch (by Kai Zhang, unpublished) with templates generated in the previous 2D classification. Local CTF was re-calculated by Gctf with the determined box files. Particles were extracted from the dose-weighted, summed micrographs in Relion2.0 and then imported into CryoSparc34 for 2D classification, ab initio modeling, heterogeneous refinement, homogenous refinement and local resolution calculation.
For images obtained with a Volta Phase Plate, following preprocessing the CTF and phase-shift values of the summed-micrographs were determined using Gctf and then applied to dose-weighted, summed micrographs for further processing. Parameters in the EM analysis for the five CasX samples have been summarized in Supplementary Table 1.
Cross-linking and mass spectrometry
CasX samples in HEPES buffer were crossed-linked using 1mM bis-sulfosuccinimidyl-suberate (BS3) at 30 °C for 30 mins. The reactions were stopped by adding 50 mM Tris (final concentration). Cross-linked samples were then digested by trypsin and purified for mass spectrometry analysis. Cross-linked peptides were identified using an upgraded version of pLink35. In pLink, parameter of cross-linker was set to BS3. Parameter of enzyme was set to trypsin with up to three missed cleavages. Precursor mass tolerance and fragment mass tolerance were both set to 20 ppm. At least 6 amino acids were required for each peptide chain. Carbamidomethylation on cysteine was searched as a fixed modification. Oxidation on methionine was searched as a variable modification. Search results were filtered by requiring False Discovery Rate (FDR) < 5% at the spectral level. Further inspection of MS/MS spectra were performed using pLabel36.
Atomic model building
For the CasX ternary complex containing a 30 bp target DNA, the cryo-EM density of State I at 3.7Å resolution was used for secondary structure search in PHENIX with the “Find Helices and Strands” program 37. The protein main chain was manually traced in Coot 38. After main chain building, side chains were assigned manually based on the EM map in Coot and then were further improved using the cryo-EM map of State I with the full R-loop at a resolution of 3.2 Å. The DNA substrates and gRNA were manually built ab initio in Coot based on the cryo-EM density. To improve backbone geometry, the atomic model was subjected to PHENIX real space refinement (global minimization and ADP refinement) with secondary structure, Ramachandran, rotamer, and nucleic-acid restraints. The final model was validated using Molprobity39 and cross-linking MS data. The atomic models of State II were obtained by running flexible fitting on the State I atomic model against the State II cryo-EM map (4.2 Å resolution) with secondary structure restrains in MDFF40. PHENIX real space refinement was further used to improve backbone geometry. This State II atomic model was directly adopted for structural interpretation of the CasX ternary complex State II with full R-loop DNA and shortened non-target strand DNA.
For the CasX-ternary complex containing a full R-loop DNA, the atomic model of CasX-ternary complex State I with 30 bp target DNA was fitted into the State I cryo-EM map of CasX-ternary complex containing full R-loop DNA (resolution of 3.2 Å) using UCSF-Chimera41. Additional DNA nucleotides were manually built in Coot. The atomic model was subjected to PHENIX real space refinement against the cryo-EM map and validated using Molprobity.
For the CasX-ternary complex containing a shortened non-target strand DNA, the atomic model of CasX-ternary complex State I with 30bp target DNA was fitted into the State I EM map of CasX-ternary complex containing the shortened non-target strand DNA (resolution 4.5 Å) using Chimera. DNA nucleotides were manually modified in Coot. The atomic model was subjected to PHENIX real space refinement against the cryo-EM map and validated using Molprobity.
X-ray fluorescence elemental analysis
Targeted proteins at the concentration around 1ug/ul were mixed with 4 volumes of acetone and incubated at −20°C for 1 hour. Precipitated proteins were collected by centrifugation. The protein pellets were smashed in the buffer containing 50 mM Tris, 150 mM NaCl, and 0.03% dodecyl maltoside at pH 7.5, loaded on to a nylon loop and then flash frozen in place on a goniometer using a nitrogen stream. The samples were excited with a 14keV X-ray beam and fluorescence spectrums were collected. Elements in the sample were identified based on characteristic emission energies.
RaxML Maximum Likelihood phylogenetic tree
Cas12 proteins8,11,42 were aligned with TnpB representatives using MAFFT43. Alignment columns were trimmed using the trimAl -gappyout method, and a maximum-likelihood phylogenetic tree was constructed using RAxML44 with PROTGAMMALG as the substitution model and 100 bootstrap samplings. The family tree was visualized using iTOL v345.
Sequence identity pairwise comparison
A non-redundant set of Cas9 orthologs (Type II-A) was compiled by clustering proteins with ≥ 90% identity using CD-HIT46. Cas9, Cpf1, and CasX proteins were each aligned separately using MAFFT and the RuvC domains for each protein in the alignment was inferred from their known crystal structures. A multiple sequence alignment of the resulting RuvC domains was used to extract the percent identity of each pair of orthologs and generate a heatmap illustrating the pairwise comparisons of the RuvC domains. Histograms of the frequency of occurrence of each identity value in pairwise comparisons of Cpf1, Cas9, and CasX were plotted from the heatmap.
Reagent, software and protocol availability
All the reagents used in this work are commercialized. All the protocols have been described in detail above. All the software used in this work have been noted with references and available for academic usage. Further information request should be addressed to J.A.D. (doudna@berkeley.edu) and E.N. (enogales@lbl.gov).
Reporting Summary
Further information on research design is available in the Nature Reporting Summary linked to this paper.
Extended Data
Extended Data Figure 1. CasX purification and substrate cleavage.
a, RaxML Maximum Likelihood phylogenetic tree of type V effector proteins with TnpB nucleases. Triangle denotes collapsed branches. Bootstrap values are indicated as percentage points; values above 88 are shown between the major branches. b, Percent sequence identity pairwise comparisons between the conserved RuvC domains of Class 2 effectors Cas9 (type II-A), Cpf1 (type V-A), and CasX (type V-E) inferred from MAFFT alignment, depicted in an all vs all fashion. High identity is shown in blue with low identity shown in red. Histograms representing interfamily and intrafamily sequence identity value distributions are shown along the edge. c, DNA cleavage site comparison among Cas12a, Cas12b and CasX. 5 repeats with consistent results. d, DNA cleavage activity of DpbCasX mutations (n =3, mean ± s.d.). e, Schematic cartoon of GFP gene. Target regions for guide 1 to 9 are marked a long the gene. CasX guide screening by GFP disruption (n >2, mean ± s.d.) f, CRISPRi efficiency for CasX active site mutations. The Cas proteins and guide RNAs used in each assay are marked. Cas9-ng indicates non-targeting RNA guide of Streptococcus pyogenes Cas9 (SpyCas9), Cas9-g indicates the targeting RNA guide of SpyCas9. CasX-ng indicates non-targeting RNA guide of DpbCasX. CasX-g indicates targeting RNA guide of DpbCasX. GFP Disruption efficiency of targeting guide is shown by GFP signal/OD compared to the non-targeting guide control. (n = 4, mean ± s.d.) g, Purification of ApoCasX, CasX-gRNA binary complex and CasX-gRNA-DNA ternary complex with three DNA designs by size exclusion chromatography. The representative S200 size exclusion traces by UV280 absorbance are shown. Samples were taken from the labeled peaks and analyzed with urea-PAGE with sybrGold. sgRNA indicates the single-guide RNA. NTS indicates the non-target strand from target DNA. TS indicates the target strand from target DNA. All the reconstitutions have been repeated for more than 3 times with consistent results.
Extended Data Figure 2. Mammalian cell editing by CasX.
a, PlmCasX T7E1 gene editing validation of the mammalian cell GFP disruption assay from Figure 2g. b, PlmCasX T7EI quantification of a (n =3, mean ± s.d.). c, PlmCasX GFP disruption dose response (n =3, mean ± s.d.). The Cas proteins and guide RNAs used in each assay are marked. Cas9-ng indicates non-targeting RNA guide of Streptococcus pyogenes Cas9 (SpyCas9), Cas9-g indicates the targeting RNA guide of SpyCas9. CasX-ng indicates non-targeting RNA guide of DpbCasX. CasX-g indicates targeting RNA guide of DpbCasX. In the human assays, CasX-g2 & CasX-g3 are GFP targeting guides to the template and non-template strand respectively, and the GFP targeting guide of Cas9 (Cas9-g) which is not expected to direct CasX activity is used as the negative control for CasX-g2 and CasX-g3. d, EGFP disruption of clonal EGFP HEK293T cell lines with PlmCasX & various doses of plasmid (n =3, mean ± s.d.). Raw FACS data is plotted GFP on the X axis and FSC on the y axis with gates drawn to demonstrate how GFP negative cells are gated. e, Indels of GFP generated by PlmCasX cleavage as analyzed by sub-cloning and sanger sequencing of 20 clones. 3 repeats with consistent results f, Map of depicting the target sites for each of the CasX & Cas9 guides on the EGFP coding sequence for Figure 2h.
Extended Data Figure 3. EM analysis of CasX-gRNA-DNA ternary complex with a 30bp target DNA.
a, Target DNA sequence in this complex. b, EM analysis pipeline. 1,698,815 particles were picked from 7,500 drift-corrected micrographs and then used for 2D classification. By 2D based manual screening, 713,219 good particles were selected for 3D classification into 4 classes. 363,431 particles from the class that shows the most intact architecture were further used for heterogeneous refinement, which generated two reconstructions, State I and State II, with 71% and 29% of the particles, respectively. State I and State II were then independently refined to 3.8 Å and 4.2 Å. c, Euler angle distribution of the refined particles belonging to State I and State II. d, Fourier shell correlation (FSC) curve calculated using two independent half maps. e, The density maps for both states, colored by local resolution as calculated in Cryopsarc. Resolution ranges from 3Å to 7 Å. Panels c and d are directly taken from the standard output of Cryosparc.
Extended Data Figure 4. EM analysis of CasX-gRNA-DNA ternary complex with full R-loop (45bp target DNA).
a, Target DNA sequence in this complex. b, Cryo-EM analysis pipeline. 1,135,443 particles were picked from 5,000 drift-corrected micrographs and then used for 2D classification. By 2D based manual screening, 485,163 good particles were selected for 3D classification into 4 classes. 222,927 particles from the class showing better structure preservation were further used for heterogeneous refinement, which generated two models, State I and State II, with 67% and 33% of the particles, respectively. State I and State II were then independently refined to 3.2 Å and 5.2 Å. c, The Euler angle distribution for State I and State II. d, FSC curve calculated using two independent half maps. e, Cryo-EM structures of State I and State II colored by local resolution as calculated in Cryopsarc. Resolution ranges from 3 to 7 Å. Panels c and d are standard outputs of Cryosparc.
Extended Data Figure 5. Atomic model building of CasX ternary complexes for State I and State II.
Atomic models and cryo-EM maps (shown with a threshold of 8σ or 9σ) for the CasX ternary complex with 30bp DNA in State I (a) and State II (b), and for State I of the CasX ternary complex with full R-loop (45bp DNA) c, Representative regions of the cryo-EM density for different secondary structure regions are shown. d, Map against model FSCs. e and f, Zoomed views of atomic models fitted in EM densities. GLY917/ GLN920 and the DNA residues within 4 angstrom distance are linked by dashed lines.
Extended Data Figure 6. Structural comparison of CRISPR effectors.
a, OBD (WED) domains are shown in aquamarine, Helical-I (REC1) domains are shown in yellow, Helical-II (REC2) domains are shown in orange, RuvC domains are shown in green, Nuc (TSL) domains are shown in pink, Bridge Helixes are shown in blue. NTSB domain in CasX is shown in red, PI domain of LbCas12a is shown in purple. Guide RNA and target DNA are shown in gray. Two orientations are presented for each model. b, Overall structure and individual domains of CasX were analyzed using Dali server against the full PDB. The protein hit with highest Z-score for each target is shown in left panel. The hits are marked with protein name and PDB code. The similarity scores between CasX overall structure/domains and AscCas12b are pulled out from Dali full PDB analysis and shown in middle panel. The similarity scores between CasX overall structure/domains and AscCas12a are pulled out from Dali full PDB analysis and shown in left panel. Z-score above 8 indicates a high degree of similarity. Z-score below 8 but above 2 indicates moderate similarity (usually irrelevant random match). Z-score below 2 indicates noise. c, TSL domain and full R-loop structures are subtracted from the ternary complex. Zinc ribbon residues are colored in blue. The primary sequence across TSL-loop is shown. Tyrosines are marked with teal circles. Positive charged residues are marked with red circles. d, Zinc finger validation by X-ray fluorescence elemental analysis. Bovine erythrocyte carbonic anhydrase that contains zinc in the active site was used as a positive control. Representative Zinc peaks appeared in the purified CasX sample but not in the purified Cas9 sample. e, Atomic models of DpbcasX, AacCas12b, LbCas12a and SpyCas9 binary complexes are shown by surface representation. Protein parts are colored in cyan, and nucleic acid in dark gray. CasX, AacCas12b and SpyCas9 require both crRNA and tracrRNA (or a fused single guide RNA), while LbCas12a uses only crRNA. Guide RNAs are subtracted out from the complexes and shown as ribbons in bottom panels, independently. Mass ratio of protein and guide RNA is shown in the right. Values of relative mass occupancy for protein and guide RNA within the three binary complexes (protein+guide RNA) are shown. Protein mass occupancies are colored in cyan, and guide RNA in dark gray. f, CRISPRi efficiency by guide RNA mutation (n = 3, mean ± s.d.). Sequence for the fused single guide RNA is shown. tracrRNA, the joint loop, crRNA and spacer region are marked respectively. The sequences for mutated guide RNA are aligned with the original guide RNA sequence and shown. Cas9 is used for positive control. (+) indicates a targeting guide (−) indicates a non-targeting guide for negative control. NC indicates the non-complementary CasX guide. WT indicates the complementary wild type guide for CasX. GFP Disruption efficiency of targeting guide is shown by GFP signal/OD compared to the non-targeting guide control.
Extended Data Figure 7. Structural comparison of apo, binary and ternary CasX samples.
a, Drift-corrected image of apoCasX obtained with a 70o phase shift and defocus of 0.5μm. The scale bar is 50nm. b, Drift-corrected image of CasX-gRNA complex with a defocus of −1.5μm. c, Drift-corrected image of CasX-gRNA-DNA complex with a defocus of −1.5μm. Representative reference-free 2D class-averages are shown on the bottom panels for the three samples. The scale bar is 20nm.d, Cryo-EM reconstruction of apoCasX. 3 representative orientations are shown with colored domains. OBD colored by aquamarine, NTSB by red, Helical-I by yellow, Helical-II by orange, RuvC by dark green, TSL by light pink and the bridge helix by blue. e, BS3 cross-linking signals revealed by mass spectrometry for the apoCasX sample. The two lysine within a cross-linked pair are connected with purple curve. f, g, As d and e for CasX-gRNA binary complex. h, i, As d and e for CasX-gRNA-DNA ternary complex. j, k, Accessibility of target strand DNA by the RuvC domain in State I and State II. Distance between the TS DNA cleavage region and RuvC active site as calculated using Pymol is 43.8 Å for State I (j) and 10.9 Å for State II (k).
Extended Data Figure 8. EM analysis of CasX-gRNA-DNA ternary complex with shortened NTS (20nt NTS and 45nt TS).
a, Target DNA sequence in this complex. b, Cryo-EM analysis pipeline. 801,927 particles were picked from 3,500 drift-corrected micrographs and then used for 2D classification. By 2D based manual screening, 369,430 good particles were selected for 3D classification into 4 classes. 181,009 particles from the class class showing better structure preservation were further used for heterogeneous refinement, which generated two models, state I and state II, with 33.6% and 66.4% of the particles, respectively. State I and State II were then independently refined to 4.5 Å and 4.4 Å by homogenous reconstruction. c, The Euler angle distribution of refined particles belong to State I and State II. d, FSC curve calculated using two independent half maps, indicating an overall resolution of 4.5Å for state I and 4.4 Å for state II. e, Cryo-EM structures of State I and State II colored by local resolution as calculated in Cryopsarc. Resolution ranges from 3Å to 7 Å. Panels c and d are directly adopted from the standard outputs of Cryosparc.
Extended Data Figure 9. CasX ΔNTSBD purification and substrate cleavage.
a, The representative S200 size exclusion traces by UV280 absorbance for wt CasX and for CasX with NTSB domain truncation. SDS-PAGE of wt CasX protein and CasX protein with NTSB domain truncation by Coomassie brilliant blue staining is shown on the up-right panel. b, Comparison of the cleavage activities of wt CasX and CasX with NTSB domain truncation on an unwound probe (only the PAM region is base-paired, the rest of the probe is mismatched) and on just a single target DNA strand. All the assays have been repeated for 3 times with consistent results.
Extended Data Figure 10. Proposed model for sequential CasX activation for DNA cleavage.
a, Proposed overall architecture of apoCasX. The different protein domains are colored as in Figure 3. b, Cryo-EM map of the gRNA-bound CasX. Upon gRNA binding, CasX undergoes a domain rearrangement (gRNA is shown as a gray solid surface). c, Cryo-EM map of the CasX ternary complex in the NTS-loading state (State I). Upon target dsDNA recognition and unwinding by the CasX-gRNA complex, the non-target strand is preferentially positioned into the RuvC active site for cleavage. d, Cryo-EM map of the CasX ternary complex in the TS-loading state (State II). After non-target strand cleavage, the entire RNA-DNA duplex is bent by the TSL domain, thus positioning the target strand into RuvC active site. e, Cryo-EM of the CasX ternary complex mimicking a hypothetical Trans-active state. After target strand DNA cleavage, the tension within the bent RNA-DNA duplex favors the return of the CasX ternary complex to State I, thus enabling the RuvC domain to cut any accessible single strand DNA. The model shown here corresponds to the CasX ternary complex with a short NTS DNA in State I to mimic the trans-ssDNA cleavage state (the 5’ overhang of TS DNA which folds back to RuvC domain is colored by blue).
Supplementary Material
Acknowledgements
EM data were collected at the Bay Area Cryo-EM (BACEM) facility located at UC Berkeley. We thank D. B. Toso and P. Grob for expert electron microscopy assistance, and A. Chintangal and P. Tobias for computational support. We thank D. Savage, J. Cofsky, and A.V. Wright for comments on the manuscript. This project was funded by NSF grant no. 1244557 (J.A.D.); NIH grant no. P50GM082250 (J.A.D.) and NIH grant no. P01GM051487 (J.A.D. and E.N.). J.A.D. and E.N. are Howard Hughes Medical Institute Investigators.
The authors declare the following competing interests
J.A.D., B.L.O, L.B.H, J.J.L, & N.O. have filed a related patent on CasX mutations and guide RNAs described herein with the United States Patent and Trademark Office. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics and Mammoth Biosciences, and a Director of Johnson & Johnson. J.A.D is a scientific advisor to Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Metagenomi and Inari. B.L.O is a co-founder of Scribe Therapeutics.
Footnotes
Data availability
All data to support the conclusions can be found in the figures and the source data file. The cryo-EM structural models and electron density maps have been deposited in the Protein Data Bank under the codes of 6E5O, 6E7A and 6E79 and the Electron Microscopy Data Bank under the codes of EMD-8987, EMD-8988, EMD-8980, EMD-8994, EMD-8996, EMD-8991, EMD-8989 and EMD-8890. More information is summarized in Supplementary Table 1. All the plasmids and oligo sequences used in this study are summarized in Supplementary Table 2.
References
- 1.Marraffini LA CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61, doi: 10.1038/nature15386 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wright AV, Nunez JK & Doudna JA Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 164, 29–44, doi: 10.1016/j.cell.2015.12.035 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R & Doudna JA Applications of CRISPR technologies in research and beyond. Nature biotechnology 34, 933 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Strutt SC, Torrez RM, Kaya E, Negrete OA & Doudna JA RNA-dependent RNA targeting by CRISPR-Cas9. Elife 7, e32724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koonin EV, Makarova KS & Zhang F Diversity, classification and evolution of CRISPR-Cas systems. Current opinion in microbiology 37, 67–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science, 1231143 (2013).
- 7.Jinek M et al. RNA-programmed genome editing in human cells. elife 2, e00471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burstein D et al. New CRISPR–Cas systems from uncultivated microbes. Nature 542, 237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamano T et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H, Gao P, Rajashankar KR & Patel DJ PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell 167, 1814–1828. e1812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JS et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swarts D & Jinek M Mechanistic Insights into the Cis-and Trans-acting Deoxyribonuclease Activities of Cas12a. bioRxiv, 353748 (2018). [DOI] [PMC free article] [PubMed]
- 14.Oakes BL, Nadler DC & Savage DF Protein engineering of Cas9 for enhanced function. Methods Enzymol 546, 491–511, doi: 10.1016/B978-0-12-801185-0.00024-6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakes BL et al. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nature biotechnology 34, 646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’connell MR et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X et al. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Scientific reports 4, 6420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamano T et al. Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1. Mol Cell 67, 633–645 e633, doi: 10.1016/j.molcel.2017.06.035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm L & Laakso LM Dali server update. Nucleic acids research 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamano T et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 165, 949–962, doi: 10.1016/j.cell.2016.04.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moolenaar GF, Hoglund L & Goosen N Clue to damage recognition by UvrB: residues in the beta-hairpin structure prevent binding to non-damaged DNA. EMBO J 20, 6140–6149, doi: 10.1093/emboj/20.21.6140 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen J, Gai D, Patrick A, Greenleaf WB & Chen XS The roles of the residues on the channel beta-hairpin and loop structures of simian virus 40 hexameric helicase. Proc Natl Acad Sci U S A 102, 11248–11253, doi: 10.1073/pnas.0409646102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castella S, Bingham G & Sanders CM Common determinants in DNA melting and helicase-catalysed DNA unwinding by papillomavirus replication protein E1. Nucleic Acids Res 34, 3008–3019, doi: 10.1093/nar/gkl384 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn S & Roberts S The zinc ribbon domains of the general transcription factors TFIIB and Brf: conserved functional surfaces but different roles in transcription initiation. Genes & development 14, 719–730 (2000). [PMC free article] [PubMed] [Google Scholar]
- 26.Okuda M et al. A novel zinc finger structure in the large subunit of human general transcription factor TFIIE. Journal of Biological Chemistry 279, 51395–51403 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Pan H & Wigley DB Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure 8, 231–239 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Larson MH et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nature protocols 8, 2180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mastronarde DN SerialEM: a program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microscopy and Microanalysis 9, 1182–1183 (2003). [Google Scholar]
- 30.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K Gctf: Real-time CTF determination and correction. Journal of structural biology 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang G et al. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Kimanius D, Forsberg BO, Scheres SH & Lindahl E Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature methods 14, 290 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Yang B et al. Identification of cross-linked peptides from complex samples. Nature methods 9, 904 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Asara JM, Christofk HR, Freimark LM & Cantley LC A label‐free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics 8, 994–999 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trabuco LG, Villa E, Schreiner E, Harrison CB & Schulten K Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49, 174–180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Shmakov S et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Molecular cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K & Standley DM MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic I & Bork P Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic acids research 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu L, Niu B, Zhu Z, Wu S & Li W CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.