Abstract
Keratins are the main cytoskeletal proteins of epithelial cells and changes in the expression of keratins have contributed to the evolutionary adaptation of epithelia to different environments. Keratin K24 was proposed to be a differentiation marker of epidermal keratinocytes but the significance of K24 expression in the epidermis versus other tissues has remained elusive. Here, we show by RT-PCR, western blot, and immunofluorescence analyses that K24 is highly expressed in the epithelium of the cornea whereas its expression levels are significantly lower in other stratified epithelia including in the epidermis. To investigate the evolutionary history of K24, we screened the genome sequences of vertebrates for orthologs of the human KRT24 gene. The results of this comparative genomics study suggested that KRT24 originated in a common ancestor of amniotes and that it was lost independently in three clades of mammals, i.e. camels, cetaceans, and a subclade of pinnipeds comprising eared seals and the walrus. Together, the results of this study identify K24 as component of the cytoskeleton in the human corneal epithelium and reveal previously unknown differences of keratin gene content among mammalian species.
Subject terms: Molecular evolution, Gene expression
Introduction
Keratins are intermediate filament (IF) proteins that form the cytoskeleton of epithelial cells1–4. Human epithelia contain 28 type I and 26 type II keratins that have a conserved central α-helical domain but differ in several molecular properties such as the presence or absence of glycine and serine-rich sequences in the amino- and carboxy-terminal domains5. The 54 keratin genes in the human genome have evolved from a common precursor and acquired specific transcription-regulatory elements during their diversification6–8. Accordingly, all type I and all type II keratins have unique expression patterns in the various epithelia of the body whereby many, but not all, type I keratins are co-expressed with a specific type II keratin9–14. For instance, keratins K5 and K14 are co-expressed in the basal layer of stratified epithelia, and K1 and K10 are co-expressed in the suprabasal layers of the epidermis to form heterodimers. However, K2 is also expressed in human suprabasal epidermis15 so that the presence of both K1:K10 and K2:K10 dimers in the same cells causes complexity in the cytoskeleton of differentiated keratinocytes.
Keratin K24 is encoded by the KRT24 gene that is located at one end of the type I keratin gene cluster where it is flanked by KRT222, which encodes an as-yet uncharacterized keratin-like protein, and KRT2516–18. KRT24 and KRT25 belong to a type I keratin gene subcluster that is characterized by expression in differentiated cells within the suprabasal layers of stratified epithelia or epithelial appendages (Fig. 1). The other members of this subcluster are KRT26, KRT27, and KRT28, which like KRT25 are expressed in the inner root sheath (IRS) of hair follicles19, KRT10 which is expressed in the epidermis, and KRT12 which is expressed in the epithelium of the cornea. Recently, K24 was reported, based on immunodetection, to be expressed in the suprabasal layers of human epidermis and in epidermal in vitro models20,21. Another article reported expression of K24 mRNA in the corneal epithelium22.
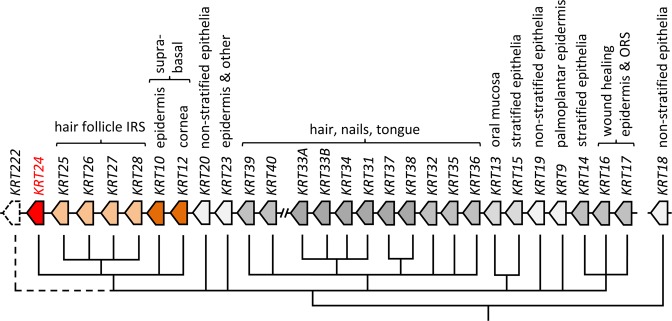
Figure 1.
Loci and phylogenetic relationships human type I keratin genes. The schematic shows the loci of KRT24 and other human type I keratin genes and the phylogenetic relationships of the genes27. Branches without significant bootstrap support are collapsed. Arrows indicate the genes with arrow tips pointing in the direction of gene transcription. The expression pattern of each keratin in human tissues is indicated above the genes. With the exception of KRT18 (chromosome 12q13), all type I keratin genes are clustered on chromosome 17q21. KRTAP genes, located between KRT40 and KRT33A, are not shown. Note that KRT222 (indicated by an arrow drawn with a broken line) encodes a keratin-like protein of unknown function. IRS, inner root sheath; ORS, outer root sheath.
In the present study, we compared the expression levels of KRT24 in the epithelia of the cornea and the skin and investigated the KRT24 gene locus in phylogenetically diverse amniotes. We provide evidence for a predominant expression of human KRT24 in differentiated corneal epithelial cells and for differential conservation and loss of KRT24 orthologs among mammals.
Results
KRT24 is expressed at high levels in the human cornea
The expression levels of KRT24 in human tissues and cell types were analyzed in the Genevestigator database, a comprehensive collection of public microarray and RNA-Seq study results23. The highest levels of KRT24 mRNA were found in the cornea, followed by the amniotic fluid, conjunctiva, oral epithelia, and the esophagus (Fig. 2a). Only very low amounts of KRT24 mRNA were found in the epidermis (Fig. 2a), although previous reports had suggested a role of KRT24 in differentiation of epidermal keratinocytes.
Figure 2.
KRT24 mRNA levels are high in the cornea. (a) KRT24 mRNA levels in human tissues and cell types were extracted from the Genevestigator database. Expression values in arbitrary units (a.u.) according to Genevestigator23 are shown. Bars indicate means and error bars show standard deviations. The number (n) of samples per tissue or cell type is indicated below the bars. P-values were calculated with the two-sided t-test. (b–e) Quantitative RT-PCR analysis of mRNAs of selected keratin genes in human cornea (n = 8) and epidermis (n = 5). Expression levels of KRT24 (b), KRT3 (c), KRT12 (d), and KRT10 (e) were calculated relative to that of GAPDH and normalized to the highest value (defined as 10 arbitrary units, a.u.) among the samples investigated. Bars indicate means and error bars show standard deviations. P-values (Mann-Whitney U Test) are shown above the graphs.
Next, we compared KRT24 mRNA amounts in cornea and epidermis by quantitative RT-PCR analysis. In agreement with the Genevestigator database (Fig. 2a), mRNA levels of KRT24 were significantly higher in the cornea than in the epidermis (Fig. 2b). The expression of KRT24 resembled that of KRT3 (Fig. 2c) and KRT12 (Fig. 2d), i.e. the established marker keratins of the differentiated corneal epithelium24,25, and was opposite to the expression of KRT10, a marker of epidermal differentiation (Fig. 2e).
K24 protein is present in the superficial cells of the corneal epithelium
To determine the distribution of K24 protein in human tissues, we generated an antibody against an epitope within the tail domain of K24. In western blot analysis, this antibody detected a band at the expected size of 55 kD (Fig. 3a). Only low amounts of K24 were present in the epidermis and conjunctiva, whereas a prominent band of K24 was detected in lysates obtained from human corneas (Fig. 3a).
Figure 3.
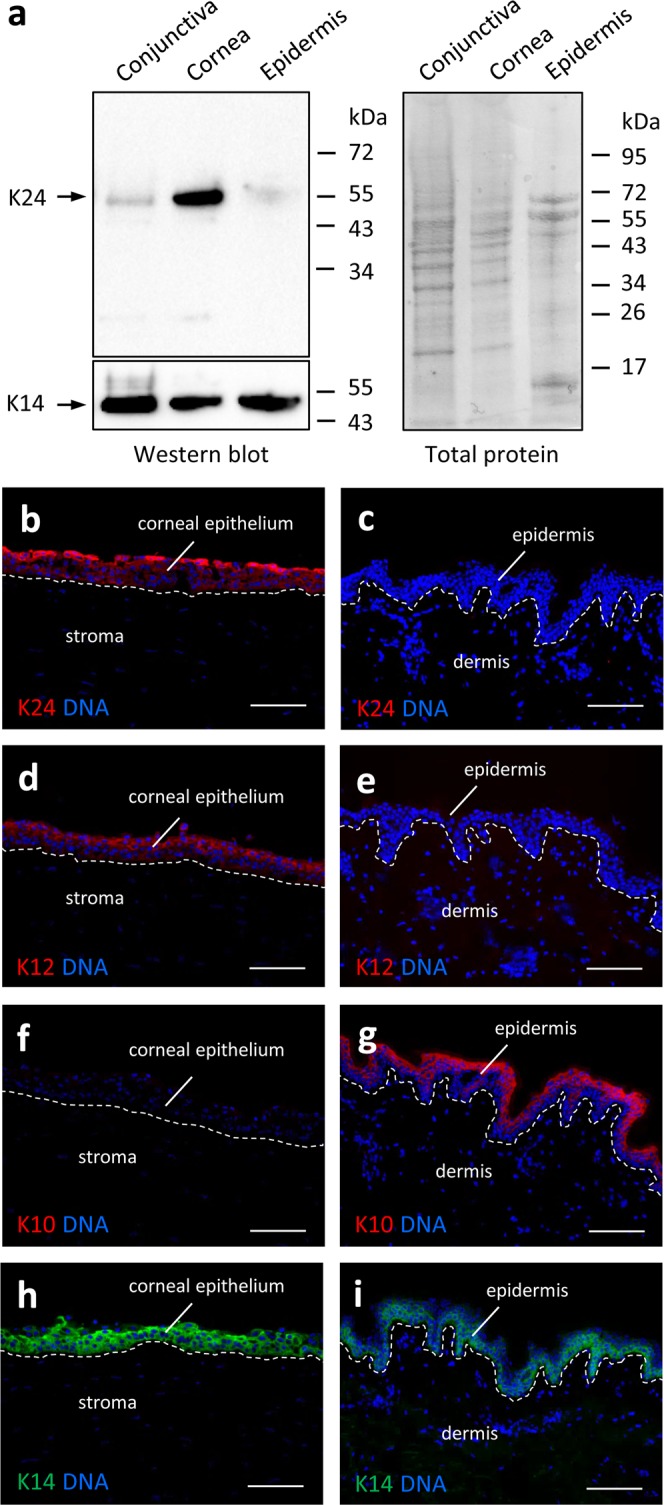
Immunodetection of K24 protein in the human cornea. (a) Western blot analysis of K24 and K14 in human conjunctiva, cornea and epidermis. The total protein blotted onto the membrane was visualized by Ponceau staining prior to incubation with blocking buffer and probing with antibodies. Results are representative for n ≥ 3 samples of each tissue. Positions of size markers are shown on the right. kDa, kilo-Dalton. (b) Immunofluorescence labeling of K24, K12, K10 (red) and K14 (green) in human cornea and skin. DNA was labeled with Hoechst 33258 dye (blue). The junctions between epithelia and mesenchymal tissues are indicated by broken lines. Results are representative for at least 3 samples of each tissue. Scale bars: 100 µm.
Immunofluorescence analysis revealed expression of K24 in the superficial layer of the corneal epithelium (Fig. 3b) whereas no significant labeling was detected in the epidermis (Fig. 3c). K12 showed a similar distribution but was also expressed in lower suprabasal corneal epithelial cells (Fig. 3d,e). K10 was absent in the cornea and abundant in the epidermis (Fig. 3e,f). K14 was expressed in the basal layer of both corneal epithelium and epidermis (Fig. 3h,i). Thus, KRT24 mRNA and K24 protein distribution patterns were highly consistent and provided solid evidence for predominant expression of KRT24 in the corneal epithelium.
Besides the cornea of the eye and the skin, we performed immunofluorescence analysis of the esophagus, representing one of the organs that contain low but significant levels KRT24 mRNA (Fig. 2)26. K24 was detected in suprabasal epithelial cells of the esophagus with immunofluorescence signal intensities weaker than in the cornea (Suppl. Fig. S1).
K24 is an evolutionary ancient keratin that has been differentially conserved or lost in different clades of mammals
Epithelia of the body surface are subjected to environment-dependent evolutionary constraints that affect their molecular composition, including proteins of the cytoskeleton. Therefore, we hypothesized that the locus of the KRT24 gene might have undergone differential evolution in species with different lifestyles. To test this hypothesis, we performed a comparative genomics study and determined the presence or absence of KRT24 orthologs in phylogenetically diverse vertebrates. The investigation was focused on the locus bordered by KRT222 and KRT12 (Fig. 4). In line with results of previous studies27,28, we identified Krt24-like genes but not 1:1 orthologs of Krt24 besides Krt222 in fish and amphibians. In the alligator a Krt24-like gene is located between Krt222 and Krt10, and three Krt24-like genes are present in the platypus, representing basal mammals (Supplementary Tables S1–S7; Suppl. Fig. S2; Fig. 4). By contrast, no Krt24 homolog is present in the chicken (Fig. 4). Krt24 orthologs are conserved in the majority of mammals with a few notable exceptions that will be discussed below.
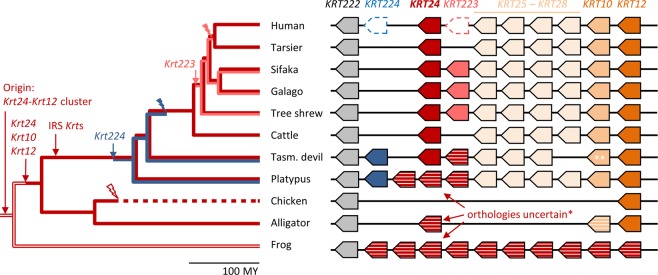
Figure 4.
Evolution of the KRT24 gene locus. A model for the evolutionary history of KRT24 and neighboring keratin genes was developed from the comparison of corresponding gene loci in tetrapods. Genes are indicated by arrows that point from the 5′-end to the 3′-end of the coding sequence. A timed phylogenetic tree46 is depicted besides the gene loci. Gene gain and loss events were inferred by the principle of maximum parsimony from the species distribution of the genes. The presence or absence of KRT24, KRT223, and KRT224 genes in ancestors of modern species is indicated by lines of the same color as the corresponding gene in the schematic on the right. The scale bar below the phylogenetic tree indicates a period of 100 million years (MY). Note that KRT223 and KRT224 are pseudogenes in the human genome and that orthology relationships could not be faithfully resolved by molecular phylogenetics for Krt24-like genes of several species (*). Two copies of the Krt10 gene are present in marsupials but only one copy is shown in this schematic (**). IRS Krts, inner root sheath keratin genes (KRT25-28).
Interestingly, we found diversity in the genes immediately flanking Krt24 in different mammals. Krt223 is located on the 5′-side of Krt24 in primates of the suborder Strepsirrhini (galago and sifaka) and the tree shrew whereas only a remnant of this gene (KRT223P, keratin 223 pseudogene) is present in the human genome (Fig. 4). Similarly, Krt224 is located on the 3′-side of Krt24 in the platypus and marsupials (Tasmanian devil) but not in the placental mammals investigated, such as human and other primates, tree shrew, and cattle (Fig. 4). A pseudogene corresponding to Krt224 was identified in the human genome (Suppl. Fig. S3). By mapping these genes and pseudogenes onto the phylogenetic tree of mammals (Fig. 4), we inferred that the evolution of the human set of type I keratins involved the loss of at least two genes, i.e. Krt223 and Krt224, that were retained in some other extant mammals.
The analysis of publicly available genome sequences showed that Krt24 was conserved in most mammals but not in cetaceans18, a subclade of pinnipeds, and camels (Fig. 5, Suppl. Fig. S4). Krt24 was lost in cetaceans after their evolutionary divergence from the lineage leading to their next extant relative, the hippopotamus (Suppl. Fig. S4). Krt24 gene remnants were found in the genome of several cetaceans, including the bottlenose dolphin (Tursiops truncatus). Amplification and sequencing confirmed that the open reading frame of Krt24 is disrupted in the dolphin (Suppl. Fig. S5). Within pinnipeds, the walrus (Odobenus rosmarus divergens), the northern fur seal (Callorhinus ursinus) and the California sea lion (Zalophus californianus), together representing the clade Otaroidea, lack a functional Krt24 gene (Suppl. Fig. S4). Within the family Camelidae, Krt24 has been inactivated in the genus Camelus including C. dromedarius, C. bactrianus, and C. ferus, whereas Krt24 is intact in the alpaca (Vicugna pacos) (Suppl. Fig. S5). These cases of gene loss indicate that Krt24 had played a redundant or even disadvantageous role in ancestors of several extant mammals. It will be interesting to determine whether loss of Krt24 correlates with adaptions of the corneal epithelium or other epithelia in these species.
Figure 5.
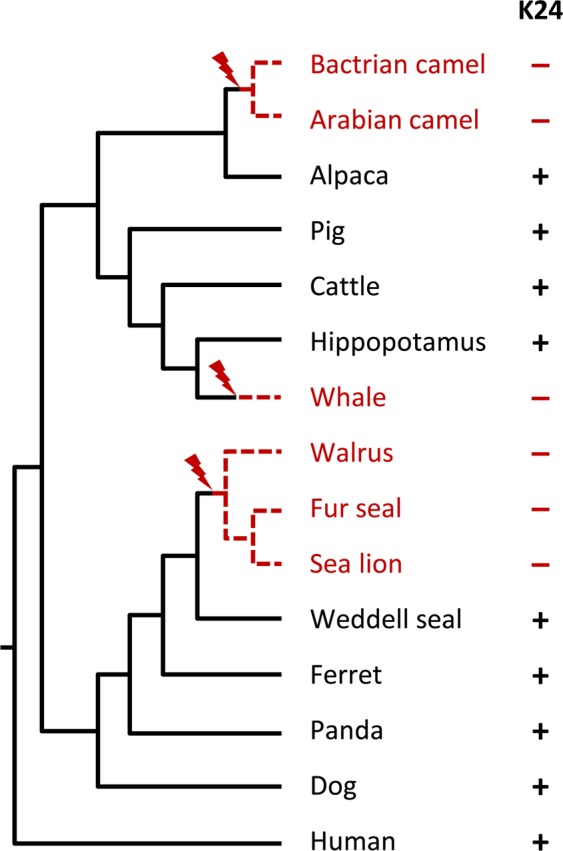
Loss of Krt24 in different clades of mammals. The presence (+) or absence (−) of functional Krt24 genes was mapped onto a phylogenetic tree of mammals. Only Krt24-deficient species and close relatives are shown.
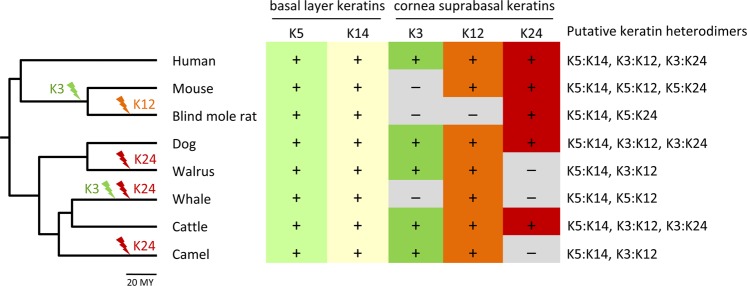
Finally, we compared the pattern of conservation and loss of K24 with that of other keratins. Neither did the pattern of K24 conservation among species correlate with that of any other keratin nor did the corneal keratins K3 and K12 show strict co-evolution (Fig. 6). Thus, the functions of each K24, K12, and K3 can be replaced by other keratins or have become dispensable in distinct evolutionary lineages of mammals.
Figure 6.
Evolution of corneal epithelial keratins in mammals. The presence (+) or absence (−) of functional orthologs of human corneal epithelial keratins K5, K14, K3, K12, and K24 was mapped onto a phylogenetic tree of mammals. Gene loss events were inferred from the distribution of functional genes and known relationships of species. Putative keratin heterodimers in the cornea are listed for each species. The scale bar beneath the timed phylogenetic tree indicates a period of 20 million years (MY).
Discussion
The results of this study provide important new insights into the expression pattern and evolution of the KRT24 gene. KRT24 is predominantly expressed in the corneal epithelium and, within this epithelium, in terminally differentiated cells on the surface. With regard to this epithelial cell differentiation-associated expression, KRT24 resembles the other members of the type I keratin subcluster also comprising suprabasal epidermal keratin KRT10, suprabasal corneal keratin KRT12, and the IRS keratins KRT25, KRT26, KRT27, and KRT28 (Fig. 1). It is therefore likely that a suprabasally expressed type I keratin gene was the molecular ancestor of all these genes including KRT24 and that duplications and modifications of promoter and coding sequences have led to the diversification and specification of expression patterns and protein properties.
KRT24 differs from the other members of the suprabasal type I keratin gene cluster because it has a relatively broad tissue expression pattern (Fig. 2a). While KRT10, KRT12, and the IRS keratins are hardly expressed outside of their predominant sites, i.e., the suprabasal epidermis, the corneal epithelium and the differentiated keratinocytes of the IRS, respectively, KRT24 mRNA levels are significant in the cornea, oral epithelium, and esophagus. This tissue distribution indicates that K24 contributes to the cytoskeleton of stratified squamous epithelia that, unlike the epidermis, do not form a cornified layer on the surface. It is also interesting to note that all K24-positive epithelia contain at least one further suprabasal type I keratin and K24 is probably a quantitatively minor keratin at all these sites.
Type I keratin K24 is coexpressed with different type II keratins in different epithelia. At its predominant expression site, the suprabasal corneal epithelium, K3 is the main type II keratin and therefore the prime candidate for heterodimerization with K24. In other human epithelia and in the corneal epithelium of the mouse29, K3 is missing whereas other type II keratins such as K4 and K5 are abundant. Therefore, K24 appears to be a promiscuous heterodimerization partner of type II keratins, however, coexpression experiments and ultrastructural localization studies will be necessary to test this hypothesis. The distribution of K24 in the corneal epithelium differs from that of K3 and K5 because K24 is confined to the most superficial cells. This distribution and our demonstration of absence of K24 in several species suggest that K24 is not indispensable for the formation of a cytoskeleton in corneal epithelial cells. Similar to the start of expression of K2, besides expression of K1 and K10, in terminally differentiating keratinocytes of the epidermis15, the increase of expression of K24 in terminally differentiating cells is likely to add new components to the cytoskeleton, thereby modifying its properties. Targeted deletion of the Krt24 gene in mice will help to define the roles of K24 in the corneal epithelium in future studies.
Importantly, our data indicate that expression of KRT24 in the epidermis, as it was reported recently20, is smaller by almost 2 orders of magnitude than that in the cornea. Accordingly, we recommend K24 not to be considered an epidermal differentiation marker. Our side-by-side comparison of cornea and epidermis by RT-PCR, western blot and immunofluorescence analysis showed consistently that mRNA and protein levels of K24 in the epidermis were very low as compared to the cornea. The identity of KRT24 RT-PCR products was confirmed by sequencing, K24 protein was only detected in the tissue that contained high amounts of KRT24 mRNA, and our anti-K24 antibody yielded only a single western blot band at the expected size of K24. Another anti-K24 antibody (ab180486) that was reported to yield strong signals in the granular and cornified layers of the epidermis20,21 was not distributed by the manufacturer when the present manuscript was written (March 2019). Yet another antibody (HPA022978) that was tested in the Human Protein Atlas project yielded staining signals in several tissues that do not contain KRT24 mRNA26. The reasons for the discrepancies in the immunoreactivities of different antibodies is not known but likely involves cross-reactivities with other antigens. Based on our RT-PCR, western blot and immunofluorescence data, we can conclude that the abundance of K24 in the epidermis, if it is present there, is considerably lower than in the cornea.
KRT24 originated in amphibious or terrestrial vertebrates and was conserved in most but not all amniotes. It is presently unknown whether Krt24 is expressed in the corneal epithelium of species other than human. The expressed sequence tag (EST) profile of murine Krt24 in GenBank (https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Mm.46378, last accessed 11 March 2019), shows expression in “eye” besides “tongue” and “embryonic tissue”, indicating that conserved expression of Krt24 in the cornea is likely. A quantitative comparison of K24 expression in the cornea and the tongue was not possible in this study because tongue samples were not available. However, KRT24 mRNA is also present in the esophagus (Fig. 2a) and immunolabeling of formalin-fixed and paraffin-embedded esophagus confirmed expression of K24, though at lower levels than in the cornea (Suppl. Fig. S1). These data suggest that Krt24 has additional roles in extra-corneal tissues of both mice and humans. In line with the notion of Krt24 function(s) outside of the cornea, the blind mole rat (Spalax galili) has retained an intact Krt24 gene (Fig. 6) despite degeneration of the corneal epithelium during the evolution of this species30,31.
Although Krt24 has been conserved in phylogenetically diverse mammals, we have also found evidence for loss of Krt24 in three clades of mammals, i.e. cetaceans, Otaroidea and camels. The premature stop codons identified in the Krt24 genes of these mammals are predicted to cause nonsense-mediated decay of mRNA32 as they are located before the last exon. Even if mRNA is translated, the protein would lack a part or the complete intermediate filament domain which is required for the function of K24 as a component of the cytoskeleton. Mutations in camels and a representative cetacean were confirmed by our sequencing of genomic DNA. A premature stop in Krt24 genes of walrus and sea lion was located at a conserved position, suggesting that this stop codon was inherited from a common ancestor. It is noteworthy that a Krt24-like gene is present in the alligator but not in birds (Fig. 4), possibly indicating loss of this gene during the evolution of birds. In mammals, Krt24 is flanked by the genes Krt25 through Krt28, which have originated after the divergence of mammals from reptiles. Krt25 through Krt28 are expressed specifically in hair follicles and were lost when hair follicles degenerated during the evolution of cetaceans18.
The identified cases of evolutionary loss of Krt24 indicate that this gene is dispensable in three clades of mammals. Furthermore, these data raise the question as to whether loss of Krt24 was associated with phenotypic changes. Cetaceans have many adaptations to aquatic life that include thickening of the epidermis18,33,34 and perhaps also changes in the cornea and internal epithelia. In the present study we show that the next relative of cetaceans, i.e. the hippopotamus does not have inactivating mutations in Krt24, whereas other largely aquatic mammals, i.e. members of a subclade (eared seals + walrus) of pinnipeds, lack functional Krt24. Yet other aquatic mammals such as sirenians and earless seals (Phocidae) have intact Krt24 genes, suggesting that loss of Krt24 is not an obligatory adaptation to aquatic life. Interestingly, Krt24 was inactivated in camels but not in their next relative, the alpaca. Camels have a thicker corneal epithelium than most or all other mammals35, possibly representing an adaptation to the low humidity of their environment or to mechanical abrasion of superficial cells during exposure to sand and dust particles. It will be interesting to determine links between loss of Krt24 and specific features of epithelia in the aforementioned species.
Taken together, Krt24 has a complex evolutionary history in amniotes and its expression pattern in humans suggests that it contributes to the keratin cytoskeleton of several stratified epithelia. High levels of K24 expression in the superficial layer of the corneal epithelium point to a predominant role of human K24 in the cornea. Accordingly, possible aberrations of K24 function should be tested in future studies using samples from normal and diseased corneas displaying impaired differentiation.
Materials and Methods
Human tissues
The cornea bank at the Medical University of Vienna procured postmortem human corneal tissues (ethics approval number EK1578/2013). Human skin was obtained from plastic surgery (ethics approval number EK2011/1149). All donors provided written informed consent.
Comparative genomics
The genome sequences of frog (Xenopus tropicalis)36, Chinese alligator (Alligator sinensis)37, platypus (Ornithorhynchus anatinus)38, Tasmanian devil (Sarcophilus harrisii)39, cattle (Bos taurus), tree shrew (Tupaia chinensis)40, mouse (Mus musculus), galago (Otolemur garnetti), sifaka (Propithecus coquereli), tarsier (Carlito syrichta), and human (Homo sapiens) were screened for the presence or absence and sequence integrity of Krt24 and neighbouring genes. In addition, corneal epithelium-specific keratin genes Krt3 and Krt12 were screened with the same criteria in blind mole rate (Spalax galili)30, sperm whale (Physeter catodon), dolphin (T. truncatus) and camels (Camelus dromedarius and Camelus bactrianus). The genome sequences of other species were used for sequence comparisons. The sequences were retrieved from the GenBank database of the National Center for Biotechnology Information (NCBI), USA (http://www.ncbi.nlm.nih.gov/). Local sequence similarity was obtained using the Basic Local Alignment Search Tool (BLAST)41. The conservation of blocks of order of genetic elements (synteny) was tested by manual alignment of gene maps including conserved genes on each side of the gene(s) of interest. Gene orthologies between species were inferred from shared synteny and phylogenetic analysis.
Sequence alignments and phylogenetic analysis
Alignments were performed with Multiple Sequence Comparison by Log-Expectation (MUSCLE)42, only the unambiguously aligned intermediate filament structures of the keratins were used for subsequent phylogenetic analyses. Maximum likelihood estimation was reconstructed using the Seaview platform43, using a JTT model with 100 bootstrap replicates to assess clade support. A mixed amino acid model with gamma correction for rate heterogeneity and invariable sites was used for estimating Bayesian posterior probabilities in MrBayes 3.2.644. Two parallel Markov chain Monte Carlo (MCMC) runs of four chains each were performed, with a length of 3,500,000 generations, a sampling frequency of 1 per 1,000 generations. To define the burnin and check the convergence of the MCMC runs, Tracer v1.7.1 was used45. Trees were visualized with FigTree v1.4.2. Phylogenetic relationships and divergence times were obtained from the Timetree website (www.timetree.org)46.
Sequence analysis of Krt24 genes
DNA from two camel species (Camelus dromedarius and Camelus bactrianus)47, transferred from the Austrian Science Fund project P1084-B17 (P. Burger), was investigated by PCRs using the following primers annealing to the Krt24 gene of both C. dromedarius (GeneID:105031793) and C. bactrianus (GeneID:105073329): camel-K24-s (5′-TCCCCTCCTTCAGTGTCCTA-3′) and camel-K24-a (5′-GAAGCAGCTTGTGTGTGACC-3′). Twenty-five ng template DNA were used for PCR, involving an annealing temperature of 59 °C and 38 cycles. DNA of bottlenose dolphin (T. truncatus)31 was subjected to PCR amplification with the primer pair dolphin-K24-s (5′-CCCCACGTCCATTCTATGACAGT-3′) and dolphin-K24-a (5′-TGATGAGATGAGCGGAAGTGGC-3′) using an annealing temperature of 62 °C and 35 cycles. Sequencing of PCR products was conducted by Microsynth AG, 6961 Wolfurt, Austria.
Quantification of mRNAs in tissues
The Genevestigator database23 was used to determine mRNA levels of KRT24 in human tissues and cell types. The expression values in Genevestigator are calculated using standard normalization methods for different microarray platforms and scaled between experiments to make the expression values comparable23.
For quantititative reverse transcription-polymerase chain reaction (RT-PCR), RNA was purified from human cornea and epidermis using the Precellys system (VWR International, Radnor, PA) and TriFast (VWR International) according to the manufacturers’ instructions. 500 ng RNA were reverse-transcribed to cDNA using the IscriptTM Kit (Biorad, Hercules, CA). Quantitative PCRs were performed using the LightCycler 480 DNA SYBR Green I Master Kit (Roche Applied Science) and the LightCycler® technology (LC480). The primer pairs K24-s (5′-GGAGGTGGTTCTAGTTTTGCA-3′) and K24-a (5′-GACGAGACAACCTTGCCATC-3′), K12-s (5′- GCTCGCCATGAAGAAATCCC-3′) and K12-a (5′-CTTTCCTCCAAACCATCACCTT-3′), K3-s (5′-ATCGAGGGTGTCAAGAAGCA-3′) and K3-a (5′-GACATCCTGTACTCCTCGCC-3′), K10-s (5′-GTGGGCGAGTCTTCATCTAA-3′) and K10-a (5′-GAGACTCTTTCCTCTTGATGCA-3′), and GAPDH-s (5′-CAGTCAGCCGCATCTTCTTTTG-3′) and GAPDH-a (5′-CGCCCAATACGACCAAATCC-3′) were used for the amplification of human K24, K12, K3, K10, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), respectively. mRNA levels were normalized to the GAPDH level of each sample. A published mathematical model was used for calculating the quantities of target relative to reference transcripts48. Statistical analyses were performed using a two-tailed Mann-Whitney U Test with GraphPad Prism version 5.01 (GraphPad Software, San Diego, CA, USA).
Immunofluorescence analysis
Cornea and epidermis samples were embedded in optimal cutting temperature (OCT) medium and frozen. The tissues were sectioned at 4 µm thickness and fixed in acetone at −20 °C. In other experiments, cornea was fixed in 4.5% phosphate-buffered formalin, embedded in paraffin according to a published protocol15, sectioned at 4 µm thickness and subjected to antigen retrieval with citrate buffer at pH 6. Paraffin-embedded esophagus from another study49 was investigated in parallel. The sections were incubated with rabbit anti-K10 (Covance, PRB159P, 1:1000), guinea pig anti-K12 (Progen, GP-K12, 1:1000), mouse monoclonal anti-K14 (Abcam, ab7800, 1:200), guinea pig anti-K72 (Progen, GP-K6irs2, 1:1000) as a negative control, and guinea pig anti-K24 (1:1000). The latter antiserum was raised against the synthetic peptide C-SGSVNMGSEDLVSGD, corresponding to amino acid residues 466–480 of human K24, coupled to keyhole limpet protein.
The fluorescence-labelled secondary antibodies, raised in goat, were anti-guinea pig immunoglobulin G (IgG) conjugated to Alexa Fluor 594 (Invitrogen, A-11076, 1:500), anti-rabbit IgG conjugated to Alexa Fluor 546 (Invitrogen, A-11010, 1:500), and anti-mouse IgG conjugated to Alexa Fluor 488 (Invitrogen, A11001, 1:500). 10% goat serum (DAKO) was added to the secondary antibody to suppress unspecific binding. Nuclei were counterstained with Hoechst 33258 solution (bisBenzimid H 33258, Sigma Aldrich, B2883).
Western blot analysis
Proteins from cornea and epidermis samples were homogenized with the Precellys system (VWR International, Radnor, PA) in Laemmli extraction buffer containing 2% SDS. Protein concentrations were determined with the Micro BCA protein assay kit (Thermo Fisher Scientific). Thirty µg of total protein per lane were electrophoresed through an ExcelGel SDS 8–18% polyacrylamide gradient gel (GE Healthcare Life Sciences, Chicago) and blotted onto a nitrocellulose membrane. The membrane was incubated with guinea pig anti-K24 (dilution 1:1000) overnight at 4 °C. After washing, the membrane was incubated for 1 h at room temperature with goat anti-guinea pig IgG (Abnova, AB10611, 1:10000) coupled to horseradish peroxidase. Bands were visualized using the enhanced chemiluminescence system (SuperSignal West Dura Extended Duration Substrate, Thermo Fisher Scientific). The membrane was reincubated with monoclonal mouse anti-K14 (Abcam, ab7800, 0.33 µg/ml) and sheep anti-mouse IgG (GE Healthcare, NXA931V, 1:10000) coupled to horseradish peroxidase as a secondary antibody.
Ethical approval and informed consent
This research has been approved by the ethics committee at the Medical University of Vienna (ethics approval numbers EK1578/2013 and EK2011/1149). All procedures involving human tissues were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments. This research has been approved by the ethics committee at the Medical University of Vienna. Postmortem cornea samples were procured from the cornea bank at the Medical University of Vienna (ethics approval number EK1578/2013). Human skin was obtained from plastic surgery (ethics approval number EK2011/1149). All donors provided written informed consent. No tissues were procured from prisoners.
Supplementary information
Acknowledgements
We thank Heinz Fischer, Silke Praetzel-Wunder, Johannes Pammer, Michael Mildner, Maria Buchberger, Marie Sophie Narzt, Julia Lachner, and Barbara Lengauer for technical support and helpful advice. The authors are grateful to Gerda Gassner from the First Camel Riding School Austria and Helmut Burgsteiner for their agreement to use previously collected samples from camels for research. Newly-reported nucleotide sequences were submitted to GenBank (Accession numbers MK422443, MK422444, MN037795). This work was supported by the Austrian Science Fund (FWF): P23801, P28004.
Author Contributions
F.E. and L.E. conceived the study, F.E. and M.L. performed experimental studies, F.E. and L.E. performed genome and transcriptome sequence studies, F.E., L.L., P.B., A.P., E.T. and L.E. analyzed the results, F.E., M.L., L.L., E.T., and L.E. wrote the manuscript. All authors reviewed the manuscript.
Data Availability
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lutz Langbein is retired.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47422-y.
References
- 1.Fuchs E, Cleveland DW. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- 2.Lane EB, McLean WH. Keratins and skin disorders. J. Pathol. 2004;204:355–366. doi: 10.1002/path.1643. [DOI] [PubMed] [Google Scholar]
- 3.Simpson CL, Patel DM, Green KJ. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat. Rev. Mol. Cell. Biol. 2011;12:565–580. doi: 10.1038/nrm3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacob JT, Coulombe PA, Kwan R, Omary MB. Types I and II keratin intermediate filaments. Cold Spring Harb. Perspect. Biol. 2018;10:pii:a018275. doi: 10.1101/cshperspect.a018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schweizer J, et al. New consensus nomenclature for mammalian keratins. J. Cell Biol. 2006;174:169–174. doi: 10.1083/jcb.200603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hesse M, Zimek A, Weber K, Magin TM. Comprehensive analysis of keratin gene clusters in humans and rodents. Eur. J. Cell Biol. 2004;83:19–26. doi: 10.1078/0171-9335-00354. [DOI] [PubMed] [Google Scholar]
- 7.Homberg M, Magin TM. Beyond expectations: novel insights into epidermal keratin function and regulation. Int. Rev. Cell. Mol. Biol. 2014;311:265–306. doi: 10.1016/B978-0-12-800179-0.00007-6. [DOI] [PubMed] [Google Scholar]
- 8.Eckhart, L. & Ehrlich, F. Evolution of trichocyte keratins. In The Hair Fibre: Proteins, Structure and Development (eds Plowman, J. E., Harland, D. P. & Deb-Choudhury, S.) 33–45 (2018). [DOI] [PubMed]
- 9.Langbein L, Schweizer J. Keratins of the human hair follicle. Int. Rev. Cytol. 2005;243:1–78. doi: 10.1016/S0074-7696(05)43001-6. [DOI] [PubMed] [Google Scholar]
- 10.Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem. Cell Biol. 2008;129:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bragulla HH, Homberger DG. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 2009;214:516–559. doi: 10.1111/j.1469-7580.2009.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langbein L, Eckhart L, Rogers MA, Praetzel-Wunder S, Schweizer J. Against the rules: The human keratin K80 - Two functional alternative splice variants, K80 and K80.1, with special cellular localization in a wide range of epithelia. J. Biol. Chem. 2010;285:36909–36921. doi: 10.1074/jbc.M110.161745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langbein L, et al. New facets of keratin K77: Interspecies variations of expression and different intracellular location in embryonic and adult skin of humans and mice. Cell Tissue Res. 2013;354:793–812. doi: 10.1007/s00441-013-1716-5. [DOI] [PubMed] [Google Scholar]
- 14.Langbein L, et al. Localisation of keratin K78 in the basal layer and the first suprabasal layers of stratified epithelia completes the expression catalog of type II keratins and provides new insights into sequential keratin expression. Cell Tissue Res. 2016;363:735–750. doi: 10.1007/s00441-015-2278-5. [DOI] [PubMed] [Google Scholar]
- 15.Fischer H, et al. Loss of keratin K2 expression causes aberrant aggregation of K10, hyperkeratosis and inflammation. J. Invest. Dermatol. 2014;134:2579–2588. doi: 10.1038/jid.2014.197. [DOI] [PubMed] [Google Scholar]
- 16.Sprecher E, et al. Refined mapping of Naegeli-Franceschetti-Jadassohn syndrome to a 6 cM interval on chromosome 17q11.2-q21 and investigation of candidate genes. J. Invest. Dermatol. 2002;119:692–698. doi: 10.1046/j.1523-1747.2002.01855.x. [DOI] [PubMed] [Google Scholar]
- 17.Rogers MA, Winter H, Langbein L, Bleiler R, Schweizer J. The human type I keratin gene family: characterization of new hair follicle specific members and evaluation of the chromosome 17q21.2 gene domain. Differentiation. 2004;72:527–540. doi: 10.1111/j.1432-0436.2004.07209006.x. [DOI] [PubMed] [Google Scholar]
- 18.Ehrlich F, et al. Differential evolution of the epidermal keratin cytoskeleton in terrestrial and aquatic mammals. Mol. Biol. Evol. 2019;36:328–340. doi: 10.1093/molbev/msy214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langbein, L. & Schweizer, J. The keratins of the human hair follicle. In International Review of Cytology Vol 243 (eds Bourne, G. H., Danielli, J. F., Jeon, K. W., Friedlander, M. & Jarvik, J.) 1–78 (2018). [DOI] [PubMed]
- 20.Min M, et al. Role of keratin 24 in human epidermal keratinocytes. PLoS One. 2017;12:e0174626. doi: 10.1371/journal.pone.0174626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klar AS, et al. The expression pattern of keratin 24 in tissue-engineered dermo-epidermal human skin substitutes in an in vivo model. Pediatr. Surg. Int. 2018;34:237–244. doi: 10.1007/s00383-017-4198-9. [DOI] [PubMed] [Google Scholar]
- 22.Hashmani K, et al. Characterization of corneal stromal stem cells with the potential for epithelial transdifferentiation. Stem Cell Res. Ther. 2013;4:75. doi: 10.1186/scrt226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hruz T, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaloin-Dufau C, Pavitt I, Delorme P, Dhouailly D. Identification of keratins 3 and 12 in corneal epithelium of vertebrates. Epithelial. Cell Biol. 1993;2:120–125. [PubMed] [Google Scholar]
- 25.Dhouailly D, Pearton DJ, Michon F. The vertebrate corneal epithelium: from early specification to constant renewal. Dev. Dyn. 2014;243:1226–1241. doi: 10.1002/dvdy.24179. [DOI] [PubMed] [Google Scholar]
- 26.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandebergh W, Bossuyt F. Radiation and functional diversification of alpha keratins during early vertebrate evolution. Mol. Biol. Evol. 2012;29:995–1004. doi: 10.1093/molbev/msr269. [DOI] [PubMed] [Google Scholar]
- 28.Vandebergh W, Maex M, Bossuyt F, Van Bocxlaer I. Recurrent functional divergence of early tetrapod keratins in amphibian toe pads and mammalian hair. Biol. Lett. 2013;9:20130051. doi: 10.1098/rsbl.2013.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu H, et al. Keratin 5 knockout mice reveal plasticity of keratin expression in the corneal epithelium. Eur. J. Cell Biol. 2006;85:803–811. doi: 10.1016/j.ejcb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Fang X, et al. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat. Commun. 2014;5:3966. doi: 10.1038/ncomms4966. [DOI] [PubMed] [Google Scholar]
- 31.Partha R, et al. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. Elife. 2017;6:pii:e25884. doi: 10.7554/eLife.25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 33.Yim HS, et al. Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 2014;46:88–92. doi: 10.1038/ng.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strasser B, Mlitz V, Fischer H, Tschachler E, Eckhart L. Comparative genomics reveals conservation of filaggrin and loss of caspase-14 in dolphins. Exp. Dermatol. 2015;24:365–369. doi: 10.1111/exd.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almubrad T, Akhtar S. Ultrastructure features of camel cornea-collagen fibril and proteoglycans. Vet. Ophthalmol. 2012;15:36–41. doi: 10.1111/j.1463-5224.2011.00918.x. [DOI] [PubMed] [Google Scholar]
- 36.Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan QH, et al. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 2013;23:1091–1105. doi: 10.1038/cr.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murchison EP, et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell. 2012;148:780–791. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan Y, et al. Genome of the Chinese tree shrew. Nat. Commun. 2013;4:1426. doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;7:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 44.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burger PA, Palmieri N. Estimating the population mutation rate from a de novo assembled Bactrian camel genome and cross-species comparison with dromedary ESTs. J. Hered. 2014;105:839–846. doi: 10.1093/jhered/est005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner T, et al. The differentiation-associated keratinocyte protein cornifelin contributes to cell-cell adhesion of epidermal and mucosal keratinocytes. J. Invest. Dermatol. 2019 doi: 10.1016/j.jid.2019.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.