Abstract
Mel4 is a novel cationic peptide with potent activity against Gram-positive bacteria. The current study examined the anti-staphylococcal mechanism of action of Mel4 and its precursor peptide melimine. The interaction of peptides with lipoteichoic acid (LTA) and with the cytoplasmic membrane using DiSC(3)-5, Sytox green, Syto-9 and PI dyes were studied. Release of ATP and DNA/RNA from cells exposed to the peptides were determined. Bacteriolysis and autolysin-activated cell death were determined by measuring decreases in OD620nm and killing of Micrococcus lysodeikticus cells by cell-free media. Both peptides bound to LTA and rapidly dissipated the membrane potential (within 30 seconds) without affecting bacterial viability. Disturbance of the membrane potential was followed by the release of ATP (50% of total cellular ATP) by melimine and by Mel4 (20%) after 2 minutes exposure (p<0.001). Mel4 resulted in staphylococcal cells taking up PI with 3.9% cells predominantly stained after 150 min exposure, whereas melimine showed 34% staining. Unlike melimine, Mel4 did not release DNA/RNA. Cell-free media from Mel4 treated cells hydrolysed peptidoglycan and produced greater zones of inhibition against M. lysodeikticus lawn than melimine treated samples. These findings suggest that pore formation is unlikely to be involved in Mel4-mediated membrane destabilization for staphylococci, since there was no significant Mel4-induced PI staining and DNA/RNA leakage. It is likely that the S. aureus killing mechanism of Mel4 involves the release of autolysins followed by cell death. Whereas, membrane interaction is the primary bactericidal activity of melimine, which includes membrane depolarization, pore formation, release of cellular contents leading to cell death.
Introduction
S. aureus is a major cause of infections in both health care and community settings which can produce high levels of mortality and have a high economic burden on society [1, 2]. S. aureus can reside on the skin and in the nasal cavity of 20–50% humans [3, 4] and this poses a risk for subsequent infections [5]. S. aureus is also a major cause of infections of medical devices and can cause 30–40% bacteraemia, surgical wound and implant-related infections [4, 6]. Methicillin-resistant S. aureus (MRSA) now causes over 50% of skin and soft tissue infections [7]. The mortality rate of S. aureus bacteraemia can reach up to 40% [8]. S. aureus associated infections are difficult to treat with currently available antibiotics [9] partly due to the increase in MRSA which are often very resistant to many different classes of antibiotics [6, 10]. To overcome these problems, new antimicrobials are needed which have unique modes of action and limited potential for resistance development.
Antimicrobial peptides (AMPs) exhibit broad spectrum antimicrobial activity against a wide range of microorganisms including bacteria, fungi, parasites and enveloped viruses at low concentrations [11–13]. AMPs are usually cationic in nature and have a varying number (from five to over a hundred) of amino acids. AMPs possess multiple modes of action, rapid bacterial killing kinetics and little toxicity toward human cells [14, 15]. Owing to the fact that these molecules exhibit multifaceted modes of action and rapidly kill bacteria, development of resistance in bacteria against AMPs is relatively rare [16, 17]. The mechanism of action of AMPs is believed to start by interacting with the negatively charged lipoteichoic acid (LTA) or teichoic acid of Gram-positive bacteria [18], through the negatively charge phosphate groups in LTA [19, 20].
The interaction with LTA is then believed to facilitate penetration of AMPs through the thick peptidoglycan layer, perhaps by the LTA acting as ladder, allowing the AMPs to reach and act on the cytoplasmic membrane [18]. AMPs disrupt phospholipid lipid bilayers of bacterial membranes by forming pores by various mechanisms called “barrel stave” or “toroidal pore” or through disintegration of lipids via the “carpet model” [21]. Cytoplasmic membrane collapse can result in leakage of cellular contents such as potassium ions, ATP and DNA/RNA which in turn may lead to cell death [22, 23]. Some AMPs translocate across the cell membrane and inhibit DNA/RNA or protein synthesis [24, 25]. AMPs can also kill Gram positive bacteria by activating cell wall bound autolytic enzymes known as autolysins [26]. LTAs, which are anchored in the cell envelope, control autolysin activity [27]. The interaction of AMPs with LTA can result in loss of regulation of autolysins which then cause autolysis by hydrolysing peptidoglycan chains and peptide bridges of murein [27].
Melimine is a cationic hybrid peptide of melittin and protamine [28]. It shows broad spectrum antimicrobial activity against Gram-negative and Gram-positive bacteria (including MRSA), fungi and protozoan such as Acanthamoeba without inducing resistance in bacteria [28, 29]. Melimine destroys the membrane potential of S. aureus [30]. Melimine, similarly to some other antimicrobial peptides such as Esculentin-1a and LL-37 [31], when covalently bound to polymers such as contact lenses inhibits initial bacterial attachment and can kill microbes [28, 29]. A shorter sequence of melimine, called Mel4, has a relatively low minimum inhibitory concentration against S. aureus (53.2 μM) and kills S. aureus when immobilized on surfaces [32]. It is non-cytotoxic to mammalian cells in vitro [28, 29]. Mel4 has improved ocular compatibility compared to melimine when bound to contact lenses [33]. Mel4 lacks the amino acids tryptophan, leucine and isoleucine that are present in the sequence of melimine. Tryptophan is a highly lipophilic amino acid [34] and its presence is often an important part of the activity to AMPs [35–37]. Leucine and isoleucine are hydrophobic residues which promote strong α-helix formation in AMPs which in turn results a higher degree of membrane disruption [38, 39]. Although various studies have shown that Mel4 has high bactericidal activity against staphylococci, the mode of action of Mel4 is not yet understood. Given the shorter peptide length and the unique amino acid sequence compared to melimine, an investigation of the killing mechanism of Mel4 against Gram-positive bacteria is necessary. Hence, this study was designed to evaluate the mode of action of Mel4 in comparison with melimine against S. aureus.
Materials and methods
Synthesis of peptides
Melimine (amino acid sequence: TLISWIKNKRKQRPRVSRRRRRRGGRRRR; molecular mass 3786.6) and Mel4 (KNKRKRRRRRRGGRRRR; 2347.8) were synthesized by conventional solid-phase peptide protocols [40, 41] and procured from the Auspep Peptide Company (Tullamarine, Victoria, Australia). The purity of the purchased peptides was ≥90%.
Bacterial strains and growth conditions
S. aureus 31 (contact lens induced peripheral ulcer isolate) [42], S. aureus 38 (microbial keratitis isolate) [43], and the type strain of S. aureus, ATCC 6538 (isolated from a “human lesion” according to the ATCC website) [44] were used in the current study. Bacteria were grown from frozen stocks overnight at 37 oC to mid-log phase in Tryptic Soy Broth (TSB; Oxoid, Basingstoke, UK) and cells were harvested following washing with phosphate buffer saline (PBS, NaCl 8 g/L, KCl 0.2 g/L, Na2HPO4 1.4 g/L, KH2PO4 0.24 g/L; pH 7.4) then diluted in the PBS containing 1/1000 TSB to OD600nm 0.05–0.06 (1× 107 colony forming units (CFU)/ml confirmed upon retrospective plate counts on TS agar (Oxoid)). Cells prepared this way were used in most experiments except for assessing the minimum inhibitory and bactericidal concentrations, measuring the release of DiSC3-5 from cells, and the autolytic activity assay.
Minimum inhibitory and minimum bactericidal concentration
The minimum inhibitory and minimum bactericidal concentrations of melimine and Mel4 were determined for all strains using a modified version of the Clinical Laboratory and Standard Institute (CLSI) broth microdilution method as described previously [45]. Bacterial suspensions were prepared to a final concentration of 5×105 CFU ml−1 in Muller Hinton Broth (MHB, Oxoid) and added to wells in a sterile 96-well microtiter plate containing two-fold dilutions of each peptide with 0.01% v/v acetic acid (Sigma Aldrich, St Louis, MO, USA) and 0.2% w/v bovine serum albumin (Sigma Aldrich). The plate was incubated at 37°C for 18 h-24 h. The MIC was set as the lowest concentration of peptides that reduced bacterial growth by ≥ 90% while the MBC was set as the lowest concentration of peptides that reduced bacterial growth by >99.99% after enumeration of viable bacteria by plate counts compared to bacteria grown without either AMP.
Interaction with LTA
Two experiments, bacterial growth in the presence of LTA and the amount of LTA in solution measured by ELISA, were performed to determine the interaction of melimine and Mel4 with LTA. Growth of S. aureus 38 was conducted in the presence of melimine or Mel4 following a method of Yang and Yousef [46]. S. aureus LTA (Sigma Aldrich; 100 μg/ml) was dissolved in MHB in wells of a 96-well microtiter plate containing S. aureus (5 ×105 CFU/ml) followed by addition of 100 μl of melimine or Mel4 to achieve their MICs. The OD600nm was determined over time during incubation at 37 OC. A growth curve was constructed over 12 h. Controls were bacteria grown in the presence of LTA alone and bacteria grown in MHB alone (a measure of maximum bacterial growth).
A competitive ELISA using kit (My BioSource, San Diego, CA, USA) was performed to evaluate whether melimine and Mel4 could bind and neutralize LTA of S. aureus. LTA (200 ng/ml) was dissolved in endotoxin free water (Sigma Aldrich) with melimine or Mel4 at their MICs or MBCs as final concentrations and incubated for 1–2 h at 37 oC. Interaction of LTA with melimine and Mel4 was assessed as an increase in OD450nm compared to negative control without peptides according to manufacturer instructions.
Cytoplasmic membrane disruption
The effect of melimine and Mel4 on the cytoplasmic membrane of S. aureus was assessed in three experiments. To examine membrane depolarization, the release of the membrane potential sensitive dye DiSC3-5 was measured. Sytox Green and Propidium Iodide (PI) dyes were used to determine whether the peptides could damage the cytoplasmic membranes and allow the stains to penetrate and interact to intracellular nucleic acids. Sytox Green has a molecular mass of 213.954 g/mol and a topological polar surface area of 28.7 A2 [47] whereas PI has a molecular mass of 668.087 g/mol and a topological polar surface area of 55.9 A2 [48]. Penetration of these two dyes through compromised membrane occurs through pores and differences may indicate different pore structures/sizes created by peptides.
Cytoplasmic membrane depolarization was performed as described by Rasul et al., [30]. Aliquots 100 μl bacteria labelled with 4 μM DiSC3-5 (Sigma Aldrich, St Louis, MO, USA) in HEPES and 100 μl of melimine and Mel4 at their MICs and MBCs (as final concentrations) were added to wells of 96 well microtiter plates. Increases in fluorescence were determined at 30 second intervals at an excitation wavelength of 622 nm and an emission wavelength of 670 nm over five minutes. Aliquots of bacteria were withdrawn over the time course and serially diluted in Dey-Engley (D/E neutralizing broth (Remel, Lenexa, KS, USA)) then plated on Tryptic Soy Agar (Oxoid, Basingstoke, UK) containing phosphatidylcholine (0.7 g /L) and Tween 80 (5ml/L) then incubated at 37 oC overnight. The number of live bacteria were enumerated and expressed as CFU/ml. A positive control of dimethyl sulfoxide (DMSO) (Merck, Billerica, MA, USA) HEPES (100 μl) was used to disrupt the cytoplasmic membrane potential of bacteria [49]. Initially a range of DMSO concentrations (1%, 2%, 5% and 20% (v/v)) were tested to determine which concentration gave maximum deporization. 20% DMSO gave maximum concentration and used in the assay.
Sytox green assay was performed following the established protocol of Li et al.,[50]. The protocol was modified as to use TSB media instead of LB media, as TSB was used in all other experiments. Bacterial cells in PBS (100 μl) were dispensed into wells of 96-well plates along with 5μM Sytox green (Invitrogen, Eugene, Oregon, USA). Plates were incubated for 15 minutes in the dark at room temperature and then 100 μl of melimine or Mel4 were added so as to achieve their MICs and MBCs. Fluorescence was measured spectrophotometrically (at an excitation wavelength of 480 nm and an emission wavelength of 522 nm) every 5 minutes up to 30 minutes, and then after 150 minutes. Triton X-100 1% (v/v) (Sigma Aldrich, St Louis, MO, USA) in PBS with 1/1000 TSB (100 μl) was used as positive control to disrupt the cytoplasmic membrane of bacteria.
Flow cytometry was used to determine the ability of melimine and Mel4 to permeabilize the cytoplasmic membrane of S. aureus 38 only. Bacteria were incubated simultaneously with SYTO9 (7.5 μM) and PI (30 μM; Invitrogen, Eugene, Oregon, USA) and incubated at room temperature for 15 min. Fluorescence intensities were recorded with a LSRFortessa SORP Flow cytometer after addition of melimine or Mel4 at their MIC at different time points. The wavelength of green fluorescence was (525/550 nm) for SYTO9 and a red fluorescence (610/20 nm) for PI [51]. Data were acquired and analysed using Flowjo software (version 10.5.0, Oregon, USA). A minimum 20000 events were recorded for each sample.
Leakage of intracellular contents
Two assays, ATP release and loss of DNA/RNA. were performed to determine the leakage of intracellular contents. Bacterial suspensions (100 μl) were added to melimine and Mel4 at their MICs and MBCs and incubated at 37°C for 10 minutes. Samples were withdrawn at two minutes intervals and centrifuged at 9000 × g for five minutes, and the supernatant removed and kept on ice until further analysis. For determination of total ATP, the bacterial pellet was resuspended in boiling 100 mM Tris, 4 mM EDTA pH (7.4) and further incubated for 2 mins at 100°C to lyse all the cells. The lysed cells were centrifuged at 9600 × g for two minutes and the supernatant was kept on ice until further use [52]. Subsequently, both total and released ATP were determined using an ATP bioluminescence kit (Invitrogen, Eugene, Oregon, USA) according to the manufacturer’s instructions.
The loss of DNA/RNA was determined by following a method of Carson et al., [53]. Bacteria (100 μl) were incubated at 37 oC with melimine and Mel4 at their MICs and MBCs. Samples were withdrawn at 5 minutes intervals, diluted (1:10) and filtered through 0.22 μm pores (Merck, Tullagreen, Ireland). The OD260nm of the filtrates was measured in a UV-star plate (Greiner Bio-one GmbH, Frickenhausen, Germany). The results were expressed as the ratio to the initial OD260nm.
Lysis of bacteria
The bacteria-lytic potential of the two peptides was evaluated using two different bacterial inoculums 1× 108 CFU/ml (OD660 0.1) and 3 × 1010 CFU/ml. The smaller inoculum size was used to see whether OD620nm was measurable at the concentration of cells used in all other assays. However, no measurable optical density change at 620nm was obtained, and so a larger inoculum of 3 × 1010 CFU/ml was used. The larger inoculum was obtained by adjusting OD620nm to 0.3, and bacterial numbers (CFU/ml) were confirmed upon retrospective plate count. Melimine and Mel4 were added at their MICs and MBCs. Bacterial cultures were immediately mixed and then diluted 1:100 in (1/1000 TSB). The OD620nm was measured and additional readings were taken at 30, 60, 90, 120 minutes, 6.5 and 24 h after incubating at room temperature. Peptides at their respective concentrations in 1/1000 TSB were used as blanks. The results were recorded as a ratio of OD620nm at each time point compared to the OD620nm at 0 minutes [53].
Time kill assay
Aliquots of bacterial cells (100 μl; 1× 107 CFU/ml) were added to melimine or Mel4 at their MICs and MBCs (as final concentration) in 96 well plates and incubated at 37 °C with gentle shaking. Aliquots (100 μl) were withdrawn at specified time points, serially diluted into D/E neutralizing broth and plated onto Tryptic Soy agar containing phosphatidylcholine (0.7 g /L) and Tween 80 (5ml/L). Bacterial colonies were counted after 24 h of incubation at 37 oC. Viable bacteria were reported as log10 CFU/ml. A control of bacterial suspensions without peptide was performed under the same conditions. The assay was performed in triplicate.
Release of autolysins
The presence of autolysins in supernatants of strains (S. aureus 31 and S. aureus 38) grown in the presence of melimine and Mel4 was determined spectrophotometrically by the ability of the supernatants to decrease the optical density of Micrococcus luteus peptidoglycan (PGN; Sigma Aldrich, St Louis, MO, USA) following a published method [54]. Bacteria were grown in MHB and harvested by washing three times with PBS. Bacteria were resuspended in the same PBS to an OD660nm of 0.1 (1× 108 CFU/ml). The bacterial suspension (100 μl) was incubated at 4X MIC of melimine or Mel4 for 5 h at 37 oC. Following centrifugation at 2000 × g for 10 minutes at 4 oC, the supernatant was recovered and filtered through 0.2 μm filter (Merck, Tullagreen, Ireland). The filtrate was saturated with 75% ammonium sulphate at 4 oC with continuous mixing for 1 h. This mixture was centrifuged at 12000 × g at 4 oC for 30 min, the supernatant was discarded, and the pellet was resuspended in 0.1 M sodium acetate buffer (pH. 6) and dialysed against four changes of buffer using Amicon ultra centrifugal filters (Merck, Millipore, Tullagreen, Carrigtwohill, IRL). The retentate was recovered and incubated with 100 μl of Micrococcus luteus peptidoglycan (1 mg/ml) at room temperature for 3 h. OD570 nm was initially determined at 0 min and then every 30 min intervals until 3 h. Results were presented as a ratio of OD570 nm at each time point versus OD570nm at 0 minutes (in percentage). The retentate from the above assay was also used to detect the autolytic activity against a lawn of Micrococcus lysodeikticus ATCC 4698. Briefly, 10 μL of each retentate from the above assay was spotted onto a pre-seeded lawn of M. lysodeikticus (108 CFU/ml) on nutrient agar. Following incubation for 24 h at 37°C, the size of the zone of inhibition was measured. Lysozyme (5 mg/ml) used as a positive control for maximum growth inhibition. Melimine or Mel4 dissolved in PBS at 4X MIC was treated in exactly the same way but in the absence of bacteria, to test whether any of the inhibitory activity in both assays (PGN and lawn of bacteria) was due to the presence of these peptides.
Lysis of horse red blood cells
The hemolytic activities of both the peptides were determined using horse red blood cells (HRBCs; Sigma) as described previously [28]. The HRBCs were washed three times with PBS at 470 × g for 5 minutes. Peptides (16.50 μM to 3407.36 μM, in PBS) were added to the washed HRBCs and incubated at 37°C for 4 h. After incubation, the cells were pelleted at 1057 × g for 5 minutes, and supernatant was removed to assess the release of haemoglobin by measuring OD540nm. HRBCs in PBS and HRBCs in distilled water were used as negative and positive controls, to achieve 0% and 100% lysis respectively. The relative OD of HRBCs treated with each peptide were compared to that treated with distilled water to determine the relative percentage of hemolysis. The therapeutic index of the peptides was calculated by the ratio of hemolytic activity and minimum inhibitory concentration.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 7.02 software (GraphPad Software, La Jolla, CA, USA). The effect of the different concentrations of peptides in each assay compared to controls was analysed using two-way ANOVA with Tukey’s test of multiple comparisons. Correlations between release of extracellular ATP and bacterial death were examined using Pearson correlation test. Oneway ANOVA with Bonferroni test was used to analyse the hemolytic effect on horse red blood cells. Statistical significance was set as p<0.05.
Results
Inhibitory concentrations of the two peptides
The MICs and MBCs for melimine or Mel4 against S. aureus are shown in Table 1. Both melimine and Mel4 were most active against S. aureus ATCC 6538. Melimine had lower MIC and MBC compared to Mel4 for all strains.
Table 1. MIC and MBC values of melimine and Mel4 against S. aureus.
| Bacterial strains | Melimine | Mel4 | ||
|---|---|---|---|---|
| MIC (μM) | MBC (μM) | MIC (μM) | MBC (μM)) | |
| S. aureus 31 | 33.01 | 66.02 | 106.48 | 212.96 |
| S. aureus 38 | 33.01 | 66.02 | 106.48 | 212.96 |
| S. aureus ATCC 6538 | 16.50 | 16.50 | 53.24 | 53.24 |
MIC = minimum inhibitory concentration that kills ≥90% of the positive control
MBC = minimum bactericidal concentration that kills ≥99.99% of bacteria.
In all subsequent experiments data of the assays for S. aureus 38 is presented. Data for all other strains are presented in the supporting information (S1–S8 Tables). Both the AMPs displayed similar modes of action against all the strains of S. aureus.
Interaction with LTA
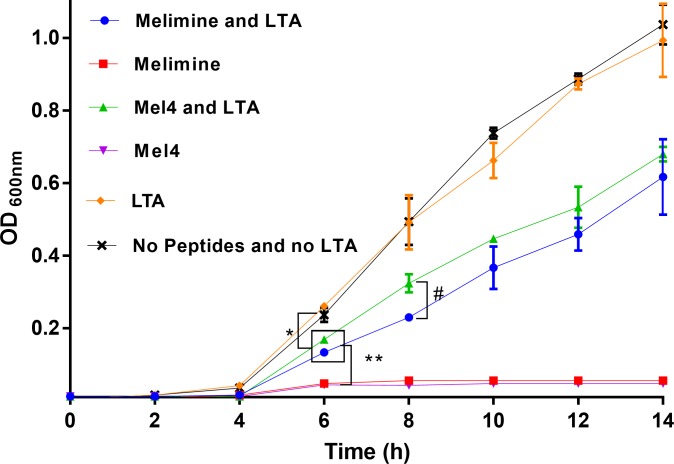
Melimine at its MIC (33.01 μM) and Mel4 at its MIC (66.02 μM) inhibited the growth of S. aureus (Fig 1) with OD600 nm<0.2 after 14 hours of growth. However, in the presence of LTA S. aureus could grow with significant increases in OD600nm starting after 6 h (p<0.001; Fig 1) with both peptides. At 6 h, the OD600nm was not different between melimine-LTA and Mel4-LTA (p = 0.601). However, at 8 h, the OD600nm with Mel4-LTA was significantly higher (p = 0.005) than melimine-LTA. The presence of LTA alone did not affect the growth of S. aureus and growth pattern was similar as in MHB alone.
Fig 1. Growth of S. aureus in the presence and absence of LTA.
The antibacterial effect of melimine and Mel4 was significantly reduced by addition of LTA compared to peptides alone after 6 h of incubation. The LTA alone had higher effect than LTA with any peptides at 6 h. Mel4-LTA increased OD600 nm more than melimine-LTA at 8 h. Error bars represents the means (±SD) of three independent experiments performed in triplicate. * represent p≤0.001, ** p≤0.0001 and # p = 0.005.
In the ELISA assay at their MICs, melimine neutralized 1.2±0.1 ng LTA/nmol and Mel4 neutralized 1.1±0.1 ng LTA/nmol (p>0.999). Similarly, at their bactericidal concentrations they neutralized 0.8±0.1 ng/nmol and 0.6±0.1 ng/nmol respectively (p>0.777).
Membrane disruption
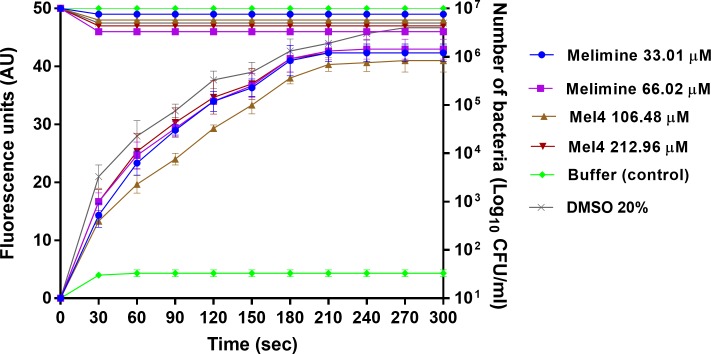
Melimine depolarized the cytoplasmic membrane of S. aureus in a predominantly concentration independent manner. An increase in fluorescence intensity was detected as early 30 seconds after addition of either peptide to all strains of S. aureus. The increase in fluorescence was continuous until at 180 sec and became constant after this time until the last observation at 300 sec (Fig 2). The fluorescein intensity was not statistically different at the MIC and MBC of melimine (p>0.999). However, at its MBC Mel4 yielded a significantly higher fluorescence than at its MIC at 60 to 90 sec (p<0.05) and after this time the degree of depolarization was similar for both the concentrations. This depolarization of the cytoplasmic membrane was not directly associated with the bacterial death as only < 0.5 log10 bacteria were affected with both the peptides at either concentration (Fig 2).
Fig 2. Cell membrane depolarization.
Cell membrane depolarization of S. aureus 38 as assessed with the release of membrane potential-sensitive dye DiSC3-(5) during treatment with peptides, measured spectroscopically at 622nm to 670nm excitation and emission wavelengths, and corresponding bacterial death as determined by plate counts. Each symbol represents data curve for increase in fluorescence (on the left y axis) and for reduction in bacterial count CFU/ml (on the right y axis). Data are presented as means (±SD) of three independent repeats in triplicate. DMSO (20%) was used as a positive control and buffer used as a negative control.
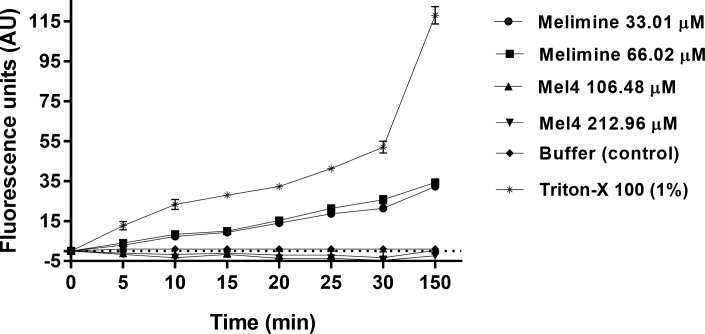
The fluorescence of Sytox green increased over time only with melimine and no fluorescence was detected with Mel4. Melimine at its MIC or MBC permeabilized the cell membrane and the fluorescence increased significantly after 10 minutes of exposure compared to buffer treated controls (p<0.026) (Fig 3). The intensity of fluorescence gradually increased over 150 minutes for both the concentrations of melimine against all strains, but no significant difference was observed between the MIC and MBC (p>0.999). Treatment with the positive control Triton-X 100 resulted emission of higher Sytox green fluorescence compared to either peptides within 10 minutes (p<0.001).
Fig 3. Cell membrane permeabilization.
Cell membrane permeability of S. aureus 38 was assessed with as the increase in fluorescence intensity due to interaction of Sytox green dye with DNA (measured spectroscopically at 480nm excitation and 522nm emission wavelengths) at their MICs and MBCs of melimine and Mel4. Data are presented as means (±SD) of three independent repeats in triplicate. Triton-X 100 (1%) was used as a positive control and buffer only as a negative control.
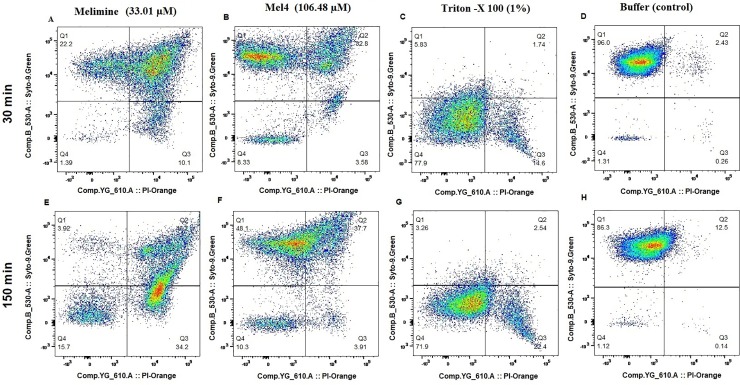
The membrane damaging effect of these peptides was also assessed with S. aureus 38 only by flow cytometry in the presence of the DNA intercalating dye PI and the membrane permeable dye Syto-9 (Fig 4). After 30 minutes exposure to melimine at its MIC, 22.2% of cells had high levels of Syto-9 staining but low levels of PI, 66.31% of cells had high levels of both Syto-9 and PI staining and 10.1% of cells had high levels of PI but low levels of Syto-9 staining. The total percentage of cells with high levels of PI staining after 30 minutes exposure to melimine was 76.41%. Whilst after 150 minutes the total number of PI stained cells remained approximately the same at 80.4%, the proportions had changed with 34.2% of cells having high levels of PI staining and low levels of Syto-9 staining and 46.2% having high levels of both Syto-9 and PI staining. The time course of staining with Mel4 at its MIC was different to melimine. After 30 minutes incubation, 55.3% of cells had high levels of Syto-9 staining but low levels of PI, 32.8% of cells had high levels of both Syto-9 and PI staining and 3.58% of cells had high levels of PI but low levels of Syto-9 staining. The total percentage of cells with high levels of PI staining after 30 minutes exposure to melimine was 36.4%. After 150 minutes the total number of PI stained cells remained approximately the same at 41.6%, as did the relative proportions with 3.91% of cells having high levels of PI staining and low levels of Syto-9 staining and 37.7% having high levels of both Syto-9 and PI staining. However, when looking at the flow cytometry readout (Fig 4) there did appear to be a general increase in the percentage of double stained cells, but the gating did not bin cells with sufficiently fine detail. There was a difference in the kinetics of cell death with the positive control Triton-X 100 (1%), which after 30 minutes stained 16.34% cells with high PI and increased up to 24.94% after 150 min incubation. With buffer treated (negative control) cells > 86% were stained with high levels of Syto-9 and low levels of PI even after 150 minutes incubation (Fig 4).
Fig 4. Flow cytometry.
Cell membrane permeabilization of S. aureus 38 as determined with Syto-9 (membrane permeable) and Propidium Iodide (membrane impermeable stains) dyes using Flow cytometry.
The release of cytoplasmic contents
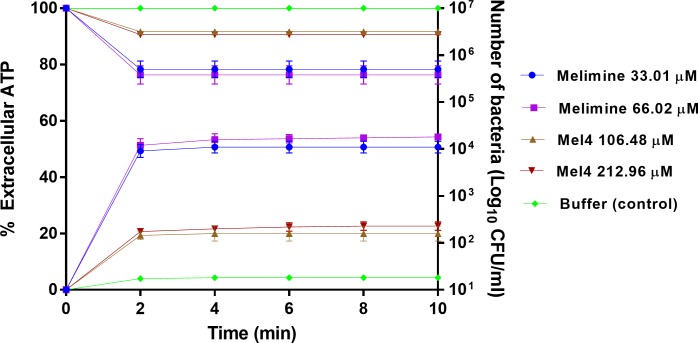
Melimine at its MIC or MBC released 49% and 51% of total cellular ATP respectively after 2 minutes of exposure (p<0.001) (Fig 5) and this amount of ATP did not increase after the initial burst at 2 minutes. Within the first two minutes, melimine decreased live bacteria by approximately 1.4 log10 at its MIC and 1.5 at its MBC, although this was not statistically different to each other and there was no further reduction in bacterial numbers to the end of the assay period (10 minutes). Compared with melimine, Mel4 released significantly (p>0.999) less ATP at its MIC (19%) and MBC (21%) after 2 min (Fig 5), with no increase up to 10 minutes. There were < 0.5 log10 viable bacteria after 2 minutes incubation with Mel4 at its MIC or MBC, with no further cell death over the remaining 8 minutes of the assay. Incubation with melimine released higher amounts of ATP and produced more bacterial death (p<0.001) than Mel4 at the MICs or MBCs.
Fig 5. Leakage of ATP.
The leakage of cellular ATP (in percentage) from S. aureus 38 after treatment with MIC or MBC of each peptide shown at X axis. The corresponding change in the number of live cells (CFU/ml) estimated through viable count displayed at Y axis. Each symbol represents data curve for increase in fluorescence (on the left y axis) and for reduction in bacterial count CFU/ml (on the right y axis). Errors bars represent the means (±SD) of three independent experiments in triplicate compared with buffer-treated control.
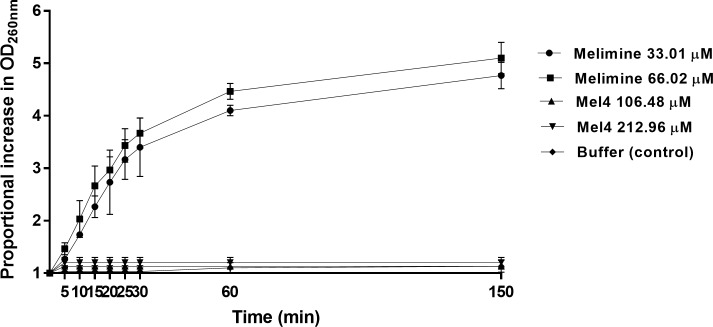
Release of DNA/RNA (260nm absorbing materials) is shown in Fig 6. Incubation with melimine resulted in release of DNA/RNA from S. aureus in time dependent but dose independent manner. Although increase in optical density first happened at 5 minutes with melimine but it was not significant (p≥0.083). A significant increase in the optical density happened after 10 minutes exposure with increases of 1.7 times and 2.0 times at MIC and MBC respectively (p<0.001). Moreover, optical density increased 4.8 times at MIC and 5.1 times at MBC after 150 minutes. On the hand, incubation with Mel4 did not increase optical density at its MICs or MBCs over the course of 150 minutes of the assay (p≥0.9945).
Fig 6. Release of DNA/RNA.
The release of UV absorbing materials (DNA/RNA) from S. aureus 38 determined spectroscopically at OD260nm.after incubation with MIC or MBC of melimine and Mel4. Data are presented as means (±SD) of three independent repeats in triplicate compared with buffer-treated control.
Bacterial lysis
At low bacterial concentrations (1× 108 CFU/ml), both the peptides showed no effect on optical density. This was probably due to bacterial concentration was too low to detect any change in optical density. However, when higher inoculum size of 1× 1010 CFU/ml was used and treated with various concentration of peptides, a significant reduction in optical density was observed at 6.5 hours (Fig 7). At this time point, melimine reduced optical density by 21±1% at its MIC and 31±4% at its MBC compared to buffer treated control (p<0.001) (Fig 7). Melimine lysed more than 42% cells after 24 h. A similar trend was observed for Mel4 which reduced optical density by 22±3% and 25±2% at its MICs and MBCs respectively after 6.5 h (Fig 7). After 24 h of incubation, Mel4 caused lysis of more than 40% cells. Over 24 hours the bacterial-lytic efficiency of both melimine and Mel4 was similar and no significant difference was observed (p≥0.851). The optical density of control cells (buffer treated) remained unchanged over the 24 h of the experiment.
Fig 7. Bacterial lysis.
Bacterial lysis of S. aureus 38 was determined spectroscopically as decrease in OD620nm after incubation with MIC and MBC of melimine and Mel4. Data are shown as means (±SD) of three independent repeats in triplicate compared with buffer-treated control. * represent p<0.001 and ** p<0.0001.
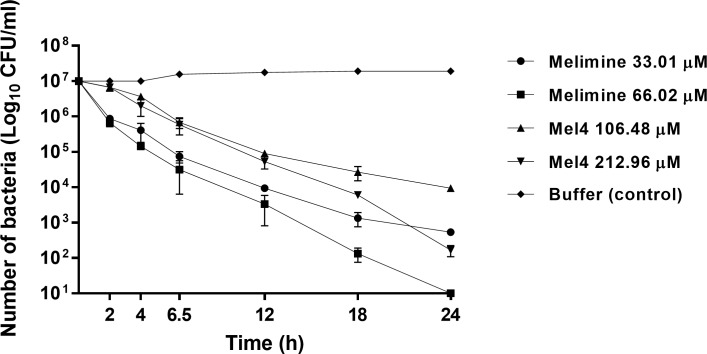
The killing kinetics of AMPs was determined by exposing S. aureus to their MIC and MBC. Melimine reduced the number of cells by 1.1 log10 at its MIC and 1.2 log10 CFU/ml at its MBC after 2 h of incubation (Fig 8). At its MIC, after 2 hours melimine continued to reduce the numbers of live cells, but the rate of reduction diminished after 12 hours. The initial rate of reduction from 0–2 h was 6.6 CFU log10/ml/hour, from 2–12 hours was 5.9 CFU log10/ml/hour and from 12–24 h was 2.9 CFU log10/ml/hour. At its MBC, the initial rate of reduction was 6.7 CFU log10/ml/hour, then from 2–24 h the rate remained constant at 5.7 CFU log10/ml/hour. Mel4 at its MIC did not significantly reduce bacterial viability within the first 2 h. The rate of reduction from 2–12 h was 5.6 CFU log10/ml/hour and 3.8 CFU log10/ml/hour from 12–24 h. At its MBC the rate of reduction was also negligible initially (0–2 h), however, from 2–24 h the rate of reduction increased to 5.5 CFU log10/ml/hour. Killing rate of melimine from 2–24 was significantly higher than Mel4 (p<0.001). The bacterial viability remained unaffected in control (buffer treated) over 24 h of the experiment.
Fig 8. Bacterial killing kinetics.
Time kill kinetics of melimine and Mel4 against S. aureus 38. Aliquotes analyzed at different time intervals for viable count showed that peptides decrease the bacterial viability (log10 CFU/ml) with different rates compared to buffer treated (negative control).
Autolytic activity
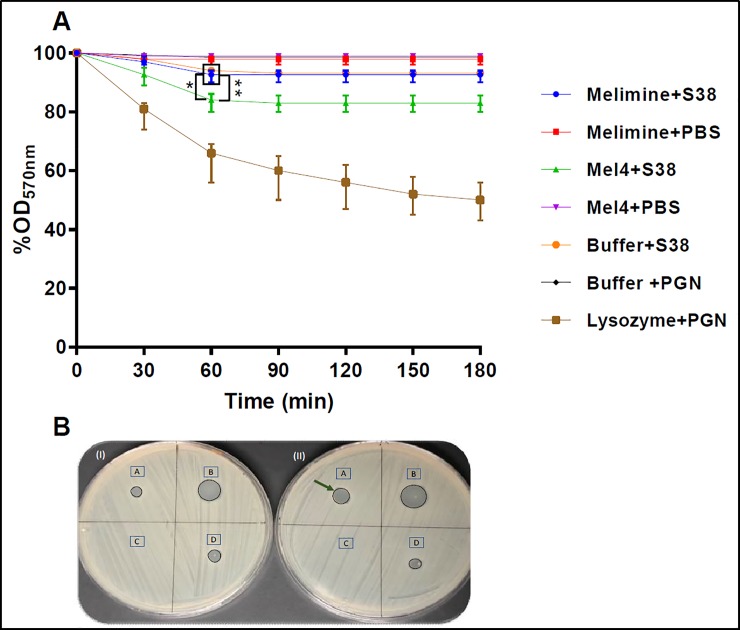
Autolytic activity of peptide-treated S. aureus supernatants was tested using PGN as the substrate. The supernatant of Mel4 treated S. aureus had a higher reduction of OD570nm of PGN suspension than melimine treated or controls (bacteria grown in the absence of peptides) (Fig 9A). The supernatants of Mel4 treated S. aureus resulted in a decrease in PGN density of 17±3% which was significantly more compared with melimine treated (8±2%; p = 0.004) and controls (7±1%, p<0.001) at 60 min ((Fig 9A). The reduction in PGN density caused by melimine or buffer supernatants was not significantly different at each time point (p = 0.999). The rate of PGN degradation for Mel4 was 0.28% OD570/min up to 60 mins and then there was no more activity from 60–180 minutes. The rate for lysozyme was 0.7% OD570/min within first 30 minutes. Lysozyme produced higher PGN hydrolysis, decreasing OD 21±4% at 30 min. The autolysis of culture supernatants was also examined using a pre-seeded lawn of Micrococcus lysodeikticus. Melimine or Mel4 suspended in buffer and treated in the same way as culture supernatants of S. aureus did not produce any zone of inhibition, showing that the peptides must either have been destroyed or removed during processing. The supernatant from Mel4 treated cells produced a zone of inhibition of 8±1 mm (Fig 9IIA) whereas for melimine and buffer treated cells smaller zones of inhibtion of 5±1 mm were produced (Fig 9IA). The positive control lysozyme formed largest 12±2 mm zone of inhibition.
Fig 9. Detection of autolysins release.
(A) Release of autolysins was determined by measuring decrease in OD570nm of PGN suspensions following treatment with cell-free supernatants of melimine and Mel4 treated S. aureus 38 (S38). Cell-free supernatants from Mel4 treated S. aureus resulted in a greater degradation of PGN than those from melimine or buffer treated S. aureus. Data presented as means (±SD) of three independent repeats in triplicate. * represent p≤0.004 compared to melimine treated bacteria and ** p<0.001 compared to buffer control (B) Release of autolysins was also monitored by determining the zone of inhibition (arrow) of cell-free supernatants against Micrococcus lysodeikticus ATCC 4698 after (I) melimine and (II) Mel4 treatment. 1A = supernatant from melimine treated S. aureus 38, IIA supernatant from Mel4 treated S. aureus, IB and IIB zones produced by lysozyme (5 mg/ml), IC no zone produced when melimine was incubated with buffer alone, IIC Mel4 incubated with buffer alone, ID and IID S. aureus incubated without addition of melimine or Mel4.
Lysis of horse red blood cells
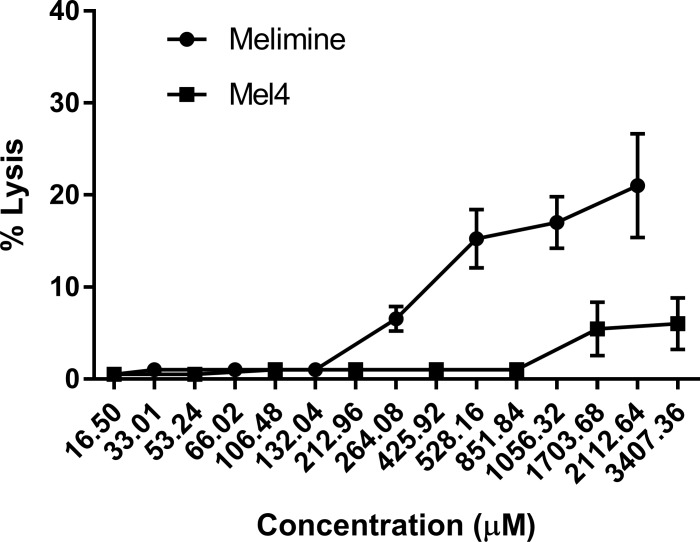
Melimine and Mel4 exhibited dose dependent hemolytic effects against HRBCs. Melimine started to hemolyse HRBCs (6%) at 264.08 μM. Significant lysis (17%) occurred at higher 518.16 μM concentrate.on (p<0.001; Fig 10). Compared with melimine, Mel4 showed less hemolysis at their corresponding MICs. The first appreciable hemolysis of HRBCs by Mel4 (5%) was observed at 1703.68 μM (p<0.041). However, exposure to 3407.36 μM resulted in slightly higher lysis (6%) of HRBCs (p<0.022). The therapeutic index (that is hemolytic concentration/MIC) was 16 for both melimine and Mel4.
Fig 10. Hemolysis of horse erythrocytes.
Hemolysis was determined as increase in OD540nm after incubation with increasing concentration of the melimine and Mel4. Errors bars represents the means (±SD) of three independent experiments performed at three separate occasion in triplicate.
Discussion
The current study demonstrated that the mode of action of S. aureus killing by Mel4 and its precursor melimine were different. A previous study had shown that 30 minutes incubation with melimine can distort S. aureus cells, induce bleb formation and accummulation of cell debris [30], as well as being able to permeabilise the cytoplasmic membrane of S. aureus in a concentration independent manner [30]. The previous study also showed that melimine was only able to produce <1 log10 reduction in numbers of S. aureus cells when incubated for up to five minutes. The current study demonstrated similar effects, but extended this to demonstrate that melimine could interact with LTA of S. aureus and produce beakage in the cytoplasmic membrane of S. aureus that resulted in leakage of intracellular contents (ATP and DNA/RNA) eventually leading to cell lysis and death. Melimine was also able to liberate a small amount of autolysins from the staphylococcal cells. Whilst Mel4 could also interact with LTA and depolarise the cytoplasmic membrane. It had a weaker effect on producing pores which was evidenced by negligible Sytox green intake by cells, minimum disruption to cytoplasmic membrane compared to melimine and minimum propidium iodide staining. Based on the flow cytometry data it is suggested that it can cause transient cell membrane permeability. In addition, less ATP was released from Mel4-treated cells and no DNA/RNA was released over 150 minutes. However, both the peptides eventually resulted in cell lysis and death after 24 hours exposure. Mel4-treated cells released greater amount of autolysins compared to melimine-treated ones. It is probable that the main killing mechanism of Mel4 was release of autolysins from LTA in cell walls along with membrane depolarization. Live bacteria produce a proton gradient outside the cell membrane, and the protons can accumulate in the cell walls and protonate the D-Alanine ester linkages of LTA [55]. The protonation of D-Ala ester linkages lowers the pH of the cell wall which in turn suppress the activity of autolysins bound to LTA [55]. Upon dissipation of the membrane protentional, deprotonation of the D-Ala ester linkages of LTA raises the pH of the cell wall resulting in activation of autolysins [55, 56]. Moreover, the membrane embedded glycolipid anchor of LTA plays an important role in its inhibitory effect on autolysins [57]. Antimicrobial peptides cause the delocalization of the glycolipid anchor and subsequently release autolysins by disrupting the cell membrane [57]. Therefore, the membrane depolarization caused by melimine and Mel4 may leads to LTA deprotonation and release of LTA which in turn liberates autolysins.
There are substantial differences in the amino acid compositions of Mel4 and melimine which contains 17 and 29 amino acids respectively. The lack of tryptophan in the sequence of Mel4 may be important in the differences in modes of action. Tryptophan (Trp) is known to interact with lipid bilayers, can enhance peptide-membrane interactions and facilitate insertion of peptides into the membrane [35, 58]. Trp and Arginine (Arg) together are known to promote stable and deeper insertion into the cell wall of Gram-positive bacteria [59]. Trp facilitates the insertion of Arg into hydrophobic regions of membranes via cation–π interactions causing rapid membrane disruption [35]. May be due to the presence of Trp, melimine can adopt a partial α helix in bio-membrane mimetic environment [30]. The antimicrobial activity of AMPs is often attributed to this helical conformation [60]. Mel4 lacks tryptophan and also has a low hydrophobic moment (0.039), meaning that it is less likely to be attracted within lipid bilayers [61]. Mel4 is unable to interact with lipid spheroids composed of 1-oleoyl-2-hydroxy-sn-glycero-3-phosphocholine (PC 18:1) or tethered lipid bilayers composed of 70% zwitterionic C20 diphytanyl-glycero-phosphatidylcholine lipid and 30% C20 diphytanyl-diglyceride but melimine can interact with these lipid layers [62].
Both melimine and Mel4 could interact with LTA. This interaction is likely due to their positive charge and the negative charge of LTA given by its phosphate groups. Melimine has an overall positive charge of +15, whilst Mel4 has +14 [32], and this small difference may partly explain some of the different effects of their interactions with LTA. At their MICs, there were 3 times more molecules of Mel4 present than melimine yet the inhibition of bacterial growth was not different, perhaps indicating a reduced ability of Mel4 to interact with LTA. In the ELISA assay at their MICs or MBCs melimine neutralized approximately the same number of LTA molecules as Mel4, further indicating a reduced Mel4-LTA interaction. However, this was not the case with the amount of autolysis observed after incubation of S. aureus cell supernatants with Micrococcus cells. LTA and teichoic acids bind to staphylococci cell wall and regulate the autolysins activity [63, 64], and addition of AMPs liberate autolysins [65].
A cytoplasmic membrane potential is essential for bacterial replication and ATP synthesis [66]. Dissipation of the membrane potential can increase membrane permeability resulting in loss of ATP [67]. Depletion of intracellular ATP can result in loss of viability of bacteria [66]. ATP leakage was associated with a increase of 1.5 log10 S. aureus bacterial death within 2 min with melimine. However, release of ATP did not correlate with bacterial death with Mel4. Melimine reduced bacterial viability in the ATP assay but not in the membrane depolarization assay at same time point. This effect may be due to the presence of 100 mM KCl in the bacterial suspension media of the membrane depolarization assay. KCl can increase the MIC of AMPs against S. aureus [68]. The amount of ATP released by Mel4 plateaued after four minutes exposure and never reached to the level of melimine until 10 minutes exposure. ATP may be depleted in the supernatant of Mel4 treated cells possibly by hydrolysis at their cell surfaces. Mel4 released small amounts of ATP without permeabilizing the cell membrane. ATP being a smaller molecule needs only a pore size of 1.5 nm to leak out from compromised membranes [69]. Thus, pores size produced by Mel4 in the cell membrane are enough for ATP release as they do not need well defined pores in the membrane to escape from bacteria.
Both melimine and Mel4 exhibited low hemolytic activity. The concentration at which both the peptides started lysis of HRBCs was 16 times higher than their MICs. These results are comparable with our previous published data where melimine produced 9% hemolysis of sheep red blood cells at 20 times higher concentration than its MIC [28]. Compared to Mel4, the slightly higher hemolytic activity of melimine at similar concentration may be related to higher hydrophobicity, amphipathcity and helicity [70]. Melimine contains the hydrophobic amino acids leucine, Isoleucine and tryptophan that can cause hemolysis of erythrocyte [71], whereas Mel4 lacks all these hydrophobic amino acids is less hydrophobic in turn showed less hemolytic activity against horse erythrocytes.
Conclusions
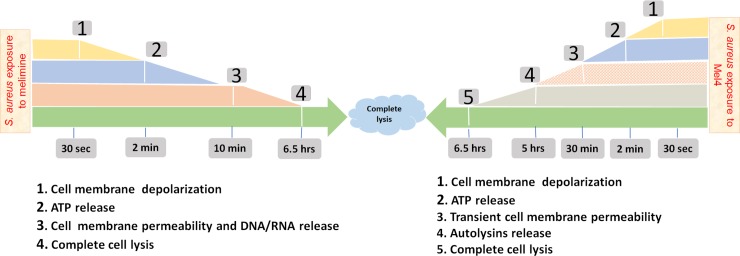
It is likely that the amphipathic characteristics of melimine allowed disruption of the cell membranes and pore formation that resulted in ATP and DNA release and ultimately cell death. However, Mel4 showed less interaction with cell membranes and its killing of S. aureus was more likely due to activation autolysins along with minimum membrane disruption. A comprehensive timeline of the mechanism of action of melimine and Mel4 against S. aureus is summarized in Fig 11.
Fig 11. Timeline of melimine or Mel4 interacting with S. aureus.
Both AMPs induced cell membrane depolarization after 30 seconds exposure to the staphylococci followed by release of ATP after 2 minutes. Cell membrane permeabilization occurred with melimine only after 10 minutes with near simultaneous release of DNA/RNA. Mel4 caused transient cell membrane after 30 minutes with no release of DNA/RNA. Mel4 showed autolysins induced killing after 5 hours. Complete bacterial lysis started after 6.5 hours of incubation with both the peptides.
Supporting information
Cell membrane depolarization as assessed by the release of the membrane potential sensitive dye DiSC3-5, measured spectroscopically at 622nm excitation and 670nm emission wavelengths. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
The decrease in the number of viable bacteria (CFU/ml) that occurred at the same time as measuring changes to their membrane depolarization. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
Cell membrane permeabilization was assessed as the increase in fluorescence intensity due to interaction of Sytox green dye with DNA (measured spectroscopically at 480nm excitation and 522nm emission wavelengths). Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
The leakage of cellular ATP (in percentage) from bacteria after treatment with 1X and 2X MIC of each peptide. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
The release of UV absorbing materials (DNA/RNA) was determined spectroscopically at OD260nm after treatment with 1X and 2X MIC of peptides. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
Bacterial lysis was determined as the decrease in OD620nm (in percentage) after treatment with 1X and 2X MIC of peptides. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
The hydrolysis of peptidoglycan (PGN) was determined by measuring decrease in OD570nm. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
Acknowledgments
The first author is grateful to Higher Education Commission (HEC) of Pakistan and the University of New South Wales, Australia for provision of Ph.D scholarship. The authors are also grateful to Christopher Brownlee of the Biological Resources Imaging Lab (BRIL) at the University of New South Wales, Australia for helping in Flow Cytometry analysis.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Prof Mark Willcox received the funding to accomplish this project. Australian Research Council was the funding body who granted fund under project number DP160101664 to conduct this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Uhlemann AC, Otto M, Lowy FD, DeLeo FR. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infection, genetics and evolution: J Mol Epi Evo Genetics Infect Dis. 2014;21:563–74. 10.1016/j.meegid.2013.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO AR. Global Report on Surveillance. Antimicrobial Resistance, Global Report on Surveillance. 2014. [Google Scholar]
- 3.Cespedes C, Gordon RJ, Lowy FD, Lo S-H, Miller M, Vavagiakis P, et al. The Clonality of Staphylococcus aureus Nasal Carriage. J Infect Dis. 2005;191(3):444–52. 10.1086/427240 [DOI] [PubMed] [Google Scholar]
- 4.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr., Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clinic Microbiol Rev. 2015;28(3):603–61. 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. New Eng J Med. 2001;344(1):11–6. 10.1056/NEJM200101043440102 [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Cheng LI, Helfer DR, Ashbaugh AG, Miller RJ, Tzomides AJ, et al. Mouse model of hematogenous implant-related Staphylococcus aureus biofilm infection reveals therapeutic targets. Proceeding of the National Academy of Sciences USA. 2017;114(26): 10.1073/pnas.1703427114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guillamet CV, Kollef MH. How to stratify patients at risk for resistant bugs in skin and soft tissue infections? Curr Opin Infect Dis. 2016;29(2):116–23. 10.1097/QCO.0000000000000244 [DOI] [PubMed] [Google Scholar]
- 8.Mylotte JM, McDermott C, Spooner JA. Prospective study of 114 consecutive episodes of Staphylococcus aureus bacteremia. Rev Infect Dis. 1987;9(5):891–907. [DOI] [PubMed] [Google Scholar]
- 9.Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clinic Invest. 2003;111(9):1265–73. 10.1172/JCI18535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Hal SJ, Fowler VG Jr. Is it time to replace vancomycin in the treatment of methicillin-resistant Staphylococcus aureus infections? Clinic Infect Dis: 2013;56(12):1779–88. 10.1093/cid/cit178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals. 2013;6(12):1543–75. 10.3390/ph6121543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3(9):710 10.1038/nri1180 [DOI] [PubMed] [Google Scholar]
- 13.Yasir M, Willcox M, Dutta D. Action of Antimicrobial Peptides against Bacterial Biofilms. Materials. 2018;11(12):2468. pmid: 11122468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim Biophys Acta-Biomembranes. 2009;1788(8):1687–92. [DOI] [PubMed] [Google Scholar]
- 15.Lee J-K, Park S-C, Hahm K-S, Park Y. Antimicrobial HPA3NT3 peptide analogs: placement of aromatic rings and positive charges are key determinants for cell selectivity and mechanism of action. Biochim Biophys Acta-Biomembranes. 2013;1828(2):443–54. [DOI] [PubMed] [Google Scholar]
- 16.Altman H, Steinberg D, Porat Y, Mor A, Fridman D, Friedman M, et al. In vitro assessment of antimicrobial peptides as potential agents against several oral bacteria. J Antimicrob Chemother. 2006;58(1):198–201. 10.1093/jac/dkl181 [DOI] [PubMed] [Google Scholar]
- 17.Fjell CD, Hiss JA, Hancock RE, Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov. 2011;11(1):37–51. 10.1038/nrd3591 [DOI] [PubMed] [Google Scholar]
- 18.Malanovic N, Lohner K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals. 2016;9(3):59. PMC5039512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bucki R, Janmey PA. Interaction of the Gelsolin-Derived Antibacterial PBP 10 Peptide with Lipid Bilayers and Cell Membranes. Antimicrob Agents Chemother. 2006;50(9):2932–40. 10.1128/AAC.00134-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Zhang S, Zhang J, Liu M, Liu Z. Vitellogenin is a cidal factor capable of killing bacteria via interaction with lipopolysaccharide and lipoteichoic acid. Mol Immunol. 2009;46(16):3232–9. 10.1016/j.molimm.2009.08.006 [DOI] [PubMed] [Google Scholar]
- 21.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462(1–2):55–70. 10.1016/s0005-2736(99)00200-x [DOI] [PubMed] [Google Scholar]
- 22.Abee T. Pore-forming bacteriocins of Gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol Lett. 1995;129(1):1–9. 10.1016/0378-1097(95)00137-T [DOI] [PubMed] [Google Scholar]
- 23.Pag U, Oedenkoven M, Sass V, Shai Y, Shamova O, Antcheva N, et al. Analysis of in vitro activities and modes of action of synthetic antimicrobial peptides derived from an α-helical ‘sequence template’. J Antimicrob Chemother. 2008;61(2):341–52. 10.1093/jac/dkm479 [DOI] [PubMed] [Google Scholar]
- 24.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–50 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- 25.Straus SK, Hancock RE. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta. 2006;1758(9):1215–23. 10.1016/j.bbamem.2006.02.009 [DOI] [PubMed] [Google Scholar]
- 26.Wilmes M, Stockem M, Bierbaum G, Schlag M, Götz F, Tran DQ, et al. Killing of staphylococci by θ-defensins involves membrane impairment and activation of autolytic enzymes. Antibiotics. 2014;3(4):617–31. 10.3390/antibiotics3040617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bierbaum G, Sahl H-G. Induction of autolysis of staphylococci by the basic peptide antibiotics Pep 5 and nisin and their influence on the activity of autolytic enzymes. Archives Microbiol. 1985;141(3):249–54. [DOI] [PubMed] [Google Scholar]
- 28.Willcox M, Hume E, Aliwarga Y, Kumar N, Cole N. A novel cationic‐peptide coating for the prevention of microbial colonization on contact lenses. J Appl Microbiol. 2008;105(6):1817–25. 10.1111/j.1365-2672.2008.03942.x [DOI] [PubMed] [Google Scholar]
- 29.Dutta D, Cole N, Kumar N, Willcox MDP. Broad Spectrum Antimicrobial Activity of Melimine Covalently Bound to Contact Lenses. Invest Ophthalmol Visual Sci. 2013;54(1):175–82. [DOI] [PubMed] [Google Scholar]
- 30.Rasul R, Cole N, Balasubramanian D, Chen R, Kumar N, Willcox MDP. Interaction of the antimicrobial peptide melimine with bacterial membranes. Int J Antimicrob Agents. 2010;35(6):566–72. 10.1016/j.ijantimicag.2010.02.005 [DOI] [PubMed] [Google Scholar]
- 31.Casciaro B, Dutta D, Loffredo MR, Marcheggiani S, McDermott AM, Willcox MD, et al. Esculentin-1a derived peptides kill Pseudomonas aeruginosa biofilm on soft contact lenses and retain antibacterial activity upon immobilization to the lens surface. Biopolymers. 2017. Epub 2017/11/01. 10.1002/bip.23074 . [DOI] [PubMed] [Google Scholar]
- 32.Dutta D, Kumar N, DP Willcox M. Antimicrobial activity of four cationic peptides immobilised to poly-hydroxyethylmethacrylate. Biofouling. 2016;32(4):429–38. 10.1080/08927014.2015.1129533 [DOI] [PubMed] [Google Scholar]
- 33.Dutta D, Zhao T, Cheah KB, Holmlund L, Willcox MDP. Activity of a melimine derived peptide Mel4 against Stenotrophomonas, Delftia, Elizabethkingia, Burkholderia and biocompatibility as a contact lens coating. Contact Lens Anterior Eye. 2017;40(3):175–83. 10.1016/j.clae.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 34.Mishra AK, Choi J, Moon E, Baek K-H. Tryptophan-Rich and Proline-Rich Antimicrobial Peptides. Molecules. 2018;23(4):815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan DI, Prenner EJ, Vogel HJ. Tryptophan-and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim Biophys Acta-Biomembranes. 2006;1758(9):1184–202. [DOI] [PubMed] [Google Scholar]
- 36.Bi X, Wang C, Ma L, Sun Y, Shang D. Investigation of the role of tryptophan residues in cationic antimicrobial peptides to determine the mechanism of antimicrobial action. J Appl Microbiol. 2013;115(3):663–72. 10.1111/jam.12262 [DOI] [PubMed] [Google Scholar]
- 37.Zhu X, Ma Z, Wang J, Chou S, Shan A. Importance of tryptophan in transforming an amphipathic peptide into a Pseudomonas aeruginosa-targeted antimicrobial peptide. PLos One. 2014;9(12):e114605 10.1371/journal.pone.0114605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saint Jean KD, Henderson KD, Chrom CL, Abiuso LE, Renn LM, Caputo GA. Effects of Hydrophobic Amino Acid Substitutions on Antimicrobial Peptide Behavior. Probiotics and Antimicrobial Proteins. 2018;10(3):408–19. 10.1007/s12602-017-9345-z [DOI] [PubMed] [Google Scholar]
- 39.Chou PY, Fasman GD. Conformational parameters for amino acids in helical, β-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13(2):211–22. 10.1021/bi00699a001 [DOI] [PubMed] [Google Scholar]
- 40.Behrendt R, White P, Offer J. Advances in Fmoc solid-phase peptide synthesis. J Pept Sci: 2016;22(1):4–27. 10.1002/psc.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Góngora-Benítez M, Tulla-Puche J, Albericio F. Handles for Fmoc Solid-Phase Synthesis of Protected Peptides. ACS Combin Sci. 2013;15(5):217–28. [DOI] [PubMed] [Google Scholar]
- 42.Jalbert I, Willcox MD, Sweeney DF. Isolation of Staphylococcus aureus from a contact lens at the time of a contact lens–induced peripheral ulcer: case report. Cornea. 2000;19(1):116–20. [DOI] [PubMed] [Google Scholar]
- 43.Hume EB, Cole N, Khan S, Garthwaite LL, Aliwarga Y, Schubert TL, et al. A Staphylococcus aureus mouse keratitis topical infection model: cytokine balance in different strains of mice. Immunol Cell Biol. 2005;83(3):294–300. 10.1111/j.1440-1711.2005.01326.x [DOI] [PubMed] [Google Scholar]
- 44.Baranska-Rybak W, Cirioni O, Dawgul M, Sokolowska-Wojdylo M, Naumiuk L, Szczerkowska-Dobosz A, et al. Activity of Antimicrobial Peptides and Conventional Antibiotics against Superantigen Positive Staphylococcus aureus Isolated from the Patients with Neoplastic and Inflammatory Erythrodermia. Chemotherapy Research and Practice. 2011;2011:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–75. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- 46.Yang X, Huang E, Yousef AE. Brevibacillin, a cationic lipopeptide that binds to lipoteichoic acid and subsequently disrupts cytoplasmic membrane of Staphylococcus aureus. Microbiol Res. 2017;195:18–23. 10.1016/j.micres.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 47.https://pubchem.ncbi.nlm.nih.gov/compound/104981. National Center for Biotechnology Information. PubChem Compound Database; CID = 104981(accessed Nov. 5, 2018).
- 48.https://pubchem.ncbi.nlm.nih.gov/compound/46863923. National Center for Biotechnology Information. PubChem Compound Database; CID = 46863923 (accessed Nov. 5, 2018).
- 49.te Winkel JD, Gray DA, Seistrup KH, Hamoen LW, Strahl H. Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front Cell Develop Biol. 2016;4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Shi Y, Su G, Le G. Selectivity for and destruction of Salmonella typhimurium via a membrane damage mechanism of a cell-penetrating peptide ppTG20 analogue. Int J Antimicrob Agents. 2012;40(4):337–43. 10.1016/j.ijantimicag.2012.05.026 [DOI] [PubMed] [Google Scholar]
- 51.Orman MA, Brynildsen MP. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob Agents and Chemother. 2013;57(9):4398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veldhuizen EJ, Schneider VA, Agustiandari H, Van Dijk A, Tjeerdsma-van Bokhoven JL, Bikker FJ, et al. Antimicrobial and immunomodulatory activities of PR-39 derived peptides. PLoS One. 2014;9(4): e95939 10.1371/journal.pone.0095939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carson CF, Mee BJ, Riley TV. Mechanism of action of Melaleuca alternifolia (tea tree) oil on Staphylococcus aureus determined by time-kill, lysis, leakage, and salt tolerance assays and electron microscopy. Antimicrob Agents and Chemother. 2002;46(6):1914–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leitch EC, Willcox MD. Elucidation of the antistaphylococcal action of lactoferrin and lysozyme. J Med Microbiol. 1999;48(9):867–71. 10.1099/00222615-48-9-867 [DOI] [PubMed] [Google Scholar]
- 55.Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiology and Molecular Biology Reviews: MMBR. 2008;72(1):85–109. 10.1128/MMBR.00030-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biswas R, Martinez RE, Göhring N, Schlag M, Josten M, Xia G, et al. Proton-binding capacity of Staphylococcus aureus wall teichoic acid and its role in controlling autolysin activity. Plos One. 2012;7(7):e41415 10.1371/journal.pone.0041415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer W, Rösel P, Koch H. Effect of alanine ester substitution and other structural features of lipoteichoic acids on their inhibitory activity against autolysins of Staphylococcus aureus. J Bacteriol. 1981;146(2):467–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37(42):14713–8. 10.1021/bi980809c [DOI] [PubMed] [Google Scholar]
- 59.Torcato IM, Huang Y-H, Franquelim HG, Gaspar D, Craik DJ, Castanho MA, et al. Design and characterization of novel antimicrobial peptides, R-BP100 and RW-BP100, with activity against Gram-negative and Gram-positive bacteria. Biochim Biophys Acta-Biomembr. 2013;1828(3):944–55. [DOI] [PubMed] [Google Scholar]
- 60.Blondelle SE, Houghten RA. Hemolytic and antimicrobial activities of the twenty-four individual omission analogs of melittin. Biochemistry. 1991;30(19):4671–8. 10.1021/bi00233a006 [DOI] [PubMed] [Google Scholar]
- 61.Kleinschmidt JH. Membrane protein folding on the example of outer membrane protein A of Escherichia coli. Cell Mol Life Sci. 2003;60(8):1547–58. 10.1007/s00018-003-3170-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berry T, Dutta D, Chen R, Leong A, Wang H, Donald WA, et al. The lipid membrane interactions of the cationic antimicrobial peptide chimeras melimine and cys-melimine. Langmuir. 2018. [DOI] [PubMed] [Google Scholar]
- 63.Brown S, Santa Maria JP Jr, Walker S. Wall teichoic acids of gram-positive bacteria. Ann Rev Microbiol. 2013;67:313–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneewind O, Missiakas D. Lipoteichoic acids, phosphate-containing polymers in the envelope of gram-positive bacteria. J Bacteriol. 2014;196(6):1133–42. 10.1128/JB.01155-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Netz DJA, Bastos MdCdF, Sahl H-G. Mode of Action of the Antimicrobial Peptide Aureocin A53 from Staphylococcus aureus. App and Environ Microbiol. 2002;68(11):5274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suh SJ, Shuman J, Carroll LP, Silo-Suh L. BEEP: An assay to detect bio-energetic and envelope permeability alterations in Pseudomonas aeruginosa. J Microbiol Methods. 2016;125:81–6. 10.1016/j.mimet.2016.04.009 . [DOI] [PubMed] [Google Scholar]
- 67.Nan YH, Bang J-K, Shin SY. Design of novel indolicidin-derived antimicrobial peptides with enhanced cell specificity and potent anti-inflammatory activity. Peptides. 2009;30(5):832–8. 10.1016/j.peptides.2009.01.015 [DOI] [PubMed] [Google Scholar]
- 68.Friedrich CL, Moyles D, Beveridge TJ, Hancock RE. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. Antimicrob Agents Chemother. 2000;44(8):2086–92. 10.1128/aac.44.8.2086-2092.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okuda K, Zendo T, Sugimoto S, Iwase T, Tajima A, Yamada S, et al. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob Agents Chemother. 2013;57(11):5572–9. 10.1128/AAC.00888-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Y, Mant CT, Farmer SW, Hancock RE, Vasil ML, Hodges RS. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J Bio Chem. 2005;280(13):12316–29. 10.1074/jbc.M413406200 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother. 2007;51(4):1398–406. 10.1128/AAC.00925-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell membrane depolarization as assessed by the release of the membrane potential sensitive dye DiSC3-5, measured spectroscopically at 622nm excitation and 670nm emission wavelengths. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
The decrease in the number of viable bacteria (CFU/ml) that occurred at the same time as measuring changes to their membrane depolarization. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
Cell membrane permeabilization was assessed as the increase in fluorescence intensity due to interaction of Sytox green dye with DNA (measured spectroscopically at 480nm excitation and 522nm emission wavelengths). Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
The leakage of cellular ATP (in percentage) from bacteria after treatment with 1X and 2X MIC of each peptide. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
The release of UV absorbing materials (DNA/RNA) was determined spectroscopically at OD260nm after treatment with 1X and 2X MIC of peptides. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
Bacterial lysis was determined as the decrease in OD620nm (in percentage) after treatment with 1X and 2X MIC of peptides. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
The hydrolysis of peptidoglycan (PGN) was determined by measuring decrease in OD570nm. Data are presented as means (±SD) of three independent repeats performed in triplicate.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.