Abstract
Advanced breast cancer, especially advanced triple-negative breast cancer, is typically more aggressive and more difficult to treat than other breast cancer phenotypes. There is currently no curable option for breast cancer patients with advanced diseases, highlighting the urgent need for novel treatment strategies. We have recently discovered that the nuclear factor of activated T cells 1 (NFAT1) activates the murine double minute 2 (MDM2) oncogene. Both MDM2 and NFAT1 are overexpressed ancconstitutively activated in breast cancer, particularly in advanced breast cancer, anc contribute to its initiation, progression, and metastasis. MDM2 regulates cancer cell proliferation, cell cycle progression, apoptosis, migration, and invasion through both p53-dependent and-independent mechanisms. We have proposed to target the NFAT1–MDM2–p53 pathway for the treatment of human cancers, especially breast cancer. We have recently identified NFAT1 and MDM2 dual inhibitors that have shown excellent in vitro and in vivo activities against breast cancer, including triple-negative breast cancer. Herein, we summarize recent advances made in the understanding of the oncogenic functions of MDM2 and NFAT1 in breast cancer, as well as current targeting strategies and representative inhibitors. We also propose several strategies for inhibiting the NFAT1–MDM2–p53 pathway, which could be useful for developing more specific and effective inhibitors for breast cancer therapy.
1. INTRODUCTION
Breast cancer remains the most common cancer and the principal cause of cancer-related death among women worldwide.1 Among various breast cancer phenotypes, triple-negative breast cancer (TNBC), which lacks the expression of estrogen receptors (ER), progesterone receptors (PR), and human epidermal growth factor receptor 2 (HER2), is the most aggressive form and represents approximately 15% of breast cancer.2–4 Advanced breast cancer, comprising the most serious stages, stage 3 (locally advanced disease) and stage 4 (metastatic stage) of breast cancer, is generally considered as a treatable but still incurable disease because of its high rates of recurrence and resistance to chemotherapy.5,6 Due to recent progress in understanding, diagnosing, and treating advanced breast cancer, particularly the HER2-positive and Luminal-like subtypes, overall survival for patients with advanced diseases has improved.7,8 However, long-term survival for patients with advanced breast cancer, especially for patients with advanced TNBC remains poor,2,3,8 emphasizing the unmet need for novel treatment strategies for these advanced diseases.
The murine double minute 2 (MDM2) oncogene has been identified as a master regulator of the p53 tumor suppressor.9,10 The deregulation of MDM2–p53 pathway owing to the amplification and overexpression of MDM2 and the mutation and deletion of p53 has been implicated in the initiation, progression, and metastasis of breast cancer.11,12 We have recently discovered that the nuclear factor of activated T cells 1 (NFAT1) transcription factor activates the transcription of MDM2.13 NFAT1 is also highly expressed and constitutively activated in breast cancer and contributes to the onset and metastatic progression of this disease.14 The activation of the NFAT1–MDM2–p53 pathway has been further observed in breast cancer, including TNBC; this pathway may be a promising molecular target for developing effective treatments for advanced breast cancer.13 In this article, we review recent progress in our understanding of the functions of MDM2 and NFAT1 in breast cancer at different stages, the current targeting approaches and inhibitors of the NFAT1–MDM2–p53 pathway, and their potentials for advanced breast cancer treatment.
2. THE MDM2 ONCOGENE IN BREAST CANCER
2.1. The MDM2 Oncogene
More than 2 decades ago, MDM2 was identified as an oncogene, as its overexpression induces tumorigenicity in mouse 3T3 cells.15 MDM2 amplification and overexpression are highly prevalent in human cancers,16–18 including breast cancer,19 and play pivotal roles in cancer onset, progression, metastasis, and response to therapy via both p53-dependent and -independent mechanisms.20,21 Due to significant advances in our understanding of MDM2 biology, MDM2 has been found to be a complicated and highly regulated protein. MDM2 consists ofmultiple functional domains, including the N-terminal p53-binding domain, the centrally located acidic domain, the zinc finger domain, and the C-terminal RING finger domain.20,22 The p53-binding domain is essential for the binding of MDM2 to p53, as well as MDM2’s inhibitory effect on p53 transcriptional activity.23 This domain is also involved in the interaction between MDM2 and other proteins,22 for example, Ring1-and YY1-binding protein (RYBP).24,25 The acidic domain of MDM2 has been demonstrated to play an important role in p53 ubiquitination and degradation.26,27 This domain also aids in interactions with ribosomal proteins S7 and S25.28,29 The linker region between the p53-binding domain and the acidic domain contains a nuclear localization signal (NLS) and a nuclear export signal (NES), which are essential for the subcellular localization of MDM2.30 There is also a cryptic nucleolar localization signal in the C-terminal domain of MDM2.31 The zinc finger domain plays a vital role in the binding of MDM2 with various proteins, including the tumor suppressor p14ARF 32,33 and several ribosomal proteins.34 The RING finger domain is critical for MDM2’s E3 ligase activity, which controls the ubiquitination of p53 and MDM2 itself.35 In addition, the RING finger domain contributes to MDM2 homodimerization and MDM2–MDMX heterodimerization.36
2.2. The MDM2–p53 Feedback Loop in Breast Cancer
MDM2 is a major negative regulator of p53 in normal cells and cancer cells. 9,10,23 MDM2 forms a tight complex with p53 and promotes p53 ubiquitination through its E3 ligase activity,9,10 enabling the recognition and interaction of p53 with the proteasome.37 Subsequently, the ubiquitinated p53 is degraded by the proteasome. In addition, MDM2 also promotes the interaction of the SUMO E3 ligase PIASy with p53 and induces the sumoylation and nuclear export of p53, 38,39 consequently inhibiting the binding ofp53 to transcriptional coactivators and decreasing p53-mediated transcription of the downstream target genes, including MDM2.40 On the other hand, p53 directly binds to the p53-responsive P2 promoter of MDM2 and mediates transcription of the MDM2 gene.41 The increased MDM2 protein, in turn, binds to p53 and promotes its degradation and inhibits its transcriptional activity, therefore forming the MDM2–p53 negative feedback loop (Fig. 1). This feedback loop is critical for controlling the cellular levels of p53 in normal cells because the high expression level of p53 is harmful for normal cell growth and differentiation. However, the amplification and overexpression of MDM2 are commonly observed in cancer cells due to deregulation of the MDM2–p53 pathway.16–19 In addition, several modulatory proteins have been reported to regulate the MDM2–p53 interaction, such as RYBP,24,25 ribosomal protein S7 and S25,28,29,34 and PA28γ,42 which have been reviewed in the recent papers.22,30
Fig. 1.
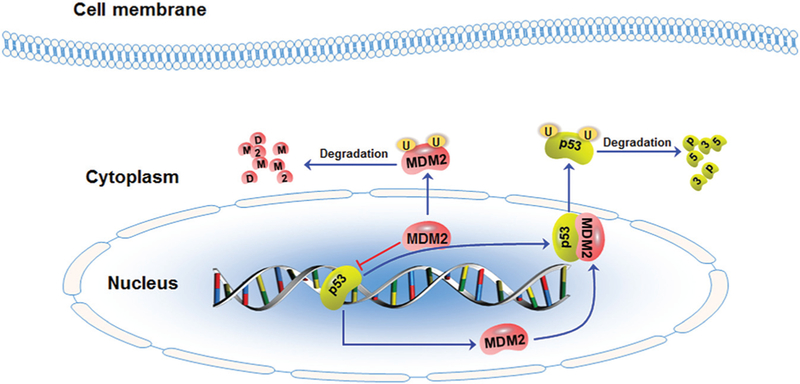
The schematic diagram illustrates the murine double minute 2 (MDM2)–p53 feedback loop in breast cancer. MDM2 oncoprotein forms a tight complex with the p53 tumor suppressor and shuttles p53 out of the nucleus, inhibiting p53-mediated transcription and promoting p53 ubiquitination and proteasomal degradation. On the other hand, p53 binds to the MDM2 P2 promoter and activates MDM2 transcription. In addition, MDM2 also targets itself for ubiquitination and degradation.
2.3. p53-Independent Functions of MDM2 in Breast Cancer
MDM2 is amplified and overexpressed in human cancer with p53 mutations and deletions, which promotes cancer initiation and progression through p53-independent mechanisms.11,12,43,44 It has been observed that the overexpression of MDM2 splice variants without the binding ability to p53 still can induce tumor formation in transgenic mice via a p53-independent mechanism.45,46 In breast cancer cells, estrogen was found to promote cell proliferation by activating MDM2, independent of p53.47 Furthermore, MDM2 promotes cell cycle progression and inhibits cell apoptosis by regulating several important mediators in a p53-independent manner. Previously, we discovered that MDM2 inhibition by an antisense antihuman MDM2 mixed-backbone oligonucleotide increased the expression levels of p21, Bax, and hypophosphorylated retinoblastoma (pRb) and reduced the levels of hyperphosphorylated pRb (ppRb) and E2F1 in p53-null cancer cells.48 Based on these observations, MDM2 has been further characterized as a negative regulator of the cyclin-dependent kinase inhibitor p21 by two laboratories independently.49–51 It has been found that MDM2 interacts with p21 and promotes its degradation in an ubiquitin-independent proteasomal degradation pathway. MDM2 also induces the proteasomal degradation of the tumor suppressor retinoblastoma (Rb) in both ubiquitin-dependent and-independent manners.52,53 We have found that MDM2 stabilizes the transcription factor E2F1 by inhibiting its ubiquitination,54 thereby activating the transcription of E2F1’s target genes related to cell cycle progression, for example, cyclin E.55 MDM2 also interacts with the Forkhead box O (FOXO) transcription factors, including FOXO3a and FOXO4, and promotes their degradation via an MDM2-mediated ubiquitin–proteasome pathway.56,57 In addition, MDM2 increases the translation of X-linked inhibitor of apoptosis protein (XIAP) by interacting with the internal ribosome entry segment (IRES) of the XIAP 5′-UTR and upregulating XIAP IRES activity.58
MDM2 has also been demonstrated to have a p53-independent role in regulating the epithelial–mesenchymal transition (EMT). In breast cancer cells, MDM2 increases cell motility and invasiveness by interacting with E-cadherin (one of the EMT markers) and promoting its ubiquitination and degradation via an endosome pathway.59 This negative regulation of E-cadherin by MDM2 has been further validated using tumor specimens from breast cancer patients with lymph node metastasis.59 In addition, MDM2 plays a crucial role in DNA replication and DNA repair. The MDM2 expression has been implicated in the chromosome instability, which has been discussed in a recent review paper.44 All these p53-independent functions of MDM2 further strengthen the potential of inhibiting MDM2 for cancer prevention and therapy.
2.4. Inhibiting MDM2 for Advanced Breast Cancer Therapy
The MDM2 oncogene has been demonstrated as a promising molecular target for cancer therapy.22,30,60,61 Several MDM2-targeting approaches have been developed: (1) inhibiting MDM2–p53 interaction, (2) blocking MDM2 E3 ligase activity, (3) destabilizing MDM2 protein, and (4) down-regulating MDM2 expression. Currently, the majority of MDM2 inhibitors have been developed to inhibit the MDM2–p53 interaction, releasing p53 from MDM2-mediated degradation and activating the transcription of p53 target genes.62 Several MDM2–p53 binding inhibitors have shown potent efficacy against human cancers harboring wild-type p53, and are undergoing further evaluation in clinical trials, for example, RG7112,63 HDM201,64 AMG 232,65 and NVP-CGM097.66 However, due to frequent mutation and deletion of p53 in advanced breast cancer, especially advanced TNBC, the MDM2–p53 binding inhibitors often show limited efficacy.12 Many MDM2 inhibitors have also been developed to block the E3 ligase activity of MDM2, activating the wild-type p53, such as HLI98C67 and MEL23.68 JNJ-26854165 (serdemetan), a novel inhibitor of the MDM2 E3 ligase activity, has been evaluated in a phase I trial performed in patients with advanced solid tumors, including seven patients with advanced breast cancer.69 A dose-and exposure-dependent p53 induction was observed in some of the patients. However, only one of seven patients with advanced breast cancer showed a partial response to JNJ-26854165. Recent studies have indicated that JNJ-26854165 also exerts its anticancer activity in p53-mutant cancer cells through p53-independent mechanisms.70
We have been focusing on the MDM2-targeting therapeutics that can directly inhibit MDM2 and induce cancer cell death, regardless ofp53 status, and have developed several MDM2 inhibitors, including antisense oligonucleotides andsiRNA,48,71–76 genistein,77 curcumin,78 ginsenosides,79–81 and makaluvamine analogs.82,83 These MDM2 inhibitors have shown excellent efficacy against human cancers, including breast cancer, regardless of the p53 status in the cancer cells. We have recently proposed the development of small-molecule inhibitors that can directly bind to MDM2 and induce its degradation, resulting in the discovery of a novel MDM2 inhibitor, termed SP141.84,85 SP141 can directly bind to the p53-binding domain ofMDM2, enhancing MDM2 autoubiquitination and proteasomal degradation.84 In human breast cancer cell lines and animal models, SP141 inhibits cancer cell growth and proliferation and induces cell cycle arrest and apoptosis in vitro and suppresses xenograft tumor growth in vivo, independent of p53.84 Furthermore, SP141 also inhibits breast cancer cell migration in vitro and prevents lung metastasis of breast cancer in vivo by modulating the expression ofEMT markers.84 The pharmacokinetic profile ofSP141 has also been investigated in our recent studies86,87; although further studies should be performed using more clinically relevant models of advanced breast cancer.
MDM2’s oncogenic functions, MDM2-targeting strategies, and the MDM2 inhibitors under development have been comprehensively discussed in recent review papers.22,30,60,61 Despite the significant advances in this field, challenges remain in the pursuit of clinically applicable MDM2 inhibitors for cancer therapy, including low specificity, poor bioavailability, limited efficacy, and potential toxicity. As MDM2 overexpression can trigger both p53-dependent and-independent pathways, inhibiting one of the MDM2 pathways could only have limited efficacy in human cancers. For cancers containing mutant p53, inhibitors that target MDM2–p53 binding may not be as effective as those that directly inhibit MDM2 expression. Moreover, p53 activation may lead to unexpected adverse effects to normal tissues. Thus, strategies to specifically and effectively inhibit MDM2 and prevent unnecessary toxicity are critically needed.
3. NFAT1, AN ONCOGENIC TRANSCRIPTION FACTOR IN BREAST CANCER
3.1. The NFAT, a Family of Transcription Factors
The NFAT family of transcription factors consist of five members, including four calcium-regulated isoforms, NFAT1 (NFATc2 or NFATp), NFAT2 (NFATc1 or NFATc), NFAT3 (NFATc4), and NFAT4 (NFATc3 or NFATx) and a tonicity-responsive enhancer-binding protein (TonEBP or NFAT5).14,88 All NFAT proteins have an N-terminal NFAT homology domain (NHD), a highly conserved Rel homology domain (RHD), and a C-terminal domain, except for NFAT5.14,88 All the calcium-responsive NFAT members, NFAT1–NFAT4 contain the NHD, which possesses a potent transactivation domain and a calcineurin docking site.89 However, NFAT5 lacks the calcineurin docking site and is completely insensitive to calcium and calcineurin.90 Indeed, NFAT5 can be activated in response to osmotic stress.91 The NHD also contains NLS1 and 2 and NES, which control NFAT subcellular localization through phosphorylation and dephos-phorylation of the NFAT proteins at the serine-rich regions (SRR).92,93 The RHD is also known as the DNA-binding domain (DBD) of NFAT proteins, which shows a structural similarity to the DBD of nuclear factor-KB (NF-kB).94 The calcineurin–NFAT signaling pathway, the posttranslational modifications of NFAT proteins, including phosphorylation by various kinases, as well as the transcriptional and posttranscriptional regulation of NFAT isoforms have been comprehensively discussed in the recent review papers.14,88 Despite significant progress in the understanding of NFAT biology, the role of these domains in the functions of NFAT proteins has not yet been completely understood.
3.2. NFAT1 as an Oncogenic Transcription Factor in Breast Cancer
Since the discovery of NFAT, as an inducible transcription factor that is capable of transactivating interleukin-2 (IL-2) during early T-cell activation,95,96 its expression and functions have been implicated in numerous aspects of vertebrate development.97,98 NFAT proteins have also been reported to play critical roles in malignant phenotypes and tumor development and progression.14,99,100 The major NFAT isoform, NFAT1 is highly expressed and aberrantly activated in various human cancers, including breast,101–104 pancreatic,105–108 lung,109–111 cervical,112 and colon cancer,113 as well as melanoma,114 and contributes to the initiation, progression, and metastasis to these diseases. This section will focus on the role of NFAT1 in breast cancer, especially advanced breast cancer.
Jauliac et al. have found that NFAT1 is highly expressed in tumor tissues from patients with grade III invasive ductal carcinoma and may play a pivotal role in invasive and metastatic properties of breast cancer.101 A tissue microarray study has been recently performed and indicated a significant increase of NFAT1 expression in invasive ductal carcinoma with lymph node metastasis, in comparison to normal adjacent tissue and primary tumor.102 In a recent paper, Kaunisto et al. have further reported that NFAT1 expression level is significantly higher in tumor-invasive front epithelium than in the central epithelium or normal breast epithelium.103 In addition, the stromal NFAT1 is also expressed at higher levels in the central and invasive front tumor stroma than in normal breast stroma.103 More importantly, in comparison with other subtypes of breast cancer, TNBC shows a higher activation rate of the calcineurin/NFATl pathway, which is essential to tumorigenesis and metastasis.104
NFAT1 regulates cell cycle progression, apoptosis, migration, invasion, angiogenesis, and resistance to chemotherapy by transactivating its downstream target genes.14,99 Specific to breast cancer, NFAT1 has been discovered to promote breast cancer cell migration and invasion through the induction of cyclooxygenase-2 (COX-2), Glypican-6 (GPC-6), and autotaxin, as well as the synthesis of prostaglandins.115–117 NFAT1 stabilizes SnoN and mediates transforming growth factor-β (TGF-β)–induced EMT in breast cancer cells by upregulating N-cadherin expression and downregulating E-cadherin expression.118 NFAT1 also cooperates with Smad2 and promotes breast cancer progression by upregulating the cyclin-dependent kinase 2 (CDK2), cyclin-dependent kinase 4 (CDK4), and cyclin E.119 The NFAT1-induced expression of IL-8 promotes intratumoral neutrophil infiltration in breast cancer.103 In addition, the negative cross talk between NFAT1 and signal transducer and activator of transcription 5 (Stat5) signaling cascades has been implicated in the growth and metastasis of breast cancer.102
Several mediators have been involved in the regulation of NFAT1 in breast cancer cells. The α6β4 integrin promotes breast carcinoma invasion by upregulating and activating NFAT1.101,117 The tumor suppressor FOXP1 binds to NFAT1 on DNA and suppresses NFAT1 transcriptional activity, resulting in enhanced breast cancer cell migration.120 Plasma membrane calcium ATPase isoform 4 (PMCA4) interacts with calcineurin and inhibits the calcineurin/NFAT1 pathway, suppressing vascular endothelial growth factor (VEGF)–mediated angiogenesis,121 while the PMCA2–calcineurin binding inhibits the calcineurin/NFAT1 pathway, leading to the inhibition of breast cancer cell apoptosis.122 Akt blocks breast cancer cell migration and invasion by inducing MDM2-mediated NFAT1 ubiquitination and proteasomal degradation, 123,124 whereas GM3 synthase promotes breast cancer lung metastasis by inhibiting phosphoinositide-3 kinase (PI3K)/Akt pathway and activating NFAT1.125 In addition, Wnt5a and Secreted frizzle-related protein 2 (SFRP2) have also been found to induce breast cancer metastasis and angiogenesis by activating NFAT1.126–128
3.3. Inhibiting NFAT1 for Advanced Breast Cancer Therapy
Pharmacological inhibition of NFAT1 signaling has been demonstrated as a promising therapeutic strategy for cancer therapy.14 Several inhibitors of calcineurin/NFATl pathway, such as cyclosporine A (CsA) have also shown promising efficacy against human cancer in vitro and in vivo.129 A calcineurin inhibitor, tacrolimus (also known as FK506), directly binds to the immunophilin FKBPB12 and inhibits the calcineurin’s phosphatase activity, leading to dephosphorylation of NFAT proteins, has shown potent antibreast cancer activity in vitro and in vivo.127,128 Tacrolimus has been found to inhibit SFRP2-induced angiogenesis and the growth of angiosarcoma xenografts in mice in vivo.128 In the MMTV-neu transgenic mice, treatment with tacrolimus markedly decreases the tumor growth rate by reducing the tumor microvascular density.127 Quercetin, a well-known anticancer flavonoid, inhibits tumor growth, induces tumor necrosis, and suppresses oncocyte proliferation in mice bearing breast cancer xenograft tumors.130 Mechanistically, quercetin suppresses the calcineurin/NFAT pathway, resulting in the inhibition of the expression of VEGF and VEGFR2.130 Moreover, the human herpesvirus 6B (HHV-6B) U54 tegument protein is reported to inhibit breast cancer cell proliferation through the inhibition of NFAT activation.131
As NFAT1 transcription factor plays a crucial role in the immune response through the regulation the transcription of cytokine genes, the long-term use of NFAT1 inhibitors may result in a reduction of immunosurveillance in tumor microenvironments, subsequently causing an increase in cancer development.132,133 Although the potential adverse effects of the current NFAT1 inhibitors are yet to be determined, novel targeting approaches for specific inhibition of oncogenic NFAT1 are still urgently needed.
4. TARGETING NFAT1-MDM2-P53 PATHWAY FOR y ADVANCED BREAST CANCER THERAPY
4.1. The NFAT1-MDM2-p53 Pathway
In our continued efforts to develop specific and potent MDM2 inhibitors, genistein has been found to inhibit MDM2 at both transcriptional and posttranslational levels.77 The inhibition of MDM2 transcription by genistein (1) requires the NFAT transcription site in the MDM2 P2 promoter. This is an intriguing observation with an obvious explanation that NFAT is involved in regulating MDM2 transcription. In this regard, our recent findings have demonstrated that NFAT1 is the predominant NFAT isoform regulating MDM2 transcription. It binds to the MDM2 P2 promoter and activates MDM2 expression in a p53-independent manner (Fig. 2).13 The NFAT1–MDM2–p53 signaling pathway is subsequently validated in human cancer cells, including breast cancer cells in vitro and in human carcinoma tissues in vivo.13 Considering these findings, it is important to ask whether targeting NFAT1–MDM2–p53 pathway could be the–MDM2–p53 pathway could be a useful approach for cancer therapy.
Fig. 2.
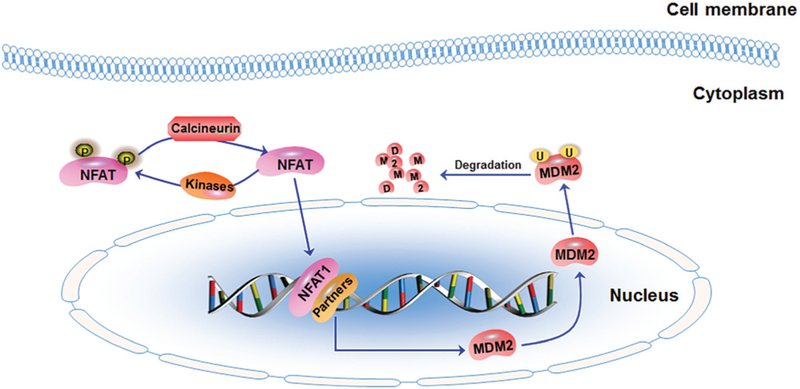
The schematic diagram illustrates the nuclear factor of activated T cells 1 (NFAT1)-MDM2 pathway in breast cancer. In the cytoplasm, calcineurin dephosphorylates NFAT1 and induces its nuclear translocation. NFAT1 can also be rephosphorylated by various kinases. In the nucleus, the dephosphorylated NFAT1 interacts with multiple transcriptional partners to activate the transcription of MDM2. MDM2 targets itself for ubiquitination and degradation.
We first wanted to know if the known NFAT 1 inhibitors could be used to target the NFAT1–MDM2–p53 pathway. Despite the structural similarities of NFAT isoforms, there is increasing evidence that these NFAT isoforms are not functionally equivalent but promote distinct signaling outputs in human cancers.14 Of note, NFAT1 and NFAT2 are frequently overexpressed and activated in human cancers, contributing to cancer development, progression, and metastasis,134–136 whereas NFAT3 functions as a tumor suppressor in breast cancer by inhibiting Lipocalin 2 expression and reducing cell motility.137 However, the majority of NFAT inhibitors, such as CsA and tacrolimus, have been developed to target the calcineurin–NFAT pathway or other upstream regulators, leading to an unwanted impact on all NFAT isoforms.14 Therefore, these NFAT inhibitors may not be useful in inhibiting the NFAT1–MDM2–p53 pathway for cancer therapy. Although genistein (1) has shown significant inhibitory effects on NFAT1–MDM2 interaction, more specific small-molecule inhibitors need to be developed for cancer therapy.
4.2. Representative Small-Molecule Inhibitors of NFAT1-MDM2-p53 Pathway
We have recently identified several small molecules (Fig. 3) that can effectively inhibit NFAT1 and MDM2 and show potent anticancer activity in vitro and in vivo. Two novel ginsenosides, 25-OCH3-PPD (2) and 25-OH-PPD (3) were initially identified as MDM2 inhibitors,79,80 and exhibited potent anticancer activity against various types of human cancer, including prostate,138–140 pancreatic,141 lung,142 and breast cancer.143 Both compounds have been found to inhibit cell viability and proliferation and induce cell cycle arrest and apoptosis in vitro. They also suppress xenograft tumor growth and sensitize tumors to chemotherapy in vivo. It has been further discovered that 25-OCH3-PPD inhibits breast cancer cell migration in vitro and prevents the lung metastasis of breast cancer in vivo.143 These activities have been attributed to its inhibitory effects on MDM2 expression and the modulatory effects on the expression of proteins related to cell growth, cell cycle progression, apoptosis, and EMT. Indeed, 25-OCH3-PPD (2) inhibits MDM2 at both transcriptional and posttranslational levels.143 The treatment of 25-OCH3-PPD enhances the MDM2 ubiquitination and degradation in breast cancer cells, independent of p53. Furthermore, 25-OCH3-PPD suppresses MDM2 transcription by inhibiting NFAT1 expression (unpublished data).
Fig. 3.
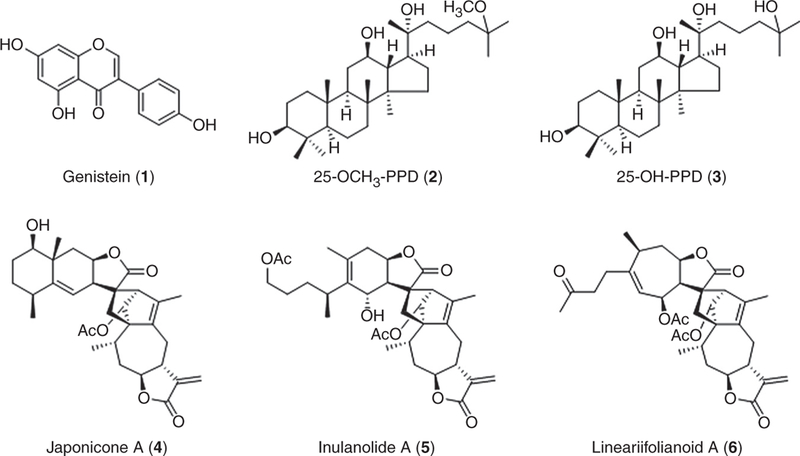
Structures ofrepresentativesmall-moleculeinhibitorsoftheNFAT1–MDM2–p53 pathway.
In our recent studies, we shed new light on the development of NFAT1–MDM2–p53 inhibitors. In a computational structure-based screening, a novel class of natural product MDM2 inhibitors, including japonicone A (JapA, 4), inulanolide A (InuA, 5), and lineariifolianoid A (LinA, 6) have been identified.144 Our results have demonstrated that JapA exerts its antibreast cancer activity via a dual-targeting mechanism. JapA directly binds to the p53-binding domain of the MDM2 protein and promotes MDM2 autoubiquitination and degradation, independent of p53.144 This compound also inhibits NFAT1 and NFAT1-mediated MDM2 transcription by suppressing NFAT1 nuclear localization, its binding to the MDM2 P2 promoter, and the induction NFAT1 ubiquitination and degradation.145 Our in vitro and in vivo studies have shown the potent antibreast cancer activity and safety profile of JapA, indicating that targeting the NFAT1–MDM2–p53 pathway is a promising approach for treating breast cancer, especially advanced breast cancer. More importantly, our data highlight the importance of developing dual NFAT1–MDM2 inhibitors instead of “mono” inhibitors for breast cancer therapy. It has been further discovered that both InuA and LinA suppress breast cancer growth and metastasis in vitro and in vivo by dually inhibiting NFAT1 and MDM2.146,147 Based on these results, we have proposed several targeting strategies for developing NFAT1-MDM2 inhibitors for the treatment of breast cancer, especially advanced breast cancer (Fig. 4).
Fig. 4.
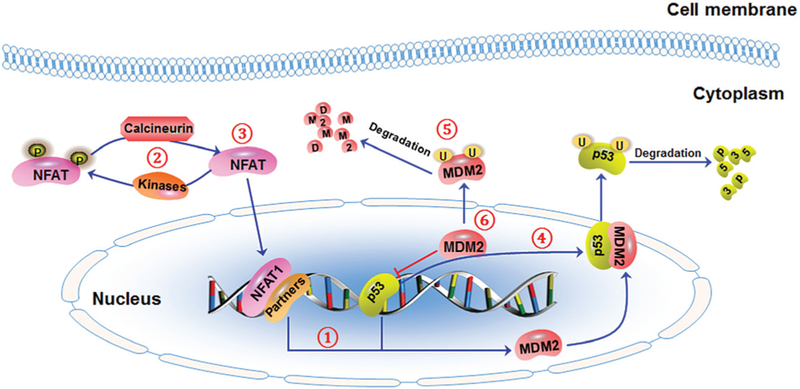
The schematic diagram illustrates targeting strategies for the NFAT1-MDM2-p53 pathway. To effectively inhibit NFAT1’s expression and activity, three targeting strategies have been proposed: (1) blocking NFAT1-MDM2 DNA binding, (2) promoting NFAT1 phosphorylation and/or inhibiting NFAT1 dephosphorylation, and (3) directly destabilizing NFAT1 protein. To effectively inhibit MDM2’s expression and activity, three targeting strategies have been proposed: (4) blocking MDM2-p53 interaction, (5) promoting MDM2 autoubiquitination and proteasomal degradation, and (6) directly inhibiting MDM2 expression. Small molecules that can dually inhibit NFAT1-MDM2 may be developed using these targeting strategies.
5. CONCLUSIONS
The overexpression and constitutive activation ofMDM2 and NFAT1 are common events in breast cancer and have been linked with the progression and metastasis of advanced breast cancer, especially advanced TNBC. Previous studies have shown that the “mono” small-molecule inhibitors of MDM2 and NFAT1 can inhibit breast tumor growth and metastasis, but may not be able to cause complete tumor regression due to inherent redundancy or acquired resistance. We have recently demonstrated that NFAT1 activates MDM2 in a p53-independent manner, and validated that the NFAT1–MDM2–p53 pathway is activated in breast cancer cell lines and carcinoma tissues. We, therefore, have proposed that targeting NFAT1–MDM2–p53 pathway is a promising strategy for developing novel and effective anticancer therapeutics, leading to the discovery of several small-molecule inhibitors, as well as their evaluation in preclinical models of breast cancer, including TNBC in vitro and in vivo.
Although the dual inhibitors of NFAT1 and MDM2 show great promise in our preliminary studies, concerns regarding its potential immune toxicity should be addressed before clinical trials are performed. Furthermore, the clinical relevance of the NFAT1–MDM2–p53 pathway in advanced breast cancer, especially advanced TNBC should be further investigated. It will be tempting to search for more specific NFAT1–MDM2 dual inhibitors. However, two questions should be answered. (1) How should specific and potent inhibitors for both NFAT1 and MDM2 be identified? (2) How should the adverse effects of these inhibitors on normal functioning NFAT1 be minimized? Both questions need to be addressed in future studies.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) grant R01 CA186662 (to R.Z.). The content is solely the responsibility of the authors, and do not necessarily represent the official views of the National Institutes of Health. W.W. was supported by the American Cancer Society (ACS) grant RSG-15–009-01-CDD.
REFERENCES
- 1.Brenner DR, Brockton NT, Kotsopoulos J, et al. Breast cancer survival among young women: a review of the role of modifiable lifestyle factors. Cancer Causes Control. 2016;27:459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szekely B, Silber AL, Pusztai L. New therapeutic strategies for triple-negative breast cancer. Oncology. 2017;31:130–137. [PubMed] [Google Scholar]
- 3.Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389:2430–2442. [DOI] [PubMed] [Google Scholar]
- 4.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liedtke C, Kolberg HC. Systemic therapy of advanced/metastatic breast cancer—current evidence and future concepts. Breast Care. 2016;11:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnenblick A, Ponde N, Piccart M. Metastatic breast cancer: the Odyssey of personalization. MolOncol. 2016;10:1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardoso F, Costa A, Norton L, et al. ESO-ESMO 2nd international consensus guidelines for advanced breast cancer (advanced breast cancer2). Ann Oncol. 2014;25: 1871–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puliti D, Bucchi L, Mancini S, et al. Advanced breast cancer rates in the epoch of service screening: the 400,000 women cohort study from Italy. EurJCancer. 2017;75:109–116. [DOI] [PubMed] [Google Scholar]
- 9.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. [DOI] [PubMed] [Google Scholar]
- 10.Kubbutat MH, Jones SN, Vousden KH. Regulation ofp53 stability by Mdm2. Nature. 1997;387:299–303. [DOI] [PubMed] [Google Scholar]
- 11.Haupt S, Vijayakumaran R, Panimaya J, Burgess A, Lim E, Haupt Y. The role ofMDM2 andMDM4 in breast cancer development and prevention. J Mol Cell Biol. 2017;9:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner N, Moretti E, Siclari O, et al. Targeting triple negative breast cancer: is p53 the answer? Cancer Treat Rev. 2013;39:541–550. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Zhang Z, Cheng J, et al. Transcription factor NFAT1 activates the mdm2 oncogene independent of p53. J Biol Chem. 2012;287:30468–30476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin JJ, Nag S, Wang W, et al. NFAT as cancer target: mission possible? BiochimBiophys Acta. 2014;1846:297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cahilly-Snyder L, Yang-Feng T, Francke U, George DL. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. SomatCellMol Genet. 1987;13:235–244. [DOI] [PubMed] [Google Scholar]
- 16.Ware PL, Snow AN, Gvalani M, Pettenati MJ, Qasem SA. MDM2 copy numbers in well-differentiated and dedifferentiated liposarcoma: characterizing progression to high-grade tumors. Am J Clin Pathol. 2014;141:334–341. [DOI] [PubMed] [Google Scholar]
- 17.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol CancerRes. 2004;2:1–8. [PubMed] [Google Scholar]
- 19.Yu Q, Li Y, Mu K, et al. Amplification of Mdmx and overexpression of MDM2 contribute to mammary carcinogenesis by substituting for p53 mutations. Diagn Pathol. 2014;9:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karni-Schmidt O, Lokshin M, Prives C. The roles of MDM2 and MDMX in cancer. Annu Rev Pathol. 2016;11:617–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nag S, Qin J, Srivenugopal KS, Wang M, Zhang R. The MDM2-p53 pathway revisited. J Biomed Res. 2013;27:254–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, Zhang J, Li M, Rayburn ER, Wang H, Zhang R. RYBP stabilizes p53 by modulating MDM2. EMBORep. 2009;10:166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W, QinJ J, Voruganti S, Nag S, Zhou J, Zhang R . Polycomb group (PcG) proteins and human cancers: multifaceted functions and therapeutic implications. Med Res Rev. 2015;35:1220–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol CellBiol. 2003;23:4939–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meulmeester E, Frenk R, Stad R, et al. Critical role for a central part ofMdm2 in the ubiquitylation of p53. Mol CellBiol. 2003;23:4929–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D, Zhang Z, Li M, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Wang W, Wang H, Wang MH, Xu W, Zhang R. Identification of ribosomal protein S25 (RPS25)-MDM2-p53 regulatory feedback loop. Oncogene. 2013;32: 2782–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.QinJ J, Nag S, Voruganti S, Wang W, Zhang R. Natural product MDM2 inhibitors: anticancer activity and mechanisms of action. Curr Med Chem. 2012;19:5705–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat CellBiol. 2000;2:179–181. [DOI] [PubMed] [Google Scholar]
- 32.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. ProcNatlAcad SciUSA. 1998;95:8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBOJ. 1998;17:5001–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, Nag S, Zhang X, et al. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev. 2015;35:225–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275: 8945–8951. [DOI] [PubMed] [Google Scholar]
- 36.Leslie PL, Ke H, Zhang Y. The MDM2 RING domain and central acidic domain play distinct roles in MDM2 protein homodimerization and MDM2-MDMX protein het-erodimerization. J Biol Chem. 2015;290:12941–12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulikov R, Letienne J, Kaur M, Grossman SR, Arts J, Blattner C. Mdm2 facilitates the association of p53 with the proteasome. Proc Natl Acad Sci USA. 2010;107: 10038–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat CellBiol. 2007;9:428–435. [DOI] [PubMed] [Google Scholar]
- 39.Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat CellBiol. 2000;2:569–573. [DOI] [PubMed] [Google Scholar]
- 40.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. [DOI] [PubMed] [Google Scholar]
- 41.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. GenesDev. 1994;8:1739–1749. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Zhang R. Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBOJ. 2008;27:852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Zhang R. p53-independent activities ofMDM2 and their relevance to cancer therapy. Curr CancerDrugTargets. 2005;5:9–20. [DOI] [PubMed] [Google Scholar]
- 44.Bouska A, Eischen CM. Murine double minute 2: p53-independent roads lead to genome instability or death. TrendsBiochem Sci. 2009;34:279–286. [DOI] [PubMed] [Google Scholar]
- 45.Fridman JS, Hernando E, Hemann MT, de Stanchina E, Cordon-Cardo C, Lowe SW. Tumor promotion by Mdm2 splice variants unable to bind p53. Cancer Res. 2003;63:5703–5706. [PubMed] [Google Scholar]
- 46.Steinman HA, Burstein E, Lengner C, et al. An alternative splice form of Mdm2 induces p53-independent cell growth and tumorigenesis. J Biol Chem. 2004;279: 4877–4886. [DOI] [PubMed] [Google Scholar]
- 47.Brekman A, Singh KE, Polotskaia A, Kundu N, Bargonetti J. A p53-independent role of Mdm2 in estrogen-mediated activation of breast cancer cell proliferation. Breast Cancer Res. 2011;13:R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, Li M, Wang H, Agrawal S, Zhang R. Antisense therapy targeting MDM2 oncogene in prostate cancer: effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc Natl Acad Sci USA. 2003;100:11636–11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Wang H, Li M, Agrawal S, Chen X, Zhang R. MDM2 is a negative regulator of p21WAF1/CIP1, independent ofp53. J Biol Chem. 2004;279:16000–16006. [DOI] [PubMed] [Google Scholar]
- 50.Jin Y, Lee H, Zeng SX, Dai MS, Lu H. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBOJ. 2003;22:6365–6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu H, Zhang Z, Li M, Zhang R. MDM2 promotes proteasomal degradation of p21Waf1 via a conformation change. J Biol Chem. 2010;285:18407–18414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uchida C, Miwa S, Kitagawa K, et al. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBOJ. 2005;24:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sdek P, Ying H, Chang DL, et al. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. MolCell. 2005;20:699–708. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z, Wang H, Li M, Rayburn ER, Agrawal S, Zhang R. Stabilization of E2F1 protein by MDM2 through the E2F1 ubiquitination pathway. Oncogene. 2005;24: 7238–7247. [DOI] [PubMed] [Google Scholar]
- 55.Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang JY, Zong CS, Xia W, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenkman AB, de Keizer PL, van den Broek NJ, Jochemsen AG, BurgeringBM. Mdm2 induces mono-ubiquitination of FOXO4. PLoS One. 2008;3:e2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 2009;15:363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang JY, Zong CS, Xia W, et al. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. MolCellBiol. 2006;26:7269–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E. Clinical overview of MDM2/X-targeted therapies. Front Oncol. 2016;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemos A, Leao M, Soares J, et al. Medicinal chemistry strategies to disrupt the p53-MDM2/MDMX interaction. MedResRev. 2016;36:789–844. [DOI] [PubMed] [Google Scholar]
- 62.Zhang B, Golding BT, Hardcastle IR. Small-molecule MDM2-p53 inhibitors: recent advances. Future Med Chem. 2015;7:631–645. [DOI] [PubMed] [Google Scholar]
- 63.AndreeffM Kelly KR, Yee K, et al. Results of the phase I trial of RG7112, a small-molecule MDM2 antagonist in leukemia. Clin Cancer Res. 2016;22:868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Furet P, Masuya K, Kallen J, et al. Discovery of a novel class of highly potent inhibitors of the p53-MDM2 interaction by structure-based design starting from a conformational argument. BioorgMedChemLett. 2016;26:4837–4841. [DOI] [PubMed] [Google Scholar]
- 65.Rew Y, Sun D. Discovery of a small molecule MDM2 inhibitor (AMG 232) for treating cancer. JMed Chem. 2014;57:6332–6341. [DOI] [PubMed] [Google Scholar]
- 66.Reuther C, Heinzle V, Nolting S, et al. The HDM2 (MDM2) inhibitor NVP-CGM097 inhibits tumor cell proliferation and shows additive effects with 5-Fluorouracil on the p53-p21-Rb-E2F1 cascade in the p53wildtype neuroendocrine tumor cell line GOT1. Neuroendocrinology. 2016. [Nov 21]. [DOI] [PubMed] [Google Scholar]
- 67.Yang Y, Ludwig RL, Jensen JP, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. CancerCell. 2005;7:547–559. [DOI] [PubMed] [Google Scholar]
- 68.Herman AG, Hayano M, Poyurovsky MV, et al. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. CancerDiscov. 2011;1:312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tabernero J, Dirix L, Schoffski P, et al. A phase I first-in-human pharmacokinetic and pharmacodynamic study of serdemetan in patients with advanced solid tumors. Clin CancerRes. 2011;17:6313–6321. [DOI] [PubMed] [Google Scholar]
- 70.You L, Liu H, Huang J, et al. The novel anticancer agent JNJ-26854165 is active in chronic myeloid leukemic cells with unmutated BCR/ABL and T315I mutant BCR/ ABL through promoting proteosomal degradation of BCR/ABL proteins. Oncotarget. 2017;8:7777–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen L, Agrawal S, Zhou W, Zhang R, Chen J. Synergistic activation of p53 by inhibition of MDM2 expression and DNA damage. Proc Natl AcadSci USA. 1998;95: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen L, Lu W, Agrawal S, Zhou W, Zhang R, Chen J. Ubiquitous induction of p53 in tumor cells by antisense inhibition ofMDM2 expression. MolMed. 1999;5:21–34. [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H, Nan L, Yu D, Agrawal S, Zhang R. Antisense anti-MDM2 oligonucleotides as a novel therapeutic approach to human breast cancer: in vitro and in vivo activities and mechanisms. Clin CancerRes. 2001;7:3613–3624. [PubMed] [Google Scholar]
- 74.Wang H, Wang S, Nan L, Yu D, Agrawal S, Zhang R. Antisense anti-MDM2 mixed-backbone oligonucleotides enhance therapeutic efficacy of topoisomerase I inhibitor irinotecan in nude mice bearing human cancer xenografts: in vivo activity and mechanisms. IntJ Oncol. 2002;20:745–752. [PubMed] [Google Scholar]
- 75.Wang H, Nan L, Yu D, Lindsey JR, Agrawal S, Zhang R. Anti-tumor efficacy of a novel antisense anti-MDM2 mixed-backbone oligonucleotide in human colon cancer models: p53-dependent and p53-independent mechanisms. MolMed. 2002;8:185–199. [PMC free article] [PubMed] [Google Scholar]
- 76.Wang H, Yu D, Agrawal S, Zhang R. Experimental therapy of human prostate cancer by inhibiting MDM2 expression with novel mixed-backbone antisense oligonucleotides: in vitro and in vivo activities and mechanisms. Prostate. 2003;54:194–205. [DOI] [PubMed] [Google Scholar]
- 77.Li M, Zhang Z, Hill DL, Chen X, Wang H, Zhang R. Genistein, a dietary isoflavone, down-regulates the MDM2 oncogene at both transcriptional and posttranslational levels. CancerRes. 2005;65:8200–8208. [DOI] [PubMed] [Google Scholar]
- 78.Li M, Zhang Z, Hill DL, Wang H, Zhang R. Curcumin, a dietary component, has anticancer, chemosensitization, and radiosensitization effects by down-regulating the MDM2 oncogene through the PI3K/mTOR/ETS2 pathway. Cancer Res. 2007;67: 1988–1996. [DOI] [PubMed] [Google Scholar]
- 79.Zhao Y, Wang W, Han L, et al. Isolation, structural determination, and evaluation of the biological activity of 20(S)-25-methoxyl-dammarane-3beta, 12beta, 20-triol [20(S)-25-OCH3-PPD], a novel natural product from Panax notoginseng. Med Chem. 2007;3:51–60. [DOI] [PubMed] [Google Scholar]
- 80.Wang W, Zhao Y, Rayburn ER, Hill DL, Wang H, Zhang R. In vitro anti-cancer activity and structure-activity relationships of natural products isolated from fruits of Panax ginseng. CancerChemotherPharmacol. 2007;59:589–601. [DOI] [PubMed] [Google Scholar]
- 81.Nag SA, Qin JJ, Wang W, Wang MH, Wang H, Zhang R. Ginsenosides as anticancer agents: in vitro and in vivo activities, structure-activity relationships, and molecular mechanisms of action. Front Pharmacol. 2012;3:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang W, Rayburn ER, Velu SE, Nadkarni DH, Murugesan S, Zhang R. In vitro and in vivo anticancer activity of novel synthetic makaluvamine analogues. Clin Cancer Res. 2009;15:3511–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang W, Rayburn ER, Velu SE, et al. A novel synthetic iminoquinone, BA-TPQ, as an anti-breast cancer agent: in vitro and in vivo activity and mechanisms of action. Breast Cancer ResTreat. 2010;123:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang W, Qin JJ, Voruganti S, et al. The pyrido[b]indole MDM2 inhibitor SP-141 exerts potent therapeutic effects in breast cancer models. Nat Commun. 2014;5:5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang W, Qin JJ, Voruganti S, et al. Identification of a new class of MDM2 inhibitor that inhibits growth of orthotopic pancreatic tumors in mice. Gastroenterology. 2014;147: 893–902e892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nag S, Qin JJ, Patil S, et al. A quantitative LC-MS/MS method for determination of SP-141, a novel pyrido[b]indole anticancer agent, and its application to a mouse PK study. J ChromatogrB. 2014;969:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nag S, Qin JJ, Voruganti S, et al. Development and validation of a rapid HPLC method for quantitation of SP-141, a novel pyrido[b]indole anticancer agent, and an initial pharmacokinetic study in mice. Biomed Chromatogr. 2015;29:654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pan MG, Xiong Y, Chen F. NFAT gene family in inflammation and cancer. CurrMol Med. 2013;13:543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luo C, Shaw KT, Raghavan A, et al. Interaction of calcineurin with a domain of the transcription factor NFAT1 that controls nuclear import. Proc Natl Acad Sci USA. 1996;93:8907–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lopez-Rodriguez C, Aramburu J, Rakeman AS, Rao A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci USA. 1999;96:7214–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. ProcNatlAcadSciUSA. 1999;96:2538–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okamura H, Aramburu J, Garcia-Rodriguez C, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. MolCell. 2000;6:539–550. [DOI] [PubMed] [Google Scholar]
- 93.Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. [DOI] [PubMed] [Google Scholar]
- 94.Nolan GP. NF-AT-AP-1 and Rel-bZIP: hybrid vigor and binding under the influence. Cell. 1994;77:795–798. [DOI] [PubMed] [Google Scholar]
- 95.Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Identification of a putative regulator of early T cell activation genes. Science. 1988;241:202–205. [PubMed] [Google Scholar]
- 96.Durand DB, Shaw JP, Bush MR, Replogle RE, Belagaje R, Crabtree GR. Characterization of antigen receptor response elements within the interleukin-2 enhancer. MolCell Biol. 1988;8:1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Minami T Calcineurin-NFAT activation and DSCR-1 auto-inhibitory loop: how is homoeostasis regulated? JBiochem. 2014;155:217–226. [DOI] [PubMed] [Google Scholar]
- 98.Fric J, Zelante T, Wong AY, Mertes A, Yu HB, Ricciardi-Castagnoli P. NFAT control of innate immunity. Blood. 2012;120:1380–1389. [DOI] [PubMed] [Google Scholar]
- 99.Mognol GP, Carneiro FR, Robbs BK, Faget DV, Viola JP. Cell cycle and apoptosis regulation by NFAT transcription factors: new roles for an old player. Cell Death Dis. 2016;7:e2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shou J, Jing J, Xie J, et al. Nuclear factor of activated T cells in cancer development and treatment. CancerLett. 2015;361:174–184. [DOI] [PubMed] [Google Scholar]
- 101.Jauliac S, Lopez-Rodriguez C, Shaw LM, Brown LF, Rao A, Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4:540–544. [DOI] [PubMed] [Google Scholar]
- 102.Zheng J, Fang F, Zeng X, Medler TR, Fiorillo AA, Clevenger CV Negative cross talk between NFAT1 and Stat5 signaling in breast cancer. Mol Endocrinol. 2011;25: 2054–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaunisto A, Henry WS, Montaser-Kouhsari L, et al. NFAT1 promotes intratumoral neutrophil infiltration by regulating IL8 expression in breast cancer. Mol Oncol. 2015;9:1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Quang CT, Leboucher S, Passaro D, et al. The calcineurin/NFAT pathway is activated in diagnostic breast cancer cases and is essential to survival and metastasis of mammary cancer cells. CellDeathDis. 2015;6:e1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Holzmann K, Kohlhammer H, Schwaenen C, et al. Genomic DNA-chip hybridization reveals a higher incidence ofgenomic amplifications in pancreatic cancer than conventional comparative genomic hybridization and leads to the identification of novel candidate genes. CancerRes. 2004;64:4428–4433. [DOI] [PubMed] [Google Scholar]
- 106.Baumgart S, Glesel E, Singh G, et al. Restricted heterochromatin formation links NFATc2 repressor activity with growth promotion in pancreatic cancer. Gastroenterology. 2012;142:388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Griesmann H, Ripka S, Pralle M, et al. WNT5A-NFAT signaling mediates resistance to apoptosis in pancreatic cancer. Neoplasia. 2013;15:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Baumgart S, Chen NM, Zhang JS, et al. GSK-3beta governs inflammation-induced NFATc2 signaling hubs to promote pancreatic cancer progression. Mol CancerTher. 2016;15:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang K, Li N, Chen Z, et al. High expression of nuclear factor of activated T cells in Chinese primary non-small cell lung cancer tissues. IntJBiolMarkers. 2007;22:221–225. [DOI] [PubMed] [Google Scholar]
- 110.Chen ZL, Zhao SH, Wang Z, et al. Expression and unique functions of four nuclear factor of activated T cells isoforms in non-small cell lung cancer. Chin J Cancer. 2011;30:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.LiuJF Zhao SH, Wu SS. Depleting NFAT1 expression inhibits the ability of invasion and migration of human lung cancer cells. CancerCellInt. 2013;13:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Noordhuis MG, Fehrmann RS, Wisman GB, et al. Involvement of the TGF-beta and beta-catenin pathways in pelvic lymph node metastasis in early-stage cervical cancer. Clin CancerRes. 2011;17:1317–1330. [DOI] [PubMed] [Google Scholar]
- 113.Gerlach K, Daniel C, Lehr HA, et al. Transcription factor NFATc2 controls the emergence of colon cancer associated with IL-6-dependent colitis. Cancer Res. 2012;72:4340–4350. [DOI] [PubMed] [Google Scholar]
- 114.Shoshan E, Braeuer RR, Kamiya T, et al. NFAT1 directly regulates IL8 and MMP3 to promote melanoma tumor growth and metastasis. CancerRes. 2016;76:3145–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yiu GK, Toker A. NFAT induces breast cancer cell invasion by promoting the induction of cyclooxygenase-2. JBiolChem. 2006;281:12210–12217. [DOI] [PubMed] [Google Scholar]
- 116.Yiu GK, Kaunisto A, Chin YR, Toker A. NFAT promotes carcinoma invasive migration through glypican-6. BiochemJ. 2011;440:157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen M, O’Connor KL. Integrin alpha6beta4 promotes expression of autotaxin/ ENPP2 autocrine motility factor in breast carcinoma cells. Oncogene. 2005;24: 5125–5130. [DOI] [PubMed] [Google Scholar]
- 118.Sengupta S, Jana S, Biswas S, Mandal PK, Bhattacharyya A. Cooperative involvement of NFAT and SnoN mediates transforming growth factor-beta (TGF-beta) induced EMT in metastatic breast cancer (MDA-MB 231) cells. ClinExpMetastasis. 2013;30: 1019–1031. [DOI] [PubMed] [Google Scholar]
- 119.Sengupta S, Jana S, Bhattacharyya A. TGF-beta-Smad2 dependent activation of CDC 25A plays an important role in cell proliferation through NFAT activation in metastatic breast cancer cells. Cell Signal. 2014;26:240–252. [DOI] [PubMed] [Google Scholar]
- 120.Oskay Halacli S FOXP1 enhances tumor cell migration by repression of NFAT1 transcriptional activity in MDA-MB-231 cells. CellBiolInt. 2017;41:102–110. [DOI] [PubMed] [Google Scholar]
- 121.Baggott RR, Alfranca A, Lopez-Maderuelo D, et al. Plasma membrane calcium ATPase isoform 4 inhibits vascular endothelial growth factor-mediated angiogenesis through interaction with calcineurin. ArteriosclerThrombVascBiol. 2014;34:2310–2320. [DOI] [PubMed] [Google Scholar]
- 122.Baggott RR, Mohamed TM, Oceandy D, et al. Disruption of the interaction between PMCA2 and calcineurin triggers apoptosis and enhances paclitaxel-induced cytotoxicity in breast cancer cells. Carcinogenesis. 2012;33:2362–2368. [DOI] [PubMed] [Google Scholar]
- 123.Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20:539–550. [DOI] [PubMed] [Google Scholar]
- 124.Yoeli-Lerner M, Chin YR, Hansen CK, Toker A. Akt/protein kinase b and glycogen synthase kinase-3beta signaling pathway regulates cell migration through the NFAT1 transcription factor. Mol Cancer Res. 2009;7:425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gu Y, Zhang J, Mi W, et al. Silencing of GM3 synthase suppresses lung metastasis of murine breast cancer cells. Breast Cancer Res. 2008;10:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dejmek J, Safholm A, Kamp Nielsen C, Andersson T, Leandersson K. Wnt-5a/Ca2 +-induced NFAT activity is counteracted by Wnt-5a/Yes-Cdc42-casein kinase 1alpha signaling in human mammary epithelial cells. Mol Cell Biol. 2006;26: 6024–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Siamakpour-Reihani S, Caster J, Bandhu Nepal D, et al. The role of calcineurin/NFAT in SFRP2 induced angiogenesis—a rationale for breast cancer treatment with the calcineurin inhibitor tacrolimus. PLoSOne. 2011;6:e20412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Courtwright A, Siamakpour-Reihani S, Arbiser JL, et al. Secreted frizzle-related protein 2 stimulates angiogenesis via a calcineurin/NFAT signaling pathway. Cancer Res. 2009;69:4621–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee CR, Chun JN, Kim SY, et al. Cyclosporin A suppresses prostate cancer cell growth through CaMKKbeta/AMPK-mediated inhibition of mTORC1 signaling. Biochem Pharmacol. 2012;84:425–431. [DOI] [PubMed] [Google Scholar]
- 130.Zhao X, Wang Q, Yang S, et al. Quercetin inhibits angiogenesis by targeting calcineurin in the xenograft model of human breast cancer. EurJPharmacol. 2016;781:60–68. [DOI] [PubMed] [Google Scholar]
- 131.IampietroM GravelA, Flamand L. Inhibition ofbreast cancer cell proliferation through disturbance of the calcineurin/NFAT pathway by human herpesvirus 6B U54 tegument protein. JVirol. 2014;88:12910–12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hogan PG. Calcium-NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium. 2017;63:66–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Muller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–656. [DOI] [PubMed] [Google Scholar]
- 134.Tie X, Han S, Meng L, Wang Y, Wu A. NFAT1 is highly expressed in, and regulates the invasion of, glioblastoma multiforme cells. PLoS One. 2013;8:e66008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Baumgart S, Chen NM, Siveke JT, et al. Inflammation-induced NFATc1-STAT3 transcription complex promotes pancreatic cancer initiation by KrasG12D. Cancer Discov. 2014;4:688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Oikawa T, Nakamura A, Onishi N, Yamada T, Matsuo K, Saya H. Acquired expression of NFATc1 downregulates E-cadherin and promotes cancer cell invasion. Cancer Res. 2013;73:5100–5109. [DOI] [PubMed] [Google Scholar]
- 137.Fougere M, Gaudineau B, Barbier J, et al. NFAT3 transcription factor inhibits breast cancer cell motility by targeting the Lipocalin 2 gene. Oncogene. 2010;29: 2292–2301. [DOI] [PubMed] [Google Scholar]
- 138.Wang W, Wang H, Rayburn ER, Zhao Y, Hill DL, Zhang R. 20(S)-25-methoxyl-dammarane-3beta, 12beta, 20-triol, a novel natural product for prostate cancer therapy: activity in vitro and in vivo and mechanisms of action. BrJ Cancer. 2008;98:792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang W, Rayburn ER, Hao M, et al. Experimental therapy of prostate cancer with novel natural product anti-cancer ginsenosides. Prostate. 2008;68:809–819. [DOI] [PubMed] [Google Scholar]
- 140.Voruganti S, Qin JJ, Sarkar S, et al. Oral nano-delivery of anticancer ginsenoside 25-OCH3-PPD, a natural inhibitor ofthe MDM2 oncogene: nanoparticle preparation, characterization, in vitro and in vivo anti-prostate cancer activity, and mechanisms of action. Oncotarget. 2015;6:21379–21394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang W, Rayburn ER, Zhao Y, Wang H, Zhang R. Novel ginsenosides 25-OH-PPD and 25-OCH3-PPD as experimental therapy for pancreatic cancer: anticancer activity and mechanisms of action. CancerLett. 2009;278:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang W, Rayburn ER, Hang J, Zhao Y, Wang H, Zhang R. Anti-lung cancer effects of novel ginsenoside 25-OCH(3)-PPD. Lung Cancer. 2009;65:306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang W, Zhang X, Qin JJ, et al. Natural product ginsenoside 25-OCH3-PPD inhibits breast cancer growth and metastasis through down-regulating MDM2. PLoS One. 2012;7:e41586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Qin JJ, Wang W, Voruganti S, Wang H, Zhang WD, Zhang R. Identification of a new class of natural product MDM2 inhibitor: in vitro and in vivo anti-breast cancer activities and target validation. Oncotarget. 2015;6:2623–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qin JJ, Wang W, Voruganti S, Wang H, Zhang WD, Zhang R. Inhibiting NFAT1 for breast cancer therapy: new insights into the mechanism of action of MDM2 inhibitor JapA. Oncotarget. 2015;6:33106–33119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Qin JJ, Wang W, Sarkar S, Voruganti S, Agarwal R, Zhang R, Inulanolide A. as a new dual inhibitor of NFAT1-MDM2 pathway for breast cancer therapy. Oncotarget. 2016;7:32566–32578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Qin JJ, Sarkar S, Voruganti S, Agarwal R, Wang W, Zhang R. Identification of linear-iifolianoid A as a novel dual NFAT1 and MDM2 inhibitor for human cancer therapy. JBiomedRes. 2016;30:322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]


